Abstract
The Golgi-localized, γ-ear-containing, Arf (ADP-ribosylation factor)-binding (GGA) proteins are clathrin adaptors that mediate the sorting of transmembrane-cargo molecules at the trans-Golgi network and endosomes. Cargo proteins can be directed into the GGA pathway by at least two different types of sorting signals: acidic cluster–dileucine motifs and covalent modification by ubiquitin. The latter modification is recognized by the GGAs through binding to their GAT [GGA and TOM (target of Myb)] domain. Here we report the crystal structure of the GAT domain of human GGA3 in a 1:1 complex with ubiquitin at 2.8-Å resolution. Ubiquitin binds to a hydrophobic and acidic patch on helices α1 and α2 of the GAT three-helix bundle that includes Asn-223, Leu-227, Glu-230, Met-231, Asp-244, Glu-246, Leu-247, Glu-250, and Leu-251. The GAT-binding surface on ubiquitin is a hydrophobic patch centered on Ile-44 that is also responsible for binding most other ubiquitin effectors. The ubiquitin-binding site observed in the crystal is distinct from the Rabaptin-5-binding site on helices α2 and α3 of the GAT domain. Mutational analysis and modeling of the ubiquitin–Rabaptin-5–GAT ternary complex indicates that ubiquitin and Rabaptin-5 can bind to the GAT domain at two different sites without any steric conflict. This ability highlights the GAT domain as a hub for interactions with multiple partners in trafficking.
Keywords: protein–protein interactions, structural biology, trafficking
The Golgi-localized, γ-ear-containing, Arf (ADP-ribosylation factor)-binding (GGA) proteins are clathrin adaptors that bind to membranes in an Arf–GTP-dependent fashion and sort specific transmembrane proteins at the trans-Golgi network (TGN) (1, 2). Three GGAs (i.e., GGA1, GGA2, and GGA3) encoded by different genes have been described in humans, and one to three have been described in most other eukaryotes. All GGAs have a conserved modular organization consisting of three folded domains, VHS (Vps27, Hrs, and Stam), GAT [GGAs and target of Myb (TOM)], and GAE (γ-adaptin ear), which are connected by two largely unstructured sequences. The mammalian VHS domain binds DXXLL-type sorting signals (X is any amino acid) that are present in the cytosolic domains of various transmembrane proteins. The GAT domain is responsible for the interaction with the GTP-bound form of Arf proteins, which recruits GGAs to membranes. The hinge that connects the GAT and GAE domains harbors clathrin-box-like motifs that interact with the terminal domain of the clathrin heavy-chain and autoinhibitory DXXLL-type motifs that bind to the VHS domain. Finally, the GAE domain binds a set of so-called “accessory proteins” that share a ΨG(P/D/E)(Ψ/L/M) consensus motif (Ψ is an aromatic residue). These interactions enable the GGAs to function as sorting adaptors at the TGN.
Recent studies have added to the already large number of known interactions in which the GGAs are engaged. The GGA–GAT domain has been shown to bind Rabaptin-5 (3), ubiquitin (4–6), and tumor-susceptibility gene (TSG) 101 (4, 7). Rabaptin-5 is a divalent effector of the Rab4 and Rab5 small GTP-binding proteins and exists as part of a complex with Rabex-5, a guanine-nucleotide exchanger for Rab5. Interactions of the Rabaptin-5/Rabex-5 complex with the GGAs are thought to promote tethering or fusion of TGN-derived coated vesicles with endosomes. TSG101 itself binds ubiquitin and is a component of the machinery involved in the sorting of transmembrane proteins to the intraluminal vesicles of multivesicular bodies (MVBs). The interaction of the GGAs with TSG101 might link the recognition of ubiquitinated transmembrane proteins to their targeting to MVBs. A growing number of trafficking proteins have been found to function as adaptors for sorting ubiquitinated proteins (8–11). The GGAs are one of the latest additions to this class, and the GAT–ubiquitin interaction endows the GGAs with the ability to sort ubiquitinated transmembrane proteins at both the TGN and endosomes.
