Abstract
High levels of inbreeding cause populations to become composed of homozygous, inbred lines. High levels of homozygosity limit the effectiveness of recombination, and therefore, retard the rate of decay of linkage (gametic phase) disequilibrium (LD) among mutations. Inbreeding and recombination interact to shape the expected pattern of LD. The actual extent of nucleotide sequence level LD within inbreeding species has only been studied in Arabidopsis, a weedy species whose global range has recently expanded. In the present study, we examine the levels of LD within and between 18 nuclear genes in 25 accessions from across the geographic range of wild barley, a species with a selfing rate of ≈98%. In addition to examination of intralocus LD, we employ a resampling method to determine whether interlocus LD exceeds expectations. We demonstrate that, for the majority of wild barley loci, intralocus LD decays rapidly, i.e., at a rate similar to that observed in the outcrossing species, Zea mays (maize). Excess interlocus LD is observed at 15% of two-locus combinations; almost all interlocus LD involves loci with significant geographic structuring of mutational variation.
Keywords: nucleotide polymorphism, population structure, Wall's B, interlocus linkage disequilibrium, inbreeding
Under recurrent self-fertilization, the level of heterozygosity decays at the rate of one-half per locus, per generation (1). Thus, within a few generations a self-fertilizing population is expected to be entirely composed of a collection of homozygous lines. An important consequence of this mating system is an extreme reduction in the rate of effective recombination. Consequently, the decay of linkage disequilibrium (LD) will be arrested. This expectation has generally been borne out in studies of natural populations; within-population levels of LD for isozyme polymorphisms are generally higher in populations of self-fertilizing plants than in outcrossers (2, 3). Many plant species have a mixed mating system with a mixture of outcrossing and self-fertilization, where occasional outcross events produce new heterozygous lines, that within a few generations, sort out into homozygous lines (4). Under this scenario, LD decays at a rate that is a function not only of recombination distance but also the level of outcrossing (5–7).
It has long been argued that the evolutionary potential of predominantly self-fertilizing species is limited by both reduced genetic diversity and a reduction in potential for effective recombination (e.g., refs. 8 and 9, reviewed in ref. 10). Recombinational potential is important because linkage drag, where selection acts on the net fitness of advantageous and disadvantageous mutations that are in LD with one another, both retards the rate of fixation of advantageous mutations and leads to the fixation of deleterious mutations. Despite the theoretical possibility of linkage drag, many of our most important crops, such as wheat, barley, beans, and tomatoes, are predominantly self-fertilizing species. Therefore, the empirical measurement of the actual extent of LD domains in predominantly self-fertilizing species is an important and largely unresolved issue. Recent studies of Arabidopsis thaliana (11) suggest that LD domains may be of the order of 200 kb, which translates into ≈1 map unit (cM), but little is known about the progenitors of major crop plants.
In this article, we examine the magnitude of LD in wild barley (Hordeum vulgare ssp. spontaneum), the progenitor of cultivated barley, and a species with a rate of self-fertilization of ≈98% (12). The sample investigated is comprised of 25 individuals from across the geographic range of wild barley. We characterize LD at the sequence level for 18 loci where haplotypes have been fully resolved. The majority of loci are mapped, so it is possible to compare LD at known physical or genetic map distances. If the mutations at a locus do not have a random geographic distribution, but instead, vary locally among geographic regions, LD can be elevated both within and between loci (13). Thus, we also examine the contribution of geographic structuring of haplotype variation to inter- and intralocus LD. The resulting analyses of wild barley data reveal levels of intralocus LD that are only slightly greater than observed in maize, an outcrossing species.
Materials and Methods
Plant Materials. Individuals sampled were drawn from across the range of wild barley, an annual grass native to southwest Asia. The accession numbers and geographic origins of samples are shown in Table 3, which is published as supporting information on the PNAS web site; see also refs. 14 and 15).
Sampled Loci. Analyzed data includes nucleotide sequences from nine previously reported loci (14–17) and nine loci reported here, referred to as: 5′Pepc, Cbf3, Dhn1, Dhn4, Dhn7, Faldh, ORF1, Stk, and Vrn1. The sequence from the enzymatic Pepc locus was reported in Morrell et al. (17); 5′Pepc is an additional 2,017 bp adjacent to, and immediately upstream from, that region. C-repeat binding factor 3 (Cbf3) is a functional, nonenzymatic locus that shows increased expression when plants are exposed to low temperatures (18). The dehydrin loci, Dhn1, Dhn4, and Dhn7, all show increased expression during dehydration (19). Faldh is an enzymatic locus, glutathione-dependent formaldehyde dehydrogenase, also classified as an alcohol dehydrogenase class III. ORF1 is an ORF, identified as a putative cleavage stimulation factor 1; Stk is a putative serine/threonine kinase (20). Vernalization 1 (Vrn1) is a MADS-box (transcription factor) gene involved in controlling flowering time in relation to a period at low temperatures (21, 22).
