Abstract
SsrA is a versatile RNA molecule found in all bacteria that functions as both a tRNA and an mRNA. SsrA rescues ribosomes stalled on damaged mRNAs and directs the tagging and degradation of their aberrant protein products. Small protein B (SmpB) is required for all known activities of SsrA. The two known functions of SmpB are binding SsrA RNA and promoting stable association of the SmpB·SsrA complex with 70S ribosomes. Using mutational analysis and biochemical experiments, we have discovered a previously uncharacterized SmpB function. This function is required for a step in the tagging process downstream of SsrA binding and ribosome association but before transpeptidation of the SsrA-linked alanine and establishment of the SsrA reading frame. Our results clearly demonstrate that residues in the C-terminal tail of SmpB confer a hitherto unrevealed function that is essential for trans-translation. Based on these results, we propose that upon binding stalled ribosomes, the unstructured C-terminal tail of SmpB acquires contacts that are critical for productive accommodation of SsrA into the ribosomal A site.
Keywords: SsrA, translation
Bacteria use a unique translational quality control system whose main components are small protein B (SmpB) protein and SsrA RNA [also known as transfer messenger RNA (tmRNA) or 10Sa RNA]. SsrA possesses a tRNA-like domain and an mRNA-like coding sequence that jointly enable it to function as both a tRNA and an mRNA (1–4). The SmpB-SsrA quality control surveillance system rescues ribosomes stalled on incomplete or damaged mRNAs and directs the addition of a proteolysis tag to the C termini of aberrant protein products to facilitate their degradation, a process known as trans-translation (1–3, 5, 6). SmpB is essential for all known SsrA functions; however, its exact mechanistic function in trans-translation is unclear. The two known functions of SmpB are specific binding to SsrA RNA and promoting its stable association with 70S ribosomes (3, 6, 7). Although not strictly required, SmpB may also stabilize SsrA RNA and promote its efficient charging by alanine tRNA synthetase (7, 8).
Recently, a great deal of structural information regarding SmpB and SsrA has become available. NMR solution structures of SmpB from Aquifex aeolicus (9) and Thermus thermophilus (10), along with a cocrystal structure of A. aeolicus SmpB in complex with the tRNA-like domain of SsrA (11), have been solved. The core SmpB structure is quite similar in all three structural models; however, none of the models were able to discern the structure of the protein's C-terminal extension. Thus, the C-terminal tail of SmpB appears to be unstructured.
In this study, we directly test the functional relevance of the C-terminal tail of the SmpB protein. We have identified residues in the C-terminal tail of SmpB that are critical in supporting SsrA tagging activity. Most interestingly, mutations in these residues do not affect the ability of SmpB to bind SsrA RNA in vivo or in vitro, nor do they affect its ability to support stable association of SsrA RNA with stalled ribosomes. We propose that the SmpB C-terminal extension gains structure within the context of the ribosome and acquires contacts that are required to support a previously undefined function of the SmpB protein. This function is crucial for events that are downstream of ribosome association but before transpeptidation and establishment of the SsrA-encoded reading frame.
Materials and Methods
Nomenclature. SmpB truncation mutants are named by using the number of the C-terminal residue of the protein. For example, SmpB155 includes amino acids 1–155 of the wild-type SmpB protein. SmpBDE is I154D/M155E, SmpBRK is I154R/M155K, SmpBAA is I154A/M155A, SmpBLI is I154L/M155I, and SmpBQQ is I154Q/M155Q. SmpB variants carrying substitution mutations of residues 137, 138, and 139 are labeled with the substitution in superscript. For a detailed description of all methods used in the study, see Supporting Text, which is published as supporting information on the PNAS web site.
Functional Assays. The endogenous tagging assay protocol was modified from Karzai and coworkers (12). The total signal in each lane of the tagging assay was quantified and compared to that of SmpBWT to generate a percent of wild type activity. Gel mobility-shift assays were performed by using 3′ end-labeled SsrA113 RNA (≈100 pM). The reaction buffer contained 50 mM Tris (pH7.5), 10 mM MgCl2, 300 mM KCl, 2 mM 2-mercaptoethanol, 100 μg/ml BSA, 0.01% Nonidet P-40 (vol/vol), 5% glycerol (vol/vol), and 100 nM total Escherichia coli tRNA. Reactions were incubated at 4°C for 1 hr. The data analysis was performed according to Berggrun and Sauer (13). Briefly, we measured the fraction of the primary bound species at each SmpB concentration and determined the apparent equilibrium dissociation constant by curve fitting using the equation: Θeq = C/(1 + Kd/[SmpB]i), where Θeq is the fraction of RNA bound at equilibrium, C is a constant representing the maximum fraction bound of the specific bound species, and [SmpB]i is the initial concentration of SmpB. Ribosomes were purified from ΔB (DE3)/pETBA or pETBAH6, as described (14).
