Abstract
The development of kidney disease is influenced by both genetic and environmental factors. Searching for models of glomerulopathy that display strong gene–environment interaction, we examined the determinants of anthracycline-induced nephropathy, a classic, strain-dependent experimental model applied to rodents in the past four decades. We produced three crosses derived from mice with contrasting susceptibility to doxorubicin (DOX) nephropathy and, surprisingly, we found that this widely studied model segregates as a single-gene defect with recessive inheritance. By genome-wide analysis of linkage, we mapped the trait locus to chromosome 16A1-B1 (DOXNPH locus) in all three crosses [peak logarithm of odds (lod) score of 92.7, P = 1 × 10-65]; this interval represents a susceptibility locus for nephropathy. Gene expression analysis indicated that susceptibility alleles at the DOXNPH locus are associated with blunted expression of protein arginine methyltransferase 7 (Prmt7) on chromosome 8, a protein previously implicated in cellular sensitivity to chemotherapeutic agents (lod = 12.4, P = 0.0001). Therefore, Prmt7 expression serves as a molecular marker for susceptibility to DOX nephropathy. Finally, increased variation in the severity of kidney disease among affected mice motivated a second genome-wide search, identifying a locus on chromosome 9 that influences the severity and progression of nephropathy (DOXmod, peak lod score 4.3, P = 0.0018). These data provide genetic and molecular characterization of a previously unrecognized Mendelian trait. Elucidation of DOX nephropathy may simultaneously provide insight into the pathogenesis of renal failure and mechanisms of cytotoxicity induced by chemotherapeutic agents.
Keywords: genetics
Diseases of the glomerulus, the filtration unit of the kidney, constitute a major cause of nephropathy and end-stage renal failure worldwide (1). Glomerulopathies have complex determination, encompassing idiopathic diseases, hereditary syndromes, or secondary disorders that develop consequent to systemic illness, exposure to infectious agents, or xenobiotics (1). Recently, genetic studies in humans and animal models have begun to clarify the pathogenesis of glomerulopathies, implicating defects in proteins that are predominantly expressed by the glomerular visceral epithelial cells, the podocytes (2, 3). Glomerular podocytes are essential to the integrity of the glomerular filtration barrier: genetic or acquired defects in these cells cause the nephrotic syndrome, a common cause of end-stage renal failure characterized by fusion of podocyte foot processes and clinically associated with the development of massive proteinuria and glomerulosclerosis (2, 3).
The occurrence of podocyte injury and nephropathy varies considerably among individuals exposed to the same systemic illness (e.g., diabetes) or environmental triggers (e.g., lithium or HIV infection). These observations are recapitulated by many experimental models of disease, where the development of podocyte injury due to toxins or immune stimuli depends on the use of specific strains or species (4). These data, together with the finding of increased familial aggregation in humans (5), suggest that genetic predisposition is an important requirement for expression of secondary nephropathies. Genetic and environmental components can be best studied in animal models, where they can be more easily controlled and distinguished. Hence, to investigate the pathogenesis of glomerular diseases, we revisited classic experimental models that exhibit strain dependence and would therefore be amenable to genetic analysis.
Among these models, nephropathy induced by anthracycline antibiotics such as daunorubicin or doxorubicin stands out because it has been the prototypical experimental model of primary focal segmental glomerulosclerosis, one of the major causes of end-stage renal failure (6–11). First described in 1970, anthracycline-mediated nephropathy has been applied in >400 studies cited in Medline (4, 6). Because it is reproducible and enables precise timing of the onset of renal injury, this model has been widely used to dissect the mechanisms that promote initiation and progression of nephropathy and test the efficacy of various therapies in preventing the deterioration of renal function (4, 6). This trait is relevant to investigations of glomerulopathies because numerous studies indicate that glomerular podocytes are the site of the initial insult in this model (6–11). Moreover, rare cases of proteinuria and nephropathy have been reported in association with anthracycline therapy in humans, suggesting that the susceptibility observed in some rodent strains has a human counterpart (12, 13).
In susceptible strains of rats and mice, a single i.v. injection of an anthracycline produces proteinuria and progressive renal disease within 5–7 days of exposure; histopathologically, animals exhibit early podocyte foot process fusion, followed by the development of typical lesions of focal segmental glomerulosclerosis, and progressive global sclerosis and interstitial fibrosis (6–11). Direct exposure of the kidneys to anthracyclines during i.v. administration is a requirement for the development of nephropathy, suggesting that this trait is independent of extrarenal drug metabolism (7, 14). Although many mechanisms have been suggested, the primary determinants of renal injury in anthracycline nephropathy have not been identified (4, 15, 16). Thus, elucidation of this trait may provide insight into pathways mediating podocyte injury and nephropathy. Moreover, this model has the advantage of allowing simultaneous investigation into the mechanisms of cytotoxicity mediated by anthracyclines.
We took advantage of contrasting susceptibility to doxorubicin (Adriamycin, DOX) nephropathy between mouse strains to produce segregating crosses that can identify loci imparting susceptibility to this trait (11). Herein, we demonstrate that genetic analysis of this old model constitutes a powerful approach for gaining insight into the pathogenesis of end-stage renal failure and understanding defense pathways against xenobiotics.
