Abstract
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Our laboratory has previously demonstrated that high-level HCV replication during acute infection of chimpanzees is associated with the modulation of multiple genes involved in lipid metabolism, and that drugs that regulate cholesterol and fatty acid biosynthesis regulate the replication of the subgenomic HCV replicon in Huh-7 cells. In this article, we demonstrate that Huh-7 cells harboring replicating, full-length HCV RNAs express elevated levels of ATP citrate lyase and acetyl-CoA synthetase genes, both of which are involved in cholesterol and fatty acid biosynthesis. Further, we confirm that the cholesterol-biosynthetic pathway controls HCV RNA replication by regulating the cellular levels of geranylgeranyl pyrophosphate, we demonstrate that the impact of geranylgeranylation depends on the fatty acid content of the cell, and we show that fatty acids can either stimulate or inhibit HCV replication, depending on their degree of saturation. These results illustrate a complex cellular-regulatory network that controls HCV RNA replication, presumably by modulating the trafficking and association of cellular and/or viral proteins with cellular membranes, suggesting that pharmacologic manipulation of these pathways may have a therapeutic effect in chronic HCV infection.
Keywords: cholesterol, replicon
Hepatitis C virus (HCV), a member of the Flaviviridae family of viruses, is a major cause of chronic hepatitis and hepatocellular carcinoma (1, 2). The HCV genome is a positive-stranded ≈9.6-kb RNA molecule consisting of a single ORF, which is flanked by 5′ and 3′ untranslated regions (UTR). The HCV 5′ UTR contains a highly structured internal ribosome entry site (3–8), and the 3′ UTR is essential for replication (9, 10). The HCV ORF encodes a single 3,008- to 3,037-aa polyprotein that is posttranslationally processed to produce ≥10 different proteins: core protein, envelope proteins E1 and E2, p7, and nonstructural proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B (1, 8, 11). Our understanding of the biology of HCV RNA replication has been greatly facilitated by the development of subgenomic and full-length HCV replicons that express HCV proteins and replicate their RNA in stably transfected human hepatoma-cell-derived Huh-7 cells.
Our laboratory has previously demonstrated (12) that multiple cellular genes involved in lipid metabolism are differentially regulated during viral spread in acutely infected chimpanzees, and that ATP citrate lyase, which is required for cholesterol and fatty acid biosynthesis, is induced during the initial rise of high-level HCV replication during acute infection in chimpanzees. There is considerable evidence suggesting that the cholesterol and fatty-acid-biosynthesis pathways may play a role in HCV replication and infection. Steatosis, i.e., the formation of hepatocellular lipid droplets, is a well documented histological characteristic of HCV infection in humans, chimpanzees, and mouse models of HCV core-protein expression (13–15). Furthermore, HCV core and NS5A proteins associate with lipid droplets, and NS5A has been reported to associate with apolipoproteins A1 and A2 (16, 17). These data strongly suggest that HCV may directly affect one or more steps in cholesterol and/or fatty acid biosynthesis. This is not surprising because cholesterol, fatty acids, and lipid rafts have been demonstrated to be critical for efficient replication and/or infection of RNA and many DNA viruses (18–27).
Genes encoding enzymes involved in cholesterol and fatty acid biosynthesis are transcriptionally regulated by sterol regulatory element-binding proteins (SREBPs), which are members of a family of the basic helix–loop–helix leucine-zipper family of transcription factors (reviewed in ref. 28). Whereas SREBP-2 plays a critical role in cholesterol biosynthesis, SREBP-1c regulates the expression of enzymes involved in fatty acid biosynthesis in the liver, e.g., fatty-acid synthase (FAS) and stearoyl-CoA desaturase-1 (29, 30). SREBPs are transcriptionally controlled by liver X receptors (LXRs) α and β, which are members of a family of nuclear hormone receptors (31). In addition to the synthesis of sterols, enzymes involved in cholesterol biosynthesis also generate the isoprenoids, farnesyl pyrophosphate and geranylgeranyl pyrophosphate, which are covalently attached to proteins and essential for their membrane association (reviewed in ref. 32). Saturated and monounsaturated fatty acids are a major component of biological membranes. In addition, a third class of fatty acids, polyunsaturated fatty acids (PUFAs), which are generated from dietary linoleate and linolenate by Δ5- and Δ6-desaturase enzymes, are involved in multiple cellular processes such as regulation of gene expression, defense against certain pathogens, interference with immune cell function, and regulation of activities of membrane-associated enzymes (33–38). In this article, we demonstrate that HCV RNA replication in Huh-7 cells is strongly regulated by pharmacological manipulation of the cholesterol and fatty-acid-synthetic pathways. Furthermore, we demonstrate that, whereas saturated and monounsaturated fatty acids are required for HCV RNA replication, PUFAs inhibit HCV replication by a mechanism that is independent of their ability to suppress lipogenic gene expression.
Materials and Methods
Tissue Culture and Generation of the Stable HCV Full-Length Replicon Cell Line. The SfiI HCV full-length replicon (genotype 1b) was obtained from Ralf Bartenschlager (University of Heidelberg, Heidelberg). An Huh-7 cell clone that stably replicates the full-length HCV RNA (called SfiI in this article) was used in all experiments and passaged as described in refs. 39–41. SfiI cells were treated with IFNα, as described in ref. 42, to eliminate self-replicating full-length SfiI replicon RNA (designated as IFN “cured” in this article). SfiI cells were cultured in complete DMEM supplemented with 10% FCS, 100 units/ml penicillin, 100 mg/ml streptomycin, 2 mM l-glutamine, and 500 μg/ml geneticin (Invitrogen). For all drug treatments, SfiI cells (≈8 × 104) were seeded into 60-mm tissue-culture dishes in complete DMEM without geneticin. Two days later, cells were treated with respective small molecules in medium containing 5% FCS. In all drug treatments, cells were treated with vehicle alone (designated as mock-treated cells), which never exceeded 0.1% of the total volume.
