Abstract
Insights into the host factors and mechanisms mediating the primary host responses after pathogen presentation remain limited, due in part to the complexity and genetic intractability of host systems. Here, we employ the model Drosophila melanogaster to dissect and identify early host responses that function in the initiation and progression of Pseudomonas aeruginosa pathogenesis. First, we use immune potentiation and genetic studies to demonstrate that flies mount a heightened defense against the highly virulent P. aeruginosa strain PA14 when first inoculated with strain CF5, which is avirulent in flies; this effect is mediated via the Imd and Toll signaling pathways. Second, we use whole-genome expression profiling to assess and compare the Drosophila early defense responses triggered by the PA14 vs. CF5 strains to identify genes whose expression patterns are different in susceptible vs. resistant host–pathogen interactions, respectively. Our results identify pathogenesis- and defense-specific genes and uncover a previously undescribed mechanism used by P. aeruginosa in the initial stages of its host interaction: suppression of Drosophila defense responses by limiting antimicrobial peptide gene expression. These results provide insights into the genetic factors that mediate or restrict pathogenesis during the early stages of the bacterial–host interaction to advance our understanding of P. aeruginosa-human infections.
Keywords: innate immunity, pathogenesis, Drosophila melanogaster, immune potentiation
The innate immunity system, which first evolved in lower animals, is ancestral and, unlike adaptive immunity, occurs throughout invertebrate and vertebrate species. Current knowledge of this system remains limited, especially with regard to the host defense mechanisms used upon initial pathogen presentation. Innate immunity must act quickly to mount a first line of defense to hold the pathogen in check before the adaptive response matures (1). Although defense strategies are diverse for different pathogens, many of them are evolutionarily conserved, including production of an array of antimicrobial peptides (AMPs), activation of phagocytic cells, and production of toxic metabolites. AMPs, the best-studied defense effectors, are rapidly elicited after microbe presentation. These ancient weapons play crucial roles in combating microbial infections in invertebrates, vertebrates, and plants (2).
Drosophila has emerged as an ideal model organism to study the genetic control of immune recognition and response, because of the high degree of conservation between the fly and mammalian innate immune systems (3), along with its genetic tractability and simplicity. The Drosophila genome contains at least 15 AMP genes that encode both broad-spectrum antibiotic peptides and more specialized activities against Gram-negative bacteria or fungi (4). Expression of many of these defense effectors is mediated via activation of the Toll and/or Imd innate immunity signaling pathways (5).
We (6) and others (7–9) have used Drosophila to explore the mechanisms used by hosts to restrict infections by the human pathogen Pseudomonas aeruginosa. This versatile and ubiquitous bacterium is the quintessential opportunistic pathogen, because it infects a broad range of hosts, including plants, insects, and humans. Furthermore, it is a major human health-care problem (10), because it is the principal agent of lethal infections in cystic fibrosis patients and the principal cause of multidrug-resistant infections in immune-suppressed individuals. Although a considerable body of research has shown that P. aeruginosa virulence in disparate hosts is mediated by means of a powerful repertoire of factors (11), the host responses triggered by this pathogen, especially during the initial stages of infection that can dictate its outcome, remain poorly understood.
Here, we use Drosophila melanogaster to uncover host genes that can promote or limit the initiation and progression of infection, with the ultimate goal to better understand and control human-bacterial pathogenesis. We profile and compare the host responses that underlie susceptible vs. nonsusceptible D. melanogaster–P. aeruginosa interactions by using PA14 and CF5, two human P. aeruginosa isolates that are highly virulent and nonvirulent in flies, respectively. We identify pathogenesis- and defense-specific genes with putative roles in pathogen detection, activation of immunity signal transduction pathways, and defense as well as nonimmunity activities. Moreover, our results indicate that AMPs are involved in fighting P. aeruginosa, reveal the signaling pathways that potentiate the Drosophila defense responses, and advance our knowledge of human infections by uncovering in vivo multicellular host responses that may promote or restrict P. aeruginosa pathogenesis.
Materials and Methods
Bacterial Strains and Growth. The P. aeruginosa PA14 and CF5 human isolates are described in refs. 11–13. The central 169 codons of the 253-codon PA14-PA1814 gene, which encodes a putative S-adenosyl-methionine-dependent methyltransferase, was deleted to generate the in-frame deletion mutant D12. The PA1814 borders were PCR-amplified from target PA14 genomic DNA and cloned into pNPTS138 to produce the replacement plasmid used to generate D12 by means of homologous recombination (14), as confirmed by hybridization and sequencing. All strains were grown at 37°C in LB medium and exhibited essentially identical growth.
