Abstract
Krüppel-like factors (KLFs), such as KLF4, KLF2, KLF5 and KLF15, have been extensively investigated in multi-cancers. However, KLF16, a member of KLFs, hasn’t been well identified in cancer, especially in gastric cancer (GC). Here, we investigated the roles of KLF16 in GC. In present study, we found that KLF16 expression levels were significantly up-regulated in GC tissues compared to adjacent normal tissues both in protein and mRNA levels by using immunohistochemistry assays (IHC) and real-time quantitative PCR (qPCR). And KLF16 expression levels were positively correlated to tumor size, invasion depth, lymphatic metastasis and TNM stage. Furthermore, KLF16 expression also could predict prognosis in patients with GC. Moreover, the knock-down of KLF16 could significantly suppress proliferation via increasing p21 expression and decreasing CDK4 expression in GC cell lines. In summary, these findings demonstrate that KLF16 plays a significant role in GC progression and could be a new therapeutic target for GC patients.
Keywords: KLF16, gastric cancer, prognosis, proliferation, p21, CDK4
Introduction
Gastric cancer (GC) has been one of the most frequent diseases which contributes to millions of deaths all over the world [1,2]. It was estimated that there were about 679,100 people who were diagnosed with GC and almost 498,000 people died from GC in China in 2015 [3]. Most of patients were too late to receive surgical treatment when they were first diagnosed with GC, and the clinical benefits of treatment for unrespectable or advanced GC patients have great room for improvement [4,5]. Therefore, it is extremely urgent for clinicians and scientists to develop novel biomarkers and targets for diagnosis and treatment of GC patients. Molecular diagnosis and targeted treatment have been a flourishing field with the high-speed development of genomics and molecular biology [6-8]. Hence, it’s necessary to identify tumor-related candidate genes and further clarify their functions and mechanisms in GC development.
The Krüppel-like factors (KLFs), one type of transcription factors, have been extensively investigated in various diseases [9-16]. KLFs can bind different GC-rich DNA elements where they can regulate transcription in a cell cycle and promoter-dependent manner [17]. Most members of KLFs function as suppressors which can inhibit proliferation, migration and invasion and induce apoptosis [15,18-23], while several members of KLF family act as oncogenic roles [24,25]. Their function and mechanism in tumor progression have been commonly recognized in cancer [26-29]. We previously revealed that KLF2 could repress tumor proliferation by up-regulating p15 and p21 expression [27]. Besides, KLF4 also displays its anti-oncogenic role in diverse cancers [30-34]. KLF5 could promote cell survival in lung cancer and GC [26,35,36]. And KLF6 promotes cell cycle arrest via regulating cyclin D1, thereby disrupting the phosphorylation of retinoblastoma protein [37].
KLF16 has been reported to be involved in metabolism and endocrinology [38]. And, KLF16 can suppress neurite outgrowth and enhance growth cone collapse in response to exogenous ephrinA5 ligands in retinal ganglion cells [39]. Besides, KLF16 inhibits cell growth, suppresses transformation mediated by oncogenic KRAS and increases apoptosis in KRAS oncogenic-mutant cancer cells [40]. So far, however, the expression and function of KLF16 have not been elucidated in GC. Hence, we measured the protein and mRNA expression of KLF16 in gastric cancer tissues and explored the essential function and underlying mechanism of KLF16 in GC.
Material and methods
Patients and specimens
All gastric adenocarcinoma tissue and adjacent normal tissue samples were obtained from the First Affiliated Hospital of Nanjing Medical University between 2013 and 2014 and all patients have written informed consent. The research was approved by the Research Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (Nanjing, China).
Cell lines and cell culture
The human normal gastric epithelial cell line GES-1 and the human gastric adenocarcinoma cell lines BGC-823, SGC-7901, MGC-803, AGS and HGC-27 were obtained from the Chinese Academy of Science Committee on Type Culture Collection Cell Bank (Shanghai, China). Cells were cultured at 37°C in an atmosphere of 5% CO2 in Dulbecco’s modified Eagle’s medium (Gibco, USA) or PRIM 1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum, and 100 IU/ml penicillin, and 100 μg/ml streptomycin (Gibco, USA).
Immunohistochemistry
Gastric adenocarcinoma tissue and adjacent normal tissue samples were immunostained for KLF16 (1:50, ab175892, abcam) and Ki-67 (1:50, ab15580, abcam). Expression was considered to be positive when 50% or more cancer cells were stained.
Quantitative real-time PCR
Total RNA was extracted from cultured cells using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions, and a total of 500 ng RNA was reversely transcribed in a final volume of 10 μl using PrimeScript RT Reagent Kit (Takara, Dalian, China) according to the manufacturer’s instructions. The GAPDH was used as internal control. The primers used for qRT-PCR were shown in Table 3. The quantitative Real-time PCR (qRT-PCR) and data collection were acquired on ABI 7900 HT real-time using SYBR Green Reagent (Vazyme, Nanjing, China) following the manufacturer’s instructions.
Table 3.