Cholesterol and Fatty-Acid-Biosynthesis Inhibitors. Lovastatin (Calbiochem) was activated as described in refs. 43 and 44. For mock-treated cells, the activation procedure was performed in the absence of lovastatin. L-659,699 and zaragozic acid A (ZA) (Merck) were dissolved in DMSO (Sigma) at concentrations of 50 mg/ml and 5 mg/ml. 5-(Tetradecyloxy)-2-furoic acid (TOFA) (obtained from Craig Townsend, The Johns Hopkins University, Baltimore) was dissolved in DMSO at a final concentration of 5 mg/ml. The LXR agonist T0901317 (Cayman Chemicals, Ann Arbor, MI) was dissolved at a concentration of 5 mM.
Generation of Ribonuclease-Protection Assay (RPA) Probes for Genes Involved in Cholesterol and Fatty Acid Biosynthesis. RPA probes were generated by PCR using gene-specific primers. Each antisense primer had a unique HindIII restriction site added to linearize the template. Total RNA was isolated as described above and RPA was performed as described in ref. 45. The sequences for each primer are as follows: ATP citrate lyase (sense) 5′-CTCCGATGAGACCATCTACATT-3′ and (antisense) 5′-ATCAAGCTTGTACTCTTGCCAGTTCATTGAGA-3′; acetyl-CoA synthetase (sense) 5′-AAGGATGCCCGGCTATGATTGGT-3′ and (antisense) 5′-ATCAAGCTTCGAGTGGTGTGATGCCGAGGAC-3′; FAS (sense) 5′-TGTCGTTGGTGCTCATCGTC-3′ and (antisense) 5′-ATCAAGCTTTGGTCTTGAGAGATGGCTTGA-3′; hydroxymethylglutaryl (HMG)-CoA synthase (sense) 5′-ATCAAGCTTTATGCCCTGGTAGTTGCAGGAG-3′ and (antisense) 5′-T TGCCTCT T TCTGCCACTGG-3′; HMG-CoA reductase (sense) 5′-ATCAAGCTTTACCATGTCAGGGGTACGTC-3′ and (antisense) 5′-CAAGCCTAGAGACATAATCATC-3′; squalene synthase (sense) 5′-AAAAACTCTGCCATCCCAATGC-3′ and (antisense) 5′-ATCAAGCTTAAGAAGGTCCCGCTGTTACACAA-3′; L32 (sense) 5′-GCGATCTCGGCACAGTAAGATTT-3′ and (antisense) 5′-ATCAAGCTTCTTGATGCCCAACATTGGTTATG-3′. Gels were dried down, and bands were visualized and quantitated as described in refs. 46–48.
Fatty Acids. Saturated (lauric, myristic, and palmitic), monounsaturated (oleic), and polyunsaturated [arachidonic, eicosapentaenoic (EPA), and docosahexaenoic (DHA)] fatty acids (Sigma) were complexed to fatty-acid-free BSA (Sigma) as described in ref. 49. Mock-treated cells were treated with equivalent amounts of BSA in the absence of any complexed fatty acids.
Northern and Western Blot Analyses. Total RNA was extracted from transfected cells at various times posttreatment and 7–8 μg of total RNA was loaded onto the gel. Generation of the probe and hybridization for positive-stranded HCV and GAPDH transcripts was performed as described in ref. 45. Western blot analysis was performed on 40–60 μg of total protein lysates by using antibodies to NS3, NS5A, and NS5B, as described in ref. 45. Relative quantitation of protein bands (normalized to GAPDH protein-expression levels) was performed by using optiquant imaging software (Packard).
Reverse Transcription and Real-Time PCR Analysis. Reverse transcription reactions and real-time PCR analyses were performed as described in ref. 45. Primers to detect gene expression of GAPDH and SREBP-1c, -1a, and -2 isoforms were generated by using sequences described in refs. 45 and 50. Analysis of relative gene expression of SREBP isoforms was performed as described in refs. 45 and 51 by normalizing to GAPDH expression levels.
Statistical Analyses. All statistical analyses were performed by using Microsoft excel software. All graphs represent the mean ± SD. P values for real-time PCR data were determined by using a paired t test.
Results
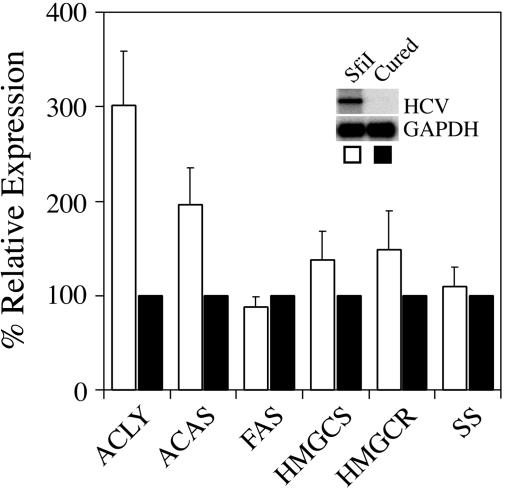
ATP Citrate Lyase and Acetyl-CoA Synthetase Gene Expression Is Induced in Huh-7 Cells That Replicate a Full-Length HCV Replicon. Because multiple lipid-metabolism genes were regulated during high-level HCV replication in acutely infected chimpanzees (12), we wanted to determine whether lipogenic gene expression was altered in Huh-7 cells containing the full-length genotype 1b HCV replicon (designated as SfiI). Using RPA analysis, we determined that ATP citrate lyase and acetyl-CoA synthetase gene expression was induced ≈3.01-fold (P = 0.023) and ≈1.96-fold (P = 0.042), respectively, in SfiI cells (Fig. 1), as compared with IFN-cured replicon cells (described in Materials and Methods), which were negative for HCV RNA by Northern blot analysis (Fig. 1 Inset) or real-time RT-PCR (data not shown). Gene expression in parental Huh-7 was similar to that seen in IFN-cured cells (data not shown). In contrast, no significant differences were detected in expression levels of other enzymes involved in cholesterol or fatty acid biosynthesis (Fig. 1). These data, along with previous gene-expression profiling analyses in acutely infected chimpanzees (12), suggest that HCV may directly or indirectly induce cholesterol and/or fatty-acid-biosynthetic gene expression.
