Abstract
Peptides of the corticotropin-releasing factor (CRF) family signal through the activation of two receptors, CRF receptor type 1 (CRFR1) and type 2 (CRFR2), both of which exist as multiple splice variants. We have identified a cDNA from mouse brain encoding a splice variant, soluble CRFR2α (sCRFR2α), in which exon 6 is deleted from the gene encoding CRFR2α. Translation of this isoform produces a predicted 143-aa soluble protein. The translated protein includes a majority of the first extracellular domain of the CRFR2α followed by a unique 38-aa hydrophilic C terminus resulting from a frame shift produced by deletion of exon 6. By using RT-PCR and Southern hybridization, the relative mRNA expression levels of full-length (seven transmembrane domains) CRFR2α and the soluble form (sCRFR2α) in the mouse brain were measured with a single reaction. The results demonstrate high levels of expression of sCRFR2α in the olfactory bulb, cortex, and midbrain regions. A rabbit antiserum raised against a synthetic peptide fragment encoding the unique C terminus revealed specific sCRFR2α immunoreactivity in mouse brain slices by immunohistochemistry and in extracts of brain regions by RIA. Interestingly, the sCRFR2α immunoreactivity distribution closely approximated that of CRFR1 expression in rodent brain. A protein corresponding to sCRFR2α, expressed and purified from either mammalian or bacterial cell systems, binds several CRF family ligands with low nanomolar affinities. Furthermore, the purified sCRFR2α protein inhibits cellular responses to CRF and urocortin 1. These data support a potential role of the sCRFR2α protein as a possible biological modulator of CRF family ligands.
Keywords: urocortins, binding protein, decoy receptor
The hypothalamic hypophysiotropic peptide corticotropin releasing factor (CRF), originally isolated from the hypothalamus (1), plays an important role in the regulation of the hypothalamo–pituitary–adrenal axis under basal and stress conditions (2, 3). Furthermore, CRF acts to integrate endocrine, autonomic and behavioral responses to stressors (2–4). The mammalian CRF peptide family comprises urocortin (Ucn) 1 (5) and the peptides Ucn 2 and Ucn 3, which are also known as stresscopin-related peptide (6, 7) and stresscopin (7, 8), respectively.
The effects of CRF-related peptides are mediated through the activation of two high-affinity membrane receptors, CRF receptors (CRFRs) 1 (9–11) and 2 (12–16), which belong to the B1 subfamily of seven-transmembrane-domain receptors that signal by coupling to G proteins. One functional variant of the CRFR1 gene is expressed in humans and rodents, along with several nonfunctional variants, which are produced by differential splicing of various exons (17, 18). The CRFR2 has three functional splice variants in human (α, β, and γ) and two rodent variants (α and β) that are produced by the use of alternate 5′ exons (12–16, 18, 19).
CRFR1 mRNA is widely expressed in mammalian brain and pituitary, with high levels found in the anterior pituitary, cerebral cortex, cerebellum, amygdala, hippocampus, and olfactory bulb (20). In the periphery, CRFR1 is expressed in testes, ovary, skin, and spleen. CRFR2 mRNA is expressed in a discrete pattern in the brain with the highest densities in the lateral septal nucleus, bed nucleus of stria terminalis, ventromedial hypothalamic nucleus, olfactory bulb, and mesencephalic raphe nuclei (20). CRFR type 2α is the major splice variant expressed in the rodent brain (21), whereas CRFR type 2β is expressed in peripheral tissues, with the highest levels in the skeletal muscle, heart, and skin (12).
The distributions of CRFR1 and CRFR2 are distinct and imply diverse physiological functions, as demonstrated by the divergent phenotypes of the CRFR1- or CRFR2-null mice. Mice deficient for CRFR1 display decreased anxiety-like behavior and have an impaired stress response (22, 23), whereas the CRFR2-null mice have increased anxiety-like behaviors and an exaggerated hypothalamo–pituitary–adrenal response to stress (24–26). However, the responses to administration of CRFR2 agonists and antagonists into specific brain regions reveal anxiolytic and anxiogenic roles for CRFR2 (27).
Radioreceptor and functional assays have demonstrated that CRFR1 and CRFR2 differ pharmacologically: Ucn 1 has equal affinities for both receptors and is more potent than CRF on CRFR2, whereas Ucn 2 and Ucn 3 appear to be selective for CRFR2 (5, 6, 8).
The activation of specific CRFRs in distinct tissues or cell types by receptor-selective CRF peptides initiates a variety of signaling pathways, including coupling to different G proteins, stimulation of protein kinase B, PKC, intracellular calcium, and mitogen-activated protein kinase (for reviews, see refs. 27–29).
Soluble proteins related to membrane receptors can be generated by enzymatic truncation of membrane-bound receptors as suggested for the growth hormone-releasing hormone receptor (30), dopamine D3 receptor (31), and calcitonin receptor (32), or by alternative splicing in the case of the glutamate receptors (33–35). Splice variants containing only the extracellular region of G-protein-coupled receptors have been reported (17–19, 33–39). In the majority of cases, these proteins act as binding, nonsignaling molecules also referred to as decoy receptors. Two partial cDNA fragments (CRFR1e and CRFR1h) comprising deletion of exons 3 and 4 and addition of a cryptic exon in CRFR1 were identified in human skin and predicted to exist as soluble proteins (17). One of these fragments, CRFR1e, exhibited dominant negative effects when cotransfected with wild-type CRFR1.
Here we present data on the cloning and pharmacological and anatomical characterization of a splice variant of the mouse CRFR type 2α (CRFR2α) gene encoding a putative soluble CRFR-related protein [soluble CRFR type 2α (sCRFR2α)]. In addition, we describe its effects on CRFR signaling.