The crystal structure of the GGA1-GAT domain (12–15) has begun to explain how the domain interacts with so many binding partners. The GGA1-GAT domain consists of an N-terminal helix–loop–helix extension (N-GAT; helices α0 and α1) that binds Arf–GTP, and a conserved C-terminal three-helix-bundle domain (sometimes referred to as C-GAT and herein referred to simply as GAT; helices α1, α2, and α3) that binds Rabaptin-5, ubiquitin, and TSG101. The GAT domains of three related proteins, TOM1, TOM1-like 1 (TOM1-L1) and TOM1-like 2 lack the N-GAT extension and do not bind Arf–GTP, but they do contain the core GAT domain and they bind ubiquitin (16, 17). Crystallographic analyses have shown that a homodimeric, parallel coiled-coil segment of Rabaptin-5 binds to a single three-helix bundle of GGA1-GAT (18). The GGA1-GAT residues involved in interactions with Rabaptin-5 comprise a hydrophobic surface patch surrounding Phe-264, Leu-277, and Leu-281 on helices α2 and α3 (18). This surface patch is ≈35 Å away and on the opposite side from the Arf–GTP-binding site (7, 15, 18). Mutation of some of these residues impairs interactions not only with Rabaptin-5 but also with ubiquitin (e.g., Leu-277 and Leu-281) and TSG101 (e.g., Leu-277) (4, 6, 7). Mutation of other residues (e.g., Phe-264 and Asn-284), however, abolishes interactions of GGA1-GAT with Rabaptin-5 without affecting interactions with ubiquitin or TSG101 (7). Recent NMR chemical-shift perturbation studies and mutational analysis showed that the GAT domain of GGA3 contains two binding sites for ubiquitin (19). One of these, site 2, centers on Leu-276 and Leu-280 and is analogous to the GGA1-binding site shared with Rabaptin-5 and TSG101. The other, site 1, centers on Leu-227 (α1 helix) and appears to be exclusively for ubiquitin. Both sites contribute to the recognition of ubiquitin by GGA3, but site 1 has a higher affinity for ubiquitin than does site 2 (19). To clarify the ubiquitin-recognition mechanism of GGA3, we determined the crystal structure of the GGA3-GAT domain in complex with ubiquitin at 2.8-Å resolution.
Materials and Methods
Cloning, Protein Expression, and Purification. For crystallization, the human GGA3-GAT domain (residues 208–301) was subcloned into the His-parallel 3 vector (20). Escherichia coli BL21(DE3) Rosetta cells harboring the plasmid were induced at OD600 > 2 at 20°C with 0.2 mM isopropyl β-d-thiogalactoside for 18 h, harvested, and treated with lysozyme in buffer A [150 mM NaCl/50 mM Tris·HCl (pH 8.0)] supplemented with 4-(2-aminoethyl)benzene sulfonyl fluoride at 4°C. The lysate was sonicated and centrifuged at 14,000 × g. The protein was purified by using Ni-nitrilotriacetate (NTA) chromatography. His6-tobacco etch virus (TEV) protease was added to the eluate, and the solution was dialyzed against buffer A with 1 mM DTT and 0.5 mM EDTA at 4°C. The cleaved His tag and the TEV protease were removed by a second Ni-NTA column. Bovine ubiquitin (Sigma) was added to form a 1:1 complex and concentrated to 38 mg/ml. For isothermal titration calorimetry (ITC) studies, GST–GGA3-GAT-208–301 constructs were generated and expressed as above. The GST-fusion proteins were purified by incubation with glutathione-Sepharose beads (Pharmacia Biotech) and eluted with 20 mM glutathione in 0.1 M NaCl/0.1 M Tris·HCl (pH 8).
ITC. GST-tagged GGA3-GAT proteins were dialyzed against PBS supplemented with 1 mM DTT. The same buffer was used to dissolve bovine ubiquitin. Final concentrations of the ubiquitin (injectant; 5–8 mM range) and GST-tagged GGA3-GAT-208–301 fragments (0.23–0.32 mM range placed in the sample cell) were calculated by measuring absorbance at 280 nm and by using extinction coefficients of 1,814 and 49,304 M-1·cm-1 for ubiquitin and GST-GAT-208–301, respectively. ITC was carried out at 30°C by using a Microcal (Studio City, CA) VP ITC instrument. Data were fitted by using the origin software (OriginLab, Northampton, MA).
Site-Directed Mutagenesis. The R259E and L276A substitutions in GGA3-GAT were introduced by using the QuikChange site-directed mutagenesis kit (Stratagene) and pGST-parallel 3-human GGA3-GAT-208–301 as a template. Mutations were confirmed by DNA sequencing.
Crystallization and Data Collection. Crystals were initially obtained in the SM3 screen (Nextal Biotechnologies, Montreal), and optimal growth was achieved in 22.5% polyethylene glycol-3350/150 mM sodium acetate (pH 4.8) at 21°C for 3 months. Data collection was performed with a laboratory Cu Kα rotating anode and an R-Axis IV image plate (MSC, The Woodlands, TX). Images were collected in 1° oscillations from a crystal cryoprotected in 18% glycerol at 95 K and processed by using HKL2000 (HKL Research, Charlottesville, VA).
