Abstract
Background and Purpose:
Candida species are the most opportunistic fungi affecting the nails and resulting in onychomycosis. In this study, we identified and evaluated in-vitro susceptibility of the recovered isolates against fluconazole (FLC), voriconazole (VRC), and clotrimazole (CLT) using the Clinical and Laboratory Standards Institute (CLSI) M27-A3 document.
Materials and Methods:
From patients with either clinically or mycologically proven onychomycosis, 97 isolates comprising of seven Candida species were isolated, which were identified by both conventional and molecular techniques such as polymerase chain reaction-restriction fragment length polymorphism. In addition, Candida dubliniensis was confirmed by restriction endonuclease analysis. Antifungal susceptibility of each isolate against the three azoles applied in this study was determined using the CLSI microdilution reference method M27-A3.
Results:
Candida parapsilosis (C. parapsilosis) was the most frequently isolated species (n=44), followed by C. albicans (n=23), C. tropicalis (n=13), C. glabrata (n=7), C. krusei (n=6), C. guilliermondii (n=3), and C. dubliniensis (n=1). All the isolates were susceptible to CLT. VRC had lower minimum inhibitory concentration (MIC) values for the isolates compared to FLC. Geometric mean MIC values of VRC, FLC, and CLT for C. parapsilosis isolates were 0.07 μg/ml, 0.8 μg/ml, and 0.35 μg/ml, respectively. Collectively, all species exhibited greater susceptibility to VRC in comparison to C. albicans (P≤0.001).
Conclusion:
This study showed that non-albicans Candida species were the most common etiologic agents of non-dermatophyte onychomycosis. The major antifungal agents used in clinics to empirically treat yeast onychomycosis are FLC and CLT. Our data suggested that CLT is a better choice for the treatment of Candida onychomycosis, especially in drug resistant cases.
Key Words: Candida albicans, Candidiasis/microbiology, Candidiasis/pathogenicity, Clotrimazole, Fluconazole, Onychomycosis, Voriconazole
Introduction
The genus Candida includes approximately 200 different species, of which only a few were adequately documented as human opportunistic pathogens. Candida infections, which may be superficial or invasive (or a combination of both), usually occur in immunocompromised patients [1]. Superficial infections that can be managed successfully with topical antifungal agents usually involve the skin, nails, or mucous membranes [2].
The azoles are the most effective antifungal agents for the management of yeast infections, which inhibit lanosterol biosynthesis from disrupting the function of the yeast cell membrane. They include imidazoles (e.g., miconazole, econazole, ketoconazole, and clotrimazole [CLT]) and triazoles (e.g., flu-conazole [FLC], itraconazole, and voriconazole [VRC]), which were used successfully to treat yeast infections [2, 3].
Even though there may be some controversy regarding the correlation between in-vitro susceptibility testing data and clinical outcomes, in-vitro antifungal data can be beneficial for predicting potential outcomes and drawbacks such as resistance [3]. The CLSI microdilution reference method is a testing platform used across the globe to obtain standardized results that can be compared with other studies.
Candida onychomycosis should to be distinguished from onychomycoses caused by either dermatophytes or a number of different filamentous fungi [4-9], as choosing the appropriate treatment without culture can result in non-responsive cases. Owing to the fact that onychomycoses may require months to resolve, providing the appropriate treatment during the early treatment phases seems to be imperative. Several groundbreaking studies, performed in Iran, identified the etiologic agents of onychomycosis, and then based upon in-vitro susceptibility studies evaluated the potential outcomes using different antifungal agents [9-13].
With the advent of molecular tools allowing for rapid genomic studies, determination of phylogenetic relationships as a way to identify yeast taxa was accentuated. This approach has resulted in several yeast taxa being redefined and reclassified. The separation of C. dubliniensis from C. albicans based on differences in their phylogeny is but an example.
Owing to the revision of many yeast species, especially in the Candida genus, the application of molecular methods has become mandatory for their accurate identification. Among the more novel molecular tools, polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) is one of the most accurate methods for the identification of Candida species [14, 15].
Material and Methods
Isolates
Patients with clinically or mycologically confirmed onychomycosis were enrolled in this study. A total of 97 Candida isolates were isolated from fingernail specimens, collected using the standard aseptic techniques, and were placed on Sabouraud dextrose agar (Merck, Germany) and incubated at 30°C. The yeast colonies were purified using the standard methods to insure only one yeast, which is devoid of potential contaminating bacteria, was present.