Fig. 1.
Induction of ATP citrate lyase and acetyl-CoA synthetase gene expression in Huh-7 cells that replicate the full-length SfiI replicon. RPA was performed by using probes specific to ATP citrate lyase (ACLY), acetyl-CoA synthetase (ACAS), fatty acid synthase (FAS), HMG-CoA synthase (HMGCS), HMG-CoA reductase (HMGCR), and squalene synthase (SS), and expression levels in SfiI cells (white bars) were compared with IFN-cured cells (black bars). Representative data (mean + SD) from three independent experiments are shown. The Inset represents Northern blot analysis to detect HCV RNA.
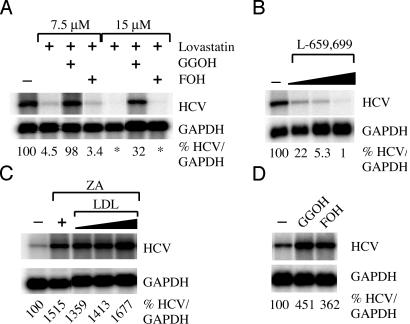
Inhibition of Geranylgeranylation Decreases HCV RNA Replication and HCV Protein Expression. We (12) and others (52) have reported that HCV RNA replication is positively and negatively regulated by activators and inhibitors, respectively, of the cholesterol biosynthetic pathway, and that the inhibitory effect can be overcome by the addition of geranylgeraniol (52). To confirm and extend these observations, we showed that lovastatin, a potent inhibitor of HMG-CoA reductase, inhibited HCV RNA replication up to ≈22-fold (Fig. 2A), and that this inhibition could be blocked by the addition of geranylgeraniol but not farnesol, as described in ref. 52. Furthermore, we showed that, under baseline conditions, the addition of both geranylgeraniol and farnesol induced HCV replication ≈4.5- and ≈3.6-fold, respectively (Fig. 2D). Collectively, these results suggest not only that geranylgeranylation of a host and/or viral protein(s) regulates HCV RNA replication under baseline conditions but also that the enhancement of these functions in response to the induction of lipogenic gene expression may promote viral replication during HCV infection. To confirm that the antiviral effect of lovastatin was not due to inhibition of cell-cycle progression, we analyzed cell-cycle progression in carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled SfiI cells treated with lovastatin. No effect on cell-cycle progression was observed (data not shown).
Fig. 2.
Geranylgeranylation is required for HCV RNA replication. Northern blot analyses were performed on total RNA harvested from SfiI cells treated with various small-molecule inhibitors. (A) SfiI cells were treated with either 7.5 or 15 μM lovastatin alone or in the presence of 10 μM geranylgeraniol or farnesol. (B) SfiI cells were treated with 5, 10, or 20 μg/ml L-659,699, and total RNA was harvested at 5 days posttreatment. (C) SfiI cells were treated with 20 μg/ml ZA alone or in the presence or absence of low-density lipoprotein (LDL). (D) SfiI cells were treated with 10 μM geranylgeraniol (GGOH) or farnesol (FOH), and total RNA was harvested on day 5 posttreatment. In all cases, relative levels of HCV RNA are shown as percentages normalized to GAPDH RNA levels. *, Transcript levels were undetectable.
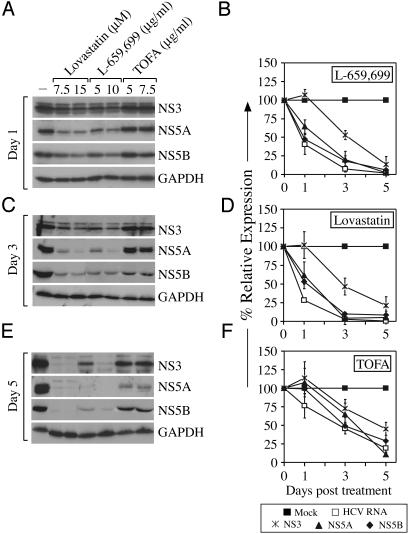
Because lovastatin has adverse effects on many host cellular processes (53, 54), we also treated SfiI cells with L-659,699 and ZA, which are specific inhibitors of HMG-CoA synthase (55, 56) and squalene synthase (57), respectively. Treatment of SfiI cells with L-659,699 also inhibited HCV RNA replication in a dose-dependent manner [≈4.5- and ≈18.9-fold at 5 and 10 μg/ml doses, respectively, at 3 days posttreatment (Fig. 2B)] without any detectable effect on cell-cycle progression (data not shown). Whereas treatment with lovastatin and L-659,699 resulted in a dose-dependent decrease in expression of HCV nonstructural proteins NS3, NS5A, and NS5B (Fig. 3 A, C, and E), the kinetics of decrease of NS5A and NS5B more closely mimicked the decay of HCV RNA (Fig. 3 B and D). Treatment of SfiI cells with ZA resulted in a ≈15-fold induction of HCV replication (Fig. 2C). Because ZA treatment has been demonstrated to induce the accumulation of upstream intermediates and/or upstream enzyme gene expression (57, 58), we wanted to confirm that the induction in HCV RNA replication was due to an accumulation of upstream intermediates (i.e., geranylgeranyl pyrophosphate) and not due to the lack of downstream products. Therefore, we incubated ZA-treated cells in the presence of low-density lipoprotein (LDL) because it has been demonstrated that cholesterol needs can be met by exogenous supplementation via the LDL-receptor system (59). Treatment of SfiI cells with LDL did not prevent the induction of HCV RNA caused by ZA treatment (Fig. 2C). Taken together, these data confirm that inhibition of geranylgeranylation, rather than the synthesis of cholesterol itself, is responsible for the inhibition of HCV RNA replication.