Results
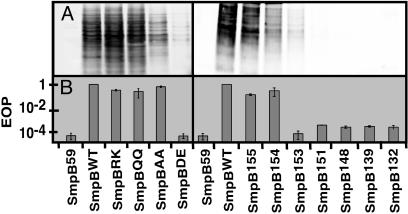
SmpB C-Terminal Tail Mutants Are Defective in Supporting SsrA-Mediated Tagging. We set out to identify functionally relevant residues in the C-terminal tail of the SmpB protein by introducing premature termination codons at various positions in the SmpB coding sequence and assessing the ability of the mutant proteins to support SsrA-mediated endogenous tagging. Endogenous tagging activity was assayed by using a plasmid that expresses SmpB and a SsrA RNA variant in which the mRNA segment is mutated to code for six histidine residues (changing the SsrA tag sequence from ANDENYALAA to ANDEHHHHHH). SmpB mutants were generated in this construct, and endogenous tagging activity was assayed in a W3110ΔsmpB1 deletion strain (hereafter called ΔB). We found that deletion of five amino acid residues from the unstructured C-terminal tail of SmpB (SmpB155) had no adverse effect on SsrA-mediated endogenous tagging activity. Indeed, a subtle increase in activity was observed with SmpB155. In contrast, removal of two additional amino acids (SmpB153) severely reduced tagging activity, whereas removal of one additional amino acid (SmpB154) had a moderate effect on endogenous tagging, reducing it to the level of wild-type protein (Fig. 1 and Table 1, which is published as supporting information on the PNAS web site). Further truncations in the C-terminal tail, including SmpB151, SmpB148, SmpB144, SmpB139, SmpB133, or SmpB59, entirely abolished the ability of the protein to support SsrAH6-mediated endogenous tagging (Fig. 1, Table 1, and data not shown).
Fig. 1.
Endogenous tagging phenotypes. (A) Western blot analysis using IR800-conjugated anti-his6 antibody, with the pattern of proteins tagged by SsrAH6 in cells expressing different SmpB variants. (B) Analysis of λimmP22 hybrid phage induction supported by different SmpB mutants. Data are presented as efficiency of plating (EOP), where the number of plaques formed when SmpBWT was expressed is taken as EOP = 1. We have used SmpBWT as a positive control and SmpB59 (an SmpB truncation mutant with only residues 1–59) as a negative control throughout these experiments.
Stepwise deletion of residues in the unstructured C-terminal tail of SmpB revealed that loss of residues 155 and 154 led to a gradual decrease in SsrA-mediated endogenous tagging activity (i.e., SmpB155 > SmpB154 > SmpB153). We therefore hypothesized that residues I154 and M155 may be important for SmpB function. To investigate the functional importance of this region, we generated substitution mutations of these residues in the context of the full-length protein and assayed the ability of mutant proteins to support endogenous SsrAH6 tagging activity. Single amino acid substitutions of residues 154 or 155 to alanine, glutamine, or glutamic acid had little effect on endogenous tagging activity (Table 1). Consequently, we explored double-substitution mutations of residues I154 and M155. Conservative substitution of both residues (I154L/M155I, hereafter called SmpBLI), as well as substitution to positively charged residues (I154R/M155K, or SmpBRK) or to polar residues (I154Q/M155Q, or SmpBQQ) had no adverse effect on endogenous tagging activity (Fig. 1 and Table 1). Conversely, when negatively charged residues were introduced (I154D/M155E, or SmpBDE), endogenous tagging activity was nearly abolished. Introduction of alanine at both positions (I154A/M155A, or SmpBAA) yielded a moderate reduction in tagging activity (Fig. 1 and Table 1). It is possible that some SmpB mutations may lead to a change in expression level or stability of the protein within cells. To control for this possibility, we performed Western analysis on S30 extracts from cells expressing each SmpB variant. We found the amount of soluble SmpB to be similar for all SmpB variants studied and not related to tagging efficiency (data not shown).
SsrA and SmpB are required for induction of the lytic cycle in the λimmP22 dis c2–5 hybrid phage (6, 15). We assessed the ability of SmpB mutants to support lytic development of this phage. Data are summarized in Fig. 1 and Table 1. As expected, the λimmP22 phage phenotypes largely mirror the observed endogenous tagging phenotypes. SmpB mutants defective in SsrAH6-mediated endogenous tagging (i.e., SmpBDE, SmpB153, and all smaller truncation mutants studied) were also severely impaired in supporting lytic growth of the hybrid phage. However, the modest loss of SsrA tagging activity supported by SmpBAA was not observed in phage assays; also, a very subtle decrease in efficiency of plating was observed with SmpB155 and SmpB154 (Fig. 1 and Table 1). These phage induction assay data support the conclusion that SmpBDE and SmpB truncation mutants are defective in supporting SmpB·SsrA-mediated tagging activity.
SmpB Mutants Are Fully Competent in Binding SsrA RNA. Next, we set out to identify the specific biochemical defect in SmpB function resulting from these mutations. The two known functions of SmpB are binding to SsrA RNA and promoting its stable association with ribosomes (3, 6, 7). First, we assessed the ability of mutant SmpB proteins to bind SsrA RNA, both in vivo and in vitro. We purified the SmpB·SsrA complex under near physiological conditions by affinity chromatography (over Ni2+-NTA beads) followed by an ion exchange step using a FPLC monoQ (HR 10/10) column. Under these conditions, SmpB protein and SsrA RNA coelute as a single peak around 600 mM KCl. The chromatographic behavior of the defective mutants SmpBDE, SmpB153, and SmpB148 was indistinguishable from SmpBWT in this regard. That is, SmpB153, SmpB148 and SmpBDE remained bound to SsrA RNA through both steps of the purification process (not shown). These results suggest that the SmpB mutants that fail to support endogenous SsrAH6 tagging are not defective in binding SsrA RNA in vivo.