Materials and Methods
Isolation of the Mouse Soluble CRFR2α cDNA. The soluble CRFR2α splice variant was isolated in parallel with that of the mouse CRFR2α ortholog (16). PCR primers were designed based on the homology between known mammalian CRFR2 genes. Oligonucleotide primers 5′ CCCCGAAGCTGCCCGACTGG 3′ (sense) and 5′ GGAAGGCTGTAAAGGATGGAGAAG 3′ (antisense) were used to screen cDNA prepared from mouse whole-brain poly(A)+ RNA, which was reverse-transcribed by using oligo(dT) or random primers. PCR was performed at 62°C for 35 cycles with 90-sec extension at 72°C. The amplified fragments were subcloned into pCRIITOPO vector (Invitrogen), sequenced, and found to encode the full-length CRFR2α (16) and a splice variant lacking exon 6, sCRFR2α.
Semiquantitative RT-PCR and Southern Analysis. The following mouse peripheral and CNS tissues were dissected and directly subjected to total RNA isolation as described in ref. 16: total brain, olfactory bulb, hypothalamus, cortex, cerebellum, hippocampus, midbrain, pons/medulla oblongata, spinal cord, and pituitary. The cDNA products were used as templates for semiquantitative and RT-PCR analysis by using specific primers for CRFR2α, sCRFR2α, and the ribosomal protein S16. The locations of the oligonuleotide primers at exons 3 and 7 result in the amplification of two products of 418 and 309 bp corresponding to CRFR2α and sCRFR2α, respectively. Oligonucleotide primer sequences and PCR conditions can be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Extracellular Receptor Kinase 1/2 (ERK1/2) Assay. CATH.a cells were equilibrated with DMEM supplemented with 1% (wt/vol) BSA for 6 h and then stimulated with 0.1% DMEM/BSA (vehicle) or 10 nM Ucn 1 in the presence or absence of 0.4 or 4 nM sCRFR2α diluted in 0.1% DMEM/BSA. Cells were harvested immediately and analyzed for phosphorylated ERK1/2-p42, 44, as described in ref. 16.
Transient Transfections and Luciferase Assay. The HEK293T cells were transfected with a luciferase reporter containing a fragment of the EVX1 gene containing a potent cAMP response element site. The cells were harvested and the luciferase reporter activity was assayed as described in ref. 16. Twenty hours after transfection, cells were treated for 4 h with vehicle, CRF, or Ucn 1 (0.0001–100 nM) in the presence or absence of 0.1 nM sCRFR2α.
RIA. Antisera were raised in rabbits immunized with a synthetic peptide fragment encoding the unique C-terminal tail (amino acids 113–143) of mouse sCRFR2α conjugated to keyhole limpet hemocyanin by using a protocol described for inhibin subunits (40). The analog Tyr113 sCRFR2α (113–143) was radiolabeled with Na 125Iby using chloramine-T and purified by HPLC (40) for use as tracer in the RIA. The procedure for sCRFR2α RIA was similar to that described in detail for inhibin subunits (40). Briefly, anti-sCRFR2α (rabbit no. 6864, Peptide Biology Laboratories, The Salk Research Institute) was used at a 1:300,000 final dilution, and synthetic sCRFR2α (113–143) was used as standard. Murine tissues were acid-extracted and partially purified with octadecyl silica cartridges as described in ref. 40. Lyophilized samples were tested at three to seven dose levels. Free tracer was separated from tracer bound to antibody by the addition of sheep anti-rabbit γ-globulins and 10% (wt/vol) polyethylene glycol. The EC50 and minimum detectable dose for sCRFR2α were ≈5 and 100 pg per tube, respectively.
Immunohistochemistry. Adult male C57B6J mice (The Jackson Laboratory) and Sprague–Dawley albino rats were anesthetized with chloral hydrate (350 mg/kg i.p.) and perfused with Zamboni's fixative (41), followed by 3 h of postfixation. Regularly spaced (one-in-four) series of 30-μm-thick frontal sections throughout the brain were prepared for nickel-enhanced avidin-biotin-immunoperoxidase localization of sCRFR2α immunoreactivity (ir) by using Vectastain Elite reagents (Vector Laboratories, Burlingame, CA) (see ref. 41). Primary sCRFR2α antisera were adsorbed against the carrier, affinity-purified, and used at a dilution of 1:2,000. Specificity of immunostaining was evaluated by using primary antisera preincubated overnight at 4°C with 0–300 μM synthetic immunogen. Labeling was also evaluated in mutant mice deficient in either or both CRFRs (22, 24). Detailed description of the fluorescence immunocytochemical analysis of COSM6 cells transfected with sCRFR2α can be found in the Supporting Materials and Methods.
Protein Expression and Purification. Mammalian expression of sCRFR2α. A cDNA corresponding to amino acids 1–143, modified by PCR to include a FLAG epitope following amino acid 143, was subcloned into pSec-Tag2 HygroA (Invitrogen, Carlsbad, CA) and used for transfection of COSM6 cells as described (42). After 4 days, the media were collected, and sCRFR2α was enriched by immunoaffinity chromatography with FLAG-agarose (Sigma). The protein was detected by immunoblot analysis with either the anti-FLAG antibody or the antibody generated to the unique sCRFR2α C terminus.
Bacterial Expression of sCRFR2α. A cDNA corresponding to amino acids 20–143 was generated by PCR with sCRFR2α as the template. The cDNA was subcloned into pET-32a(+) (Novagen, La Jolla, CA) and the protein purified by S-protein-affinity chromatography as described in ref. 42. The protein was detected by immunoblot analysis with the antibody generated to the unique sCRFR2α C terminus.
Radioreceptor Assays. The soluble protein, purified either from COS M6 cell media or Escherichia coli was incubated in triplicate wells with [125I-DTyr0]-astressin and an increasing concentration of unlabeled peptides, as described (43).