Structure Determination. A model of the GGA3-GAT domain was generated by using the CaspR server at http://igs-server.cnrsmrs.fr/Caspr/(21), based on structures of the GGA1 GAT domain (12–15). The GAT domains of GGA1 and GGA3 have 66% sequence identity. Mobile surface side chains were removed manually. An initial molecular-replacement solution was found in the apparent space group C2221 with the program como (22) by using bovine ubiquitin and the GGA3 GAT homology model to search. One ubiquitin and one GAT domain were located in the asymmetric unit. The R factor for the initial solution was 45.4% after rigid-body refinement. The quality of the initial electron-density map was insufficient to trace missing residues, and the difference between R and Rfree (12%) was unreasonable. These observations led us to hypothesize that a noncrystallographic twofold axis in the space group C2 might be in almost the same orientation with the twofold rotational axis in the space group C2221. The structure was placed in the C2 unit cell by using the partially refined model of 1:1 complex in C2221 to search. Two complexes in the asymmetric unit were located. The molecular-replacement solution in C2 had R and Rfree values of 34% and 36%, respectively. The C2 solution led to an electron-density map sufficient to locate the section from residues 237–246, which comprises the entire α1-α2 linker and the first turn of α2 and was absent in the search model. Pseudomerohedral twinning is a possibility in C2 when β = 90°. We checked for this possibility by using the twinning operator (-h, -k, l) and determined that the crystals were twinned with a twinning fraction of 0.46 estimated based on intensity statistics by using the parameter H (23). At this stage, a new Rfree test set was chosen such that twinning operator-related reflections were selected together, and simulated annealing was used to remove bias toward reflections in the new test set before continuing refinement. Implementing the detwinning operation resulted in an immediate drop of 2% in Rfree. Because the twinning fraction was estimated to be ≈0.5, and a reasonable structural model was available, the data set was detwinned according to the algorithm for perfect twinning in cns (24, 25). All subsequent calculations were done on the detwinned dataset. Because of the low parameter-to-reflection ratio, noncrystallographic symmetry restraints were applied throughout the refinement process. The model was refined by using cns (24) and manually improved with the program o (26). The model contains no residues in disallowed regions of the Ramachandran plot.
Results
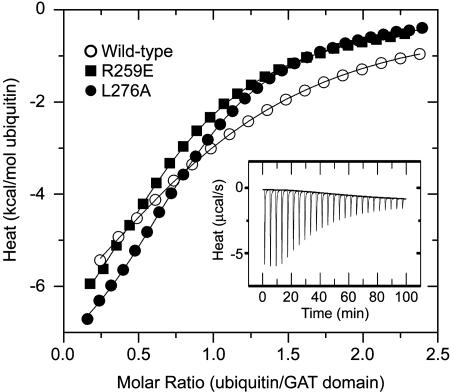
Analysis of GAT–Ubiquitin-Binding by ITC. The affinity of ubiquitin for the GAT domain of GGA3 (Kd = 181 ± 39 μM; n = 3) was measured by ITC. Mutation of Arg-259 and Leu-276 in ubiquitin-binding site 2 had no significant effect on the interaction with ubiquitin measured by ITC (Fig. 1). Similar results were obtained upon excision of the GST tag from the GST–GGA3-GAT-208–301 construct with tobacco etch virus protease (data not shown). These observations are consistent with the existence of a high-affinity ubiquitin-binding site distinct from site 2 (19).
Fig. 1.
ITC analysis of the binding of ubiquitin to the GGA3-GAT domain in vitro. The Inset shows the raw heat change elicited by successive injections of ubiquitin into a solution of wild-type GGA3-GAT, whereas the main figure depicts the normalized integration data (kcal/mol of ubiquitin as a function of the molar ratio of ubiquitin to the GST–GGA3-GAT-208–301 fragment), as well as the fitting to a one-site model.
