Abstract
The yeast cyclin-dependent kinase Srb10 phosphorylates various transcriptional activators as they activate transcription, and acidic transcriptional activating domains found on several activators directly bind Srb10. Here we show that the interaction between Srb10 (with its associated cyclin Srb11) and each of several different activating regions, in vitro, leads to the phosphorylation of peptide sequences attached to but outside of the activating regions themselves. In some cases, residues within the activating regions are also phosphorylated. The results define a mechanism by which a kinase is recruited to alternate substrates with diverse physiological consequences.
Keywords: kinase-substrate targeting, proteolysis, Srb10/11, transcriptional regulation
Five different yeast activators are phosphorylated, as they activate transcription, by Srb10, a cyclin-dependent kinase (Cdk) that associates with the cyclin Srb11. The effect of the modification is unique to each case. For example, phosphorylation of Gal4 increases the efficiency of induction of the galactose-responsive genes; that of Gcn4 and Ste12 leads to their proteolysis; that of Msn2 leads to its export from the nucleus; and that of Sip4 increases the efficiency of induction of glucose-responsive genes (1–5). Srb10/11 also phosphorylates a heptapeptide (YSPTSPS), 26 tandem copies of which comprise the carboxyl-terminal domain (CTD) of the largest subunit of RNA polymerase II (6–9). Other defined targets of Srb10/11 in the transcriptional machinery include two subunits of the transcription factor IID complex (10, 11).
Srb10/11 associates with the mediator, a multiprotein complex that itself associates with RNA polymerase II (6, 8, 10, 12–15). In a previous article, we showed that each of an array of transcriptional acidic activating regions (yeast, mammalian, and artificial) binds Srb10 (5). The interaction, detected in vivo, was also observed in vitro with purified Srb10 alone (but not with Srb11 alone), with Srb10/11, and with Srb10/11 as part of the mediator complex (5).
The activator–Srb10 interactions, like those between activators and other components of the transcriptional machinery, may help recruit that machinery to a nearby gene. Here we show a different (and/or additional) role played by various activator–Srb10/11 interactions: in each case the interaction recruits the kinase to a specific potential substrate. Thus, for five cases tested here, interaction of the activating region with the kinase leads to phosphorylation of a peptide sequence attached to the activating region, and in two of these cases, residues within the activating region itself are also phosphorylated.
Materials and Methods
Srb10/11 Purification. Recombinant Srb10 and Srb11 were expressed and purified from a baculovirus-expression system. Purifications of Srb10/11 from insect cells were performed as described by Koh et al. (5, 16).
Srb10 bearing a carboxyl-terminal 3× hemagglutinin tag was expressed in SKY-559 yeast cells. Cells were grown to midlog phase and lysed in buffer A [50 mM Tris, pH 7.5/5 mM EDTA/50 mM NaF/250 mM NaCl/0.1% Nonidet P-40/1 mM benzamidine/1 mM chymostatin/1 mM pepstatin/1 mM PMSF/1× protease inhibitor mix (Roche Molecular Biochemicals, CA 1697498)]. Cell lysate then was incubated with hemagglutinin antibody (Santa Cruz Biotechnology, 7392), and the enzyme was precipitated by using protein A-Sepharose.
Gst-Fusion Proteins. Gst-fused proteins were purified as described (5). Gst-fusion proteins were expressed in BL21 (DE3) pLysS bacterial strain. Cells were grown at 37°C until they reached midlog phase and then were induced for 3 h with 1 mM isopropyl β-d-thiogalactoside. Cells were lysed in buffer B (PBS with 10% glycerol and 0.01% Nonidet P-40 along with 1 mM PMSF and 1× protease inhibitor mix). After centrifugation (15 min at 10,000 × g), the protein was purified by using glutathione-Sepharose 4B (Amersham Pharmacia). Gst-fused proteins were eluted by using 50 mM glutathione in buffer C (5 mM Hepes, pH 7.4/10 mM K-glutamate/10 mM magnesium acetate/0.5 mM EGTA, pH 8.0/10% glycerol/0.1% Nonidet P-40 with 1 mM PMSF and 1× protease inhibitor mix). The relevant fractions were concentrated by using centriprep-10 concentrators (Amicon).