Primers used in qRT-PCR
| Primer Name | Primer Sequences (5’-3’) |
|---|---|
| KLF16 Forward | CAAGTCCTCGCACCTAAAGTC |
| KLF16 Reverse | AGCGGGCGAACTTCTTGTC |
| CDNK1A/p21 Forward | TGTCCGTCAGAACCCATGC |
| CDNK1A/p21 Reverse | AAAGTCGAAGTTCCATCGCTC |
| CDK4 Forward | ATGGCTACCTCTCGATATGAGC |
| CDK4 Reverse | CATTGGGGACTCTCACACTCT |
| GAPDH Forward | GCTCTCTGCTCCTCCTGTTC |
| GAPDH Reverse | CCAAATCCGTTGACTC |
Plasmid construction and cell transfection
We obtained KLF16 short hairpin RNA (shRNA) plasmid and empty vector from Genechem (Shanghai, China). KLF16 shRNA target sequences were shown as follows: sh-KLF16 1#: 5’-AGCGCTTCACCCGCAGTGA-3’; sh-KLF16 2#: 5’-CGCACCTAAAGTCGCACCT-3’; sh-KLF16 3#: 5’-GTGCTCATGGCCATCTCTT-3’. Plasmid vector for transfection were prepared using DNA Midiprep kits (Qiagen, Germany), and 2.5 μg plasmids were transfected into cells using X-tremeGENE HP DNA Transfection reagent (Roche, Switzerland) according to the manufacturer’s instruction. Transfected cell lines were further selected with puromycin (1 μg/ml) for 4 weeks. The stably interfering cell lines were identified using real-time PCR and western blotting.
Cell proliferation
The cell proliferation assay was carried out with Cell Counting Kit-8 (CCK8, Biotool, China) according to the manufacturer’s instruction. A total of 2×103 per well transfected cells were seed into 96-well plates in 200 μl of medium supplemented with 10% FBS and incubated at 37°C in an atmosphere of humidified air 5% CO2 incubator. Cell viability was assessed ever 24 h following the manufacturer’s protocol. For colony formation assay, a certain number (n=500) of transfected cells were placed into 6-well plates and cultured in medium supplemented with 10% FBS for 14 days, and medium was replaced every 4 days. Colonies were fixed by methanol and stained with 0.05% crystal violet (Sigma). Each experiment was performed in triplicated and repeated three times.
Flow cytometry
MGC-803 and SGC-7901 cells transfected with sh-KLF16 were harvested 48 h after transfection by trypsinisation. After double staining with FITC-Annexin V and propidium iodide (PI) using the FITC Annexin V Apoptosis Detection Kit (BD Biosciences) according to the manufacturer’s recommendations, the cells were analyzed by flow cytometry (FACScan) equipped with Cell Quest software (BD Biosciences). Cells were classified as viable, dead, early apoptotic, and apoptotic, then the relative number of early apoptotic cells was compared with that in cells transfected with control plasmids. Cells for cell cycle analysis were stained with PI using the CycleTEST™ Plus DNA Reagent Kit (BD Biosciences) following the protocol, and analyzed by FACScan. The percentage of cells in G0/G1, S, and G2/M phase were counted and compared.
Western blotting
The cells were lysed using protein extraction reagent RIPA (Beyotime, China) supplemented with protease inhibitors cocktail (Roche) and PMSF (Beyotime, China). The concentration of proteins was calculated by the Bio-Rad protein assay kit. Thirty micrograms of the protein were separated by 10% SDS polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred to 0.22 μm PVDF membranes (Roche) and incubated with specific antibodies. ECL chromogenic substrate was used to visualize the bands, and the intensity of bands was quantified by densitometry (Quantity One software, Bio-Rad). GAPDH antibody was used as internal control. GAPDH, p21 and CDK4 were provided by Cell Signaling Technology (2118, 2947, 12790). Anti-KLF16 antibodies were purchased from abcam (ab175892).
Statistical analysis
All statistical analyses were performed by SPSS 22.0 software (IBM, USA). The significance of differences between groups was estimated by Student’s t test, χ2 test, or Wilcoxon test, as appropriate. DFS rates were calculated by the Kaplan-Meier method with the log-rank test applied for comparison. Survival data were evaluated using univariate and multivariate Cox proportional hazards model. Variables with a value of p<0.05 in univariate analysis were used in subsequent multivariate analysis on the basis of Cox regression analyses. Two-sided p values were calculated, and a probability level of 0.05 was chosen for statistical significance.
Results
KLF16 was up-regulated in gastric cancer (GC) tissues and cell lines
We found KLF16 was up-regulated in gastric tissues compared with the adjacent normal tissues in TCGA datasets by using bioinformatics software Cancer RNA-SeqNexus (CRN, http://syslab4.nchu.edu.tw/) [41] (data not shown). We determined that KLF16 expression was obviously higher in the gastric tumors than in the adjacent normal tissues by using immunochemistry assays (Figure 1A). Besides, we also assessed the mRNA expression of KLF16 in 60 GC tissues and their paired adjacent normal tissues. The qPCR results showed that KLF16 was dramatically up-regulated in GC tissues compared to paired normal tissues (P<0.001; Figure 1B). KLF16 expression was also detected in the GC cell lines, including BGC-823, SGC-7901, MGC-803, AGS and HGC-27, and the normal gastric epithelial cell line GES-1. KLF16 expression was found distinctly increased in MGC-803 (P=0.0003), SGC-7901 (P=0.005) and BGC-823 (P=0.0006) compared with that in GES-1 (Figure 2A).