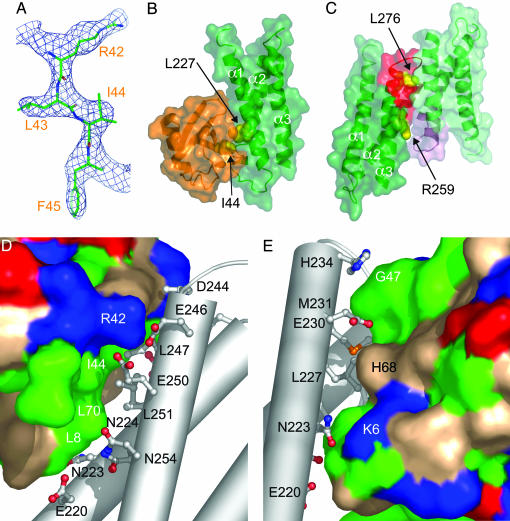
Structure of the GAT Domain of GGA3. To determine the mode of ubiquitin binding to the GGA3-GAT domain, we solved the crystal structure of the GGA3-GAT domain in complex with bovine ubiquitin at 2.8 Å (Fig. 2 A and B; Table 1). GGA3-GAT consists of a three-helix bundle (α1, α2, and α3) very similar to that of GGA1-GAT. Seventy-three Cα positions superimpose with an rms deviation of 1.1 Å. The N terminus of α1 in GGA1 packs against the α2 and α3 loop, but in GGA3, the N terminus of α1 is pulled away from its position in GGA1 by 5 Å. The loop from residues 236–240 connecting helices α1 and α2 also differs in the GGA3 vs. GGA1. The hydrophobic patch between helices α2 and α3, which is the binding site for Rabaptin-5 (18) as well as the ubiquitin-binding site 2 (4, 6, 7), is present in GGA3. Residues in this patch include Arg-259, Phe-263, Leu-276, and Leu-280. This site is not occupied by ubiquitin in the crystal. Instead, two GAT-domain molecules related by noncrystallographic twofold symmetry contact each other at this site (Fig. 2C). The GGA3 GAT domain is a monomer in solution as judged by gel-filtration analysis at a concentration of 1 mg/ml (data not shown), suggesting that these contacts between GAT domains occur only in the crystal.
Fig. 2.
Crystal structure of the GGA3-GAT–ubiquitin complex. (A) A (2Fo - Fc) omit map showing the region around Ile-44 of ubiquitin, contoured at 0.9σ. Residues 42–45 of ubiquitin were omitted and the structure was subjected to simulated annealing and individual B-factor refinement before map calculation. The refined structure is shown with atoms colored as follows: green, carbon; red, oxygen; blue, nitrogen. (B) The GAT–ubiquitin complex interacts via site 1 (green, the GAT domain; orange, ubiquitin). Helices of the GAT domain are labeled. GGA3 Leu-227 and ubiquitin Ile-44 are shown in a yellow Corey–Pauling–Koltun model. (C) Crystal packing in the GAT dimer blocks site 2. One GAT monomer is colored green overall except with red for site 2 (residues 259–280). The second GAT monomer is colored light green overall, except with light purple for site 2. (D and E) Interactions in the site 1 interface. Ubiquitin is shown in a surface representation (green, hydrophobic residues; blue, basic; red, acidic; beige, uncharged polar). The GAT-domain helices are shown as gray cylinders, and residues that interact with ubiquitin are shown in ball-and-stick representations. Views in parts D and E differ by a 180° rotation about the y axis.
Table 1. Crystallographic data and refinement statistics.
| Space group | C2 |
| Unit cell, Å | a = 47.9, b = 97.3, c = 66.5 |
| Angles, ° | α = γ = 90.00, β = 90.03 |
| Resolution, Å | 66.5-2.8 (2.9-2.8)* |
| Unique reflections | 7,207 (605)* |
| Completeness, % | 95.4 (81.1)* |
| Redundancy | 2.7 |
| 〈I〉/〈σ〉 | 12.1 (2.6)* |
| Rmerge,† % | 8.0 (36.1)* |
| R‡/Rfree,§ % | 23.0/30.5 |
| rms deviation | |
| Bonds, Å | 0.0087 |
| Angles, ° | 1.271 |
| Average B factor, Å2 | 46.5 |
| Allowed ϕ-ψ angles, % | 100 |
Statistics in parentheses are for the highest resolution shell (Å)
Rmerge = Σ|I(k) - 〈I(k)〉|/ΣI(k)
R = Σ|Fobs - kFcalc|/Σ|Fobs|
Rfree is the R value calculated for a test set of reflections, comprising a randomly selected 10% of the data, not used during refinement
Structure of the GGA3-GAT–Ubiquitin Complex. The crystal structure contains two copies of a 1:1 complex between GGA3-GAT and ubiquitin related to each other by noncrystallographic symmetry. The lattice is tightly packed, and the GAT and ubiquitin molecules make contact with each other at multiple interfaces, two of which are more extensive than the others. One interface surrounds the Ile-44 of ubiquitin and the Leu-227 of GGA3 (Fig. 2B, D, and E). This contact involves residues from GGA3 previously identified as ubiquitin-binding site 1 (19). The Ile-44 region was also previously shown to participate in GAT binding (19). The shape-complementarity score calculated with the Collaborative Computational Program Number 4 (CCP4) program sc is 0.72. A value of >0.7 is comparable to that of specific protein–protein-inhibitor complexes (27), consistent with a functional role for the interface. The other contact surrounds Ile-36 of ubiquitin and Leu-233 of GGA3. In contrast to ubiquitin-binding site 1, Leu-233 and surrounding residues are conserved poorly or not at all among the GGAs, and NMR chemical-shift perturbation studies do not highlight any of the residues in this region as potential interactors (19). This contact has a poor shape-complementarity score of 0.56. The Ile-36/Leu-233 site is likely to be a crystal-packing artifact rather than a functional binding site and therefore will not be described further.