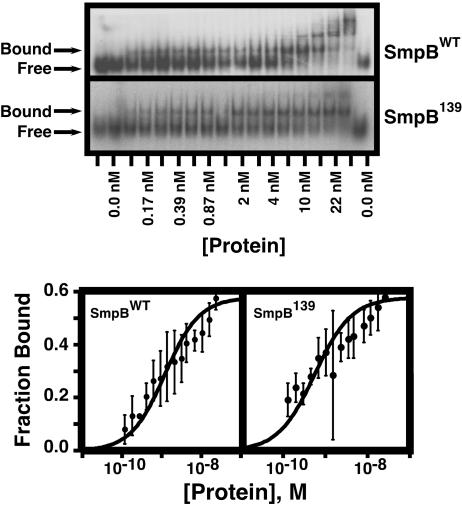
Binding of one SmpB molecule to one SsrA molecule (presumably the most functionally relevant interaction) produces only a small and difficult-to-discern band shift (not shown). A large body of evidence, including enzymatic footprinting, mutational analysis, and a recent cocrystal structure model, suggests that the tRNA-like domain of SsrA RNA is necessary and sufficient for specific binding of one molecule of SmpB protein (8, 11, 16, 17). Therefore, we used a synthetic SsrA variant (SsrA113, that includes the tRNA-like domain of SsrA without the pseudoknots or the mRNA sequence) to assess the SsrA-binding affinities of SmpBWT and the severely defective mutants, SmpBDE, SmpB153, and SmpB139. In vivo, SmpB must recognize SsrA RNA in the presence of a large excess of cellular RNAs. Therefore, to ensure that only specific interactions of SmpB protein with SsrA RNA were detected, we conducted our gel mobility-shift assays under high-stringency binding conditions. Binding reactions were carried out in the presence of 300 mM KCl and 100 nM total E. coli tRNA (a 1,000-fold molar excess of the structurally related competitor tRNA over SsrA). The gel mobility-shift assay data include multiple bound species (Fig. 2). Cocrystal and cryo-EM structural data both suggest that the functional SmpB·SsrA complex contains one molecule of each binding partner (11, 26). Hence, for the purposes of data analysis, we have treated the primary bound species as the specific bound complex and the additional bound species, which appear at higher SmpB concentrations, as products of additional protein–protein and protein–RNA interactions (see Materials and Methods and ref. 13). The binding data are very complex, because multiple bound species appear at SmpB concentrations where the free RNA is >0. We attempted several methods of data analysis but were unable to obtain a perfect fit. The curve fit analysis presented is meant to be an estimate of SsrA-binding affinity used to compare different SmpB variants.
Fig. 2.
SsrA-binding assays. (Upper) Gel mobility-shift assays of the SsrA-binding propensity of SmpBWT and SmpB139. (Lower) Curve-fit analysis used to determine the apparent equilibrium dissociation constants (Kd) of SsrA113–SmpB interactions.
Most significantly, regardless of what data analysis method was used, we did not observe any difference in the affinities of SmpBWT, SmpBDE, SmpB153, and SmpB139 for SsrA RNA (Fig. 2 and Table 1). The apparent equilibrium dissociation constants, Kd, of all mutants studied were similar to SmpBWT (Kd for SmpBWT, SmpBDE, SmpB153, and SmpB139 were calculated to be 1.17 ± 0.18 nM, 0.623 ± 0.13 nM, 0.848 ± 0.22 nM, and 0.519 ± 0.095 nM, respectively). Furthermore, we observed no difference in the binding affinity of SmpBWT and the SmpB153 truncation mutant to full-length SsrA (not shown). Taken together, these data clearly show that mutations to the unstructured C-terminal tail of SmpB affect neither the ability of the protein to bind full-length SsrA RNA in vivo nor the affinity of SmpB interactions with SsrA under stringent in vitro conditions. Therefore, we conclude that SmpBDE, SmpB153, and SmpB139, although defective in endogenous tagging, are fully capable of binding SsrA RNA with high affinity and specificity both in vivo and in vitro.