Materials and Methods
Animal Breeding. We purchased the following 6- to 8-week-old mice from Jackson Laboratories: BALB/cJ, C57BL6/J, FVB/NJ, 129/SvJ, and CAST/EiJ mice (BALB, B6, FVB, 129, and CAST hereafter). F1 hybrids were produced by intercrossing the parent strains. We produced two backcrosses [(BALB × B6) F1 × BALB] and [(BALB × FVB) F1 × BALB] and one intercross [(BALB × B6) F1 × (BALB × 129) F1]. DOX nephropathy was produced by injecting DOX 10 mg/kg by tail vein at 8 weeks of age. We tested 8–20 mice (equal number of males and females) from each inbred strain and F1 hybrid; two saline injected animals matched for strain served as the control. Fifteen days after DOX injection, animals were killed for histologic analysis of kidneys, spontaneously voided urine was collected for urinalysis, and blood was collected by cardiac puncture for determination of blood urea nitrogen (BUN). Animals that were moribund were killed before the 15-day end point. The protocol was approved by the Institutional Animal Care and Use Committee at Columbia University.
Phenotypic Characterization. Proteinuria was measured by spot urine dipsticks (Roche Diagnostics), which quantitated levels as 0, 0.5 (trace), 1+, 2+, and 3+. Normal range in saline-treated mice was 0–2+. Kidneys sections were stained with hematoxylin/eosin and periodic acid/Schiff reagent. Renal histology was scored independently by two investigators (A.G.G. and K.A.F.) blinded to genetic background and genotype; discrepancies between scores were resolved by a third expert pathologist (V.D.D.). On each slide, we examined at least 100 glomeruli and scored four traits: glomerular injury, tubular cyst/cast formation, podocyte hyperplasia, and interstitial inflammation. We used the following semiquantitative scale: 0, no disease; 1, 1–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100% of tissue affected. In our first genome screen, we applied dichotomous criteria to define affection status: mice with at least 3+ proteinuria and histologic evidence of ≥5% glomerulosclerosis at the time of death were classified as affected. Mice with <3+ proteinuria and normal histology were classified as unaffected. Application of these phenotypic criteria yielded a clear dichotomous distribution in the backcross, distinguishing affected from unaffected mice. Of 312 backcross mice bred, only four fell into an indeterminate category. Because indeterminate results may have been due to a suboptimal DOX injection, these four mice were not used in genetic analyses. In the second genome screen to search for modifier loci, we used the numerical average of the four histology scores (total histology score) and BUN as primary phenotypes for qtl mapping (see below). Phenotypic data were adjusted for the interval spanning DOX injection and the day of killing (computation of residuals). This adjustment was necessary because some animals died before the 15-day end point or were killed early to avoid pain and distress (range 9–15 days), reducing the time interval separating DOX injection to killing date and, therefore, providing less opportunity for disease progression.
Genotyping and Analysis of Linkage. Genotyping was performed by using informative microsatellite markers distributed across the mouse genome; fluorescent primers were used to direct PCR from genomic DNA, and products were analyzed on a capillary sequencer. Marker positions were obtained from the Broad Institute, Mouse Genome Informatics (The Jackson Laboratory), and National Center for Biotechnology Information web sites and the Mouse single-nucleotide polymorphism (SNP) database (http://mousesnp.roche.com).
We conducted a first genome-wide screen with 62 informative markers to detect the major DOX nephropathy susceptibility locus. To search for this Mendelian locus, we applied the dichotomous, affection status criteria described above. For linkage analysis, we followed homozygosity for BALB alleles at each marker among affected and unaffected mice and calculated P values from Fisher's exact tests; pairwise logarithm of odds (lod) scores were manually calculated by counting recombinants and confirmed by using the Map Manager qtx program (17).
A second genome-wide screen with 112 informative markers was conducted to detect QTLs that modify the severity of disease in affected animals. For this second genome screen, primary phenotypes included BUN and the total renal histology score as quantitative traits. Pairwise and multipoint lod scores and the proportion of the variance of each trait explained by each trait locus were calculated by using Map Manager qtx under an additive model. Additional genotyping was performed for intervals that showed lod scores of ≥1; thus, we typed a total of 134 informative markers in this second screen. Traditional thresholds for significance in the backcross were used (lod score > 3.3) (18). We also conducted permutation tests in which the observed sets of phenotypes for each animal were randomized on the genotypes; 10,000 replicates were performed to determine the empirical significance of the linkage findings. In secondary analyses, we performed composite interval mapping and two-locus interaction analyses in an attempt to detect additional trait loci.
Microarray Analysis. We used the Affymetrix 430A array to analyze gene expression in whole kidneys from the B6 and BALB mice: three DOX-treated mice of each strain were compared with two saline-treated counterparts (two-way factorial design, total of 10 arrays). Kidneys were profiled 3 days after DOX injection (before development of proteinuria and any histological changes in BALB mice) to ensure that observed differences were not attributable to proteinuria or tissue sclerosis. Sample preparation, labeling, and hybridization were performed according to Affymetrix-recommended procedures (www.affymetrix-.com). Gene expression data were compared by two-way analysis of variance by using the r statistical language, specifically searching for differentially expressed genes that demonstrate interaction between strain and DOX treatment.
