Abstract
The macrocyclic antiviral drug xylyl-bicyclam blocks entry of HIV into cells by targeting the CXCR4 coreceptor, a seven-helix transmembrane G-protein-coupled receptor. Its affinity for CXCR4 is enhanced by binding to Cu2+, Ni2+, or Zn2+. Metallocyclams have a rich configurational chemistry and proteins may bind selectively to specific metallocyclam configurations. Our studies of lysozyme reveal structural details of protein–metallocyclam interactions that are important for receptor recognition. Solution NMR studies show that Cu-cyclam interacts with specific tryptophan residues of lysozyme (Trp-62, Trp-63, and Trp-123). Two major binding sites for both Cu-cyclam and Cu2-xylyl-bicyclam were detected by x-ray crystallography. In the first site, Cu2+ in one cyclam ring of Cu2-xylyl-bicyclam adopts a trans configuration and is coordinated to a carboxylate oxygen of Asp-101, whereas for Cu-cyclam two ring NH groups form H bonds to the carboxylate oxygens of Asp-101, stabilizing an unusual cis (folded) cyclam configuration. For both complexes in this site, a cyclam ring is sandwiched between the indole side chains of two tryptophan residues (Trp-62 and Trp-63). In the second site, a trans cyclam ring is stacked on Trp-123 and H bonded to the backbone carbonyl of Gly-117. We show that there is a pocket in a model of the human CXCR4 coreceptor in which trans and cis configurations of metallobicyclam can bind by direct metal coordination to carboxylate side chains, cyclam-NH···carboxylate H bonding, together with hydrophobic interactions with tryptophan residues. These studies provide a structural basis for the design of macrocycles that bind stereospecifically to G-coupled and other protein receptors.
Keywords: zinc, copper, x-ray crystallography, NMR, CXCR4 coreceptor
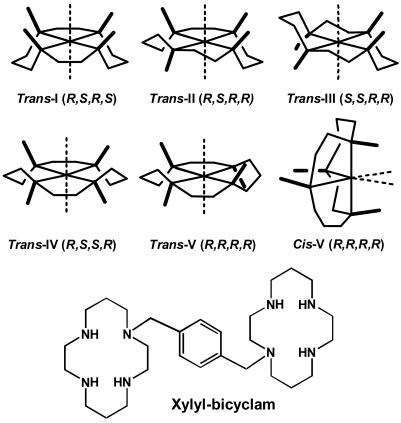
Cyclam (1,4,8,11-tetraazacyclotetradecane; Fig. 1) derivatives block entry of HIV into cells by binding specifically to the CXCR4 coreceptor (1), a G-protein-coupled, seven-helix transmembrane protein. An example is xylyl-bicyclam (AMD3100), which has two cyclam rings connected by a p-phenylenebis(methylene) linker (Fig. 1) and is a highly potent anti-HIV drug (2, 3). AMD3100 has recently been on clinical trial for the treatment of AIDS. Additional medical interest in cyclams arises from their ability to mobilize stem cells and to carry diagnostic and therapeutic radionucleotides (4), and from their potential use in transplant, inflammation, and cancer therapy (5). Metal ions such as Zn2+ and Cu2+ bind to cyclam strongly (log K values ≈15 and 27, respectively (6)) and relatively rapidly (7), and it seems likely that metal complexation by xylyl-bicyclam is involved in the mechanism of action of the drug in vivo. For metal-bicyclam complexes, there is a close correlation (8) between antiviral activity and binding to the coreceptor CXCR4 as monitored by inhibition of 12G5 mAb binding and the intracellular Ca2+ signal induced by SDF-1α chemokine, the order of decreasing activity being Zn2+ ≈ Ni2+ > Cu2+ >> Co3+ >> Pd2+. The affinity of AMD3100 for the CXCR4 receptor is enhanced by factors of 7, 36, and 50 by incorporation of Cu2+, Zn2+, or Ni2+, respectively, into the cyclam rings (9), and a similar metal-induced enhancement in CXCR4 affinity is observed for cyclam itself, although the affinity for the receptor is much less. Aspartate carboxylate groups of CXCR4 (especially Asp-171 and Asp 262) have been implicated in the bicyclam binding site (9, 10), and models of CXCR4 containing (carboxylate)O···HN(cyclam) H bonding and carboxylate(O)-metal(cyclam) coordination have been built (9–11).
Fig. 1.
Structures of the major configurations of a metal cyclam and of xylyl-bicyclam (the hydrochloride salt is the drug AMD3100).
