Abstract
The aim of this study was to investigate the association of circulating miRNAs profile with the risk of knee osteoarthritis (OA), and evaluate their correlation with clinical characteristics. This study was divided into two parts: exploration stage and validation stage. In exploration stage, 8 knee OA patients and 8 age and gender highly matched health controls (HCs) were recruited, and plasma sample were collected for microarray examination. Differentially expressed miRNAs and enrichment analysis were subsequently performed. In validation stage, 100 knee OA patients and 100 age and gender matched HCs were enrolled, and Top 8 differentially expressed miRNAs in microarray were selected for further validation by qPCR. In exploration stage, 41 up-regulated miRNAs and 29 down-regulated miRNAs were identified by microarray, and enrichment analysis disclosed these miRNAs were involved in inflammation- and immunity- related process. Top 8 differentially expressed miRNAs in microarray were determined in the validation stage, and miR-19b-3p, miR-92a-3p, miR-122-5p, miR-486-5p and miR-320b expression were increased in knee OA. Univariate and multivariate logistic analysis showed only miR-19b-3p, miR-122-5p and miR-486-5p were independent factors for knee OA risk, and ROC curve showed combination of miR-19b-3p, miR-122-5p and miR-486-5p has a great diagnostic value for knee OA. Besides, miR-19b-3p and miR-486-5p positively correlates with disease severity. This study revealed that circulating miRNA profiles played a key role in knee OA diagnosis, and combined measurement of miR-19b-3p, miR-122-5p and miR-486-5p could be served as a novel and promising biomarker for diagnosis and disease severity of knee OA.
Keywords: Knee osteoarthritis, biomarker, miR-19b-3p, miR-122-5p, miR-486-5p
Introduction
Osteoarthritis (OA) is common progressive joint disease characterized by degenerative changes in articular cartilage and subchondral bone with synovial inflammation and bone remodeling [1]. It is estimated that approximately 26 million adults were affected by OA during 2010-2012 [2], and 67 million people will be suffered by OA in 2030 in the US [3]. As a dominant type of OA, knee OA is characterized by degeneration of knee joint, pain, swelling as well as snapping joints, especially in elderly patients [4]. Unfortunately, Knee OA is usually only detected in late stage of disease progression by insensitive clinical symptom and radiographic findings. Obviously, there is a huge challenge of identifying patients with early-stage knee OA damages. To timely diagnose and treat, effective and reliable biomarkers of knee OA are deadly needed. A number of investigators have proposed biochemical markers based on cartilage, synovium or bone for evaluating diagnosis and prognosis of OA [5], but limited convincing biomarkers for early diagnosis of knee OA were discovered to facilitate timely treatment.
MicroRNA (miRNAs), endogenous RNAs with about 23 nucleotides, can regulate protein expression by pairing to the mRNAs of protein-coding genes and guiding their post-transcriptional repression [6], and play critical roles in regulating cell differentiation, progression and apoptosis [7]. MicroRNAs have been reported to play a serious of roles in the development and progression OA, and more than 30 miRNAs are abnormally expressed in OA samples [8]. Cartilage degradation has been reported to associate with deregulation of miR-25, miR-145, MiR-455-3p, miR-483-5p and miR-675; while inflammation was correlated with miR-203 and miR-483 [9]. Several deregulated miRNAs have also been proposed as biomarkers of important pathological inflammation or skeletal damage to evaluate disease progression or treatment response [10-13]. In addition to studies on miRNAs expressions from tissues, miRNAs have been investigated in some other pathology in circulation, which demonstrated the associations of their circulating level and tissue expression in different disease states [14-17]. However, limited studies have revealed association between knee OA and aberrant-expressed plasma miRNAs, which might be developed to be a reliable and sensitive biomarker for knee OA.
The aim of this study was to investigate the association of circulating miRNAs profile with the risk of knee OA, and evaluate their correlation with clinical characteristics, subsequently identify convincing biomarkers for diagnosis and disease severity in knee OA patients.
Materials and methods
Participants
100 patients with knee OA according to 1986 classification of osteoarthritis of knee in diagnostic criteria of the American Rheumatism Association [18], from May 2016 to Nov 2016, at the department of Rheumatology & Immunology in Changhai Hospital were recruited in this study. In the meanwhile, 100 age and gender matched health volunteers at the department of Physical Examination were also enrolled as health controls (HCs). All the participants provided written informed consents. This study was approved by Ethics Committee of Changhai Hospital.
Study design
This study was divided into two parts: exploration stage and validation stage (Figure 1). In exploration stage, 8 knee OA patients and 8 age as well as gender highly matched HCs were selected from the total participants, and miRNA profiles were detected by microarray. Differentially expressed miRNAs were determined subsequently and enrichment analysis on the disease (HMDD2), GO BP, KEGG Pathway, Organ, miRNA family and clusterwas performed. In validation stage, top 8 differentially expressed miRNAs were selected to further validate in total participants (100 knee OA patients and 100 HCs) by qPCR. And the association of candidate circulating miRNAs with OA risk and clinical features of OA was determined.
Figure 1.
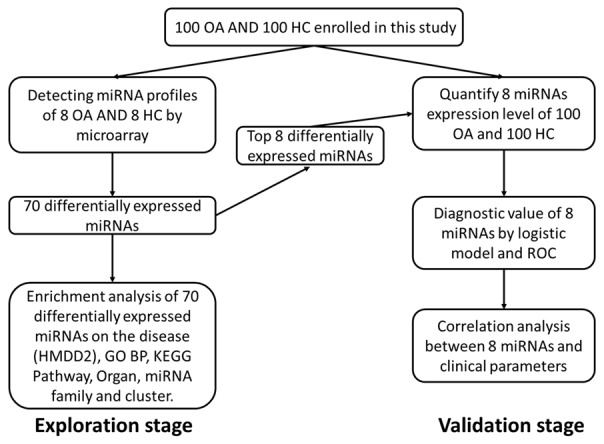
Study flow. This study was divided into two parts: exploration stage and validation stage. In the exploration stage, plasma sample from 8 knee OA patients and 8 HCs were obtained, and microarray were performed (Left). Top 8 differentially expressed miRNAs in microarray was subsequently determined in 100 knee OA patients and 100 HCs for further validation (Right).
Sample acquisition and handling
8 mL aliquot of blood was obtained from all participants directly into sodium citrate tubes. The whole blood was allowed to stand for ~3 h at -4°C before centrifuging at 1,500 g for 10 min at room temperature. The resultant plasma was aliquoted into Eppendorf tubes and stored at -70°C.