Measurement of Gene Expression by Real-Time PCR. Total kidney RNA was extracted by using TRIzol reagent (Invitrogen) and further purified by using the RNeasy Mini Kit with the RNase-Free DNase Set (Qiagen, Valencia, CA), and cDNA was generated with the Omniscript kit (Qiagen). We quantitated gene expression in triplicate by real-time PCR on a Bio-Rad MyiQ Thermal Cycler with SYBR Green (Bio-Rad) reagent. Primer sequences were as follows: Prmt7-sense TTGCGGTGACTGCGAAGG, Prmt7-antisense: GAGGCTTGGAGAGGCTTCTG; syndecan4-sense GTATCCATGTCCAGCACTG, syndecan4-antisense ATGCGTAGAACTCATTGGTG; β-actin-sense: CACCACACCTTCTACAATGA, β-actin-antisense: AGCCTGGATGGCTACGTACAC. We generated standard curves by using serial 10-fold dilutions of kidney cDNA to measure amplification efficiencies and calculate the starting amounts of target gene and β-actin transcript in test samples. Calculation of gene expression level was performed by using both the standard curve and the ΔΔCt methods; these methods gave similar results because reaction efficiencies were equal between target genes and β-actin. Target gene expression, normalized to β-actin expression, is reported as fold change over the same saline-treated B6 kidney sample that was included in each run. Gene expression in time course experiments was analyzed by global ANOVA, followed by post hoc comparisons between strains and time points (Dunnett's C test). We also used Prmt7 gene expression as a quantitative phenotype for analysis among a subset of backcross mice for which both genotyping data and gene expression data were available.
In Situ Hybridization. In situ hybridization was performed as previously described in developing (embryonic day 15.5) and adult kidneys by using digoxigenin-labeled riboprobes (19). cDNA encoding a fragment from mouse Prmt7 (GenBank entry no. AY673972, position 95–1395) was amplified by PCR and cloned into pBluescript KS (+) by directional cloning with linkers. The resulting plasmid was linearized with SacI, and T3 was used to produce riboprobes, which were subsequently fragmented by alkaline hydrolysis.
Results
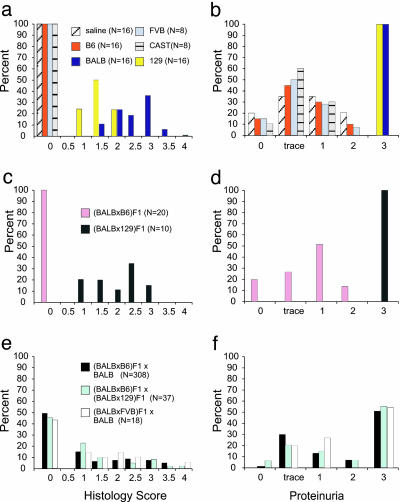
DOX Nephropathy Is a Mendelian Trait That Maps to Chromosome 16A1-B1. We first tested five inbred strains for susceptibility to DOX nephropathy. BALB and 129 mice demonstrate severe susceptibility with the onset of proteinuria (3+) ≈5 days after injection and progression to significant glomerulosclerosis within 14 days of injections (Figs. 1 and 2), similar to previous descriptions in BALB (11). DOX injury was specific to the kidneys because these susceptible strains showed no histologic evidence of tissue injury in the lungs, livers, or hearts in the tested interval. In contrast, B6, FVB, and CAST mice are completely resistant to DOX nephropathy with renal histology and proteinuria indistinguishable from saline-treated controls (Figs. 1 and 2); these protected strains did not demonstrate any evidence of renal injury when examined up to 8 weeks after injection. Presented with these clear contrasting phenotypes, we produced F1 hybrids by breeding the susceptible BALB to the resistant B6 strain to study the dominance pattern. Similar to B6, (BALB × B6) F1 hybrids were completely resistant to DOX nephropathy (Fig. 2 c and d). These data suggested that BALB alleles are recessive to B6 alleles and that a (BALB × B6) F1 × BALB backcross would enable the estimation of the number of BALB loci contributing to the trait. We therefore generated 308 (BALB × B6) F1 × BALB backcross mice and tested them for the development of DOX nephropathy. To our surprise, we found that 157 backcross mice (50.9%) were affected, developing 3+ proteinuria and at least 5% glomerulosclerosis with tubular casts. In contrast, the remaining backcross mice were phenotypically indistinguishable from saline-treated controls (Fig. 2 e and f). This bimodal distribution conforms to expectations under autosomal recessive inheritance of a single gene and suggests that DOX susceptibility in mice is a Mendelian trait.
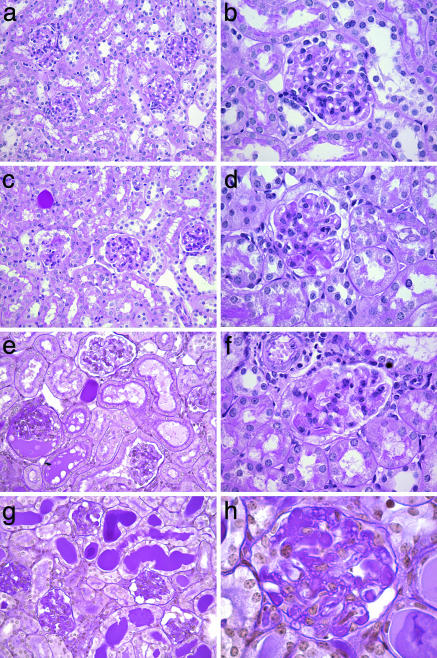
Fig. 1.