Fig. 3.
Inhibition of HCV protein synthesis by small-molecule inhibitors of cholesterol and fatty acid biosynthesis. (A, C, and E) SfiI cells were mock-treated or treated with increasing doses of lovastatin, L-659,699, or TOFA. Total cell lysates were harvested at various times posttreatment, and SDS/PAGE analyses were performed by using antibodies specific to NS3, NS5A, NS5B, or GAPDH. (B, D, and F) Kinetic analysis of HCV protein expression of SfiI cells treated with L-659,699 (B), lovastatin (D), or TOFA (F) compared with HCV RNA levels. Representative data (mean ± SD) from at least three independent experiments are shown.
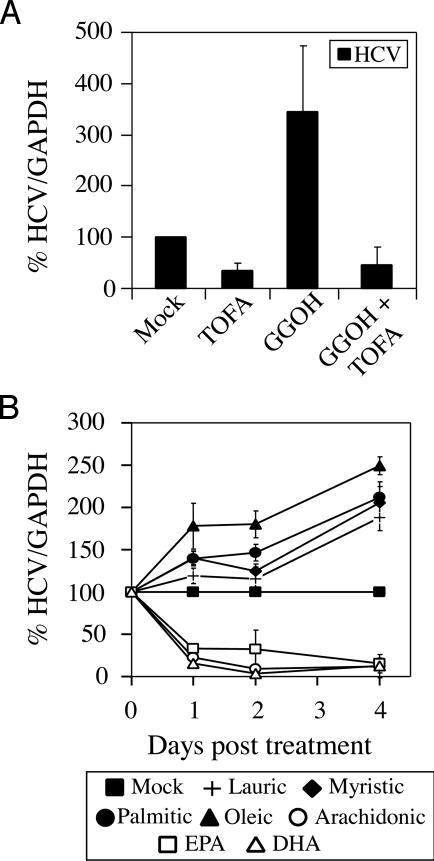
Inhibition of Fatty Acid Biosynthesis Inhibits HCV RNA Replication and HCV Protein Expression. To determine whether the fatty acid biosynthetic pathway plays a role in regulating HCV replication, we inhibited the enzyme acetyl-CoA carboxylase by using TOFA (60, 61). Treatment of SfiI cells with TOFA resulted in a ≈3-fold inhibition of HCV RNA replication (Fig. 4A), consistent with previous published results from our laboratory (12) using a FAS inhibitor. As expected, treatment of SfiI cells with TOFA also resulted in decreased HCV protein expression (Fig. 3). Because TOFA and cerulenin inhibit global fatty acid synthesis (61, 62), we studied the impact of representative saturated (lauric, myristic, and palmitic) and monounsaturated (oleic) fatty acids on HCV RNA replication in SfiI cells. As shown in Fig. 4B, lauric, myristic, palmitic, and oleic acids induced HCV RNA replication [≈1.9-fold (P = 0.0174), ≈2.1-fold (P = 0.018), ≈2.1-fold (P = 0.0184), and ≈2.5-fold (P = 0.0029), respectively, at 4 days posttreatment; Fig. 4B], demonstrating that fatty acid synthesis and, in particular, these saturated and monounsaturated fatty acids were required for HCV RNA replication.
Fig. 4.
Inhibition of basal and geranylgeraniol-induced HCV RNA replication by TOFA. (A) SfiI cells were mock-treated or treated with 5 μg/ml TOFA, 10 μM geranylgeraniol (GGOH), or TOFA and GGOH. (B) SfiI cells were treated with 50 μM fatty acids complexed to fatty-acid-free BSA. Total RNA was harvested at various times posttreatment and Northern blot analysis was performed. Levels of HCV RNA are represented as a percentage normalized to GAPDH RNA levels. Representative data (mean + SD) from three independent experiments are shown.
Because the addition of geranylgeranyl isoprenoids to certain proteins facilitates their interactions with membrane bilayers that contain fatty acids as a major component (32, 63), we proposed that deregulation of fatty acid synthesis may affect membrane integrity, and, thus, affect the localization and/or function of prenylated proteins. Therefore, we examined whether the prenylation and fatty-acid-synthetic pathways worked together in regulating HCV RNA replication. To test this effect, we incubated geranylgeraniol-treated SfiI cells in the presence or absence of TOFA. Whereas the addition of geranylgeraniol induced HCV RNA replication by ≈3.5-fold, this increase could be completely blocked by the fatty-acid-synthesis inhibitor TOFA (Fig. 4A), suggesting that both these pathways are required for efficient HCV RNA replication to occur.
Exogenous Addition of PUFAs Inhibits HCV RNA Replication. It is noteworthy that the inhibitors of acetyl-CoA carboxylase (Fig. 4A) and FAS (12) that suppress HCV RNA replication have no effect on PUFA synthesis. In contrast to saturated and monounsaturated fatty acids, PUFAs have long been known for their antilipogenic effects (64–66). Thus, we hypothesized that PUFAs might suppress HCV RNA replication. Indeed, whereas saturated (lauric, myristic, and palmitic) and monounsaturated (oleic) fatty acids induced HCV RNA replication at 4 days posttreatment (Fig. 4B), PUFAs, such as arachidonic acid, EPA, and DHA, inhibited HCV RNA replication at even 1 day posttreatment [≈4.5-fold (P = 0.0005), ≈3-fold (P = 0.0047), and ≈6.4-fold (P = 0.0002) for arachidonic acid, EPA, and DHA, respectively (Fig. 4B)]. Treatment with PUFAs had no detectable effect on cell-cycle progression (data not shown).
