Abstract
An increasing number of deregulated long non-coding RNAs (lncRNA) have been implicated in cancer in humans, suggesting that lncRNAs may be involved in tumorigenesis or tumor progression. In previous investigations, lncRNA, AFAP1-AS1 has been found to be associated with several cancers, including nasopharyngeal carcinoma, lung cancer and esophageal adenocarcinoma. However, the function of AFAP1-AS1 in lung cancer has not been reported. In our present study, we found that AFAP1-AS1 was overexpressed in lung adenocarcinoma and associated with survival time. The low expression of AFAP1-AS1 was an independent predictor for disease-free survival in patients with lung adenocarcinoma. Additionally, we transfected AFAP1-AS1 siRNA into H1975 and HCC827, both lung adenocarcinoma cancer cells, and found that cell growth was suppressed, apoptosis induced and invasion inhibited. Taken together, AFAP1-AS1 down-regulation exerts growth suppression and apoptosis induction in lung adenocarcinoma cells, suggested it may participate in turmorigenesis and be a therapeutic target for treating lung adenocarcinoma in future.
Keywords: AFAP1-AS1, lung cancer, growth, apoptosis, invasion
Introduction
Lung cancer is the leading cause of cancer-associated mortality worldwide. In 2010, the mortality rate from lung cancer approached 1,500,000, accounting for 19% of all cases of cancer related death that year [1]. With increase in environmental pollution and weak tobacco control in China, the incidence of lung cancer in China has surged in last 10 years [2]. Of all patients with lung cancer, more than half are diagnosed with locally advanced or metastatic disease. Despite continuous improvements in surgical resection, radiation therapy technologies and chemotherapy or target drugs, patients with lung cancer are vulnerable to relapse and associated mortality [3]. The cure rate for lung cancer is low, with a 5-year survival rate of less than 15% [4]. Certain cases of lung cancer have been found to be associated with tobacco consumption [5]. However, the mechanisms underlying the development of lung cancer remain to be fully elucidated. Therefore, the investigation of lung cancer tumorigenesis and development remains important.
Previous studies have indicated that only 1.5-2% of mammalian genomic sequences code for proteins, and it has been shown that mammalian genomes are largely transcribed, comprising numerous non-coding RNAs [6]. Non-coding RNAs are classified into two groups, represented by short RNAs, which are generally less than 200 bp in length and include microRNA (miRNAs), and long non-coding RNAs (lncRNAs), which are more than 200 bp in length [7]. MiRNAs have been intensively investigated in lung cancer, and a number of specific miRNAs have been reported as oncogenes, tumor suppressors and regulators of drug-resistance in lung cancer [8,9]. It has also become increasingly apparent that lncRNAs are critical in cellular processes, including cell growth, differentiation and apoptosis, via transcriptional and/or post-transcriptional regulation of genes [10]. However, the detailed characterization of the effects of lncRNAs on tumorigenesis and the development of lung cancer require further elucidation.
In lung cancer, a number of studies have investigated the role of lncRNAs. The lncRNA expression profile of human lung adenocarcinoma compared to normal lung samples have been investigated and analyzed using microarray. And 2,420 lncRNAs have been identified as being differentially expressed, suggesting certain specific lncRNAs may be important in tumorigenesis and development of lung cancer [11]. In addition, our previous study found that MALAT1 promotes brain metastasis by inducing epithelial to mesenchymal transition in lung cancer [12]. HOTAIR has been found to be overexpressed in metastatic lung cancer, and has subsequently been found to be involved in motility and invasion of lung cancer [13]. MEG3 has been found to inhibit non-squamous cell lung carcinoma (NSCLC) cell proliferation and induce apoptosis by affecting the expression of p53 [14]. AFAP1-AS1 has been shown to exert biological functions in esophageal adenocarcinoma, pancreatic ductal adenocarcinoma and lung cancer [15-17]. Therefore, the present study aimed to investigate the biological functions of AFAP1-AS1 in lung adenocarcinoma from clinical and cellular aspects.
Methods and materials
Ethics
All patients signed informed consent and human samples used in this study were approved by the Committee for Ethical Review of Research Involving Human Subjects in Drum Tower Hospital, Affiliated to Medical School of Nanjing University.
