Abstract
The suppressors of cytokine signaling (SOCS) proteins are a family of SH2 domain-containing intracellular inhibitors of cytokine signal transduction that act by several different mechanisms. Recent evidence suggests that the action of the SOCS proteins may extend beyond the cytokine receptors to signaling initiated by members of the tyrosine kinase receptor family. In this study, the ability of SOCS-5 to negatively regulate signaling cascades downstream of the epidermal growth factor receptor (EGF-R) has been examined by using an EGF-responsive cell line engineered to constitutively express the EGF-R and SOCS-5 or SOCS-5 mutants. SOCS-5 associated with the EGF-R complex in an EGF-independent manner, and the mitogenic response to EGF of all SOCS-5-expressing cell lines was dramatically inhibited when compared with control cell lines. Furthermore, this effect was abrogated after deletion of the SOCS-5 SOCS box. This result suggests that the inhibition of signaling occurs through enhanced proteasomal degradation of the EGF-R through SOCS box recruitment of E3 ubiquitin ligase activity.
Cytokines control a wide variety of cellular responses by binding to cell surface receptor proteins and inducing activation of an intracellular signaling cascade. Cytokines initiate signaling through activation of a receptor-associated family of cytoplasmic protein tyrosine kinases, the Janus kinases (JAKs). Once activated, JAKs then phosphorylate both receptor subunits and cytoplasmic proteins recruited to the receptor complex (1, 2). In contrast, receptor tyrosine kinases contain an intrinsic tyrosine kinase domain but again initiate signaling by crossphosphorylation and recruitment of second messengers (3). Several mechanisms for negative regulation of signaling have been described, including dephosphorylation of critical residues by protein phosphatases, ubiquitination, and targeting of signaling proteins for degradation through the proteasome, receptor internalization, and interaction of signaling molecules with specific regulatory molecules.
The SOCS proteins are a family of negative regulators. The first member of this family, cytokine-inducible SH2-containing protein, was discovered in a screen for cytokine-inducible genes (4). Three groups discovered SOCS-1 independently as a suppressor of cytokine signaling (SOCS) (5), a JAK-binding protein (6), or based on antigenic similarity with the signal transducers and activators of transcription (STAT) SH2 domain (STAT-induced STAT inhibitor; SSI) (7). There are now eight SOCS family members (cytokine-inducible SH2-containing protein and SOCS-1–SOCS-7) characterized by a central SH2 domain, an N-terminal region of variable length and composition and a C-terminal 40-aa motif, termed the “SOCS box” (4, 8), which has been shown to couple the SOCS proteins to an E3 ubiquitin ligase complex (9, 10). Based on amino acid identity, the SOCS proteins can be grouped into pairs; SOCS-1 and SOCS-3, cytokine-inducible SH2-containing protein and SOCS-2, SOCS-4 and SOCS-5 (sharing 49% amino acid identity), and SOCS-6 and SOCS-7 (8).
Through studies of mice engineered to lack particular SOCS genes, SOCS-1 has been shown to be a critical regulator of IFN-γ signaling during postnatal development (11, 12), and SOCS-2 appears to regulate the growth hormone and insulin-like growth factor I pathways (13). SOCS-3 has a critical role in placental development (14) and more recently has been shown to negatively regulate IL-6 signaling (15, 16). When overexpressed, SOCS-5 has been shown to partially inhibit IL-6 and leukemia inhibitory factor signaling in vitro but less potently than SOCS-1 or SOCS-3 (17). More recently, Seki and colleagues (18) have proposed a role for SOCS-5 in regulation of the Th1/Th2 T cell balance through interaction with the IL-4 receptor.
Interestingly, SOCS-1 has also been shown to inhibit the mitogenic response to stem cell factor, which utilizes the tyrosine kinase receptor, c-kit (19). In addition, SOCS-3 has been shown to inhibit platelet-derived growth factor signaling (20), and several SOCS proteins have been shown to interact with the insulin-like growth factor I receptor (21, 22). SOCS family proteins have also been implicated in signaling from the epidermal growth factor receptor (EGF-R) with SOCS-1, SOCS-3 (23), and SOCS-7 (24) shown to be capable of interacting with the EGF-R cytoplasmic domain. In Drosophila, the homologue of the mammalian SOCS-5 gene, SOCS36E has been implicated in both the JAK/STAT pathway and EGF-R signaling (25). This study has investigated the potential of mammalian SOCS-5 to negatively regulate EGF-R signaling.
Methods
SOCS Expression Vectors. The cDNA-encoding SOCS-5 was described in ref. 8. Constructs encoding SOCS proteins with an N-terminal Flag epitope tag (DYKDDDDK) were generated by PCR to give fragments with in-frame MluI restriction enzyme sites at both N and C termini and subcloned into the mammalian expression vector pEF-FLAG-I, a derivative of the mammalian expression vector pEF-BOS (26). SOCS-5 deletion mutants lacking either the N terminus (amino acids 370–536; ΔNT) or the SOCS box (amino acids 2–480; ΔSB) were generated by PCR to give fragments with in-frame AscI and MluI restriction enzyme sites at the N and C termini, respectively. The SOCS-5 SH2 mutant in which the invariant arginine was replaced by lysine (R406K; mSH2) and mutations in the SOCS-5 SOCS box to eliminate elongin C binding (L484P, C488F; mSB) were generated by using the PCR-based technique, splicing by overlap extension (27). PCR fragments with in-frame AscI and MluI restriction sites at the N and C termini, respectively, were then subcloned into pEF-FLAG-I to encode proteins with an N-terminal Flag epitope tag. PCR fragments encoding SOCS-5 with either a deleted or mutated SOCS box were subcloned into a bicistronic pEFBOS vector containing an internal ribosome entry site, followed by sequence-encoding GFP. All constructs were sequenced in their entirety before use.