The new subcultures were transferred to Sabouraud dextrose agar after which small portions of the purified colonies were added to microtubes containing distilled water and were maintained at -20C. For the evaluation of conventional and molecular methods, both were applied for the identification of each isolate.
The conventional method
The isolates were initially identified according to the results of chlamydoconidia formation in Corn Meal Agar with Tween 80 (Merck, Germany) incubated at 30°C, germ tube formation in fresh serum incubated for up to three hours at 37°C, and colony color on the chromogenic medium of CHROMagar Candida (Biomeriux, France) incubated at 30°C. CHROMagar is used for presumptive identification of some yeasts and differentiation of C. albicans, C. tropicalis, and C. krusei.
The molecular method (PCR-RFLP)
We used a PCR-RFLP technique as described before [14, 15]. DNA extraction was performed using phenol-chloroform method [16]. A loop full of fresh yeast was harvested and suspended in 300 μl of lysis buffer (10 mM Tris, 1 mM ethylenediaminetetraacetic acid [EDTA] pH 8, 1% sodium dodecyl sulfate [SDS], 100 mM NaCl, and 2% Triton X-100) with phenol-chloroform and glass beads, and then was vortexed. Total DNA was precipitated with 2-propanol, washed with 70% ethanol, air-dried, and suspended in 50 μl of Tris–EDTA buffer (10 mM Tris, 1 mM EDTA), and was kept at -20°C before use.
Internal transcribe spacer (ITS) region of ITS1-5.8S-ITS2 segment of the ribosomal DNA gene was amplified [14]. A set of universal primers (ITS1, 5-TCCGTAGGTGAACCTGCGG and ITS4, 5-TCCTCCGCTTATTGATATGC) (Meta-bion International, Martinsried, Germany) were employed for amplification. PCR amplification was carried out in a final volume of 50 μl. Each reaction contained 1 μl of template DNA, 0.5 μM of each primer, and 0.20 mM of each deoxynucleoside triphosphate (dNTP), 5 μl of 10× PCR buffer, and 1.25 U of Taq polymerase (Roche Molecular Biochemicals, Mannheim, Germany). An initial denaturation step at 94°C for five minutes was followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 56°C for 45 seconds, and extended at 72°C for one minute, with a final extension step at 72°C for seven minutes. The PCR product was electrophoresed on 1.2% agarose gel and stained with ethidium bromide.
For differentiation between Candida species, amplified PCR products were digested with Msp1 restriction endonuclease to achieve the best species-specific pattern, [14] and additional enzyme of B1nI (AvrII) was applied to differentiate C. dubliniensis from C. albicans [15]. Restriction fragments were separated by 1.8% agarose gel electrophoresis in Tris-Borate-EDTA (TBE) buffer for approximately one hour at 100 V and visualized by staining with ethidium bromide.
Antifungal susceptibility testing
Antifungal susceptibility testing of FLC, VRC, and CLT was performed according to the Clinical and Laboratory Standards Institute (CLSI) using micro-dilution method with minor modification [17, 18]. RPMI 1640 (with L-glutamine and phenol red, without bicarbonate; Sigma, USA) was prepared and buffered at pH 7.0 with 0.165 mol of 3-N-morpholino propanesulfonic acid (MOPS) (Sigma- Aldrich, Germany).
Serial dilutions of the drugs were prepared in 96-well microtiter trays using RPMI 1640 media buffered with MOPS (Sigma, St. Louis, USA). Stock inocula were prepared by suspending three colonies of each isolate in 5 ml sterile 0.85% NaCl and adjusting the turbidity to a 0.5 McFarland standard at 530 nm wavelength to achieve 1–5×106 cells/ml density.
Working suspensions were prepared by making a 1/1000 dilution with RPMI of the stock suspension for each Candida species. The trays were incubated at 35°C for 24-48 hours in humid atmosphere. The growth in each well was compared with control wells. Minimum inhibitory concentrations (MICs) were visually determined and defined as the lowest concentration of the drugs that produced no visible growth. Each experiment was performed in duplicate.
Candida albicans (ATCC10261) and C. parapsilosis (ATCC 4344) were used as controls. For FLC, MIC for susceptible, susceptible dose dependent, and resistant were ≤8 μg/ml, 16-32 μg/ml, and ≥64 μg/ml, respectively. For VRC, MIC for susceptible, susceptible dose dependent, and resistant were ≤1 μg/ml, 2 μg/ml, and ≥4 μg/ml, respectively [18].