Renal pathology in DOX nephropathy backcross mice. Periodic acid/Schiff reagent-stained kidney sections of (BALB × B6) F1 × BALB backcross mice with varying severity of disease are shown. (Left) Low power views. (Right) Higher power magnification of glomeruli from the same mouse. a and b are from a mouse with no detectable disease, and c–f are from mice with progressively increasing severity of histologic renal disease. Histology scores: c and d = 1; e and f = 2.1; g and h = 3. The affected kidneys show microcystic tubular dilatation, tubular casts, and focal segmental glomerulosclerosis. Glomerular damage progresses from segmental sclerosis (in d and f) to global glomerulosclerosis.
Fig. 2.
Distribution of phenotypes in DOX-treated mice. Histograms depicting the distribution of phenotypic scores among inbred strains (a and b), F1 hybrids (c and d), and mapping cohorts (e and f). Left shows the histology scores (range 0–4); Right shows proteinuria (range 0–3). The bars show the percent of mice in each category from each strain. The saline-treated mice are two each from inbred and F1 hybrid strains. The number (N) of mice in each group is indicated in parentheses.
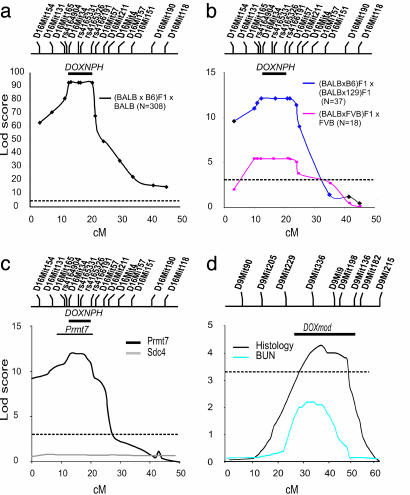
To map this putative Mendelian locus, we performed a genome-wide scan in 42 backcross mice (31 affected and 11 unaffected) by using 62 informative markers distributed across the mouse genome. Linkage was initially detected based on strong departure from the null distribution at D16Mit154, on the telomeric portion of chromosome 16, with homozygosity for BALB alleles in 29 of 31 affected mice but none of the unaffected mice. Additional genotyping confirmed linkage to the 26-centimorgan interval between D16mit154 and D16Mit157 in these mice (lod score = 9.2, Fisher exact P value < 1 × 10-5). We found no evidence of linkage elsewhere in the genome, with the next highest lod score observed of 0.9 at D12Mit28. We further genotyped 15 loci in the interval and identified five informative markers. Genotyping of informative markers in all 308 (BALB × B6) F1 × BALB backcross mice confirmed the initial localization of the DOX nephropathy gene, yielding a peak lod score of 92.7 (P = 7.2 × 10-69) on chromosome 16A1-B1 (Fig. 3a and Table 1, which is published as supporting information on the PNAS web site). These data are robust to quantitative analysis of the DOX nephropathy phenotype: analyzed as quantitative traits, the renal histology score and proteinuria yield lod scores of 153 and 170, respectively, at this interval, accounting for ≈50% of the variance in each phenotype. These linkage data confirm that susceptibility to DOX nephropathy in the mouse is a Mendelian trait with recessive inheritance; we have named this chromosome 16 locus DOX-NPH. Analysis of haplotypes in recombinant mice refined the linkage to the ≈14-Mb interval between D16Mit165 and rs4166191 (Fig. 2a).
Fig. 3.
lod plots in mapping cohorts. Map distance is presented in centimorgans. The positions of informative markers genotyped are shown above the plot. The dashed horizontal bars indicate the threshold for genome-wide significance for a complex trait (lod = 3.3). The number (N) of animals in each cross is indicated in parentheses. The thick solid bar above the lod plots in a–c indicates the minimal recombinant interval combined from the three crosses. (a) Single-point lod score plot for linkage of DOX nephropathy to chromosome 16A1-B1 in the (BALB × B6) F1 × BALB backcross. (b) Single-point lod score plot for the (129 × BALB) F1 × (BALB × B6) F1 intercross and (BALB × FVB) F1 × BALB backcross mice chromosome 16A1-B1. (c) Multipoint lod score plots on chromosome 16 for renal Prmt7 and sdc4 expression in 149 (BALB × B6) F1 × BALB backcross mice. The thin solid bar above the lod plot indicates the 95% confidence interval for Prmt7 expression computed by bootstrap resampling. (d) Mapping a modifier gene for DOX nephropathy to chromosome 9. Multipoint lod score plots for histology score and BUN on chromosomes 9 for 157 affected (BALB × B6) F1 × BALB backcross mice are shown. The solid bar above the chromosome 9 lod plot indicates the 95% confidence interval for DOXmod computed by bootstrap resampling (histology score).
We next produced additional crosses to determine whether resistance or susceptibility in other tested strains were attributable to variation at the DOXNPH locus. If the resistant phenotype in FVB is also due to resistance alleles at the DOXNPH locus, crosses between BALB and FVB should demonstrate linkage to DOXNPH. We therefore used the FVB as the counterstrain to generate a (BALB × FVB) F1 × BALB backcross. This cross also demonstrated a bimodal distribution in phenotype, with inheritance of FVB alleles at the DOXNPH interval providing protection from DOX nephropathy (lod = 5.4, P = 2 × 10-5; Figs. 2 e and f and 3b). These data demonstrate that protection from DOX nephropathy in both FVB and B6 strains are due to resistance alleles at the DOXNPH locus.
