Abstract
The hepatitis C virus (HCV) is a major cause of liver disease worldwide. The understanding of the viral life cycle has been hampered by the lack of a satisfactory cell culture system. The development of the HCV replicon system has been a major advance, but the system does not produce virions. In this study, we constructed an infectious HCV genotype 1b cDNA between two ribozymes that are designed to generate the exact 5′ and 3′ ends of HCV. A second construct with a mutation in the active site of the viral RNA-dependent RNA polymerase (RdRp) was generated as a control. The HCV-ribozyme expression construct was transfected into Huh7 cells. Both HCV structural and nonstructural proteins were detected by immunofluorescence and Western blot. RNase protection assays showed positive- and negative-strand HCV RNA. Sequence analysis of the 5′ and 3′ ends provided further evidence of viral replication. Sucrose density gradient centrifugation of the culture medium revealed colocalization of HCV RNA and structural proteins in a fraction with the density of 1.16 g/ml, the putative density of HCV virions. Electron microscopy showed viral particles of ≈50 nm in diameter. The level of HCV RNA in the culture medium was as high as 10 million copies per milliliter. The HCV-ribozyme construct with the inactivating mutation in the RdRp did not show evidence of viral replication, assembly, and release. This system supports the production and secretion of high-level HCV virions and extends the repertoire of tools available for the study of HCV biology.
Keywords: assembly, cell culture, infection, ribozyme, viral replication
The hepatitis C virus (HCV) is an important cause of human illness worldwide (1). Although it has proven to be a difficult public health problem, it has been no easier to study in the laboratory. A major impediment has been the lack of robust model systems to study the complete viral life cycle. HCV is a member of the Flaviviridae family of ≈9.6 kb, and it has a central ORF flanked by the 5′ and 3′ noncoding regions. The ORF is divided into the coding sequences for the structural proteins at the 5′ end and the nonstructural proteins at the 3′ end. Study of the biology of hepatitis C at a molecular level focused initially on expression and manipulation of individual viral proteins in tissue culture.
The development of the subgenomic and genomic replicons is a major breakthrough to understanding viral replication and viral-cell interactions and provides a means to test therapeutic targets (2, 3). However, as yet, none of these systems produce viral particles, nor do they produce infectious virions. Although some infectious tissue culture systems have been described; in general, these systems have not been robust enough to study the complete viral life cycle (4, 5).
Why virion production has been such an elusive goal remains unclear; however, the promise of a system that produces authentic virions is clear. Not only would more of the biology of the virus become accessible for study, but also such a system would provide a means to screen a wider range of potential therapeutic compounds. There is evidence for an inverse relationship between viral replication in tissue culture and virulence in the host organism. This relationship is true for hepatitis A, and there is evidence that it may be true for HCV as well (6, 7). Regardless of the reason for this difficulty, there is an urgent need to establish such a system if improved therapies are to be developed, particularly given the absence of a simple small-animal model of HCV infection. This need is especially true for genotype 1, given that this genotype is the major genotype of human infections worldwide and is the type most resistant to current therapies (8, 9).
In this study, we describe an in vitro HCV replication system that is capable of producing viral particles in the culture medium. A full-length HCV construct, CG1b of genotype 1b, known to be infectious (10), was placed between two ribozymes designed to generate the exact 5′ and 3′ ends of HCV when cleaved. By using this system, we showed that HCV proteins and positive and negative RNA strands were produced intracellularly, and viral particles that resemble authentic HCV virions were produced and secreted into the culture medium. This system provides a unique opportunity to further study the life cycle and biology of HCV and to test potential therapeutic targets.