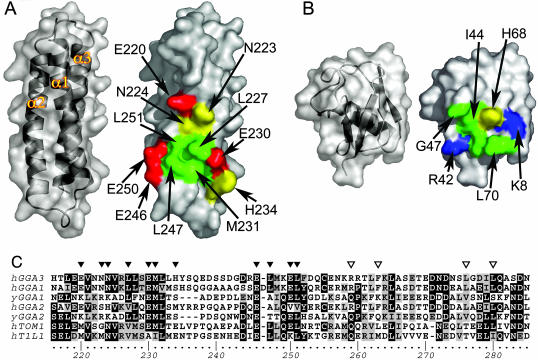
On the GAT domain, the functional contact centers on the hydrophobic residues Leu-227, Met-231, Leu-247, and Leu-251 (Fig. 3A). These hydrophobic residues are surrounded by a ring of polar-contact residues that interact with ubiquitin by polar interactions and nonpolar interactions with the aliphatic portions of their side chains. These include Glu-220, Asn-223, Asn-224, Glu-230, His-234, Asp-244, Glu-246, Glu-250, and Gln-254. On ubiquitin, the hydrophobic contact site centers on the residues Leu-8, Ile-44, Gly-47, and Val-70 (Fig. 3B). Contacts are also made by the basic residues Lys-6, Arg-42, and His-68 (Fig. 2 D and E; Fig. 3B). Glu-246 and Glu-250 of the GAT domain form a salt-bridge network with Arg-42 of ubiquitin, highlighting a role for both charged and hydrophobic interactions in the interface (Fig. 2 D and E). Lys-48, the most common site of conjugation in polyubiquitin chains, does not make direct contact with GAT but is in a sterically constricted site within 5 Å of His-234.
Fig. 3.
Recognition surfaces on GGA3-GAT and ubiquitin. (A) The surface of GGA3-GAT as viewed directly into the ubiquitin-binding site. Interacting residues are colored as follows: red, acidic; yellow, polar and uncharged at neutral pH; and green, hydrophobic. Noninteracting surface regions are gray. (B) The surface of ubiquitin as viewed directly into the GAT-binding site, colored as in A, except that surfaces of residues positively charged at neutral pH are colored blue. (C) GAT sequences aligned in the region of sites 1 and 2. Residues in site 1 are marked by filled triangles, and residues in site 2 are marked by open triangles.
Discussion
Functional Ubiquitin-Binding Sites. NMR chemical-shift perturbation and mutational analyses highlighted two sites on the GAT domain that interact with a single region on ubiquitin (19). Site 1 identified in the chemical-shift perturbation study corresponds to the binding site we observe in the crystal structure. A quadruple mutation in site 1 severely reduces the affinity of the GGA3-GAT domain for ubiquitin. A quadruple mutation in site 2 also reduces binding, but to a lesser degree (19). ITC data presented here show that single mutations in site 2 lead to no measurable impairment of in vitro binding. Taken together, the crystallographic and binding analyses indicate that site 1 is the primary binding site for ubiquitin.