Sequencing and Data Assembly. PCR amplification, sequencing, and fragment assembly follow the methods described by Morrell et al. (17); i.e., primarily direct sequencing of PCR products with abi big dye v. 3.1 followed by assembly of sequence fragments with phred/phrap/consed (23–25), using polyphred (26) for polymorphism detection. Multiple sequence alignment was performed with clustalw (27).
Data Analysis. compute and descpoly (28) were used to estimate sequence diversity, including Watterson's θ (29) (denoted here as ΘW) and Tajima's π (30), and to calculate the LD measure, Wall's B (31). Permutation tests (with 1,000 replicates) of the product moment correlation between distance and r2, the squared value of Pearson's correlation coefficient for diallelic nucleotide states were performed with rsq (28). Fisher's exact test (FET), as implemented in dnasp v. 4.06 (32), was used to test for LD.
Wall's B is a summary statistic for nucleotide sequence data from a population genetic sample. It is calculated based on pairs of segregating sites. The method considers each of the possible pairs of adjacent segregating sites. In some cases, the adjacent sites have the same pattern of variation among the individuals within the sample. In these cases, the two adjacent sites are in complete disequilibrium. Such pairs of sites are called congruent pairs. Wall's B is the proportion of pairs that are congruent. Wall's B can be considered a measure of LD with values approaching 1 indicating extensive congruence among adjacent segregating sites.
The P values for FET resulting from pairwise comparisons of nucleotide site produce a distribution that is very similar to that obtained from comparing sites with the more frequently used correlation coefficient, r2. One advantage of using P values from the FET is to provide a more direct means of identifying a threshold for significant LD. FET P values were calculated for combinations of nucleotide polymorphisms for which each polymorphic state occurred at least twice in the sample (frequency of ≈8% or greater). Six segregating sites with more than two nucleotide states were excluded.
If some degree of LD is observed within a locus, then a conservative test of interlocus LD must control for the nonindependence of linked sites. Maintaining haplotypes as a sampling unit provides a direct means of determining if observed interlocus LD exceeds that expected under random combinations of haplotypes. We have implemented a simple resampling method that produces 1,000 randomizations of the sample order of haplotypes at the first of two loci. LD statistics (in this case, r2) for interlocus pairwise comparsions are then calculated for each randomized sample order. As applied here, the procedure tests whether the observed median r2 values between all interlocus pairs of parsimony informative sites differ from what would be expected under the null hypothesis of no association between the haplotypes at the two loci. The P value is the proportion of median r2 values from replicates that fall at or above the observed value, implying the presence of significant LD between loci.
The extent of geographic structure at individual loci between the three major geographic regions within the range of wild barley [as defined in Morrell et al. (17)] was estimated by using the Kst* and Snn methods of Hudson (33, 34) as implemented in dnasp (32), using 1,000 replicates for each test.
The randomization test for interlocus haplotype configurations, plots of LD relative to physical distance, and counts of haplotypes and their frequency were implemented in the r statistical package and programming language (which can be accessed at www.r-project.org).
Results
The sequence data reported here, in combination with data from the same sample of 25 individuals reported in previous studies (14–17), includes 25,030 bp of aligned sequenced length, 23,962 bp of unaligned length (i.e., excluding indel polymorphisms), with an average length per locus of 1,363.1 bp. A total of 699 mutations were identified, 418 of these occur at least twice in the sample.
For the majority of loci, alignment is unambiguous; clustalw was used for aligning Dhn1, Dhn4, and Dhn7, where indel polymorphism is extensive. For Dhn4, two deeply divergent haplotypes cannot be aligned through most of the intron between exons 1 and 2. We have chosen a conservative alignment that minimizes the number of segregating sites in the region.