Fly Infections. Oregon-R-S wild-type and spz197 mutant flies were obtained from the Bloomington Fly Stock Center (Bloomington, IN). imd1, relE20, spzrm7, and imd1;spzrm7 mutant flies were provided by B. Lemaitre (Centre de Genetique Moleculaire, Gif-sur-Yvette, France). imd1, relE20, and imd1;spzrm7 flies were used as homozygotes, and spz197 and spzrm7 flies were used as transheterozygotes. For AMP overexpression, flies overexpressing Gal4 under the ubiquitous promoter daughterless (P{Gal4-da.G32} UH1; Bloomington Stock Center) were crossed to transgenic strains carrying AMP coding sequences under the control of upstream activating sequence enhancer elements (U-Drom, U-CecA, U-Att, U-Def, U-Dipt, and U-Drc) (15). Fly maintenance, inoculations, survival kinetics, and assessment of number of bacteria were performed as described in ref. 6. Mock-injured flies served as mock-inoculated controls. To minimize effects of circadian rhythms on fly responses after infection, PA14 or CF5 infections were performed sequentially within 1 h at the same time of day. Each survival kinetic experiment used a minimum of 50 flies. All experiments were performed at least twice, and independent experiments gave reproducible differences between the control and experimental conditions. Comparisons in each set were made by using isogenic flies. The Kaplan–Meier (16) and Cox (17) statistical models were used to analyze fly survival kinetics.
RNA Extraction, Labeling, and Scanning. RNA was extracted by using the RNAeasy Kit (Qiagen, Valencia, CA) from 40 1- to 4-day-old male flies infected with CF5, PA14, or D12. EnzoBioArray High Yield Transcript Labeling and GeneChip Sample Cleanup Module kits (Affymetrix) were used for labeling. Fluidics and scanning were performed by using Affymetrix protocols, with all experiments performed in duplicate.
Data Analysis. The raw scanned image files were processed by using data normalization, quality assurance and control, filtering, and clustering. Stable invariant-set normalization and perfect match model-based expression values were generated by using the dchip program (www.biostat.harvard.edu/complab/dchip) (18). All arrays passed quality control by dchip. A probe set was eliminated if it showed small variation (SD/mean <0.2) across samples (after pooling replicate arrays) or an overall signal intensity at or below background [4 × rawQ (a measure of noise value, representing the degree of pixel to pixel variation of probe cells on a GeneChip array)]. A dual filter was applied to identify differentially expressed genes in flies infected with PA14 or CF5 by the following criteria: (i) each gene had at least a 1.5-fold change in its expression value between either PA14 and injury or CF5 and injury for at least one time point; and (ii) the gene expression value had at least a 1.5-fold change between PA14 and CF5 for at least one time point. Data for 241 filtered genes were analyzed by supervised hierarchical clustering using dchip. The color visualization and fold change were generated by using the guigraph software (http://genetics.mgh.harvard-.edu/RahmeWeb/guigraph/index.html) and are presented as expression ratios in Tables 1–3, which are published as supporting information on the PNAS web site.
Gene Categorization and Classification. Only genes with >1.5-fold positive or negative expression change were deemed up- or down-regulated. These genes group into two major classes: defense (Class I) and pathogenesis (Class II), with expression of a defense gene (DG) either higher or lower in CF5-inoculated vs. both mock- and PA14-inoculated flies and of a pathogenesis gene (PG) either higher or lower in PA14-inoculated vs. mock- and CF5-inoculated flies.
Results
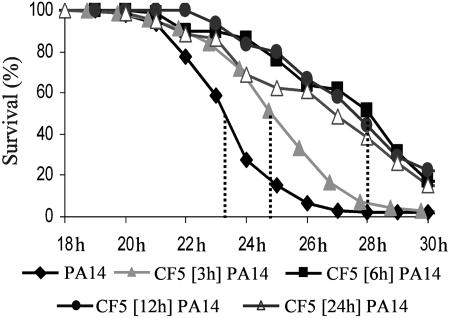
Nonvirulent P. aeruginosa Bacteria Restrict Subsequent Infection by Virulent P. aeruginosa. The fly defense systems fail to prevent infection by the highly virulent P. aeruginosa human isolate PA14, which proliferates to high titers in fly tissues, kills flies in hours, and has ≈100% mortality in both flies and mice (ref. 6; see also Fig. 6, which is published as supporting information on the PNAS web site). Conversely, the CF5 human isolate proliferates weakly in fly tissues, has very limited fly mortality (Fig. 6), and is avirulent in mice (data not shown), suggesting that it either lacks virulence factors and/or is susceptible to or activates fly defenses. To this end, we asked whether CF5 could potentiate the fly innate immune response to limit subsequent infection by PA14. We preinoculated flies in the dorsal thorax with CF5 at ≈100 cells per fly and then secondarily inoculated these flies with PA14 at 3, 6, 12, or 24 h.
Fig. 1 and Table 4, which is published as supporting information on the PNAS web site, show that CF5 triggers host responses that protect against subsequent infection, because PA14 mortality was significantly delayed in preinoculated flies, with a >4-h delay to 50% mortality and ≈25% fly survivors at 30 h vs. the nonpreinoculated flies. This effect also was produced to a lesser degree by preinoculation with avirulent Escherichia coli, whereas preinfection with avirulent Staphylococcus aureus cells, or with Cryptococcus neoformans fungal cells, was similar to sterile injury (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 1.
Immune potentiation elicited by initial inoculation with CF5 cells protects flies from subsequent infection with virulent PA14 cells. Infection with CF5 at 3, 6, 12, or 24 h before PA14 infection delays mortality and increases fly survival. This protection maximizes at 6 h after primary inoculation and lasts for >24 h; P < 0.0001 (Table 4).