Results
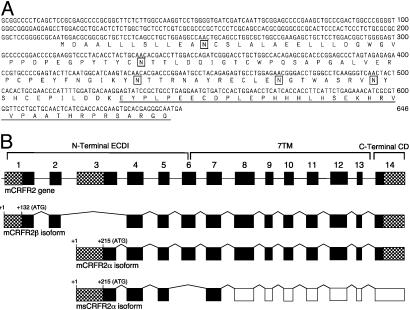
A cDNA transcript of smaller size (≈100 bp) was observed during the isolation of the mouse CRFR2α (16). This smaller fragment was isolated and found to encode a variant of CRFR2α bearing a deletion of exon 6. Translation of the variant transcript predicts a 143-aa protein, sCRFR2α, comprising the majority of the first extracellular domain (ECD1) of CRFR2α followed by a unique 38-aa C terminus (Fig. 1A). Screening of GenBank showed homology of the C terminus to no other protein. The genomic arrangement of the sCRFR2α is shown in Fig. 1B.
Fig. 1.
Sequence of sCRFR2α and genomic structure CRFR2. (A) Nucleotide and translated amino acid sequence of the sCRFR2α. Underlined amino acids indicate the unique C-terminal tail. Boxed residues indicate putative N-linked glycosylation sites. (B) Schematic representation of the structure of the mouse CRFR2 gene (first scheme), the two known functional transcripts in mouse, β and α (second and third schemes, respectively), and the sCRFR2α splice variant (fourth scheme). The locations of the translation start sites (ATG) are indicated. Exons coding for the N-terminal ECD (ECD1), the seven transmembrane domains (7TM), and the C-terminal cytoplasmic domain (CD) are indicated. 5′ and 3′ UTRs are indicated by hatched boxes. Black boxes represent coding regions, and white boxes represent exons downstream to the stop codon.
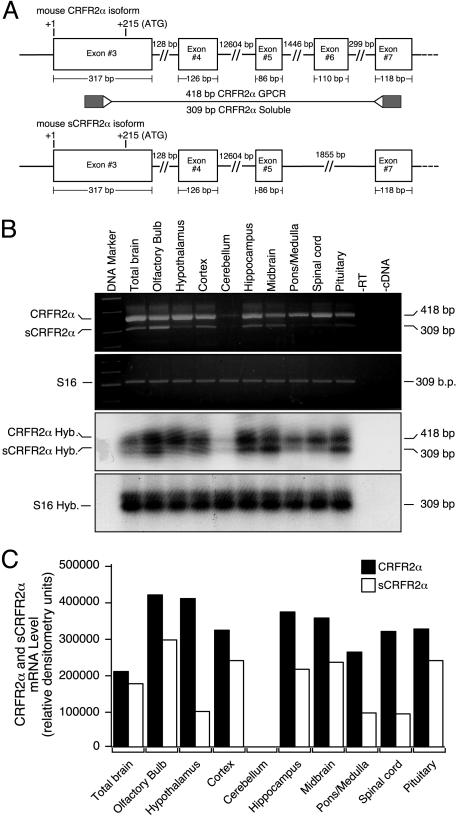
If the sCRFR2α mRNA is merely a product of splicing errors, it should be much less abundant than the correctly spliced RNA. To examine this question, semiquantitative RT-PCR followed by Southern hybridization analysis was used to compare the relative abundance of CRFR2α and sCRFR2α mRNA in several brain regions. Total RNA prepared from mouse tissues was reverse-transcribed to generate cDNAs that were used as templates for semiquantitative RT-PCR analysis, followed by Southern hybridization by using specific primers and probes for CRFR2α and sCRFR2α (Fig. 2). The oligonucleotide primer pair (located in exons 3 and 7) allowed the simultaneous amplification of both the soluble form and the full-length membrane bound receptor in a single reaction (Fig. 2A). The sCRFR2α is highly expressed in the olfactory bulb, cortex, midbrainm and the pituitary (Figs. 2 B and C). Lower levels of expression were found in the hippocampus, hypothalamus, pons, medulla, and spinal cord (Figs. 2 B and C). As shown in Fig. 2, the abundance of sCRFR2α mRNA is lower but comparable with that of CRFR2α mRNA. The sequences of cDNA fragments from RT-PCR were found to encode a splice variant of the mouse CRFR2α gene (Fig. 1A).
Fig. 2.
Expression of CRFR2α and sCRFR2α mRNA in mouse brain and pituitary. (A) Schematic representation and the oligonucleotide primer locations of the amplified portion of mouse CRFR2α (Upper) and sCRFR2α (Lower) transcripts. The locations of the oligonuleotide primers at exons 3 and 7, which result in the amplification of two products of 418 and 309 bp corresponding to CRFR2α and sCRFR2α, respectively, are indicated. GPCR, G-protein-coupled receptor. (B) Representative image of electrophoretic analysis of the semiquantitative RT-PCR for mCRFR2α and sCRFR2α mRNA and the ribosomal protein S16 mRNA (first and second gels). Southern blot hybridization (Hyb.) of amplified mCRFR2α and sCRFR2α cDNA and the ribosomal protein S16 cDNA fragments were also performed (third and fourth gels). RT, reverse transcriptase. (C) The radioactive bands were quantified by PhosphorImager, and normalized values (relative to S16 expression) are presented as relative densitometry units.
