Abstract
The geochemical energy budgets for high-temperature microbial ecosystems such as occur at Yellowstone National Park have been unclear. To address the relative contributions of different geochemistries to the energy demands of these ecosystems, we draw together three lines of inference. We studied the phylogenetic compositions of high-temperature (>70°C) communities in Yellowstone hot springs with distinct chemistries, conducted parallel chemical analyses, and carried out thermodynamic modeling. Results of extensive molecular analyses, taken with previous results, show that most microbial biomass in these systems, as reflected by rRNA gene abundance, is comprised of organisms of the kinds that derive energy for primary productivity from the oxidation of molecular hydrogen, H2. The apparent dominance by H2-metabolizing organisms indicates that H2 is the main source of energy for primary production in the Yellowstone high-temperature ecosystem. Hydrogen concentrations in the hot springs were measured and found to range up to >300 nM, consistent with this hypothesis. Thermodynamic modeling with environmental concentrations of potential energy sources also is consistent with the proposed microaerophilic, hydrogen-based energy economy for this geothermal ecosystem, even in the presence of high concentrations of sulfide.
Keywords: geothermal springs, phylogenetic study, primary productivity, Yellowstone National Park, hydrogen metabolism
Microbial communities associated with volcanic hot springs have attracted broad interest because of the unique thermophilic properties of the constituent organisms. However, little attention has been given to hot spring communities as whole microbial ecosystems. One fundamental consideration in understanding any ecosystem is the energy budget: the relative contributions of different energy sources that fuel primary productivity, the conversion of carbon dioxide into biomass. Most of Earth's biomass is considered to be the product of photosynthesis. However, at temperatures higher than ≈70°C, photosynthesis is not known to occur,§ but thermophilic microbial communities develop well beyond that temperature (1–5). Consequently, high-temperature primary productivity must derive from chemosynthesis based on the oxidation of reduced inorganic or organic sources. A variety of lithotrophic microorganisms (which use inorganic energy sources) and heterotrophic organisms (which use reduced carbon) have been cultured from hot spring communities (6–11). However, the relative contributions of different potential energy sources to particular communities have not been systematically addressed.
Previous chemical analyses of Yellowstone hot springs have not provided satisfactory explanations of the energy sources that fuel the communities. Potential energy sources detected in different hot springs included sulfide, CH4 and other short-chain hydrocarbons, and reduced metals such as As[III], Fe[II], and Mn[II] (12, 13). However, none of these chemicals is ubiquitous in the hot springs, and robust microbial communities occur in some hot springs with little or none of these potential energy sources.
We propose that one way to gain insight into the relative contributions of potential energy sources available to microbial habitats is to assess the relative abundances of organisms that make up the communities. Microorganisms that engage in primary productivity are expected to be conspicuous in an autotrophic system. If the relative abundances of particular physiological types of organisms are taken to reflect the relative amounts of different energy sources that are drawn on for primary productivity, then a census of the physiologies that comprise a microbial community would correspond to a biological assessment of the energy demands of the particular ecosystem. The phenotypes of different microbes often are revealed by their phylogenetic associations, so a phylogenetic survey of the organisms that comprise a microbial community is expected to yield information on the community bioenergetics. Such a survey cannot be achieved with traditional culture-based methods, because most naturally occurring microbes resist cultivation with standard techniques (14). With the advent of molecular methods for the phylogenetic identification of organisms without the requirement for culture, the relative abundances of microbial community constituents can be estimated (15).
In the most commonly used culture-independent analysis of microbial community composition, small subunit rRNA genes are amplified by PCR from natural community DNA and then cloned and sequenced for phylogenetic identifications. The collection of rRNA gene sequences is a census of the phylogenetic types of organisms that comprise the community. If the organisms indicated by the sequences fall into relatedness groups with predictable forms of energy metabolism, based on cultured representatives, then the probable energy sources for the environmental organisms can be inferred. Microbial communities associated with high-temperature hot springs in Yellowstone National Park and elsewhere have been analyzed to some extent using these culture-independent methods (10, 11, 16–22). One finding of all studies has been the abundant occurrence of microorganisms from the Aquificales bacterial phylogenetic division (8, 10, 18, 20, 21, 23). All known representatives of Aquificales exclusively or preferentially use molecular hydrogen, H2, as an energy source. This dominance by Aquificales members suggested that H2 could be a main energy source in these hot spring ecosystems. However, the occurrence of H2 in Yellowstone hot springs had not been documented, and the few communities previously analyzed were from settings with limited variation in chemical composition.
To explore further the bioenergetics that underpin Yellowstone hot spring communities, we conducted extensive additional characterizations of microbial communities that thrive at >70°C in different chemical regimes. In parallel, we determined the chemical compositions of the hot springs, including the first systematic measurements of aqueous molecular hydrogen in the Yellowstone geothermal system. We then used thermodynamic modeling based on the hot spring chemistries to evaluate the bioenergetic potentials of the available fuels. The results collectively provide new perspective on the energetics of high-temperature ecosystems.