We next performed a complementation test to determine whether the susceptible 129 strain shares the same susceptibility gene(s) as BALB. We therefore produced (BALB × 129) F1 hybrids: these mice are expected to develop DOX nephropathy if parental strains share the same susceptibility loci or, alternatively, demonstrate resistance if susceptibility loci differ between the parents. We found that (BALB × 129) F1 hybrids develop DOX nephropathy similar to the BALB and 129 strains (Fig. 2 c and d), providing evidence for noncomplementation and shared susceptibility gene(s) between parental strains. We next confirmed linkage to the DOXNPH in the 129 strain by using an intercross between the susceptible (BALB × 129) F1 and the resistant (BALB × B6) F1 hybrids (Fig. 2 e and f). As predicted by the phenotype of the (BALB × 129) F1 hybrid, we again observed Mendelian distribution of the trait; linkage confirmed that the BALB/BALB and BALB/129 genotypes at the DOX-NPH locus confer susceptibility to DOX nephropathy (lod = 11.4, Fisher exact P = 3 × 10-11; Fig. 3b). Together, these data provide unequivocal evidence of linkage at the DOXNPH locus in the BALB and 129 strains and demonstrate that the trait is likely to be genetically homogeneous for these two susceptible strains.
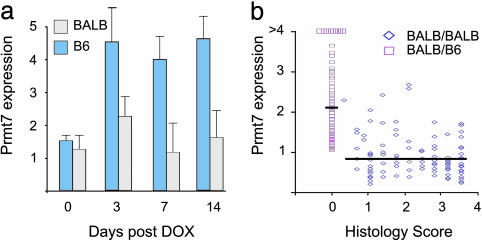
DOXNPH Alleles Predict Expression of Prmt7 in DOX Nephropathy. To gain better insight into molecular pathways characterizing genetic susceptibility to DOX nephropathy, we performed gene expression analysis of whole mouse kidneys in mice with contrasting susceptibility. We found 15 genes with significant evidence of interaction between strain and DOX treatment (Table 2, which is published as supporting information on the PNAS web site), but none of these genes localize to our linkage intervals. However, we further characterized one of these genes, protein arginine methyltransferase 7 (Prmt7, located on chromosome 8) because it had been identified in genetic screens for susceptibility to chemotherapeutic cytotoxicity and might therefore participate in the pathogenesis of disease upstream or downstream from the DOXNPH gene (20). In developing kidneys (embryonic day 15.5), Prmt7 expression is confined to the nephrogenic zone in the cortical region, where the tips of the ureteric bud induce de novo formation of epithelia in the metanephric mesenchyme and early glomeruli (Fig. 5, which is published as supporting information on the PNAS web site). Prmt7 expression was ≈8-fold lower in adult kidneys, resulting in suboptimal staining by in situ hybridization; therefore, we quantified Prmt7 expression in the adult by real-time PCR. Time course experiments show that Prmt7 expression is up-regulated in B6 after DOX exposure but remains unchanged in BALB (Fig. 4a; ANOVA P = 0.0001, post hoc P < 0.005 between DOX-treated B6 and BALB groups). These expression differences are observed before the development of proteinuria and histologic abnormalities that occur at days 5–7, suggesting the development of tissue injury and cell death are not responsible for blunted Prmt7 expression in BALB. These data are further confirmed in (BALB × B6) F1 × BALB backcross mice (n = 149; Fig. 4b): by using Prmt7 expression as a quantitative phenotype, we performed a genome-wide analysis of linkage in the backcross and found strong evidence of linkage of Prmt7 expression to the DOXNPH locus on chromosome 16 (peak lod = 12.4 at D16Mit34, 37% of variance explained; Fig. 3c). Thus, Prmt7 expression in response to DOX injection is predicted by the genotype at the DOXNPH locus and serves as a marker for DOX nephropathy in this cross. We found no other loci significantly contributing to Prmt7 expression, including the murine Prmt7 locus on chromosome 8. We also measured the expression of Sdc4, another differentially expressed gene previously implicated in experimental nephropathy (21), but found no linkage to the DOXNPH locus (Fig. 3c; maximum lod = 0.3) or elsewhere in the genome in backcross mice. These data show multifactorial determination of Sdc4 expression and argue in favor of the specificity of the association of Prmt7 expression with DOX nephropathy.
Fig. 4.
Prmt7 gene expression. Whole kidney Prmt7 expression was measured by real-time RT-PCR and is presented as fold expression relative to a baseline B6 mouse. (a) Time course for Prmt7 gene expression in BALB and B6 mice treated at day 0 with DOX. Each bar represents the average expression in six mice measured in triplicate. ANOVA, P = 0.0001; *, P < 0.005 B6 vs. BALB by post hoc Dunnett's C test. (b) Distribution of renal Prmt7 expression vs. histology score in 149 (BALB × B6) F1 × BALB backcross mice. Mice with BALB/B6 genotype (□) at the DOXNPH locus (rs4165326) have significantly higher Prmt7 expression values than mice with BALB/BALB genotypes (⋄); P = 3 × 10-8 by two-sided t test. The horizontal lines represent the mean values for each genotypic class.