RNA isolation
Total RNA was isolated with LeukoLOCK kit (Ambion) according to the specific protocol to capture small RNAs. Quality, quantity and integrity of RNA were measured by a NanoDrop spectrophotometer (ND-1000, Nanodrop Technologies) and Ry gel electrophoresis, respectively.
Microarray hybridization
Total RNAs (500 ng) from 8 Knee OA patients and 8 HCs were labeled using the FlashTag Biotin labeling kit (Genisphere) and hybridized to the GeneChip miRNA 4.0 Array (Affymetrix) respectively, which covered 2578 human mature miRNA, 2025 pre-miRNA and 1996 snoRNA. In brief, RNA molecules were first polyadenylated and followed by a ligation step with a biotin-labeled DNA molecule attached, and then labeled RNA was hybridized to the array, finally washed and stained in a GeneChip Fluidics Station 450 and scanned in a GeneChip Scanner 7G (Affymetrix).
Data preprocessing and differentially expressed miRNAs screening in microarray
Considering the intrinsic background of different chips might have impact on the calculating the expression values, the raw data of each chip were normalized via the Geoquery package (version 2.34.0) in R language [19]. When data have been transformed to log2 expression, the Limma package in R language was used to identify the differentially expressed miRNAs in knee OA and HC samples. P values were adjusted by method of Benjamini and Hochberg procedure to control false discovery rate [20]. The statistical significance was defined as P<0.05, and the clinical significance was defined as a difference of at least 1.5 folds, abs (log2 (fold change)) >1.5.
For illustrate the pattern of differentially expressed miRNAs, Principal Components Analysis (PCA) plot and hierarchical clustering analysis were performed by R and heatmap package (version 1.0.2, available at http://cran.r-project.org/web/packages/pheatmap/index.html).
Enrichment analysis of differentially expressed miRNAs
miEAA database was annotated to differentially expressed miRNAs [21], which including miRNA-related disease (HMDD), KEGG pathway, GO BP and Organ. The enrichment analysis of differentially expressed miRNAs and its precursor were completed by the Fisher’s exact test to identify overrepresented miRNA-related items.
qPCR validation
Eight miRNAs showing highest fold changes in keen OA in the microarray experiment were selected for further validation: miR-19b-3p, miR-92a-3p, miR-122-5p, miR-486-5p, miR-663a, miR-320b, miR-887-5p and miR-1180-3p.
Samples of 100 knee OA patients and 100 HCs were used for qPCR validation by TaqMan miRNA assays. After RNA (100 ng) reserve transcribed to cDNA with the TaqMan microRNA Reverse Transcription kit (Lif Technologies) under the instruction of manufacturer’s protocol, cNDA (15 ng) was amplified in thermal cycler (7900 HT) according to the manufacturer’s protocol. U6 was employed as endogenous controls due to its wide usage and limited variance. The 2-∆∆CT method was used to express the levels of miRNAs.
Statistics
Data were analyzed using the SPSS (version 20.0, SPSS, Inc., Chicago, Illinois). The Kolmogorov-Smirnov test was used to assess the normality of continuous data. To compare the expression of miRNAs between groups, a Mann-Whitney U test was performed. Categorical variables were presented as counts and proportions and were compared using Chi-square test or Fisher’s exact test. To determine the correlation of the miRNA expression and diagnostic value, univariate and multivariate logistic regression analysis were used. Receiver operating characteristic (ROC) curve was used to assess the diagnostic value of miRNAs for OA. Spearman’s correlation coefficient was used to test the correlation between the expression of miRNAs and clinical index. P<0.05 was considered significant.
Results
Characteristics of participants in exploration stage
8 knee OA patients and 8 HCs were included in the exploration stage, and miRNA profiles were detected in plasma by microarray (Figure 1). The characteristics were presented in Table 1. There were 62.5% female in knee OA group with age 51.13±2.03 years, 62.5% female in HC group with age 50.75±1.67 years, which showed no difference between groups.
Table 1.
Demographic characteristic of Knee OA patients and HCs in exploration stage
| Parameters | Knee OA | HC | P value |
|---|---|---|---|
| Number | 8 | 8 | |
| Age (Years) | 51.13±2.03 | 50.75±1.67 | 0.6930 |
| Gender (Female) | 5 (62.5%) | 5 (62.5%) | - |
Data distribution was described by mean ± standard deviation for continues variables and count (percentage) for categorical variables. Significance of the comparison was determined by t test for continues variables or Chi-squared test for categorical variables. P valve <0.05 was considered significant. OA, osteoarthritis; HC, health control.
Differentially expressed miRNAs in microarray
The Principal Components Analysis (PCA) plot of the eight pairs of samples in microarray was shown in Figure 2, demonstrating the first component vs. the second component (Figure 2A) and the first component vs. the third component (Figure 2B). This revealed that the miRNA profiles of knee OA and HC samples clustered irrespectively and relatively independent, which indicated the similarity of these eight pairs of samples and feasibility to differentiate disease and healthy based on miRNA patterns.
Figure 2.
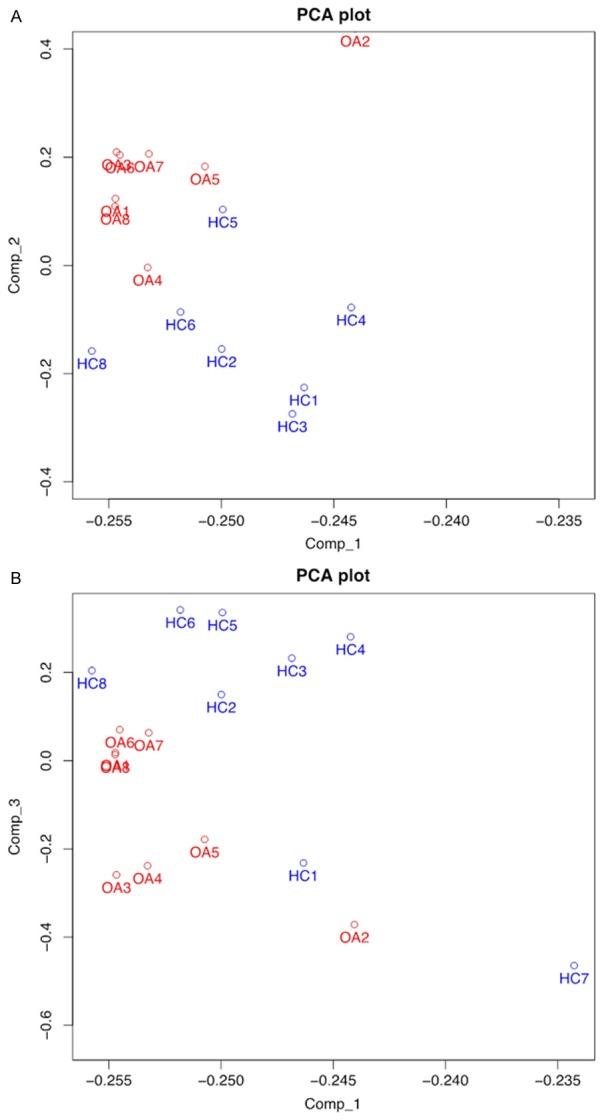
Principal Components Analysis (PCA) of microarray. The result of first component vs. second component (A) and first component vs. the third component (B) revealed that the miRNA profiles of knee OA and HC samples clustered irrespectively and relatively independent, which implied knee OA patients and HCs could be differentiated by the miRNA patterns.