Materials and Methods
Molecular Phylogenetic Analyses. Microbial communities associated with hot springs with high H2 concentrations were surveyed with molecular phylogenetic methods previously described (19). Sediment samples were collected from Washburn Spring 1 (WB1), Washburn Spring 3 (WB3), and Cinder Pool (CPC) and frozen immediately on liquid nitrogen. In springs with little or no sediment, we collected and froze biomass that colonized glass growth slides placed in hot springs for periods of time from 48 h [Obsidian Pool Prime (OPP) Mud Volcano region] to 2 months [West Thumb Pool (WTP), 44°25′26′′, W 110°34′36′′]. Community DNA was extracted from ≈1 g of sample by bead beating (24), which yielded an average of ≈18 μg of DNA per g of sample. For OPP and WB1, we used the UltraClean fecal DNA extraction kit (MoBio Laboratories, Carlsbad, CA); for Cinder Pool, we used the 10-g UltraClean Mega Soil DNA extraction kit (MoBio Laboratories).
PCR primers used in this study included 515F, 1391R, 27F, 1492R, 4Fa, 333Fa (25), 360Fe, 82Fe, and 1391Re (26). PCRs were incubated at 94°C for 2 min followed by 29 cycles at 94°C for 30 sec, 55.5°C for 1 min, and 72°C for 1.5 min, followed by a single 72°C step for 12 min. Each 30-μl reaction contained 1× PCR buffer, 0.2 mM each dNTP, 2.5 mM MgCl2, ≈0.2 μM each primer, 1 mg/ml BSA, 1 M betaine, 0.5 units of Taq polymerase, and ≈200 ng of template. PCR products were gel-purified with the QIAquick Gel Extraction Kit (Qiagen) and cloned with the TOPO TA Cloning Kit for Sequencing (Invitrogen). Unique clones were identified by restriction fragment-length polymorphism (24) and sequenced on a MegaBACE 1000 DNA Sequencer (Amersham Pharmacia). Sequences were aligned and analyzed with the arb software package (www.mikro.biologie.tu-muenchen.de).
Nucleotide sequences have been deposited in the GenBank database under accession nos. AY861719–AY862082. Clone sequences deposited are coded for site (CPC, OPP, WB1, and WB3), PCR primer pair used (A, 515F and 1391R; B, 8F and 1492R; C, 333Fa and 1391R; D, 4Fa and 1391R; E, 360Fe and 1391R; F, 82Fe and 1391R), and three-digit clone number (e.g., OPPD012 = Obsidian Pool Prime, archaeal primer pair 4Fa&1391R, clone number 12).
Hydrogen and Water Chemistry. We measured aqueous H2 concentrations in Yellowstone waters [hot springs, streams, geothermal vents, and a well (27)] with a modified bubble-stripping method (28). Source waters were pumped with a 12-V portable peristaltic pump through insulated, H2-impermeable polyethylene tubing for 20 min at a flow rate of 200 ml/min, through a 250-ml glass bottle bubble stripping device (28). Twenty milliliters of atmospheric air was introduced into the water-filled bottle. The temperature of the bubble was measured with a thermister. Bubbles were collected with an air-tight syringe and transferred to nitrogen-charged, H2-impermeable, glass septum vials and sent to Microseeps (Pittsburgh) for analysis of H2,CH4, CO2, and light hydrocarbons on a RGA3 reduction gas analyzer (Trace Analytical, Newark, DE) (28).
To determine the actual H2 concentration in hot springs, we adjusted the measured values to account for the solubility of H2 at high temperatures with Henry's law: CW = CgHc where CW is the concentration of the gas in the water, Cg is the concentration (partial pressure) of the gas in the bubble, and Hc is Henry's constant for that gas (the solubility of a gas at a given temperature). The temperature effect on Henry's constant is relatively small for most gases. However, for hydrogen, Henry's constant decreases by 28% from 0 to 100°C. Values of Hc at high temperature were estimated with Ostwald's expression: Hc = PH2/RTCW, where PH2 is 1.22 × 10-5 atm (1 atm = 101.3 kPa), R is the gas constant (0.0821 liters atm mol-1 K-1), T is the measured bubble temperature (K), and CW is 1 × 10-8 mol/liter.
Sulfide measurements were conducted with a colorimetric assay (CHEMetrics, Calverton, VA). Samples for water chemistry were filtered by syringe through a 0.2-μm filter, acidified with ultrapure nitric acid, and stored at 4°C until analysis. Anions, cations, and elemental analyses were conducted on a Series 4500I IC (Dionex, Sunnyvale, CA), an ARL 3410+ ICP-AES (Thermo Electron), and a Varian ICP-MS (Varian, Palo Alto, CA).
Thermodynamic Modeling. The amounts of chemical energy available from lithoautotrophic reactions were quantified with the Gibbs free energy equation: 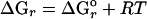 ln Q, where ΔGr is the change in free energy of the reaction,
ln Q, where ΔGr is the change in free energy of the reaction, 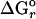 is the standard Gibbs free energy, and Q is the activity quotient of compounds involved in the reaction. Values of
is the standard Gibbs free energy, and Q is the activity quotient of compounds involved in the reaction. Values of 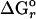 were calculated with the computer program supcrt92 and thermodynamic data contained therein (29). Values of Q were determined with the measured composition of hot spring fluids. Because these are dilute solutions, activity coefficients were assumed to equal one for all dissolved compounds. Distributions of dissolved CO2 and sulfide were calculated from the measured concentrations of these compounds, appropriate dissociation constants, and the measured pH, assuming the species were in equilibrium.
were calculated with the computer program supcrt92 and thermodynamic data contained therein (29). Values of Q were determined with the measured composition of hot spring fluids. Because these are dilute solutions, activity coefficients were assumed to equal one for all dissolved compounds. Distributions of dissolved CO2 and sulfide were calculated from the measured concentrations of these compounds, appropriate dissociation constants, and the measured pH, assuming the species were in equilibrium.
