Abstract
The outer membrane protein Ail (Adhesion invasion locus) is one of the most abundant proteins on the cell surface of Yersinia pestis during human infection. Its functions are expressed through interactions with a variety of human host proteins, and are essential for microbial virulence. Structures of Ail have been determined by X-ray diffraction and solution NMR spectroscopy, but those samples contained detergents that interfere with functionality, thus, precluding analysis of the structural basis for Ail’s biological activity. Here, we demonstrate that high-resolution solid-state NMR spectra can be obtained from samples of Ail in detergent-free phospholipid liposomes, prepared with a lipid to protein molar ratio of 100. The spectra, obtained with 13C or 1H detection, have very narrow line widths (0.40–0.60 ppm for 13C, 0.11–0.15 ppm for 1H, and 0.46–0.64 ppm for 15N) that are consistent with a high level of sample homogeneity. The spectra enable resonance assignments to be obtained for N, CO, CA and CB atomic sites from 75 out of 156 residues in the sequence of Ail, including 80% of the transmembrane region. The 1H-detected solid-state NMR 1H/15N correlation spectra obtained for Ail in liposomes compare very favorably with the solution NMR 1H/15N TROSY spectra obtained for Ail in nanodiscs prepared with a similar lipid to protein molar ratio. These results set the stage for studies of the molecular basis of the functional interactions of Ail with its protein partners from human host cells, as well as the development of drugs targeting Ail.
Keywords: Membrane protein, Solid-state NMR, Magic angle spinning, Ail, Yersinia pestis
The importance of the lipid bilayer membrane for supporting the structure and function of membrane proteins has motivated the development of samples for structural studies by X-ray diffraction, electron microscopy (EM) and NMR spectroscopy, that resemble native membranes as closely as possible (Cross et al. 2014; De Zorzi et al. 2016; Maslennikov and Choe 2013; Moraes et al. 2014). NMR has the unique advantage of being compatible with samples that are similar to the physiological protein environments (Zhou and Cross 2013). NMR signals provide accurate structural restraints as well as information about global and local dynamics. Moreover, since they are highly sensitive to the local molecular environment NMR signals are useful for characterizing even weak ligand binding through chemical shift changes, enabling structure–activity correlations to be made for binding events.
The long correlation times of large protein-lipid assemblies pose a limitation on the types of membrane proteins that can be studied by solution NMR, although advances are being made in the development of experimental methods (Takeuchi et al. 2015) and the preparation of small, detergent-free lipid nanodiscs (Hagn et al. 2013). Protein-containing phospholipid bilayers are, however, well suited for solid-state NMR studies, where the correlation time is not a limiting factor. Advances in magic angle spinning (MAS) and oriented sample (OS) solid-state NMR methods have enabled structural studies of a number of membrane proteins in near-native lipid membranes (Baker et al. 2015; Brown and Ladizhansky 2015).
High quality solid-state NMR spectra have been reported for samples of membrane proteins in proteolipid 2D crystals, precipitates, and microcrystals (Andreas et al. 2015; Barbet-Massin et al. 2014; Eddy et al. 2012; Li et al. 2007; Linser et al. 2011; Saurel et al. 2017; Shahid et al. 2012). These samples, prepared with lipid to protein molecular ratios in the range of 6:1 and 25:1, have the advantage that large quantities of protein can be packed in the NMR rotor for enhanced sensitivity. Such low lipid content, however, is well below that of most biological membranes (Gennis 1989; Lee 2003, 2011). Although some specialized membranes do have very low lipid to protein ratios, this is not generally the case, and even rod outer segment disc membranes, which are highly enriched in rhodopsin, have a lipid to protein molar ratio of ~40:1 (Devaux and Seigneuret 1985). Analysis of the crowded membranes of synaptic vesicles estimates that about 12 molecules of lipid and 9 molecules of cholesterol are present for every protein transmembrane helix (Takamori et al. 2006). For a protein with a single transmembrane helix, this translates to a lipid:protein molar ratio of 12:1 and total protein mole fraction of ~4.5%, when cholesterol is also taken into account; for a protein with four transmembrane helices, the lipid:protein molar ratio is 48:1, and the total protein mole fraction in a lipid-cholesterol membrane is 1.2%.
Low lipid content can compromise protein stability and induce the formation of non-native contacts, including contacts between protein molecules with opposite transmembrane orientations (Andreas et al. 2015; Behlau et al. 2001; Dolder et al. 1999). These samples may need to be diluted into liposomes to reconstitute activity (Eddy et al. 2012), indicating that bulk lipid is important for function. Furthermore, 2D crystals can suffer from structural heterogeneity (Behlau et al. 2001). This is not necessarily problematic for cryo-EM, where individual crystals can be selected for analysis, but poses a challenge for NMR, where the signals reflect ensemble properties of the bulk sample, and heterogeneity manifests as broader line widths, which decrease spectral resolution.
An important advantage of NMR is that neither long- nor short-range molecular order is required to obtain high-resolution spectra, thus bypassing the burden of preparing either 2D or 3D crystalline samples. The only prerequisite for reducing inhomogeneous line broadening and obtaining single, narrow resonance lines, is that the protein adopts a homogeneous conformational ensemble state on the time-scale determined by the frequency span of spin interactions. For solid-state NMR studies of membrane proteins, this can be achieved by using proteolipid samples with a total lipid mole fraction high enough to prevent the formation of non-native protein conformations and non-native contacts in the membrane. Here, we show that the outer membrane protein Ail (Adhesion invasion locus) yields very high resolution NMR spectra when incorporated in lipid bilayers with 100 lipid molecules for each protein.
Ail is one of the most highly expressed proteins on the cell surface of Yersinia pestis during human infection. Y. pestis, the causative agent of plague, is an invasive blood stream pathogen that poses very high risk to public health because it is highly pathogenic, can be easily disseminated, and causes high mortality. The interactions of Ail with human host proteins are critical for promoting the survival of Y. pestis bacteria in serum, and are targets for therapy development. Two structures of Ail have been determined: by X-ray diffraction (Yamashita et al. 2011) for the protein crystallized in tetra-ethylene glycol monooctyl ether (C8E4), and by solution NMR (Marassi et al. 2015) for the protein in decyl-phosphocholine (DePC) micelles. Ail adopts the same eight-stranded β-barrel fold in both cases, but the protein’s activity is compromised by detergent (Ding et al. 2015), which is present in both the crystal and micelle samples, underscoring the need to perform structure/function studies in detergent-free membranes. Well-resolved NMR spectra of Ail in lipid bilayers provide access to experimental approaches for determining its functional structure, understanding its functions, and NMR-based drug discovery.
Materials and methods
Protein preparation
Ail was cloned into the E. coli plasmid pET-30b, and then expressed and purified as described previously (Ding et al. 2015). For uniform 15N and 13C labeling (u-15N, u-13C), bacteria were grown in M9 medium containing 1 g/L of (99% 15N)-ammonium sulfate and 2 g/L of (99% 13C)-glucose (Cambridge Isotope Laboratories). For additional 2H labeling, the growth medium was prepared with (99.99% 2H)-water to obtain fractional (~70%) 2H labeling (f-2H), or with both (99.99% 2H)-water and (98% 2H, 99% 13C)-glucose for uniform 2H labeling (u-2H) The 2H atoms at exchangeable sites were replaced with 1H during Ail purification under denaturing conditions.