Figure 1.
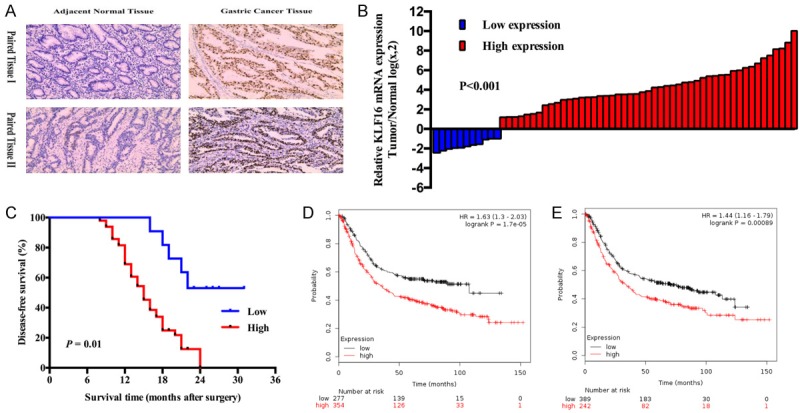
KLF16 expression in patients with GC was significantly up-regulated and positively associated with prognosis. A. Representative KLF16 protein expression levels in GC tissues and adjacent normal tissues were analyzed by immunohistochemistry. B. KLF16 mRNA expression levels in GC tissues and adjacent normal tissues were analyzed by qPCR. C. Kaplan-Meier analysis of Disease-free survival based on KLF16 expression in all the 60 patients. D, E. Online Kaplan-Meier plotter analysis of OS and first-progression survival (FPS) in patients with GC from TCGA and GEO datasets.
Figure 2.
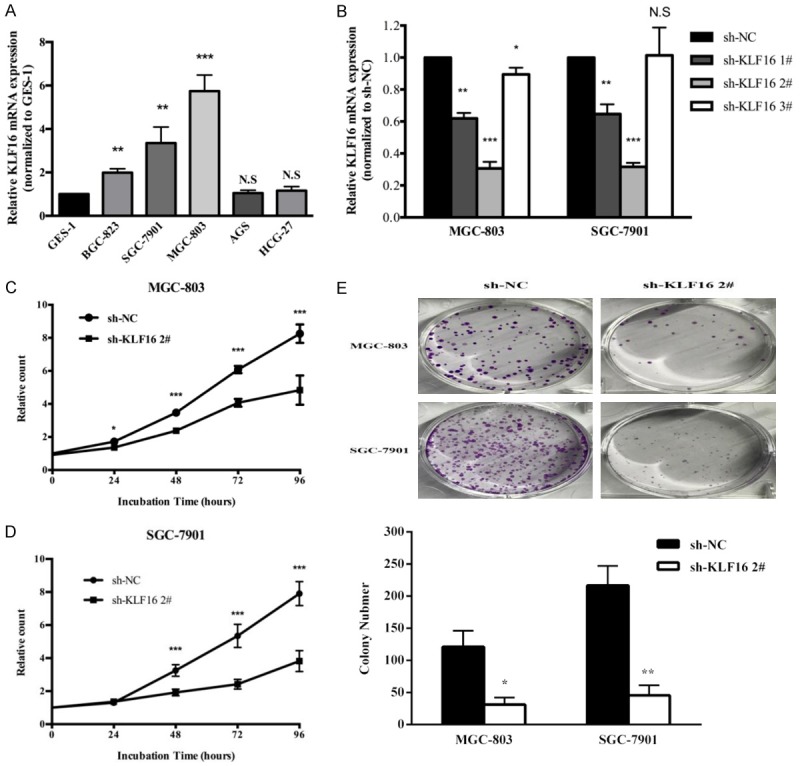
KLF16 knockdown significantly inhibited GC cell proliferation. A. qPCR analysis of KLF16 expression in the normal gastric epithelium cell line (GES-1) and GC cells. Bars: SD. B. qPCR analysis of KLF16 expression transfected with KLF16 shRNA in MGC-803 and SGC-7901. Bars: SD. C, D. CCK8 assays were conducted to determine the cell viability of MGC-803 and SGC-7901 after knockdown expression of KLF16. Experiments were performed in triplicate. Bars: SD. E. Colony-formation growth assays were conducted to determine the cell viability of MGC-803 and SGC-7901 after knockdown expression of KLF16. Experiments were performed in triplicate. Bars: SD.