Results
Chemistry of Yellowstone Hot Springs. To provide a chemical context for interpretation of the results of microbiological studies, we conducted chemical analyses of selected hot springs in geologically distinct areas of Yellowstone with evidence of significant biomass (Fig. 1). The results of these analyses and other available data are presented in Tables 1 and 2. Collectively, the sites analyzed are representative of geothermal springs worldwide. Hot springs in Upper Geyser Basin, for instance, contain little sulfide and tend toward alkalinity (pH 8–9), with high concentrations of silica. Hot springs in Norris Geyser Basin and the Mud Volcano area contain relatively high concentrations of sulfides and low-to-neutral pH.
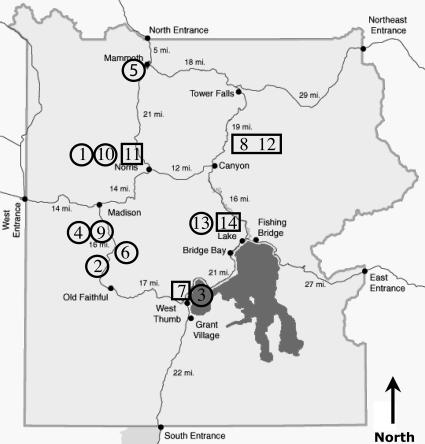
Fig. 1.
Site locations. A map of Yellowstone National Park shows locations of hydrogen measurements indicated by site number (Tables 1 and 2). Boxed numbers identify sites with associated phylogenetic analyses.
Table 1. Hydrogen in Yellowstone National Park hot springs.
| Location | Site | Temp., °C | pH | Eh, V | D.O., mg/liter | SSU* source | Sulfide, μM | Sulfate, μM | H2, nM | CH4, μM | CO2, mM | CnHn, ng/liter |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2X Distilled Water (Control)† | - | 23 | - | 0 | 0 | 2.1 ± 0.00 | 0.0 ± 0.00 | 0.0 ± 0.00 | 0 | |||
| Dragon Pool, (Norris Basin) | 1 | 72 | 3.1 | 21 | 5.8 | ND | 2.9 ± 0.17 | 1.1 ± 0.14 | 22.5 ± 2.0 | 164 | ||
| Well Y-7, Biscuit Basin | 2 | 55 | 7 | 27 | ND | ND | 3.8 ± 0.15 | 8.6 ± 1.10 | 2.7 ± 0.36 | 673 | ||
| Yellowstone Lake | 3 | 9 | 7 | Several | 0 | ND | 4.3 ± 0.00 | 0.1 ± 0.00 | 0.1 ± 0.00 | 20 | ||
| Canary Spring, Mammoth | 5 | 68 | 8.5 | ND | ND | 11.0 ± 3.80 | 0.0 ± 0.00 | 8.1 ± 0.39 | 19 | |||
| Octopus Spring | ||||||||||||
| Fall 2000 | 6 | 92 | 8.5 | 0.018‡ | 0.92‡ | 44 | <0.47‡ | 0.22‡ | 15.0 ± 0.28 | 1.9 ± 0.15 | 1.5 ± 0.08 | 175 |
| Summer 2001 | 92 | 8.5 | 14.0 ± 0.25 | 1.7 ± 0.15 | 1.5 ± 0.08 | 183 | ||||||
| West Thumb Pool | 7 | 89 | 7.3 | This study | 0 | 0.25 | 15.5 ± 0.00 | 7.8 ± 0.41 | 7.9 ± 0.27 | 524 | ||
| Washburn Spring #3 | 8 | 86 | 6.2 | 0.223§ | ND§ | This study | 167§ | 44§ | 18.5 ± 0.60 | 5.8 ± 0.30 | 9.75 ± 0.54 | 436 |
| Queen's Laundry | ||||||||||||
| Fall 2000 | 9 | 89 | 8 | 18 | 2.2¶ | 0.042¶ | 28.0 ± 0.59 | 0.73 ± 0.03 | 1.7 ± 0.21 | 604 | ||
| Summer 2001 | 89 | 8 | 30.4 ± 0.40 | 0.95 ± 0.04 | 2.2 ± 0.27 | 525 | ||||||
| Cinder Pool | ||||||||||||
| Fall 2000 | 11 | 88 | 4.2 | 0.022§ | 0.5§ | This study | 47§ | 1.0§ | 77.6 ± 27.8 | 1.2 ± 0.21 | 16.6 ± 3.80 | 241 |
| Summer 2001 | 88 | 4.3 | 13.7 ± 0.23 | 1.85 ± 0.18 | 24.5 ± 3.18 | 362 | ||||||
| Washburn Spring 1 | 12 | 76 | 6.7 | 0.067§ | 0.3§ | This study | 235§ | 32.5§ | 103.1 ± 1.10 | 8.3 ± 0.36 | 17.2 ± 0.40 | 436 |
| Obsidian Pool | 13 | 80 | 6.5 | 19 | 17.6 | 0.33 | 133.2 ± 5.80 | 0.1 ± 0.00 | 14.9 ± 0.66 | 63 | ||
| Obsidian Pool Prime | 14 | 74 | 5.7 | This study | 17.6 | 0.52 | 325.3 ± 40.0 | 0.1 ± 0.01 | 12.8 ± 0.13 | 21 |
Table 2. Limited water chemistry for springs examined phylogenetically.