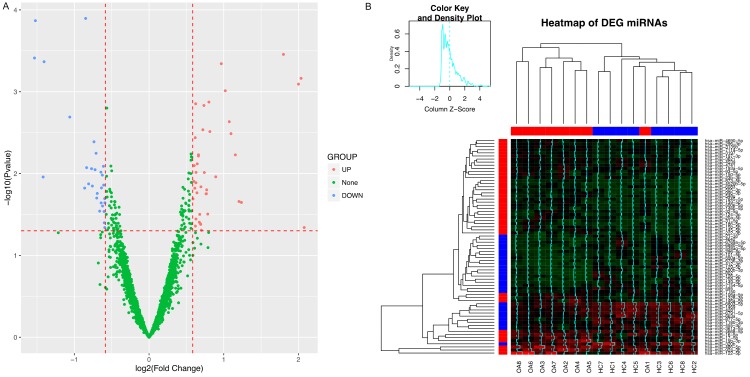
Comparing the miRNA profiles between knee OA and HC by limma package, 70 differentially expressed miRNAs were identified according to the standard in method section. The volcano plot of the differentially expressing miRNAs demonstrated 41 up-regulated miRNAs and 29 down-regulated miRNAs (Figure 3A). The heatmap disclosed differentially expressed miRNAs were aggregated in blocks (Figure 3B) (Row: upregulation in red, downregulation in blue; column: disease in red, control in blue), then disease and control could be distinguished with the exception of knee OA.
Figure 3.
Differentially expressing miRNAs analysis of microarray. A. 41 up-regulated miRNAs and 29 down-regulated miRNAs were observed in microarray. B. Heatmap. Upregulation was presented in red, while downregulationshown in blue. Heat map disclosed that differentially expressed miRNAs were aggregated in blocks.
Enrichment analysis
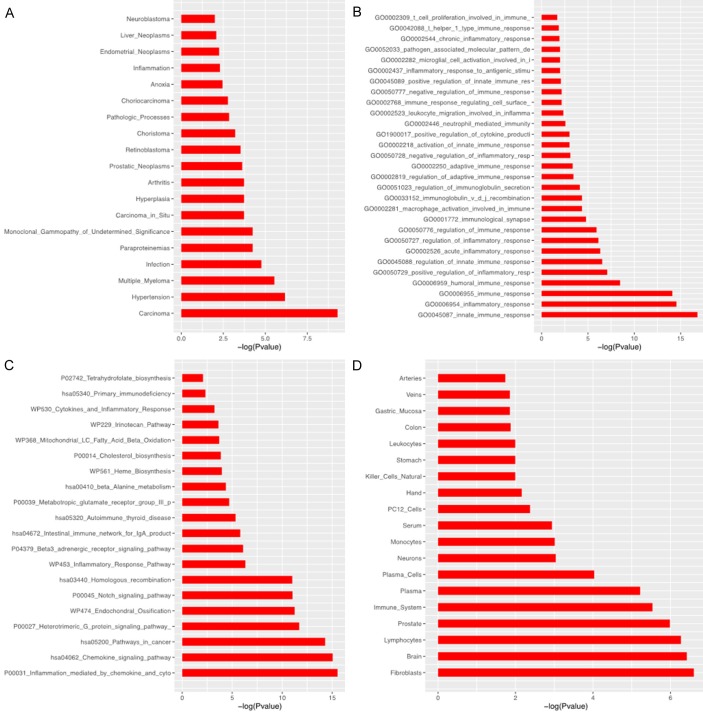
The enrichment analysis of 70 differentially expressed miRNAs was performed including disease (HMDD2), biology process (GO), pathway (KEGG) and organ (Figure 4). The result indicated these differentially expressed miRNAs were prone to express in blood or joints and were involved in inflammation- and immunity-related process. Based on these 70 mature miRNAs, 78 corresponding precursors were identified, among which miR-17 (17.85 folds) and miR-15 (21.42 folds) clusters and families of the differential expressing miRNAs were identified (Table 2).
Figure 4.
The enrichment analysis of differentially expressing miRNAs. HMDD2 analysis (A) presented that differentially expressing miRNAs correlated with arthritis; Biology process (GO) and pathway (KEGG) analysis showed differentially expressing miRNAs were involved in inflammation- and immunity-related process (B) and pathways (C). Besides, they were prone to express in blood or joints (D).
Table 2.
Enrichment analysis in miRNA clusters and families of the differential expressing miRNAs by Fisher’s exact test
| Items | Group | Nseta | Fold change | P value | Hitb |
|---|---|---|---|---|---|
| chr13_91351314_91351391 | Cluster | 6 | 23.79 | 1.18E-08 | mir-17; mir-20a; mir-19b-1; mir-92a-1 |
| chr3_160404745_160404825 | Cluster | 2 | 35.69 | 5.62E-06 | mir-15b; mir-16-2 |
| chrX_134169378_134169452 | Cluster | 6 | 17.85 | 9.22E-06 | mir-92a-2; mir-19b-2; mir-106a |
| mir-17 | Family | 8 | 17.85 | 2.59E-07 | mir-17; mir-20a; mir-93; mir-106a |
| mir-15 | Family | 5 | 21.42 | 2.28E-06 | mir-16-1; mir-15b; mir-16-2 |
Data was presented by Items, Group, Nset, Fold Change, P-value and hit. The enrichment analysis in miRNA clusters and families of the differential expressing miRNA was determined by Fisher’s exact test. P value <0.05 was considered significant.