Statistical analysis
To analyze the data, Chi-square test was performed, using SPSS version 15. Geometric mean of MICs was calculated, as well.
Results
The number of Candida species, as identified by the conventional and molecular methods, is summarized in Table 1. PCR-RFLP using MspI restriction endonuclease enzyme was able to distinguish seven Candida species as shown in Table 2 and Figure 1.
Table 1.
Identification of Candida species by conventional and molecular methods
| Candida species | Germ tube | Chlamidoconidia | Colony color on CHROMagar | Polymerase chain reaction- restriction fragment length polymorphism |
|---|---|---|---|---|
| C. parapsilosis (44) | 1 | 0 | White-pink(38) | 44 |
| C. ablicans (23) | 21 | 22 | Green (23) | 23 |
| C. tropicalis (13) | 4 | 1 | Blue (13) | 13 |
| C. glabrata (7) | 0 | 0 | Pink–purple (5) | 7 |
| C. krusei (6) | 0 | 0 | Pink–purple (6) | 6 |
| C. guilliermondii (3) | 0 | 0 | Blue-purple (3) | 3 |
| C. dubliniensis | 1 | 1 | Green (1) | 1 |
| Total | 97 |
Table 2.
Frequency of Candida species isolated from onychomycosis
| Candida species | Polymerase chain reaction- restriction fragment length polymorphism |
|
|---|---|---|
| No. | Percent | |
| C. parapsilosis | 44 | 45.3 |
| C. ablicans | 23 | 23.7 |
| C. tropicalis | 13 | 13.4 |
| C. glabrata | 7 | 7.2 |
| C. krusei | 6 | 6.1 |
| C. guilliermondii | 3 | 3.09 |
| C. dubliniensis | 1 | 1.03 |
| Total | 97 | 100 |
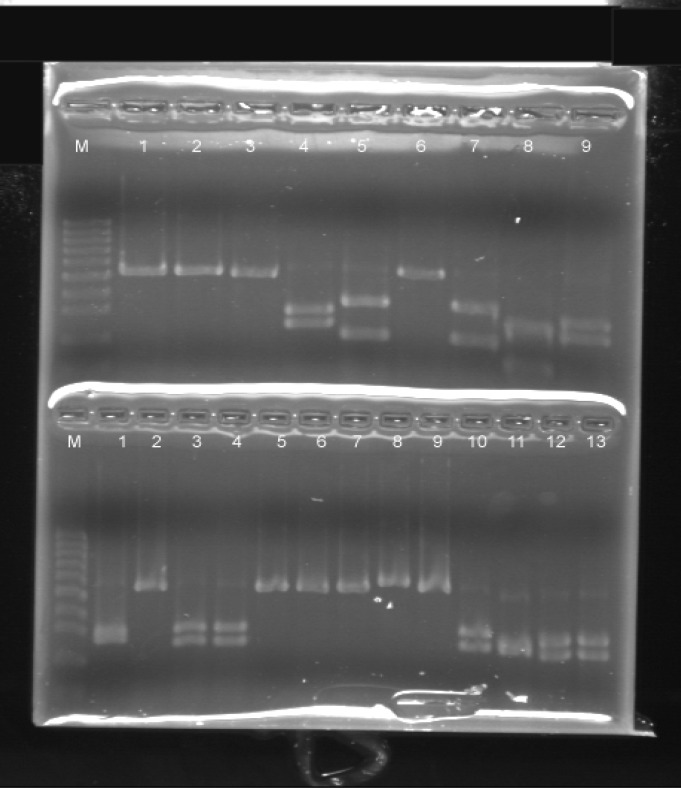
Figure 1.
Polymerase chain reaction- restriction fragment length polymorphism profile of some isolates: C. parapsilosis (up 1-3, 6 and down 2, 5-9), C. albicans (up 4, 9 and down 3, 4, 10, 12, 13), C.tropicalis (up 5, 7) and C. krusei (up 8 and down 11)
C. parapsilosis was the most predominant species among the isolates (45.3%), whereas C. albicans was the second (23.7%) most common taxon. When B1nI (AvrII) restriction endonuclease enzyme was employed, one of the C. albicans isolates was re-identified as C. dubliniensis.