Generation of Cell Lines. All Ba/F3 cell lines were maintained in DMEM containing 10% (vol/vol) bovine calf serum and 10% (vol/vol) conditioned medium from WEHI-3B D- cells as a source of murine IL-3. The construction of the Ba/F3 cells expressing the wild-type EGF-R was described in ref. 28. The BaF/ERX line was derived by continuous growth in EGF (F.W., unpublished data). BaF/ERX cell derivatives were generated by electroporation with the pEF-FLAG-I expression vector carrying cDNAs encoding either Flag-tagged SOCS-5 or SOCS-5 mutant protein and the pPGKPuropA expression vector. Cells were selected in 20 μg/ml puromycin (Sigma). SOCS expression was determined by immunoprecipitation with anti-Flag antibody (M2; Eastman Kodak) and Western blot with rat anti-Flag antibody. Maintenance of EGF-R expression was confirmed by flow cytometric analysis by using monoclonal antibody 528, and independently derived clonal cell lines were generated from individual colonies grown in soft agar. SOCS box deleted or mutated lines were screened by FACS analysis for GFP expression and cloned by sorting to give one cell per well. Protein expression was then confirmed by immunoprecipitation and Western blot with anti-Flag antibodies. Control cell lines were generated by electroporation with the pPGKPuropA vector alone. A number of independently derived control cell lines were selected and maintained in 20 μg/ml puromycin.
Antibodies and Growth Factors. EGF was prepared from mouse submaxillary glands according to a published procedure (mouse EGF) (29). Murine IL-3 was purchased from PeproTech (Rocky Hill, NJ). Monoclonal anti-EGF-R antibody 528 (30) and monoclonal anti-EGF-R antibody 806 were produced and purified at the Ludwig Institute from hybridoma culture medium, and these antibodies recognize different extracellular epitopes of the EGF-R. Polyclonal rat anti-Flag antibody was a gift of D. Huang & L. O'Reilly (The Walter and Eliza Hall Institute of Medical Research).
Flow Cytometric Analysis. To examine EGF-R expression, BaF/ERX cell lines were incubated with 25 μg/ml monoclonal antibody 528 for 30 min at 4°C. Antibody binding was detected by incubation with fluorescein-conjugated rabbit anti-mouse Ig (Silenus, Melbourne) for 30 min at 4°C and analyzed by using a FACScan flow cytometer (Becton Dickinson). Parental Ba/F3 cells lacking the EGF-R were used as a negative control. To determine changes in EGF-R surface expression, cells were washed twice and incubated at 37°C in the presence of 50 ng/ml mouse EGF. Cell aliquots (1 × 106) were removed at 0 and 4 h incubation for determination of available EGF receptors on the cell surface by using anti-EGF-R antibody 528.
Survival and Proliferation Assays. Microwell assays were performed in 60-well plates as described in ref. 31. Briefly, cells were washed and dispensed into plates at 200 cells per well in the presence of increasing concentrations of either EGF or IL-3. After 3 days of incubation, the number of viable cells were counted by using an inverted microscope. Alternatively, 1 × 105 cells were washed and dispensed into 24-well plates (Nunc) in the presence of 100 ng/ml mouse EGF. After 94 h, viable cells were counted by using an inverted microscope. After 48 h, cells were subcultured to prevent overgrowth.
Immunoprecipitation and Analysis of Western Blots. BaF/ERX cell lines were washed and incubated in serum-free media for 1–2 h before incubation with 50 ng/ml EGF at 37°C unless otherwise indicated. Cells were then centrifuged at 4°C and lysed in KALB lysis buffer (32), containing protease inhibitors (Complete Mixture tablets, Roche Applied Science, Indianapolis), 1 mM Na3VO4, and 1 mM NaF. Proteins were immunoprecipitated with anti-EGF-R monoclonal antibody 528 and protein G Sepharose (Amersham Pharmacia). Alternatively, proteins were immunoprecipitated by using anti-Flag antibody conjugated to Sepharose (M2; Eastman Kodak). Proteins were separated by SDS/PAGE under reducing conditions and electrophoretically transferred to BioTrace poly(vinylidene difluoride) membranes (Pall). Membranes were blocked overnight in 10% wt/vol skim milk and incubated with primary antibody for 2 h. Antibody binding was visualized with either peroxidase-conjugated goat anti-rat Ig (Southern Biotechnology Associates) or peroxidase conjugated anti-mouse IgFc, which specifically recognizes the Ig heavy chain (Jackson ImmunoResearch), and the enhanced chemiluminescence system (Amersham Pharmacia). To reblot, the membrane was first stripped of antibodies in either 0.1 M glycine (pH 2.9) or 62.5 mM Tris·HCl (pH 6.7), containing 2% (wt/vol) SDS and 100 mM 2-mercaptoethanol.