Sample sizes for some loci are <25 individuals because it was not possible to obtain complete sequences for all accessions. In most cases, the problem of incomplete sequence was encountered because of interruption by transposable elements. Dhn1 has 23 complete sequences, Dhn4 has 24, and (as reported in ref. 17) Dhn5 has 23. For Vrn1, the sample size is 19, the locus could not be amplified in six individuals even after six additional amplification primers were tested in combination with internal primers. For Dhn1, sample 39 contains a 562-bp insertion. This insertion is not included in the aligned length for Dhn1. Also, a number of heterozygous individuals were identified, specifically: sample 9 for Cbf3, samples 12 and 28 for Dhn7, sample 28 for G3pdh, samples 12 and 28 for ORF1, sample 12 for Stk, and samples 6 and 28 for Waxy. Haplotype configurations and frequencies for all parsimony informative mutations are shown in Fig. 4, which is published as supporting information on the PNAS web site.
Diversity Estimates. Diversity estimates for the 18 loci are shown in Table 1. Diversity per locus ranged from ΘW = 0.0007–0.0200 with a mean of 0.0081. Table 4, which is published as supporting information on the PNAS web site, reports levels of diversity, the number of haplotypes, and the number of segregating sites identified in the three principal geographic regions within the range of wild barley, previously identified as the Western, Zagros, and Eastern regions; i.e., areas of the Middle East to the east and west of the Zagros Mountains, and the mountainous region itself (17). Mean diversity (as estimated from average values of the pairwise diversity estimate, π) is greatest in the Western portion of the sample, ≈15% greater than in the Zagros and 25% greater than in the Eastern region (Table 4).
Table 1. Estimates of nucleotide sequence diversity, Tajima's T (commonly reported as Tajima's D test), and Wall's B, for a common set of 25 samples at 18 loci in wild barley.
| Gene | Length, bp | Ungapped length, bp | Θw × 103 | π × 103 | T | Wall's B |
|---|---|---|---|---|---|---|
| Adh1 | 1,362 | 1,359 | 2.73 (±1.11) | 2.07 (±0.17) | -0.926 | 0.154 |
| Adh2 | 1,980 | 1,971 | 4.84 (±1.72) | 3.19 (±0.31) | -1.289 | 0.057 |
| Adh3 | 1,873 | 1,803 | 15.42 (±5.11) | 22.42 (±1.15) | 1.734 | 0.423 |
| α-amy1 | 856 | 856 | 3.10 (±1.36) | 1.27 (±0.63) | -1.948 | 0.222 |
| Cbf3 | 1,514 | 1,477 | 4.61 (±1.69) | 4.38 (±0.52) | -0.183 | 0.160 |
| Dhn1 | 1,538 | 1,034 | 18.87 (±6.47) | 13.22 (±0.73) | -1.187 | 0.070 |
| Dhn4 | 1,074 | 815 | 14.13 (±4.97) | 17.18 (±2.00) | 0.831 | 0.381 |
| Dhn5 | 1,088 | 1,076 | 13.35 (±4.66) | 11.31 (±1.11) | -0.130 | 0.269 |
| Dhn7 | 1,400 | 1,322 | 20.02 (±6.35) | 15.02 (±1.48) | -0.971 | 0.158 |
| Dhn9 | 1,011 | 1,011 | 4.90 (±1.91) | 3.91 (±0.43) | -0.725 | 0.167 |
| Faldh | 1,092 | 1,075 | 5.91 (±2.12) | 5.71 (±0.38) | -0.125 | 0.381 |
| G3pdh | 2,010 | 1,992 | 7.76 (±2.54) | 9.90 (±1.74) | 0.823 | 0.536 |
| ORF1 | 1,533 | 1,516 | 6.16 (±2.16) | 5.18 (±0.61) | -0.592 | 0.143 |
| 5'Pepc | 2,019 | 2,017 | 0.66 (±0.35) | 0.23 (±0.08) | -1.841 | — |
| Pepc | 1,154 | 1,154 | 1.15 (±0.61) | 1.14 (±0.12) | -0.023 | — |
| Stk | 1,057 | 1,044 | 9.29 (±3.27) | 6.77 (±0.64) | -1.019 | 0.111 |
| Vrn1 | 1,262 | 1,208 | 3.79 (±1.48) | 3.57 (±0.35) | -0.216 | 0.077 |
| Waxy | 1,232 | 1,232 | 9.27 (±1.05) | 7.89 (±0.57) | -0.615 | 0.233 |
For Θw, SD is shown, based on no recombination.