The results of antifungal activity of VRC, FLC, and CLT against the Candida species are presented in Table 3. CLT had potent activity against the isolates tested with MIC ranging from 0.03 µg/ml to 16 µg/ml. MIC50 and MIC90 of the isolates for CLT, VRC, and FLC were 0.5 mg/L and 0.06 mg/L, 0.5 mg/L and 5.6mg/L , as well as 8 mg/L and 16 mg/L, respectively.
Table 3.
Antifungal susceptibility of Candida species isolated from onychomycosis, S (Susceptible), SDD (Susceptible Dose Dependent), and R (resistant), GM (Geometric mean
| Species | Voriconazole |
Fluconazole |
Clotrimazole |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S | SDD | R | GM (μg/ml) |
S | SDD | R | GM (μg/ml) |
S | R | GM (μg/ml) |
|
| C.parapsilosis | 42 | 0 | 2 | 0.07 | 41 | 2 | 1 | 0.8 | 44 | 0 | 0.35 |
| C. ablicans | 13 | 0 | 10 | 0.57 | 22 | 0 | 1 | 0.9 | 23 | 0 | 0.25 |
| C. tropicalis | 6 | 0 | 7 | 1.06 | 8 | 1 | 4 | 8.9 | 13 | 0 | 1.8 |
| C. glabrata | 3 | 0 | 0 | 0.14 | 3 | 0 | 0 | 1.8 | 3 | 0 | 2 |
| C. krusei | 6 | 0 | 0 | 0.06 | 6 | 0 | 0 | 0.71 | 6 | 0 | 0.35 |
| C.guilliermondii | 7 | 0 | 0 | 0.5 | 5 | 2 | 0 | 1.41 | 7 | 0 | 0.14 |
| C.dubliniensis | 0 | 1 | 0 | 0.5 | 0 | 0 | 1 | 16 | 1 | 0 | 2 |
| Total (%) | 77 (79.3%) | 1 (1%) | 19 (19.5%) | 85 (87.6%) | 5 (5.1%) | 7 (7.2%) | 97 (100%) | 0 (0%) | |||
Geometric mean MICs for CLT were 0.25 μg/ml for C. albicans, 0.35 μg/ml for C. parapsilosis, 1.8 μg/ml for C. tropicalis, 2 μg/ml for C. glabrata, 0.35 μg/ml for C. krusei, 0.14 μg/ml for C. guilliermondii, and 2 μg/ml for C. dubliniensis.
Furthermore, 19.5% and 7.2% of the isolates had low susceptibility to VRC and FLC, respectively. VRC was active against 79.3% of the isolates with geometric mean MIC values of 0.57 μg/ml for C. albicans, 0.07 μg/ml for C. parapsilosis, 1.06 μg/ml for C. tropicalis, 0.14 μg/ml for C. glabrata, 0.06 μg/ml for C. krusei, 0.5 μg/ml for C. guilliermondii, and 0.5 μg/ml for C. dubliniensis.
FLC geometric mean MIC values for Candida species are exhibited in Table 3. FLC was more effective compared to VRC (P≤0.001). There was a significant difference between C. albicans and non-C. albicans regarding sensitivity to VRC (P=0.001).
Discussion
Candida species are the principal non-dermatophytic etiologic agents of onycho-mycosis in patients living in Shiraz, Iran. Identification of Candida spp. isolated from patients with onychomycosis using molecular tools was an important contribution to understanding their epidemiology in the city of Shiraz, southern Iran. PCR-RFLP is an ideal method for distinguishing Candida species from each other, especially C. dubliniensis [14, 15].
Using B1nI restriction enzymes, we were able to distinguish C. dubliniensis from among the C. albicans isolates. This is the first instance in which C. dubliniensis was demonstrated to be an etiologic agent of onychomycosis in Iran. Typical for this taxon and C. albicans, it produced germ tubes and chlamydoconidia, and developed green colored colonies on CHROMagar. C. dubliniensis was sensitive to CLT and VRC at 2 μg/ml and 0.5 μg/ml concentrations, respectively, but had a low MIC of 16 μg/ml to FLU.
Zomorodian et al. [19] recovered 16 isolates of C. dubliniensis from denture-related stomatitis from Iranian patients, whereas Ghahri et al. [20] used PCR-RFLP with MboI endonuclease restriction enzyme and could not distinguish C. dubliniensis from among 67 isolates of C. albicans recovered from onychomycosis in Tehran, Iran. This discrepancy in results might be pertinent to the type of restricted enzyme they used.