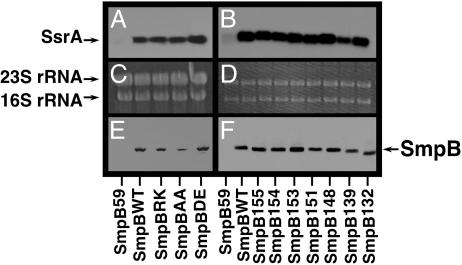
C-Terminal Tail Mutants Support SsrA Association with Ribosomes. Next, we hypothesized that mutations in the C-terminal tail of SmpB might impair the protein's ability to stably associate with ribosomes and/or to support the association of SsrA RNA with ribosomes. To test this hypothesis, we purified tight coupled 70S ribosomes from cells expressing SmpBWT and the various mutants using a method modified from Vila-Sanjurjo et al. (14) and analyzed them for the presence of SmpB protein and SsrA RNA. We used SmpBWT as a positive control and SmpB59 (a SmpB truncation mutant with only residues 1–59, which does not support stable association of SsrA with ribosomes) as a negative control throughout these experiments. We found by Western blot analysis that SmpBWT, SmpBRK, SmpBAA, and SmpBDE all stably associate with ribosomes (Fig. 3E). Northern blot analysis of these samples, using SsrA RNA specific probes, revealed that all of the aforementioned SmpB mutants also support stable association of SsrA RNA with ribosomes (Fig. 3A). The SmpB truncation mutants SmpB155, SmpB154, SmpB153, SmpB151, SmpB148, SmpB139, and SmpB132 also displayed no defects in this regard (i.e., these SmpB truncation mutants associate with ribosomes and are fully proficient in promoting stable association of SsrA RNA with ribosomes) (Fig. 3 B and F). Ethidium bromide staining of the gels used for Northern blot (Fig. 3 C and D) is shown to demonstrate that a similar amount of ribosome preparation was loaded onto each lane.
Fig. 3.
Ribosome association. (A and B) Northern blot analysis using an SsrA-specific probe to detect SsrA RNA in purified ribosome preparations. (C and D) Ethidium bromide staining of the same gel as in A and B, shown to demonstrate that similar amounts of ribosomal RNA were loaded in each lane. (E and F) Western blot analysis using anti-his6 antibody to detect his6-tagged SmpB protein in the same purified ribosome preparations used in A–D. The SmpB variant expressed in the cells from which the ribosomes were purified is indicated on the horizontal axis.
Taken together, these data directly demonstrate that SmpB variants carrying specific mutations near their C termini are fully capable of performing the two known functions of SmpB (i.e., specific binding of SsrA RNA and supporting its stable association with 70S ribosomes); nonetheless, these mutants fail to support SsrA tagging activity.
SmpB Mutants Fail to Support Transpeptidation and Partial Tagging. Having identified tagging-deficient SmpB mutants that retain the ability to bind SsrA and promote stable association of the SmpB·SsrA complex with stalled ribosomes, we set out to further define the mechanistic time frame of the defect. We wanted to know whether the defective SmpB mutants were capable of adding any part of the tag sequence (addition of the SsrA-charged alanine or any portion of the SsrA encoded tag) to the C-terminal end of incompletely synthesized protein fragments (i.e., partial tagging).
To assay for partial tagging, we used λ-N-trpAt, a synthetic gene construct that contains the N-terminal 93 residues of the λ cI repressor followed by a His-6 epitope and a trpAt transcriptional terminator (6, 18). Transcription of this gene yields a nonstop mRNA that leads to ribosome stalling and subsequent tagging by the SmpB·SsrA system. We coexpressed the λ-N-trpAt protein (hereafter referred to as λ-N) along with SsrAH6 and the SmpB C-terminal tail mutants, then purified the λ-N protein and analyzed the product for the presence or absence of any SsrA-encoded tag sequence by MALDI-TOF MS (Fig. 4 and Fig. 6, which is published as supporting information on the PNAS web site). In the absence of functional SmpB·SsrA, a series of peaks is observed, the largest of which represents the major translation product from the λ-N-trpAt gene. The smaller flanking peaks represent minor translation products, most likely arising from λ-N-trpAt transcript degradation (Fig. 6 and ref. 18). In the presence of functional SmpB·SsrAH6, a second set of peaks appear, corresponding to the major and minor translation products with the SsrAH6 encoded tag sequence at their C termini (Figs. 4 and 6 and ref. 18). If only the SsrA-linked alanine is added to the λ-N protein product, one would expect a peak corresponding to the mass of the major untagged translation product plus the mass of an alanine residue.
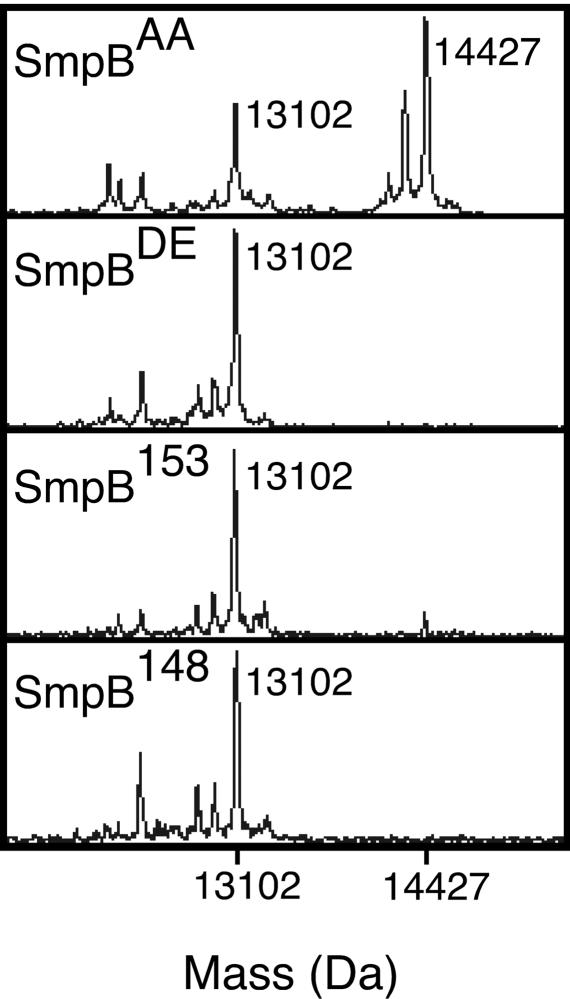
Fig. 4.