Tissue samples and cell lines
Lung adenocarcinoma samples (n = 36) and matched normal tissues (n = 36) were collected at Nanjing Drum Tower Hospital (Nanjing, China). The lung adenocarcinoma cell lines-H1915 and HCC827 were obtained from American Type Culture Collection (ATCC) and cultured in RPMI-1640 medium (Invitrogen, CA) with 10% Fetal bovine serum (FBS) and 100 U/mL penicillin/streptomycin (Sigma, St Louis, MO).
Quantitative Real-Time PCR (qRT-PCR)
Total RNA from tissues or cell lines was extracted using TRIzol reagent (Invitrogen, CA). The concentration of isolated total RNA was measured by NanoDrop ND-1000 Spectrophotometer (Agilent, CA). For mRNA detection, the total RNA was reversely transcribed by using SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, CA). The qPCR was performed by using SsoFastTMEvaGreen® Supermix (Bio-Rad). The primers of AFAP1-AS1 were 5’-TCGCTCAATGGAGTGACGGCA-3’ and 5’-CGGCTGAGACCGCTGAGAACT-3’ (Reverse) [18]. And primers for HPRT1 were TGACACTGGCAAAACAATGCA (Forward) and GGTCCTTTTCACCAGCAAGCT (Reverse). Amplification was done on a Bio-Rad CFX96 system in 20 uL. HPRT1 internal control was used as endogenous controls, and fold changes were calculated via relative quantification 2-ΔCt [19].
Cell proliferation assay
Cells were detached by treatment with 0.25% trypsin-EDTA (Invitrogen, Carlsbad, CA, USA) and seeded into 96-well plates at a density of 5×103 per well in 100 µl. For the MTS assay, the CellTiter 96®AQueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI, USA) was used following the manufacturer’s instruction. Briefly, 2 h before each of the desired time points (12 h, 24 h and 48 h), 20 µl of the MTS reagent was added to each well and cells were incubated at 37°C for 3 h. The absorbance was detected at 490 nm using a Wallac Victor 1420 Multi-label plate reader. All of the experiments were repeated three times.
Cell apoptosis assay
The Cells were harvested using 40 μm cell strainers (BD Biosciences, San Jose, CA, USA). The cells were detached by treatment with 0.25% trypsin-EDTA (Invitrogen, Carlsbad, CA, USA) and washed once with phosphate-buffered saline. The cells were then resuspended in 100 μl Staining Buffer (eBioscience, San Diego, CA, USA) containing 1% fetal bovine serum (FBS; Gibco BRL, Grand Island, NY, USA) and place on ice for 20 min to block Fc receptors. After incubating with primary phycoerythrin (PE) for another 45 min on ice in dark, the cells were washed twice with 1 ml ice-cold Staining Buffer and centrifuged (400×g) for 5 min at 4°C. Then Cells were resuspended in 0.5 ml 2% formaldehyde fixation buffer and analysed using a FACSCalibur flow cytometer and CellQuest software (BD Biosciences, San Jose, CA, USA). All flow cytometry results were obtained from two independent experiments performed in triplicate.
In vitro Matrigel invasion assay
The cell invasiveness was assessed with the use of BioCoatMatrigel Invasion Chambers (BD Biosciences, Bedford, MA). Medium with 10% FBS was added to the lower chamber as chemoattractant. Then equal numbers of H1975, HCC827 or stable transfected with siRNA of ASAP1-AS1 or siControl were resuspended in 500 μl serum-free medium and seeded into the rehydrated insert. After 24 h of incubation at 37°C, non-invading cells on the upper surface of the Matrigel membrane were gently removed with a cotton-tipped swab. The cells were then fixed with 100% methanol and stained with 1% toluidine blue (Sigma). The stained invasive cells on the lower surface of the membrane were photographed under an inverted light microscope (40× objective) and quantified by manual counting in three randomly selected areas. This experiment was performed in duplicate in three independent experiments.
Statistical analysis
The experimental data are presented as the mean ± standard deviation (SD). All statistical analyses were performed using ANOVA or a two-tailed Student’s t test (SPSS 22.0 SPSS Inc, Chicago, IL, USA). Disease-free survival (DFS) was measured from the date of hepatic resection to the date of death or the last follow-up. The survival curves were calculated using the Kaplan-Meier method and statistically compared using a log-rank test. Differences were considered statistically significant when the P-values were less than 0.05.