In the current study, C. parapsilosis was the most common isolate recovered from our patients. Our results are not congruent with the findings of other Iranian studies [21, 22]. Hashemi et al. identified C. albicans (41%) as the main etiologic agent of onychomycosis in Tehran, Iran [10]. Chadeganipour et al. reported C. albicans as the most prevalent yeast causing onychomycosis in Isfahan, Iran [11].
Moreover, Farasat et al., using an ITS sequencing method, identified C. pulcherrima as the causative agent of nail lesions [12]. Our findings are in line with those of the study by Segal et al. [23] and other studies performed in Spain and Hong Kong, which reported C. parapsilosis as the predominant species [24, 25]. The inconsistency among the results may be associated with the use of different identification tools and demographic groups as well as the fact that C. parapsilosis is a complex of closely related species, which require further studies. We purposefully chose our identification methods to avoid such problems
The CLSI broth microdilution method is recommended for evaluation of antifungal activity of Candida species [17, 18]. In our former study, we assessed antifungal activity among clinical Candida isolates and reported 96.6% susceptibility to FLC [19]. Khosravi et al. in a study conducted in Tehran, Iran, reported that 85.7% of onychomycosis isolates belonged to Candida species and that the isolates were susceptible to FLC [13].
In our study, MICs of non-albicans isolates were significantly lower than the C. albicans isolates (P=0.001). All the isolates had diverse geometric mean MICs to CLT; 19.5% of the isolates were resistant to VRC, and C. albicans were the most resistant isolates. VRC was employed for the treatment of disseminated candidiasis; however, it is not commonly used for the treatment of onychomycosis [26-34]. In Iran, VRC is used for highly resistant cases when the imidazoles are ineffective due to resistance or possible non-compliance. FLC is the most common drug of choice, used for the treatment of onychomycosis, primarily in combination with other antifungal agents [3, 26]. In our study, 87.6% of the isolates were sensitive to FLC; thus, it could be the best choice for combination therapy with CLT.
Conclusion
In summary, this study showed that PCR-RFLP is an efficient method for identification of Candida species and that FLC is more effective than the other agents tested against the isolates. We were unable to correlate in vitro antifungal data to in vivo response according to 90/60 rule [32] and it was not in the scope of this study to attempt to correlate in-vitro antifungal data to in-vivo response. The data can aid physicians to choose an effective potential drug for treating onychomycosis patients.
Acknowledgments
This study was extracted from an MD thesis by Alireza Zakaei and was funded by Deputy of Research and Technology of Shiraz University of Medical Sciences, Shiraz, Iran (Grant No. 6000). Our special thanks go to professor Emeritus Michael R. McGinnis from Medical University of Texas for his editorial comments.
Authors’ Contributions
K. Pakshir contributed to study concept and design, drafted and revised the manuscript, and analyzed and interpreted the data. K. Zomorodian contributed to study concept and design, drafted and revised the manuscript, and interpreted the data. A. Zakaei contributed to sample collection and laboratory examination. M. Motamedi cooperated with sample collection and laboratory examination, interpreted the data, revised the manuscript, and performed statistical analysis. M. Rahimi Ghiasi and M. Karamitalab contributed to sample collection and laboratory examination. P. Jafari performed statistical analysis.
Conflicts of Interest:
The authors declare no conflicts of interest.
Financial Disclosure
Dr Keyvan Pakshir reported receiving research grants from Shiraz University of Medical Sciencesfor this research (grant number 6000).
Authors have no financial interests related to the material in the manuscript.