MALDI-TOF MS spectra of purified λ-N protein from cells expressing the SmpBAA, SmpBDE, SmpB153, and SmpB148 variants. The species with m/z = 13,102 is the major untagged λ-N protein product, whereas the peak with m/z = 14,427 corresponds to the major product with the full SsrAH6 encoded tag sequence. For details of the experiment and results, see Fig. 6.
The observed peaks in the MALDI-TOF spectra of λ-N protein purified from cells expressing the SmpBDE, SmpB153, and SmpB148 mutants correspond to protein products encoded solely by the λ-N-trpAt construct; the major peak in each profile corresponds exactly to the predicted mass identity of untagged λ-N protein (Figs. 4 and 6). We did not observe any peaks corresponding to SsrA-tagged products. These results agree with the endogenous tagging and hybrid phage phenotypes of these mutants, demonstrating that the SmpBDE, SmpB153, and SmpB148 mutants are unable to support the addition of the SsrA encoded tag sequence to nascent polypeptides in response to ribosome stalling. Furthermore, we did not observe any peaks corresponding to partially tagged λ-N protein products that could result from the addition of either alanine alone or any portion of the tag sequence arising from the mRNA-like function of SsrA (Figs. 4 and 6). Therefore, the MALDI-TOF analysis demonstrates that the SmpBDE, SmpB153, and SmpB148 mutants are unable to support either the tRNA- or the mRNA-like function of SsrA RNA.
As expected, the MALDI-TOF spectrum of λ-N protein purified from cells expressing SmpBAA includes the set of mass peaks corresponding to untagged products, along with an additional set of peaks corresponding to products containing the full SsrAH6 encoded tag sequence (Figs. 4 and 6). Once again, no partially tagged products were observed with SmpBAA. The absence of such products implies that SmpBAA is partly defective in supporting both the tRNA- and mRNA-like functions of SsrA.
Although the MALDI-TOF spectra in all experiments were of sufficient resolution to detect several previously characterized minor translation products (Figs. 4 and 6, data not shown, and ref. 18), we did not observe mass peaks in any spectrum that corresponded to partially tagged proteins. One possible explanation for this observation is that addition of an alanine to the C terminus of the λ-N-trpAt protein renders it unstable within cells. To control for this possibility we expressed a variant of the λ-N-trpAt gene that codes for a C-terminal alanine followed by a termination codon. We found this product to be stable and soluble when expressed (data not shown). Another possibility is that the MALDI-TOF-MS signal of a partially tagged product is masked by other proteins in the sample. The clearly defined mass peaks corresponding to minor translation products seen here and by Williams et al. (18) argue against this possibility. It is also formally possible that a partially tagged product is not released from the ribosome; however, considering the labile nature of the ester linkage between SsrA and alanine, we consider this to be unlikely. One would expect that a partially tagged protein bound to SsrA would be released via the same mechanism that permits the release of the untagged species from P site tRNA in the absence of SsrA. The most likely explanation is that the SmpBDE, SmpB153, and SmpB148 mutants do not support either full or partial tagging, and that the decreased tagging activity of SmpBAA mutant is due not to partial tagging activity but rather to an overall reduction in the tagging proficiency of this mutant. Thus, the unstructured C-terminal tail of SmpB plays a crucial role after association of the SmpB·SsrA complex with ribosomes but before transpeptidation of the SsrA linked alanine and establishment of the SsrA reading frame.
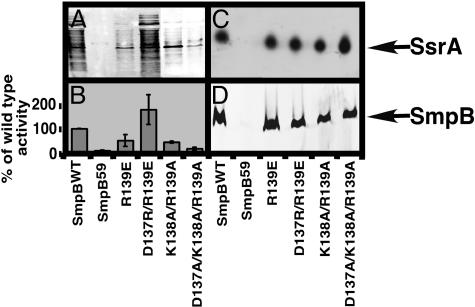
Mutation of Conserved C-Terminal Tail Residues Yields the Same Phenotypes. The unstructured C-terminal tail of SmpB contains a number of invariant or highly conserved amino acids. For instance, residues D137, K138, and R139, near the proximal base of the SmpB C-terminal tail, show a high degree of conservation among bacterial species (the three residues are 100%, 98%, and 100% conserved among the 130 known SmpB sequences). As such, we hypothesized that these C-terminal tail residues should also play an analogous and critical role in SmpB-SsrA-mediated tagging. Single point mutation of each invariant residue to alanine had little effect on SsrAH6-mediated endogenous tagging (not shown). Replacing the positively charged R139 with a negatively charged glutamic acid (SmpBR139E) led to a moderate tagging defect. Interestingly, this defect was rescued by substitution of the negatively charged D137 with arginine (SmpBD137R/R139E) (Fig. 5 and Table 1). A double alanine substitution mutation, SmpB K138A/R139A, caused a moderate defect in endogenous tagging. More notably, the triple alanine substitution, SmpBD137A/K138A/R139A, nearly abolished tagging activity (Fig. 5 and Table 1). These data support the conclusion that these residues in the C-terminal tail of SmpB play a critical role in trans-translation.