Metal cyclam complexes have a potentially rich configurational chemistry (4, 12). Each of the coordinated N atoms is chiral. The metal ion can commonly be 4-, 5-, or 6-coordinate, and symmetrical trans configurations can fold to give cis structures, as illustrated for trans-V in Fig. 1. In view of the targeting of anti-HIV cyclam derivatives to CXCR4, it is important to understand the nature of the interactions that determine their binding sites on proteins and in particular whether proteins can recognize different configurations of metal cyclams selectively. However, there appear to be no reported structures of protein adducts of metal cyclams. In particular, we have suggested (11) that carboxylate groups in a protein positioned above the faces of a metallocyclam might stabilize the unusual cis-V configuration. To explore the structures of protein–cyclam adducts, we have studied the binding of metal–cyclam and metal–bicyclam complexes to lysozyme, chosen as a model protein because it is readily crystallized (e.g., ref. 13) and the active site contains carboxylates from Glu-35 and Asp-52 in close proximity. These amino acid side chains are known to bind to metal ions such as Gd3+ (14). Our data provide structural insight into specific polar and nonpolar interactions involved in the recognition of metallocyclams by proteins, features that can be found in a binding pocket for anti-HIV metallocyclams on the human CXCR4 coreceptor.
Experimental Section
Materials. Hen egg white lysozyme (HEWL) and cyclam were purchased from Sigma. Xylyl-bicyclam and Zn2(xylyl-bicyclam)-(ClO4)2 were prepared as previously described (11). (15N)Cyclam was synthesized by using the route shown in Scheme 1 with (15N)phthalimide (98 atom % 15N) and (15N)glycine (99 atom % 15N), both from Aldrich, as sources of 15N, and the Zn complex was prepared and characterized as described in ref. 15.
[Cu(cyclam)(H2O)2](OAc)2 (1). Cyclam (400.7 mg, 2 mmol) was dissolved in methanol (50 ml), and Cu(OAc)2 (363.5 mg, 2 mmol) was added. The reaction mixture was heated to reflux, stirred for 2 h, and then filtered to give a clear, purple solution. The solvent was removed in vacuo, and the purple solid was recrystallized by slow diffusion of methanol into ether to give purple needle crystals of 1·2MeOH (m/z = 322.2 [CuC12N4H27O2]+, 262.0 [CuC10N4H23]+). Samples of this complex used in NMR studies did not contain MeOH, which is readily lost on drying the complex.
[Cu2(xylyl-bicyclam)(OAc)4] (2). Xylyl-bicyclam (50.2 mg, 0.1 mmol) was dissolved in methanol (5 ml) and Cu(OAc)2 (36.9 mg, 0.2 mmol) was added. The dark blue solution was heated under reflux for 2 h, filtered, and concentrated on a rotary evaporator to give a dark blue crystalline material, which was recrystallized from methanol to give dark blue crystals (m/z = 805.1, [Cu2C34N8O6H63]+).
Lysozyme Crystallization. The hanging-drop method was used. The reservoir solution contained 100 μl of 50 mM acetate buffer, pH 4.5, 200 μl of saturated NaCl solution, and 700 μl of distilled water, and the hanging drop contained 2.5 μl of HEWL (50 mg·ml-1 in acetate buffer) and 2.5 μl of the reservoir solution. Crystals suitable for x-ray diffraction grew at 277 K within a week. Attempts to cocrystallize adducts of HEWL with complex 2 or with the Zn analogue were unsuccessful.
Crystal Soaking. Soaking was carried out for 5 days at 288 K with HEWL crystals in drops to which either solid 1 had been added (to saturation) or 2 had been added as a saturated solution in the well solution. Soaked crystals became purple, and they were removed in a cryoloop and frozen in liquid nitrogen by using type B immersion oil as a cryoprotectant.
X-Ray Crystallography. Diffraction data for complex 1 were collected with Mo-Kα radiation at 150 K on a Bruker SMART APEX charge-coupled device diffractometer equipped with an Oxford Cryosystems low-temperature device. Systematic errors were treated with sadabs (16). The structure was solved by Patterson methods (dirdif (17)) and refined by least squares against F2 (crystals (18)). H atoms were placed on C in calculated positions, but those attached to N and O were located in difference maps and refined. Crystal data: Orthorhombic, space group Pbca, a = 15.9421(7) Å, b = 7.0819(3) Å, and c = 20.9833(10) Å. The final conventional R factor was 0.0341; other data have been deposited in the Cambridge Structural Database.