KLF16 expression was associated with clinicopathological features in GC
KLF16 expression levels in tumor tissues were categorized as low or high depending on whether there were 50% cancer cells that were positively stained with anti-KLF16 antibody compared with the corresponding adjacent noncancerous tissue samples. We analyzed the clinicopathological features in the high- and low-KLF16 expression groups. As shown in Table 1, the high KLF16 group (n=49) showed a greater tumor size (P=0.030), a greater depth of invasion (P=0.004), more lymphatic metastasis (P=0.034) and more advanced TNM stages (P=0.049) than the low KLF16 group (n=11). However, there was no significant correlation between KLF16 expression and other clinicopathological factors, such as gender and age (P>0.05).
Table 1.
Correlation between KLF16 expression and clinicopathological characteristics of gastric cancer cohort (n=60)
| Clinical parameter | KLF16 expression | χ2-test P-value | |
|---|---|---|---|
|
| |||
| High (n=49) | Low (n=11) | ||
| Gender | 1.000 | ||
| Male | 35 | 8 | |
| Female | 14 | 3 | |
| Age (years) | 0.990 | ||
| <50 | 15 | 4 | |
| ≥50 | 34 | 7 | |
| Tumor size | 0.030a | ||
| <5 cm | 12 | 7 | |
| ≥5 cm | 37 | 4 | |
| Histologic differentiation | 0.508 | ||
| Well | 11 | 3 | |
| Moderate | 30 | 5 | |
| Poor | 5 | 1 | |
| Undifferentiated | 3 | 2 | |
| Invasion depth | 0.004a | ||
| T1 | 2 | 3 | |
| T2 | 10 | 4 | |
| T3 | 18 | 3 | |
| T4 | 19 | 1 | |
| Lymphatic metastasis | 0.034a | ||
| Yes | 40 | 5 | |
| No | 9 | 6 | |
| TNM stages | 0.049a | ||
| I | 6 | 5 | |
| II | 9 | 4 | |
| III | 34 | 2 | |
Overall P<0.05.
High KLF16 expression is associated with poor prognosis in GC patients
We used Kaplan-Meier analysis and log-rank test to evaluate the effects of KLF16 expression and clinicopathological features on disease-free survival (DFS). Our results showed that the high KLF16 expression patients had shorter disease-free time (median DFS: 15.698 months) than the low group (median DFS: 25.5 months, P=0.01; Figure 1C). We also found that KLF16 could exert clear influence on patients’ overall survival and progression survival through analyzing GEO, EGA and TCGA datasets from Kaplan-Meier Plotter (http://kmplot.com/analysis/) [42]. Online Kaplan-Meier Plotter results which were consistent with our data indicated that GC patients with higher expression of KLF16 would have a shorter OS and free-progression survival (Figure 1D, 1E). In Table 2, the results of univariate analyses and multivariate Cox proportional hazards model showed that KLF16 expression, together with TNM stage was negatively associated with the DFS in our cohort. The results showed that KLF16 could be an independent prognostic biomarker for predicting DFS (HR=0.221, 95% CI: 0.084-0.580; P=0.02).
Table 2.
Univariate and multivariate Cox regression analyses of KLF16 for DFS of patients in study cohort (n=60)
| Variables | DFS | ||
|---|---|---|---|
|
| |||
| HR | 95% CI | P value | |
| Univariate analysis | |||
| Gender (female vs. male) | 1.233 | 0.643-2.365 | 0.529 |
| Age (<50 years vs. ≥50 years) | 0.851 | 0.522-3.282 | 0.618 |
| Tumor size (<5 cm vs. ≥5 cm) | 0.701 | 0.372 -1.322 | 0.273 |
| Histological differentiation (poor + undifferentiated vs. well + moderate) | 1.395 | 0.645-3.013 | 0.398 |
| Invasion depth (T3 + T4 vs. T1 + T2) | 4.157 | 1.915-9.024 | <0.001a |
| Lymphatic metastasis (Yes vs. No) | 3.402 | 1.431-8.089 | 0.006a |
| TNM stage (III vs. I + II) | 5.144 | 2.436-10.862 | <0.001a |
| KLF16 expression (Low vs. High) | 0.221 | 0.084-0.580 | 0.002a |
| Multivariate analysis | |||
| Invasion depth (T3 + T4 vs. T1 + T2) | 1.728 | 0.601-4.972 | 0.310 |
| Lymphatic metastasis (Yes vs. No) | 0.854 | 0.267-2.728 | 0.790 |
| TNM stage (III vs. I + II) | 2.965 | 1.031-8.523 | 0.044a |
| KLF16 expression (Low vs. High) | 0.369 | 0.358-2.747 | 0.060 |
Overall P<0.05;
HR: hazard ratio; CI: confidence interval.
KLF16 promotes GC cells proliferation
In order to investigate the function of KLF16 in GC cell lines, we chose MGC-803 and SGC-7901 cell lines for research which had a relative higher expression of KLF16. The expression of KLF16 in GC cells and the knock-down efficiency of KLF16 shRNAs were confirmed by qRT-PCR (Figure 2B). We chose KLF16 shRNA 2# for research in vitro because it had more significant knock-down efficiency of KLF16. We performed CCK8 assays to determine the effects of KLF16 on cell proliferation. As shown in Figure 2C, the knock-down of KLF16 could significantly inhibit cell viability. And colony formation assays authenticated the same results as CCK8 assays provided (Figure 2D).