Fig. 5.
Phenotypes of mutations to the 137–139 region. (A) Western analysis of endogenous tagging activity. (B) Bar graph depicting the mean and standard deviation of the percent of wild-type endogenous tagging signals for three separate experiments. (C) Northern blot using an SsrA-specific probe showing copurification of SsrA RNA with ribosomes. (D) Western blot showing the presence of SmpB in the same ribosome preparation as in C. The samples in C and D were normalized by A260 to ensure that a similar amount of ribosomes was loaded into each lane (data not shown).
For further biochemical analysis of their defect, we purified the defective SmpB mutants (SmpBR139E, SmpBK138A/R139A, and SmpBD137A/K138A/R139A) and analyzed their SsrA-binding affinities by gel mobility-shift assay. We observed no defect in the SsrA-binding affinities of these mutants. The equilibrium dissociation constants for interactions of SmpBR139E, SmpBK138A/R139A, and SmpBD137A/K138A/R139A with SsrA113 were very similar to that of SmpBWT (0.411 ± 0.05 nM for SmpBR139E, 0.877 ± 0.14 nM for SmpBK138A/R139A, and 0.688 ± 0.07 nM for SmpBD137A/K138A/R139A) (Table 1). Similarly, none of the mutations to these residues affected the association of the SmpB·SsrA complex with ribosomes (Fig. 5). Thus, substitution mutations of the conserved D137, K138, and R139 residues lead to the same set of phenotypes observed for other C-terminal tail mutants (full competence in supporting SsrA-binding and SmpB·SsrA ribosome association but loss of ability to support SsrA-mediated endogenous tagging). Once again, these data strongly support the conclusion that the C-terminal tail of SmpB confers a hitherto unknown function essential for trans-translation.
Discussion
Our results demonstrate that the C terminus of SmpB is involved in supporting a previously uncharacterized function of the protein in trans-translation. We have shown that mutations to specific C-terminal residues of SmpB protein impair the protein's ability to support endogenous SsrA-mediated tagging, as well as induction of lytic development of a λimmP22 hybrid phage (Fig. 1 and Table 1). Significantly, these mutants are not defective in binding SsrA RNA in vivo or in vitro nor are they impaired in their ability to promote stable association of the SmpB·SsrA complex with tight-coupled 70S ribosomes (Figs. 2 and 3). Nonetheless, the mutant proteins are unable to support addition of the SsrA-linked alanine to incompletely synthesized polypeptides (Figs. 4 and 6). Thus, the newly identified function of the C-terminal tail of SmpB is required for events in trans-translation downstream of SmpB·SsrA complex formation and its stable association with ribosomes but before addition of the SsrA-linked alanine to incomplete polypeptides.
Residues I154 and M155 are important in supporting this function. Interestingly, whereas the length of the C-terminal extension varies among bacterial SmpB genes, all known SmpB C-terminal tails include at least the amino acid that aligns with I154 in E. coli. Negative charges at residues 154 and 155 are not well tolerated, because introduction of negative charge at both residues 154 and 155 renders the protein severely defective in supporting SsrA-mediated tagging (Fig. 1 and Table 1). The presence of charged amino acids as well as the net charge of the 137–139 region seems to be a key element in determining tagging efficiency. The complete loss of charged amino acids that is generated by the triple alanine substitution mutation elicits the most severe tagging defect of all of the mutations we've generated in the region (Fig. 5 and Table 1). Furthermore, mutations that confer net negative charge to this region (i.e., R139E and K138A/R139A) also produce a tagging defect, whereas mutations that confer a net positive charge (i.e., D137A, K138A, R139A, and D137A/R139A) do not adversely affect the tagging proficiency of the protein (Fig. 5, Table 1, and data not shown). The tagging defect elicited by the R139E mutation is rescued by restoring the net charge (D137R/R139E), indicating that some level of flexibility exists with regard to the specific location of charged amino acids in this region.
It is conceivable that charge–charge interaction of residues 137–139 with either rRNA phosphate backbone or other ribosomal elements anchors the structure of the SmpB C-terminal tail within the context of the ribosome. This type of interaction may be required to position other distal elements (perhaps residues 154 and 155) for functional contacts. Alternatively, charged amino acids in the 137–139 region may garner direct functional contacts. Regardless, residues 154–155 and the invariant residues in the 137–139 region are likely to be required to support the same SmpB function.