EGF-R Internalization and Degradation. Binding, internalization, and degradation of 125I-labeled EGF was performed essentially as described in refs. 33 and 34. Briefly, 125I-EGF (with or without 100-fold excess unlabeled EGF) was added to prewarmed BaF/ERX- and SOCS-5-expressing lines (2.5 × 105 cells per point). At the indicated times, aliquots were removed and centrifuged through chilled FCS to remove free 125I-EGF, the cells were resuspended in 3% acetic acid in PBS for 5 min, and the cell surface 125I-EGF (acid-released) or internalized 125I-EGF (acid-resistant) was determined. Cell-associated radioactivity was counted on a Packard Cobra γ counter. Each cell line was tested in duplicate. 125I-labeled EGF was iodinated by the iodogen method (Pierce).
Results
Most Ba/F3 cell lines expressing the EGF-R show an increased survival response to EGF but do not proliferate in the absence of IL-3 (35); however, a mitogenically responsive line was derived by selection in EGF. The resulting cell line (BaF/ERX) can be grown continuously in EGF and depends on EGF for transit through the cell cycle (F.W., unpublished data). One of the advantages of the BaF/ERX line is that the EGF-R (ErbB-1) is the only family member present, allowing analysis of this receptor subunit in the absence of heterodimeric binding partners. To examine a possible role for SOCS-5 in EGF-R signaling, we established BaF/ERX lines that stably overexpressed either SOCS-5 or SOCS-5 mutants.
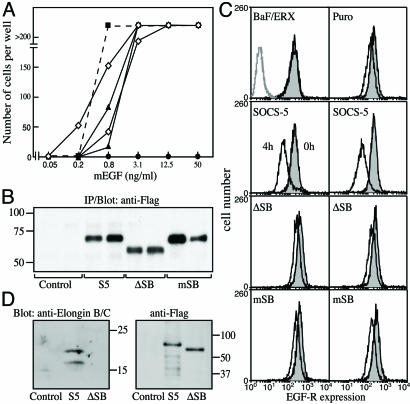
Generation of SOCS-5-Expressing Cell Lines. BaF/ERX cells were electroporated with expression plasmids for Flag-tagged SOCS-5 and SOCS-5 mutants lacking the N terminus (ΔNT) or the SOCS box (ΔSB) or containing a mutated SH2 domain (R406K; mSH2) or a mutated SOCS box (L484P, C488F; mSB). Multiple independent clones expressing each protein were obtained. As a control for variability in the parental cell line, independent lines were also derived after transfection of BaF/ERX cells with the plasmid conferring puromycin resistance alone. All lines were derived and maintained by culturing in the presence of IL-3 and puromycin. Expression of the SOCS-5 protein was confirmed in each cell line by immunoprecipitation and Western blot analysis with anti-Flag antibodies (Fig. 1B). Endogenous SOCS-5 is expressed in Ba/F3 cells at relatively low levels that are comparable with those observed in thymus or spleen and are in contrast to the high level expression in the brain, as determined by quantitative PCR (data not shown). Maintenance of EGF-R expression was confirmed by FACS analysis, although it was interesting to observe that all SOCS-5-expressing clones appeared to have slightly reduced EGF-R expression (Fig. 1 A).
Fig. 1.
Generation of SOCS-5-expressing lines. (A) FACS analysis of BaF/ERX lines showing EGF-R expression levels. Parental Ba/F3 cells, which had not been transfected with EGF-R cDNA, are shown to the left in histogram A. EGF-R levels are shown to the right of histogram A and in subsequent panels. (A Upper) Puromycin-resistant control lines. (A Lower) SOCS-5-expressing lines. (B) SOCS-5 expression levels. Equal cell numbers of BaF/ERX puromycin-resistant lines and BaF/ERX cells expressing SOCS-5 were lysed and analyzed by immunoprecipitation and Western blot by using anti-Flag antibody. Lanes: A–E, independent puromycin-resistant lines; F–J, independent SOCS-5 clones.