Kinase Assay. For Figs. 1, 2, 3, 4, kinase assays were performed by using 0.5 pmol of baculovirus-purified Srb10/11. Yeast-purified Srb10 was used in the reactions shown in Fig. 5. The yeast-purified Srb10 was also used to repeat the experiment shown in Fig. 4 to determine the concentration required for the same enzymatic activity (data not shown). Each assay was conducted in a 25-μl volume of buffer D [20 mM Hepes-KOH, pH 7.3/10% glycerol/2.5 mM EGTA/15 mM magnesium acetate/1 mM DTT/100 mM potassium acetate/100 μM ATP/5 μCi (1 Ci = 37 GBq) of [γ-32P]ATP (Amersham Pharmacia Biosciences, 6,000 Ci/mmol, 10 mCi/ml)], a combination of phosphatase inhibitors (1 mM NaN3/1 mM NaF/0.4 mM NaVO3/0.4 mM Na3VO4), 1× protease inhibitor mix, and 1 mM PMSF. Reactions were performed at 25°C and resolved on 4–20% gradient gels by SDS/PAGE.
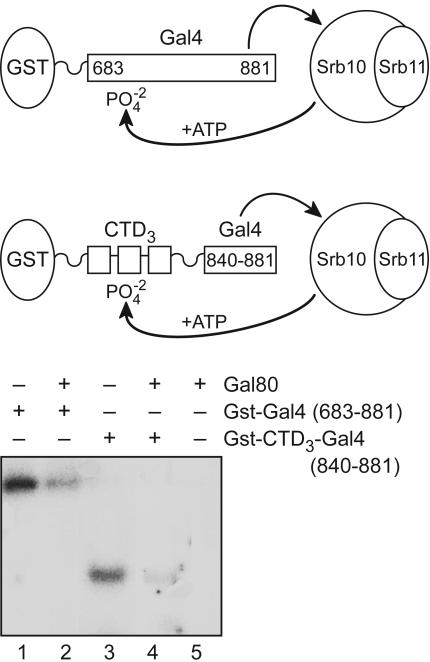
Fig. 1.
Effect of Gal80 on the phosphorylation of two substrates. Srb10/11 complex was incubated with 0.1 nmol of either Gst-Gal4 (683–881) or Gst-CTD3-Gal4 (840–881) in the presence or absence of 2-fold molar excess of Gal80. We expect that Gal80 binds stoichiometrically to the Gal4-activating region (840–874) under these conditions (17). We use three repeats of the CTD heptapeptide YSPTSPS as a substrate based on experiments of Hengartner et al. (8).
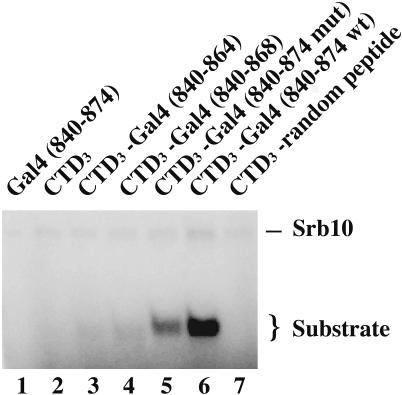
Fig. 2.
Role of activating region strength on the phosphorylation of an attached substrate. Srb10/11 complex was mixed with 0.1 nmol of the indicated Gst-fusion proteins. Lane 1 contains a Gst-Gal4 (840–874) control peptide, which is not attached to a CTD3 substrate. Lanes 2–7 contain Gst-CTD3 fused to five peptides, two of which are deletion derivatives of Gal4 and the third of which bears F869A, a point mutation in the activating region. This point mutation decreases the activating function of Gal4 (840–881) by 3- to 4-fold (16, 17). The sequence of the fifth peptide, which has no activating region, is (YSPTSPS)3-GVTWTKLRIT-RLESLQGCLI-PLQWMMYITI-YSMMKIPNRI-PGNSS.