Nanodisc preparation
The preparation of Ail nanodiscs (Ding et al. 2015) and expression and purification of the nanodisc membrane scaffold protein MSP1D1 Δh5 (Hagn et al. 2013) were performed as described previously. Briefly, Ail was assembled into nanodiscs by refolding in 170 mM n-decyl-phosphocholine (DePC; Anatrace), mixing with phospholipids (Avanti), MSP1D1 Δh5 and Na-cholate, and finally removing the detergent with Bio-Beads SM-2 (Bio-Rad) and dialysis. The nanodiscs were composed of a 75:25 molar mixture of the phospholipids (Avanti) dimyristoyl-phos-phatidylcholine (DMPC) and dimyristoylphosphatidyl-glycerol (DMPG), 2H labeled at all 54 protons of their acyl chains. The molar ratio 100:2:1 of lipid:MSP1D1 Δh5:Ail gave the highest level of nanodisc size homogeneity and the narrowest NMR linewidths (Ding et al. 2015). For solution NMR experiments, the nanodiscs were transferred into NMR buffer (25 mM Na-PO4, pH 6.5, 1 mM EDTA) supplemented with 5 mM NaCl and 10% D2O, and concentrated with a Vivaspin 500 centrifugal device to obtain a final concentration of 0.6 mM (u-15N, u-13C, u-2H) Ail in a 500 uL volume of the 5 mm NMR tube.
Proteoliposome preparation
Ail liposomes were prepared with a lipid:protein molar ratio of 100:1. A solution of refolded Ail and 170 mM DePC in NMR buffer was mixed with a solution containing phospholipids (DMPC:DMPG, 75:25 molar) and 100 mM Na-cholate in NMR buffer. Detergent was removed by dialysis against three 4 L volume changes of NMR buffer over the course of 24 h, followed by incubation with Bio-Beads SM-2 (Biorad) overnight. Proteoliposomes were harvested by ultracentrifugation at 168,000xg, 4 °C, for 16 h, and then packed into MAS rotors (Bruker). We estimate that ~90% of the final sample volume could be transferred to the MAS rotor for NMR studies. Three NMR samples were prepared each containing ~3 mg of Ail packed in a 4 mm MAS rotor with a 50 uL insert, ~3 mg of Ail packed in a 3.2 mm thin-walled rotor, or ~0.3 mg of Ail packed in a 1.3 mm rotor. The solid-state NMR spectra acquired at the start and at the end of data collection show no difference, indicating that the samples are stable over a period of time longer than 1 month.
Analysis of lipid content in the MAS rotor
The total amount of lipid contained in the MAS rotor was estimated by measuring the 1H NMR signal intensity at 1.2 ppm from the lipid acyl CH2 groups, and comparing it to the intensity at 1.2 ppm measured for a standard sample prepared by adding a known quantity of hydrated lipid directly into the rotor. The 1D 1H NMR spectra were obtained with a MAS rate of 12.5 kHz, at 27 °C. By this estimate, the final NMR sample contains ~90% of the lipid added to the initial preparation. This is consistent with our estimate of the total sample volume that we can transfer to the MAS rotor after centrifugation, and indicates that the final sample maintains the lipid:protein ratio of the initial preparation, with no significant loss of lipid. It is likely that adding Biobeads after, and not before, allowing liposomes to form by extensive detergent dialysis, helps prevent the loss of lipid through adsorption.
Differential scanning calorimetry (DSC)
DSC experiments were performed using an N-DSC II instrument (Calorimetry Sciences Corporation). Pure phospholipid and proteoliposomes samples were suspended in NMR buffer at concentrations between 0.5 and 3.0 mg/mL, and scanned at a rate of 1 K/min, under a constant pressure of 3 atm. NMR buffer served as the reference. The DSC data were analyzed with NanoAnalyze software (TA Instruments).
Activity assays
Enzyme linked immunosorbent assays (ELISA) were performed as described (Ding et al. 2015), using C-terminal His-tagged Ail, and human plasma fibronectin (Sigma; F2006) coated on 96-well plates (Nunc). Binding was detected with primary mouse anti-His monoclonal antibody (Qiagen), and secondary goat anti-mouse antibody, conjugated to horseradish peroxidase (Sigma) to develop absorbance at 490 nm upon reaction with its substrate o-phenylenediamine (Pierce).
Solution NMR experiments
Solution NMR spectra were recorded on a Bruker AVANCE III 800 MHz spectrometer equipped with a triple-resonance cryoprobe. 2D and 3D 1H-15N TROSY, TROSY-HNCA and NOESY-HSQC experiments were performed with 2H decoupling. Chemical shift assignments in the spectra of Ail nanodiscs were obtained by analysis of 3D HNCA and 3D NOESY-HSQC data sets, aided by comparison with the assigned spectra of Ail in micelles (Marassi et al. 2015).
Solid-state NMR experiments
A suite of 13C detection experiments was recorded on 500 MHz Bruker AVANCE and 750 MHz Bruker AVANCE III HD spectrometers, each equipped with a Bruker 4 mm E-free MAS probe, operating at a spinning rate of 12 kHz ± 5 Hz. Typical π/2 pulse lengths for 1H, 13C and 15N were 2.5, 2.5 and 5 μs (500 MHz), or 2.7, 3.2 and 5.6 μs (750 MHz). SPINAL64 (Fung et al. 2000), implemented with a 90 kHz RF field strength, was used for 1H decoupling during data acquisition.
1D 13C spectra were acquired using either dipolar-based cross polarization (CP) (Pines et al. 1973) or J-coupling-based polarization transfer (INEPT) (Morris and Freeman 1979). The respective delays in the INEPT and refocused INEPT blocks were set to 1/4 × J (1.7 ms) and 1/6 × J (1.1 ms), to obtain positive phase carbon signals. 2D homo-nuclear 13C–13C correlation spectra were acquired using the PDSD pulse sequence (Szeverenyi et al. 1982) with 50 ms mixing.
2D heteronuclear 13C–15N correlation spectra were acquired using SPECIFIC-CP (Baldus et al. 1998) for band selective polarization transfer, during which continuous wave (CW) 1H decoupling (100 kHz RF field strength) was applied, and a tangent ramp was applied on the 15N channel with contact times of 3 ms (500 MHz) or 4.5 ms (750 MHz) for N–CA transfers, and 3.5 ms (500 MHz) or 6 ms (750 MHz) for N–CO transfer. For NCACX and NCOCX experiments, a 50 ms PDSD mixing interval was added following the SPECIFIC-CP transfer (Pauli et al. 2001).
2D homonuclear 13C–13C correlation spectra were recorded using DARR (Takegoshi et al. 2001, 2003) with 200 ms mixing, on a 900 MHz Bruker AVANCE III HD spectrometer equipped with a Bruker 3.2 mm E-free MAS probe, at a spinning rate of 12 kHz ± 5 HzC.