The Severity of DOX Nephropathy Among Affected Mice Is Determined by a Modifier Locus on Chromosome 9. The large variation in phenotypic severity among affected (BALB × B6) F1 × BALB backcross mice compared with inbred BALB mice implied additional genetic variance attributable to the existence of loci that modify the severity or progression of renal disease (Fig. 2 e and f). To identify these modifier loci, we performed a second genome screen in affected (BALB × B6) F1 × BALB backcross mice, conditioning on the presence of the BALB/BALB genotype at the DOXNPH locus. We compared the inheritance of each segment of each chromosome with the inheritance of quantitative renal disease phenotypes and found strong evidence for linkage across chromosome 9 with a peak lod score between D9Mit336 and D9Mit9 (positioned at 34 and 42.6 centimorgans on the Whitehead Institute linkage map, respectively). The total histology scores showed the strongest evidence of linkage (lod score 4.3; Fig. 3d); however, the linkage to this chromosome segment is robust to all other component histology scores and BUN (lod 2.1; Fig. 3d). Based on published lod thresholds in a backcross, the histology score has genome-wide significance, whereas the score for BUN is suggestive. The lower lod score with BUN is explained by the finding that many severely affected mice with global glomerulosclerosis had moderately elevated BUN levels, likely secondary to poor oral intake. Permutations of phenotypes on genotypes to calculate genome-wide empiric thresholds (10,000 replicates) confirmed that the data for the total histology score are highly significant (P value = 0.0018). Evaluation of the 95% confidence interval, determined by bootstrap resampling, localizes this trait locus to a 30-centimorgans interval (≈52 Mb) delimited by D9Mit229 to D9Mit182. At this modifier locus, which we named DOXmod, homozygosity for BALB alleles confers ≈30% increase in the histology score. Secondary analyses did not reveal evidence for other loci modifying the progression of renal disease.
Discussion
Genetic factors are implicated in the development of nephropathy and susceptibility to cytotoxicity induced by chemotherapeutic agents (5, 22, 23). Both of these traits are generally considered to have multifactorial determination, complicating efforts to distinguish contributions of single genes. Anthracycline-induced nephropathy represents an interesting and unusual combination of these phenotypes, revealing a unique vulnerability to anthracycline toxicity in renal podocytes of susceptible rodent strains and some humans (6–13). Prior data suggest that this trait is likely autonomous to the kidney: podocyte damage and glomerulosclerosis can be prevented by transient clipping of the renal artery during drug injection, suggesting that initial exposure to the anthracycline, rather than its metabolites, causes nephropathy (7, 14). Moreover, interstrain and interspecies variations in drug levels do not correlate with susceptibility to nephrosis, further suggesting that variation in extrarenal drug metabolism is not a major contributor to renal injury (24, 25). Because anthracyclines are naturally occurring antibiotic agents, we reasoned that this trait can serve as an excellent model for genetic investigation of podocyte susceptibility to environmental damage and that its elucidation would enable insight into pathways leading to nephropathy. We examined five inbred mouse strains and their F1 hybrids, which disclosed clear contrasting susceptibilities. To our surprise, mapping crosses demonstrated that this widely studied model segregates as a Mendelian trait with recessive inheritance; a genome-wide search provided unequivocal evidence of linkage to the same segment of chromosome 16 in the two susceptible strains identified. Our linkage data can now be pursued by additional meiotic mapping to refine the trait locus before proceeding with further annotation and sequence analysis of positional candidates. Such positional cloning approaches may require production of thousands of mice (26). However, recent data indicate that laboratory mice have few independent alleles, suggesting that traits mapped to the same interval in different strains can be identified by searching for shared chromosomal segments within the linkage interval (27). Hence, the finding of linkage of DOX nephropathy to the same interval in BALB and 129 strains strongly suggests the presence of an ancestral mutation predisposing to this trait in the laboratory mouse and implies the feasibility of linkage disequilibrium mapping for prioritizing positional candidates and accelerating gene identification efforts.
We found that expression of Prmt7, a recently characterized member of the protein arginine methyltransferases is increased in mice protected from DOX nephropathy. Differences in Prmt7 expression precedes clinical or histologic evidence of renal disease in BALB, suggesting that this impaired up-regulation does not occur secondary to the development of kidney injury. Prmt7 response to DOX injection is strongly predicted by the genotype at the DOXNPH locus (lod = 12.4), suggesting that Prmt7 may act downstream of the DOXNPH gene. However, this finding may still represent a secondary compensation without causal relationship. Thus, the mechanisms underlying the association between Prmt7 and DOX nephropathy remain to be established. Arginine methylation of proteins participates in many cellular processes such as protein–protein interaction, signal transduction, transcription, and RNA processing (28, 29). Methylation of doxorubicin has also been reported to decrease its cytotoxic potential (30), raising the possibility that direct methylation of doxorubicin by Prmt7 may mediate protection from kidney injury. The biological role and the natural substrate(s) for Prmt7 are yet unknown, but available data suggest that it is a type II methyltransferase (31, 32). Prmt7 has conserved orthologs in many species, including Drosophila menalogaster and Caenorhabditis elegans, and was recently identified in a screen of genes conferring sensitivity to many DNA damaging agents, suggesting a role in cellular response to xenobiotics (20). Interestingly, expression of Prmt5, the other known type II methyltransferase, is up-regulated in tubular epithelium in a model of ischemic renal injury (33), further suggesting a role for these enzymes in nephropathy. The role of arginine methylation in DOX nephropathy will likely be better clarified with the identification of the DOXNPH gene.