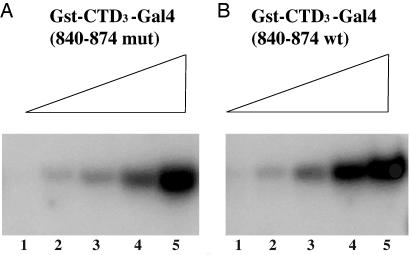
Fig. 3.
The effect of increasing concentrations of two substrates. In both cases [Gst-CTD3-Gal4 (840–874) wild type and mutant], the substrate concentration increases at 3.3-fold increments, with lane 5 containing 6.6 nmol.
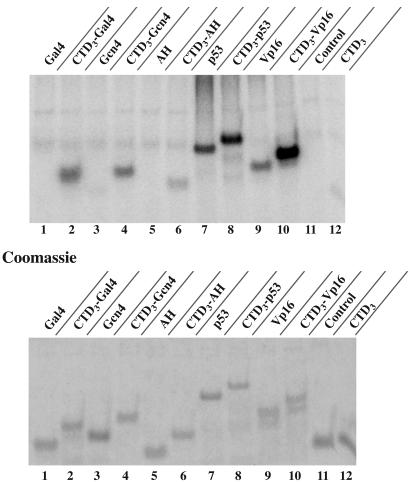
Fig. 4.
The effect of various activating regions on the targeting of Srb10/11 to an attached CTD3 substrate. The activating regions used were AH (PGIELQELQELQALLQQQ), Gal4 (840–874), Gcn4 (107–144), p53 (1–97), and Vp16 (420–490). Each reaction contained Srb10/11 complex and equimolar amounts of each substrate, as demonstrated in the Coomassie-blue-stained gel.
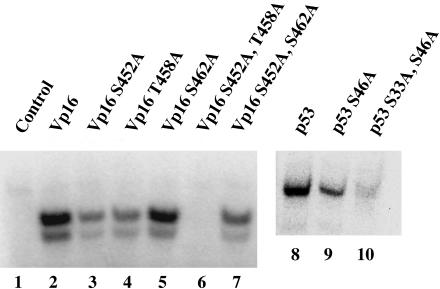
Fig. 5.
Mapping the phosphorylation sites of two mammalian activating regions. Srb10 was incubated separately with 17.5 pmol of Vp16 and p53 wild type and with the same quantity of their SP/TP mutant forms.
Results
Fig. 1 shows phosphorylation of the following two proteins by purified Srb10/11: (i) a fragment of Gal4 (683–881) that contains an activating region (residues 840–881) as well as a region bearing three residues (S690, S696, and S699) reported by Hirst et al. (2) to be phosphorylated by Srb10 in vivo (lanes 1 and 2) and (ii) a fusion peptide bearing an activating region (Gal4 residues 840–881) attached to three heptad repeats (CTD3) from the CTD (lanes 3 and 4). In both cases, phosphorylation was severely attenuated by addition of Gal80, the inhibitor that binds to and masks the activating region of Gal4 (lanes 2 and 4). Thus, at the concentrations of reactants used here, phosphorylation requires the unmasked Gal4-activating region. The sites phosphorylated in each case must lie outside the activating region (residues 840–881), because the activating region bears no required target sites of Srb10/11 [i.e., no Ser-Pro nor Thr-Pro sites]. Gal80 was not phosphorylated by Srb10/11 (lane 5).