Fast MAS 1H detection experiments were performed on a 900 MHz Bruker AVANCE III HD spectrometer, using a Bruker 1.3 mm MAS probe at a spinning rate of 60 kHz ± 15 Hz. 2D CP-HSQC (Barbet-Massin et al. 2014) spectra were obtained with 1.5 ms of 1H–15N CP mixing for the initial transfer of magnetization from 1H to 15N. During 15N evolution, a 13C π pulse was employed for one bond JN−CA and JN−CO decoupling, and a low power (20 kHz RF field strength) XiX (Ernst et al. 2003) scheme was used for 1H decoupling. After 15N evolution, magnetization was transferred from 15N back to 1H with 300 μs of CP mixing. Water suppression was performed using MISSISSIPPI (Zhou and Rienstra 2008), implemented with a saturation pulse train of 300 ms, prior to 1H detection. WALTZ-16 (Shaka et al. 1983) (10 kHz RF field strength) was used for 15N heteronuclear decoupling during data acquisition. The sample temperature at each spinning frequency was measured using the temperature-dependent 1H chemical shift of water.
Results and discussion
In solid-state NMR experiments, line broadening from heteronuclear dipolar and scalar couplings can be reduced by performing experiments at higher magnetic field, increasing the MAS rate, optimizing decoupling schemes and spectroscopic editing. Sample homogeneity, however, is also critical for obtaining high-resolution NMR spectra. Line broadening due to the presence of multiple conformational states reduces spectral resolution and sensitivity, and must be avoided as much as possible by sample optimization.
Membrane proteins require surrounding lipids to maintain native function, dynamics and structure; without sufficient lipid they can oligomerize, adopt non-native conformations, and exhibit altered global and local dynamics. For example, the structure of OmpX is destabilized in nanodiscs with lipid to protein ratios below 40:1 (Hagn et al. 2013). In earlier studies, we experimented with refolding both Ail and OmpX in liposomes, large bicelles (q = 3.2), and nanodiscs with a range of lipid and protein compositions (Ding et al. 2015, 2013; Mahalakshmi et al. 2007; Mahalakshmi and Marassi 2008; Plesniak et al. 2011; Yao et al. 2013). We found that lipid to protein molar ratios greater than 100 yield the most homogeneous reconstitutions, with minimal protein aggregation and the narrowest NMR line widths. For example, we obtained high resolution OS solid-state NMR spectra of OmpX and Ail in magnetically aligned bicelles prepared with 190 lipids per protein molecule, and optimal solution NMR spectra of Ail nanodiscs with 100 lipids per protein. Based on this experience, we focused the present study on Ail liposomes containing 100 lipids (DMPC:DMPG, 75:25 molar) per molecule of Ail.
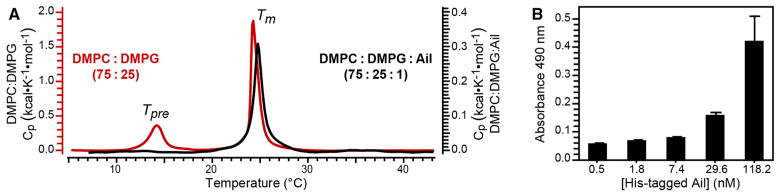
DSC analysis shows that these preparations form proper lipid bilayers with properties that are virtually indistinguishable from those of pure lipids. The DSC endotherms (Fig. 1a, black) measured for Ail-containing bilayers display a sharp, symmetric main transition (Tm) at 24.7 °C, only slightly higher than that observed at 24.2 °C for a binary, 75:25 molar mixture of DMPC and DMPG (Fig. 1a, red). This behavior is typical of the highly cooperative transformation of lipid bilayers from a lamellar gel phase (Lβ′), where the lipid acyl chains are tilted and in an all-trans conformation, to a lamellar liquid crystalline phase (Lα), where the lipid chains are disordered and fluid (Janiak et al. 1976; Rand et al. 1975; Tardieu et al. 1973). Binary mixtures of DMPC and DMPG exhibit nearly ideal mixing in both the gel and fluid phases, over the whole composition range (Findlay and Barton 1978). The observation of a single, sharp, symmetric transition for Ail-lipid bilayers indicates that they also exhibit nearly ideal mixing behavior, with no evidence of phase separation in either the gel or fluid phase.
Fig. 1.
Phase and activity properties of Ail proteoliposomes. a DSC endotherms measured for a binary 75:25 molar mixture of DMPC:DMPG (red), and the ternary 75:25:1 molar mixture of DMPC:DMPG:Ail (black) used for NMR studies. Note different vertical scales for heat capacity (Cp) for either pure lipids (left, red) or Ail-lipids (right, black). The main transition (Tm) to Lα and the pre-transition (Tpre) from Lβ′ to Pβ′, are marked. b ELISA detection of the Fibronectin binding activity of Ail in liposomes. His-tagged Ail in liposomes was added at increasing concentrations to Fibronectin-coated plates and incubated overnight. Binding was detected with a mouse anti-His antibody. Each data point represents the average of three experiments
As expected, the pure DMPC:DMPG lipid bilayers also undergo a pretransition (Tpre) at 14.2 °C, from the lamellar gel phase (Lβ′) to a rippled gel phase (Pβ′), where the lipid chain tilt is reduced. The enthalpy of the pretransition is known to be modulated by pH, salts and phospholipid headgroup interactions, and is often reduced beyond detection in mixed membranes. This is, indeed, observed for the Ail-lipid bilayers. The DSC data also show that less heat is needed for melting Ail-containing bilayers, as the heat capacity of the main gel to fluid transition is ~5.5 times lower than that of pure lipids. This indicates that Ail destabilizes the gel phase, potentially by interfering with lipid–lipid headgroup or acyl chain interactions, even at this relatively low concentration.
Activity assays performed with ELISA and Ail liposomes (Fig. 1b) show that Ail binds its human ligand, Fibronectin, in a dose-dependent manner. Additional evidence for the activity of Ail in these samples was presented previously (Ding et al. 2015), using an assay that probes the interactions of Ail’s extracellular loop 2 (EL2) with a specific anti-Ail-EL2 antibody. Thus, taken together, the DSC and ELISA data demonstrate that Ail liposomes prepared with 100 lipids per protein molecule, form homogeneous lipid bilayers, that are fluid above ~25 °C and support the ligand binding activity of the protein.