Diffraction data for HEWL complexes were collected at Station 14.2 at the Daresbury Synchrotron Radiation Source and processed by using the programs mosf lm and scala (19). The initial structure was solved by using a reported lysozyme structure (ref. 13, PDB code 193L). Refinement was performed by using the program refmac (20) with waters being added by arp/warp, and manual checking and correction were performed with the program o (21). Data collection and refinement are summarized in Table 1. The positions of the metal ions in the structures of the adducts were confirmed in anomalous difference maps produced from data on HEWL crystals soaked with Ni-cyclam at a wavelength of 1.488 Å (Daresbury Synchrotron Radiation Source Station 14.1; data not shown).
Table 1. Crystallographic data and refinement statistics for lysozyme adducts.
| Cu-cyclam | Cu2-bicyclam | |
|---|---|---|
| Data statistics | ||
| Space group | P43212 | P43212 |
| Unit cell | ||
| a, Å | 77.9064 | 78.258 |
| b, Å | 77.9064 | 78.258 |
| c, Å | 37.7624 | 37.900 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 90 |
| Resolution range,* Å | 39-1.75 (1.84-1.75) | 39-1.6 (1.69-1.6) |
| Molecules per asymmetric unit | 1 | 1 |
| Observed reflections | 81,871 | 111,324 |
| Unique reflections | 12,234 | 16,097 |
| I/σ* | 6.1 (2.0) | 8.5 (2.0) |
| Completeness,* % | 99.9 (99.9) | 100 (100) |
| Multiplicity* | 6.7 (6.8) | 6.9 (6.9) |
| Rsym,* % | 8.1 (34) | 6.1 (36.2) |
| Refinement | ||
| R/Rfree, % | 17.6/23.3 | 17.7/22.3 |
| rms bonds, Å | 0.015 | 0.014 |
| rms angles, ° | 1.577 | 1.55 |
| Average isotropic B, Å2 | 19.55 | 17.63 |
Numbers in parentheses refer to high shell
NMR Spectroscopy. 1H NMR data were acquired over a range of 15 ppm at 298 K on a Bruker Avance 600 (1H = 599.92 MHz) NMR spectrometer using a TXI [1H, 13C, 15N] cryoprobe, equipped with z-field gradients. 1H chemical shifts were referenced internally to dioxane (3.75 ppm). All NMR experiments were performed in 90% H2O/10% D2O. The water resonance was suppressed by presaturation. Standard pulse sequences were used for 2D total correlation spectroscopy (TOCSY) and correlation spectroscopy (COSY) (mixing times of 60 and 80 ms, respectively).
The pH of 5 mM solutions of HEWL in 90% H2O/10% D2O used for NMR titrations was adjusted to 4.6 with 0.1 M HCl. Titrations were carried out by adding microliter aliquots of aqueous Cu-cyclam (125 mM, pH 4.6). The assignments for 1H NMR resonances of HEWL are based on those given by Noda et al. (22).
Mass Spectrometry. Positive-ion electrospray ionization mass spectra were acquired on a Micromass Platform II mass spectrometer, collision energy 4 eV, source temperature 353 K, capillary voltage 1.5 kV, cone voltage 25 V (Cu complexes 1 and 2) or 90 V (protein). Aqueous solutions were diluted 1:1 with acetonitrile before injection.
Supporting Information. Scheme 1 showing the synthetic route to (15N)cyclam and Figs. 4–10 showing 1D and 2D NMR spectra, x-ray crystal structure of complex 1·2MeOH, lysozyme crystals, crystal packing, contact distances, and comparison of tryptophan sandwiches are published as supporting information on the PNAS web site. Crystallographic data have also been deposited as follows: complex 1 in the Cambridge Crystallographic Data Centre CCDC 260045, Cu-cyclam-HEWL in the Protein Data Bank 1YIK, and Cu2-bicyclam-HEWL in the Protein Data Bank 1YIL.