Geographic Structure. With sampled individuals divided among the Western, Eastern, and Zagros regions, Kst* values for six loci exceed that expected for random distribution of haplotypes, (Kst*, P < 0.01), Adh3, Cbf3, Faldh, G3pdh, Pepc, and Vrn1 (Table 4). Snn indicates significant geographic structure (P < 0.05) for four additional loci (α-amy1, Dhn7, Dhn9, and ORF1). Snn values with P < 0.01 were found for six loci, Adh3, Cbf3, Dhn7, Faldh, ORF1, Vrn1. For the Dhn4 locus, which has extensive indel polymorphism, inclusion of indels in the analysis as a fifth nucleotide state results in a significant Snn value (P < 0.05). Thus, 11 of the 18 sampled loci demonstrate significant geographic structuring of haplotype diversity. At Dhn4, deeply divergent sequence types predominate in the Eastern and Western portions of the species range, a situation similar to that reported for Adh3 (15) and G3pdh (17).
Intralocus LD. The majority of loci do not show evidence of extensive LD. Based on permutation tests of the product moment correlation between physical distance and r2, 10 of the 18 sampled loci show a significant, negative association of physical distance and r2, as would be expected if mutations are disassociated by recombination.
Wall's B per locus varied from 0.070 to 0.536, with a mean of 0.222. The largest values of Wall's B were observed at three loci, Adh3, Dhn4, and G3pdh, which have a distinctive geographic distribution of haplotype polymorphism relative to all other sampled loci (Table 1). All three loci have two predominant sequence types that differ at the vast majority of segregating sites (Fig. 4). They are also exceptional in having positive values of Tajima's T (T is negative at all other loci) (Table 1).
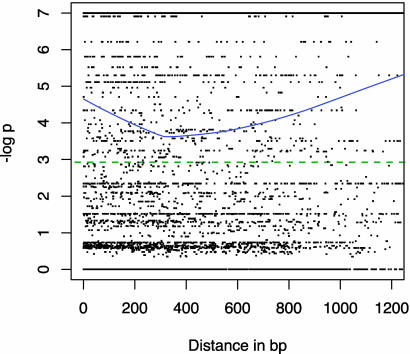
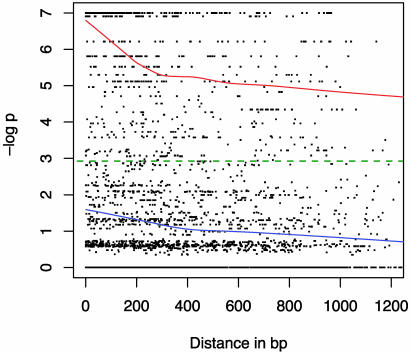
The LD for all pairs of diallelic sites within each locus, plotted against the distance in base pairs between sites is shown in Fig. 1. In Fig. 2, Adh3, Dhn4, and G3pdh loci have been excluded because the very strong geographic structure evident at these loci contributes a large number of points and obscures the pattern of LD at all other loci. Plotted values are the negative log of the P value of each FET. The significance threshold of P < 0.05 is shown as a green dotted line. The blue curve is the lowess approximation (35, 36) of the mean value of LD for all points, and the red curve is the lowess approximation for the subset of points with significant LD. No decline in LD with distance is evident in Fig. 1. In Fig. 2, significant LD declines rapidly for the first 300 bp, and a gradual decay is evident out to 1,200 bp.
Fig. 1.
The decay of LD at all 18 sampled wild barley loci. Plotted values are the negative log of FET P values versus distance in base pairs. The significance threshold of P ≤ 0.05 is shown with a green dotted line. The blue curve is the lowess approximation of mean LD for all comparisons.
Fig. 2.
The decay of LD within 13 wild barley loci. Plotted values are the negative log of FET P values versus distance in base pairs. The significance threshold of P ≤ 0.05 is shown with a green dotted line. The blue curve is the lowess approximation of mean LD for all comparisons and the red curve is the lowess approximation for significant values only.
Interlocus LD. For the comparison of LD among loci, there are 136 interlocus pairwise comparisons (the two portions of Pepc are combined in this analysis). Of these comparisons, 14.7% have a median value of r2 significantly larger (P < 0.05) than expected (Table 5, which is published as supporting information on the PNAS web site). The majority of interlocus comparisons with an excess of LD involve either closely linked loci, or two-locus pairs where one or more of the loci show evidence of geographic structure. The proportion of two-locus pairs with significant LD is impacted by the power of detection. As with previously reported methods of detecting interlocus LD (37, 38), both the number of individuals sampled and allele frequencies impact the power for detecting significant associations. In the present method, power is also impacted by the number of parsimony-informative sites that make up observed haplotypes at each locus. With a small number of sites at either of the loci in a two-locus comparison, there is little power to detect significant interlocus LD because the distribution of r2 values is constructed from a limited number of pairwise comparisons.