Results
AFAP1-AS1 is increased in lung cancer tissues and is associated with patients’ survival
To investigate the potential role of AFAP1-AS1 in lung cancer, the level of AFAP1-AS1 mRNA in 36 lung adenocarcinoma samples, which was divided to 3 groups and 12 cases were in each group, and 36 normal matched tissues, was compared by qRT-PCR. The relative level of AFAP1-AS1 was significantly higher in lung tumor tissues compared with matched normal tissues (P < 0.0001, paired Student’s t-test; Figure 1A). Moreover, we analyzed the association between AFAP1-AS1 and survival of patients. The high level of AFAP1-AS1 was significantly correlated with shorter disease-free survival (DFS) of patients with lung adenocarcinoma (P = 0.0382; Figure 1B). These results indicate that AFAP1-AS1 level is increased in lung adenocarcinoma patients and associated with patients’ survival.
Figure 1.
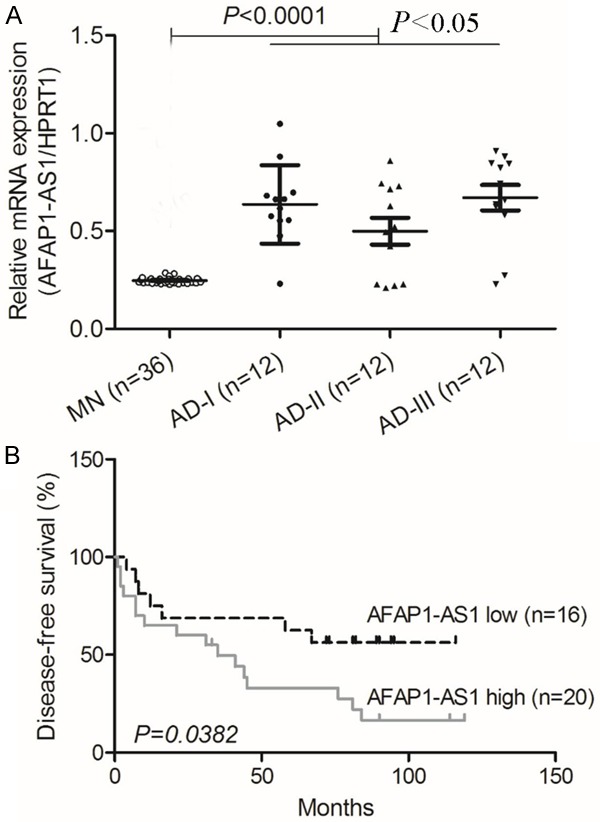
AFAP1-AS1 is over-expressed in lung adenocarcinoma cancer and is associated with patient survival. A: The level of AFAP1-AS1 in 36 pairs of lung adenocarcinoma cancer tissue samples was analyzed by qRT-PCR. Lung adenocarcinoma cancer patients were divided to 3 groups and the average level of AFAP1-AS1 in each group was higher than that in matched normal tissues (P < 0.0001). B: The Kaplan-Meier method was used to analyze survival in patients with lung adenocarcinoma cancer. The probability of patient survival: AFAP1-AS1 high, n = 20; AFAP1-AS1 low, n = 16 (P = 0.0382).
Down-regulation of AFAP1-AS1 leads to growth inhibition in lung adenocarcinoma cells
To reveal the potential role of AFAP1-AS1 in lung adenocarcinoma, we transfected lung adenocarcinoma cells-HCC827 and H1975 with synthetic oligo nucleotides to knock-down AFAP1-AS1 expression. Then we investigated whether AFAP1-AS1 affect lung adenocarcinoma cell growth. The two-step quantitative RT-PCR results showed that transfection of AFAP1-AS1 siRNA could significantly decrease the level of AFAP1-AS1 in HCC827 or H1975 cells, as compared to their respective control groups (P < 0.05, Figure 2A, 2B). Subsequently, MTS assay showed that down-regulation of AFAP1-AS1 significantly inhibited cell growth of both HCC827 and H1975 cells (Figure 2C, 2D).