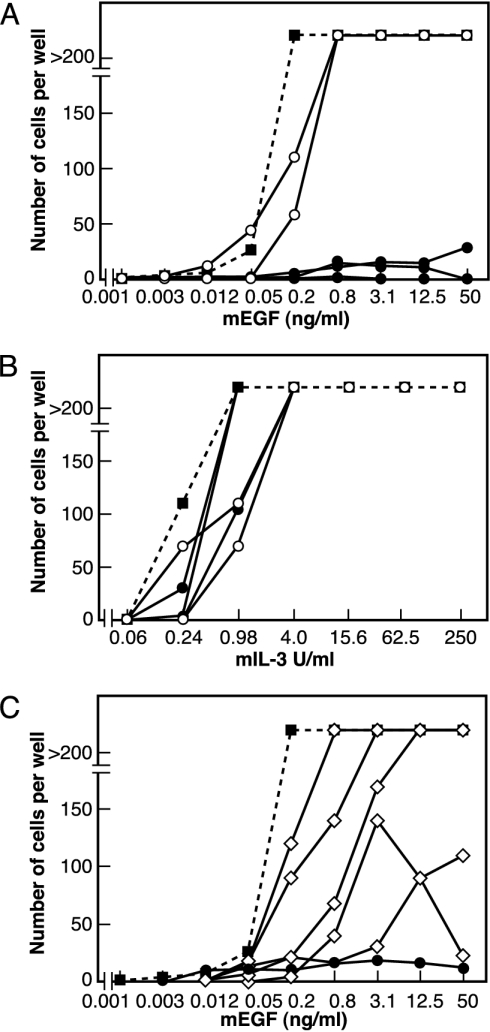
SOCS-5 Inhibits EGF Survival and Mitogenic Responses. Five independent SOCS-5-expressing clones were examined. All had a dramatically inhibited proliferative response to EGF when compared with puromycin-resistant control lines and the parental BaF/ERX line. Three representative lines are shown in Fig. 2A. To confirm that this effect was specific to EGF signaling, the same cell lines were analyzed for their proliferative response to IL-3. All five SOCS-5 clones and the puromycin-resistant lines demonstrated similar responsiveness to IL-3 (Fig. 2B). To examine the EGF response in more detail, total cell numbers were determined after incubation with 100 ng/ml EGF for 94 h. The SOCS-5-expressing lines were again unable to respond, indicating that EGF-stimulated proliferation was severely impaired (puromycin-resistant BaF/ERX clones, n = 4, 13.78 ± 6.99 × 105 cells; SOCS-5-expressing clones, n = 3, 0.61 ± 0.38 × 105 cells). To determine whether the N-terminal region was required for the proliferative response, we examined five independent clones expressing SOCS-5 lacking the N terminus. Three clones showed a clear response to EGF, whereas two clones gave a partial response (Fig. 2C). Protein levels of the N-terminally truncated mutant were comparable with levels of full-length SOCS-5 protein as determined by immunoprecipitation and Western blot. EGF-R levels were also comparable with BaF/ERX cells (data not shown).
Fig. 2.
SOCS-5 inhibits EGF proliferative responses. (A) A representative experiment showing the number of cells obtained per well of the parental BaF/ERX line (▪), two puromycin-resistant lines (○), and three SOCS-5 clones (•) in response to a 3-day incubation with increasing concentrations of EGF. (B) A representative experiment showing the number of cells obtained per well in response to a 3-day incubation with increasing concentrations of IL-3. (C) A representative experiment showing the number of cells obtained per well of the parental BaF/ERX line (▪), a SOCS-5 clone (•), and five clones expressing SOCS-5 lacking the N terminus (⋄) in response to a 3-day incubation with increasing concentrations of EGF.
SOCS-5 Associates with the EGF-R Complex. The EGF-R is highly tyrosine phosphorylated after EGF stimulation, recruiting a wave of secondary signaling molecules through their SH2 domains. We therefore examined the ability of SOCS-5 to associate with the EGF-R complex by immunoprecipitation of EGF-R proteins and Western blot analysis for the presence of Flag-tagged SOCS-5 or SOCS-5 mutants. SOCS-5 and SOCS-5 lacking the SOCS box (S5ΔSB) were observed to associate with the EGF-R complex. Both the N-terminal region and the SH2 domain appear to be involved in recruitment to the receptor complex as mutation of the SOCS-5-SH2 domain (R406K; S5mSH2) decreased the association, whereas SOCS-5 lacking the N-terminal region (S5ΔNT) was unable to associate with the complex. Interestingly, SOCS-5 was present in the receptor complex before stimulation with EGF, suggesting that this interaction occurs in an EGF-independent manner (Fig. 3A). EGF-R levels were apparently decreased after EGF stimulation (Fig. 3C). This finding is unlikely to result from receptor internalization because cells were stimulated at 4°C and, most probably, results from a reduction in the ability of the antibody to recognize phosphorylated receptor. Overexpression of SOCS-5 did not inhibit either EGF-R tyrosine phosphorylation (Fig. 3B) or kinase activity as measured by phosphorylation of a substrate peptide (data not shown). Expression levels of SOCS-5 and mutated SOCS-5 proteins are shown in Fig. 3D.
Fig. 3.
Association of SOCS-5 with the EGF-R complex. BaF/ERX cells stably expressing Flag-tagged SOCS-5 (S5), SOCS-5 lacking the N terminus (ΔNT), and SOCS-5 with a mutated SH2 domain (mSH2) or with a deleted SOCS box (ΔSB) were incubated with (+) or without (-) 50 ng/ml EGF for 15 min on ice and lysed. EGF-R proteins were immunoprecipitated by using monoclonal antibody 528, and associated Flag-tagged SOCS proteins were detected by Western analysis with rat anti-Flag antibody (A). Total cell lysates were analyzed by Western blot with anti-phosphotyrosine antibodies (B). Western blots were reprobed by using anti-EGF-R monoclonal antibody 806 (C). Levels of SOCS-5 protein were determined by immunoprecipitation with anti-Flag antibodies conjugated to agarose (M2-beads) and Western blot with rat anti-Flag antibodies (D).