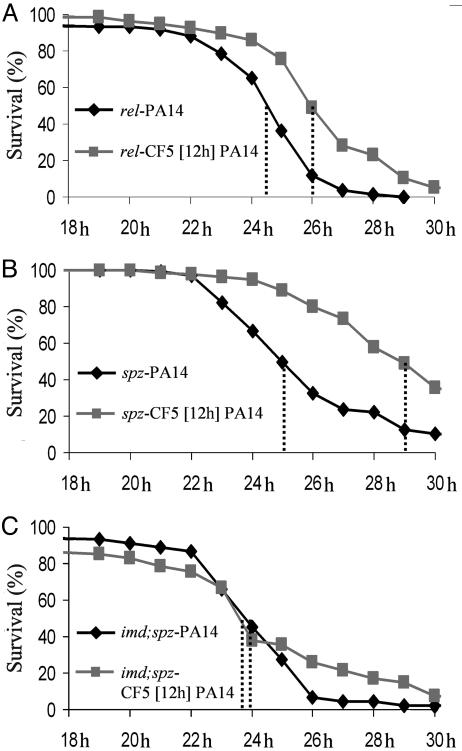
The CF5-Potentiated Defense Response Requires the Toll and Imd Pathways. Many fly immune defenses are under the control of the Imd and Toll innate immunity signaling pathways (5), which are both essential to restrict P. aeruginosa infection (6, 7). Indeed, mutations in the Imd and Toll pathway components limit the induction of most AMP genes (5). To determine whether Imd and Toll are relevant in CF5-triggered immune potentiation, we preinoculated rel- and spz- mutant flies with CF5. Fig. 2 A and B and Table 4 show that these flies are significantly less susceptible to PA14 infection after CF5 infection. Similar results were obtained with imd- flies (Table 4 and data not shown). In contrast, imd-;spz- double mutant flies, in which both innate immunity signaling pathways are impaired, are equally susceptible to PA14 with or without CF5 preinoculation (Fig. 2C), suggesting that the Imd and Toll pathways act synergistically in CF5-triggered innate immune potentiation.
Fig. 2.
P. aeruginosa-triggered immune potentiation requires both the Imd and Toll signaling pathways. Fly survival after preinoculation with CF5 of rel- (A) and spz- (B) mutant flies and of a imd-;spz- double mutant (C). P < 0.0001 for A and B and P > 0.2 for C (Table 4).
P. aeruginosa-Triggered Gene Expression. We performed whole-genome expression studies on CF5-infected vs. PA14-infected flies to dissect the CF5-triggered immune potentiation, vs. the host responses to PA14 that favor infection. RNA was isolated from flies at 1, 6, and 12 h after CF5 or PA14 inoculation, with untreated and mock-inoculated flies as controls. By using the criteria presented in Materials and Methods, we identified 241 genes differentially expressed in CF5-infected vs. PA14-infected flies (Tables 1 and 2). These genes group into two major classes: the Class I DGs, whose expression is altered only by CF5 inoculation (defense-specific genes; DSGs) or primarily altered by CF5 (Table 1), and the Class II PGs, whose expression is altered only by PA14 inoculation (pathogenesis-specific genes; PSGs) or primarily altered by PA14 (Table 2).
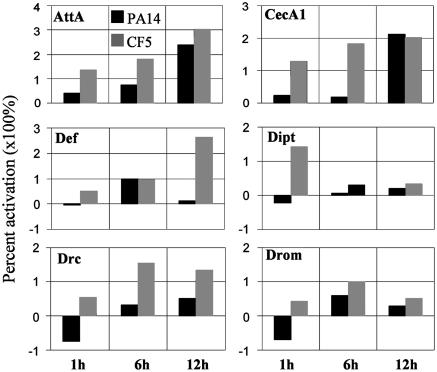
Class I: DGs. DGs include 133 up-regulated and 80 down-regulated genes (Table 1). Several immunity-related functions, including most of the AMPs (Fig. 3 and Table 1), are predominantly up-regulated by CF5. Other immunity-related genes predominately expressed after CF5 inoculation are the pattern recognition receptor PGRP-SB1 (19); relish, the Imd pathway transcriptional activator of most AMP genes in our list; the γ- and θ-trypsin genes (20); the phagocytosis-related gene TepII (21); and windbeutel, whose product activates the Toll receptor during dorsoventral axis determination (22).
Fig. 3.
Relative AMP levels induced in PA14- and CF5-infected flies over time. Values for selected AMPs correspond to PA14 vs. mock injury (black bars) and CF5 vs. mock injury (gray bars) as percent activation of expression levels at 1, 6, and 12 h after inoculation. Several differences between PA14 and CF5 infection are seen: Attacin A (AttA) and Cecropin A1 (CecA1) show prominent differences at 1 and 6 h; Diptericin (Dipt) and Drosomycin (Drom) at 1 h; Drosocin (Drc) at all three time points; and Defensin (Def) at 1 and 12 h. Note the negative percent change in expression levels of Drc and Drom at 1 h after PA14 infection, suggesting they are suppressed by PA14.