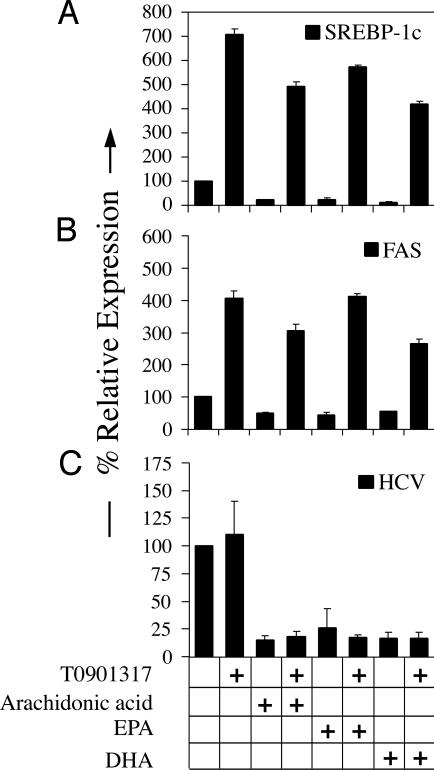
The LXR–SREBP-1c Pathway Does Not Play a Role in PUFA-Mediated Suppression of HCV Replication. Using the fatty acid synthesis inhibitors TOFA (Fig. 3A) and cerulenin (12), we have demonstrated that HCV RNA replication requires the synthesis of fatty acids. PUFAs have been demonstrated to inhibit de novo fatty acid and triglyceride synthesis, in part, by acting as antagonists of the nuclear receptor LXRα, and, thus, inhibiting transcription of lipogenic genes such as SREBP-1c, FAS, and stearoyl-CoA desaturase (64–68). Therefore, we wanted to determine whether the PUFA-mediated inhibition of HCV RNA replication was mediated through antagonism of the LXR–SREBP-1c pathway. As expected, PUFAs, but not the saturated or monounsaturated fatty acids, inhibited gene expression of SREBP-1c and FAS (Fig. 6 B and C, which is published as supporting information on the PNAS web site), whereas expression of SREBP-2, which is preferentially involved in inducing cholesterol-biosynthetic enzyme gene expression, was not significantly changed (Fig. 6D). Thus, PUFAs specifically inhibited SREBP-1c and FAS gene expression in Huh-7 cells replicating the SfiI HCV replicon, consistent with the previously published (64, 69) role of PUFAs in hepatocytes. To determine whether PUFA-mediated inhibition of LXRα activation was the mechanism by which HCV RNA replication was being inhibited, we treated SfiI cells with PUFAs in the presence or absence of an LXR agonist T0901317, which has been previously reported (70, 71) to induce expression of SREBP-1c, FAS, and stearoyl-COA desaturase. T0901317 not only induced the expression of SREBP-1c and FAS in the SfiI cells, it also rescued the inhibitory effect of arachidonic acid, EPA, and DHA on SREBP-1c and FAS expression (Fig. 5). However, T0901317 had no effect on baseline HCV RNA replication, and it did not reverse the antagonistic effects of PUFAs (Fig. 5C). These results suggest that the PUFAs inhibit HCV RNA replication by a mechanism that is independent of their ability to inhibit lipogenic gene expression by antagonizing LXRα.
Fig. 5.
PUFA-mediated inhibition of HCV RNA replication is not mediated through antagonizing LXRα–SREBP-1c activation. SfiI cells were treated with 50 μM arachidonic acid, EPA, or DHA alone or in the presence of the LXR agonist T0901317. Total RNA was harvested at 2 days posttreatment, and real-time RT-PCR analyses were performed to determine expression levels of SREBP-1c (A) and FAS (B). Northern blot analyses were performed to determine HCV RNA levels (C). Representative data (mean + SD) from three independent experiments are shown.
Discussion
We have demonstrated that elements of the cholesterol and fatty-acid-biosynthetic pathways are required for HCV RNA replication in Huh-7 cells that stably replicate the full-length SfiI HCV replicon. Using small-molecule inhibitors that interfere with cholesterol biosynthesis, we demonstrated that geranylgeranylation is essential for HCV RNA replication, as described by Ye et al. (52). Whereas the large delta antigen of the hepatitis delta virus contains a CXXX motif and is farnesylated (59), no consensus CAAX motifs are present in the HCV polypeptide sequence. This observation led Ye et al. (52) to suggest that one or more host geranylgeranylated proteins are needed for HCV RNA replication to occur. In the current study, we have shown that geranylgeraniol and farnesol actually stimulate HCV RNA replication, suggesting that they may exert a proviral effect as a result of the induction of lipogenic gene expression in replicon-containing Huh-7 cells (Fig. 1) and in acutely infected chimpanzees (12). Our results further demonstrate that the proviral effect of geranylgeraniol is antagonized by the acetyl-CoA carboxylase inhibitor, TOFA, suggesting that geranylgeranylation alone does not mediate efficient HCV RNA replication, but that fatty acid synthesis is required as well, perhaps to help form the fatty-acid-containing cellular membranes to which the putative geranylgeranylated protein(s) are most likely targeted. Furthermore, Western blot data suggest that NS5A and NS5B proteins seem to disappear faster than does NS3 after treatment with cholesterol-synthesis inhibitors (Fig. 3). Whereas the actual half-lives of these proteins in replicon-containing cells is unknown, it is possible that NS5A and/or NS5B localization to the intracellular membrane and HCV-replication complex may directly or indirectly depend on the geranylgeranylated protein(s). Further experiments will be needed to address this issue.