Materials and Methods
Plasmid Construction. The ribozymes were constructed by means of three pairs of overlapping primers that were based on a described ribozyme pair that was functional in hepatocytes (11). The innermost set (5′-CGG TAC CCG GTA CCG TCG CCA GCC CCC GA and 3′-ACG GAT CTA GAT CCG TCA CAT GAT CTG CA) was used to amplify pHCVGFP2. The pH-CVGFP2 was derived from an infectious full-length HCV CG1b clone (10) and was constructed by replacing the HCV sequences between nucleotide 709 (ClaI) and 8935 (BglII) by the sequence coding for the GFP. The middle (5′-TCC GTG AGG ACG AAA CGG TAC CCG GT and 3′-CAC GGA CTC ATC AGG ACG GAT CTA GA) and outermost (5′-GGC TGG CCT GAT GAG TCC GTG AGG A and 3′-GAT CAT GTT CGT CCT CAC GGA CTC A) sets were then added on to this sequence by PCR. This fragment was cloned into the SrfI site of pCMV-Script (Stratagene) and in turn subcloned into pcDNA3.1 (Invitrogen) by using NotI and HindIII sites to generate the pHt plasmid. pcDNA has both a CMV and a T7 promoter. The GFP was then removed, and the missing part of the HCV sequence was reinserted to generate the pHr plasmid. The pHr was used to generate the HCV-ribozyme RNA by T7 polymerase to assess the efficiency of the ribozymes. The HCV-ribozyme fragment was subcloned into pTRE2hyg+ (Clontech) under the control of a tetracycline-responsive promoter. This construct was named pTHr. In all the experiments described in this study, pTHr transfection always refers to cotransfection with pTet-Off (Clontech) expressing the tetracycline-responsive transactivator. A mutation in the GDD motif of the polymerase (GDD→GND) was introduced into this construct, and the mutated construct was named pTHrGND. The plasmid pTREhyg2+, without any insert, was also used as a control and is hereon referred to as pTRE.
Tissue Culture and Transfection and RNase Protection Assay. A human hepatoma cell line (Huh7) was maintained at 37°C in Dulbecco's modified Eagle's medium containing 10% FBS with 5% CO2. Transfection was carried out by using Lipofectamine (Invitrogen) according to the manufacturer's instructions. RPA 111 ribonuclease protection assay kits (Ambion) were used according to the manufacturer's directions. The probe used was transcribed from a construct containing the core region from nucleotide 342 to nucleotide 707 of HCV CG1b strain flanked by the T3 and T7 promoters.
Immunofluorescence and Western Blot. Huh7 cells were grown on glass coverslips and transfected as described. Cells were fixed with acetone/methanol on ice at different time points after transfection. Cells were washed with PBS three times, incubated with primary antibody for 1 h, washed with PBS, incubated with secondary antibody, and washed again with PBS. Monoclonal antibodies against the core (C1) and E1 (A4) were from H. Greenberg (Stanford Medical School, Palo Alto, CA) (12). The anti-E2 monoclonal antibodies AP33 and ALP98 were from A. Patel (Medical Research Council, Glasgow, Scotland) (13). The NS5A monoclonal antibody was obtained from J. Lau (ICN). The Cy3-labeled donkey anti-mouse IgG was obtained from Kirkegaard & Perry Laboratories. The same primary antibodies were used for Western blotting. The peroxidase-labeled goat anti-mouse IgG used as the secondary antibody was obtained from Kirkegaard & Perry Laboratories.
Sucrose Gradient Density Centrifugation. The tissue culture medium was centrifuged to remove cellular debris, and the supernatant was pelleted over a 30% sucrose cushion. The pellet was resuspended in TNC buffer (10 mM Tris·HCl, pH 7.4/1 mM CaCl2/150 mM NaCl) with EDTA-free protease inhibitors (Roche Applied Science) and applied onto a 20–60% sucrose gradient (10.5-ml volume) in SW41 tubes (Beckman Coulter) and centrifuged at 100,000 × g for 16 h at 4°C. We collected 1-ml fractions from the top of the gradient. The fractions were tested for HCV proteins and viral RNA as described below. Cryoelectron microscopy was performed by using standard techniques.
HCV RNA, Protein Quantitation, and RACE. HCV RNA level was quantitated by using the TaqMan real-time PCR method as described in ref. 10. RNA was extracted from 100 μl of the sucrose gradient fractions or tissue culture media by using TRIzol (Invitrogen) and resuspended in 20 μl of double-filtered RNase-free water. Samples were tested in duplicate. The core protein was quantitated by using the HCV core ELISA kits, which were provided by S. Yagi (Advanced Life Technology, Saitama, Japan) and used as described in ref. 14. Samples were tested in 50- or 100-μl aliquots. RNA was extracted by using TRIzol (Invitrogen), reverse-transcribed, and amplified by RNA ligase-mediated RACE (RLM-RACE, Ambion). The 5′ and 3′ RACE procedure was performed as described in ref. 15.