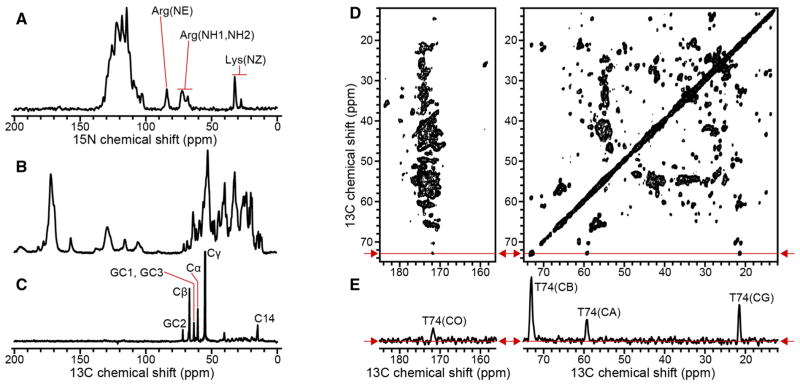
To further assess the extent of sample homogeneity and overall protein dynamics, we acquired a series of 1D and 2D MAS NMR spectra (Fig. 2). The 1D 15N and 13C NMR spectra obtained with CP (Fig. 2a, b) display high resolution for a 156-residue integral membrane protein in lipid bilayers. Several signals can be detected and resolved, including those from Arg and Lys side chain nitrogens, indicating that these basic side chains are sufficiently immobilized on the time scale for CP detection (10−6 s), and adopt a unique conformation, due to specific hydrogen bond and electrostatic interactions.
Fig. 2.
15N and 13C NMR spectra of (u-15N, u-13C) Ail in liposomes. Spectra were obtained at a 1H resonance frequency of 750 MHz at 10 °C. a, b 1D CP spectra acquired with 256 scans. c 1D INEPT 13 C spectrum acquired with 8192 scans. The delay of the refocused INEPT block was chosen so that all 13C signals are positive. d 2D PDSD 13C-13C spectrum acquired with 50 ms mixing time and 64 scans. e 1D slice taken from the 2D PDSD spectrum at a 13C chemical shift of 72.3 ppm, as indicated by the red arrows and red lines
INEPT pulse sequences, based on the transfer of polarization via through-bond scalar couplings, have been used to detect fast motions of proteins and lipids (Andronesi et al. 2005; Warschawski and Devaux 2005). The 13C spectrum (Fig. 2c), obtained with INEPT at 10 °C, is dominated by signals from the natural abundance 13C in the phospholipids, with no detectable signals from the protein. The lipid signals in the INEPT spectrum come primarily from carbons in the lipid headgroup (GC1–GC3, Cα, Cβ, Cγ) and the hydrocarbon tail termini (C14), which remain relatively mobile at 10 °C, compared to the rest of hydrocarbon chain. This is consistent with the DSC data showing that the lipids are in the ordered gel phase at this temperature, with hydrocarbon chains fully extended, closely packed, and rigid. By contrast, the lack of protein signals show that the global and local motions of the protein are sufficiently slowed at 10 °C, to prevent detection with INEPT transfer. The principal motions that account for INEPT signals from the lipids are uniaxial rotation and segmental trans-gauche isomerization (Warschawski and Devaux 2005), which occur on a time scale between 10−8 and 10−12 s (Leftin and Brown 2011), thus, we conclude that any motions present in Ail, including motions in its extracellular loops, must be slower than this time range.
The 2D 13C–13C exchange spectrum (Fig. 2d) of Ail liposomes obtained with 50 ms PDSD mixing displays excellent resolution. The line widths measured in the direct dimension, for well-resolved 13C signals, such as those from the CA, CB and CG atoms of T74 (Fig. 2e), are in the range of 0.4–0.6 ppm (80–110 Hz at 750 MHz). Taking into account that the one bond, homonuclear C–C and heteronuclear C–N scalar J couplings are of the order of tens of Hz (JCC = 35 Hz; JCACO = 55 Hz; JNCA = 11 Hz; JNCO = 15 Hz), and were not removed in these experiments, the observed line widths are quite narrow, consistent with a high level of sample homogeneity for Ail in lipid bilayers.
Narrow 13C line widths have been reported for other β-barrel membrane proteins in 2D crystals, including OmpG (0.8–1.2 ppm; 80–120 Hz at 400 MHz) and VDAC (0.3–0.6 ppm; 60–90 Hz at 750 MHz) (Eddy et al. 2015; Hiller et al. 2005). Solid-state NMR line widths are independent of protein size and reflect the extent of inhomogeneous line broadening from the samples. The line widths in the spectra of Ail liposomes are similar to or narrower than those reported for similar proteins in 2D crystals. Lipid content is a major difference between these types of samples, with 2D crystals containing significantly less lipid. Solvation in a proper lipid bilayer with properties dominated by bulk lipid, may contribute to the conformational homogeneity of Ail liposomes.
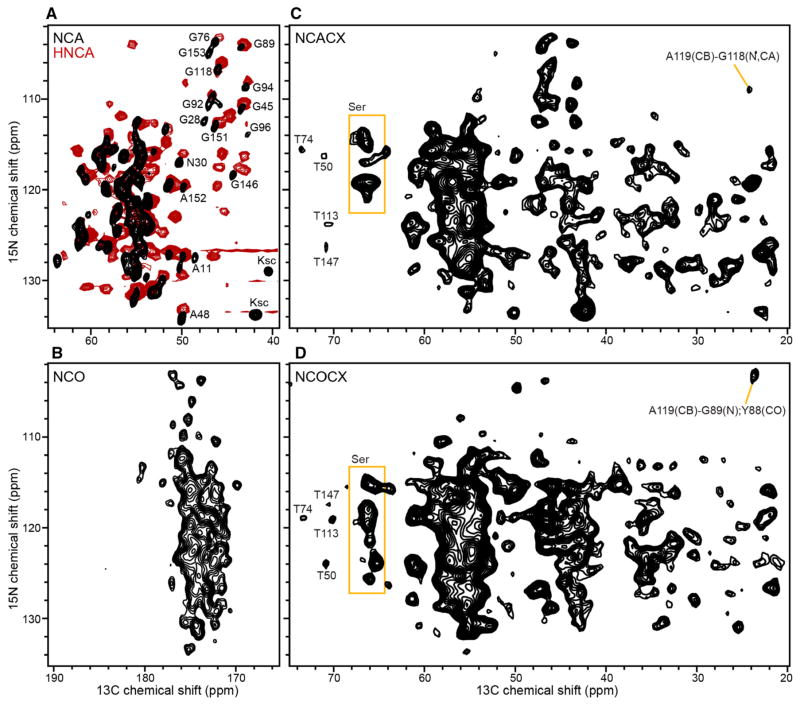
2D 13C/15N NCA and NCO correlation spectra serve as the building blocks of 3D experiments that establish intra-residue (NiCAi) and inter-residue (NiCOi−1) connectivities. The spectra (Fig. 3a, b) obtained with less than 10 h of signal averaging at 750 MHz display excellent signal-to-noise ratios and resolution. The efficiencies of the selective heteronuclear polarization transfer were 35% for NCA and >50% for NCO, consistent with high sample homogeneity and minimal local motions for Ail in lipid bilayers.
Fig. 3.