Inhibition of fatty acid synthesis by inhibiting acetyl-CoA carboxylase led to a decrease in HCV replication, consistent with our previously published data (12), which demonstrated that inhibition of FAS by using the drug cerulenin inhibited replication of the subgenomic HCV RNA replicon. We have further demonstrated that saturated and monounsaturated fatty acids can enhance HCV RNA replication, consistent with the requirement of the fatty-acid-biosynthetic pathway in HCV RNA replication. Our data suggest that saturated and monounsaturated fatty acids are required for efficient HCV replication, most likely, in part, by maintaining optimal membrane structure. As HCV replication has been demonstrated to occur in an HCV-induced “membranous web,” consisting of modified lipid-containing intracellular vesicles (72, 73), it is plausible to propose that fatty acids may be needed for proper architecture of this structure that is required for HCV RNA replication to occur. However, further experiments are warranted to fully determine the role(s) of saturated and monounsaturated fatty acids in HCV replication.
We also demonstrated that PUFAs inhibit HCV RNA replication by a mechanism(s) that is independent of their ability to antagonize LXR activation. PUFAs dramatically inhibited HCV RNA replication (Fig. 4B), which is consistent with recent data published by Leu et al. (74). PUFAs have long been known to suppress hepatic lipogenesis, reduce hepatic triglyceride levels, and induce fatty-acid oxidation and degradation (75–77). Because we have previously demonstrated that HCV RNA replication requires fatty-acid synthesis (12), it was interesting that the inhibitory effect on HCV replication by PUFAs is not mediated by an inhibition of lipogenesis (Fig. 5), suggesting that PUFAs inhibit HCV RNA replication by some other mechanism. In addition to their roles in regulating lipogenesis, PUFAs have been demonstrated to interfere with membrane fluidity (38), and certain PUFAs, such as arachidonic acid, can be metabolized into short-lived prostaglandins, leukotrienes, and thromboxanes, which serve as ligands for G-protein-coupled receptors (78). Evidence in the literature suggests that PUFAs can have both positive and negative effects on the host antiviral response (reviewed in ref. 79). Thus, further experiments are needed to determine how PUFAs inhibit HCV RNA replication in replicon-containing cells and also whether, and to what extent, PUFAs can inhibit HCV replication in vivo.
In conclusion, we have demonstrated that HCV requires elements of the cholesterol- and fatty-acid-biosynthetic pathways for efficient replication. Further experiments to determine the precise mechanism(s) responsible for the antiviral effects of lovastatin and PUFAs may lead to the development of novel therapeutic agents for the treatment of chronic HCV infection.
Supplementary Material
Acknowledgments
We thank Dr. Ralf Bartenschlager for providing the HCV full-length (SfiI) replicon; Dr. Darius Moradpour (University of Freiburg, Freiburg, Germany) for supplying the monoclonal antibodies to NS3, NS5A, and NS5B proteins; and Drs. Stefan Wieland, Alan McLachlan, and Pablo Gastaminza for helpful comments and discussions. This work was supported by National Institutes of Health Grants CA A1053988 and CA76403 (to F.V.C.) and an American Cancer Society Gloria Rosen Postdoctoral Research Fellowship (to S.B.K.).
Author contributions: S.B.K. and F.V.C. designed research; S.B.K. performed research; S.B.K. and F.V.C. analyzed data; and S.B.K. and F.V.C. wrote the paper.
Abbreviations: HCV, hepatitis C virus; SREBP, sterol regulatory element-binding protein; FAS, fatty-acid synthase; HMG, hydroxymethylglutaryl; LXRs, liver X receptors; PUFAs, polyunsaturated fatty acids; ZA, zaragozic acid A; TOFA, 5-(tetradecyloxy)-2-furoic acid; RPA, ribonuclease-protection assay; EPA, eicosapentaenoic acid; DHA, docosahexaenoic acid.
References
- 1.Bradley, D. W. (2000) Curr. Top. Microbiol. Immunol. 242, 1-23. [DOI] [PubMed] [Google Scholar]
- 2.Alter, M. J., Margolis, H. S., Krawczynski, K., Judson, F. N., Mares, A., Alexander, W. J., Hu, P. Y., Miller, J. K., Gerber, M. A., Sampliner, R. E., et al. (1992) N. Engl. J. Med. 327, 1899-1905. [DOI] [PubMed] [Google Scholar]
- 3.McCaffrey, A. P., Ohashi, K., Meuse, L., Shen, S., Lancaster, A. M., Lukavsky, P. J., Sarnow, P. & Kay, M. A. (2002) Mol. Ther. 5, 676-684. [DOI] [PubMed] [Google Scholar]
- 4.Trowbridge, R. & Gowans, E. J. (1998) J. Viral Hepat. 5, 95-98. [DOI] [PubMed] [Google Scholar]
- 5.Wang, C., Sarnow, P. & Siddiqui, A. (1993) J. Virol. 67, 3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsukiyama-Kohara, K., Iizuka, N., Kohara, M. & Nomoto, A. (1992) J. Virol. 66, 1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blight, K. J. & Rice, C. M. (1997) J. Virol. 71, 7345-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reed, K. E. & Rice, C. M. (2000) Curr. Top. Microbiol. Immunol. 242, 55-84. [DOI] [PubMed] [Google Scholar]