Results
Ribozyme Activity. To prove that the ribozymes function properly in the context of HCV genome, the HCV-ribozyme RNA was generated by in vitro transcription of pHr and analyzed by formamide gel electrophoresis. The results are shown in Fig. 1B. A band corresponding to the full-length HCV genome of ≈9,587 nt was detected. Also seen were bands corresponding to the vector (5,400 nt), a 150-nt fragment corresponding to the RNA between the T7 transcription initiation and the cleavage site of the 5′ ribozyme, and other molecular weight fragments probably representing uncleaved or prematurely terminated transcripts. A similarly expected pattern of cleavage was also observed with the pHt, which is the precursor construct of the pHr and contains the GFP sequence in place of the HCV polyprotein sequence (data not shown). Further proof of the ribozymes cleaving correctly is discussed later with the RACE results.
Fig. 1.
Construction of HCV-ribozyme plasmid. (A) The design of the construct is shown with the positions and sequences of the ribozymes (Rbz) flanking the 5′ and 3′ ends of the HCV CG1B sequence. The cleavage sites are indicated by arrows. The boxes shown 5′ and 3′ to the construct represent the promoter sequence (5′ end) and the simian virus 40 small T antigen intron and polyadenylation signal (3′ end). (B) An RNA gel with in vitro transcription products from pHr. The first lane shows molecular weight (MW) markers, and the second lane shows a sense transcript beginning at the 5′ end under the control of the T7 promoter. (Upper) The expected fragments at ≈9,500 and 5,400 nucleotides are indicated by arrows. The third lane shows an antisense transcript from the 3′ end under the control of a T3 promoter showing bands representing the full length of the plasmid and a population of RNA ≈1,400 bp long that possibly represents a termination sequence or difficult secondary structure at that region. (Lower) The expected 150-nt fragment can be seen on this gel with longer exposure (both lanes labeled T7).
HCV RNA and Protein Production in Transfected Cells. Both positive- and negative-strand HCV RNAs were detected in cells transfected with pTHr (Fig. 2). The level of positive-strand HCV RNA was at least 10-fold higher than the level of negative-strand HCV RNA in multiple experiments. The GND mutant pTHrGND produced a small amount of positive-strand RNA but did not produce any detectable negative-strand RNA. The positive-strand RNA produced with the GND mutant was less than that produced with pTHr. No viral RNA was detected in cell lysates transfected with pTRE.
Fig. 2.
Detection of HCV positive- and negative-strand RNAs. (Upper) The experiment. Shown are the total cellular RNA probed for the HCV core sequence, either positive or negative strand, and the findings when cellular RNA from pTRE-, pTHr-, or pTHrGND-transfected cells were probed for either positive- or negative-strand core sequence. (Lower) The control. Shown is the total cellular RNA probed for GAPDH messenger RNA. Note that the amounts are roughly comparable in the three lanes.
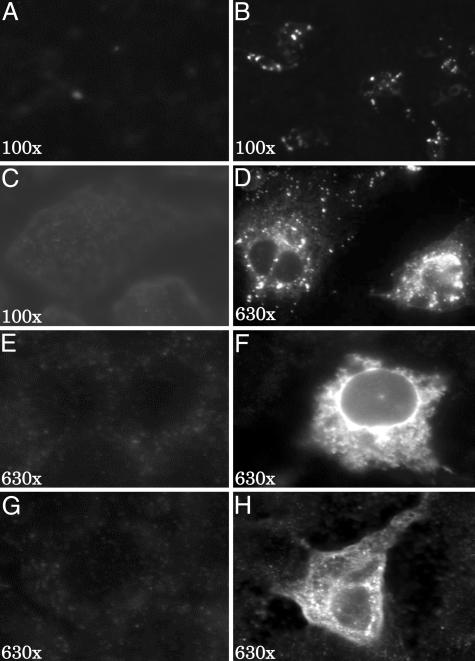
Cells transfected with pTHr or the control plasmid pTRE were analyzed by immunofluorescence with monoclonal antibodies directed against the core, E2, and NS5A. A granular cytoplasmic staining was seen with antibodies against all three proteins (Fig. 3). A time-course experiment showed peak protein expression on day 2 and a significant decrease on day 4 after transfection (data not shown). The percentage of cells with fluorescence was ≈10%, despite the transfection efficiency of ≈50% with a GFP-containing plasmid (data not shown). No immunofluorescence was seen in the cells transfected with pTRE.