Strictly speaking, it is also conceivable that the observed ribosome association phenotypes do not reflect functional binding of SmpB·SsrA·EF-Tu·GTP quarternary complexes with the ribosomal A site. SmpB and SsrA may be copurifying with ribosomes due to nonspecific protein–RNA or RNA–RNA interactions. That the ribosomes are purified under stringent salt conditions (0.5 M NH4Cl) argues against this possibility. The observation that SsrA fails to copurify with ribosomes when the truncated SmpB variant SmpB59 is expressed indicates even more strongly that the observed in vivo ribosome association results reflect specific functional binding of quarternary complexes to ribosomes. We have performed ribosome purifications by using three different high-stringency protocols. All SmpB C-terminal tail mutants tested support association of the SmpB·SsrA complex with ribosomes regardless of the purification protocol used. Even when ribosomes are purified by sedimenting tight-coupled 70S ribosomes through a high salt sucrose cushion (0.5 M NH4Cl), followed by isolation of the 70S peak through a linear sucrose gradient in high salt (0.3 M NH4Cl), the mutant SmpB·SsrA complexes remain bound to the ribosome (data not shown). This provides strong evidence that the observed ribosome association phenotypes reflect specific interactions with the ribosome.
When a ternary complex of cognate tRNA with EF-Tu·GTP is brought to the ribosomal A site, proper codon–anticodon interactions somehow trigger conformational changes that activate the GTPase domain of EF-Tu. Rapid GTP hydrolysis is followed by release of EF-Tu·GDP and accommodation of the tRNA acceptor stem into the peptidyl transferase center (19–21). Considering the structural and mechanistic similarities between tRNAs and the tRNA-like domain of SsrA, one would presume that accommodation of SsrA would proceed much like that of tRNAs. However, SsrA lacks a traditional anticodon stem loop, and thus codon–anticodon interactions are necessarily absent from the SsrA accommodation step (2, 3, 22). It is unclear what mechanistic events trigger GTP hydrolysis by EF-Tu when a cognate tRNA is presented to the ribosomal A site. Likewise, it is unclear what mechanistic events trigger GTP hydrolysis by EF-Tu when SsrA RNA is presented to the ribosomal A site. It is possible that the C-terminal tail region of SmpB plays a direct or indirect role in GTPase activation. Karzai et al. (6) first suggested the hypothesis that SmpB might serve as a tRNA anticodon stem loop mimic in a manner analogous to domain IV of EF-G. Our results demonstrate that the C-terminal tail of SmpB is not required for initial binding of the SsrA-EF-Tu-GTP complex to stalled ribosomes. However, this region might gain structure in the context of the ribosome to mediate specific contacts with ribosomal elements necessary for proper positioning or accommodation of SsrA RNA into the A site. Indeed, there is extensive precedent in the literature to support the idea that a ribosome-associated protein may contain extensions that are unstructured in solution but gain structure within the context of the ribosome (23–25). Deletion of residues I154 and M155, introduction of negative charges at these positions, or substitution of the invariant C-terminal tail amino acids D137, K138, and K139 may destabilize interactions of the SmpB C-terminal tail with rRNA or ribosomal proteins near the decoding center, explaining the observed loss of SmpB·SsrA-mediated tagging activity of these mutants (Fig. 1 and Table 1). Support for this hypothesis comes from the cocrystal structure model of SmpB·SsrA tRNA-like domain. Docking SmpB·SsrA into the ribosomal A site, Gutman et al. (11) orient SmpB provocatively toward the decoding center but not in close enough proximity to make direct contacts. Consistent with our observation, the authors postulate that the positively charged SmpB C terminus may extend into the decoding center and act as an anticodon mimic (11). The cryo-EM reconstruction of SmpB·SsrA interactions with the ribosomal A site before accommodation suggests another attractive possibility. At the current level of resolution, this model does not permit prediction of specific interactions; however, SmpB is shown to be in close proximity to helix 69 of 23S rRNA. Valle et al. (26) postulate that interactions between the D stem of traditional tRNAs and the 50S subunit are mediated through SmpB protein when SsrA interacts with the ribosome. That is, SmpB, bound to SsrA, makes contacts with 23S rRNA analogous to those made by traditional tRNAs (26). Differences between these two models are expected, because they represent different states of SsrA–A site interactions. Hence, one possible explanation for the observed phenotype of the SmpB C-terminal tail mutants is that they are defective in properly engaging ribosomal elements responsible for eliciting EF-Tu GTPase activity.
It is also possible that mutations to the SmpB C terminus affect the protein's ability to support a step in trans-translation downstream of GTP hydrolysis. Contacts made by the SmpB C-terminal tail may be required for proper positioning of the SsrA acceptor stem in the peptidyl transferase center during accommodation. Alternatively, these contacts could play a role in the establishment of the SsrA-encoded reading frame. The precise mechanistic role of the SmpB C-terminal tail remains to be elucidated; however, the results presented in this study clearly demonstrate that SmpB performs an essential function in trans-translation that extends beyond specific association and transport of SsrA to the ribosomal A site.
The results of this study demonstrate a previously uncharacterized role for SmpB in the transtranslation mechanism, a role beyond binding SsrA RNA, delivering it to the ribosome, and promoting its stable association. Taken together with previous structural data, our results support the notion that the SmpB C-terminal extension plays a crucial role in proper engagement and accommodation of SsrA RNA in the ribosomal A site.