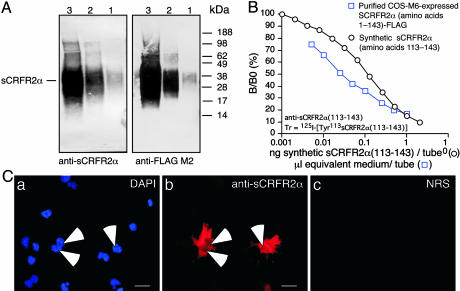
Computer analysis of the sequence predicted that the first 19 amino acids serve as a putative signal peptide. Because the sequence contains no obvious sites for membrane attachment, the protein is hypothesized to be secreted as a soluble form. To explore this hypothesis, the protein was expressed in COS M6 cells. After purification from the media, a protein band of ≈30 kDa was visualized by immunoblot analyses with either anti-FLAG antiserum or the anti-sCRFR2α, an antiserum raised against a synthetic peptide fragment encoding the unique C-terminal tail of sCRFR2α protein (amino acids 113–143) (Fig. 3A). The larger size of the protein compared with that predicted from the cDNA is probably a result of glycosylation.
Fig. 3.
A highly specific antiserum raised in rabbit by using a synthetic peptide fragment encoding the unique C-terminal tail of mouse sCRFR2α protein (amino acids 113–143) was used to develop a sCRFR2α RIA, which was used for immunoblot analysis and for immunocytochemistry. (A) Western immunoblot of mouse sCRFR2α protein isolated from the medium of COS-M6 cells transiently transfected with sCRFR2α-FLAG construct reacted with anti-sCRFR2α (amino acids 113–143) serum (Left) or monoclonal M2 anti-FLAG (Right). Lanes 1, 2, and 3 correspond to 0.1, 1, and 10 μl of sCRFR2α-FLAG extract, respectively. (B) Displacement of [125I]Tyr113 sCRFR2α (amino acids 113–143) binding to rabbit anti-sCRFR2α (amino acids 113–143) by synthetic sCRFR2α (amino acids 113–143) and by purified COS-M6-expressed sCRFR2α (amino acids 1–143)-FLAG. (C) (b) Immunofluorescence staining of COS-M6 cells transiently transfected with mouse sCRFR2α construct visualized with the anti-sCRFR2α (amino acids 113–143) serum followed by a Cy3-conjugated secondary antibody. (a) The slides were counterstained with DAPI to visualize transfected and nontransfected cells. (c) Cells incubated with normal rabbit serum (NRS) as negative control followed by a Cy3-conjugated secondary antibody did not show any staining. B/B0, (Bound to maximum bound) ×100.
To obtain a larger quantity of sCRFR2α, a protein lacking the putative signal peptide was expressed as a fusion protein in Escherichia coli (42). After cleavage and purification, the protein was visualized (by using the anti-sCRFR2α serum) by immunoblot analysis as a narrow band of size ≈20 kDa. The anti-sCRFR2α serum detects the sCRFR2α proteins in RIA (Fig. 3B) and immunocytochemistry (Fig. 3C).
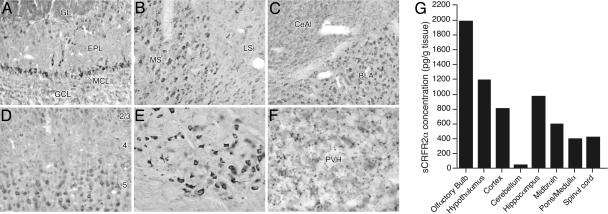
Immunohistochemical studies with anti-sCRFR2α serum revealed the distribution of sCRFR2α-ir in rodent brain. The cellular distribution of immunolabeling for sCRFR2α-ir was widespread and conformed more closely to the location of CRFR1 mRNA expression pattern than to that of CRFR2 (Fig. 4 A–F). The results described are from studies in mice; a similar pattern of labeling was observed in rats. Major sites of cellular expression include mitral and tufted cells of the olfactory bulb, the medial septal/diagonal band complex, piriform cortex, substantia nigra, red nucleus, basolateral amygdaloid, deep cerebellar, and dorsal column nuclei, all of which are prominent sites of CRFR1 expression. Similar to CRFR1, sCRFR2α-ir cell bodies are numerous throughout isocortex, although the laminar distributions are only partly overlapping. Thus, although CRFR1- and sCRFR2α-expressing cell bodies are numerous in layer 2/3, the dominant cortical seat of CRFR1 expression is in layer 4, whereas that of sCRFR2α is in layer 5. Major sites of CRFR2 expression, including the lateral septal, midbrain raphe, ventromedial hypothalamic, and medial amygdaloid nuclei, were all lacking in sCRFR2α-stained cell bodies, although, interestingly, the latter two sites were among the few invested with labeled varicosities that we take to be representative of sCRFR2α-ir terminal fields. The paraventricular nucleus of the hypothalamus also contained a presumed sCRFR2α-ir terminal field of moderate density.
Fig. 4.
Presence of sCRFR2α-like ir in the mouse brain by using immunohistochemistry and RIA. (A–F) Immunoperoxidase staining for sCRFR2α in select mouse brain regions. (A–E) Major sites of cellular expression included the principal output neurons of the olfactory bulb (A); the medial septal nucleus (B); the basolateral (BLA) but not the central (CeA) nucleus of the amygdala (C); cerebral cortex, where stained cells were localized mainly in layers 5 and 2/3 (D); and red nucleus (E). GL, glomerular layer; EPL, external plexiform layer; MCL, mitral cell layer; GCL, granule cell layer; MS, medial septal nucleus; LSi, intermediate lateral septal nucleus. In each of these sites, the pattern of cellular labeling was similar although not necessarily identical to that of CRFR1 mRNA expression. (F) Immunolabeled fibers and varicosities were restricted to a handful of cell groups, including the paraventricular nucleus of the hypothalamus (PVH). (G) sCRFR2α-like ir in acid-extracted and partially purified tissue isolated from mouse brain was measured by RIA. Tissue extracts were tested at five to seven dose levels and displaced 125I-labeled Tyr113 sCRFR2α (amino acids 113–143) binding to rabbit anti-sCRFR2α (amino acids 113–143) in a dose-dependent manner.