| Location | Site | Al, μg/liter | As, μg/liter | Cl, mg/liter | Li, μg/liter | Mn, μg/liter | Fe(II), mg/liter |
|---|---|---|---|---|---|---|---|
| Octopus Spring* | 6 | 512 | 1,380 | 262 | 3,420 | 3.4 | 0.0014† |
| West Thumb Pool | 7 | 66 | 1,265 | 153 | 1,384 | 13.2 | DL |
| Washburn Spring 3‡ | 8 | 68,000 | <1 | 6.7 | <70 | 340 | 65 |
| Queen's Laundry | 9 | 282 | 1,313 | 239 | 1,996 | 0.87 | DL |
| Cinder Pool‡ | 11 | 1,130 | 2,400 | 601 | 4,700 | <6 | 0.088 |
| Washburn Spring 1‡ | 12 | 34,000 | <1 | <10 | 50 | 510 | 23.6 |
| Obsidian Pool | 13 | 349 | DL | 25 | 199 | 427 | 0.11 |
| Obsidian Pool Prime | 14 | 206 | 526 | 305 | 1,171 | 50 | 0.26 |
Concentrations of potential energy sources other than H2, such as sulfide and reduced metals, are highly variable in different hot springs (12, 13). Particularly notable, however, is our finding of ubiquitous H2 at concentrations appropriate for energy metabolism, >5–10 nM (30–33). H2 concentrations ranged to > 300 nM and were spring-dependent but seasonally constant in three springs tested (Queens Laundry, Octopus Spring, and Cinder Pool) (see Tables 1 and 2). Other potential energy sources, such as Fe[II], Mn[II], and NH4, occur variably (Table 2 and refs. 12 and 13). However, the energy yield from microbial oxidation of such compounds is low relative to other sources, so they probably do not contribute substantially to the overall energy budget of these communities. Moreover, deposits of iron and manganese oxide/hydroxide minerals, the products of microbial oxidation of Fe[II] and Mn[II], although sometimes present in the hot springs, are not conspicuous.
Microbiological Analyses. We determined the composition of microbial communities from hot springs >70°C with high H2 concentrations. To test the impact of reduced sulfur compounds on community composition, we examined hot springs with a range of sulfide concentrations (Table 1). The presence or absence of sulfide might influence the composition of a community if significant in the energy budget of that community. Hydrogen concentrations varied among the springs, generally with higher concentrations in springs with higher concentrations of Fe[II] and sulfide (Tables 1 and 2). To determine the composition of microbial communities associated with these chemical settings, we amplified, cloned, and sequenced rRNA genes from crust and sediment communities as well as pioneer communities scraped from glass slides incubated in hot springs. Overall, ≈2,500 randomly chosen rRNA gene clones were surveyed by restriction fragment-length polymorphism, and ≈400 new sequences were determined and submitted to the GenBank database.
To determine the phylogenetic types of organisms present, we compared the sequences to sequences of known organisms in public databases. We also compared the compositions of the communities. Although the detail of compositions varied, all of the communities contained sequences representative of the same phylogenetic groups. Samples obtained on artificial growth surfaces generally overlapped with the environmental sediment samples. Fig. 2 summarizes the census results.
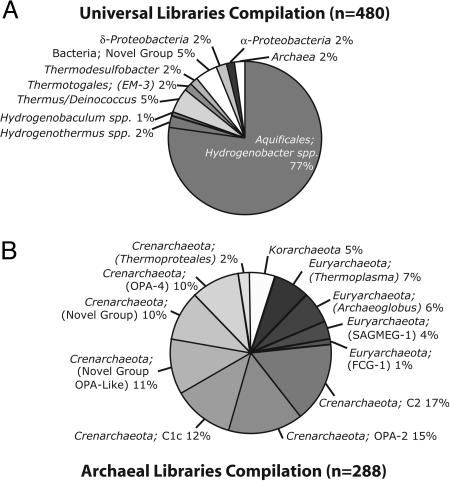
Fig. 2.
Cumulative rRNA gene analyses. (A) Distribution of sequences by phylogenetic group as identified with arb. Universal PCR primers (515F and 1391R) were used with environmental DNA templates from five hot springs, and resultant sequences were compiled for the assemblage. Five percent of the sequences are from one potentially new candidate bacterial division encountered in this study. (B) Distribution of archaeal sequences in three hot springs with two archaeal-specific PCR primer pairs. The majority, 77% of the sequences, are identified as crenarchaeotes. Eighteen percent fall within Euryarchaeota, and 5% fall within Korarchaeota. OPA-2, OPA-4, and OPA-Like represent environmental DNA sequences from a previous study of Obsidian Pool (16); FCG-1 represents sequences from marine/hydrothermal vent benthic archaea; and SEGMEG-1 represents sequences from deep South African mines.