Prior studies, including the original observations of DOX nephropathy among outbred rats, have suggested that the mechanisms of initiation of renal disease are distinct from those influencing progression (10). Consistent with these data, we found that the development of nephrosis requires the presence of susceptibility alleles at DOXNPH, but the severity and progression of disease is further influenced by a modifier gene on chromosome 9. The effect of the DOXmod modifier was striking, resulting in nearly a 30% increase in the severity of renal injury in mice homozygous for BALB alleles. Interestingly, DOXmod overlaps completely with a progression locus in a murine model of Alport syndrome in which 129 alleles increase risk and B6 alleles confer protection against nephropathy (34). In addition, DOXNPH is syntenic to a renal progression locus identified in a rat model of polycystic kidney disease (35). This convergence of mapping data from three independent models raises the possibility that these loci may be identical and that DOXmod represents a progression locus for many forms of renal disease. The DOXmod interval is quite broad, requiring the development of congenic lines for further refinement. Nevertheless, the similarity in phenotypes between BALB, 129, and their F1 hybrids suggest that they share the same susceptibility alleles at DOXmod and that efforts to identify this modifier gene may also benefit from sequence comparison across multiple strains.
In summary, our data suggest that genetic approaches in the DOX nephropathy model may significantly extend our understanding of the mechanisms of initiation and progression of renal disease. Because cells often use similar molecular defenses against xenobiotics, elucidation of this model may also provide insight into biological pathways relevant to anthracycline cardiotoxicity and multidrug resistance in cancer, the main clinical problems associated with the use of anthracyclines in humans. Finally, these data motivate a careful evaluation of rare patients who develop renal disease in association with anthracycline therapy because this presentation may be a manifestation of underlying genetic susceptibility.
Supplementary Material
Acknowledgments
We thank Richard Lifton and Qais Al-Awqati for their insightful comments. A.G.G. was supported by a young investigator award from the Emerald Foundation, National Institutes of Health Grant K08 DK02610-01, and the Irving Clinical Scholar Program. This study was also supported by the Columbia Diabetes and Endocrinology Research Center (National Institutes of Health Grant P30-DK63608).
Author contributions: Z.Z. and A.G.G. designed research; Z.Z., K.M.S.-O., S.C., K.A.F., R.Z.F., P.P., J.B., V.D.D.A., and A.G.G. performed research; K.M.S.-O., S.C., and J.B. contributed new reagents/analytic tools; Z.Z., K.M.S.-O., K.A.F., P.P., J.B., V.D.D.A., and A.G.G. analyzed data; and Z.Z., K.M.S.-O., S.C., and V.D.D.A. wrote the paper.
Abbreviations: BUN, blood urea nitrogen; DOX, doxorubicin; lod, logarithm of odds.
References
- 1.U.S. Renal Data System (2004) USRDS 2004 Annual Data Report: Atlas of End-Stage Renal Disease in the United States (Nat. Inst. Health, Bethesda, MD).
- 2.Pollak, M. R. (2003) Semin. Nephrol. 23, 141-146. [DOI] [PubMed] [Google Scholar]
- 3.Mundel, P. & Shankland, S. J. (2002) J. Am. Soc. Nephrol. 13, 3005-3015. [DOI] [PubMed] [Google Scholar]
- 4.Fogo, A. B. (2003) Semin. Nephrol. 23, 161-171. [DOI] [PubMed] [Google Scholar]
- 5.Freedman, B. I., Soucie, J. M. & McClellan, W. M. (1997) J. Am. Soc. Nephrol. 8, 1942-1945. [DOI] [PubMed] [Google Scholar]
- 6.Sternberg, S. S. (1970) Lab. Invest. 23, 39-51. [PubMed] [Google Scholar]
- 7.Bertani, T., Cutillo, F., Zoja, C., Broggini, M. & Remuzzi, G. (1986) Kidney Int. 30, 488-496. [DOI] [PubMed] [Google Scholar]
- 8.Bertani, T., Poggi, A., Pozzoni, R., Delaini, F., Sacchi, G., Thoua, Y., Mecca, G., Remuzzi, G. & Donati, M. B. (1982) Lab. Invest. 46, 16-23. [PubMed] [Google Scholar]
- 9.Bertani, T., Remuzzi, G., Rocchi, G., Delaini, F., Sacchi, G., Falchetti, M. & Donati, M. B. (1984) Appl. Pathol. 2, 32-38. [PubMed] [Google Scholar]