Labeling throughout the brain was blocked by preincubation of the antiserum with low micromolar concentrations (≥30 μM) of the sCRFR2α (amino acids 113–143) peptide used as immunogen; competition with the corresponding peptide predicted from the CRFR1 sequence did not interfere with immunolabeling at concentrations as high as 3 mM. Further support for the specificity of labeling are observations that all immunolocalizations persisted in CRFR1 and/or CRFR2-deleted mice; note that the targeting construct used for generating each of the existing receptor knockout lines would be expected to spare the sCRFR2α coding region (22–24).
To determine the presence of sCRFR2α-like ir in brain, a highly specific RIA was developed with anti-sCRFR2α and [125I-Tyr113]sCRFR2α (amino acids 113–143) as the tracer. Tissue from mouse brain was acid-extracted, partially purified on C18 cartridges, and assayed at multiple doses in the RIA. The tissue extracts displaced [125I-Tyr113]sCRFR2α (amino acids 113–143) bound to anti-sCRFR2α in a dose-dependent manner (Fig. 4G). The highest levels of expression were found in the olfactory bulb, hypothalamus, cortex, and midbrain, all of which correlate with the presence of ir cells and fibers, determined by the immunohistochemical studies (Fig. 4). A putative soluble form of CRFR1 (generated by deletion of exon 5) would comprise a different unique C-terminal sequence. A protein corresponding to that sequence did not displace [125I-Tyr113]sCRFR2α (amino acids 113–143) in the RIA. These results further confirm the existence of sCRFR2α protein in rodent CNS.
The interactions of the sCRFR2α with CRF family ligands were assessed by radioreceptor assay by using competitive displacement of [125I-DTyr0]-astressin bound to sCRFR2α. The soluble proteins, secreted by COS M6 cells or produced in bacteria, bind the agonists (Ucn 1 and CRF) and the antagonist (astressin) with nanomolar affinities, whereas the affinities for Ucn 2 and Ucn 3 are much lower (Table 1).
Table 1. Inhibitory binding constants, Ki (nM), for CRF ligands binding to sCRFR2α proteins.
| Protein | CRF | rUcn 1 | mUcn 2 | mUcn 3 | Astressin |
|---|---|---|---|---|---|
| Mam sCRFR2α | 23 (14-39) | 6.6 (3.5-12) | 113 (68-190) | >200 | 6.7 (3.6-12) |
| Bact sCRFR2α | 14.8 (9.2-24) | 5.8 (2.5-13.3) | 116 (85-158) | >200 | 10 (7.9-12.5) |
Binding of CRF family members to sCRFR2α proteins purified from either COS M6 cell media [mammalian (mam) sCRFR2α] or E. coli [bacterial (bact) sCRFR2α]. See Materials and Methods for details. rUcn 1, rat Ucn 1; mUcn 2/3, mouse Ucn 2/3.
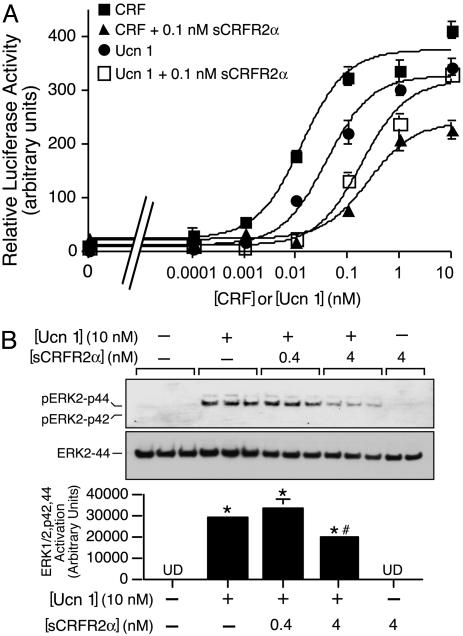
To delineate the possible functions of sCRFR2α, we studied its effects on signaling by CRF family ligands. The mammalian-expressed sCRFR2α and the bacterially-expressed sCRFR2α proteins inhibit, in a dose-dependent manner, the cAMP response to Ucn 1 and CRF in HEK293T cells transfected with mouse CRFR2α as measured by cAMP response element-luciferase activity of the EVX1 gene (Fig. 5A). Because the Ucns activate mitogen-activated protein kinase signaling (29), we measured the ability of sCRFR2α to inhibit the activation by Ucn 1 of ERK1/2-p42,44 in CATH.a cells, which endogenously express CRFR1 and CRFR2α. The sCRFR2α inhibits the induction of phosphorylated ERK by Ucn 1 in CATH.a cells (Fig. 5B).
Fig. 5.
sCRFR2α protein interferes with the induction of cAMP and mitogen-activated protein kinase signaling mediated by Ucn1 or CRF. (A) Activation of cAMP response element-luciferase reporter by Ucn1 or CRF with or without sCRFR2α preincubation in 293T cells transiently transfected with mouse CRFR2α. Luciferase reporter containing a fragment of the cAMP response element promoter of the EVX1 gene was cotransfected into 293T cells with CRFR2α expression vectors. Luciferase activity was measured after 4-h treatment with 0.0001–100 nM Ucn1 or CRF, in the presence or absence of 0.1 nM sCRFR2α. Assays were normalized to cotransfected β-galactosidase activity. The mean of six replicates from one experiment are shown in the graph. (B) Equilibrated CATH.a cells were treated with 10 nM Ucn1 with or without 0.4 or 4 nM sCRFR2α. After 5 min of receptor stimulation, cell lysates were harvested and subjected to SDS/PAGE immunoblot analysis by using phosphorylated ERK1/2-p42,44 antibody and ERK2-p44 antibody. ERK activation was calculated by normalizing the levels of phosphorylated ERK1/2-p42,44 to total ERK2-p44. The means of triplicates from one experiment are shown in the graph. *, P < 0.05 vs. vehicle treatment; #, P < 0.05 vs. Ucn 1 treatment. UD, undetected.