Non-immunity-related genes up-regulated by CF5 encode cell-proliferative, metabolic, neuronal, transport, signaling, and structural functions, along with putative proteases and several unknown proteins (Table 1). Thirteen of 28 of the structural protein genes are defense-specific and only induced by CF5. Although tissue reconstruction is stimulated as thoracic structures are injured during bacterial inoculation, expression of these genes is not significantly increased by PA14-inoculation. Other up-regulated activities that might also mediate such reconstruction include the centaurin γ1A and Rgk2 GTPases, related to cytoskeleton organization; the tissue growth signal transducer cubitus interruptus (23); the Activin-Like Protein at 23B; PAK, POSH, Abelson-Interacting Protein, and menin, which potentially mediate developmental and tissue repair morphogenetic movements; and the cell proliferation gene pan gu (24).
Prominent functions predominantly down-regulated by CF5 include the stress-related protein genes, Frost, Hsp22, Hsp26, Hsp68, Hsp70Bbb, Hsp70Bc, and Hsp20-like (GenBank accession no. CG7409); and the detoxification genes, Cyp6d5, Cyp9b2, Cyp6a2, Cyp6g1, Cyp6w1, urate oxidase, and GenBank no. CG6045, predicted to encode a xanthine dehydrogenase (Table 1). In addition, immunity-related factors, drosomysin-5; GenBank no. CG12780, which encodes a putative recognition Gram-negative binding protein-like molecule proposed to function in antifungal defense (25); the hemocyte-specific and putatitive pattern recognition receptor PGRP-LA (26); a serpin (GenBank no. CG6289); a cathepsin L (GenBank no. CG6357); two serine-type proteases (GenBank nos. CG11037 and CG3739); and circadian and neuronal factors are down-regulated by CF5 (Table 1). Metabolic enzymes involved in glucose catabolism, GenBank nos. CG6484, CG11909, CG11669, and CG8690, LvpH, and Sodh-1, are also down-regulated (Table 1). Hsp-like CG7409, Cyp6d5, Cyp6a2, Cyp9b2, xanthine dehydrogenase (GenBank no. CG6045), drosomysin-5, and PGRP-LA are all DSGs (Table 1).
Class II: PGs. PGs include 16 up-regulated and 12 down-regulated genes in PA14-inoculated vs. CF5-inoculated flies (Table 2), 20 of which are PSGs, because their expression is solely altered by PA14, with 15 up-regulated and 5 down-regulated. Up-regulated PSGs include the stress-related genes methuselah-like 6 (27) and sesB (28); the macrophage marker Sap-r (29); the hemocyte proliferation-related gene GenBank no. CG14557 (30); two putative catabolism genes, GenBank nos. CG8685 and CG3699; and the V-ATPases genes, GenBank nos. CG12403 and CG7007, predicted to regulate proton transport (31).
Down-regulated PSGs include GenBank no. CG6639, predicted to encode a serine protease (20); the thoracic tissue morphogenesis gene Beadex; the putative olfactory genes with GenBank nos. CG4202 and CG10274; and a putative G-protein coupled receptor, GenBank no. CG13579. Immunity-related PGs with lower expression in PA14-inoculated vs. CF5-inoculated flies include AttC; the humoral immunity gene totM (32); GenBank no. CG9162, which contains putative NFκB-like dorsal binding sites (33); and the hemocyte-expressed gene twins (34) (Table 2).
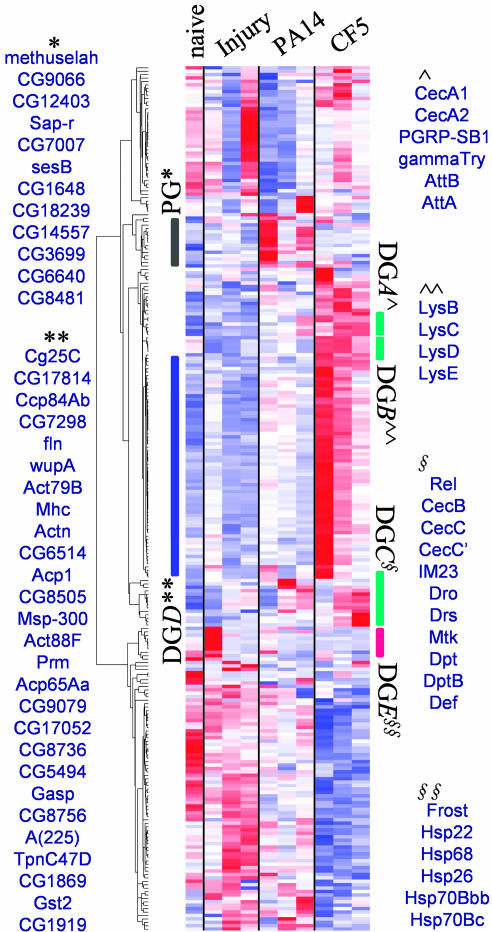
Clustering Analysis. Genes sharing a unique pattern of expression might pinpoint functionally related responses having pivotal roles in the infection process. To identify such coregulated gene profiles, we subjected the 241 Class I and II genes to hierarchical cluster analysis. Fig. 4 shows that this analysis identifies several gene clusters, five of which are mainly comprised of DGs (DG A–E) and one comprised of PGs. All members of the DG A–E clusters are up-regulated by 1.5- to 4-fold as early as 1 h after CF5 inoculation (Table 3).