Nset, the size of given miRNA cluster or family;
hit, differential expressing miRNAs in given cluster or family.
Characteristics of participants in validation stage
100 knee OA patients and 100 HCs were included in the validation stage (Figure 1). The demographic and clinical characteristics of knee OA patients and HCs were listed in Table 3. There were no significant differences between groups in age (51.69±8.78 vs. 51.09±9.23, P=0.6384) or gender (69% vs. 61% female, P=0.2356).
Table 3.
Demographic and clinical characteristic of Knee OA patients and HC in validation stage
| Parameters | Knee OA | HC | P value |
|---|---|---|---|
| Number | 100 | 100 | |
| Age (Years) | 51.69±8.78 | 51.09±9.23 | 0.6384 |
| Gender (Female) | 69 (69%) | 61 (61%) | 0.2356 |
| Disease duration (months) | 33.19±14.08 | ||
| VAS pain rest | 3.28±1.83 | ||
| VAS pain walking | 3.5±1.67 | ||
| WOMAC pain | 10.74±3.49 | ||
| WOMAC stiffness | 5.11±1.29 | ||
| WOMAC function | 38.02±12.14 | ||
| WOMAC total | 53.87±14.12 |
Data distribution was described by mean ± standard deviation for continues variables and count (percentage) for categorical variables. Significance of the comparison was determined by t test for continues variables or Chi-squared test for categorical variables. P valve<0.05 was considered significant. OA, osteoarthritis; HC, health control; VAS, visual analogue scale; WOMAC, Western Ontario and McMaster Universities Osteoarthritis Index.
Expressions of candidate miRNAs in qPCR
Top 8 differentially expressing miRNAs in microarray were selected as candidate for qPCR validation which listed in Table 4. As presented in Figure 5, the expression of miR-19b-3p, miR-92a-3p, miR-122-5p, miR-486-5p were up-regulated in knee OA patients compared with HCs, which was consistent with the results from microarray. And miR-320b level was increased as well in opposite to the result of microarray. Besides, no difference in miR-663a, miR-887-5p and miR-1180-3p was observed between two groups which disagreed with data from microarray.
Table 4.
Top 8 differentially expressing miRNAs in microarray
| MiRNA | LogFC | AveExpr | P value | Sig |
|---|---|---|---|---|
| miR-122-5p | 2.08 | 4.59 | 0.0456 | UP |
| miR-92a-3p | 2.03 | 3.84 | 0.0007 | UP |
| miR-19b-3p | 2.00 | 2.54 | 0.0008 | UP |
| miR-486-5p | 1.80 | 4.14 | 0.0003 | UP |
| miR-877-5p | -1.53 | 2.40 | 0.0004 | DOWN |
| miR-1180-3p | -1.52 | 2.20 | 0.0001 | DOWN |
| miR-320b | -1.42 | 3.96 | 0.0110 | DOWN |
| miR-663a | -1.41 | 2.72 | 0.0004 | DOWN |
Top 8 differentially expressing miRNAs (according to abs (logFC)) were presented by the logFC, AveExpr, P-value and Sig. Significance of the comparison was completed by limma package in R. P value <0.05 was considered significant. LogFC, log2 (fold change); AveExpr, average of expression level; UP, Up-regulated; Down, Down-regulated.
Figure 5.
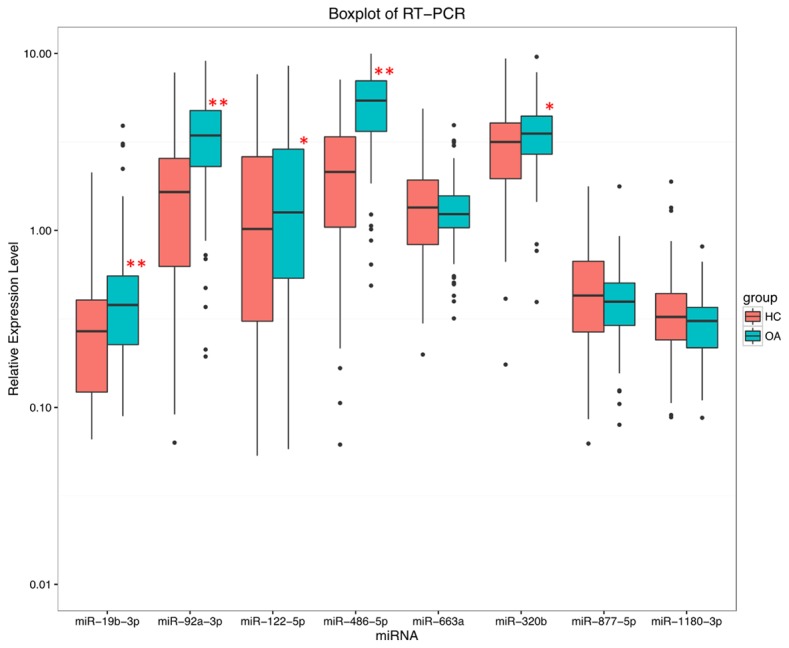
Expressions of candidate eight miRNAs by qPCR. Five miRNAs were observed to be dysregulated in knee OA patients compared to HCs by qPCR, among which miR-19b-3p, miR-92a-3p, miR-122-5p, miR-486-5p and miR-320b were all up-regulated. No difference was discovered in miR-663a, miR-877-5p, miR-1180-3p between knee OA patients and HCs.
Diagnostic value of candidate miRNAs for knee OA
In order to investigate the diagnostic value of the candidate miRNAs for OA, logistic regression analysis was performed. The univariate logistic regression found that miR-19b-3p (P=0.001), miR-92a-3p (P<0.001), miR-486-5p (P<0.001) and miR-320b (P=0.019) expressions increases the risk of OA (Table 5). All factors with a P value <0.1 were subsequently included in the multivariate logistic regression (stepwise) analysis, which demonstrated that only miR-19b-3p (P<0.001), miR-122-5p (P=0.037) and miR-486-5p (P<0.001) levels were independent factors for OA risk. The independent predicting miRNAs were further analyzed by ROC curve to build a model of circulating miRNAs for OA diagnosis. As presented in Figure 6, combination of miR-19b-3p, miR-122-5p and miR-486-5p expressions could diagnose knee OA with dramatically high area under curve (AUC): 0.926, 95% CI 0.885-0.967, with sensitivity 80.0% and specificity 88.0% at the best cut-off point.
Table 5.