The phylogenetic distribution of rRNA genes amplified with the universal PCR primers (Fig. 2 A) provides some perspective on the overall microbial composition of the Yellowstone geothermal ecosystem. Communities were dominated by bacterial rRNA genes. Archaea are considered common in geothermal and other “extreme” environments, but these and all previous surveys indicate that such organisms are not more abundant than bacteria (19). Most of the archaeal sequences encountered were related to environmental crenarchaeote sequences previously observed in Obsidian Pool (16, 17), none with a specific relationship to a cultured organism (Fig. 2B).
Fig. 3 shows the main phylogenetic groups identified in springs with five different chemical compositions. Although several hundred unique bacterial sequences were determined, these fell into only a few phylogenetic groups. Sequences representative of Aquificales were most abundant in the communities, and sequences representative of Thermotogales, Thermus/Deinococcus, and Thermodesulfobacteria also were common.
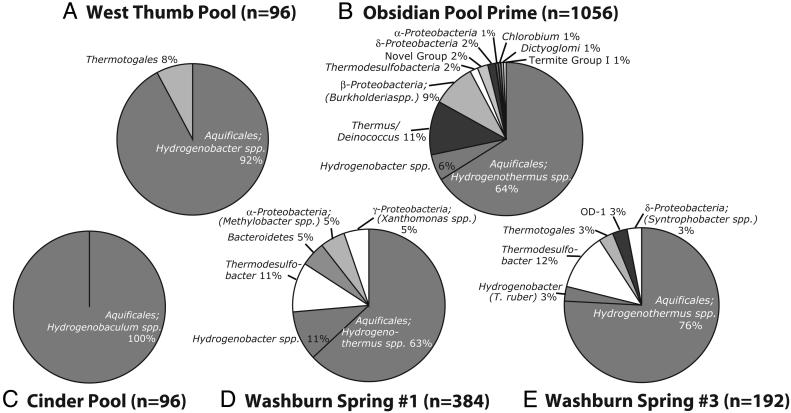
Fig. 3.
Bacterial rRNA gene clone libraries. Bacterial sequences (arb phylogenetic assignment) for five previously unexamined hot springs are shown as pie charts. At least two PCR primer pairs and as many as eight (Obsidian Pool Prime) were used to determine the compositions for each hot spring.
Collectively, ≈90% of sequences obtained were representatives of these phylogenetic groups. These results are consistent with earlier findings from more limited studies (16–20). Most Aquificales sequences were closely related to cultured organisms that rely on H2 as an energy source, including Thermocrinus ruber (8), Hydrogenobacter spp. (34), Hydrogenobaculum spp. (35), and Hydrogenothermus spp. (36). Because representatives of a relatedness group are expected to have properties that are uniformly present in known members of the group, the environmental hot spring organisms represented by the dominant sequences are predicted to engage in hydrogen oxidation.
Comparison of community compositions in low- and high-sulfide samples (Fig. 4) indicates that organisms recognized for sulfur oxidation, such as relatives of Thiobacillus spp., do not dominate. Instead, δ-proteobacterial sequences emerge in communities with higher sulfide concentrations. Many of these sequences are specifically related to δ-proteobacteria known for sulfate reduction and commonly use H2 as a reductant. Sulfate is often present in the hot springs (Table 1). Our results suggest that, when sulfate is present, sulfate-reducing bacteria can contribute significantly to the energy budget of the community.
Fig. 4.
Low- and high-sulfide communities compilation. (A) The phylogenetic distribution of rRNA gene sequences obtained from the two low-sulfide springs of this study (West Thumb Pool and Obsidian Pool Prime) combined with the five low-sulfide springs studied by Blank et al. in ref. 18 (Octopus Spring, Queens Laundry, Eclipse Geyser, Spindle Spring, and Boulder Spring). (B) The phylogenetic distribution of rRNA gene sequences obtained from the three high-sulfide springs in this study (Cinder Pool, Washburn Spring 1, and Washburn Spring 3).
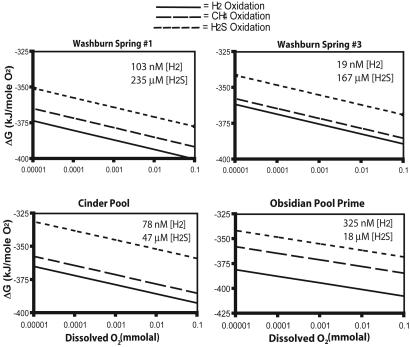
Thermodynamic Modeling. We modeled the potential energy available to the microbial communities in hot springs. Because photosynthesis does not occur above ≈70°C,§ most microbes in Yellowstone hot spring environments must obtain their energy from reduced compounds in geothermal fluids. Dissolved sulfides, CH4, and H2 are the principal potential energy sources available to these communities. The relative potential energy yields of available energy sources can be estimated from the measured chemical compositions of the hot springs. This potential energy depends on available O2, which is difficult to measure but is low because of the reduced nature of hot springs and the poor solubility of O2 in hot water (12, 13). Therefore, we modeled potential energy available in four springs for oxygen-consuming lithoautotrophic reactions over a range of O2 concentrations (Fig. 5) (37). Results show that H2 oxidation was favored under oxygen-limited conditions observed in hot springs, which is consistent with the apparent dominance of putative hydrogen metabolizers in this study. The dominant abundance of predicted hydrogen-oxidizing organisms occurs in hot springs with high sulfide concentrations (e.g., Washburn Spring 1, site 12; see Fig. 1) and low sulfide concentrations (e.g., Obsidian Pool Prime, site 14, and West Thumb Pool, site 7).