KLF16 promotes proliferation by regulating cyclin-dependent kinases
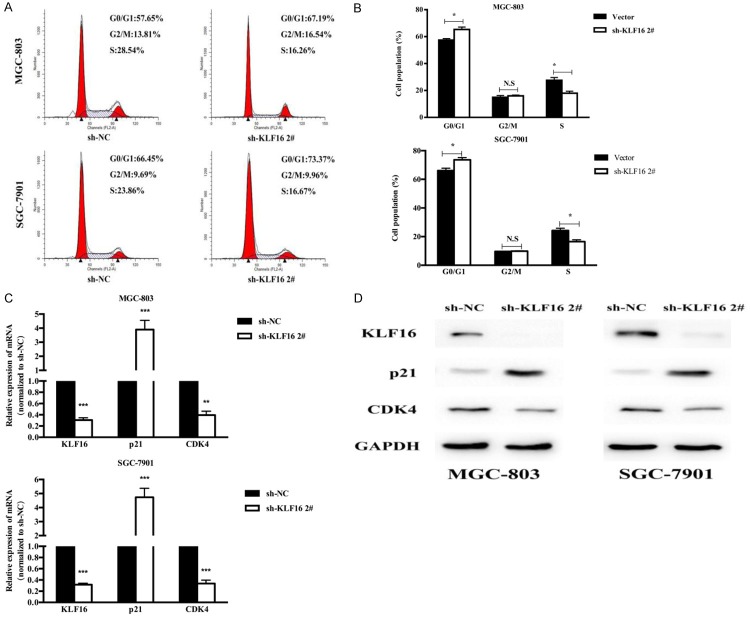
We further observed whether KLF16 promoted proliferation by regulating cell cycle. We used flow cyctometric assays to identify the changes after knock-down of KLF16 in GC cells. The results showed that knock-down of KLF16 could induce cell cycle arrest at G0/G1 phase both in MGC-803 and SGC-7901 cell lines (Figure 3A, 3B). In order to reveal the mechanism of KLF16 which was involved in the induction of G0/G1 arrest, we then assessed the mRNA expression of genes related to cell cycle G0/G1 phase. The qPCR results showed that the mRNA expressions of p21 and CDK4 dramatically decreased after KLF16 silencing (Figure 3C). Moreover, the WB assays confirmed these findings (Figure 3D).
Figure 3.
KLF16 promoted cell proliferation by regulating G0/G1 phase cell cycle transition. A, B. Cell cycle of MGC-803 and SGC-7901 cells were analyzed by flow cytometry after transfection with KLF16 shRNA. C. qPCR analysis of p21 and CDK4 mRNA expression levels following the treatment of KLF16 shRNA in MGC-803 and SGC-7901 cell lines. D. Western blotting analysis of p21 and CDK4 protein expression levels following the transfection of KLF16 shRNA in MGC-803 and SGC-7901 cell lines.
Discussion
The Krüppel-like factors (KLFs) are an essential subfamily of zinc finger-containing transcription factors in eukaryotes that regulate transcription in a cell cycle and promoter-dependent manner. To our best knowledge, KLFs display varied roles in tumor process. For example, KLF2 acts as a tumor suppressor via regulating p15 and p21 in lung cancer [27]. KLF4 can suppress cell survival in gastric cancer, breast cancer and other cancers [10,31,43]. Besides, KLF15 has been reported to function as a tumor suppressor in breast cancer and diverse cancer cell lines [44].
KLF16 has been reported to act as a regulator involved in metabolism and endocrinology [38], and it could suppress neurite outgrowth and enhance growth cone collapse in response to exogenous ephrinA5 ligands in retinal ganglion cells [9]. In addition, KLF16 also could inhibit adipogenesis via decreasing PPARγ expression [45]. Moreover, KLF16 inhibited cellular proliferation, suppressed transformation mediated by oncogenic KRAS and increased apoptosis in KRAS oncogenic-mutant cancer cells [40].
So far, however, the expression pattern and biological function of KLF16 in GC remain unclear. Our study was the first to investigate the expression, prognostic role of KLF16 in patients with GC and potential biological roles in GC cells. We found that KLF16 was highly expressed in GC tissues compared to paired adjacent normal tissues both in protein and mRNA expression levels. Moreover, KLF16 expression was positively related to tumor size, depth of invasion and TNM stage. Moreover, our data also demonstrated that GC patients with higher expression levels of KLF16 also would suffer from a poor prognosis. The Kaplan-Meier analysis showed that DFS was significantly shorter among patients with higher KLF16 expression than in those with lower expression, which was consistent with online Kaplan-Meier Plotter data. Besides, the online Kaplan-Meier Plotter analyses also showed that GC patients with higher KLF16 expression have decreased overall survival than those patients with lower KLF16 expression. Therefore, KLF16 could be used as a novel biomarker for predicting prognosis in GC patients.