The experiment shown in Fig. 2 examines the efficiency of phosphorylation of a series of substrates bearing activating regions of various strengths. These strengths were determined by the degree to which each activating region activated transcription in vivo when tethered to DNA (data not shown). In the experiments shown here, each activating region, a deletion derivative of that used for Fig. 1, was attached to CTD3. The results show that, at subsaturating concentrations of substrate (see below), phosphorylation of the CTD3 increased as the strength of the attached activating region increased (lanes 3–6). Thus, for example, the longest (and strongest) activating region (Gal4 residues 840–874) promoted approximately five times as much phosphorylation as did the shortest (and weakest) activating region [Gal4 (840–864) (compare lanes 6 and 3)]. A point mutation (F869A) in that longer activating region, which is known to decrease activation in vivo (17), also decreased phosphorylation in the depicted reaction (compare lanes 5 and 6). In an experiment not shown, we failed to detect any effect of a Ga14 activating region on the catalytic activity of Srb10/11.
Fig. 3 shows that the different efficiencies of phosphorylation of two substrates [one bearing a strong activating region (B) and another the same activating region damaged by the F869A point mutation (A)] can be overcome by increasing substrate concentration. Thus, as in the experiment shown in Fig. 2, at low substrate concentration, wild-type Gal4 was phosphorylated more efficiently than was the point mutant (compare lane 4 in A and B). However, at higher concentration, both the wild-type and mutant Gal4-fused substrates were phosphorylated approximately equally efficiently (compare lane 5 in A and B).
The experiments described thus far have illustrated the ability of the activating region of Gal4 to appose an attached substrate with Srb10/11. Fig. 4 extends the finding to the following additional activating regions: a peptide excised from the yeast activator Gcn4 (residues 107–144); the 18-residue artificial activating region, AH (18); and peptides from the mammalian activators p53 (residues 1–97) and VP16 (residues 420–490) (19). The latter two activating regions contain Ser-Pro and Thr-Pro residues that could serve as potential sites of phosphorylation. As the figure shows, Gal4 (840–874), Gcn4 (107–144), and AH each enhanced phosphorylation of its attached CTD3 (lanes 2, 4, and 6). Moreover, the relative efficiencies of phosphorylation (lanes 2, 4, and 6) were consistent with their relative activating strengths in vivo and their relative affinities for Srb10/11 in vitro (AH < Gcn4 < Gal4) (5, 18, 19). The p53- and VP16-activating region fragments also increased phosphorylation of attached CTD3 (compare lane 7 with lane 8 and lane 9 with lane 10). These two mammalian activating regions were themselves phosphorylated, a reaction observed with these regions unattached to CTD3 (lanes 7 and 9). Phosphorylation within the activation domain of the yeast activator Ste12 has been reported recently (3).
The experiment of Fig. 5 maps the phosphorylation sites (either Ser-Pro or Thr-Pro) within these two mammalian activating regions to at least two sites in p53 and to two sites in VP16. In the case of VP16, changing either one of the potential phosphorylation sites (S452A or T458A) diminished phosphorylation and substituting both residues with alanine abolished phosphorylation altogether. As a control, changing S462A (in an SAP site) had no effect on the phosphorylation of the VP16-activating peptide by Srb10/11. Fig. 5 also shows that the p53 single-point mutant S46A diminished phosphorylation and with the additional mutation S33A nearly abolished phosphorylation of p53. In parallel binding experiments, we found that the mutated, unphosphorylatable p53 and VP16 derivatives bound Srb10 as efficiently as did the wild-type peptides (data not shown).