Fig. 3.
Detection of HCV proteins by immunofluorescence. (A) Low-power view of cells transfected with pTHr and stained without primary antibody but with the secondary antibody. No fluorescence was seen. (B) Low-power view of cells transfected with pTHr and stained with anti-core. Multiple cells with fluorescence can be seen. (C) Low-power view of cells transfected with the control pTRE and stained with anti-core. There was no fluorescence. (D) High-power view of B. (E and F) High-power views of cells stained with anti-E2. Cells were transfected with pTRE (E) or pTHr (F). (G and H) High-power views of cells transfected with pTRE (G) and pTHr (H) and stained with anti-NS5A.
Western blot of cell lysates transfected with pTRE or pTHr showed the presence of core, E2, and NS5A in cells transfected with pTHr but not in cells transfected with pTRE (Fig. 4). As expected, viral protein was not detected in the presence of doxycycline (data not shown). Furthermore, little or no HCV protein was detected in pTHrGND-transfected cells, suggesting that viral replication is required for efficient protein production in this system (data not shown).
Fig. 4.
Detection of HCV proteins by Western blot. In each blot, the first lane shows cells transfected with pTHr and the second lane shows cells transfected with pTRE. The molecular weights are shown on the left of the blots. (Left) Blot was probed with anti-core. (Center) Blot probed with anti-E2. (Right) Blot probed with anti-NS5A.
HCV Virion Production and Secretion. To assess the possibility of HCV particle production, culture medium of the pTHr- and pTHrGND-transfected cells was subjected to sucrose density gradient centrifugation. The fractions were analyzed for two HCV structural proteins, core and E2, and HCV RNA. These results are shown in Fig. 5A. In the culture medium from cells transfected with pTHr, a peak of HCV proteins and RNA coincided in fraction 5, which has the density of 1.16 g/ml. This density is consistent with the published density of free HCV virions (16). Viral particles were visualized by electron microscopy only in fraction 5 (Fig. 5B). These particles are heterogeneous in appearance and have at least two sizes (≈50 and 100 nm in diameter) with the 50 nm being the major form. This heterogeneity has been described in ref. 17. Viral particles are double-shelled and appear to have spike-like projections from their surface. Shown in Fig. 5A are the results for pTHrGND-transfected cells. The HCV protein and RNA levels are at least 10-fold less than those of the pTHr-transfected cells.
Fig. 5.
Sucrose density gradient analysis of culture medium of HCV-transfected cells. (A) (Lower) Results of the sucrose gradient for pTHr (solid lines) and pTHrGND (dotted lines) transfections. The buoyant density of the sucrose is plotted with the levels of HCV RNA measured by TaqMan PCR and HCV core protein measured by core ELISA. (Upper) Western blot for the E2 protein in the fractions of the sucrose gradient of the pTHr transfection. Each lane corresponds to the fraction number below it on the x axis of the graph. Three hundred microliters of each fraction was spun at 100,000 × g for 90 min, and the pellet was resuspended in loading buffer and used for the Western blot. (B) Cryoelectron microscopy of fraction 5. (Bar, 100 nm.)
RACE. RACE was used to ensure the exact cleavage of the 5′ and 3′ ends of HCV by the ribozymes. In vitro-transcribed RNA from pHr and RNA from the culture medium of pTHr-transfected cells were analyzed by RACE. The 5′ end of the in vitro-transcribed RNA, as expected, had the same sequence as the cDNA construct (Fig. 6A). However the 3′ end of the in vitro transcript could not be amplified by RACE, possibly because of a less efficient cleavage by the 3′ ribozyme and subsequent difficulty in amplifying a heterogeneous population of the 3′ ends. Both the 5′ and 3′ ends of HCV RNA from the culture medium were successfully determined. Interestingly, a change in the most 5′ nucleotide from G to A was noted; this change has been frequently observed in HCV RNA replicons and circulating HCV RNA in infected humans (15). In the 3′ end, two nucleotide changes in the stem loop region were noted: U→A and A→U. These changes preserved the stem loop structure (Fig. 6B). Such changes have also been reported in HCV RNA from infected individuals (18). The RNA levels in the medium of the GND-transfected cells were not adequate to perform RACE.