In biological functional study, we found that GC cell proliferation was greatly attenuated following KLF16 knock-down by shRNA. Cell cycle analyses demonstrated that knock-down of KLF16 inhibited cell growth by suppressing G0/G1 phase transition. To confirm the molecular mechanism by which KLF16 promoted proliferation, we screened potential targets involved in cell cycle. And it was identified that KLF16 knock-down could significantly decrease the expression of CDK4 both in mRNA and protein levels.
Generally, our research firstly revealed that KLF16 was dramatically up-regulated in GC tissues and related cell lines. In addition, KLF16 was positively associated with aggressive clinical characteristics, including larger tumor size, deeper invasion, advanced TNM stage, shorter DFS and OS. More importantly, KLF16 knock-down inhibited cell proliferation and KLF16 mediated oncogenic effects occurred partially through up-regulating CDK4 expression and down-regulating p21 expression. Conclusively, KLF16 could be a new therapeutic target and prognosis predictor for GC. In future, we will carry out more studies to exploit the specific regulating pathway of KLF16.
Acknowledgements
We acknowledge the support from the National Natural Science Foundation of China (Grant Numbers 81672896, 81302012) and the Priority Academic Program Development of Jiangsu Higher Education Institutions. The authors are thankful to the National Natural Science Foundation of China and the Priority Academic Program Development of Jiangsu Higher Education Institutions for supporting this study.
Informed consent was obtained from all individual participants included in the study.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 5.Ohtsu A. Chemotherapy for metastatic gastric cancer: past, present, and future. J Gastroenterol. 2008;43:256–264. doi: 10.1007/s00535-008-2177-6. [DOI] [PubMed] [Google Scholar]
- 6.Fenech M. The genome health clinic and genome health nutrigenomics concepts: diagnosis and nutritional treatment of genome and epigenome damage on an individual basis. Mutagenesis. 2005;20:255–269. doi: 10.1093/mutage/gei040. [DOI] [PubMed] [Google Scholar]
- 7.Udler M, Maia AT, Cebrian A, Brown C, Greenberg D, Shah M, Caldas C, Dunning A, Easton D, Ponder B, Pharoah P. Common germline genetic variation in antioxidant defense genes and survival after diagnosis of breast cancer. J. Clin. Oncol. 2007;25:3015–3023. doi: 10.1200/JCO.2006.10.0099. [DOI] [PubMed] [Google Scholar]
- 8.Komori T, Takemasa I, Yamasaki M, Motoori M, Kato T, Kikkawa N, Kawaguchi N, Ikeda M, Yamamoto H, Sekimoto M, Matsubara K, Matsuura N, Monden M. Gene expression of colorectal cancer: preoperative genetic diagnosis using endoscopic biopsies. Int J Oncol. 2008;32:367–375. [PubMed] [Google Scholar]
- 9.Wang J, Galvao J, Beach KM, Luo W, Urrutia RA, Goldberg JL, Otteson DC. Novel roles and mechanism for Kruppel-like factor 16 (KLF16) regulation of neurite outgrowth and ephrin receptor A5 (EphA5) expression in retinal ganglion cells. J Biol Chem. 2016;291:21422. doi: 10.1074/jbc.A116.732339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei D, Kanai M, Jia Z, Le X, Xie K. Kruppellike factor 4 induces p27Kip1 expression in and suppresses the growth and metastasis of human pancreatic cancer cells. Cancer Res. 2008;68:4631–4639. doi: 10.1158/0008-5472.CAN-07-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Sun L, Xiao X, Xie R, Liu C, Wang Y, Wei Y, Zhang H, Liu L. Kruppel-like factor 8 contributes to hypoxia-induced MDR in gastric cancer cells. Cancer Sci. 2014;105:1109–1115. doi: 10.1111/cas.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Hamza MS, Leong HS, Lim CB, Pan YF, Cheung E, Soo KC, Iyer NG. Kruppel-like factor 5 modulates p53-independent apoptosis through Pim1 survival kinase in cancer cells. Oncogene. 2008;27:1–8. doi: 10.1038/sj.onc.1210625. [DOI] [PubMed] [Google Scholar]
- 13.Dang DT, Mahatan CS, Dang LH, Agboola IA, Yang VW. Expression of the gut-enriched Kruppel-like factor (Kruppel-like factor 4) gene in the human colon cancer cell line RKO is dependent on CDX2. Oncogene. 2001;20:4884–4890. doi: 10.1038/sj.onc.1204645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown AR, Simmen RC, Raj VR, Van TT, MacLeod SL, Simmen FA. Kruppel-like factor 9 (KLF9) prevents colorectal cancer through inhibition of interferon-related signaling. Carcinogenesis. 2015;36:946–955. doi: 10.1093/carcin/bgv104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao QQ, Chen JJ, Dong L, Zhong L, Sun X. Kruppel-like factor 2 suppresses growth and invasion of gastric cancer cells in vitro and in vivo. J Biol Regul Homeost Agents. 2016;30:703–712. [PubMed] [Google Scholar]