Figure 2.
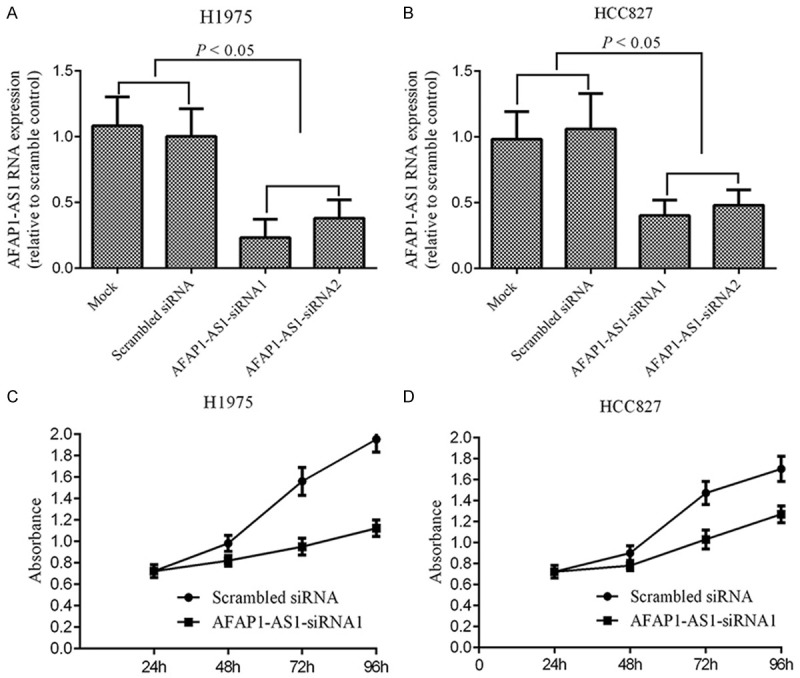
Silencing of AFAP1-AS1 inhibits lung adenoncarcinoma cancer cell growth. (A, B) The level of AFAP1-AS1 in H1975 (A) and HCC827 (B) cells transfected with siRNA or scramble siRNA as control by qRT-PCR analysis. (C, D) The cell growth rate was detected in H1975 and HCC827 cells transfected with siRNA or scramble siRNA by MTS assay at different time points (24 h, 48 h, 72 h and 96 h). Down-regulation of AFAP1-AS1 inhibits lung adenoncarcinoma cancer cell growth significantly (P < 0.05).
Inhibition of AFAP1-AS1 promoted apoptosis in lung adenocarcinoma cells
Since down-regulation of AFAP1-AS1 has been shown to inhibit the growth of lung adenocarcinoma cells, we next examined whether knockdown of AFAP1-AS1 is able to induce apoptosis of lung adenocarcinoma cells. HCC827 and H1975 cells were transfected with AFAP1-AS1 siRNA. The cells transfected with scramble siRNA were used as control. Following 72 hours of transfection, flow cytometry analysis revealed that knockdown of AFAP1-AS1 induced approximately 23% apoptosis in HCC827 cells compared to 0.6% induced by scramble siRNA, and almost 37% in H1975 cells compared to 3.8% by scramble siRNA (Figure 3A, 3B). Collectively, these data indicate that AFAP1-AS1 is one of apoptosis inducers in lung adenocarcinoma cells.
Figure 3.