Fig. 6.
Sequences of the 5′ and 3′ ends of HCV RNA. (A) The cDNA sequence for the 5′ end of the CG1B strain (a) and the RACE results for the 5′ ends of in vitro-transcribed RNA (b) and of the HCV RNA from the culture medium (c). (B) The cDNA sequences and the stem-loop structures of the 3′ ends of the CG1b strain (Left) and the HCV RNA from the medium (Right). Nucleotide changes are boxed.
Discussion
Since the discovery of HCV in 1989, working with HCV has proven to be difficult, mostly because of the lack of model systems (19). Each aspect of the life cycle has been difficult to reproduce in vitro. The infectious clone was developed after multiple attempts and had to be demonstrated in a chimpanzee (20, 21). Other small-animal models require complicated systems (22, 23). In vitro, virus obtained from infected individuals can replicate only in certain B cell lines and primary human hepatocytes but only at a low level (4, 5). Until the development of the replicon, most model systems have been difficult to work with (2, 24). Development of virus-like particles and pseudovirus have allowed study of viral entry into the cell but do not model other aspects of the viral life cycle (25–28). Therefore, a model system with viral replication, assembly, and release is urgently needed. Furthermore, genotype 1, the most prevalent form of HCV and the most difficult to treat, was chosen for this model.
By engineering two hammerhead ribozyme sequences, one at the 5′ end and the other at the 3′ end of an infectious HCV cDNA clone, we generated a DNA expression construct for the production of HCV virions. An important initial consideration was to ensure that the ribozymes are indeed functional. This functionality was demonstrated by in vitro-translation and RACE. Transfection of this HCV-ribozyme construct into Huh7 cells demonstrated the production of structural and nonstructural proteins by immunofluorescence and Western blot. Both positive- and negative-strand RNAs could be detected intracellularly. As expected, the positive strand is much more abundant than the negative strand.
The GND mutant was constructed as a control to determine the extent of replication in this model. Evidence for replication was derived from a number of results. The simplest evidence was the presence of negative-strand viral RNA in pTHr-transfected cells and the lack of negative strand in pTHrGND-transfected cells. A >10-fold difference in the relative amounts of the positive-strand viral RNA between the wild-type and GND constructs provided additional evidence. This observation can be explained by the lack of amplification as a result of defective replication. The positive strand seen with the GND mutant was generated from transcription of the cDNA plasmid. This difference in product was also evident in the culture medium. The amounts of viral RNA and core protein on the sucrose gradients were >10-fold higher in wild-type cells than in the GND mutant-transfected cells. The final and perhaps the most interesting evidence for replication is the RACE findings. The 5′ and 3′ nucleotide changes have been described in refs. 15 and 18. The G→A switch of the initial nucleotide of HCV is associated with replication in vivo and in vitro (15). A transposition from an A–T to a T–A base pairing has also been reported (18) and represents a base pair in the putative terminal stem loop of the 3′ end of HCV. These observations provide support for the replication of viral RNA in this system.
Evidence for assembly and release was derived in a number of ways. The presence of HCV RNA in the media with the exact 5′ and 3′ ends showed that the correctly processed RNA was secreted into the culture medium. The association of viral RNA and core and E2 protein in the same fraction on the sucrose gradient with a density of 1.16g/ml (the published density of free HCV virions) supported the interpretation that viral particles are assembled and secreted into the medium. The most compelling evidence is the visualization of particles resembling virions by electron microscopy, and these particles were visualized only in fraction 5, where viral RNA and proteins are present. It is interesting that the core protein extends into fractions 6 and 7 more than the viral RNA and E2 protein. This core reactivity might represent free core particles, although they were not seen on electron microscopy (29). The production and release of HCV particles is rather robust in this system, capable of achieving >10 million copies of HCV RNA per ml in the culture medium.
Although replicons using the full-length HCV genome have been developed, particles have not been described. In those replicons where sequence coding for the neomycin is included, difficulty in packaging a longer RNA molecule might be the problem. Alternatively the block could be the result of the inhibitory effects of the replicon adaptive mutations on virion assembly and release. Both possibilities are speculative. However, in the system presented here, there is no extraneous RNA and, although mutations can and do occur (see the RACE results), the source of the RNA (the cDNA) maintains a stable sequence without adaptive mutations. This difference might partially explain why particles are seen. It may also be of importance that there is a constant RNA production inside the cells being channeled directly into the appropriate cellular machinery for assembly.