- 16.Nakamura Y, Migita T, Hosoda F, Okada N, Gotoh M, Arai Y, Fukushima M, Ohki M, Miyata S, Takeuchi K, Imoto I, Katai H, Yamaguchi T, Inazawa J, Hirohashi S, Ishikawa Y, Shibata T. Kruppel-like factor 12 plays a significant role in poorly differentiated gastric cancer progression. Int J Cancer. 2009;125:1859–1867. doi: 10.1002/ijc.24538. [DOI] [PubMed] [Google Scholar]
- 17.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito G, Uchiyama M, Kondo M, Mori S, Usami N, Maeda O, Kawabe T, Hasegawa Y, Shimokata K, Sekido Y. Kruppel-like factor 6 is frequently down-regulated and induces apoptosis in non-small cell lung cancer cells. Cancer Res. 2004;64:3838–3843. doi: 10.1158/0008-5472.CAN-04-0185. [DOI] [PubMed] [Google Scholar]
- 19.Kang L, Lu B, Xu J, Hu H, Lai M. Downregulation of Kruppel-like factor 9 in human colorectal cancer. Pathol Int. 2008;58:334–338. doi: 10.1111/j.1440-1827.2008.02233.x. [DOI] [PubMed] [Google Scholar]
- 20.Reeves HL, Narla G, Ogunbiyi O, Haq AI, Katz A, Benzeno S, Hod E, Harpaz N, Goldberg S, Tal-Kremer S, Eng FJ, Arthur MJ, Martignetti JA, Friedman SL. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology. 2004;126:1090–1103. doi: 10.1053/j.gastro.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Shi Q, Gao Y, Xu S, Du C, Li F, Tang XS, Jia J, Wang X, Chang L, He D, Guo P. Kruppel-like factor 5 promotes apoptosis triggered by tumor necrosis factor alpha in LNCaP prostate cancer cells via up-regulation of mitogen-activated protein kinase kinase 7. Urol Oncol. 2016;34:58, e11–8. doi: 10.1016/j.urolonc.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Wei D, Gong W, Kanai M, Schlunk C, Wang L, Yao JC, Wu TT, Huang S, Xie K. Drastic down-regulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746–2754. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Sun M, Zang C, Ma P, He J, Zhang M, Huang Z, Ding Y, Shu Y. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. 2016;7:e2225. doi: 10.1038/cddis.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu N, Wang Y, Zhou Y, Pang H, Zhou J, Qian P, Liu L, Zhang H. Kruppel-like factor 8 involved in hypoxia promotes the invasion and metastasis of gastric cancer via epithelial to mesenchymal transition. Oncol Rep. 2014;32:2397–2404. doi: 10.3892/or.2014.3495. [DOI] [PubMed] [Google Scholar]
- 25.Mori A, Moser C, Lang SA, Hackl C, Gottfried E, Kreutz M, Schlitt HJ, Geissler EK, Stoeltzing O. Up-regulation of Kruppel-like factor 5 in pancreatic cancer is promoted by interleukin-1beta signaling and hypoxia-inducible factor-1alpha. Mol Cancer Res. 2009;7:1390–1398. doi: 10.1158/1541-7786.MCR-08-0525. [DOI] [PubMed] [Google Scholar]
- 26.Chia NY, Deng N, Das K, Huang D, Hu L, Zhu Y, Lim KH, Lee MH, Wu J, Sam XX, Tan GS, Wan WK, Yu W, Gan A, Tan AL, Tay ST, Soo KC, Wong WK, Dominguez LT, Ng HH, Rozen S, Goh LK, Teh BT, Tan P. Regulatory crosstalk between lineage-survival oncogenes KLF5, GATA4 and GATA6 cooperatively promotes gastric cancer development. Gut. 2015;64:707–719. doi: 10.1136/gutjnl-2013-306596. [DOI] [PubMed] [Google Scholar]
- 27.Yin L, Wang JP, Xu TP, Chen WM, Huang MD, Xia R, Liu XX, Kong R, Sun M, Zhang EB, Shu YQ. Downregulation of Kruppel-like factor 2 is associated with poor prognosis for nonsmall-cell lung cancer. Tumour Biol. 2015;36:3075–3084. doi: 10.1007/s13277-014-2943-4. [DOI] [PubMed] [Google Scholar]
- 28.Xu Q, Liu M, Zhang J, Xue L, Zhang G, Hu C, Wang Z, He S, Chen L, Ma K, Liu X, Zhao Y, Lv N, Liang S, Zhu H, Xu N. Overexpression of KLF4 promotes cell senescence through microRNA-203-survivin-p21 pathway. Oncotarget. 2016;7:60290–60302. doi: 10.18632/oncotarget.11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tseng WC, Chuang CW, Yang MH, Pan CC, Tarng DC. Kruppel-like factor 4 is a novel prognostic predictor for urothelial carcinoma of bladder and it regulates TWIST1-mediated epithelial-mesenchymal transition. Urol Oncol. 2016;34:485, e15–e24. doi: 10.1016/j.urolonc.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Nagata T, Shimada Y, Sekine S, Moriyama M, Hashimoto I, Matsui K, Okumura T, Hori T, Imura J, Tsukada K. KLF4 and NANOG are prognostic biomarkers for triple-negative breast cancer. Breast Cancer. 2017;24:326–335. doi: 10.1007/s12282-016-0708-1. [DOI] [PubMed] [Google Scholar]