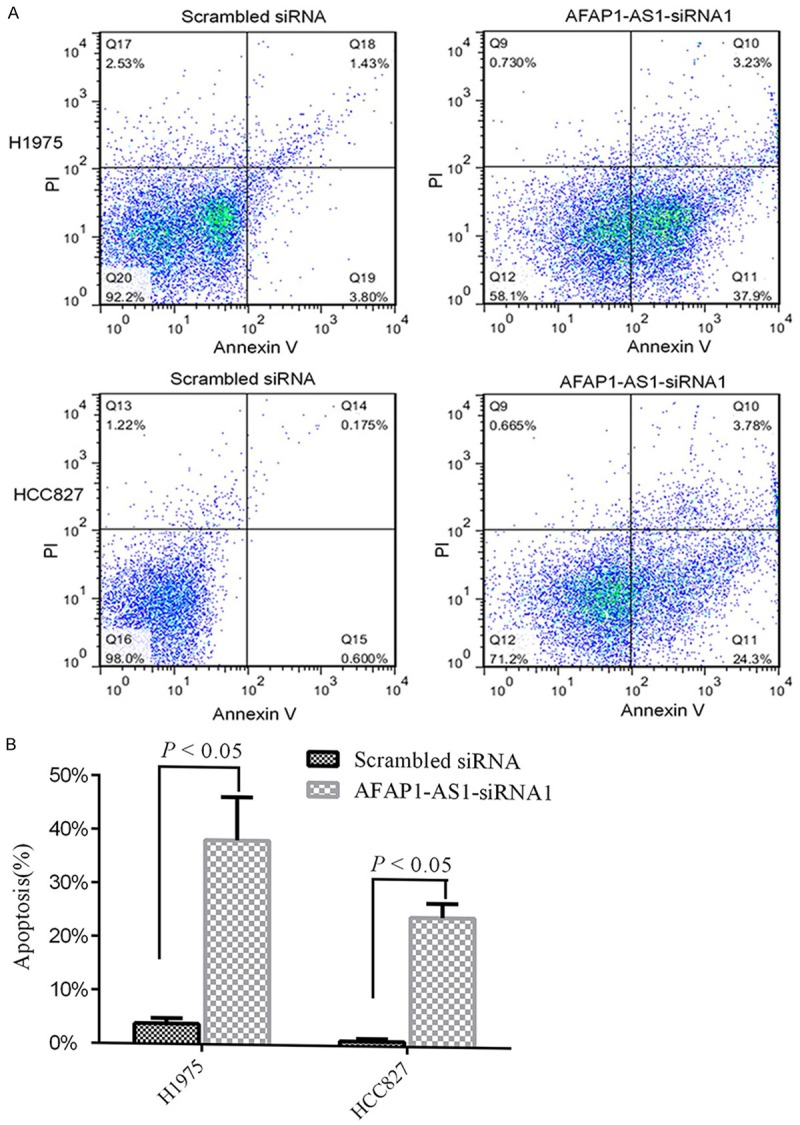
Silencing of AFAP1-AS1 induces lung cancer apoptosis in vitro. A: Flow cytometry of apoptosis in H1975 and HCC827 transfected with siRNA or scramble siRNA. Specific AFAP1-AS1 siRNA induced significant apoptosis in H1975 and HCC827 (lower right quadrant, 37.9% vs 3.80%; 24.3% vs 0.60%). B: Apoptosis rate relative to treatment with a scrambled siRNA or with a specific AFAP1-AS1 siRNA. Specific siRNA induced significant apoptosis in both H1975 and HCC827 (P < 0.05).
Inhibition of AFAP1-AS1 in lung adenocarcinoma cells leads to reduced invasion
In light of the above observation that AFAP1-AS1 is greatly affected the growth and apoptosis of lung adenocarcinoma cells, we hypothesized that it may be associated with the invasiveness of tumor cells too. We transfected HCC827 and H1975 with AFAP1-AS1 siRNA or scramble siRNA. Then following 72 hours of transfection, we investigate the invasion of HCC827 and H1975 cells. Consistently, HCC827 and H1975 transfected with AFAP1-AS1 siRNA demonstrated low invasiveness and the invasive cells almost 3 times less than that of transfected with scramble siRNA as control (P < 0.05, Figure 4A, 4B). These results suggested that AFAP1-AS1 affected the invasion of lung adenocarcinoma cells.
Figure 4.

Silencing of AFAP1-AS1 inhibits lung cancer cell invasion in vitro. A: Image of invasion in H1975 and HCC827 cells transfected with specific siRNA or scramble siRNA by transwell Matrige invasion assay. B: The average invasive cells was counted from three independent repeated wells and shown in the graph. Silencing of AFAP1-AS1 inhibits lung cancer cell invasion significantly (P < 0.05).