Fig. 4.
Hierarchical cluster analysis of the 241 differentially expressed DGs and PGs. Each gene is clustered across naive, sterile-injury, PA14-infection, and CF5-infection conditions and is presented colorwise in a single row. The darker the color, the lower (blue) or the higher (red) the expression vs. the mean expression level. Groups of similarly affected genes are defined by each cluster. DG clusters A–C (DGA–C) predominantly contain immunity genes (green bars), the DGD cluster contains predominantly tissue reconstruction genes (blue bar), and the DGE cluster (red bar) contains stress proteins. The black bar marks the PGs. The complete list of gene identities is presented in Table 3.
Immunity genes predominantly cluster in the DG A–C groups. DGA contains seven genes, including the AMPs Cecropin A1 (CecA1) and Cecropin A2 (CecA2); Attacin A (AttA) and Attacin B (AttB); NOT-serine type endopeptidase γ-trypsin; the pattern recognition receptor PGRP-SB1; and the odorant binding protein Obp56e. DGB contains the lysozymes Lys B, C, D, and E. DGC contains nine AMP genes, whose expression peaks at 6 or 12 h after CF5 inoculation (Fig. 3 and 4), which mirrors the immune potentiation timing (Fig. 1). In addition, the IM23 immune molecule and the putative lipid and amino acid catabolism activities, GenBank nos. CG6675, CG8665, and CG4757, belong to the DGC cluster (Table 3), suggesting coregulation of catabolic and immune functions.
DGD, the other CF5 up-regulated cluster, contains 55 genes, 27 of which function in tissue reconstruction (Fig. 4). DGD is part of a clade that includes DGB and -C, suggesting that immunity and tissue reconstruction genes may act together to limit P. aeruginosa infection. DGE, a fifth functional cluster, is comprised of six stress-response genes and one circadian DG (Fig. 4 and Table 1).
The sixth cluster, PG, includes 12 Class II, PA-14 up-regulated genes (Fig. 4), which encode cellular immunity, stress, metabolism, and proton balance activities. These genes are coregulated, with 11 subclassed as PSGs. PA14-down-regulated genes failed to cluster.
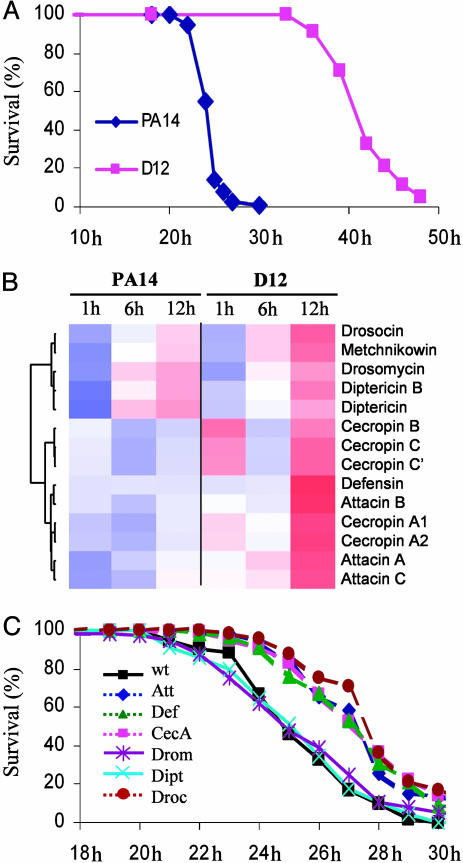
P. aeruginosa Restricts AMP Gene Expression to Cause Disease. Our genomic data show that AMP gene expression is significantly lower in PA14-infected vs. CF5-infected flies, and in some cases even lower than in the mock-injured control flies (Fig. 3), indicating that PA14 suppresses or poorly elicits their expression. To this end, we inoculated flies with the PA14-isogenic mutant D12, which has attenuated fly virulence (Fig. 5A), to ask whether the difference in AMP gene expression elicited by CF5 vs. PA14 reflects the strain “virulence-potential” vs. its general antigenicity. RNA was isolated from D12-inoculated flies at 1, 6, and 12 h after inoculation, and AMP gene expression was assessed by microarray. Fig. 5B shows that AMP gene transcription levels were higher in D12-infected vs. PA14-infected flies and similar to those elicited by CF5 (Table 1). These results further suggest that PA14 suppresses AMP expression.
Fig. 5.
Highly virulent PA14 cells suppress AMP expression. (A) Fly survival after infection with PA14 and PA14-isogenic mutant D12. (B) Relative expression of 14 AMP genes in flies infected with PA14 vs. avirulent PA14-isogenic mutant D12 cells. Cecropin C′ refers to the splice variant of the gene. Color representations are the same as in Fig. 4. (C) Overexpression of the AMP genes AttA, CecA, Def, and Drc, but not Dipt or Drom, renders flies significantly less susceptible to PA14 infection (P < 0.0001; Table 4).