In the crystal, site 2 is completely occupied by hydrophobic contacts with its own counterpart from a noncrystallographic twofold-symmetry-related GAT domain. This excludes ubiquitin binding at this site, and explains why binding at site 2 is not observed in the crystal lattice. The lack of interaction at site 2 is in apparent contradiction to previous reports by us (7) and others (6) that mutations in site 2 impair ubiquitin binding as assessed by yeast two-hybrid and pull-down experiments. However, ubiquitin binding is strongly affected by the manner of presentation, as well as by the oligomerization state of the interacting partner, the presence of other proteins that can provide bridging interactions, and other factors. To directly assess the role of site 2 in a 1:1 interaction between the purified GAT domain and ubiquitin, we determined the binding constants for wild type and two site-2 mutants in the GAT domain by ITC (Fig. 1). We found that these site-2 mutants do not detectably impair binding between isolated GAT and ubiquitin. This is consistent with the observation that site 2 is nonessential for ubiquitin binding in vitro and that mutations in site 2 are less disruptive than those in site 1 (19). However, site 2 is still capable of supporting the known biological role for the GAT–ubiquitin interaction, the trafficking of the ubiquitinated general amino-acid permease, Gap1p. Either one of the ubiquitin binding sites on GGA3 was able to support TGN to endosome trafficking of Gap1p, and mutation of both sites was required to block trafficking (19).
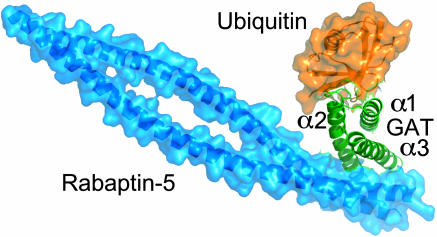
A hypothetical model of a GGA-GAT–Rabaptin-5–ubiquitin ternary complex was created to judge whether binding at the two sites is sterically feasible. The model was generated by superimposing the GGA1-GAT portion of the structure of the GGA1-GAT–Rabaptin-5 complex (18) onto the GGA3-GAT in the ubiquitin complex (Fig. 4). Rabaptin-5 and ubiquitin do not contact each other and do not approach each other within <6 Å. The crystal structure of the GGA3-GAT–ubiquitin complex shows that Rabaptin-5 and ubiquitin bind at separate sites and is consistent with the possibility that they may interact simultaneously with GGA1 in vivo (7).
Fig. 4.
Model of a ternary GAT–Rabaptin-5–ubiquitin complex. Molecules are colored as follows: blue, Rabaptin-5; green, GAT; orange, ubiquitin.
Implications for Other GAT-Domain Proteins. Human and yeast GGAs, TOM1, and TOM1-L1 protein share GAT domains. Like the GGA-GAT domains, the GAT domains of TOM1 and TOM1-L1 bind to ubiquitin (16). The hydrophobic center of the ubiquitin-binding site is conserved in the aligned sequences of all of these GAT domains (Fig. 3C). Leu-227 of GGA3 corresponds to either a Leu or Val in all sequences. Met-231 is almost completely conserved, except that an Ile replaces it in TOM1-L1. Leu-247 is well conserved but replaced by Ala in human GGA2 and by Ala or Thr in yeast GGAs. Leu-251 is almost completely conserved but is replaced by Val in human GGA2. Glu-246 is identically conserved in all sequences, and Glu-250 is conserved in most. Glu-230 is identically conserved except that it is changed to Ala in TOM1-L1. Other polar residues are less well conserved, and these sequence differences might account for the lower affinity of the GAT domain of GGA2 as compared with those of GGA1 and GGA3 (4–6).
Comparison to Other Ubiquitin-Binding Domains. The past 2 years have brought a rapid increase in structural information on complexes of ubiquitin with various effector domains. Ile-44 was identified some years ago as a ubiquitin residue essential for both the proteolytic (28) and endocytic (29) functions of ubiquitin. The coupling of unfolded-protein response to endoplasmic reticulum degradation (CUE) domains of Vps9p and Cue2p (30, 31) are small, three-helix bundles that interact with the same Ile-44 hydrophobic patch on ubiquitin as described here. The complex of the structurally related ubiquitin-associated (UBA) domain has been modeled to interact with the same patch on the basis of the Cue2p complex structure (30). The one-helix ubiquitin-interacting motif (UIM) also interacts with this patch (32). The ubiquitin-E2-variant (UEV) domains of Vps23p and TSG101 (33, 34) have a more polar interface with ubiquitin than do the other domains, but even these domains interact with Ile-44. The Npl4 zinc finger (NZF) provides yet another example of a ubiquitin-binding domain that interacts with the Ile-44 region (35). The site-1 interface in the GAT–ubiquitin complex buries 530 Å2 of solvent-accessible surface area. This is similar to the surface area buried in most other ubiquitin-binding domain–ubiquitin complexes, most of which have comparably high micromolar affinities. The structure of the site-1 interface thus fits the pattern established for five other ubiquitin-binding domains and has the hallmarks of a bona fide functional ubiquitin-recognition interface.