- 31.Zhang N, Zhang J, Shuai L, Zha L, He M, Huang Z, Wang Z. Kruppel-like factor 4 negatively regulates beta-catenin expression and inhibits the proliferation, invasion and metastasis of gastric cancer. Int J Oncol. 2012;40:2038–2048. doi: 10.3892/ijo.2012.1395. [DOI] [PubMed] [Google Scholar]
- 32.Yu F, Shi Y, Wang J, Li J, Fan D, Ai W. Deficiency of Kruppel-like factor KLF4 in mammary tumor cells inhibits tumor growth and pulmonary metastasis and is accompanied by compromised recruitment of myeloid-derived suppressor cells. Int J Cancer. 2013;133:2872–2883. doi: 10.1002/ijc.28302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu T, Chen X, Lin T, Liu J, Li M, Zhang W, Xu X, Zhao W, Liu M, Napier DL, Wang C, Evers BM, Liu C. KLF4 deletion alters gastric cell lineage and induces MUC2 expression. Cell Death Dis. 2016;7:e2255. doi: 10.1038/cddis.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Wang B, Chen LQ, Yang J, Gong ZQ, Zhao XL, Zhang CQ, Du KL. miR-10b promotes invasion by targeting KLF4 in osteosarcoma cells. Biomed Pharmacother. 2016;84:947–953. doi: 10.1016/j.biopha.2016.09.108. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Liu X, Xu Y, Liu J, Xie M, Ni W, Chen S. KLF5 promotes hypoxia-induced survival and inhibits apoptosis in non-small cell lung cancer cells via HIF-1alpha. Int J Oncol. 2014;45:1507–1514. doi: 10.3892/ijo.2014.2544. [DOI] [PubMed] [Google Scholar]
- 36.Li Q, Dong Z, Zhou F, Cai X, Gao Y, Wang LW. Kruppel-like factor 5 promotes lung tumorigenesis through upregulation of Sox4. Cell Physiol Biochem. 2014;33:1–10. doi: 10.1159/000356645. [DOI] [PubMed] [Google Scholar]
- 37.Benzeno S, Narla G, Allina J, Cheng GZ, Reeves HL, Banck MS, Odin JA, Diehl JA, Germain D, Friedman SL. Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res. 2004;64:3885–3891. doi: 10.1158/0008-5472.CAN-03-2818. [DOI] [PubMed] [Google Scholar]
- 38.Daftary GS, Lomberk GA, Buttar NS, Allen TW, Grzenda A, Zhang J, Zheng Y, Mathison AJ, Gada RP, Calvo E, Iovanna JL, Billadeau DD, Prendergast FG, Urrutia R. Detailed structural-functional analysis of the Kruppel-like factor 16 (KLF16) transcription factor reveals novel mechanisms for silencing Sp/KLF sites involved in metabolism and endocrinology. J Biol Chem. 2012;287:7010–7025. doi: 10.1074/jbc.M111.266007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Galvao J, Beach KM, Luo W, Urrutia RA, Goldberg JL, Otteson DC. Novel roles and mechanism for kruppel-like factor 16 (KLF16) regulation of neurite outgrowth and ephrin receptor A5 (EphA5) expression in retinal ganglion cells. J Biol Chem. 2016;291:18084–18095. doi: 10.1074/jbc.M116.732339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernandez-Zapico ME, Lomberk GA, Tsuji S, DeMars CJ, Bardsley MR, Lin YH, Almada LL, Han JJ, Mukhopadhyay D, Ordog T, Buttar NS, Urrutia R. A functional family-wide screening of SP/KLF proteins identifies a subset of suppressors of KRAS-mediated cell growth. Biochem J. 2011;435:529–537. doi: 10.1042/BJ20100773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li JR, Sun CH, Li W, Chao RF, Huang CC, Zhou XJ, Liu CC. Cancer RNA-Seq Nexus: a database of phenotype-specific transcriptome profiling in cancer cells. Nucleic Acids Res. 2016;44:D944–951. doi: 10.1093/nar/gkv1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szasz AM, Lanczky A, Nagy A, Forster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabo A, Gyorffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel NV, Ghaleb AM, Nandan MO, Yang VW. Expression of the tumor suppressor Kruppel-like factor 4 as a prognostic predictor for colon cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:2631–2638. doi: 10.1158/1055-9965.EPI-10-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoda T, McNamara KM, Miki Y, Onodera Y, Takagi K, Nakamura Y, Ishida T, Suzuki T, Ohuchi N, Sasano H. KLF15 in breast cancer: a novel tumor suppressor? Cell Oncol (Dordr) 2015;38:227–235. doi: 10.1007/s13402-015-0226-8. [DOI] [PubMed] [Google Scholar]
- 45.Jang MK, Lee S, Jung MH. RNA-Seq analysis reveals a negative role of KLF16 in adipogenesis. PLoS One. 2016;11:e0162238. doi: 10.1371/journal.pone.0162238. [DOI] [PMC free article] [PubMed] [Google Scholar]