Univariate and multivariate logistic regression analysis of miRNAs for predicting OA states
| miRNA | Univariate logistic | Multivariate logistic (stepwise) | ||||||
|---|---|---|---|---|---|---|---|---|
| OR |
|
|||||||
| 95% CI | P value | OR | 95% CI | P value | ||||
|
|
|
|||||||
| Lower | Higher | Lower | Higher | |||||
| miR-19b-3p | 3.91 | 2.61 | 5.86 | 0.001 | 3.83 | 2.16 | 6.78 | <0.001 |
| miR-92a-3p | 2.73 | 1.95 | 3.82 | <0.001 | - | - | - | - |
| miR-122-5p | 1.24 | 1.08 | 1.42 | 0.069 | 1.24 | 1.01 | 1.53 | 0.037 |
| miR-486-5p | 6.56 | 3.79 | 11.38 | <0.001 | 7.12 | 3.59 | 14.09 | <0.001 |
| miR-663a | 1.06 | 0.75 | 1.48 | 0.055 | - | - | - | - |
| miR-320b | 2.35 | 1.54 | 3.59 | 0.019 | - | - | - | - |
| miR-877-5p | 0.90 | 0.65 | 1.25 | 0.016 | - | - | - | - |
| miR-1180-3p | 0.46 | 0.29 | 0.73 | 0.151 | - | - | - | - |
Data was presented by P value, OR and 95% CI. The value of miRNAs to predict OA states were tested by univariate and multivariate logistic regression model. P valve <0.05 was considered significant. OA, osteoarthritis; OR, odds ratio; 95% CI, 95% confidence interval.
Figure 6.
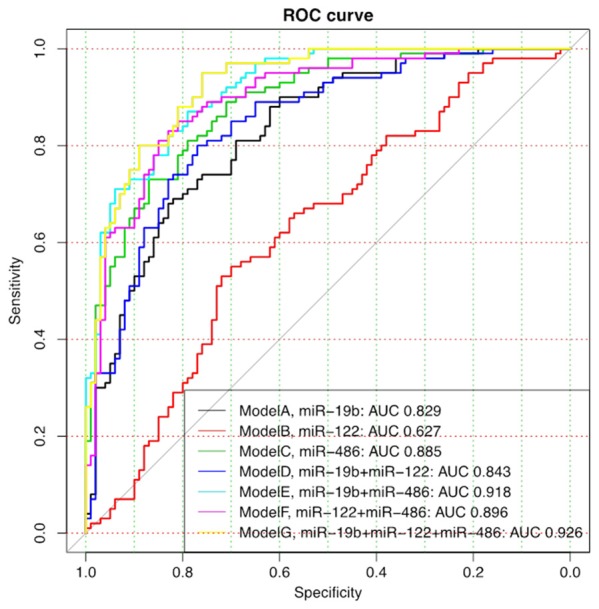
ROC curve analysis of candidate miRNAs for knee OA diagnosis. Seven models were constructed to investigate the diagnostic value of miR-19b, miR-122 and miR-486 for knee OA. miR-19b, miR-122 and miR486 were firstly analyzed alone by ROC curve for knee OA risk (Model A-C), and then combined each other for the ROC curve analysis (Model D-F). And at last, all the three miRNAs were combined to analyze the comprehensive predictive value for knee OA risk (Model G), which illuminated a great diagnostic value with AUC 0.926.
Correlation of candidate miRNAs with clinical features of knee OA
Further correlation analysis between the expression level of candidate miRNAs and clinical features of OA patients were performed. As shown in Table 6, miR-19b-3p was positively correlated with disease duration (P<0.001), pain VAS walking (P=0.010), WOMAC pain (P=0.002) and WOMAC stiffness (P=0.017) scores, while miR-122-5p was only negatively correlated with pain VAS walking score (P=0.018). As to miR-486-5p, it was positively associated with age (P<0.001), disease duration (P=0.022), pain VAS rest (P=0.004), pain VAS walking (P=0.001), WOMAC pain (P=0.002), WOMAC function (P=0.002) and WOMAC total scores (P<0.001). Other detailed correlation information was presented in Table 6.
Table 6.
The correlation analysis between the expression level of miRNAs and characteristic of knee OA patients (n=100)
| Parameters | miR-19b-3p | miR-92a-3p | miR-122-5p | miR-486-5p | miR-663a | miR-320b | miR-877-5p | miR-1180-3p | |
|---|---|---|---|---|---|---|---|---|---|
| Age | r | 0.128 | 0.254* | -0.086 | 0.423** | 0.186 | 0.036 | -0.038 | 0.040 |
| P_value | 0.204 | 0.011 | 0.397 | <0.001 | 0.304 | 0.721 | 0.706 | 0.692 | |
| Gender | r | 0.186 | 0.086 | -0.056 | 0.034 | 0.040 | -0.154 | 0.075 | -0.004 |
| P_value | 0.063 | 0.396 | 0.581 | 0.740 | 0.693 | 0.127 | 0.456 | 0.970 | |
| Disease duration | r | 0.354** | 0.150 | -0.045 | 0.222* | 0.154 | 0.069 | 0.051 | 0.085 |
| P_value | <0.001 | 0.137 | 0.654 | 0.026 | 0.127 | 0.495 | 0.615 | 0.403 | |
| VAS pain rest | r | 0.142 | 0.201 | 0.085 | 0.285** | 0.103 | 0.224* | -0.093 | 0.110 |
| P_value | 0.159 | 0.076 | 0.403 | 0.004 | 0.309 | 0.025 | 0.357 | 0.274 | |
| VAS pain walking | r | 0.258** | 0.195 | -0.236* | 0.336** | 0.352** | 0.078 | 0.026 | 0.044 |
| P_value | 0.010 | 0.115 | 0.018 | 0.001 | <0.001 | 0.438 | 0.800 | 0.666 | |
| WOMAC pain | r | 0.302** | 0.184 | 0.103 | 0.301** | 0.135 | 0.146 | 0.005 | 0.067 |
| P_value | 0.002 | 0.159 | 0.306 | 0.002 | 0.182 | 0.147 | 0.964 | 0.505 | |
| WOMAC stiffness | r | 0.239* | -0.032 | 0.080 | 0.116 | 0.185 | 0.079 | 0.261** | 0.115 |
| P_value | 0.017 | 0.754 | 0.427 | 0.250 | 0.066 | 0.434 | 0.009 | 0.256 | |
| WOMAC function | r | 0.122 | 0.202 | 0.137 | 0.310** | 0.037 | -0.064 | -0.095 | -0.076 |
| P_value | 0.226 | 0.104 | 0.174 | 0.002 | 0.717 | 0.525 | 0.349 | 0.454 | |
| WOMAC total | r | 0.191 | 0.198 | 0.141 | 0.352** | 0.088 | 0.008 | -0.022 | -0.030 |
| P_value | 0.057 | 0.184 | 0.162 | <0.001 | 0.383 | 0.938 | 0.829 | 0.771 |
Data was presented as r and P value. Correlation was determined by Spearman test.