SOCS-5 Down-Regulates EGF-R Surface Expression in a SOCS Box-Dependent Manner. Because the SOCS proteins are coupled to an E3 ubiquitin ligase complex through the SOCS box, SOCS-5 may inhibit EGF-R signaling by regulating EGF-R levels. To determine whether the inhibition of EGF-R signaling was SOCS box-dependent, BaF/ERX lines were generated that expressed SOCS-5 with either a deleted SOCS box (ΔSB) or a SOCS box with a mutated elongin C binding site (mSB). In contrast to lines expressing wild-type SOCS-5, cell lines expressing SOCS-5 protein unable to bind elongin C (ΔSB and mSB) responded to EGF in a manner comparable with the control lines (Fig. 4A). Protein expression levels are shown in Fig. 4B. EGF-R expression levels were confirmed by FACS analysis (data not shown). To determine whether the loss of inhibition correlated with changes in EGF-R levels, FACS analysis was performed to determine EGF-R surface expression after EGF stimulation. BaF/ERX lines expressing SOCS-5 displayed enhanced down-regulation of surface EGF-R after incubation with EGF for 4 h. In contrast, lines expressing SOCS-5 with either a deleted or mutated SOCS box (ΔSB and mSB) did not display enhanced down-regulation of the receptor (Fig. 4C). This finding indicates that the SOCS-5 inhibition of the EGF-R signaling is mediated by the SOCS-5 SOCS box and correlates with enhanced receptor loss from the cell surface. Fig. 4D confirms that, as with other SOCS box proteins, the SOCS-5 SOCS box binds to elongins B and C.
Fig. 4.
The SOCS-5 SOCS box is required for inhibition of EGF-R signaling. (A) Proliferation of BaF/ERX lines expressing SOCS-5 with a mutated (mSB) or deleted (ΔSB) SOCS box. The number of cells obtained per well is shown for the parental BaF/ERX line (▪), two SOCS-5 clones (•), two SOCS-5mSB clones (▴), and two SOCS-5ΔSB clones (⋄) in response to a 3-day incubation with increasing concentrations of EGF. (B) SOCS-5 expression levels. Equal cell numbers of two independent BaF/ERX lines expressing SOCS-5 or SOCS-5 with a mutated (mSB) or deleted SOCS box (ΔSB) were lysed and analyzed by immunoprecipitation and Western blot by using anti-Flag antibody. (C) SOCS-5-dependent down-regulation of EGF-R surface levels. BaF/ERX, puromycin-resistant BaF/ERX cells (Puro) and BaF/ERX cells expressing SOCS-5 or SOCS-5 with a mutated (mSB) or deleted SOCS box (ΔSB) were incubated with 50 ng/ml EGF for 4 h at 37°C and analyzed for EGF-R surface expression at 0 and 4 h (filled and unfilled histograms, respectively) by FACS with anti-EGF-R antibody 528. Data are shown for two independently derived clonal lines expressing SOCS-5, SOCS-5ΔSB, or SOCS-5mSB. Untransfected parental Ba/F3 cells are shown in gray in Upper Left. (D) Association of the SOCS-5 SOCS box with elongins B and C. 293T cells were transiently transfected with either vector alone (control) or plasmids expressing either Flag-tagged SOCS-5 or SOCS-5 with a deleted SOCS box (S5ΔSB). Flag-tagged proteins were immunoprecipitated and analyzed by SDS/PAGE and Western blot for the presence of elongins B and C (Left). Membranes were stripped and reprobed with anti-Flag antibodies (Right).
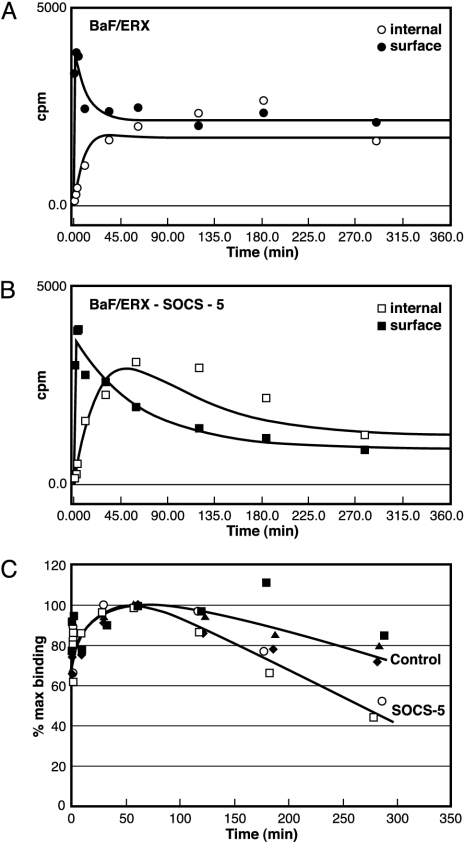
EGF-R Degradation Is Enhanced in the Presence of SOCS-5. Down-regulation of surface receptor levels can result from increased receptor endocytosis, receptor degradation, or decreases in the rate of protein synthesis. We therefore performed 125I-EGF binding assays to investigate internalization and degradation of the EGF-R in the SOCS-5-expressing lines. Cell lines were incubated with 125I-EGF at 37°C with and without unlabeled EGF, and samples were removed at intervals. Surface-bound 125I-EGF and internalized 125I-EGF were determined, and cpm were plotted versus time (Fig. 5 A and B). In contrast to the BaF/ERX line, the SOCS-5-expressing lines demonstrated an increased loss of total 125I-EGF that was apparent after 100 min of incubation with EGF (Fig. 5C). Initial rates of 125I-EGF internalization were indistinguishable between control and SOCS-5-expressing lines (Fig. 5), suggesting that the enhanced EGF-R down-regulation results from an increased rate of receptor degradation.