Tables 1–3 demonstrate that the expression of 48 genes is similarly altered by D12- and CF5-inoculation. Eighteen of these genes are immunity-related, including 13 AMP genes (Fig. 5B); their transcriptional regulator relish; and windbeutel, TotM, TepII, and PGRP-SB1 (Tables 1–3). The remaining 30 genes act in tissue reconstruction (Rgk2, ci, Abi, Alp23B, and GenBank no. CG14557), metabolism (Lsp2, Sodh-1, and GenBank nos. CG17121, CG4757, and CG17176), stress (Hsp70Bbb, Hsp70Bc, methuselah-like 6, and sesB), neuronal function (GenBank nos. CG13948 and CG9948), signaling (GenBank nos. CG17766 and CG13579), gene regulation (GenBank nos. CG15619 and CG9772), circadian rhythm (GenBank no. CG11853), detoxification (Cyp6g1), or proteolysis (GenBank no. CG6580), or have unknown functions.
What is the benefit to P. aeruginosa to suppress AMP expression? An intriguing possibility is that AMPs are essential defense molecules during P. aeruginosa early infection, and their suppression aids bacterial survival and pathogenesis in the host. To this end, we infected flies that constitutively express different AMPs and assessed mortality over time. Fig. 5C and Table 4 show that AttA, CecA, Defensin (Def), or Drosocin (Drc) overexpression renders flies significantly less susceptible to PA14 infection, because these flies showed a 2- to 3-h delay to 50% mortality and 10–20% survival at 30 h. Conversely, Diptericin (Dipt) and Drosomysin (Drom) overexpression failed to give protection (Fig. 5C). As Fig. 5B shows, DiptA and Drom are part of one the six subclusters of the AMP genes differentially expressed in PA14-infected vs. D12-infected flies.
Discussion
We have used immune potentiation, genetic, and whole genome expression experiments to gain insights into the host responses that mediate susceptible vs. nonsusceptible pathogen–host interactions and, as such, may have clinical implications for limiting P. aeruginosa-human infections. Specifically, we show that Drosophila, when challenged by nonvirulent P. aeruginosa cells, mounts a complex defense by means of its innate immunity system that includes the expression of a wide array of genes and that this initial defense, including the expression of AMPs, is suppressed by a highly pathogenic P. aeruginosa strain. Our data also reveal the host signaling pathways that mediate the immune potentiation response, identify genes that potentially favor or limit the initiation and progression of pathogenesis, and demonstrate the importance of AMPs to restrict infection. These results are discussed below in the context of host–defense and pathogen–disease mechanisms.
The avirulent CF5 strain is an informative tool to dissect the primary steps of the host response to bacterial challenge, in the absence of progressive infection, because CF5 bacteria “prime” host defenses to subsequent challenge by highly virulent PA14 cells. This immune potentiation requires both Imd and Toll, in agreement with our previous results that these signaling pathways play an essential regulatory role in the ability of the fly to initially combat P. aeruginosa infection (6). Sterile injury, albeit to a lesser degree, also primes host defenses (Fig. 7), in concert with the low induction of certain DGs after injury (Fig. 4). These data extend classic studies that first characterized the Drosophila inducible defense response (39).
Our comparison of whole genome gene expression in CF5-challenged vs. PA14-challenged flies has three major implications. First, the 241 genes identified as differentially expressed fall into two distinct classes: the Class I DGs, whose expression is altered by CF5, and the Class II PGs, whose expression is altered by PA14. Second, the altered expression of ≈50% of these genes is strain-restricted: 101 Class I and 20 Class II genes are exclusive to CF5 (DSGs) and to PA14 (PSGs), respectively (Fig. 8, which is published as supporting information on the PNAS web site). Third, many of these genes likely enhance or limit pathogenesis, because they encode recognition and phagocytosis functions, or immune pathway components and effectors. Indeed, 51 of these genes specify known or predicted immunity-related activities that are part of recognition and phagocytosis, AMP production, or stress and detoxification functions (Fig. 8).
Although many of the differentially expressed genes exhibit low-magnitude changes, this differential expression is significant, because several of them cluster together and encode immunity-related functions, including AMPs. That many of the CF5-specific genes encode immunity and tissue reconstruction functions might reflect the whole-organism nature of our analysis, which inherently overlooks underrepresented cell types and magnifies the contributions of responses specific to the fat body, the main source of AMPs, and thorax muscle, a major component of the total body mass. This skewing lends further credence to the significance of the low-magnitude gene expression changes.
That certain immunity genes are down-regulated by CF5 may suggest that these genes are dispensable against P. aeruginosa infection, and, therefore, it is beneficial for the host to down-regulate them. Conversely, immunity genes down-regulated by PA14, similar to the CF5-up-regulated genes, benefit the host so their expression is compromised during virulent infection.