Discussion
We have identified and characterized a splice variant of CRFR2α encoding a soluble protein containing a major CRF ligand-binding domain (44), i.e., the ECD1 of CRFR2α. This soluble protein is expressed in areas of the brain that express CRFR1 and is detected in tissue extracts from these regions. Furthermore, the protein binds the CRF family ligands CRF and Ucn 1 with high affinity and, thus, may act to modulate the activity of two potent agonists. These data lend support to a potential functional significance of the sCRFR2α protein as a biological modulator of CRF family ligands. Based on the gene structures of CRFR2β and CRFR1, it is possible that similar soluble splice variants may also exist for these other receptors. The unique C-terminal peptide for a putative soluble splice variant for CRFR1 displayed no cross-reactivity in the specific sCRFR2α RIA and could not abolish the sCRFR2α immunohistochemical staining.
One explanation for the existence of sCRFR2α is that it arises as the result of abnormal posttranscriptional processing (splicing errors) that lead to frameshift deletions with resultant nonfunctional mRNA. Another possibility is that the mRNA is translated and may serve physiologic roles. Although the frequency of splicing errors is unknown, it is thought that they are probably common in the splicing of premRNAs that are derived from complex genes (45). Frameshift deletions often cause premature translation termination and affect the stability of such RNAs. Therefore, erroneously spliced RNAs are thought to reflect noise in the RNA splicing system and are rapidly destroyed in the cytoplasm if they contain an ORF that is interrupted by a premature stop codon (46). The data on the relative abundance of CRFR2α and sCRFR2α mRNAs as well as the apparent conservation of sCRFR2α expression in mouse and rat suggest that a splicing error is an unlikely explanation for the existence of the soluble splice variant. Furthermore, the difference in the ratio of expression of the full-length receptor and the soluble form in the different brain regions suggests distinct regulation of the two transcripts.
Because the sCRFR2α comprises nearly all of the ECD1 of the receptor, we find it intriguing that the sCRFR2α binds CRF and Ucn 1 with higher affinity than Ucn 2, whereas a soluble protein expressing the ECD1 of CRFR2β binds Ucn 1 and Ucn 2 with higher affinity than CRF (43). Indeed, the binding characteristics of sCRFR2α are similar to those of CRFR1 rather than to those of CRFR2. Another CRF-binding protein, CRF-BP, (47) is expressed both peripherally and centrally in both human and rodent with a distribution distinct from that of sCRFR2α (48). CRF-BP also binds CRF and Ucn 1 with high affinity (47, 49).
Truncated splice variants containing only the extracellular region of transmembrane receptors have been reported for several G-protein-coupled receptors. Three alternative splice variants of the metabotropic glutamate receptor have been reported to generate soluble proteins that contain only the ECD of the receptor (33–35). These splice variants are all generated by insertion of an exon and result in an in-frame or out-of-frame stop codon. A human thyroid-stimulating hormone receptor mRNA variant encoding the extracellular ligand-binding domain but lacking the transmembrane domain was also reported (37). A truncated soluble protein isoform of the luteinizing hormone receptor that lacks the transmembrane region was isolated from turkey cDNA (38), and a putative soluble form of the follicle-stimulating hormone receptor was isolated from ovine cDNA (36). A splice variant encoding only the extracellular ligand-binding domain of the GABAB receptor subunit GABAB(1a) was reported lacking exon 11, which results in the introduction of two in-frame stop codons (39).
Two putative soluble variants of CRFR1 were isolated from human skin cDNA and were cloned as chimeric constructs (17). Cotransfection of these CRFR1 constructs (CRFR1e and CRFRh) with wild-type CRFR1 in COS cells showed that CRFR1e inhibited, whereas CRFR1h enhanced, Ucn 1-stimulated cAMP response (17). However, the proteins were not isolated nor were their ligand-binding characteristics determined (17). Unlike the sCRFR2α protein characterized in this report, the protein expression of the other G-protein-coupled receptor soluble isoforms have not demonstrated.
In conclusion, we propose that sCRFR2α protein may modulate ligand activity by either competing with the full-length membrane receptors for available ligand or by presenting the ligand or prolonging its action. Furthermore, the observations that sCRFR2α is expressed in areas of CRFR1 expression and that the protein binds CRFR1 ligands, Ucn 1, and CRF with high affinity suggest that the protein may be important to the function of this receptor. The existence of this binding protein increases the cast of characters that play roles in the action of CRFRs and CRF ligands and may help elucidate and explain the complex responses observed for the ligands.
Supplementary Material
Acknowledgments
We thank Dr. W. Fischer for helpful discussions, K.-F. Lee (The Salk Institute) for knockout mice, Dr. J. Gulyas and R. Kaiser (The Salk Institute) for peptide synthesis, and S. Guerra for assistance in the preparation of the manuscript. This work is supported by National Institute of Diabetes and Digestive and Kidney Diseases Program Project Grant DK 26741, the Robert J. and Helen C. Kleberg Foundation, the Adler Foundation, and the Foundation for Research (W.W.V.). W.W.V. is a Senior Foundation for Research Investigator.
Author contributions: A.M.C., M.H.P., and W.W.V. designed research; A.M.C., M.H.P., M.R.D., J.M.V., B.K.B., C.M.A., and P.E.S. performed research; J.M.V. and J.E.R. contributed new reagents/analytic tools; A.M.C., M.H.P., J.M.V., and K.A.L. analyzed data; and A.M.C. and M.H.P. wrote the paper.