Fig. 5.
EGF-R degradation is enhanced in the presence of SOCS-5. Approach to steady state at 37°C for 125I-EGF interacting with BaF/ERX control cells (A) and BaF/ERX cells stably expressing SOCS-5 (B) is shown. 125I-EGF was added to prewarmed cells, and, at the indicated times, aliquots of cells were removed, centrifuged through chilled FCS to remove free 125I-EGF, and cell surface (• and ▪) or internalized (○ and □) 125I-EGF was determined. The lines drawn through the experimental points are the lines of best fit according to the computer program of Myers et al. (40). (C) Total cell-associated 125I-EGF (cell surface plus internalized cpm) is expressed as a percentage of maximal binding for control lines (▪, ▴, and ♦) and two independent SOCS-5-expressing lines (○ and □). Cell lines were assayed in duplicate.
Discussion
This study has demonstrated that overexpression of SOCS-5 can dramatically inhibit EGF mitogenic signaling in BaF/ERX cells. Because BaF/ERX cells only express the Erb-B1 protein, this system has allowed us to exclusively examine the effects of SOCS-5 on EGF-activated Erb-B1 homodimeric signaling. Inhibition of EGF signaling was shown to depend on the SOCS-5 SOCS box and most likely results from enhanced receptor degradation over time. There is now compelling evidence that the SOCS box recruits E3 ubiquitin ligase activity and, in the case of SOCS-1, results in polyubiquitination of VAV, JAK2, and FAK and a decrease in the half-life of those proteins (36–39). Our data provide evidence of SOCS box-dependent degradation of a receptor protein, the EGF-R.
SOCS-5 association with the EGF-R complex required the SOCS-5 N-terminal region and occurred in the absence of a ligand, suggesting that it was independent of EGF-R phosphorylation. SOCS-5 has also been shown to interact with the IL-4 receptor in a phosphorylation-independent manner through the N-terminal region (18). It is therefore likely that SOCS-5 may have a unique mechanism of association with target proteins that occurs through the N terminus. Paradoxically, the SOCS-5 SH2 domain is also required for interaction with the EGF-R in the absence of EGF stimulation. It is not clear whether this interaction is tyrosine-dependent, but low-level tyrosine phosphorylation of the receptor in the BaF/ERX cells is observed before the addition of EGF. This interaction may be auto-phosphorylation or trans-phosphorylation by another kinase such as c-src. Potentially, SOCS-5 could also be binding through an EGF-R-interacting protein that is tyrosine-phosphorylated. Taken together, our data suggest that both the SOCS-5 N-terminal region and SH2 domain mediate the EGF-R interaction.
It is likely that a threshold level of receptors is required to maintain a biological response and, that in the presence of SOCS-5, this standard is reduced to a level that is insufficient to maintain proliferation. Deletion of the SOCS-5 N-terminal region did not fully restore EGF-induced proliferation in all clones tested. This result suggests that although SOCS-5 association with the EGF-R was apparently abrogated, some inhibition of signaling, through the SH2 domain or SOCS box, was still in effect, thus providing a further explanation for the dramatic inhibition of signaling observed with the full-length protein.
The enhanced EGF-R degradation in the presence of SOCS-5 is ligand-dependent. However, SOCS-5 is constitutively associated with the EGF-R complex in the BaF/ERX system before ligand stimulation. This result suggests that an activation step other than association with the receptor complex may be required for SOCS-5-mediated ubiquitin ligase activity. It is unclear at this stage what this might be, but ligand binding to and activation of the EGF-R complex may result in changes to the EGF-R cytoplasmic domain or SOCS-5, which allow the associated E3 ubiquitin ligase complex access to lysine residues within the receptor.
A recent publication by Kario and colleagues (40) has also described down-regulation of EGF-R in the presence of SOCS-5 and supports the findings described here. In addition, Kario et al. have demonstrated up-regulation of SOCS-4 and SOCS-5 mRNA in response to EGF stimulation (40). The EGF-R and SOCS-5 have overlapping expression patterns and are ubiquitously expressed in most tissues in the mouse (8, 41) (data not shown). This finding is consistent with endogenous SOCS-5 being involved in the regulation of EGF signaling and acting as part of a negative feedback loop in a similar manner to other SOCS proteins.
Although expression of SOCS-5 has a clear effect on EGF-induced proliferation in the BaF/ERX cells, its physiological role remains to be determined. A study in Drosophila by Callus and colleagues (25) suggests that the Drosophila homologue of SOCS-5, SOCS36E, can negatively regulate both JAK/STAT and EGF-R signaling pathways (25). Our data suggest that the interaction between SOCS-5 and the EGF pathway may be functionally conserved and may result from a direct interaction with the EGF-R complex. We have recently generated SOCS-5-null mice (41), and our initial studies indicate no obvious EGF-related phenotype in the knockout animals. SOCS-5 shares a great deal of similarity with SOCS-4 (92% amino acid identity within the SH2 domain), and both SOCS-4 and SOCS-5 can associate with the EGF receptor when overexpressed (data not shown). Similar data using overexpression systems also indicates that SOCS-1, SOCS-3, and SOCS-7 (23, 24) have the capacity to interact with the EGF-R. It is therefore possible that other SOCS proteins may compensate for the loss of SOCS-5. The question of whether EGF signaling is a physiological site of action for SOCS-5 will need to be addressed by analysis of compound SOCS knockout mice. Notwithstanding that result, the present SOCS-5-EGF-R system may prove of value in helping to establish the mechanism of SOCS-5 action.