Fig. 5.
Results of thermodynamic models. Shown are the amounts of energy available from O2-consuming metabolic reactions, expressed in terms of available energy per mole of limiting O2 for comparative purposes. The available energy is shown for a range of hypothetical O2 concentrations because accurate O2 concentrations in hot waters are difficult to assess.
Discussion
Microbiology historically has focused on single organisms, with limited attention to microbial ecosystems. Indeed, it is a challenge even to identify a microbial “ecosystem,” in the sense of an ecological unit. Microbial ecosystems are constrained not by geography or climate, but rather by local chemical and physical conditions. Innumerable microbial ecosystems collectively underpin and mold our biosphere, so it is important to strive to understand their biochemical webs.
In this study we considered a relatively simple and confined ecosystem setting, Yellowstone geothermal springs >70°C, and we posed a simple question: What is the main source of metabolic energy that drives such communities? We draw together three lines of inference to propose that the main energy source for these communities is H2.O2 and, to a lesser extent, oxidized sulfur species serve as the main terminal electron acceptors. Sulfide, long considered an important energy source for hot spring communities, seems to play a minor role. Microbial sulfide oxidation may play a more prominent role further away from hot spring sources, where cooler waters allow higher O2 solubility.
The phylogenetic composition of these communities is the first line of inference that leads us to the conclusion that hydrogen is their main energy source. These and previous molecular analyses of hot springs with varied chemistries show that the dominant rRNA genes are derived from close relatives of species known for hydrogen metabolism. The second line of inference, which corroborates the molecular results, is the finding of ubiquitous H2 in Yellowstone hot springs, at concentrations sufficient for microbial bioenergetics. Finally, thermodynamic calculations based on O2 limitation show that H2 metabolism is favored in this ecosystem.
Our conclusions are based on several assumptions about the molecular approach to microbial community analysis. In principal, rRNA clone libraries provide a snapshot of the relative proportions of phylogenetic types in a community, and some properties of those individuals can be inferred from phylogenetic information. Representatives of a relatedness group are expected to have properties that are common to the group (38). However, we acknowledge that potential experimental artifacts can bias how well clone libraries represent the actual proportions of phylogenetic types in a sample (reviewed in refs. 39 and 40). Such artifacts include variable PCR amplification due to primer selectivity and differential extraction of genomic DNA from samples. Nonetheless, comparisons of results obtained from clone libraries and other methods, such as fluorescent in situ hybridization (41) and rRNA hybridization (42), show that careful application of molecular methods accurately identifies the abundant organisms in a sample.
We endeavored to minimize potential experimental artifacts by analysis of clone libraries prepared using different suites of PCR primers with broad specificities, an approach used successfully in other studies (43). Obsidian Pool Prime, for example, was examined with eight different PCR primer pairs. Although different libraries always contained some unique sequences not seen in other libraries from the same environmental DNA, we see no significant difference in the proportions of phylogenetic groups in the libraries. Regardless of potential biases, >93% of rRNA sequences characteristic of H2-oxidizing microbes dominate both low- and high-sulfide springs. This finding provides strong evidence that such organisms constitute the main component of these communities. Also, each of the communities is probably more complex than we detect. At most, we analyzed several hundred randomly chosen clones, and in no case did we exhaust the diversity in a library. Thus, our analysis captures only the most abundant rRNA genes. Substantial diversity remains to be uncovered in these and other geothermal systems.
The Yellowstone hot spring communities are relatively simple from the perspective of rRNA gene sequences. However, microbes that have even identical rRNA sequences may not be entirely identical. Because hot spring geochemistry varies, organisms are expected to evolve adaptations to local conditions. For instance, organisms in settings with little reduced iron (e.g., Octopus Spring) may have mechanisms for the acquisition or utilization of iron that are not required in high-iron hot springs (e.g., Obsidian Pool). Previous studies have shown genetic variation among microbes with identical rRNA sequences from different hot springs, indicated by variation in sequences of rRNA internal transcribed spacers (18, 44).
The importance of H2-metabolizing organisms in environmental microbiology has long been recognized (1). The nM concentrations of H2 reported here are consistent with those reported for other oxygen-poor environments such as lake sediment (36 nM), rice paddies (28 nM), and sewage sludge (203 nM) (30). Hydrogen in Yellowstone geothermal waters is likely geochemical in origin. Sources of geochemical H2 are not well understood in general (45), but in the Yellowstone environment they probably derive from subsurface interaction of water with Fe[II] (46–51). Life in the subsurface probably is limited more by the availability of oxidant than of fuels such as H2 (52). This theme of hydrogen as a main fuel in Yellowstone hot springs likely resonates to other geohydrothermal ecosystems, where H2 probably is common in anoxic water.