- 10.Bertani, T., Rocchi, G., Sacchi, G., Mecca, G. & Remuzzi, G. (1986) Am. J. Kidney Dis. 7, 12-19. [DOI] [PubMed] [Google Scholar]
- 11.Wang, Y., Wang, Y. P., Tay, Y. C. & Harris, D. C. (2000) Kidney Int. 58, 1797-1804. [DOI] [PubMed] [Google Scholar]
- 12.Burke, J. F., Jr., Laucius, J. F., Brodovsky, H. S. & Soriano, R. Z. (1977) Arch. Intern. Med. (Moscow) 137, 385-388. [PubMed] [Google Scholar]
- 13.Sathiapalan, R. K., Velez, M. C., McWhorter, M. E., Irwin, K., Correa, H., Baliga, R. & Warrier, R. P. (1998) J. Pediatr. Hematol. Oncol. 20, 482-485. [DOI] [PubMed] [Google Scholar]
- 14.De Boer, E., Navis, G., Tiebosch, A. T., De Jong, P. E. & De Zeeuw, D. (1999) J. Am. Soc. Nephrol. 10, 2359-2366. [DOI] [PubMed] [Google Scholar]
- 15.Deman, A., Ceyssens, B., Pauwels, M., Zhang, J., Houte, K. V., Verbeelen, D. & Van den Branden, C. (2001) Nephrol. Dial. Transplant 16, 147-150. [DOI] [PubMed] [Google Scholar]
- 16.Lebrecht, D., Setzer, B., Rohrbach, R. & Walker, U. A. (2004) Nephrol Dial Transplant 19, 329-336. [DOI] [PubMed] [Google Scholar]
- 17.Manly, K. F., Cudmore, R. H., Jr., & Meer, J. M. (2001) Mamm. Genome 12, 930-932. [DOI] [PubMed] [Google Scholar]
- 18.Lander, E. & Kruglyak, L. (1995) Nat. Genet. 11, 241-247. [DOI] [PubMed] [Google Scholar]
- 19.Batourina, E., Choi, C., Paragas, N., Bello, N., Hensle, T., Costantini, F. D., Schuchardt, A., Bacallao, R. L. & Mendelsohn, C. L. (2002) Nat. Genet. 32, 109-115. [DOI] [PubMed] [Google Scholar]
- 20.Gros, L., Delaporte, C., Frey, S., Decesse, J., de Saint-Vincent, B. R., Cavarec, L., Dubart, A., Gudkov, A. V. & Jacquemin-Sablon, A. (2003) Cancer Res. 63, 164-171. [PubMed] [Google Scholar]
- 21.Ishiguro, K., Kadomatsu, K., Kojima, T., Muramatsu, H., Matsuo, S., Kusugami, K., Saito, H. & Muramatsu, T. (2001) Lab. Invest. 81, 509-516. [DOI] [PubMed] [Google Scholar]
- 22.Brown, D. M., Provoost, A. P., Daly, M. J., Lander, E. S. & Jacob, H. J. (1996) Nat. Genet. 12, 44-51. [DOI] [PubMed] [Google Scholar]
- 23.Watters, J. W., Kraja, A., Meucci, M. A., Province, M. A. & McLeod, H. L. (2004) Proc. Natl. Acad. Sci. USA 101, 11809-11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gustafson, D. L., Rastatter, J. C., Colombo, T. & Long, M. E. (2002) J. Pharm. Sci. 91, 1488-1501. [DOI] [PubMed] [Google Scholar]
- 25.Kimura, M., Takahasi, H., Ohtake, T., Sato, T., Hishida, A., Nishimura, M. & Honda, N. (1993) Nephron 63, 193-198. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, Y., Proenca, R., Maffei, M., Barone, M., Leopold, L. & Friedman, J. M. (1994) Nature 372, 425-432. [DOI] [PubMed] [Google Scholar]
- 27.Wade, C. M., Kulbokas, E. J., III, Kirby, A. W., Zody, M. C., Mullikin, J. C., Lander, E. S., Lindblad-Toh, K. & Daly, M. J. (2002) Nature 420, 574-578. [DOI] [PubMed] [Google Scholar]
- 28.Mowen, K. A., Tang, J., Zhu, W., Schurter, B. T., Shuai, K., Herschman, H. R. & David, M. (2001) Cell 104, 731-741. [DOI] [PubMed] [Google Scholar]
- 29.Mowen, K. A., Schurter, B. T., Fathman, J. W., David, M. & Glimcher, L. H. (2004) Mol. Cell 15, 559-571. [DOI] [PubMed] [Google Scholar]
- 30.Westendorf, J., Marquardt, H. & Marquardt, H. (1984) Cancer Res. 44, 5599-5604. [PubMed] [Google Scholar]
- 31.Lee, J. H., Cook, J. R., Yang, Z. H., Mirochnitchenko, O., Gunderson, S., Felix, A. M., Herth, N., Hoffmann, R. & Pestka, S. (October 19, 2004) J. Biol. Chem. 10.1074/jbc.M405295200. [DOI] [PubMed]
- 32.Miranda, T. B., Miranda, M., Frankel, A. & Clarke, S. (2004) J. Biol. Chem. 279, 22902-22907. [DOI] [PubMed] [Google Scholar]
- 33.Braun, M. C., Kelly, C. N., Prada, A. E., Mishra, J., Chand, D., Devarajan, P. & Zahedi, K. (2004) Am. J. Nephrol. 24, 250-257. [DOI] [PubMed] [Google Scholar]
- 34.Andrews, K. L., Mudd, J. L., Li, C. & Miner, J. H. (2002) Am. J. Pathol. 160, 721-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bihoreau, M. T., Megel, N., Brown, J. H., Kranzlin, B., Crombez, L., Tychinskaya, Y., Broxholme, J., Kratz, S., Bergmann, V., Hoffman, S., et al. (2002) Hum. Mol. Genet. 11, 2165-2173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.