This model system does not allow the study of viral entry and the earliest events in the HCV life cycle. In addition, whether these particles are infectious or not remains to be determined. The HCV sequence used is known to be infectious in chimpanzees. It should be noted that the sequence is genotype 1b. The results that would be obtained with other genotypes in this system is unknown. Despite these caveats, it represents a robust system to study the viral life cycle, specifically viral assembly and release. Very little is known about the assembly and release of HCV. This work might present an opportunity to better elucidate the biology of HCV as well as to develop therapeutic targets for the treatment of hepatitis C, in particular for genotype 1.
Note Added in Proof. During the preparation of this manuscript, two groups (T. Wakita, T. Takanobu, T. Date, and M. Miyamoto and T. Pietschmann, G. Koutsoudakis, S. Kallis, T. Kato, S. Foung, T. Wakita, and R. Bartenschlager) at the 11th International Symposium on HCV and Related Viruses (Heidelberg, Germany, Oct. 3–7, 2004) reported the production of infectious HCV in cell culture by transfecting a full-length HCV RNA genome.
Acknowledgments
We thank Z. Hong Zhou, Ph.D., for superb cryoelectron microscopy, Shintaro Yagi for providing the HCV core ELISA kit, and Anthony Davis for excellent technical assistance.
Author contributions: T.H., S.S., G.L., and T.J.L. designed research; T.H., S.S., J.A., T.W., T.R.M., A.J., B.C., N.J., R.S., G.L., and T.J.L. performed research; T.H., S.S., J.A., T.W., T.R.M., A.J., B.C., N.J., R.S., G.L., and T.J.L. analyzed data; and T.H. and T.J.L. wrote the paper.
This work was presented in part at the 55th Annual Meeting of the American Association for the Study of Liver Disease, Boston, October 30–November 2, 2004.
Abbreviation: HCV, hepatitis C virus.
References
- 1.Liang, T. J., Rehermann, B., Seeff, L. B. & Hoofnagle, J. H. (2000) Ann. Intern. Med. 132, 296-305. [DOI] [PubMed] [Google Scholar]
- 2.Lohmann, V., Korner, F., Koch, J., Herian, U., Theilmann, L. & Bartenschlager, R. (1999) Science 285, 110-113. [DOI] [PubMed] [Google Scholar]
- 3.Ikeda, M., Yi, M., Li, K. & Lemon, S. M. (2002) J. Virol. 76, 2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimizu, Y. K., Iwamoto, A., Hijikata, M., Purcell, R. H. & Yoshikura, H. (1992) Proc. Natl. Acad. Sci. USA 89, 5477-5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sung, V. M., Shimodaira, S., Doughty, A. L., Picchio, G. R., Can, H., Yen, T. S., Lindsay, K. L., Levine, A. M. & Lai, M. M. (2003) J. Virol. 77, 2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raychaudhuri, G., Govindarajan, S., Shapiro, M., Purcell, R. H. & Emerson, S. U. (1998) J. Virol. 72, 7467-7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukh, J., Pietschmann, T., Lohmann, V., Krieger, N., Faulk, K., Engle, R. E., Govindarajan, S., Shapiro, M., St Claire, M. & Bartenschlager, R. (2002) Proc. Natl. Acad. Sci. USA 99, 14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manns, M. P., McHutchison, J. G., Gordon, S. C., Rustgi, V. K., Shiffman, M., Reindollar, R., Goodman, Z. D., Koury, K., Ling, M. & Albrecht, J. K. (2001) Lancet 358, 958-965. [DOI] [PubMed] [Google Scholar]
- 9.Fried, M. W., Shiffman, M. L., Reddy, K. R., Smith, C., Marinos, G., Goncales, F. L., Jr., Haussinger, D., Diago, M., Carosi, G., Dhumeaux, D., et al. (2002) N. Engl. J. Med. 347, 975-982. [DOI] [PubMed] [Google Scholar]