Acknowledgments
We thank S. Mifsud and L. Di Rago for excellent technical assistance. This work was supported by the Anti-Cancer Council of Victoria, AMRAD Corp., National Health and Medical Research Council, Australia (Program Grant 257500), and the Australian Federal Government Cooperative Research Centre Program. S.E.N. was supported by an Australian Postdoctoral Fellowship (Australian Research Council).
Author contributions: S.E.N., D.M., J.-G.Z., and N.A.N. designed research; S.E.N., D.M., N.S.S., R.C., F.W., A.S., D.C., and T.A.W. performed research; F.W., D.J.H., and W.S.A. contributed new reagents/analytic tools; S.E.N., D.M., N.S.S., R.C., A.S., D.C., and N.A.N. analyzed data; and S.E.N. wrote the paper.
Abbreviations: EGF-R, EGF receptor; JAK, Janus kinase; SOCS, suppressors of cytokine signaling; STAT, signal transducers and activators of transcription.
References
- 1.Ihle, J. N., Thierfelder, W., Teglund, S., Stravapodis, D., Wang, D., Feng, J. & Parganas, E. (1998) Ann. N.Y. Acad. Sci. 865, 1-9. [DOI] [PubMed] [Google Scholar]
- 2.Leonard, W. J. & O'Shea, J. J. (1998) Annu. Rev. Immunol. 16, 293-322. [DOI] [PubMed] [Google Scholar]
- 3.Schlessinger, J. (2000) Cell 103, 211-225. [DOI] [PubMed] [Google Scholar]
- 4.Yoshimura, A., Ohkubo, T., Kiguchi, T., Jenkins, N. A., Gilbert, D. J., Copeland, N. G., Hara, T. & Miyajima, A. (1995) EMBO J. 14, 2816-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Starr, R., Willson, T. A., Viney, E. M., Murray, L. J., Rayner, J. R., Jenkins, B. J., Gonda, T. J., Alexander, W. S., Metcalf, D., Nicola, N. A. & Hilton, D. J. (1997) Nature 387, 917-921. [DOI] [PubMed] [Google Scholar]
- 6.Endo, T. A., Masuhara, M., Yokouchi, M., Suzuki, R., Sakamoto, H., Mitsui, K., Matsumoto, A., Tanimura, S., Ohtsubo, M., Misawa, H., et al. (1997) Nature 387, 921-924. [DOI] [PubMed] [Google Scholar]
- 7.Naka, T., Narazaki, M., Hirata, M., Matsumoto, T., Minamoto, S., Aono, A., Nishimoto, N., Kajita, T., Taga, T., Yoshizaki, K., et al. (1997) Nature 387, 924-929. [DOI] [PubMed] [Google Scholar]
- 8.Hilton, D. J., Richardson, R. T., Alexander, W. S., Viney, E. M., Willson, T. A., Sprigg, N. S., Starr, R., Nicholson, S. E., Metcalf, D. & Nicola, N. A. (1998) Proc. Natl. Acad. Sci. USA 95, 114-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamura, T., Sato, S., Haque, D., Liu, L., Kaelin, W. G., Jr., Conaway, R. C. & Conaway, J. W. (1998) Genes Dev. 12, 3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang, J. G., Farley, A., Nicholson, S. E., Willson, T. A., Zugaro, L. M., Simpson, R. J., Moritz, R. L., Cary, D., Richardson, R., Hausmann, G., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 2071-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexander, W. S., Starr, R., Fenner, J. E., Scott, C. L., Handman, E., Sprigg, N. S., Corbin, J. E., Cornish, A. L., Darwiche, R., Owczarek, C. M., et al. (1999) Cell 98, 597-608. [DOI] [PubMed] [Google Scholar]
- 12.Marine, J. C., Topham, D. J., McKay, C., Wang, D., Parganas, E., Stravopodis, D., Yoshimura, A. & Ihle, J. N. (1999) Cell 98, 609-616. [DOI] [PubMed] [Google Scholar]
- 13.Metcalf, D., Greenhalgh, C. J., Viney, E., Willson, T. A., Starr, R., Nicola, N. A., Hilton, D. J. & Alexander, W. S. (2000) Nature 405, 1069-1073. [DOI] [PubMed] [Google Scholar]
- 14.Roberts, A. W., Robb, L., Rakar, S., Hartley, L., Cluse, L., Nicola, N. A., Metcalf, D., Hilton, D. J. & Alexander, W. A. (2001) Proc. Natl. Acad. Sci. USA 98, 9324-9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yasukawa, H., Ohishi, M., Mori, H., Murakami, M., Chinen, T., Aki, D., Hanada, T., Takeda, K., Akira, S., Hoshijima, M., et al. (2003) Nat. Immunol. 4, 551-556. [DOI] [PubMed] [Google Scholar]