Results and Discussion
Interaction of Zn-Cyclams with Lysozyme. First we sought to demonstrate that Zn2-bicyclam binds to lysozyme. Electrospray ionization mass spectrometry of an aqueous solution containing lysozyme and Zn2(bicyclam)(ClO4)2 in a 1:1 molar ratio gave two well resolved series of charged states with masses in the transformed spectrum of 14,308 ± 1 and 14,943 ± 9 Da corresponding to lysozyme and an adduct with [Zn2(bicyclam)]4+, respectively. An interaction of Zn-cyclam with HEWL was detected by using Zn{(15N)cyclam}Cl2 and 2D [1H, 15N] NMR spectroscopy (Fig. 4). Of the three cross-peaks for the trans-I, trans-III, and cis-V configurations of Zn-cyclam, those for trans-I and cis-V shifted by 0.04 and 0.13 ppm, respectively, in the presence of HEWL {calculated from [(ΔδHN)2 + (δN × 0.17)2]1/2 (23)}, whereas that for trans-III was unaffected. These experiments show that both zinc cyclam and zinc bicyclam bind to HEWL, and thus suggest that HEWL is a useful model for identification of specific interactions relevant to the binding of metallocyclams to the CXCR4 coreceptor.
Cu2-xylyl-bicyclam also exhibits anti-HIV activity, although it is less active than Zn2-xylyl-bicyclam (8). We used Cu2+-cyclam as a paramagnetic probe to investigate interactions of metallocyclams with the model target protein HEWL in solution. The structures of Cu-cyclam (1·2MeOH, Fig. 5) and Zn-cyclam {[Zn(cyclam)(H2O)2](OAc)2 (15)} are similar.
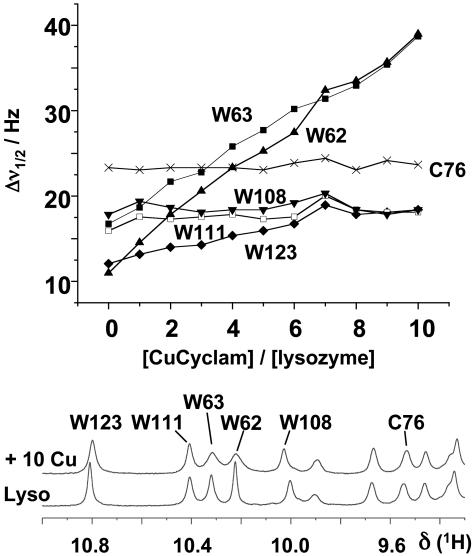
Cu-Cyclam–HEWL Interactions in Solution. Resonances from protons in the protein that are within a few ångstroms of Cu2+ in a bound Cu-cyclam would be expected to broaden significantly on account of the paramagnetism of Cu2+ [3d9; dipolar broadening ∝ r-6 (24), where r = Cu–H distance]. The main features of the 1D 1H NMR spectrum of HEWL were unchanged after addition of up to 10 mol equiv of the paramagnetic complex 1 (Fig. 6), which indicated that the interaction had little effect on the overall protein fold. However, specific Trp resonances broadened, as seen in 1D (Fig. 2) and 2D (Fig. 6) spectra. Line-broadening of tryptophan indole NH resonances decreased in the order (Fig. 2): Trp-62, Trp-63 > Trp-123 >> Trp-108, Trp-111, with Trp-108 and Trp-111 being little affected by Cu-cyclam. Cu-cyclam also had little effect on the Hδ and Hε resonances of the only His residue in lysozyme, His-15. Next we used x-ray diffraction to study the binding of Cu-cyclam and Cu2-bicyclam to crystals of HEWL.
Fig. 2.
Effect of Cu-cyclam on tryptophan indole NH resonances of lysozyme. (Lower) The low-field region of the 1H NMR spectrum of HEWL in 90% H2O/10% D2O before and after addition of 10 mol equiv of the Cu-cyclam complex 1. The pH of the solution (4.6) was chosen so as to be close to that under which lysozyme readily crystallizes. As is evident from the plot of linewidth versus Cu-cyclam concentration (Upper), there are specific paramagnetic broadening effects on Trp-62, Trp-63, and Trp-123, implying that these residues are close to the bound Cu2+. The linewidth of the backbone NH resonance of Cys-76 is also shown for comparison.
Cu-(Bi)Cyclam Binding Sites in Crystalline HEWL. Colorless HEWL crystals turned purple on soaking in solutions containing either Cu-cyclam 1 or Cu2-bicyclam 2 (Fig. 7). The x-ray structures of the adducts were solved to a resolution of 1.75 and 1.6 Å, respectively. The overall structure of lysozyme is not changed by formation of adducts with either complex 1 or complex 2. The positions of the copper atoms in both the monocyclam and bicyclam structures were unequivocally detected as strong peaks in an |Fo| - |Fc| map. In each case, this revealed two binding sites per asymmetric unit.