Discussion
Our results show that five different acidic transcriptional activating regions, by binding to the Cdk/cyclin pair Srb10/11, promote phosphorylation of the substrate CTD3 that is artificially attached to the activating regions. We imagine that this in vitro reaction mirrors the physiologically relevant phosphorylation of activators described in the Introduction. We also found that two mammalian activating regions were phosphorylated by Srb10/11, but we do not know the physiological consequences of the modification. Phosphorylation of the CTD of RNA polymerase II is believed to be crucial for efficient transcription (6–9). It is possible that the binding of activating regions to Srb10 (as well as to other targets in the transcriptional machinery) not only apposes the activators themselves with the kinase but also indirectly apposes the CTD (as part of RNA polymerase II) with the kinase to promote phosphorylation of the CTD. Thus, a series of phosphorylations would be triggered by the kind of binding reaction we have described as transcription is activated. In addition to its positive role in gene activation (2, 5–7, 20), Srb10/11 has also been characterized as required for efficient repression by Tup1/Ssn6, a protein complex that is recruited to and works at many yeast genes (12, 21–25). Suggestions as to the mechanism of this effect have been proposed, but the matter has not been fully resolved (1, 3, 8, 21, 22).
The mode of targeting by Srb10 that we have described here differs in detail from previously described mechanisms for targeting Cdks to one or another substrate in the following way. Cdks are typically recruited to target substrates by a “recruiting patch” on their associated cyclins (26–32). By associating with different cyclins, a Cdk can in this fashion be directed to different targets (26–32). In our case, targeting is achieved by direct interactions with the Cdk (Srb10) rather than its associated cyclin Srb11. It has been argued that there are few stereospecific constraints on the cyclin–target interactions leading to specific phosphorylations, and that rule seems to hold for the cases we have described as well. Our experiments thus provide another example of a strategy for imposing specificity on kinases, one that is used widely for signal transduction pathways involving many aspects of cellular function (32, 33).
Acknowledgments
We thank B. Schulman, M. Floer, Z. Zaman, S. S. Koh, R. Young, A. Gann, I. Sadowski, and W. Tansey for help. This work was supported by funds from the National Institutes of Health (to M.P., Ludwig Professor of Molecular Biology) and by University of Wisconsin (Madison) Trust Funds and the March of Dimes Foundation (to A.Z.A.).
Author contributions: A.Z.A., A.O., and M.P. designed research; A.Z.A. and A.O. performed research; A.Z.A. and A.O. contributed new reagents/analytic tools; A.Z.A., A.O., and M.P. analyzed data; and A.Z.A., A.O., and M.P. wrote the paper.
Abbreviations: Cdk, cyclin-dependent kinase; CTD, carboxyl-terminal domain.
References
- 1.Chi, Y., Huddleston, M. J., Zhang, X., Young, R. A., Annan, R. S., Carr, S. A. & Deshaies, R. J. (2001) Genes Dev. 15, 1078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirst, M., Kobor, M. S., Kuriakose, N., Greenblatt, J. & Sadowski, I. (1999) Mol. Cell 3, 673-678. [DOI] [PubMed] [Google Scholar]
- 3.Nelson, C., Goto, S., Lund, K., Hung, W. & Sadowski, I. (2003) Nature 421, 187-190. [DOI] [PubMed] [Google Scholar]