- 16.Croker, B. A., Krebs, D. L., Zhang, J. G., Wormald, S., Willson, T. A., Stanley, E. G., Robb, L., Greenhalgh, C. J., Forster, I., Clausen, B. E., et al. (2003) Nat. Immunol. 4, 540-545. [DOI] [PubMed] [Google Scholar]
- 17.Nicholson, S. E., Willson, T. A., Farley, A., Starr, R., Zhang, J. G., Baca, M., Alexander, W. S., Metcalf, D., Hilton, D. J. & Nicola, N. A. (1999) EMBO J. 18, 375-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seki, Y., Hayashi, K., Matsumoto, A., Seki, N., Tsukada, J., Ransom, J., Naka, T., Kishimoto, T., Yoshimura, A. & Kubo, M. (2002) Proc. Natl. Acad. Sci. USA 99, 13003-13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Sepulveda, P., Okkenhaug, K., Rose, J. L., Hawley, R. G., Dubreuil, P. & Rottapel, R. (1999) EMBO J. 18, 904-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cacalano, N. A., Sanden, D. & Johnston, J. A. (2001) Nat. Cell Biol. 3, 460-465. [DOI] [PubMed] [Google Scholar]
- 21.Dey, B. R., Spence, S. L., Nissley, P. & Furlanetto, R. W. (1998) J. Biol. Chem. 273, 24095-24101. [DOI] [PubMed] [Google Scholar]
- 22.Dey, B. R., Furlanetto, R. W. & Nissley, P. (2000) Biochem. Biophys. Res. Commun. 278, 38-43. [DOI] [PubMed] [Google Scholar]
- 23.Xia, L., Wang, L., Chung, A. S., Ivanov, S. S., Ling, M. Y., Dragoi, A. M., Platt, A., Gilmer, T. M., Fu, X. Y. & Chin, Y. E. (2002) J. Biol. Chem. 277, 30716-30723. [DOI] [PubMed] [Google Scholar]
- 24.Matuoka, K., Miki, H., Takahashi, K. & Takenawa, T. (1997) Biochem. Biophys. Res. Commun. 239, 488-492. [DOI] [PubMed] [Google Scholar]
- 25.Callus, B. A. & Mathey-Prevot, B. (2002) Oncogene 21, 4812-4821. [DOI] [PubMed] [Google Scholar]
- 26.Mizushima, S. & Nagata, S. (1990) Nucleic Acids Res. 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horton, R. M., Hunt, H. D., Ho, S. N., Pullen, J. K. & Pease, L. R. (1989) Gene 77, 61-68. [DOI] [PubMed] [Google Scholar]
- 28.Walker, F., Hibbs, M. L., Zhang, H. H., Gonez, L. J. & Burgess, A. W. (1998) Growth Factors 16, 53-67. [DOI] [PubMed] [Google Scholar]
- 29.Burgess, A. W., Lloyd, C. J. & Nice, E. C. (1983) EMBO J. 2, 2065-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato, J. D., Kawamoto, T., Le, A. D., Mendelsohn, J., Polikoff, J. & Sato, G. H. (1983) Mol. Biol. Med. 1, 511-529. [PubMed] [Google Scholar]
- 31.Metcalf, D., Willson, T. A., Hilton, D. J., Di Rago, L. & Mifsud, S. (1995) Leukemia 9, 1556-1564. [PubMed] [Google Scholar]
- 32.Nicholson, S. E., Novak, U., Ziegler, S. F. & Layton, J. E. (1995) Blood 86, 3698-3704. [PubMed] [Google Scholar]
- 33.Nicola, N. A., Peterson, L., Hilton, D. J. & Metcalf, D. (1988) Growth Factors 1, 41-49. [DOI] [PubMed] [Google Scholar]
- 34.Nicola, N. A. & Metcalf, D. (1988) Growth Factors 1, 29-39. [DOI] [PubMed] [Google Scholar]
- 35.Shibuya, H., Yoneyama, M., Ninomiya-Tsuji, J., Matsumoto, K. & Taniguchi, T. (1992) Cell 70, 57-67. [DOI] [PubMed] [Google Scholar]
- 36.De Sepulveda, P., Ilangumaran, S. & Rottapel, R. (2000) J. Biol. Chem. 275, 14005-14008. [DOI] [PubMed] [Google Scholar]
- 37.Liu, E., Cote, J. F. & Vuori, K. (2003) EMBO J. 22, 5036-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamizono, S., Hanada, T., Yasukawa, H., Minoguchi, S., Kato, R., Minoguchi, M., Hattori, K., Hatakeyama, S., Yada, M., Morita, S., et al. (2001) J. Biol. Chem. 276, 12530-12538. [DOI] [PubMed] [Google Scholar]
- 39.Ali, S., Nouhi, Z. & Chughtai, N. (2003) J. Biol. Chem. 278, 52021-52031. [DOI] [PubMed] [Google Scholar]
- 40.Kario, E., Marmor, M. D., Adamsky, K., Citri, A., Amit, I., Amariglio, N., Rechavi, G. & Yarden, Y. (December 7, 2004) J. Biol. Chem., 10.1074/jbc.M408575200. [DOI] [PubMed]
- 41.Brender, C., Columbus, R., Metcalf, D., Handman, E., Starr, R., Huntington, N., Tarlinton, D., Odum, N., Nicholson, S. E., Nicola, N. A., et al. (2004) Mol. Cell. Biol. 24, 6094-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]