Interlocus comparisons can be divided into four classes, those between tightly linked, loosely linked, and unlinked loci, and finally, between loci with an unknown linkage relationship. There is a significant excess of LD at four of six comparisons at tightly linked loci (Table 5), i.e., loci at a 0-cM distance in genetic mapping populations of moderate size (19, 39, 40). The Dhn4, Dhn5, and Dhn7 loci that have been mapped to the same location on chromosome 6H all show a significant association, as do Dhn1 and Vrn1, which map to the same portion of 5H barley genetic map information, can be accessed at http://wheat.pw.usda.gov/index.shtml. At loosely linked loci (in all cases, separated by ≥7 cM), 2 of 13, or 15.4%, show significant LD. Both significant comparisons involve at least one locus at which geographic structure was detected. Significant LD is detected in 12 of 87 (13.8%) of unlinked comparisons, all but two (Adh2_Dhn5 and Dhn1_Dhn5) of these comparisons involve at least one locus with geographic structure. Two of 30 (67%) exhibit significant intralocus LD, where the linkage relationship is unknown (one or more of the loci does not have a known genetic map position). Again, both cases involve at least one locus with geographic structure.
For detailed examination of interlocus LD, six two-locus comparisons are shown in Table 2. ORF1 and Stk are at the closest known distance, separated by 37,900 bp (20). As is evident in Table 2, 166 of 1,000 haplotype configurations in the simulation had median values of r2 less than or equal to that in the actual data; however, the P values for the significance tests on the mean, 0.75, and 0.95 quantiles are <0.05. Comparisons of Adh1 and Adh2, and Dhn5 and Dhn7, where the pairs of loci are linked at 0 genetic distance (16, 19, 39, 40), show very different levels of LD. The level of interlocus LD in the observed Adh1 by Adh2 haplotype configuration is not significantly different from that expected in random configurations of the haplotypes (Fig. 5, which is published as supporting information on the PNAS web site). However, only 3 of 1,000 configurations of the Dhn5 and Dhn7 haplotypes have a median r2 value as large as that observed in the empirical data set (Table 2). Dhn4 and G3pdh are also linked, although the precise genetic map location of G3pdh is not known. Segregating sites at both loci, but particularly at G3pdh, are divided among two predominant haplotypes (Fig. 4), leading to a large number of interlocus comparisons of segregating sites that produce identical r2 values and variation among haplotype configurations that produces very large differences in distribution of values in some correlation matrices but also many replicates of nearly identical matrices (Fig. 5). The median r2 value for comparison of interlocus sites in the empirical data set is the lowest of only seven unique values observed in 1,000 replicates (Fig. 5). The Adh2 by Dhn5 and Cbf3 by Dhn7 comparisons involve unlinked loci. Neither Adh2 nor Dhn5 showed evidence of geographic structure when samples were divided among the three major geographic regions. However, the median level of r2 in the empirical data are significantly greater than expected at P < 0.05. Adh2 shows significant LD with three other loci (Table 5), all of which have a significant pattern of geographic structure (Table 4). Dhn5 has a significant excess of LD with three other loci, including two loci that showed evidence of geographic structure and one that did not (Table 5). Both Cbf3 and Dhn7 showed evidence of geographic structure. The level of association between mutations at Cbf3 and Dhn7 is slightly greater than in the majority of randomizations of haplotypes from the two loci (Fig. 5).
Table 2. The proportion of 1,000 randomizations of two-locus haplotype configurations with a mean, 0.25 quantile, median, etc., greater than or equal to that in the empirical data in six interlocus comparisons.
| Locus pair | Linkage | Mean | 25th Percentile | Median | 75th Percentile | 95th Percentile |
|---|---|---|---|---|---|---|
| Adh1 and Adh2 | Tightly linked | 0.291 | 0.658 | 0.688 | 0.670 | 0.191 |
| Adh2 and Dhn5 | Unlinked | 0.037* | 0.155 | 0.023* | 0.020* | 0.103 |
| Cbf3 and Dhn7 | Unlinked | 0.014* | 0.058 | 0.032* | 0.015* | 0.022* |
| Dhn4 and G3pdh | Linked | 0.635 | 0.782 | 0.857 | 0.563 | 0.698 |
| Dhn5 and Dhn7 | Tightly linked | 0.000* | 0.008* | 0.003* | 0.003* | 0.000* |
| ORF1 and Stk | Tightly linked | 0.007* | 0.395 | 0.166 | 0.013* | 0.004* |
, P < 0.05
Comparison with Maize. Tenaillon et al. (41, 42) reported levels of diversity and the extent of LD for a species-wide sample from 25 individuals (9 inbred lines and 16 haploidized landrace samples) at 21 loci along maize chromosome 1. Although the average sequence length is considerably shorter for the maize study (average length of 724.0 bp versus 1,361.1 bp in the present study), the data sets are otherwise directly comparable. Based on 1,000 permutations of the product moment correlation between distance and r2, only 7 of the 21 maize loci show a significant correlation at P < 0.05. This result may be because of a relatively small number of parsimony-informative sites at loci that do not show a significant correlation. There are always 13 or fewer sites at such loci.