Supplementary Material
Acknowledgments
We thank Dr. Charles McHenry (University of Colorado Health Science Center, Boulder) for providing materials, Drs. Rolf Sternglanz and James Bliska for insightful comments on the manuscript, and members of the Karzai laboratory for helpful discussions. This research was supported by grants from National Institutes of Health and the Pew Scholars Award (to A.W.K.).
Author contributions: T.R.S., D.P.D., and A.W.K. designed research; T.R.S., D.P.D., H.J.C., and A.W.K. performed research; T.R.S., D.P.D., H.J.C., and A.W.K. contributed new reagents/analytic tools; T.R.S., D.P.D., and A.W.K. analyzed data; and T.R.S. and A.W.K. wrote the paper.
Abbreviations: tmRNA, transfer messenger RNA; SmpB, small protein B.
References
- 1.Keiler, K. C., Waller, P. R. & Sauer, R. T. (1996) Science 271, 990-993. [DOI] [PubMed] [Google Scholar]
- 2.Gillet, R. & Felden, B. (2001) Mol. Microbiol. 42, 879-885. [DOI] [PubMed] [Google Scholar]
- 3.Karzai, A. W., Roche, E. D. & Sauer, R. T. (2000) Nat. Struct. Biol. 7, 449-455. [DOI] [PubMed] [Google Scholar]
- 4.Withey, J. & Friedman, D. (1999) J. Bacteriol. 181, 2148-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Withey, J. H. & Friedman, D. I. (2003) Annu. Rev. Microbiol. 57, 101-123. [DOI] [PubMed] [Google Scholar]
- 6.Karzai, A. W., Susskind, M. M. & Sauer, R. T. (1999) EMBO J. 18, 3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimizu, Y. & Ueda, T. (2002) FEBS Lett. 514, 74-77. [DOI] [PubMed] [Google Scholar]
- 8.Hanawa-Suetsugu, K., Takagi, M., Inokuchi, H., Himeno, H. & Muto, A. (2002) Nucleic Acids Res. 30, 1620-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, G., Nowakowski, J. & Hoffman, D. W. (2002) EMBO J. 21, 1845-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Someya, T., Nameki, N., Hosoi, H., Suzuki, S., Hatanaka, H., Fujii, M., Terada, T., Shirouzu, M., Inoue, Y., Shibata, T., et al. (2003) FEBS Lett. 535, 94-100. [DOI] [PubMed] [Google Scholar]
- 11.Gutmann, S., Haebel, P. W., Metzinger, L., Sutter, M., Felden, B. & Ban, N. (2003) Nature 424, 699-703. [DOI] [PubMed] [Google Scholar]
- 12.Karzai, A. W. & Sauer, R. T. (2001) Proc. Natl. Acad. Sci. USA 98, 3040-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berggrun, A. & Sauer, R. T. (2000) J. Mol. Biol. 301, 959-973. [DOI] [PubMed] [Google Scholar]
- 14.Vila-Sanjurjo, A., Ridgeway, W. K., Seymaner, V., Zhang, W., Santoso, S., Yu, K. & Cate, J. H. (2003) Proc. Natl. Acad. Sci. USA 100, 8682-8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Retallack, D. M., Johnson, L. L. & Friedman, D. I. (1994) J. Bacteriol. 176, 2082-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barends, S., Karzai, A. W., Sauer, R. T., Wower, J. & Kraal, B. (2001) J. Mol. Biol. 314, 9-21. [DOI] [PubMed] [Google Scholar]
- 17.Barends, S., Bjork, K., Gultyaev, A. P., de Smit, M. H., Pleij, C. W. & Kraal, B. (2002) FEBS Lett. 514, 78-83. [DOI] [PubMed] [Google Scholar]
- 18.Williams, K. P., Martindale, K. A. & Bartel, D. P. (1999) EMBO J. 18, 5423-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark, B. F. & Nyborg, J. (1997) Curr. Opin. Struct. Biol. 7, 110-116. [DOI] [PubMed] [Google Scholar]
- 20.Ogle, J. M., Brodersen, D. E., Clemons, W. M., Jr., Tarry, M. J., Carter, A. P. & Ramakrishnan, V. (2001) Science 292, 897-902. [DOI] [PubMed] [Google Scholar]
- 21.Ramakrishnan, V. (2002) Cell 108, 557-572. [DOI] [PubMed] [Google Scholar]
- 22.Withey, J. H. & Friedman, D. I. (2002) Curr. Opin. Microbiol. 5, 154-159. [DOI] [PubMed] [Google Scholar]
- 23.Helgstrand, M., Rak, A. V., Allard, P., Davydova, N., Garber, M. B. & Hard, T. (1999) J. Mol. Biol. 292, 1071-1081. [DOI] [PubMed] [Google Scholar]
- 24.Brodersen, D. E., Clemons, W. M., Jr., Carter, A. P., Wimberly, B. T. & Ramakrishnan, V. (2002) J. Mol. Biol. 316, 725-768. [DOI] [PubMed] [Google Scholar]
- 25.Hoang, L., Fredrick, K. & Noller, H. F. (2004) Proc. Natl. Acad. Sci. USA 101, 12439-12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valle, M., Gillet, R., Kaur, S., Henne, A., Ramakrishnan, V. & Frank, J. (2003) Science 300, 127-130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.