- 4.Vincent, O. Kuchin, S., Hong, S. P., Townley, R., Vyas, V. K. & Carlson, M. (2001) Mol. Cell. Biol. 21, 5790-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ansari, A. Z., Koh, S. S., Zaman, Z., Bongards, C., Lehming, N., Young, R. A. & Ptashne, M. (2002) Proc. Natl. Acad. Sci. USA 99, 14706-14709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao, S.-M., Zhang, J., Jeffery, D. A., Koleske, A. J., Thompson, C. M., Chao, D. M., Viljoen, M., van Vuuren, H. J. & Young, R. A. (1995) Nature 374, 193-196. [DOI] [PubMed] [Google Scholar]
- 7.Kuchin, S., Yeghiayan, P. & Carlson, M. (1995) Proc. Natl. Acad. Sci. USA 92, 4006-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hengartner, C. J., Myer, V. E., Liao, S. M., Wilson, C. J., Koh, S. S. & Young, R. A. (1998) Mol. Cell 2, 43-53. [DOI] [PubMed] [Google Scholar]
- 9.Borggrefe, T., Davis, R., Erdjument-Bromage, H., Tempst, P. & Kornberg, R. D. (2002) J. Biol. Chem. 277, 44202-44207. [DOI] [PubMed] [Google Scholar]
- 10.Liu, Y., Kung, C., Fishburn, J., Ansari, A. Z., Shokat, K. M. & Hahn, S. (2004) Mol. Cell. Biol. 24, 1721-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallberg, M., Polozkov, G. V., Hu, G. Z., Beve, J., Gustafsson, C. M., Ronne, H. & Bjorklund, S. (2004) Proc. Natl. Acad. Sci. USA 101, 3370-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlson, M. (1997) Annu. Rev. Cell Dev. Biol. 13, 1-23. [DOI] [PubMed] [Google Scholar]
- 13.Lee, T. I. & Young, R. A. (2000) Annu. Rev. Genet. 34, 77-137. [DOI] [PubMed] [Google Scholar]
- 14.Malik, S. & Roeder, R. G. (2000) Trends Biochem. Sci. 25, 277-283. [DOI] [PubMed] [Google Scholar]
- 15.Myers, L. C. & Kornberg, R. D. (2000) Annu. Rev. Biochem. 69, 729-749. [DOI] [PubMed] [Google Scholar]
- 16.Koh, S. S., Ansari, A. Z., Ptashne, M. & Young, R. A. (1998) Mol. Cell 1, 895-904. [DOI] [PubMed] [Google Scholar]
- 17.Wu, Y., Reece, R. J. & Ptashne, M. (1996) EMBO J. 15, 3951-3963. [PMC free article] [PubMed] [Google Scholar]
- 18.Giniger, E. & Ptashne, M. (1987) Nature 330, 670-672. [DOI] [PubMed] [Google Scholar]
- 19.Melcher, K. (2000) J. Mol. Biol. 301, 1097-1112. [DOI] [PubMed] [Google Scholar]
- 20.Larschan, E. & Winston, F. (2005) Mol. Cell. Biol. 25, 114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green, S. R. & Johnson, A. D. (2004) Mol. Biol. Cell. 15, 4191-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaman, Z., Ansari, A. Z., Koh, S. S., Young, R. A. & Ptashne, M. (2001) Proc. Natl. Acad. Sci. USA 98, 2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mennella, T. A., Klinkenberg, L. G. & Zitomer, R. S. (2003) Eukaryot. Cell 2, 1288-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuchin, S. & Carlson, M. (1998) Mol. Cell. Biol. 18, 1163-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balciunas, D. & Ronne, H. (1995) Nucleic Acids Res. 23, 4421-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown, N. R., Noble, M. E. M., Endicott, J. A. & Johnson, L. N. (1999) Nat. Cell Biol. 1, 438-443. [DOI] [PubMed] [Google Scholar]
- 27.Endicott, J. A., Noble, M. E. & Tucker, J. A. (1999) Curr. Opin. Struct. Biol. 9, 738-744. [DOI] [PubMed] [Google Scholar]
- 28.Russo, A. A., Jeffrey, P. D. & Pavletich, N. P. (1996) Nat. Struct. Biol. 3, 696-700. [DOI] [PubMed] [Google Scholar]
- 29.Schulman, B. A., Lindstrom, D. L. & Harlow, E. (1998) Proc. Natl. Acad. Sci. USA 95, 10453-10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Byrne, M., Miller, N., Springer, M. & O'Shea, E. K. (2004) J. Mol. Biol. 335, 57-70. [DOI] [PubMed] [Google Scholar]
- 31.Roberts, J. M. (1999) Cell 98, 129-132. [DOI] [PubMed] [Google Scholar]
- 32.Ptashne, M. & Gann, A. (2002) Genes and Signals (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 33.Ptashne, M. & Gann, A. (2003) Science 299, 1025-1027. [DOI] [PubMed] [Google Scholar]