- 10.Thomson, M., Nascimbeni, M., Gonzales, S., Murthy, K. K., Rehermann, B. & Liang, T. J. (2001) Gastroenterology 121, 1226-1233. [DOI] [PubMed] [Google Scholar]
- 11.Benedict, C. M., Pan, W., Loy, S. E. & Clawson, G. A. (1998) Carcinogenesis 19, 1223-1230. [DOI] [PubMed] [Google Scholar]
- 12.Dubuisson, J., Hsu, H. H., Cheung, R. C., Greenberg, H. B., Russell, D. G. & Rice, C. M. (1994) J. Virol. 68, 6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Triyatni, M., Saunier, B., Maruvada, P., Davis, A. R., Ulianich, L., Heller, T., Patel, A., Kohn, L. D. & Liang, T. J. (2002) J. Virol. 76, 9335-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka, E., Ohue, C., Aoyagi, K., Yamaguchi, K., Yagi, S., Kiyosawa, K. & Alter, H. J. (2000) Hepatology 32, 388-393. [DOI] [PubMed] [Google Scholar]
- 15.Cai, Z., Liang, T. J. & Luo, G. (2004) J. Virol. 78, 3633-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaito, M., Watanabe, S., Tsukiyama-Kohara, K., Yamaguchi, K., Kobayashi, Y., Konishi, M., Yokoi, M., Ishida, S., Suzuki, S. & Kohara, M. (1994) J. Gen. Virol. 75, 1755-1760. [DOI] [PubMed] [Google Scholar]
- 17.Andre, P., Komurian-Pradel, F., Deforges, S., Perret, M., Berland, J. L., Sodoyer, M., Pol, S., Brechot, C., Paranhos-Baccala, G. & Lotteau, V. (2002) J. Virol. 76, 6919-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolykhalov, A. A., Feinstone, S. M. & Rice, C. M. (1996) J. Virol. 70, 3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choo, Q. L., Kuo, G., Weiner, A. J., Overby, L. R., Bradley, D. W. & Houghton, M. (1989) Science 244, 359-362. [DOI] [PubMed] [Google Scholar]
- 20.Kolykhalov, A. A., Agapov, E. V., Blight, K. J., Mihalik, K., Feinstone, S. M. & Rice, C. M. (1997) Science 277, 570-574. [DOI] [PubMed] [Google Scholar]
- 21.Yanagi, M., Purcell, R. H., Emerson, S. U. & Bukh, J. (1997) Proc. Natl. Acad. Sci. USA 94, 8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mercer, D. F., Schiller, D. E., Elliott, J. F., Douglas, D. N., Hao, C., Rinfret, A., Addison, W. R., Fischer, K. P., Churchill, T. A., Lakey, J. R., et al. (2001) Nat. Med. 7, 927-933. [DOI] [PubMed] [Google Scholar]
- 23.Labonte, P., Morin, N., Bowlin, T. & Mounir, S. (2002) J. Med. Virol. 66, 312-319. [DOI] [PubMed] [Google Scholar]
- 24.Blight, K. J., Kolykhalov, A. A. & Rice, C. M. (2000) Science 290, 1972-1974. [DOI] [PubMed] [Google Scholar]
- 25.Baumert, T. F., Ito, S., Wong, D. T. & Liang, T. J. (1998) J. Virol. 72, 3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nam, J. H., Bukh, J., Purcell, R. H. & Emerson, S. U. (2001) J. Virol. Methods 97, 113-123. [DOI] [PubMed] [Google Scholar]
- 27.Bartosch, B., Bukh, J., Meunier, J. C., Granier, C., Engle, R. E., Blackwelder, W. C., Emerson, S. U., Cosset, F. L. & Purcell, R. H. (2003) Proc. Natl. Acad. Sci. USA 100, 14199-14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Logvinoff, C., Major, M. E., Oldach, D., Heyward, S., Talal, A., Balfe, P., Feinstone, S. M., Alter, H., Rice, C. M. & McKeating, J. A. (2004) Proc. Natl. Acad. Sci. USA 101, 10149-10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maillard, P., Krawczynski, K., Nitkiewicz, J., Bronnert, C., Sidorkiewicz, M., Gounon, P., Dubuisson, J., Faure, G., Crainic, R. & Budkowska, A. (2001) J. Virol. 75, 8240-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]