- 9.Friebe, P. & Bartenschlager, R. (2002) J. Virol. 76, 5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi, M. & Lemon, S. M. (2003) J. Virol. 77, 3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartenschlager, R. & Lohmann, V. (2000) J. Gen. Virol. 81, 1631-1648. [DOI] [PubMed] [Google Scholar]
- 12.Su, A. I., Pezacki, J. P., Wodicka, L., Brideau, A. D., Supekova, L., Thimme, R., Wieland, S., Bukh, J., Purcell, R. H., Schultz, P. G. & Chisari, F. V. (2002) Proc. Natl. Acad. Sci. USA 99, 15669-15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moriya, K., Fujie, H., Shintani, Y., Yotsuyanagi, H., Tsutsumi, T., Ishibashi, K., Matsuura, Y., Kimura, S., Miyamura, T. & Koike, K. (1998) Nat. Med. 4, 1065-1067. [DOI] [PubMed] [Google Scholar]
- 14.Moriya, K., Yotsuyanagi, H., Shintani, Y., Fujie, H., Ishibashi, K., Matsuura, Y., Miyamura, T. & Koike, K. (1997) J. Gen. Virol. 78, 1527-1531. [DOI] [PubMed] [Google Scholar]
- 15.Perlemuter, G., Sabile, A., Letteron, P., Vona, G., Topilco, A., Chretien, Y., Koike, K., Pessayre, D., Chapman, J., Barba, G. & Brechot, C. (2002) FASEB J. 16, 185-194. [DOI] [PubMed] [Google Scholar]
- 16.Barba, G., Harper, F., Harada, T., Kohara, M., Goulinet, S., Matsuura, Y., Eder, G., Schaff, Z., Chapman, M. J., Miyamura, T. & Brechot, C. (1997) Proc. Natl. Acad. Sci. USA 94, 1200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi, S. T., Polyak, S. J., Tu, H., Taylor, D. R., Gretch, D. R. & Lai, M. M. (2002) Virology 292, 198-210. [DOI] [PubMed] [Google Scholar]
- 18.Simons, K. & Ehehalt, R. (2002) J. Clin. Invest. 110, 597-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu, S. X., Ahlquist, P. & Kaesberg, P. (1992) Proc. Natl. Acad. Sci. USA 89, 11136-11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, W. M., Ishikawa, M. & Ahlquist, P. (2001) J. Virol. 75, 2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guinea, R. & Carrasco, L. (1990) EMBO J. 9, 2011-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez, L. & Carrasco, L. (1991) FEBS Lett. 280, 129-133. [DOI] [PubMed] [Google Scholar]
- 23.Kushner, D. B., Lindenbach, B. D., Grdzelishvili, V. Z., Noueiry, A. O., Paul, S. M. & Ahlquist, P. (2003) Proc. Natl. Acad. Sci. USA 100, 15764-15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahlquist, P., Noueiry, A. O., Lee, W. M., Kushner, D. B. & Dye, B. T. (2003) J. Virol. 77, 8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noueiry, A. O. & Ahlquist, P. (2003) Annu. Rev. Phytopathol. 41, 77-98. [DOI] [PubMed] [Google Scholar]
- 26.Lee, W. M. & Ahlquist, P. (2003) J. Virol. 77, 12819-12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz, M., Chen, J., Lee, W. M., Janda, M. & Ahlquist, P. (2004) Proc. Natl. Acad. Sci. USA 101, 11263-11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown, M. S. & Goldstein, J. L. (1997) Cell 89, 331-340. [DOI] [PubMed] [Google Scholar]
- 29.Horton, J. D., Goldstein, J. L. & Brown, M. S. (2002) J. Clin. Invest. 109, 1125-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimano, H. (2001) Prog. Lipid Res. 40, 439-452. [DOI] [PubMed] [Google Scholar]
- 31.Janowski, B. A., Grogan, M. J., Jones, S. A., Wisely, G. B., Kliewer, S. A., Corey, E. J. & Mangelsdorf, D. J. (1999) Proc. Natl. Acad. Sci. USA 96, 266-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, F. L. & Casey, P. J. (1996) Annu. Rev. Biochem. 65, 241-269. [DOI] [PubMed] [Google Scholar]
- 33.Meves, H. (1994) Prog. Neurobiol. 43, 175-186. [DOI] [PubMed] [Google Scholar]
- 34.Goldberg, E. M. & Zidovetzki, R. (1998) Biochemistry 37, 5623-5632. [DOI] [PubMed] [Google Scholar]
- 35.Goldberg, E. M. & Zidovetzki, R. (1997) Biophys. J. 73, 2603-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsutsumi, T., Yamauchi, E., Suzuki, E., Watanabe, S., Kobayashi, T. & Okuyama, H. (1995) Biol. Pharm. Bull. 18, 664-670. [DOI] [PubMed] [Google Scholar]
- 37.Clarke, S. D. (2000) Br. J. Nutr. 83, Suppl. 1, S59-S66. [DOI] [PubMed] [Google Scholar]
- 38.Stulnig, T. M. (2003) Int. Arch. Allergy Immunol. 132, 310-321. [DOI] [PubMed] [Google Scholar]
- 39.Pietschmann, T., Lohmann, V., Kaul, A., Krieger, N., Rinck, G., Rutter, G., Strand, D. & Bartenschlager, R. (2002) J. Virol. 76, 4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blight, K. J., Kolykhalov, A. A. & Rice, C. M. (2000) Science 290, 1972-1974. [DOI] [PubMed] [Google Scholar]
- 41.Guo, J. T., Bichko, V. V. & Seeger, C. (2001) J. Virol. 75, 8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blight, K. J., McKeating, J. A. & Rice, C. M. (2002) J. Virol. 76, 13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagasawa, K., Muraki, Y., Matsuda, T., Ohnishi, N. & Yokoyama, T. (2000) J. Pharm. Sci. 89, 1594-1604. [DOI] [PubMed] [Google Scholar]
- 44.Huang, K. C., Chen, C. W., Chen, J. C. & Lin, W. W. (2003) J. Biomed. Sci. 10, 396-405. [DOI] [PubMed] [Google Scholar]