Acknowledgments
This article is dedicated to the memory of Tommy Gold, a pioneer in thought on carbon and energy sources in the Earth's crust. We gratefully acknowledge the help and assistance of the Yellowstone Center for Resources. Dr. Kirk Nordstrom (U.S. Geological Survey, Boulder, CO) provided useful insight and feedback throughout the life of the project. We thank members of the N.R.P. laboratory for collegiality, review, and thoughtful comments on the manuscript. Funds for this work have been provided by National Science Foundation Life in Extreme Environments Grant DEB-9870880 (to N.R.P.) the National Aeronautics and Space Administration Astrobiology Institute (to N.R.P.), a National Science Foundation Microbial Biology Postdoctoral fellowship (to J.R.S.), and an Agouron Institute Postdoctoral Fellowship (to J.R.S.).
Author contributions: J.R.S. designed research; J.R.S. and J.J.W. performed research; J.R.S. contributed new reagents/analytic tools; J.R.S., J.J.W., T.M.M., and N.R.P. analyzed data; J.R.S. and N.R.P. wrote the paper; J.R.S. conducted all field work.; and J.J.W. assisted with all field work.
Data deposition: Nucleotide sequences have been deposited in the GenBank database (accession nos. AY861719–AY862082).
Footnotes
Cox, A. D. & Shock, E. L., Poster B41D-0927, American Geophysical Union Annual Meeting, Dec. 8–12, 2003, San Francisco.
References
- 1.Madigan, M. T., Martinko, J. M. & Parker, J. (2003) Brock Biology of Microorganisms (Prentice Hall, Upper Saddle River, NJ).
- 2.Brock, T. D. (1967) Science 158, 1012-1019. [DOI] [PubMed] [Google Scholar]
- 3.Brock, T. D. (1978) Thermophilic Microorganisms and Life at High Temperatures (Springer, New York).
- 4.Jannasch, H. W. (1985) Proc. R. Soc. London Ser. B. 225, 277-297. [Google Scholar]
- 5.Jannasch, H. W. & Mottl, M. J. (1985) Science 229, 717-725. [DOI] [PubMed] [Google Scholar]
- 6.Huber, G., Drobner, E., Huber, H. & Stetter, K. O. (1992) Syst. Appl. Microbiol. 15, 502-504. [Google Scholar]
- 7.Huber, R., Burggraf, S., Mayer, T., Barns, S. M., Rossnagel, P. & Stetter, K. O. (1995) Nature 376, 57-58. [DOI] [PubMed] [Google Scholar]
- 8.Huber, R., Eder, W., Heldwein, S., Wanner, G., Huber, H., Rachel, R. & Stetter, K. O. (1998) Appl. Environ. Microbiol. 64, 3576-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Norris, T. B., Wraith, J. M., Castenholz, R. W. & McDermott, T. R. (2002) Appl. Environ. Microbiol. 68, 6300-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reysenbach, A.-L., Wickham, G. S. & Pace, N. R. (1994) Appl. Environ. Microbiol. 60, 2113-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reysenbach, A. L., Ehringer, M. & Hershberger, K. (2000) Extremophiles 4, 61-67. [DOI] [PubMed] [Google Scholar]
- 12.Ball, J. W., Nordstrom, D. K., Cunningham, K. M., Schoonen, M. A. A., Xu, Y. & Demonge, J. M. (1998) Water-Chemistry and On-Site Sulfur-Speciation Data for Selected Springs in Yellowstone National Park, Wyoming, 1994–1995 (U.S. Geological Survey, Boulder, CO), Open-File Report 98-574.
- 13.Ball, J. W., Nordstrom, D. K., Jenne, E. A. & Vivit, D. V. (1998) Chemical Analyses of Hot Springs, Pools, Geysers, and Surface Waters from Yellowstone National Park, Wyoming, and Vicinity, 1974–1975 (U.S. Geological Survey, Boulder, CO), Open-File Report 98-182.
- 14.Amann, R. I., Ludwig, W. & Schleifer, K. H. (1995) Microbiol. Rev. 59, 143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pace, N. R. (1997) Science 276, 734-740. [DOI] [PubMed] [Google Scholar]
- 16.Barns, S. M., Delwiche, C. F., Palmer, J. D. & Pace, N. R. (1996) Proc. Natl. Acad. Sci. USA 93, 9188-9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barns, S. M., Fundyga, R. E., Jeffries, M. W. & Pace, N. R. (1994) Proc. Natl. Acad. Sci. USA 91, 1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blank, C. E., Cady, S. L. & Pace, N. R. (2002) Appl. Environ. Microbiol. 68, 5123-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hugenholtz, P., Pitulle, C., Hershberger, K. L. & Pace, N. R. (1998) J. Bacteriol. 180, 366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugenholtz, P., Goebel, B. M. & Pace, N. R. (1998) J. Bacteriol. 180, 4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donahoe-Christiansen, J., D'Imperio, S., Jackson, C. R., Inskeep, W. P. & McDermott, T. R. (2004) Appl. Environ. Microbiol. 70, 1865-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stahl, D. A., Lane, D. J., Olsen, G. J. & Pace, N. R. (1985) Appl. Environ. Microbiol. 49, 1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skirnisdottir, S., Hreggvidsson, G. O., Hjorleifsdottir, S., Marteinsson, V. T., Petursdottir, S. K., Holst, O. & Kristjansson, J. K. (2000) Appl. Environ. Microbiol. 66, 2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dojka, M. A., Hugenholtz, P., Haack, S. K. & Pace, N. R. (1998) Appl. Environ. Microbiol. 64, 3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lane, D. J. (1991) in Nucleic Acid Techniques in Bacterial Systematics, eds. Stackebrandt, E. & Goodfellow, M. (Wiley, New York), pp. 115-175.