Wall's B for the 21 maize loci varies from 0 to 0.645 with a mean of 0.207. This value is very similar to the mean value of Wall's B of 0.222 from the 18 wild barley loci.
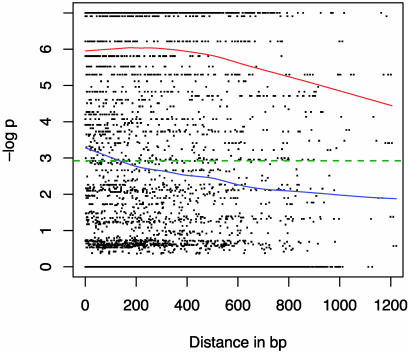
Intralocus LD at 18 of the maize loci is shown in Fig. 3; we have excluded loci that appear to have been subject to strong selection (D8, Tb1, and Ts2) (41). This results in 1,516 pairwise comparisons. Based on the lowess curves in Figs. 2 and 3, the initial level of LD is lower in wild barley than in maize. The rate of decay of significant LD is greater in the wild barley data set than in maize. The mean level of significant LD in wild barley shows the greatest decline over the first 300 bp (Fig. 1), whereas the level of significant LD in the maize data set declines most rapidly after 400 bp (Fig. 2). At 1,000 bp, the two data sets show very similar levels of significant LD.
Fig. 3.
The decay of LD within 18 maize loci. Plotted values are the negative log of FET P values versus distance in base pairs. The significance threshold of P ≤ 0.05 is shown with a green dotted line. The blue curve is the lowess approximation of mean LD for all comparisons and the red curve is the lowess approximation for significant values only.
Discussion
We have examined the level of LD within and between 18 loci from a sample of wild barley from across the species range. Wild barley has a rate of self-fertilization of ≈98%, which results in a very low level of heterozygosity; reducing dramatically the effective rate of recombination relative to random mating. Although the effective rate of recombination should be reduced, 10 of 18 sampled loci show a significant negative correlation of LD with physical distance in base pairs. The loci where a decline of LD with distance is not evident within the locus have either very low levels of polymorphism (10 or fewer segregating sites, i.e., Adh1, α-amy1, Pepc, 5′Pepc, and Vrn1) or geographic subdivision of haplotypes (i.e., Cbf3, Dhn9, and G3pdh).
Interlocus comparisons of LD were performed by using randomization of haplotypes at two-locus pairs to test for an excess of LD in the empirical data sets relative to random configurations of the haplotypes present at each locus. The majority of closely linked loci show an excess of interlocus LD. Loci linked at a distance of ≥7 cM show a level of LD of similar to that at unlinked loci.
A number of factors can contribute to excess interlocus LD, including selection, species-wide reductions in effective population size, or geographic structure. Among these factors, geographic structure provides the most plausible explanation for significant LD between unlinked loci (in the absence of rare epistatic interactions). Under selection or reduced effective population size, the decay of LD is a function of recombination rate and distance. However, LD due to geographic structure is independent of linkage. Approximately 15% of two-locus comparisons demonstrate significant interlocus LD. In the majority of cases, at least one locus in each of the two-locus pairs had been shown (based on the Kst* or Snn tests) to have significant geographic structure among the three principal geographic regions. Clearly, at the genomic level, large numbers of interlocus associations can be expected simply because of nonrandom spatial distribution of haplotypes. Multilocus associations should occur more frequently in predominantly inbreeding species, because associations among haplotypes that are generated by mutation and random genetic drift can persist in inbred and partially isolated subpopulations (or demes) (2, 43, 44)
Why is relatively low LD observed in wild barley despite an expected 40-fold reduction in the effective rate of recombination in a highly inbreeding species? There are at least three possible explanations. The first concerns the time scale spanned by the data; a species-level sample incorporates all of the history of a locus. For the wild barley Adh2 locus for example, time to most recent common ancestor (TMRCA) was estimated to be 460,000 years based on observed nucleotide substitutions and a mutation rate of 3.5 ×10-9 sites per year (16). In highly inbred species, effective recombination events are associated with outcrossing events (45). Even with only 2% outcrossing, given this time scale, a substantial number of recombination events could accumulate.