- 45.Kapadia, S. B., Brideau-Andersen, A. & Chisari, F. V. (2003) Proc. Natl. Acad. Sci. USA 100, 2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guidotti, L. G., Ishikawa, T., Hobbs, M. V., Matzke, B., Schreiber, R. & Chisari, F. V. (1996) Immunity 4, 25-36. [DOI] [PubMed] [Google Scholar]
- 47.McClary, H., Koch, R., Chisari, F. V. & Guidotti, L. G. (2000) J. Virol. 74, 2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guidotti, L. G., McClary, H., Loudis, J. M. & Chisari, F. V. (2000) J. Exp. Med. 191, 1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cousin, S. P., Hugl, S. R., Wrede, C. E., Kajio, H., Myers, M. G., Jr., & Rhodes, C. J. (2001) Endocrinology 142, 229-240. [DOI] [PubMed] [Google Scholar]
- 50.Yang, Y. A., Morin, P. J., Han, W. F., Chen, T., Bornman, D. M., Gabrielson, E. W. & Pizer, E. S. (2003) Exp. Cell Res. 282, 132-137. [DOI] [PubMed] [Google Scholar]
- 51.Livak, K. J. & Schmittgen, T. D. (2001) Methods 25, 402-408. [DOI] [PubMed] [Google Scholar]
- 52.Ye, J., Wang, C., Sumpter, R., Jr., Brown, M. S., Goldstein, J. L. & Gale, M., Jr. (2003) Proc. Natl. Acad. Sci. USA 100, 15865-15870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steiner, S., Gatlin, C. L., Lennon, J. J., McGrath, A. M., Aponte, A. M., Makusky, A. J., Rohrs, M. C. & Anderson, N. L. (2000) Electrophoresis 21, 2129-2137. [DOI] [PubMed] [Google Scholar]
- 54.Bouterfa, H. L., Sattelmeyer, V., Czub, S., Vordermark, D., Roosen, K. & Tonn, J. C. (2000) Anticancer Res. 20, 2761-2671. [PubMed] [Google Scholar]
- 55.Thupari, J. N., Pinn, M. L. & Kuhajda, F. P. (2001) Biochem. Biophys. Res. Commun. 285, 217-223. [DOI] [PubMed] [Google Scholar]
- 56.Li, J. N., Mahmoud, M. A., Han, W. F., Ripple, M. & Pizer, E. S. (2000) Exp. Cell Res. 261, 159-165. [DOI] [PubMed] [Google Scholar]
- 57.Bergstrom, J. D., Kurtz, M. M., Rew, D. J., Amend, A. M., Karkas, J. D., Bostedor, R. G., Bansal, V. S., Dufresne, C., VanMiddlesworth, F. L., Hensens, O. D., et al. (1993) Proc. Natl. Acad. Sci. USA 90, 80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaidya, S., Bostedor, R., Kurtz, M. M., Bergstrom, J. D. & Bansal, V. S. (1998) Arch. Biochem. Biophys. 355, 84-92. [DOI] [PubMed] [Google Scholar]
- 59.Glenn, J. S., Marsters, J. C., Jr., & Greenberg, H. B. (1998) J. Virol. 72, 9303-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malewiak, M. I., Griglio, S. & Le Liepvre, X. (1985) Metabolism 34, 604-611. [DOI] [PubMed] [Google Scholar]
- 61.Halvorson, D. L. & McCune, S. A. (1984) Lipids 19, 851-856. [DOI] [PubMed] [Google Scholar]
- 62.Kuhajda, F. P., Jenner, K., Wood, F. D., Hennigar, R. A., Jacobs, L. B., Dick, J. D. & Pasternack, G. R. (1994) Proc. Natl. Acad. Sci. USA 91, 6379-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edwards, P. A. & Ericsson, J. (1999) Annu. Rev. Biochem. 68, 157-185. [DOI] [PubMed] [Google Scholar]
- 64.Ou, J., Tu, H., Shan, B., Luk, A., DeBose-Boyd, R. A., Bashmakov, Y., Goldstein, J. L. & Brown, M. S. (2001) Proc. Natl. Acad. Sci. USA 98, 6027-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshikawa, T., Shimano, H., Yahagi, N., Ide, T., Amemiya-Kudo, M., Matsuzaka, T., Nakakuki, M., Tomita, S., Okazaki, H., Tamura, Y., et al. (2002) J. Biol. Chem. 277, 1705-1711. [DOI] [PubMed] [Google Scholar]
- 66.Xu, J., Nakamura, M. T., Cho, H. P. & Clarke, S. D. (1999) J. Biol. Chem. 274, 23577-23583. [DOI] [PubMed] [Google Scholar]
- 67.Kim, H. J., Takahashi, M. & Ezaki, O. (1999) J. Biol. Chem. 274, 25892-25898. [DOI] [PubMed] [Google Scholar]
- 68.Kersten, S. (2002) Proc. Nutr. Soc. 61, 371-374. [DOI] [PubMed] [Google Scholar]
- 69.Foretz, M., Foufelle, F. & Ferre, P. (1999) Biochem. J. 341, 371-376. [PMC free article] [PubMed] [Google Scholar]
- 70.Field, F. J., Born, E., Murthy, S. & Mathur, S. N. (2002) Biochem. J. 368, 855-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murthy, S., Born, E., Mathur, S. N. & Field, F. J. (2004) Biochem. J. 377, 545-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aizaki, H., Lee, K. J., Sung, V. M., Ishiko, H. & Lai, M. M. (2004) Virology 324, 450-461. [DOI] [PubMed] [Google Scholar]
- 73.Egger, D., Wolk, B., Gosert, R., Bianchi, L., Blum, H. E., Moradpour, D. & Bienz, K. (2002) J. Virol. 76, 5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leu, G. Z., Lin, T. Y. & Hsu, J. T. (2004) Biochem. Biophys. Res. Commun. 318, 275-280. [DOI] [PubMed] [Google Scholar]
- 75.Jump, D. B. & Clarke, S. D. (1999) Annu. Rev. Nutr. 19, 63-90. [DOI] [PubMed] [Google Scholar]
- 76.Nestel, P. J., Connor, W. E., Reardon, M. F., Connor, S., Wong, S. & Boston, R. (1984) J. Clin. Invest. 74, 82-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thomassen, M. S., Christiansen, E. N. & Norum, K. R. (1982) Biochem. J. 206, 195-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Funk, C. D. (2001) Science 294, 1871-1875. [DOI] [PubMed] [Google Scholar]
- 79.Anderson, M. & Fritsche, K. L. (2002) J. Nutr. 132, 3566-3576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.