P value <0.05 was considered significant;
P value <0.01 was considered extremely significant.
OA, osteoarthritis; VAS, visual analogue scale; WOMAC, Western Ontarioand McMaster Universities Osteoarthritis Index.
Discussion
In the present study, we screened plasma miRNAs with aberrant expressions in patients with keen OA compared with healthy controls by microarray and verified by quantitative PCR. Our results revealed: (1) 41 up-regulated miRNAs and 29 down-regulated miRNAs were detected by microarray, which were prone to express in blood or joints, and were involved in inflammation- and immunity- related process in enrichment analysis; (2) In validation stage, miR-19b-3p, miR-122-5p and miR-486-5p were observed to be independent factors for risk of knee OA, and their combination has a high diagnostic value with AUC 0.926 for knee OA; (3) miR-19b-3p and miR-486-5p were found to be positively correlated with comprehensive disease severity such as pain VAS scores and WOMAC scores, while miR-122-5p was only negatively correlated with pain VAS walking score.
Circulating miRNAs in various biofluids, such as plasma, sera, urine, extracellular fluid and others, have been observed in these years, and investigators speculated some of them might play an important role in mediating the endocrine or paracrine signaling between cells. Previous reports demonstrated the main sources of circulating miRNAs in plasma included inactive release from damaged tissue [22], active release from cells [23], and these miRNAs could be highly stabilized via formation of microvesicles or protein compounds [23,24]. Although the exact role of these circulating miRNAs remains to be fully understood, they have already incited great interests to employ these circulating miRNAs as biomarkers for various diseases. Given current protein biomarkers are suffered numerous technical problems, including challenges of high-affinity capture reagent and high-protein abundancy development, miRNAs are expected to overcome these limitations with their advantages: firstly, better understanding of their possible function; secondly, their stability in extracellular environment; thirdly, noninvasive detection could be used in patients refuse invasive practice; fourthly, it’s sensitive enough to make early screen practicable. Therefore, further efforts to develop appropriate miRNA biomarkers for various diseases including knee OA is critical and necessary.
Cellular components and extracellular matrix are critical for normal function of articular cartilage, by maintaining mechanical structure of articular and homeostasis of the extracellular environment [25]. Meanwhile, articular cartilage could be affected by the following inflammatory pathways, including cytokine interleukin-1β via synthesis stimulation of extracellular matrix-degrading enzymes, inhibiting gene expression for collagen type II and synthesis of aggrecan via downregulating Sox9 [26,27]. MiR-101 and miR-145 were reported to suppress Sox9 expression by binding to its mRNA, then effected IL-1β-induced chondrocyte extracellular matrix-degrading and increased the hypertrophicmarkers, respectively [28,29]. Silencing of miR-34a reduces IL-1β-induced downregulation of collagen gene Col2a and upregulation of inducible nitric oxide synthase, apoptosis, and leading to chondrolytic effects in chondrocytes [30]. Downregulation of miR-148a was found in OA cartilage, while its overexpression led to increased production of type II collagen [31]. On the other hand, matrix metalloproteinase-13 (MMP-13) is produced in OA joints and could degrade extracellular matrix, which regulates tissue modeling and repair [32]. MiR-27b, miR-488, miR-33a, miR-127-5p and miR-181b have been reported to have impact on MMP-13 expression and play a role in the pathology of knee OA [33-37].
Meanwhile, miR-9 and miR-140 play important roles in the normal cartilage development, and their aberrant expressions are involved in the pathology of knee OA [38-40]. Although these aberrant miRNAs from tissues provide some information for OA pathology, most of them need invasive detection. Noninvasive detection of circulating miRNAs might provide a practical strategy for effective diagnosis and disease severity evaluation of knee OA.
Several circulating miRNAs have been reported related to OA, including miR-155, miR-16, miR-223 and miR-146a at synovial level were lower in patients with OA compared with RA [41]; increased expression of miR-155, miR-146a, miR-223 and miR-181a at peripheral blood mononuclear cells level in patients with OA compared with control [42]; and negatively correlated let-7e expression at serum level in symptomatic OA patients with arthroplasty [13]. As for the plasma level, overexpression of a panel of 12 miRNAs (miR-20b, miR-30b, miR-16, miR-29c, miR-93, miR-146a, miR-126, miR-186, miR-234, miR-184, miR-195, and miR-885-5p) was determined in patients with OA compared with healthy controls [43]. Interestingly, the expression of miR-223 and miR-16 were decreased in both synovial fluid and plasma but without correlation of miRNA concentrations, which indicating their different origins [41]. Plasma miRNAs are promising biomarker candidates for clinical use based on its great advantage of great stability and are not degraded from the endogenous RNase. Plasma miRNAs have been reported to be stable for up to 24 h at room temperature [14]. Thus further efforts might be put on better understanding function and origin of plasma miRNAs and interpret aberrant expression of miRNAs, and finally, facilitate the development of miRNA biomarkers with clear clinical suggestions.
To our knowledge, this study is the first time to systematically explore the aberrant expression of plasma miRNAs in knee OA by microarray, and further, validate in qPCR as well as investigate their association with clinical feature of knee OA patients. We found upregulation of miR-19b-3p, miR-122-5p and miR-486-5p were independent predictors of OA, with miR-486-5p as the most valuable one. And the combination of the three miRNAs appeared a great diagnostic value in knee OA with high AUC. Aberrant expression of miR-486 have been reported to be associated with various disease: increased circulating miR-486 is associated with acute myocardial infarction. Meanwhile, downregulation of miR-486-5p expression is related to human cancers, including hepatocellular carcinoma, gastric carcinoma, and breast tumors [44-46]. Especially plasma miR-486 has been recognized as an effective non-invasive biomarker of recurrence of early-stage non-small-cell lung cancer [47], and serum miR-486-5p is expected to be used in clinical diagnosis of lung cancer [48]. Furthermore, our univariate and multivariate Cox regression analysis between the expressions level of miRNAs and characteristics of OA patients identified a significant relationship between miR-486-5p expression and VAS rest, WOMAC pain, and WOMAC function. This suggested circulating miR-486-5p could provide useful information for both early diagnostic and prognostic evaluation in OA patients. In addition to that, we demonstrated increased has-miR-19b-3p expression were associated with OA duration, VAS walking, WOMAC pain and stiffness and has-miR-122-5p was correlated with WAS rest. Philippe L and colleagues demonstrated miR-19a/b regulate IL-6 and matrix metalloproteinase 3 release by controlling TLR2 expression, and could act as negative regulators of inflammation [49]; meanwhile targeting miR-19 was reported to regulate the activity of NF-kB signaling inflammation [50]. On the other hand, previous reports demonstrated the critical role of miR-122 in liver diseases, and chronic inflammation leads to decreased expression of miR-122 which might contribute to the development of hepatocellular carcinoma [51]. These association between plasma miRNAs and clinical characteristics provide the possibility to develop not only diagnostic but also progressive biomarkers for knee OA patients.