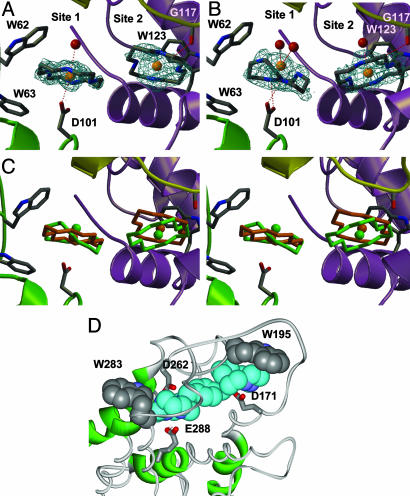
The metal binding positions are similar in the monocyclam and the bicyclam structures. Site 1 is close to the side chain of Asp-101, whereas site 2 is closest to the side chain of Trp-123 and the backbone carbonyl of Gly-117. These residues are on different regions on the protein surface. The packing of the protein molecules in the crystal brings the binding sites close to each other (Fig. 8). The crystal packing and the interaction between the cyclam rings therefore appear to play a crucial role in the formation of the binding site in the crystal.
In the structure of Cu2-bicyclam-HEWL, two cyclam rings were readily identified. Both adopt trans configurations (Fig. 3A). No electron density was present for the p-phenylenebis-(methylene) group that links the two cyclam rings. Although the rings in the two sites are close (Cu–Cu distance 8.45 Å), it was not possible to model the two cyclam rings as part of a single bicyclam unit (therefore only monocyclam rings were built and refined). The next closest cyclam ring in the crystal is too distant (Cu–Cu distance 14.87 Å) to be modeled as part of a single bicyclam. Hence it appears that the cyclam rings in sites 1 and 2 belong to two different Cu2-bicyclam molecules.
Fig. 3.
The two Cu-cyclam and Cu2-bicyclam binding sites in crystals of HEWL. (A) Cu-bicyclam-HEWL. 2|Fo| - |Fc| density is shown around the cyclams at 0.9σ. No electron density is seen for the p-phenylenebis(methylene) linker. (B) Cu-cyclam-HEWL. 2|Fo| - |Fc| density is shown around the cyclams at 0.9σ. (C) Stereoview of an overlay of the cyclam positions in lysozyme. Color code: orange, Cu2-bicyclam; green, Cu-cyclam. The sites are at the intersection of three lysozyme molecules in the crystal (Fig. 8). Molecule 1 is green and contains Asp-101, Trp-62, and Trp-63; molecule 2 is purple and contains Trp-123; and molecule 3 is yellow and contains Gly-117. (D) A model showing [Zn2(xylyl-bicyclam)]4+ bound to the human coreceptor CXCR4 (11). One of the cyclam rings in a trans configuration is stacked on Trp-195 and its Zn is bound to Asp-171. The other cyclam ring has a folded configuration (cis) and is close to Trp-283; its Zn is bound to Asp-262, and two ring NH groups are H bonded to the oxygens of Glu-288.
In Cu-cyclam-HEWL, the Cu in site 1 is in a slightly different position from that in the bicyclam structure, and the cyclam ring in this structure has strong density consistent with a cis configuration. Copper in site 2 is in a position similar to that found in the bicyclam structure, and the planarity of the cyclam ring is indicative of the presence of a trans configuration (Fig. 3B). Cu-cyclam itself in 1·2MeOH adopts the trans-III configuration even though carboxylate groups are present from acetate, the counter anion (Fig. 5). It is evident, therefore, that the protein plays an important role in stabilizing the bound cis configuration in site 1.
Metal Coordination, H Bonding, and Hydrophobic Interactions. In site 2, the cyclams in both the monocyclam and bicyclam structures adopt trans configurations. The trans-III configuration is common for octahedral metal cyclam complexes, and trans-I is common for square-planar metal cyclam complexes (4). The cyclam ring stacks on Trp-123 and a (cyclam)CH···OC(Gly-117) hydrogen bond is formed (Fig. 3). Cu2+ in the monocyclam structure is also very weakly coordinated to a water molecule (Cu–O 3.05 Å). Comparison of the positions of the cyclam in the monocyclam and bicyclam structures reveals that although there is very little movement in the Cu position (0.8 Å), there is a distinct change in the orientation of the cyclam plane (Fig. 3C). This change appears to be a consequence of the difference in binding mode for the cyclam in site 1.