Solid-state NMR (black) and solution NMR (red) spectra of (u-15N, u-13C) Ail in lipid bilayers. a, b 2D NCA and NCO solid-state NMR spectra (black) obtained at 750 MHz, at 10 °C, with 128 scans per increment. The 2D NCA plane projection (red) from the 3D solution NMR HNCA spectrum of Ail in nanodiscs was obtained at 800 MHz, at 45 °C; additional signals in the HNCA spectrum arise from inter-residue HN and CA connectivities. c, d NCACX and NCOCX spectra obtained at 500 MHz, at 10 °C, with 400 scans per increment. Examples of resolved and assigned signals are marked
Inclusion of a 50 ms PDSD mixing scheme, following the SPECIFIC-CP transfer, allowed us to obtain 2D NCACX and NCOCX spectra (Fig. 3c, d), which are essential for correlating signals from side chain carbons to the backbone CA, CO and N atomic sites. Inspection of these 2D spectra shows that multiple signals can be resolved and assigned. For example, the complete spin systems of the four Thr residues in the Ail sequence can be assigned by combining 2D PDSD, NCACX and NCOCX data sets, and by taking advantage of the characteristic 13C chemical shifts of CA and CB resonances from Thr. Furthermore, the CA and CB signals of the 16 Ser, which are highly overlapped in the 2D PDSD spectrum, are better resolved in the 2D NCACX and NCOCX spectra.
Comparison with the HNCA spectrum obtained for a sample of Ail in nanodiscs, using solution NMR (Fig. 3a, red), reveals significant similarities, notwithstanding the different temperatures (10 °C for solid-state NMR and 45 °C for solution NMR). This indicates that liposomes and nanodiscs prepared with the same lipid composition (DMPC:DMPG, 75:25 molar), and the same lipid to protein ratio (100:1 molar), provide similar environments for Ail. The solution NMR HNCA spectrum correlates the HN atom from residue i, to the CA atoms from both residues i and i−1, and therefore, has more signals. Nevertheless, the spectral similarities are evident among many of the Gly and Ala signals, which are well resolved in the NCA solid-state NMR spectrum.
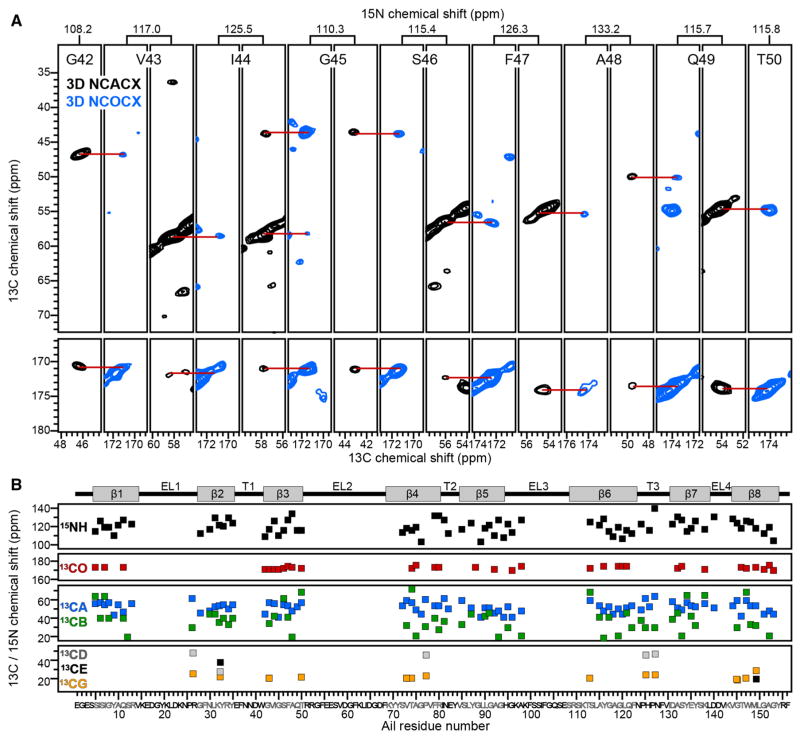
The 2D PDSD and NCA spectra display good resolution, and several assignments could be readily transferred from the solution NMR data. Nevertheless, 3D NCACX and NCOCX spectra (Fig. 4a) were needed to obtain unambiguous sequential resonance assignments. The assignment strategy is similar to that used in solution NMR. Peak lists from NCA and NCO spectra were compiled and used to construct strip plots from 3D NCACX and NCOCX data sets, which serve as the key experiments used to link residue i to its preceding neighbor, residue i−1. Amino acid types were identified on the basis of their characteristic side chain 13C chemical shifts, and then mapped to the protein primary sequence. The process was assisted by analysis of the 2D PDSD spectrum (Fig. 2d), which provides finger-print signal patterns for some amino acid types.
Fig. 4.
Examples and summary of resonance assignments. a 2D 13C–13C strips taken from 3D NCACX (black) and NCOCX (blue) spectra of (u-15N, u-13C) Ail in liposomes. Spectra were recorded at 750 MHz, 10 °C, with 256 scans per increment. Alternating sequential strips show assignments for signals from backbone atomic sites in strand β3. b Resonance assignments mapped on the amino acid sequence of Ail. The protein topology (top), with eight β strands (β1–β8), four extracellular loops (EL1–EL4) and three intracellular turns (T1–T3), is taken from the solution NMR structure in micelles
By combining data from 3D NCACX, 3D NCOCX, and 2D PDSD experiments, it was possible to assign 13C and 15N chemical shifts for 75 of the 156 residues in the sequence of Ail, including nearly 80% of the residues in the transmembrane β-strands (Fig. 4b). Assignments for extracellular loop sites are incomplete. The INEPT spectra (Fig. 2) show no evidence of protein signals and, hence, indicate that no regions of Ail, including the loops, exhibit motions faster than the lower limit of the range of lipid dynamics (~10−8 s). Missing assignments, therefore, may reflect line broadening due to conformational exchange dynamics in the extracellular loops on a slower time scale. Signal overlap is also a challenge for obtaining complete assignments, but should be alleviated through the use of alternative labeling schemes.
Using a proteoliposome sample containing (u-15N, u-13C, f-2H) Ail, we also acquired a 2D 13C/13C DARR spectrum with 200 ms mixing at 900 MHz (Fig. 5a, b). In this sample, the amide 2H atoms were back exchanged with 1H during protein purification, while the backbone and side chain carbon sites remain deuterated. Deuteration dilutes the 1H concentration and suppresses the strong homonuclear 1H–1H dipolar couplings, resulting in longer lifetimes of the 13C and 15N coherences, leading to improvements in sensitivity and resolution.
Fig. 5.