- 26.Dawson, S. C. & Pace, N. R. (2002) Proc. Natl. Acad. Sci. USA 99, 8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spear, J. R., Walker, J. J. and Pace, N. R. (2002) Yellowstone Science 10, 15-21. [Google Scholar]
- 28.Chapelle, F. H., Vroblesky, D. A., Woodward, J. C. & Lovley, D. R. (1997) Environ. Sci. Technol. 31, 2873-2877. [Google Scholar]
- 29.Johnson, J. W., Oelkers, E. H. & Helgeson, H. C. (1992) Comput. Geosci. 18, 899-947. [Google Scholar]
- 30.Zinder, S. H. (1993) in Methanogenesis: Ecology, Physiology, Biochemistry & Genetics, ed. Ferry, J. G. (Chapman & Hall, New York), pp. 128-206.
- 31.Lovley, D. R. (1985) Appl. Environ. Microbiol. 49, 1530-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovley, D. R., Dwyer, D. F. & Klug, M. J. (1982) Appl. Environ. Microbiol. 43, 1373-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovley, D. R. & Klug, M. J. (1982) Appl. Environ. Microbiol. 43, 552-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pitulle, C., Yang, Y., Marchiani, M., Moore, E. R., Siefert, J. L., Aragno, M., Jurtshuk, P., Jr., & Fox, G. E. (1994) Int. J. Syst. Bacteriol. 44, 620-626. [DOI] [PubMed] [Google Scholar]
- 35.Shima, S., Yanagi, M. & Saiki, H. (1994) FEMS Microbiol. Lett. 119, 119-122. [DOI] [PubMed] [Google Scholar]
- 36.Stohr, R., Waberski, A., Volker, H., Tindall, B. J. & Thomm, M. (2001) Int. J. Syst. Evol. Microbiol. 51, 1853-1862. [DOI] [PubMed] [Google Scholar]
- 37.McCollom, T. M. & Shock, E. L. (1997) Geochim. Cosmochim. Acta 61, 4375-7391. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt, T. M., DeLong, E. F. & Pace, N. R. (1991) J. Bacteriol. 173, 4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Wintzingerode, F., Gobel, U. B. & Stackebrandt, E. (1997) FEMS Microbiol. Rev. 21, 213-229. [DOI] [PubMed] [Google Scholar]
- 40.Kanagawa, T. (2003) J. Biosci. Bioeng. 96, 317-323. [DOI] [PubMed] [Google Scholar]
- 41.Juretschko, S., Loy, A., Lehner, A. & Wagner, M. (2002) Syst. Appl. Microbiol. 25, 84-99. [DOI] [PubMed] [Google Scholar]
- 42.Massana, R., Murray, A. E., Preston, C. M. & DeLong, E. F. (1997) Appl. Environ. Microbiol. 63, 50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frank, D. N., Spiegelman, G. B., Davis, W., Wagner, E., Lyons, E. & Pace, N. R. (2003) J. Clin. Microbiol. 41, 295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papke, R. T., Ramsing, N. B., Bateson, M. M. & Ward, D. M. (2003) Environ. Microbiol. 5, 650-659. [DOI] [PubMed] [Google Scholar]
- 45.Apps, J. A. & van de Kamp, P. C. (1993) The Future of Energy Gases (U.S. Geological Survey, Washington, DC), Vol. 1570.
- 46.Stevens, T. O. & McKinley, J. P. (1995) Science 270, 450-454. [Google Scholar]
- 47.Madsen, E. L., Lovley, D. R., Chapelle, F. H., Stevens, T. & McKinley, J. (1996) Science 272, 896-897. [Google Scholar]
- 48.Anderson, R. T., Chapelle, F. H. & Lovley, D. R. (2001) Environ. Sci. Technol. 35, 1556-1559. [DOI] [PubMed] [Google Scholar]
- 49.Anderson, R. T., Chapelle, F. H. & Lovley, D. R. (1998) Science 281, 976-977. [DOI] [PubMed] [Google Scholar]
- 50.Stevens, T. O. & McKinley, J. P. (2000) Environ. Sci. Technol. 34, 826-831. [Google Scholar]
- 51.Sleep, N. H., Meibom, A., Fridriksson, T., Coleman, R. G. & Bird, D. K. (2004) Proc. Natl. Acad. Sci. USA 101, 12818-12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gold, T. (1992) Proc. Natl. Acad. Sci. USA 89, 6045-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson, J. M. & DeMonge, J. M. (1996) Chemical Analyses of Hot Springs, Pools, and Geysers from Yellowstone National Park, Wyoming, and Vicinity, 1980–1993 (U.S. Geological Survey, Reston, VA), Open-File Report 96-68.