In site 1, the cyclam ring of Cu2-bicyclam has a trans configuration. The Cu2+ in this cyclam coordinates directly to oxygen Oδ1 of the side chain of Asp-101 (Cu–Oδ1 2.7 Å) and also very weakly to a water molecule (Cu–O 3.07 Å) on the opposite side of the ring (Fig. 9). In the monocyclam structure, the cyclam ring adopts a cis configuration. The Cu in this ring does not coordinate directly to Asp-101 and is in a significantly different position from that found in the bicyclam structure (Cu movement 1.61 Å; Fig. 3C). In this folded conformation, two NH···O hydrogen bonds are formed between the carboxylate oxygens of Asp-101 and two NH groups on one face of the cyclam ring (Oδ2–N1 2.70 Å, Oδ1–N10 2.93 Å, Fig. 9). The Cu in this ring coordinates weakly to two water molecules (Cu–O 2.76, 2.88 Å). In both the monocyclam and bicyclam structures, the cyclam in site 1 is sandwiched between the indole rings of Trp-62 and Trp-63 and stacks directly with Trp-62. This hydrophobic interaction with tryptophan would therefore appear to be important for the binding of these cyclams in both cis and trans configurations. Girard et al. (25), in their search for heavy atom derivatives useful in anomalous dispersion methods for solving protein structures, have studied adducts of HEWL with Gd-HPDO3A, the Gd3+ complex of a derivative of the smaller [12]aneN4 macrocycle cyclen (1,4,7,10-tetraazacyclododecane). They were unable to produce Gd-HPDO3A-HEWL by soaking, only by cocrystallization. Intriguingly Gd-HPDO3A binds in the same sites 1 and 2 as Cu-(bi)cyclam (Fig. 10C); the main protein interaction is between Trp-62 and the cyclen macrocycle with no direct metal coordination or involvement of H bonds with Asp-101 (which is turned away from the site).
The difference in the orientation of the cyclam plane in site 2 of the monocyclam and bicyclam structures is likely to be a consequence of the difference in conformation of the cyclam in site 1. In both structures, the cyclams interact weakly with each other (closest ring–ring distance 3.21 Å for bicyclam, 3.34 Å for monocyclam, Figs. 9 and 10). The overlay of the monocyclam and bicyclam structures (Fig. 3C) reveals that the different orientations of the plane conserve the tightness of the cyclam–cyclam interaction (which would otherwise increase to 3.62 Å). It is unclear why the cyclam in site 1 is in the trans configuration in the bicyclam structure compared to the cis configuration for monocyclam. The lack of difference between the protein structures does not provide any clues. It would therefore seem likely that this difference is related to the presence of a linker between the cyclam rings for bicyclam (although the linker is disordered).
What are the important factors in stabilization of the cis cyclam configuration? In crystals of the zinc complex of the drug xylyl-bicyclam, [Zn2(xylyl-bicyclam)(OAc2)2](OAc)2·2MeOH, one acetate is bidentate and the other forms double H bonds to the NH groups on the opposite face of the cyclam ring that adopts the folded cis-V configuration (11). In the Cu-cyclam-HEWL structure, there is no direct coordination of Cu2+ to a carboxylate, only NH hydrogen bonding, and the carboxylate oxygen coordination sites appear to be occupied by weakly bound water molecules. cis-Metallocyclams with two bound aqua ligands are unusual, although Barefield et al. (26) prepared cis-[Ni(cyclam)(H2O)2]Cl2·2H2O by displacement of a chelated ethylenediamine by water under acidic conditions. In solution, the interaction of carboxylate groups with Zn2+ cyclams readily leads to the formation of the folded cis-V configuration, equilibria being reached within 1 h at millimolar concentrations, at 298 K (11, 15).
CXCR4 Binding Sites. Previously (11) we built a model of the human CXCR4 coreceptor based on the x-ray structure of rhodopsin (27) and docked Zn2-bicyclam such that there is direct coordination of Zn2+ in one cyclam ring to the carboxylate of Asp-171, and of Zn2+ in the other to Asp-262. In the latter site, Glu-288 on the opposite side of the ring can form double H bonds to two ring NH groups, and the macrocycle folds into the cis-V configuration. In the first site, a trans-I or trans-III configuration is more likely for the cyclam ring. Inspection of the same CXCR4 model now reveals that there are tryptophan indole rings close to each of the docked cyclams (Fig. 3D). Notable is the stacking of Trp-195 on the trans-cyclam ring that contains Zn2+ bound to Asp-171. These are all features seen in the HEWL-(bi)cyclam structures. The hydrophobic interaction of the N(CH2)2N and N(CH2)3N backbone of cyclam with tryptophan indole rings has a parallel in the hydrophobic contact of the N(CH2)3CH cyclic side-chain of proline with tryptophan at some protein–protein interfaces, often as a Trp-Pro-Trp sandwich, e.g., the IgE receptor (28) (Fig. 10D). Such hydrophobic interactions may therefore play a key role in the binding of cyclam anti-HIV complexes to the CXCR4 coreceptor.