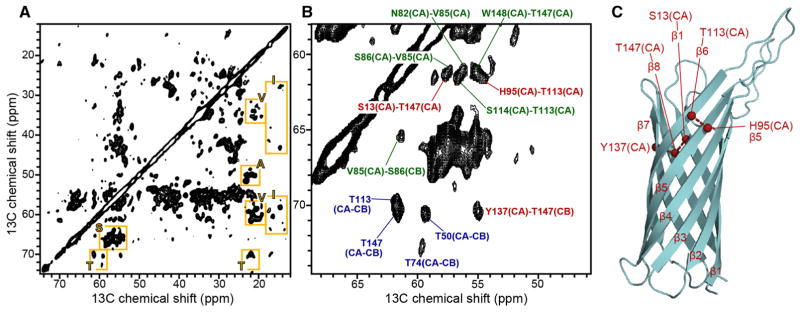
2D 13C–13C DARR spectrum of (u-15N, u-13C, f-2H) Ail in liposomes. a Fingerprint region of the spectrum showing examples of resolved signals from A, I, S, T, V residues (yellow boxes). b Expanded spectral region showing examples of short-range intra-residue connectivities (blue), short-range inter-residue connectivities (green), and long-range inter-residue connectivities (red). c Structure of Ail determined in micelles (PDB: 2N2L) showing inter-strand connectivities (red) assigned in the DARR spectrum. The spectrum was obtained at 900 MHz, at 10 °C, with 200 ms DARR mixing time, and 144 scans per increment
The 2D DARR spectrum, which was obtained in 12 h, shows excellent resolution. Signals for the Thr, Ser, Ile and Ala spin systems can be readily identified based on their characteristic chemical shifts, which reflect the β-strand conformation. A number of cross-peaks can be assigned, including several from intra-residue (Fig. 5b, blue) and inter-residue (Fig. 5b, green) connectivities, and several reflecting long-range (Fig. 5b, red) connectivities between neighboring β-strands (Fig. 5c) that provide essential input for β-barrel structure determination.
The resolution of 13C-detected spectra from deuterated proteins can be improved by suppressing 13C–2H scalar couplings with low power 2H decoupling during acquisition (Akbey et al. 2012), made possible by more advanced RF probes (Huber et al. 2012). Furthermore, deuteration at a carbon site induces an isotope effect that manifests as a chemical shift difference relative to that of proton-attached carbon (Tang et al. 2010). In our case of fractional (~70%) deuteration, broadening of the 13C line widths is expected to arise from the mixed contributions of 1H and 2H attached to each carbon site. This broadening effect is ~0.4 ppm for CA and can be as large as 1 ppm for CH3, meaning that the resolution enhancement obtained through the use of higher magnetic field and sample deuteration is partially compromised by the presence of 13C–2H scalar couplings and by broadening induced by isotope effects. Thus, further improvements should be available through the use of 2H-decoupling and protein samples with higher levels of deuteration.
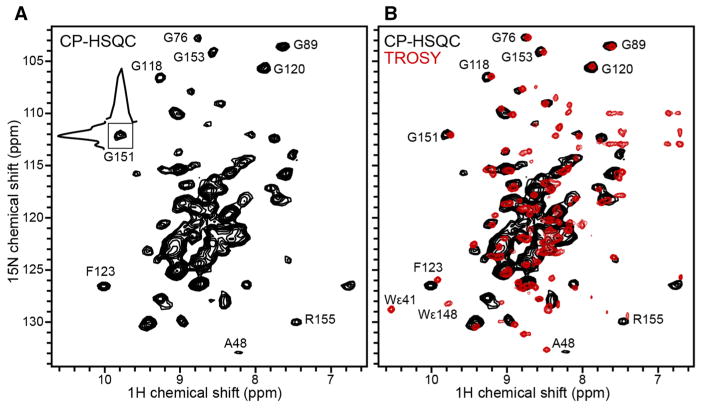
Proton detection, facilitated by fast MAS (>50 kHz) and 2H labeling to suppress the large network of strong homonuclear 1H–1H dipolar couplings, has become an important technique for enhancing sensitivity in solid-state NMR spectra. The 1H-detected 1H/15N CP-HSQC spectrum (Fig. 6) obtained for Ail (~0.3 mg) liposomes at 30 °C, is well resolved and displays the wide signal dispersion characteristic of folded β-barrel proteins. The DSC data show that at 30 °C the lipid bilayer has completed its transition from the gel to liquid crystalline phase, hence the spectrum represents Ail in fully fluid membranes. Individual resonances have line widths in the range of 0.11–0.15 ppm (107–138 Hz) for 1H, and 0.46–0.64 ppm (42–58 Hz) for 15N. These are consistent with a high level of sample homogeneity and compare very favorably with recent reports of 1H-detected spectra of β-barrel and α-helical membrane proteins in 2D crystals and other lower lipid content preparations (Barbet-Massin et al. 2014; Eddy et al. 2015).
Fig. 6.
2D 1H–15N correlation spectra of Ail in phospholipid bilayers. a Solid-state NMR 1H-detected CP-HSQC spectrum of (u-15N, u-13C, f-2H) Ail in liposomes, recorded at 900 MHz, 30 °C, with 160 scans and a MAS rate of 60 kHz. b Solution NMR 1H-detected TROSY-HSQC spectrum (red) of (u-15N, u-13C, u-2H) Ail in nanodiscs prepared with 2H labeled lipids, recorded at 800 MHz, 45 °C, with 128 scans. The solid-state NMR CP-HSQC spectrum (black) is superimposed
The solid-state NMR CP-HSQC spectrum of Ail in liposomes overlaps very well with the solution NMR TROSY spectrum (Fig. 6b, red) of Ail in nanodiscs (Ding et al. 2015), as clearly shown by the Gly signals in the region between 100 and 115 ppm. Some signals, present in the solution NMR spectrum, are not observed in the CP-HSQC spectrum; for example, signals from the two Trp (W41, W148) indole nitrogens are absent.
Conclusions
The overall backbone structures of β-barrel outer membrane proteins appear to be similar in different environments, including crystals, micelles, nanodiscs and liposomes. It is evident, however, that the environment has significant effects on both protein dynamics and function. There may be local environment-specific effects on side chain and backbone sites, and on the accessibility of protein sites to ligands, which may be occluded in one setting and unmasked in another. Here we demonstrate that MAS solid-state NMR experiments yield well resolved spectra for the Ail β-barrel embedded in liposomes, where the lipid content is sufficiently high to allow the formation of a proper bilayer.
Previous solid-state NMR studies (Ding et al. 2015, 2013; Mahalakshmi et al. 2007; Mahalakshmi and Marassi 2008; Plesniak et al. 2011; Yao et al. 2013) on Ail and OmpX in magnetically aligned bicelles, prepared with lipid to protein molar ratios of 190:1, showed that the proteins undergo rotational diffusion in the membrane, and thus, enable dipolar couplings and anisotropic chemical shifts to be measured for structural studies. Since the Ail liposomes described in this study form lamellar bilayers with phase characteristics that are very similar to those of pure lipids, we expect that the protein will be able to undergo rotational diffusion at temperatures above the gel to fluid transition (24.7 °C), and that these samples will also be suitable for experiments (Das et al. 2012) designed to measure orientation-dependent anisotropic signals under MAS.
The majority of spectra presented in this study were acquired at temperatures where the lipids are in the gel phase, to enhance spectroscopic sensitivity by dampening dynamics that interfere with magnetization transfer mediated by dipolar couplings. The 1H-detected, 1H–15N CP-HSQC spectrum, however, was obtained at 30 °C, where the lipid bilayer is completely liquid crystalline. This indicates the feasibility of performing structural studies under more physiological conditions. NMR studies with liposomes are an important first step in this direction, due to their relative ease of preparation with multiple components.