In conclusion, this study revealed that circulating miRNA profiles played a key role in knee OA diagnosis, and combined measurement of miR-19b-3p, miR-122-5p and miR-486-5p could be served as a novel and promising biomarker for diagnosis and disease severity of knee OA.
Disclosure of conflict of interest
None.
References
- 1.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum. 2012;64:1697–1707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease C and Prevention (CDC) Prevalence of doctor-diagnosed arthritis and arthritis-attributable activity limitation-United States, 2010-2012. MMWR Morb Mortal Wkly Rep. 2013;62:869–873. [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter DJ, Schofield D, Callander E. The individual and socioeconomic impact of osteoarthritis. Nat Rev Rheumatol. 2014;10:437–441. doi: 10.1038/nrrheum.2014.44. [DOI] [PubMed] [Google Scholar]
- 4.Hochberg MC. Osteoarthritis: the rheumatologist’s perspective. HSS J. 2012;8:35–36. doi: 10.1007/s11420-011-9253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Spil WE, DeGroot J, Lems WF, Oostveen JC, Lafeber FP. Serum and urinary biochemical markers for knee and hip-osteoarthritis: a systematic review applying the consensus BIPED criteria. Osteoarthritis Cartilage. 2010;18:605–612. doi: 10.1016/j.joca.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 8.Asahara H. Current status and strategy of microRNA research for cartilage development and osteoarthritis pathogenesis. J Bone Metab. 2016;23:121–127. doi: 10.11005/jbm.2016.23.3.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vicente R, Noel D, Pers YM, Apparailly F, Jorgensen C. Deregulation and therapeutic potential of microRNAs in arthritic diseases. Nat Rev Rheumatol. 2016;12:496. doi: 10.1038/nrrheum.2016.119. [DOI] [PubMed] [Google Scholar]
- 10.Duroux-Richard I, Jorgensen C, Apparailly F. What do microRNAs mean for rheumatoid arthritis? Arthritis Rheum. 2012;64:11–20. doi: 10.1002/art.30651. [DOI] [PubMed] [Google Scholar]
- 11.Duroux-Richard I, Jorgensen C, Apparailly F. miRNAs and rheumatoid arthritis-promising novel biomarkers. Swiss Med Wkly. 2011;141:w13175. doi: 10.4414/smw.2011.13175. [DOI] [PubMed] [Google Scholar]
- 12.Alevizos I, Illei GG. MicroRNAs as biomarkers in rheumatic diseases. Nat Rev Rheumatol. 2010;6:391–398. doi: 10.1038/nrrheum.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beyer C, Zampetaki A, Lin NY, Kleyer A, Perricone C, Iagnocco A, Distler A, Langley SR, Gelse K, Sesselmann S, Lorenzini R, Niemeier A, Swoboda B, Distler JH, Santer P, Egger G, Willeit J, Mayr M, Schett G, Kiechl S. Signature of circulating microRNAs in osteoarthritis. Ann Rheum Dis. 2015;74:e18. doi: 10.1136/annrheumdis-2013-204698. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nugent M, Miller N, Kerin MJ. Circulating miR-34a levels are reduced in colorectal cancer. J Surg Oncol. 2012;106:947–952. doi: 10.1002/jso.23174. [DOI] [PubMed] [Google Scholar]
- 16.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 18.Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K, Christy W, Cooke TD, Greenwald R, Hochberg M. Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and therapeutic criteria committee of the American rheumatism association. Arthritis Rheum. 1986;29:1039–1049. doi: 10.1002/art.1780290816. [DOI] [PubMed] [Google Scholar]
- 19.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 20.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- 21.Backes C, Khaleeq QT, Meese E, Keller A. miEAA: microRNA enrichment analysis and annotation. Nucleic Acids Res. 2016;44:W110–116. doi: 10.1093/nar/gkw345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009;55:1944–1949. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- 23.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 24.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 2011;7:33–42. doi: 10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- 26.Sekiya I, Tsuji K, Koopman P, Watanabe H, Yamada Y, Shinomiya K, Nifuji A, Noda M. SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J Biol Chem. 2000;275:10738–10744. doi: 10.1074/jbc.275.15.10738. [DOI] [PubMed] [Google Scholar]
- 27.Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai L, Zhang X, Hu X, Zhou C, Ao Y. Silencing of microRNA-101 prevents IL-1beta-induced extracellular matrix degradation in chondrocytes. Arthritis Res Ther. 2012;14:R268. doi: 10.1186/ar4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Sanchez A, Dudek KA, Murphy CL. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145) J Biol Chem. 2012;287:916–924. doi: 10.1074/jbc.M111.302430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanco FJ, Ochs RL, Schwarz H, Lotz M. Chondrocyte apoptosis induced by nitric oxide. Am J Pathol. 1995;146:75–85. [PMC free article] [PubMed] [Google Scholar]
- 31.Vonk LA, Kragten AH, Dhert WJ, Saris DB, Creemers LB. Overexpression of hsa-miR-148a promotes cartilage production and inhibits cartilage degradation by osteoarthritic chondrocytes. Osteoarthritis Cartilage. 2014;22:145–153. doi: 10.1016/j.joca.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Fosang AJ, Last K, Knauper V, Murphy G, Neame PJ. Degradation of cartilage aggrecan by collagenase-3 (MMP-13) FEBS Lett. 1996;380:17–20. doi: 10.1016/0014-5793(95)01539-6. [DOI] [PubMed] [Google Scholar]