A second possible explanation is that the relatively low level of LD observed in wild barley results from a recent transition in mating system from outcrossing to selfing (16). As noted by Lin et al. (16) the closely related species Hordeum bulbosum is self-incompatible. If the transition to self-fertilization was relatively recent, say within the last 100,000 years, then many recombination events that occurred before the transition may still be evident in the data, and levels of LD may be reduced relative to an equilibrium situation. A third possibility is that increased chiasmata frequencies may elevate recombination rates within self-fertilizing lineages. An increase in chiasmata frequency in inbreeding species relative to outcrossing relatives has been reported (reviewed in refs. 9 and 46).
Even by taking these possible explanations into account, it is remarkable that LD in wild barley is of essentially the same magnitude as observed in maize. Recent work in Arabidopsis (47) also suggests relatively restricted LD domains, although still larger in magnitude than observed in wild barley. It is clear that species-wide LD within selfers can be quite limited, perhaps surprisingly limited, given the high levels of LD reported within populations. The question now is whether wild barley is unusual or whether relatively low levels of LD are a common feature of inbreeding species. If wild barley and Arabidopsis are typical, then classical arguments about the evolutionary potential of predominantly self-fertilizing species will need to be revisited.
Supplementary Material
Acknowledgments
We thank T. J. Close, A. D. Long, S. J. MacDonald, and S .I. Wright for helpful discussion and A. H. D. Brown, B. S. Gaut, S. Hegde, D. B. Neale, N. Takebayashi, and B. S. Weir for comments on an earlier version of the manuscript. This work was supported by National Science Foundation Grant DEB-0129247.
Author contributions: P.L.M. and M.T.C. designed research; K.E.L. performed research; D.M.T. analyzed data; and P.L.M. and M.T.C. wrote the paper.
Abbreviations: LD, linkage disequilibrium; FET, Fisher's exact test.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY895831–AY896053).
References
- 1.Mendel, G. (1866) 4, 3-47.
- 2.Brown, A. H. D. (1979) Theor. Popul. Biol. 15, 1-42. [Google Scholar]
- 3.Hastings, A. (1990) in Plant Population Genetics, Breeding and Genetic Resources, eds. Brown, A. H. D., Clegg, M. T., Kahler, A. L. & Weir, B. S. (Sinauer, Sunderland, MA), p. 449.
- 4.Allard, R. W. (1975) Genetics 79, 115-126. [PubMed] [Google Scholar]
- 5.Weir, B. S. & Cockerham, C. C. (1973) Genet. Res. 21, 247-262. [DOI] [PubMed] [Google Scholar]
- 6.Golding, G. B. & Strobeck, C. (1980) Genetics 94, 777-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nordborg, M. (2000) Genetics 154, 923-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stebbins, G. L. (1950) Variation and Evolution in Plants (Columbia Univ. Press, New York).