Discussion
Intensive studies over the last decades have focused on the role of protein coding genes in the tumorigenesis and development of cancer. However, recent advances in technologies such as microarray and sequencing of RNA have made it possible to survey the transcripts of many organisms and diseases [20]. In fact, at least 90% of the genome is actively transcribed to non-coding RNAs, including microRNA and long non-coding RNAs. Although initially regarded as transcriptional noise, recent studies indicate that they may have a biological role in cellular development, differentiation and metabolism [21]. Furthermore, accumulating evidences of dys-regulated lncRNAs across a number of cancers suggest that aberrant lncRNA expression may be a major contributor to tumorigenesis and cancer development.
As mentioned above, lncRNAs have been implicated in a wide variety of human diseases including cancers, such as in breast, liver and lung cancer [22-24]. However, MALAT-1 has been previously associated with worse prognosis in a small group of lung cancer patients. And in our previous research, we also found that MALAT-1 was correlated with development of brain metastasis in lung cancer patients [25]. Cumulatively, these studies suggest that lncRNA might be involved in both the process of tumorigenesis as well as metastatic progression and, thus, the further investigation of the function of lncRNA is needed. In our current study, we found that the average level of AFAP1-AS1 in lung adenocarcinoma tissues was significantly higher than those in matched non-tumor tissues. The low expression level of AFAP1-AS1 in lung adenocarcinoma patients was associated with better survival and could be an independent prognostic indicator.
AFAP1-AS1 level was higher in lung adenocarcinoma compared with matched non-tumor tissues, but in other studies, it was demonstrated down-regulated in esophagus cancer [15-17]. These contrary findings are probably because lncRNAs exhibit remarkably tissue-specific expression patterns than protein-coding genes. AFAP1-AS1 too, may have a tissue-specific expression profile and exhibit important role in NSCLC development and progression.
The previous studies suggest that lncRNAs may play a very important role in tumorigenesis and development of cancer, similarly to protein coding genes. For example, long noncoding RNA HOTAIR has been found to be relevant to cellular proliferation, invasiveness, and clinical relapse in small-cell lung cancer. LncRNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) [26]. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression [27]. In our study, we demonstrated that AFAP1-AS1 involves in growth, apoptosis and progression of lung adenocarcinoma.
Recently, Zeng et al demonstrated that AFAP1-AS1 knockdown significantly inhibited the cell invasive and migration capability in lung cancer cells. And they further speculated that AFAP1-AS1 may promote cancer cell metastasis via regulation of actin filament integrity [17]. These results are similar to our experimental results in vitro, which further proved our conclusion. AFAP1-AS1 is likely to be a useful biomarker of lung adenocarcinoma. Therefore, AFAP1-AS1 may serve as a novel therapeutic application for lung adenocarcinoma patients in future. However, the observation and specific mechanism of AFAP1-AS1 are needed to elucidate in the future studies.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China; Grant Number: 81370161.
Disclosure of conflict of interest
None.
Authors’ contribution
YS Wang and HP Xia participated in the design of the study, data acquisition and analysis as well as drafting the manuscript. HY Jiang, H Li, Y Li, LY Miao, Y Zhuang, JH Dai, and Y Liu were responsible for the laboratory assay and troubleshooting. HR Cai, YL Xiao and HP Xia participated in data acquisition, analysis, and interpretation. MK Shi collected all samples and helped to draft the manuscript. All authors read and approved the final manuscript.