Conclusions
Cyclams, macrocyclic tetraamines, have a wide range of potential uses in diagnosis and therapy (4). Antiviral bicyclams target the membrane coreceptor protein CXCR4 (1), and coreceptor binding is enhanced by the presence of Ni2+, Cu2+, or Zn2+ in the cyclam ring (9). Currently there are only structural models of CXCR4–cyclam adducts (9–11). These models have emphasized the importance of aspartate carboxylate side chains of CXCR4 in forming H bonds to cyclam NH groups and in coordination to cyclam-bound metal ions. Cyclam rings in metallocyclam complexes have configurational flexibility and in one model a folded cis configuration is stabilized by interactions with carboxylate side chains (11). Although it would be desirable to carry out structural studies directly on CXCR4, such transmembrane proteins are very difficult to crystallize. To date, the only member of this class of proteins for which the x-ray crystal structure has been solved is bovine rhodopsin (27). We found that anti-HIV metallocyclams bind to HEWL, which is therefore a useful model for CXCR4.
The x-ray crystal structures of Cu-cyclam and Cu2-bicyclam complexes of lysozyme establish that protein recognition can involve a range of both polar and nonpolar interactions. These include H bonding between the cyclam ring and protein carboxylate groups, which, for the Cu-cyclam structure, anchors the cyclam ring in the unusual folded cis configuration without direct coordination of Cu2+ to the protein. The presence of an additional cyclam ring, as in a bicyclam, can influence the configuration adopted by the bound macrocycle. Intriguingly, hydrophobic interactions between the cyclam ring and the indole ring of tryptophan, reminiscent of tryptophan-proline sandwiches often found at protein–protein interfaces, occur not only in crystals of HEWL but also in aqueous solution. Such tryptophan stacking interactions are also possible in models of CXCR4 adducts of metallocyclams, which contain at the same time other features important for metallocyclam docking, including metal coordination to Asp-171 and Asp-262 (Fig. 3D). Tryptophan, an amphipathic amino acid, is often located toward the surface (interfacial regions) of membrane proteins (29), and may play a crucial role in the recognition of antiviral metallocyclams and other macrocycles by the protein coreceptor. Knowledge of these diverse interactions will aid the design of new generations of chemokine receptor antagonists.
Supplementary Material
Acknowledgments
We thank Dr. Claudia Blindauer for advice and assistance with NMR experiments and Robert Smith for his help with mass spectrometry. We thank the Wellcome Trust (U.K.) for support for the Edinburgh Protein Interaction Centre (EPIC), and the Biotechnology and Biological Sciences Research Council for a studentship for T.M.H.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: HEWL, hen egg white lysozyme.
Data deposition: The atomic coordinates and structure factors have been deposited in the Cambridge Structural Database, Cambridge Crystallographic Data Centre, Cambridge CB2 1EZ, United Kingdom (CSD reference no. 260045) and the Protein Data Bank, www.pdb.org (PDB ID codes 1YIK and 1YIL).