Abbreviations: CRF, corticotropin-releasing factor; CRFR, CRF receptor; sCRFR2α, soluble CRFR type 2α; Ucn, urocortin; ECD, extracellular domain; ir, immunoreactivity; ERK, extracellular receptor kinase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY753668).
References
- 1.Vale, W., Spiess, J., Rivier, C. & Rivier, J. (1981) Science 213, 1394-1397. [DOI] [PubMed] [Google Scholar]
- 2.Rivier, C. & Vale, W. (1983) Nature 305, 325-327. [DOI] [PubMed] [Google Scholar]
- 3.Muglia, L., Jacobson, L., Dikkes, P. & Majzoub, J. A. (1995) Nature 373, 427-432. [DOI] [PubMed] [Google Scholar]
- 4.Koob, G. F. & Heinrichs, S. C. (1999) Brain Res. 848, 141-152. [DOI] [PubMed] [Google Scholar]
- 5.Vaughan, J., Donaldson, C., Bittencourt, J., Perrin, M. H., Lewis, K., Sutton, S., Chan, R., Turnbull, A. V., Lovejoy, D., Rivier C., et al. (1995) Nature 378, 287-292. [DOI] [PubMed] [Google Scholar]
- 6.Reyes, T. M., Lewis, K., Perrin, M. H., Kunitake, K. S., Vaughan, J., Arias, C. A., Hogenesch, J. B., Gulyas, J., Rivier, J., Vale, W. W. & Sawchenko, P. E. (2001) Proc. Natl. Acad. Sci. USA 98, 2843-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu, S. Y. & Hsueh, A. J. (2001) Nat. Med. 7, 605-611. [DOI] [PubMed] [Google Scholar]
- 8.Lewis, K., Li, C., Perrin, M. H., Blount, A., Kunitake, K., Donaldson, C., Vaughan, J., Reyes, T. M., Gulyas, J., Fischer, W., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 7570-7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, R., Lewis, K. A., Perrin, M. H. & Vale, W. W. (1993) Proc. Natl. Acad. Sci. USA 90, 8967-8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vita, N., Laurent, P., Lefort, S., Chalon, P., Lelias, J. M., Kaghad, M., Le, F. G., Caput, D. & Ferrara, P. (1993) FEBS Lett. 335, 1-5. [DOI] [PubMed] [Google Scholar]
- 11.Chang, C. P., Pearse, R. V., II, O'Connell, S. & Rosenfeld, M. G. (1993) Neuron 11, 1187-1195. [DOI] [PubMed] [Google Scholar]
- 12.Perrin, M., Donaldson, C., Chen, R., Blount, A., Berggren, T., Bilezikjian, L., Sawchenko, P. & Vale, W. (1995) Proc. Natl. Acad. Sci. USA 92, 2969-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenzel, P., Kesterson, R., Yeung, W., Cone, R. D., Rittenberg, M. B. & Stenzel-Poore, M. P. (1995) Mol. Endocrinol. 9, 637-645. [DOI] [PubMed] [Google Scholar]
- 14.Kishimoto, T., Pearse, R. V., II, Lin, C. R. & Rosenfeld, M. G. (1995) Proc. Natl. Acad. Sci. USA 92, 1108-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovenberg, T. W., Liaw, C. W., Grigoriadis, D. E., Clevenger, W., Chalmers, D. T., De Souza, E. B. & Oltersdorf, T. (1995) Proc. Natl. Acad. Sci. USA 92, 836-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, A., Perrin, M., Brar, B., Li, C., Jamieson, P., Digruccio, M., Lewis, K. & Vale, W. (2005) Mol. Endocrinol. 19, 441-458. [DOI] [PubMed] [Google Scholar]
- 17.Pisarchik, A. & Slominski, A. (2004) Eur. J. Biochem. 271, 2821-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grammatopoulos, D. K., Dai, Y., Randeva, H. S., Levine, M. A., Karteris, E., Easton, A. J. & Hillhouse, E. W. (1999) Mol. Endocrinol. 13, 2189-2202. [DOI] [PubMed] [Google Scholar]
- 19.Kostich, W. A., Chen, A., Sperle, K. & Largent, B. L. (1998) Mol. Endocrinol. 12, 1077-1085. [DOI] [PubMed] [Google Scholar]
- 20.Van Pett, K., Viau, V., Bittencourt, J. C., Chan, R. K. W., Li, H. Y., Arias, C., Prins, G. S., Perrin, M., Vale, W. & Sawchenko, P. E. (2000) J. Comp. Neurol. 428, 191-212. [DOI] [PubMed] [Google Scholar]
- 21.Lovenberg, T. W., Chalmers, D. T., Liu, C. & De Souza, E. B. (1995) Endocrinology 136, 4139-4142. [DOI] [PubMed] [Google Scholar]
- 22.Smith, G. W., Aubry, J. M., Dellu, F., Contarino, A., Bilezikjian, L. M., Gold, L. H., Chen, R., Marchuk, Y., Hauser, C., Bentley, C. A., et al. (1998) Neuron 20, 1093-1102. [DOI] [PubMed] [Google Scholar]
- 23.Timpl, P., Spanagel, R., Sillaber, I., Kresse, A., Reul, J. M., Stalla, G. K., Blanquet, V., Steckler, T., Holsboer, F. & Wurst, W. (1998) Nat. Genet. 19, 162-166. [DOI] [PubMed] [Google Scholar]
- 24.Bale, T. L., Contarino, A., Smith, G. W., Chan, R., Gold, L. H., Sawchenko, P. E., Koob, G. F., Vale, W. W. & Lee, K. F. (2000) Nat. Genet. 24, 410-414. [DOI] [PubMed] [Google Scholar]
- 25.Coste, S. C., Kesterson, R. A., Heldwein, K. A., Stevens, S. L., Heard, A. D., Hollis, J. H., Murray, S. E., Hill, J. K., Pantely, G. A., Hohimer, A. R., et al. (2000) Nat. Genet. 24, 403-409. [DOI] [PubMed] [Google Scholar]
- 26.Kishimoto, T., Radulovic, J., Radulovic, M., Lin, C. R., Schrick, C., Hooshmand, F., Hermanson, O., Rosenfeld, M. G. & Spiess, J. (2000) Nat. Genet. 24, 415-419. [DOI] [PubMed] [Google Scholar]
- 27.Bale, T. L. & Vale, W. W. (2004) Annu. Rev. Pharmacol. Toxicol. 44, 525-557. [DOI] [PubMed] [Google Scholar]
- 28.Perrin, M. H. & Vale, W. W. (1999) Ann. N.Y. Acad. Sci. 885, 312-328. [DOI] [PubMed] [Google Scholar]
- 29.Brar, B. K., Vale, W. & Perrin, M. (2002) in Encyclopedia of Hormones and Related Cell Regulators, eds. Henry, H. L. & Norman, A. W. (Academic, New York), pp. 313-325.