Implications for GGA and GAT Domain-Mediated Sorting. The structure presented here highlights a role for GAT domains as hubs for multiple protein–protein interactions that occur by means of hydrophobic surface interactions. The role of trafficking domains as interaction hubs has been noted recently in another context by structural analyses showing that adaptor-protein-appendage domains have multiple peptide-binding grooves (36–38). We have been able to model a ternary ubiquitin–Rabaptin-5–GAT complex and show that the two different GAT ligands do not clash with one another, consistent with a potential role for GGAs in sorting ubiquitinated Gap1p and other ubiquitinated cargo to Rab5-positive endosomes. The two hydrophobic sites on the GAT domain are conserved throughout the family of GGA and TOM1 proteins, but the polar residues surrounding the hydrophobic patches are variable and can modulate their functions. This ability increases the remarkable versatility of GAT domains, considered as a class.
Acknowledgments
We thank Y. Im and D. Hurt for assistance with crystallography and data collection and C. Abergel for advice on molecular replacement.
Author contributions: J.S.B. and J.H.H. designed research; G.P., S.L., R.M., C.N.A., and B.M.B. performed research; S.L. contributed new reagents/analytic tools; and J.S.B. and J.H.H. wrote the paper.
Abbreviations: Arf, ADP-ribosylation factor; GGA, Golgi-localized, γ-ear-containing, Arf-binding; TGN, trans-Golgi network; TOM, target of Myb; GAT, GGA and TOM; TOM1-L1, TOM1-like 1; TSG, tumor-susceptibility gene; CUE, coupling of unfolded protein response to endoplasmic reticulum degradation; ITC, isothermal titration calorimetry.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1YD8).
References
- 1.Bonifacino, J. S. (2004) Nat. Rev. Mol. Cell Biol. 5, 23-32. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh, P. & Kornfeld, S. (2004) Eur. J. Cell Biol. 83, 257-262. [DOI] [PubMed] [Google Scholar]
- 3.Mattera, R., Arighi, C. N., Lodge, R., Zerial, M. & Bonifacino, J. S. (2003) EMBO J. 22, 78-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puertollano, R. & Bonifacino, J. S. (2004) Nat. Cell Biol. 6, 244-251. [DOI] [PubMed] [Google Scholar]
- 5.Scott, P. M., Bilodeau, P. S., Zhdankina, O., Winistorfer, S. C., Hauglund, M. J., Allaman, M. M., Kearney, W. R., Robertson, A. D., Boman, A. L. & Piper, R. C. (2004) Nat. Cell Biol. 6, 252-259. [DOI] [PubMed] [Google Scholar]
- 6.Shiba, Y., Katoh, Y., Shiba, T., Yoshino, K., Takatsu, H., Kobayashi, H., Shin, H. W., Wakatsuki, S. & Nakayama, K. (2004) J. Biol. Chem. 279, 7105-7111. [DOI] [PubMed] [Google Scholar]
- 7.Mattera, R., Puertollano, R., Smith, W. J. & Bonifacino, J. S. (2004) J. Biol. Chem. 279, 31409-31418. [DOI] [PubMed] [Google Scholar]
- 8.Hicke, L. (2001) Nat. Rev. Mol. Cell Biol. 2, 195-201. [DOI] [PubMed] [Google Scholar]
- 9.Katzmann, D. J., Odorizzi, G. & Emr, S. D. (2002) Nat. Rev. Mol. Cell Biol. 3, 893-905. [DOI] [PubMed] [Google Scholar]
- 10.Di Fiore, P. P., Polo, S. & Hofmann, K. (2003) Nat. Rev. Mol. Cell Biol. 4, 491-497. [DOI] [PubMed] [Google Scholar]
- 11.Haglund, K., Di Fiore, P. P. & Dikic, I. (2003) Trends Biochem. Sci. 28, 598-603. [DOI] [PubMed] [Google Scholar]
- 12.Collins, B. M., Watson, P. J. & Owen, D. J. (2003) Dev. Cell 4, 321-332. [DOI] [PubMed] [Google Scholar]
- 13.Suer, S., Misra, S., Saidi, L. F. & Hurley, J. H. (2003) Proc. Natl. Acad. Sci. USA 100, 4451-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiba, T., Kawasaki, M., Takatsu, H., Nogi, T., Matsugaki, N., Igarashi, N., Suzuki, M., Kato, R., Nakayama, K. & Wakatsuki, S. (2003) Nat. Struct. Biol. 10, 386-393. [DOI] [PubMed] [Google Scholar]