That the majority of the differentially expressed genes respond to CF5 suggests that the high virulence of PA14 is due in part to its ability to suppress and escape host-defense responses. To this end, the expression changes observed in flies inoculated with the highly attenuated PA14-isogenic mutant D12 vs. PA14 are primarily for immunity genes, including those encoding AMPs; and mortality is reduced and the life expectancy is increased of PA14-infected flies that overexpress single AMPs vs. wild-type flies. AMPs are important defense effectors: imd-;spz- double mutant flies, which are severely compromised in AMP expression (35), do not display immune potentiation against P. aeruginosa when preinoculated with CF5 (Fig. 2), and peak AMP induction, by means of CF5 inoculation (Fig. 4), coincides with the time required to trigger potentiation (Fig. 1). AMPs are considered to be the last line of fly defense. The cellular and humoral responses, although effective in combating microbial invasion, are unable to fully clear the hemocoel if challenged by large numbers of invading cells (36). In contrast, the AMPs are specifically released into the hemolymph where they attack bacterial and fungal cell wall components (36). Thus, the absence or low production of AMPs will impair hemocoel clearing of infectious organisms. Although known AMPs were reported ineffective against P. aeruginosa infection (15), the speedy mortality rate of this pathogen might result in the underestimation of small contributions of specific AMPs to fly defense.
Use of CF5 and D12 have enabled us to uncover a previously undescribed mechanism used by highly virulent PA14 to suppress and escape host defense mechanisms and thus increase its survival and pathogenicity. Although the function of the D12 locus is unknown, we speculate that other P. aeruginosa genes also may limit host defense responses, because D12 fails to affect many of the genes whose expression is altered by CF5 (Tables 1–3).
Our analysis also identifies fly genes whose expression has not previously been reported to be affected by bacterial challenge or infection (20, 37, 38). These differentially expressed genes encode stress, longevity, metabolic, muscle reconstruction, neuronal, olfactory, and circadian functions, suggesting that noninnate immunity functions also may mediate the enhanced resistance of flies to CF5 vs. PA14. To this end, bacterial challenge up-regulates muscle and cuticle structural constituents via Toll pathway activation (38), and aging, oxidative stress (39), circadian rhythms (40), and innate immunity are linked in Drosophila. Many of the genes identified here may function to locally or systemically enhance or restrict pathogenesis, with those that encode receptors or serine proteases, or that mediate tissue reconstruction, hemocyte proliferation, or phagocytosis, of particular interest.
This study presents, to our knowledge, the first whole-genome and whole-organism expression analysis of P. aeruginosa pathogenesis. Our data identify host-defense functions, reveal Drosophila genes that respond to bacterial challenge, and demonstrate that highly virulent P. aeruginosa cells suppress host defenses. Furthermore, the identified host-response genes may provide molecular signatures of sets of early host responses that lead to susceptible vs. nonsusceptible pathogen–host interactions. Furthermore, our results for immune potentiation and the defensive role AMPs in P. aeruginosa infection provide insights into the Drosophila–P. aeruginosa antagonistic interaction. Given the high degree of molecular and mechanistic conservation between the Drosophila and human innate immunity systems, our results also provide insights into P. aeruginosa–human pathogenesis and should help formulate future therapeutic strategies.
Supplementary Material
Acknowledgments
We thank B. Lemaitre for fly stocks, S. Stachel for comments and editing, J. Wilhelmy and G. Coelho for technical assistance, and S. Gopalan for microarray list comparisons. This work was supported in part by a grant from Aventis S.A. (to L.G.R.).
Author contributions: Y.A., M.N.M., and L.G.R. designed research; Y.A., M.N.M., G.W.L., and R.L.B. performed research; R.W.D. contributed new reagents/analytic tools; Y.A. and W.X. analyzed data; and Y.A. and L.G.R. wrote the paper.
Abbreviations: AMP, antimicrobial peptide; DG, defense gene; PG, pathogenesis gene; PSG, pathogenesis-specific gene; DSG, defense-specific gene.
References
- 1.Medzhitov, R. & Janeway, C., Jr. (2000) Immunol. Rev. 173, 89-97. [DOI] [PubMed] [Google Scholar]
- 2.Tzou, P., Ohresser, S., Ferrandon, D., Capovilla, M., Reichhart, J. M., Lemaitre, B., Hoffmann, J. A. & Imler, J. L. (2000) Immunity 13, 737-748. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann, J. A. & Reichhart, J. M. (2002) Nat. Immunol. 3, 121-126. [DOI] [PubMed] [Google Scholar]
- 4.Hetru, C., Troxler, L. & Hoffmann, J. A. (2003) J. Infect. Dis. 187, Suppl. 2, S327-S334. [DOI] [PubMed] [Google Scholar]