References
- 1.Schols, D., Esté, J. A., Henson, G. & De Clercq, E. (1997) Antiviral Res. 35, 147-156. [DOI] [PubMed] [Google Scholar]
- 2.De Clercq, E., Yamamoto, N., Pauwels, R., Baba, M., Schols, D., Nakashima, H., Balzarini, J., Debyser, Z., Murrer, B. A., Schwartz, D., et al. (1992) Proc. Natl. Acad. Sci. USA 89, 5286-5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bridger, G. J. & Skerlj, R. T. (1999) Adv. Antiviral Res. 3, 161-229. [Google Scholar]
- 4.Liang, X. & Sadler, P. J. (2004) Chem. Soc. Rev. 33, 246-266. [DOI] [PubMed] [Google Scholar]
- 5.De Clercq, E. (2003) Nat. Rev. Drug Discov. 2, 581-587. [DOI] [PubMed] [Google Scholar]
- 6.Izatt, R. M., Pawlak, K., Bradshaw, J. S. & Bruening, R. L. (1991) Chem. Rev. 91, 1721-1785. [Google Scholar]
- 7.Paisey, S. J. & Sadler, P. J. (2004) Chem. Commun. 306-307. [DOI] [PubMed]
- 8.Esté, J. A., Cabrera, C., De Clercq, E., Struyf, S., Van Damme, J., Bridger, G., Skerlj, R. T., Abrams, M. J., Henson, G., Gutierrez, A., et al. (1999) Mol. Pharmacol. 55, 67-73. [DOI] [PubMed] [Google Scholar]
- 9.Gerlach, L. O., Jakobsen, J. S., Jensen, K. P., Rosenkilde, M. R., Skerlj, R. T., Ryde, U., Bridger, G. J. & Schwartz, T. W. (2003) Biochemistry 42, 710-717. [DOI] [PubMed] [Google Scholar]
- 10.Gerlach, L. O., Skerlj, R. T., Bridger, G. J. & Schwartz, T. W. (2001) J. Biol. Chem. 276, 14153-14160. [DOI] [PubMed] [Google Scholar]
- 11.Liang, X., Parkinson, J. A., Weishäupl, M., Gould, R. O., Paisey, S. J., Park, H.-S., Hunter, T. M., Blindauer, C. A., Parsons, S. & Sadler, P. J. (2002) J. Am. Chem. Soc. 124, 9105-9112. [DOI] [PubMed] [Google Scholar]
- 12.Bosnich, B., Poon, C. K. & Tobe, M. L. (1965) Inorg. Chem. 4, 1102-1108. [Google Scholar]
- 13.Vaney, M. C., Maignan, S., Ries-Kautt, M. & Ducruix, A. (1996) Acta Crystallogr. D 52, 505-517. [DOI] [PubMed] [Google Scholar]
- 14.Perkins, S. J., Johnson, L. N., Machin, P. A. & Phillips, D. C. (1979) Biochem. J. 181, 21-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang, X., Weishäupl, M., Parkinson, J. A., Parsons, S., McGregor, P. A. & Sadler, P. J. (2003) Chem. Eur. J. 9, 4709-4717. [DOI] [PubMed] [Google Scholar]
- 16.Sheldrick, G. M. (2004) sadabs (University of Göttingen, Germany).
- 17.Beurskens, P. T., Beurskens, G., Bosman, W. P., de Gelder, R., Garcia Granda, S., Gould, R. O., Israel, R. & Smits J. M. M. (1996) dirdif(Crystallography Laboratory, University of Nijmegen, The Netherlands).
- 18.Betteridge, P. W., Carruthers, J. R., Cooper, R. I., Prout, K. & Watkin, D. J. (2003) J. Appl. Crystallogr. 36, 1487. [Google Scholar]
- 19.Leslie, A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography 26.
- 20.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 21.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 22.Noda, Y., Yokota, A., Horii, D., Tominaga, T., Tanisaka, Y., Tachibana, H. & Segawa, S.-I. (2002) Biochemistry 41, 2130-2139. [DOI] [PubMed] [Google Scholar]
- 23.Farmer, B. T., II, Constantine, K. L., Goldfarb, V., Friedrichs, M. S., Wittekind, M., Yanchunas, J., Jr., Robertson, J. G. & Mueller, L. (1996) Nat. Struct. Biol. 3, 995-997. [DOI] [PubMed] [Google Scholar]
- 24.Bertini, I. & Luchinat, C. (1996) Coord. Chem. Rev. 150, 1-292. [Google Scholar]
- 25.Girard, E., Chantalat, L., Vicat, J. & Kahn, R. (2002) Acta Crystallogr. D 58, 1-9. [DOI] [PubMed] [Google Scholar]
- 26.Barefield, E. K., Bianchi, A., Billo, E. J., Connolly, P. J., Paoletti, P., Summers, J. S. & Van Derveer, D. G. (1986) Inorg. Chem. 25, 4197-4202. [Google Scholar]
- 27.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T., Stenkamp, R. E., et al. (2000) Science 289, 739-745. [DOI] [PubMed] [Google Scholar]
- 28.Garman, S. C., Sechi, S., Kinet, J.-P. & Jardetzky, T. S. (2001) J. Mol. Biol. 311, 1049-1062. [DOI] [PubMed] [Google Scholar]
- 29.Ulmschneider, M. B. & Sansom, M. S. P. (2001) Biochim. Biophys. Acta 1512, 1-14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.