It is interesting that high resolution spectra were obtained in both the gel phase, where the hydrophobic core of the lipid bilayer is expanded, and in the liquid crystalline phase, where the hydrophobic core is thinner. It should be noted that physiological conditions for bacterial outer membrane proteins also include the presence of lipopolysaccharides, which are highly enriched in the outer leaflet of the outer membrane, and appear to have an ordering effect on the extracellular loops of β-barrels (Lee et al. 2017). Additional studies will be needed to examine the effects of membrane thickness and lipopolysaccharides on the structure and dynamics of Ail, including comparisons with spectra from protein present in native purified membranes, as described (Renault et al. 2012) for components of the E. coli outer membrane.
Importantly, the protein is functional in detergent-free lipid bilayers. As amide chemical shifts are very sensitive to the local chemical environment, 1H–15N correlation spectra are widely used to probe ligand binding in solution NMR studies of proteins. In solid-state NMR, the ability to obtain high quality CP-HSQC spectra of Ail in lipid bilayers opens an avenue to study the molecular basis for the functional interactions of Ail with its human host partners and develop therapeutic agents.
Acknowledgments
This research was supported by grants from the National Institutes of Health (GM 118186, GM 099986, and GM 066978) and by the Biotechnology Resource for Molecular Imaging of Proteins at UCSD supported by the National Institutes of Health (P41 EB 002031).
References
- Akbey U, Rossum BJ, Oschkinat H. Practical aspects of high-sensitivity multidimensional (1)(3)C MAS NMR spectroscopy of perdeuterated proteins. J Magn Reson. 2012;217:77–85. doi: 10.1016/j.jmr.2012.02.015. [DOI] [PubMed] [Google Scholar]
- Andreas LB, et al. Structure and mechanism of the influenza A M218-60 dimer of dimers. J Am Chem Soc. 2015;137:14877–14886. doi: 10.1021/jacs.5b04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J Am Chem Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- Baker LA, Folkers GE, Sinnige T, Houben K, Kaplan M, van der Cruijsen EA, Baldus M. Magic-angle-spinning solid-state NMR of membrane proteins. Methods Enzymol. 2015;557:307–328. doi: 10.1016/bs.mie.2014.12.023. [DOI] [PubMed] [Google Scholar]
- Baldus M, Petkova A, Herzfield J, Griffin R. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol Phys. 1998;95:1197–1207. [Google Scholar]
- Barbet-Massin E, et al. Rapid proton-detected NMR assignment for proteins with fast magic angle spinning. J Am Chem Soc. 2014;136:12489–12497. doi: 10.1021/ja507382j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlau M, Mills DJ, Quader H, Kuhlbrandt W, Vonck J. Projection structure of the monomeric porin OmpG at 6 A resolution. J Mol Biol. 2001;305:71–77. doi: 10.1006/jmbi.2000.4284. [DOI] [PubMed] [Google Scholar]
- Brown LS, Ladizhansky V. Membrane proteins in their native habitat as seen by solid-state NMR spectroscopy. Protein Sci. 2015;24:1333–1346. doi: 10.1002/pro.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross TA, Ekanayake V, Paulino J, Wright A. Solid state NMR: the essential technology for helical membrane protein structural characterization. J Magn Reson. 2014;239:100–109. doi: 10.1016/j.jmr.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das BB, Nothnagel HJ, Lu GJ, Son WS, Tian Y, Marassi FM, Opella SJ. Structure determination of a membrane protein in proteoliposomes. J Am Chem Soc. 2012;134:2047–2056. doi: 10.1021/ja209464f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Zorzi R, Mi W, Liao M, Walz T. Single-particle electron microscopy in the study of membrane protein structure. Microscopy. 2016;65:81–96. doi: 10.1093/jmicro/dfv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux PF, Seigneuret M. Specificity of lipid-protein interactions as determined by spectroscopic techniques. Biochim Biophys Acta. 1985;822:63–125. doi: 10.1016/0304-4157(85)90004-8. [DOI] [PubMed] [Google Scholar]
- Ding Y, Yao Y, Marassi FM. Membrane protein structure determination—in membrana. Acc Chem Res. 2013;46:2182–2190. doi: 10.1021/ar400041a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Fujimoto LM, Yao Y, Plano GV, Marassi FM. Influence of the lipid membrane environment on structure and activity of the outer membrane protein Ail from Yersinia pestis. Biochim Biophys Acta. 2015;1848:712–720. doi: 10.1016/j.bbamem.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolder M, Zeth K, Tittmann P, Gross H, Welte W, Wallimann T. Crystallization of the human, mitochondrial voltage-dependent anion-selective channel in the presence of phospholipids. J Struct Biol. 1999;127:64–71. doi: 10.1006/jsbi.1999.4141. [DOI] [PubMed] [Google Scholar]
- Eddy MT, et al. Lipid dynamics and protein-lipid interactions in 2D crystals formed with the beta-barrel integral membrane protein VDAC1. J Am Chem Soc. 2012;134:6375–6387. doi: 10.1021/ja300347v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy MT, et al. Lipid bilayer-bound conformation of an integral membrane beta barrel protein by multidimensional MAS NMR. J Biomol NMR. 2015;61:299–310. doi: 10.1007/s10858-015-9903-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst M, Samoson A, Meier BH. Low-power XiX decoupling in MAS NMR experiments. J Magn Reson. 2003;163:332–339. doi: 10.1016/s1090-7807(03)00155-1. [DOI] [PubMed] [Google Scholar]
- Findlay EJ, Barton PG. Phase behavior of synthetic phosphatidylglycerols and binary mixtures with phosphatidylcholines in the presence and absence of calcium ions. BioChemistry. 1978;17:2400–2405. doi: 10.1021/bi00605a023. [DOI] [PubMed] [Google Scholar]
- Fung BM, Khitrin AK, Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J Magn Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- Gennis RB. Biomembranes : molecular structure and function. springer advanced texts in chemistry. Springer; New York: 1989. [Google Scholar]
- Hagn F, Etzkorn M, Raschle T, Wagner G. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J Am Chem Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Krabben L, Vinothkumar KR, Castellani F, van Rossum BJ, Kuhlbrandt W, Oschkinat H. Solid-state magic-angle spinning NMR of outer-membrane protein G from Escherichia coli. Chembiochem. 2005;6:1679–1684. doi: 10.1002/cbic.200500132. [DOI] [PubMed] [Google Scholar]
- Huber M, With O, Schanda P, Verel R, Ernst M, Meier BH. A supplementary coil for (2)H decoupling with commercial HCN MAS probes. J Magn Reson. 2012;214:76–80. doi: 10.1016/j.jmr.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Janiak MJ, Small DM, Shipley GG. Nature of the Thermal pretransition of synthetic phospholipids: dimyristolyl- and dipalmitoyllecithin. BioChemistry. 1976;15:4575–4580. doi: 10.1021/bi00666a005. [DOI] [PubMed] [Google Scholar]