- 5.De Gregorio, E., Spellman, P. T., Tzou, P., Rubin, G. M. & Lemaitre, B. (2002) EMBO J. 21, 2568-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau, G. W., Goumnerov, B. C., Walendziewicz, C. L., Hewitson, J., Xiao, W., Mahajan-Miklos, S., Tompkins, R. G., Perkins, L. A. & Rahme, L. G. (2003) Infect. Immun. 71, 4059-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Argenio, D. A., Gallagher, L. A., Berg, C. A. & Manoil, C. (2001) J. Bacteriol. 183, 1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fauvarque, M. O., Bergeret, E., Chabert, J., Dacheux, D., Satre, M. & Attree, I. (2002) Microb. Pathog. 32, 287-295. [DOI] [PubMed] [Google Scholar]
- 9.Boman, H. G., Nilsson, I. & Rasmuson, B. (1972) Nature 237, 232-235. [DOI] [PubMed] [Google Scholar]
- 10.Lyczak, J. B., Cannon, C. L. & Pier, G. B. (2000) Microbes Infect. 2, 1051-1060. [DOI] [PubMed] [Google Scholar]
- 11.Rahme, L. G., Stevens, E. J., Wolfort, S. F., Shao, J., Tompkins, R. G. & Ausubel, F. M. (1995) Science 268, 1899-1902. [DOI] [PubMed] [Google Scholar]
- 12.Wolfgang, M. C., Kulasekara, B. R., Liang, X., Boyd, D., Wu, K., Yang, Q., Miyada, C. G. & Lory, S. (2003) Proc. Natl. Acad. Sci. USA 100, 8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He, J., Baldini, R. L., Deziel, E., Saucier, M., Zhang, Q., Liberati, N. T., Lee, D., Urbach, J., Goodman, H. M. & Rahme, L. G. (2004) Proc. Natl. Acad. Sci. USA 101, 2530-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai, J. W. & Alley, M. R. (2000) J. Bacteriol. 182, 504-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tzou, P., Reichhart, J. M. & Lemaitre, B. (2002) Proc. Natl. Acad. Sci. USA 99, 2152-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan, E. & Meier, P. (1958) J. Am. Stat. Assoc. 53, 562-563. [Google Scholar]
- 17.Cox, D. R. (1972) J. R. Stat. Soc. B. 34, 187-220. [Google Scholar]
- 18.Li, C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98, 31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellroth, P., Karlsson, J. & Steiner, H. (2003) J. Biol. Chem. 278, 7059-7064. [DOI] [PubMed] [Google Scholar]
- 20.De Gregorio, E., Spellman, P. T., Rubin, G. M. & Lemaitre, B. (2001) Proc. Natl. Acad. Sci. USA 98, 12590-12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagueux, M., Perrodou, E., Levashina, E. A., Capovilla, M. & Hoffmann, J. A. (2000) Proc. Natl. Acad. Sci. USA 97, 11427-11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson, K. V. (1998) Cell 95, 439-442. [DOI] [PubMed] [Google Scholar]
- 23.Lum, L. & Beachy, P. A. (2004) Science 304, 1755-1759. [DOI] [PubMed] [Google Scholar]
- 24.Fenger, D. D., Carminati, J. L., Burney-Sigman, D. L., Kashevsky, H., Dines, J. L., Elfring, L. K. & Orr-Weaver, T. L. (2000) Development (Cambridge, U.K.) 127, 4763-4774. [DOI] [PubMed] [Google Scholar]
- 25.Ferrandon, D., Imler, J. L. & Hoffmann, J. A. (2004) Semin. Immunol. 16, 43-53. [DOI] [PubMed] [Google Scholar]
- 26.Choe, K. M., Werner, T., Stoven, S., Hultmark, D. & Anderson, K. V. (2002) Science 296, 359-362. [DOI] [PubMed] [Google Scholar]
- 27.Brody, T. & Cravchik, A. (2000) J. Cell Biol. 150, F83-F85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Homyk, T., Jr., Szidonya, J. & Suzuki, D. T. (1980) Mol. Gen. Genet. 177, 553-565. [DOI] [PubMed] [Google Scholar]
- 29.Freeman, M. R., Delrow, J., Kim, J., Johnson, E. & Doe, C. Q. (2003) Neuron. 38, 567-580. [DOI] [PubMed] [Google Scholar]
- 30.Asha, H., Nagy, I., Kovacs, G., Stetson, D., Ando, I. & Dearolf, C. R. (2003) Genetics 163, 203-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dow, J. A. (1999) J. Bioenerg. Biomembr. 31, 75-83. [DOI] [PubMed] [Google Scholar]
- 32.Ekengren, S. & Hultmark, D. (2001) Biochem. Biophys. Res. Commun. 284, 998-1003. [DOI] [PubMed] [Google Scholar]
- 33.Markstein, M., Markstein, P., Markstein, V. & Levine, M. S. (2002) Proc. Natl. Acad. Sci. USA 99, 763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braun, A., Lemaitre, B., Lanot, R., Zachary, D. & Meister, M. (1997) Genetics 147, 623-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leulier, F., Rodriguez, A., Khush, R. S., Abrams, J. M. & Lemaitre, B. (2000) EMBO Rep. 1, 353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kavanagh, K. & Reeves, E. P. (2004) FEMS Microbiol. Rev. 28, 101-112. [DOI] [PubMed] [Google Scholar]
- 37.Irving, P., Troxler, L., Heuer, T. S., Belvin, M., Kopczynski, C., Reichhart, J. M., Hoffmann, J. A. & Hetru, C. (2001) Proc. Natl. Acad. Sci. USA 98, 15119-15124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boutros, M., Agaisse, H. & Perrimon, N. (2002) Dev. Cell 3, 711-722. [DOI] [PubMed] [Google Scholar]
- 39.Landis, G. N., Abdueva, D., Skvortsov, D., Yang, J., Rabin, B. E., Carrick, J., Tavare, S. & Tower, J. (2004) Proc. Natl. Acad. Sci. USA 101, 7663-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDonald, M. J. & Rosbash, M. (2001) Cell 107, 567-578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.