- 30.Rekasi, Z., Czompoly, T., Schally, A. V. & Halmos, G. (2000) Proc. Natl. Acad. Sci. USA 97, 10561-10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, K., Bergson, C., Levenson, R. & Schmauss, C. (1994) J. Biol. Chem. 269, 29220-29226. [PubMed] [Google Scholar]
- 32.Seck, T., Baron, R. & Horne, W. C. (2003) J. Biol. Chem. 278, 23085-23093. [DOI] [PubMed] [Google Scholar]
- 33.Malherbe, P., Kratzeisen, C., Lundstrom, K., Richards, J. G., Faull, R. L. & Mutel, V. (1999) Brain Res. Mol. Brain Res. 67, 201-210. [DOI] [PubMed] [Google Scholar]
- 34.Zhu, H., Ryan, K. & Chen, S. (1999) Brain Res. Mol. Brain Res. 73, 93-103. [DOI] [PubMed] [Google Scholar]
- 35.Valerio, A., Zoppi, N., Ferraboli, S., Paterlini, M., Ferrario, M., Barlati, S. & Spano, P. NeuroReport 12, 2711-2715. [DOI] [PubMed]
- 36.Khan, H., Yarney, T. A. & Sairam, M. R. (1993) Biochem. Biophys. Res. Commun. 190, 888-894. [DOI] [PubMed] [Google Scholar]
- 37.Graves, P. N., Tomer, Y. & Davies, T. F. (1992) Biochem. Biophys. Res. Commun. 187, 1135-1143. [DOI] [PubMed] [Google Scholar]
- 38.You, S., Kim, H., Hsu, C. C., El Halawani, M. E. & Foster, D. N. (2000) Biol. Reprod. 62, 108-116. [DOI] [PubMed] [Google Scholar]
- 39.Schwarz, D. A., Barry, G., Eliasof, S. D., Petroski, R. E., Conlon, P. J. & Maki, R. A. (2000) J. Biol. Chem. 275, 32174-32181. [DOI] [PubMed] [Google Scholar]
- 40.Vaughan, J. M., Rivier, J., Corrigan, A. Z., McClintock, R., Campen, C. A., Jolley, D., Voglmayr, J. K., Bardin, C. W., Rivier, C. & Vale, W. (1989) Methods Enzymol. 168, 588-617. [DOI] [PubMed] [Google Scholar]
- 41.Bittencourt, J. C., Vaughan, J., Arias, C., Rissman, R. A., Vale, W. W. & Sawchenko, P. E. (1999) J. Comp. Neurol. 415, 285-312. [PubMed] [Google Scholar]
- 42.Perrin, M. H., Fischer, W. H., Kunitake, K. S., Craig, A. G., Koerber, S. C., Cervini, L. A., Rivier, J. E., Groppe, J. C., Greenwald, J., Moller Nielsen, S. & Vale, W. W. (2001) J. Biol. Chem. 276, 31528-31534. [DOI] [PubMed] [Google Scholar]
- 43.Perrin, M. H., DiGruccio, M. R., Koerber, S. C., Rivier, J. E., Kunitake, K. S., Bain, D. L., Fischer, W. H. & Vale, W. W. (2003) J. Biol. Chem. 278, 15595-15600. [DOI] [PubMed] [Google Scholar]
- 44.Grace, C. R., Perrin, M. H., DiGruccio, M. R., Miller, C. L., Rivier, J. E., Vale, W. W. & Riek, R. (2004) Proc. Natl. Acad. Sci. USA 101, 12836-12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nigro, J. M., Cho, K. R., Fearon, E. R., Kern, S. E., Ruppert, J. M., Oliner, J. D., Kinzler, K. W. & Vogelstein, B. (1991) Cell 64, 607-613. [DOI] [PubMed] [Google Scholar]
- 46.Pulak, R. & Anderson, P. (1993) Genes Dev. 7, 1885-1897. [DOI] [PubMed] [Google Scholar]
- 47.Potter, E., Behan, D. P., Fischer, W. H., Linton, E. A., Lowry, P. J. & Vale, W. W. (1991) Nature 349, 423-426. [DOI] [PubMed] [Google Scholar]
- 48.Chan, R. K., Vale, W. W. & Sawchenko, P. E. (2000) Neuroscience 101, 115-129. [DOI] [PubMed] [Google Scholar]
- 49.Sutton, S. W., Behan, D. P., Lahrichi, S. L., Kaiser, R., Corrigan, A., Lowry, P., Potter, E., Perrin, M. H., Rivier, J. & Vale, W. W. (1995) Endocrinology 136, 1097-1102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.