- 9.Grant, V. (1958) Cold Spring Harbor Symp. Quant. Biol. 23, 337-363. [DOI] [PubMed] [Google Scholar]
- 10.Takebayashi, N. & Morrell, P. L. (2001) Am. J. Bot. 88, 1143-1150. [PubMed] [Google Scholar]
- 11.Nordborg, M., Borevitz, J. O., Bergelson, J., Berry, C. C., Chory, J., Hagenblad, J., Kreitman, M., Maloof, J. N., Noyes, T., Oefner, P. J., et al. (2002) Nat. Genet. 30, 190-193. [DOI] [PubMed] [Google Scholar]
- 12.Brown, A. H. D., Zohary, D. & Nevo, E. (1978) Heredity 41, 49-62. [Google Scholar]
- 13.Nei, M. & Li, W. H. (1973) Genetics 75, 213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cummings, M. P. & Clegg, M. T. (1998) Proc. Natl. Acad. Sci. USA 95, 5637-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin, J.-Z., Brown, A. H. D. & Clegg, M. T. (2001) Proc. Natl. Acad. Sci. USA 98, 531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, J.-Z., Morrell, P. L. & Clegg, M. T. (2002) Genetics 162, 2007-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrell, P. L., Lundy, K. E. & Clegg, M. T. (2003) Proc. Natl. Acad. Sci. USA 100, 10812-10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi, D. W., Rodriguez, E. M. & Close, T. J. (2002) Plant Physiol. 129, 1781-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi, D. W., Zhu, B. & Close, T. J. (1999) Theor. Appl. Genet. 98, 1234-1247. [Google Scholar]
- 20.Dubcovsky, J., Ramakrishna, W., SanMiguel, P. J., Busso, C. S., Yan, L. L., Shiloff, B. A. & Bennetzen, J. L. (2001) Plant Physiol. 125, 1342-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T. & Dubcovsky, J. (2003) Proc. Natl. Acad. Sci. USA 100, 6263-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan, L. L., Loukoianov, A., Blechl, A., Tranquilli, G., Ramakrishna, W., SanMiguel, P., Bennetzen, J. L., Echenique, V. & Dubcovsky, J. (2004) Science 303, 1640-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ewing, B., Hillier, L., Wendl, M. C. & Green, P. (1998) Genome Res. 8, 175-185. [DOI] [PubMed] [Google Scholar]
- 24.Ewing, B. & Green, P. (1998) Genome Res. 8, 186-194. [PubMed] [Google Scholar]
- 25.Gordon, D., Abajian, C. & Green, P. (1998) Genome Res. 8, 195-202. [DOI] [PubMed] [Google Scholar]
- 26.Nickerson, D. A., Tobe, V. O. & Taylor, S. L. (1997) Nucleic Acids Res. 25, 2745-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thornton, K. (2003) Bioinformatics 19, 2325-2327. [DOI] [PubMed] [Google Scholar]
- 29.Watterson, G. A. (1975) Theor. Popul. Biol. 7, 256-276. [DOI] [PubMed] [Google Scholar]
- 30.Tajima, F. (1983) Genetics 105, 437-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wall, J. D. (1999) Genet. Res. 74, 65-79. [Google Scholar]
- 32.Rozas, J., Sanchez-DelBarrio, J. C., Messeguer, X. & Rozas, R. (2003) Bioinformatics 19, 2496-2497. [DOI] [PubMed] [Google Scholar]
- 33.Hudson, R. R. (1992) Mol. Biol. Evol. 9, 969-969. [DOI] [PubMed] [Google Scholar]
- 34.Hudson, R. R. (2000) Genetics 155, 2011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cleveland, W. S. (1981) Am. Stat. 35, 54. [Google Scholar]
- 36.Cleveland, W. S. (1979) J. Am. Stat. Assoc. 74, 829-836. [Google Scholar]
- 37.Brown, A. H. D. (1975) Theor. Popul. Biol. 8, 184-201. [DOI] [PubMed] [Google Scholar]
- 38.Weir, B. S. (1996) Genetic Data Analysis II, Methods for Discrete Population Genetic Data (Sinauer, Sunderland, MA).
- 39.Kleinhofs, A., Kilian, A., Maroof, M. A. S., Biyashev, R. M., Hayes, P., Chen, F. Q., Lapitan, N., Fenwick, A., Blake, T. K., Kanazin, V., et al. (1993) Theor. Appl. Genet. 86, 705-712. [DOI] [PubMed] [Google Scholar]
- 40.Choi, D.-W., Koag, M. C. & Close, T. J. (2000) Theor. Appl. Genet. 101, 350-354. [Google Scholar]
- 41.Tenaillon, M. I., Sawkins, M. C., Long, A. D., Gaut, R. L., Doebley, J. F. & Gaut, B. S. (2001) Proc. Natl. Acad. Sci. USA 98, 9161-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tenaillon, M. I., Sawkins, M. C., Anderson, L. K., Stack, S. M., Doebley, J. & Gaut, B. S. (2002) Genetics 162, 1401-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown, A., Nevo, E. & Zohary, D. (1977) Nature 268, 430-431. [Google Scholar]
- 44.Charlesworth, D. (2003) Philos. Trans. R. Soc. London B 358, 1051-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nordborg, M. (1999) in Statistics in Molecular Biology and Genetics: IMS Lecture Notes-Monograph Series, ed. Seillier-Moiseiwitsch, F. (Inst. Math. Stat., Hayward, CA), Vol. 33.
- 46.Charlesworth, D., Charlesworth, B. & Strobeck, C. (1977) Genetics 86, 213-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nordborg, M., Hu, T. T., Ishino, Y., Jhaveri, J., Toomajian, C., Zheng, H., Bakker, E., Calabrese, P., Gladstone, J., Goyal, R., et al. (2005) PLoS Biol., in press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.