References
- 1.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FG, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo JP, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KM, Nasseri K, Norman P, O’Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA 3rd, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De Leon FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh PH, Yip P, Zabetian A, Zheng ZJ, Lopez AD, Murray CJ, AlMazroa MA, Memish ZA. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the global burden of disease study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen WQ, Zheng RS, Zhang SW, Zeng HM, Zou XN. The incidences and mortalities of major cancers in China, 2010. Chin J Cancer. 2014;33:402–405. doi: 10.5732/cjc.014.10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgensztern D, Ng SH, Gao F, Govindan R. Trends in stage distribution for patients with non-small cell lung cancer: a national cancer database survey. J Thorac Oncol. 2010;5:29–33. doi: 10.1097/JTO.0b013e3181c5920c. [DOI] [PubMed] [Google Scholar]
- 4.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, Zhang S, Wang J, Zhou S, Ren S, Lu S, Zhang L, Hu C, Hu C, Luo Y, Chen L, Ye M, Huang J, Zhi X, Zhang Y, Xiu Q, Ma J, Zhang L, You C. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, openlabel, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 5.Ito H, Matsuo K, Tanaka H, Koestler DC, Ombao H, Fulton J, Shibata A, Fujita M, Sugiyama H, Soda M, Sobue T, Mor V. Nonfilter and filter cigarette consumption and the incidence of lung cancer by histological type in Japan and the United States: analysis of 30-year data from population-based cancer registries. Int J Cancer. 2011;128:1918–1928. doi: 10.1002/ijc.25531. [DOI] [PubMed] [Google Scholar]
- 6.Ponting CP, Belgard TG. Transcribed dark matter: meaning or myth? Hum Mol Genet. 2010;19:R162–168. doi: 10.1093/hmg/ddq362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26:155–165. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 8.Wang YS, Wang YH, Xia HP, Zhou SW, Schmid-Bindert G, Zhou CC. MicroRNA-214 regulates the acquired resistance to gefitinib via the PTEN/AKT pathway in EGFR-mutant cell lines. Asian Pac J Cancer Prev. 2012;13:255–260. doi: 10.7314/apjcp.2012.13.1.255. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez S, Risolino M, Mandia N, Talotta F, Soini Y, Incoronato M, Condorelli G, Banfi S, Verde P. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene. 2015;34:3240–3250. doi: 10.1038/onc.2014.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Xu G, Chen J, Pan Q, Huang K, Pan J, Zhang W, Yu F, Zhou T, Wang Y. Long noncoding RNA expression profiles of lung adenocarcinoma ascertained by microarray analysis. PLoS One. 2014;9:e104044. doi: 10.1371/journal.pone.0104044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelialmesenchymal transition in lung cancer. J Neurooncol. 2015;121:101–8. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- 13.Zhao W, An Y, Liang Y, Xie XW. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur Rev Med Pharmacol Sci. 2014;18:1930–1936. [PubMed] [Google Scholar]
- 14.Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu W, Bhagat TD, Yang X, Song JH, Cheng Y, Agarwal R, Abraham JM, Ibrahim S, Bartenstein M, Hussain Z, Suzuki M, Yu Y, Chen W, Eng C, Greally J, Verma A, Meltzer SJ. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett’s esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144:956–966. e954. doi: 10.1053/j.gastro.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye Y, Chen J, Zhou Y, Fu Z, Zhou Q, Wang Y, Gao W, Zheng S, Zhao X, Chen T, Chen R. High expression of AFAP1-AS1 is associated with poor survival and short-term recurrence in pancreatic ductal adenocarcinoma. J Transl Med. 2015;13:137. doi: 10.1186/s12967-015-0490-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng Z, Bo H, Gong Z, Lian Y, Li X, Li X, Zhang W, Deng H, Zhou M, Peng S, Li G, Xiong W. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biol. 2016;37:729–737. doi: 10.1007/s13277-015-3860-x. [DOI] [PubMed] [Google Scholar]
- 18.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, Prasanth KV. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Xu X, Zhang Y, Williams J, Antoniou E, McCombie WR, Wu S, Zhu W, Davidson NO, Denoya P, Li E. Parallel comparison of Illumina RNASeq and Affymetrix microarray platforms on transcriptomic profiles generated from 5-azadeoxycytidine treated HT-29 colon cancer cells and simulated datasets. BMC Bioinformatics. 2013;14(Suppl 9):S1. doi: 10.1186/1471-2105-14-S9-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 22.Askarian-Amiri ME, Seyfoddin V, Smart CE, Wang J, Kim JE, Hansji H, Baguley BC, Finlay GJ, Leung EY. Emerging role of long non-coding RNA SOX2OT in SOX2 regulation in breast cancer. PLoS One. 2014;9:e102140. doi: 10.1371/journal.pone.0102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Hou Z, Zhao W, Zhou J, Shen L, Zhan P, Xu C, Chang C, Bi H, Zou J, Yao X, Huang R, Yu L, Yan J. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int J Biochem Cell Biol. 2014;53:380–388. doi: 10.1016/j.biocel.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 26.Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, Mo YY. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) Cell Death Dis. 2014;5:e1008. doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, Han L, Xia R, Wang KM, Yang JS, De W, Shu YQ, Wang ZX. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]