- 33.Akhtar N, Rasheed Z, Ramamurthy S, Anbazhagan AN, Voss FR, Haqqi TM. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum. 2010;62:1361–1371. doi: 10.1002/art.27329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song J, Kim D, Lee CH, Lee MS, Chun CH, Jin EJ. MicroRNA-488 regulates zinc transporter SLC39A8/ZIP8 during pathogenesis of osteoarthritis. J Biomed Sci. 2013;20:31. doi: 10.1186/1423-0127-20-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kostopoulou F, Malizos KN, Papathanasiou I, Tsezou A. MicroRNA-33a regulates cholesterol synthesis and cholesterol efflux-related genes in osteoarthritic chondrocytes. Arthritis Res Ther. 2015;17:42. doi: 10.1186/s13075-015-0556-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SJ, Cheon EJ, Lee MH, Kim HA. MicroRNA-127-5p regulates matrix metalloproteinase 13 expression and interleukin-1betainduced catabolic effects in human chondrocytes. Arthritis Rheum. 2013;65:3141–3152. doi: 10.1002/art.38188. [DOI] [PubMed] [Google Scholar]
- 37.Song J, Lee M, Kim D, Han J, Chun CH, Jin EJ. MicroRNA-181b regulates articular chondrocytes differentiation and cartilage integrity. Biochem Biophys Res Commun. 2013;431:210–214. doi: 10.1016/j.bbrc.2012.12.133. [DOI] [PubMed] [Google Scholar]
- 38.Min Z, Zhang R, Yao J, Jiang C, Guo Y, Cong F, Wang W, Tian J, Zhong N, Sun J, Ma J, Lu S. MicroRNAs associated with osteoarthritis differently expressed in bone matrix gelatin (BMG) rat model. Int J Clin Exp Med. 2015;8:1009–1017. [PMC free article] [PubMed] [Google Scholar]
- 39.Makki MS, Haseeb A, Haqqi TM. MicroRNA-9 promotion of interleukin-6 expression by inhibiting monocyte chemoattractant proteininduced protein 1 expression in interleukin-1beta-stimulated human chondrocytes. Arthritis Rheumatol. 2015;67:2117–2128. doi: 10.1002/art.39173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang ZJ, Zhuang H, Wang GX, Li Z, Zhang HT, Yu TQ, Zhang BD. MiRNA-140 is a negative feedback regulator of MMP-13 in IL-1betastimulated human articular chondrocyte C28/I2 cells. Inflamm Res. 2012;61:503–509. doi: 10.1007/s00011-012-0438-6. [DOI] [PubMed] [Google Scholar]
- 41.Murata K, Yoshitomi H, Tanida S, Ishikawa M, Nishitani K, Ito H, Nakamura T. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2010;12:R86. doi: 10.1186/ar3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okuhara A, Nakasa T, Shibuya H, Niimoto T, Adachi N, Deie M, Ochi M. Changes in microRNA expression in peripheral mononuclear cells according to the progression of osteoarthritis. Mod Rheumatol. 2012;22:446–457. doi: 10.1007/s10165-011-0536-2. [DOI] [PubMed] [Google Scholar]
- 43.Borgonio Cuadra VM, Gonzalez-Huerta NC, Romero-Cordoba S, Hidalgo-Miranda A, Miranda-Duarte A. Altered expression of circulating microRNA in plasma of patients with primary osteoarthritis and in silico analysis of their pathways. PLoS One. 2014;9:e97690. doi: 10.1371/journal.pone.0097690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun H, Cui C, Xiao F, Wang H, Xu J, Shi X, Yang Y, Zhang Q, Zheng X, Yang X, Wu C, Wang L. miR-486 regulates metastasis and chemosensitivity in hepatocellular carcinoma by targeting CLDN10 and CITRON. Hepatol Res. 2015;45:1312–1322. doi: 10.1111/hepr.12500. [DOI] [PubMed] [Google Scholar]
- 45.Oh HK, Tan AL, Das K, Ooi CH, Deng NT, Tan IB, Beillard E, Lee J, Ramnarayanan K, Rha SY, Palanisamy N, Voorhoeve PM, Tan P. Genomic loss of miR-486 regulates tumor progression and the OLFM4 antiapoptotic factor in gastric cancer. Clin Cancer Res. 2011;17:2657–2667. doi: 10.1158/1078-0432.CCR-10-3152. [DOI] [PubMed] [Google Scholar]
- 46.Tahiri A, Leivonen SK, Luders T, Steinfeld I, Ragle Aure M, Geisler J, Makela R, Nord S, Riis ML, Yakhini Z, Kleivi Sahlberg K, Borresen-Dale AL, Perala M, Bukholm IR, Kristensen VN. Deregulation of cancer-related miRNAs is a common event in both benign and malignant human breast tumors. Carcinogenesis. 2014;35:76–85. doi: 10.1093/carcin/bgt333. [DOI] [PubMed] [Google Scholar]
- 47.Li W, Wang Y, Zhang Q, Tang L, Liu X, Dai Y, Xiao L, Huang S, Chen L, Guo Z, Lu J, Yuan K. MicroRNA-486 as a biomarker for early diagnosis and recurrence of non-small cell lung cancer. PLoS One. 2015;10:e0134220. doi: 10.1371/journal.pone.0134220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Liang L, Zhang CY. Isothermally sensitive detection of serum circulating miRNAs for lung cancer diagnosis. Anal Chem. 2013;85:11174–11179. doi: 10.1021/ac403462f. [DOI] [PubMed] [Google Scholar]
- 49.Philippe L, Alsaleh G, Suffert G, Meyer A, Georgel P, Sibilia J, Wachsmann D, Pfeffer S. TLR2 expression is regulated by microRNA miR-19 in rheumatoid fibroblast-like synoviocytes. J Immunol. 2012;188:454–461. doi: 10.4049/jimmunol.1102348. [DOI] [PubMed] [Google Scholar]
- 50.Gantier MP, Stunden HJ, McCoy CE, Behlke MA, Wang D, Kaparakis-Liaskos M, Sarvestani ST, Yang YH, Xu D, Corr SC, Morand EF, Williams BR. A miR-19 regulon that controls NF-kappaB signaling. Nucleic Acids Res. 2012;40:8048–8058. doi: 10.1093/nar/gks521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li C, Deng M, Hu J, Li X, Chen L, Ju Y, Hao J, Meng S. Chronic inflammation contributes to the development of hepatocellular carcinoma by decreasing miR-122 levels. Oncotarget. 2016;7:17021–17034. doi: 10.18632/oncotarget.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]