- Lee AG. Lipid-protein interactions in biological membranes: a structural perspective. Biochim Biophys Acta. 2003;1612:1–40. doi: 10.1016/s0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- Lee AG. Biological membranes: the importance of molecular detail. Trends Biochem Sci. 2011;36:493–500. doi: 10.1016/j.tibs.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Lee J, Patel DS, Kucharska I, Tamm LK, Im W. Refinement of OprH-LPS interactions by molecular simulations. Biophys J. 2017;112:346–355. doi: 10.1016/j.bpj.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leftin A, Brown MF. An NMR database for simulations of membrane dynamics. Biochim Biophys Acta. 2011;1808:818–839. doi: 10.1016/j.bbamem.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Berthold DA, Frericks HL, Gennis RB, Rienstra CM. Partial (13)C and (15)N chemical-shift assignments of the disulfide-bond-forming enzyme DsbB by 3D magic-angle spinning NMR spectroscopy. Chembiochem. 2007;8:434–442. doi: 10.1002/cbic.200600484. [DOI] [PubMed] [Google Scholar]
- Linser R, et al. Proton-detected solid-state NMR spectroscopy of fibrillar and membrane proteins. Angew Chem Int Ed Engl. 2011;50:4508–4512. doi: 10.1002/anie.201008244. [DOI] [PubMed] [Google Scholar]
- Mahalakshmi R, Marassi FM. Orientation of the Escherichia coli outer membrane protein OmpX in phospholipid bilayer membranes determined by solid-state NMR. BioChemistry. 2008;47:6531–6538. doi: 10.1021/bi800362b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalakshmi R, Franzin CM, Choi J, Marassi FM. NMR structural studies of the bacterial outer membrane protein OmpX in oriented lipid bilayer membranes. Biochim Biophys Acta. 2007;1768:3216–3224. doi: 10.1016/j.bbamem.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marassi FM, Ding Y, Schwieters CD, Tian Y, Yao Y. Backbone structure of Yersinia pestis Ail determined in micelles by NMR-restrained simulated annealing with implicit membrane solvation. J Biomol NMR. 2015;63:59–65. doi: 10.1007/s10858-015-9963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslennikov I, Choe S. Advances in NMR structures of integral membrane proteins. Curr Opin Struct Biol. 2013;23:555–562. doi: 10.1016/j.sbi.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes I, Evans G, Sanchez-Weatherby J, Newstead S, Stewart PD. Membrane protein structure determination—the next generation. Biochim Biophys Acta. 2014;1838:78–87. doi: 10.1016/j.bbamem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GA, Freeman R. Enhancement of nuclear magnetic resonance signals by polarization transfer. J Am Chem Soc. 1979;101:760–762. doi: 10.1021/ja00497a058. [DOI] [Google Scholar]
- Pauli J, Baldus M, van Rossum B, de Groot H, Oschkinat H. Backbone and side-chain 13 C and 15 N signal assignments of the alpha-spectrin SH3 domain by magic angle spinning solid-state NMR at 17.6 T. Chembiochem. 2001;2:272–281. doi: 10.1002/1439-7633(20010401)2:4<272::AID-CBIC272>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Pines A, Gibby MG, Waugh JS. Proton-enhanced NMR of dilute spins in solids. J Chem Phys. 1973;59:569–590. doi: 10.1063/1.1680061. [DOI] [Google Scholar]
- Plesniak LA, Mahalakshmi R, Rypien C, Yang Y, Racic J, Marassi FM. Expression, refolding, and initial structural characterization of the Y. pestis Ail outer membrane protein in lipids. Biochim Biophys Acta. 2011;1808:482–489. doi: 10.1016/j.bbamem.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand RP, Chapman D, Larsson K. Tilted hydrocarbon chains of dipalmitoyl lecithin become perpendicular to the bilayer before melting. Biophys J. 1975;15:1117–1124. doi: 10.1016/S0006-3495(75)85888-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault M, Tommassen-van Boxtel R, Bos MP, Post JA, Tommassen J, Baldus M. Cellular solid-state nuclear magnetic resonance spectroscopy. Proc Natl Acad Sci USA. 2012;109:4863–4868. doi: 10.1073/pnas.1116478109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurel O, et al. Local and global dynamics in klebsiella pneumoniae outer membrane protein a in lipid bilayers probed at atomic resolution. J Am Chem Soc. 2017 doi: 10.1021/jacs.6b11565. [DOI] [PubMed] [Google Scholar]
- Shahid SA, Markovic S, Linke D, van Rossum BJ. Assignment and secondary structure of the YadA membrane protein by solid-state MAS NMR. Sci Rep. 2012;2:803. doi: 10.1038/srep00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaka AJ, Keeler J, Frenkiel T, Freeman R. An improved sequence for broadband decoupling: WALTZ-16. J Magn Reson. 1983;52:335–338. doi: 10.1016/0022-2364(83)90207-X. [DOI] [Google Scholar]
- Szeverenyi NM, Sullivan MJ, Maciel GE. Observation of spin exchange by two-dimensional fourier transform 13 C cross polarization-magic-angle spinning. J Magn Reson. 1982;47:462–475. doi: 10.1016/0022-2364(82)90213-X. [DOI] [Google Scholar]
- Takamori S, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Takegoshi K, Nakamura S, Terao T. 13 C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. doi: 10.1063/1.2364503. [DOI] [PubMed] [Google Scholar]
- Takegoshi K, Nakamura S, Terao T. 13C–1 H dipolar-driven 13C–13 C recoupling without 13 C rf irradiation in nuclear magnetic resonance of rotating solids. J Chem Phys. 2003;118:2325–2341. doi: 10.1063/1.1534105. [DOI] [Google Scholar]
- Takeuchi K, Arthanari H, Shimada I, Wagner G. Nitrogen detected TROSY at high field yields high resolution and sensitivity for protein NMR. J Biomol NMR. 2015;63:323–331. doi: 10.1007/s10858-015-9991-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Comellas G, Mueller LJ, Rienstra CM. High resolution (1)(3)C-detected solid-state NMR spectroscopy of a deuterated protein. J Biomol NMR. 2010;48:103–111. doi: 10.1007/s10858-010-9442-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu A, Luzzati V, Reman FC. Structure and polymorphism of the hydrocarbon chains of lipids: a study of lecithin-water phases. J Mol Biol. 1973;75:711–733. doi: 10.1016/0022-2836(73)90303-3. [DOI] [PubMed] [Google Scholar]
- Warschawski DE, Devaux PF. 1 H-13C polarization transfer in membranes: a tool for probing lipid dynamics and the effect of cholesterol. J Magn Reson. 2005;177:166–171. doi: 10.1016/j.jmr.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Yamashita S, et al. Structural insights into Ail-mediated adhesion in Yersinia pestis. Structure. 2011;19:1672–1682. doi: 10.1016/j.str.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Ding Y, Tian Y, Opella SJ, Marassi FM. Membrane protein structure determination: back to the membrane. Methods Mol Biol. 2013;1063:145–158. doi: 10.1007/978-1-62703-583-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX, Cross TA. Influences of membrane mimetic environments on membrane protein structures. Annu Rev Biophys. 2013;42:361–392. doi: 10.1146/annurev-biophys-083012-130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou DH, Rienstra CM. High-performance solvent suppression for proton detected solid-state NMR. J Magn Reson. 2008;192:167–172. doi: 10.1016/j.jmr.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]