- 15.Zhu, G. Y., Zhai, P., He, X. Y., Terzyan, S., Zhang, R. G., Joachimiak, A., Tang, J. & Zhang, X. J. C. (2003) Biochemistry 42, 6392-6399. [DOI] [PubMed] [Google Scholar]
- 16.Yamakami, M., Yoshimori, T. & Yokosawa, H. (2003) J. Biol. Chem. 278, 52865-52872. [DOI] [PubMed] [Google Scholar]
- 17.Katoh, Y., Shiba, Y., Mitsuhashi, H., Yanagida, Y., Takatsu, H. & Nakayama, K. (2004) J. Biol. Chem. 279, 24435-24443. [DOI] [PubMed] [Google Scholar]
- 18.Zhu, G. Y., Zhai, P., He, X. Y., Wakeham, N., Rodgers, K., Li, G. P., Tang, J. & Zhang, X. J. C. (2004) EMBO J. 23, 3909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bilodeau, P. S., Winistorfer, S. C., Allaman, M. M., Surendhran, K., Kearney, W. R., Robertson, A. D. & Piper, R. C. (2004) J. Biol. Chem. 279, 54808-54816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheffield, P., Garrard, S. & Derewenda, Z. (1999) Protein Expression Purif. 15, 34-39. [DOI] [PubMed] [Google Scholar]
- 21.Claude, J. B., Suhre, K., Notredame, C., Claverie, J. M. & Abergel, C. (2004) Nucleic Acids Res. 32, W606-W609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jogl, G., Tao, X., Xu, Y. W. & Tong, L. (2001) Acta Crystallogr. D 57, 1127-1134. [DOI] [PubMed] [Google Scholar]
- 23.Yeates, T. O. (1997) Methods Enzymol. 276, 344-358. [PubMed] [Google Scholar]
- 24.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 25.Barends, T. R. M. & Dijkstra, B. W. (2003) Acta Crystallogr. D 59, 2237-2241. [DOI] [PubMed] [Google Scholar]
- 26.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence, M. C. & Colman, P. M. (1993) J. Mol. Biol. 234, 946-950. [DOI] [PubMed] [Google Scholar]
- 28.Beal, R., Deveraux, Q., Xia, G., Rechsteiner, M. & Pickart, C. (1996) Proc. Natl. Acad. Sci. USA 93, 861-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloper-Mould, K. E., Jemc, J. C., Pickart, C. M. & Hicke, L. (2001) J. Biol. Chem. 276, 30483-30489. [DOI] [PubMed] [Google Scholar]
- 30.Kang, R. S., Daniels, C. M., Francis, S. A., Shih, S. C., Salerno, W. J., Hicke, L. & Radhakrishnan, I. (2003) Cell 113, 621-630. [DOI] [PubMed] [Google Scholar]
- 31.Prag, G., Misra, S., Jones, E. A., Ghirlando, R., Davies, B. A., Horazdovsky, B. F. & Hurley, J. H. (2003) Cell 113, 609-620. [DOI] [PubMed] [Google Scholar]
- 32.Swanson, K. A., Kang, R. S., Stamenova, S. D., Hicke, L. & Radhakrishnan, I. (2003) EMBO J. 22, 4597-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teo, H., Veprintsev, D. B. & Williams, R. L. (2004) J. Biol. Chem. 279, 28689-28696. [DOI] [PubMed] [Google Scholar]
- 34.Sundquist, W. I., Schubert, H. L., Kelly, B. N., Hill, G. C., Holton, J. M. & Hill, C. P. (2004) Mol. Cell 13, 783-789. [DOI] [PubMed] [Google Scholar]
- 35.Alam, S. L., Sun, J., Payne, M., Welch, B. D., Black, B. K., Davis, D. R., Meyer, H. H., Emr, S. D. & Sundquist, W. I. (2004) EMBO J. 23, 1411-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritter, B., Denisov, A. Y., Philie, J., Deprez, C., Tung, E. C., Gehring, K. & McPherson, P. S. (2004) EMBO J. 23, 3701-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishra, S. K., Harwryluk, M. J., Brett, T. J., Keyel, P. A., Dupin, A. L., Jha, A., Heuser, J. E., Fremont, D. H. & Traub, L. M. (2004) J. Biol. Chem. 279, 46191-46203. [DOI] [PubMed] [Google Scholar]
- 38.Praefcke, G. J., Ford, M. G. J., Schmid, E. M., Olesen, L. E., Gallop, J. L., Peak-Chew, S. Y., Vallis, Y., Babu, M. M., Mills, I. G. & McMahon, H. T. (2004) EMBO J. 23, 4371-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]