References
- 1.Lopez-Martınez R. Candidosis, a new challenge. Clin Dermatol. 2010;28(2):178–84. doi: 10.1016/j.clindermatol.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 2.pampiato C, Leonardi D. Candida infections, causes, targets, and resistance mechanisms: traditional and alternative antifungal agents. Biomed Res Int. 2013;2013:204237. doi: 10.1155/2013/204237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X, Hiruma M, Shivaki Y, Ogawa H. Combination therapy of once-weekly fluconazole (100, 150, or 300 mg) with topical application of ketoconazole cream in the treatment of onychomycosis. Jpn J Infect Dis. 2004;57(6):260–3. [PubMed] [Google Scholar]
- 4.Ilkit M. Onychomycosis in Adana, Turkey: a 5-year study. Int J Dermatol. 2005;44(10):851–4. doi: 10.1111/j.1365-4632.2005.02265.x. [DOI] [PubMed] [Google Scholar]
- 5.Souse LK, Fernondes OF, Passos XS, Costa CR, Lemos JA, Silva MR. Epidemiological and mycological data of onychomycosis in Goiania, Briazil. Mycoses. 2010;53(1):68–71. doi: 10.1111/j.1439-0507.2008.01663.x. [DOI] [PubMed] [Google Scholar]
- 6.Dorko E, Jautora J, Tkacikova L, Wantrubova A. The frequeney of Candida species in onychomycosis. Folia Microbil. 2002;47(6):727–31. doi: 10.1007/BF02818679. [DOI] [PubMed] [Google Scholar]
- 7.Swinne D, Watelle M, Nolard N. In vitro activities of voriconazole, fluconazole, itraconazole and amphotericin B against non Candida albicans yeast isolates. Rev Iberoam Micol. 2005;22(1):24–8. doi: 10.1016/s1130-1406(05)70002-4. [DOI] [PubMed] [Google Scholar]
- 8.Figueiredo VT, de Assis Santos D, Resende MA, Hamdan JS. Identification and in vitro antifungal susceptibility testing of 200 clinical isolated of Candida spp responsible for fingernail infection. Mycopathologia. 2007;164(1):27–33. doi: 10.1007/s11046-007-9027-6. [DOI] [PubMed] [Google Scholar]
- 9.Shokohi T, Hashemi Soteh MB, Saltanat Pouri ZS, Hedayati MT, Mayahi S. Identification of Candida species using PCR-RFLP in cancer patients in Iran. Indian J Med Microbiol. 2010;28(2):147–51. doi: 10.4103/0255-0857.62493. [DOI] [PubMed] [Google Scholar]
- 10.Hashemi SJ, Gerami M, Zibafar E, Daei M, Moazeni M, Nasrollahi A. Onychomycosis in Tehran: mycological study of 504 patients. Mycoses. 2010;53(3):251–5. doi: 10.1111/j.1439-0507.2009.01703.x. [DOI] [PubMed] [Google Scholar]
- 11.Chadeganipour M, Nilipour S, Ahmadi G. Study of onychomycosis in Isfahan, Iran. Mycoses. 2010;53(2):153–7. doi: 10.1111/j.1439-0507.2008.01679.x. [DOI] [PubMed] [Google Scholar]
- 12.Farasat A, Ghahri M, Mirhendi H, Beiraghi S. Morphological and Molecular characteristics of Candida pulcherrima, an opportunistic yeast, isolated from nail lesions in Iran. Adv Stud Biol. 2012;4(6):297–306. [Google Scholar]
- 13.Khosravi AR, Shokria H, Mansourib P, Katiraeea F, Ziglaria T. Candida species isolated from nails and their in vitro susceptibility to antifungal drugs in the department of dermatology (University of Tehran, Iran) J Mycol Med. 2008;18(4):210–5. [Google Scholar]
- 14.Mirhendi H, Makimura K, Khoramizadeh M, Yamaguchi H. A one-enzyme PCR-RFLP assay for identification of six medically important Candida species. Nihon Ishinkin Gakkai Zasshi. 2006;47(3):225–9. doi: 10.3314/jjmm.47.225. [DOI] [PubMed] [Google Scholar]
- 15.Mirhendi H, Makimura K, Zomorodian K, Maeda N, Ohshima T, Yamaguchi H. Differentiation of Candida albicans and Candida dubliniensis using a single-enzyme PCR-RFLP method. Jpn J Infect Dis. 2005;58(4):235–7. [PubMed] [Google Scholar]
- 16.Yamada Y, Makimura K, Mirhendi H, Ueda K, Nishiyama Y, Yamaguchi H, et al. Comparison of different methods for extraction of mitochondrial DNA from human pathogenic yeasts. Jpn J Infect Dis. 2002;55(4):122–5. [PubMed] [Google Scholar]
- 17.Johan HR, Barbara DA, Andes D, Arthington-Skaggs B, Brown SD, Chaturvedi V, et al. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standards-Third edition. Clin Lab Stand Inst. 2008;28(14):13–25. [Google Scholar]
- 18.Schwalbe R, Steele-Moore L, Goodwin AC. Antimicrobial susceptibility testing protocols. Boca Raton: CRC press; 2007. pp. 173–208. [Google Scholar]
- 19.Zomorodian K, Haghighi NN, Rajaee N, Pakshir K, Tarazooie B, Vojdani M, et al. Assessment of Candida species colonization and denture-related stomatitis in complete denture wearers. Med Mycol. 2011;49(2):208–11. doi: 10.3109/13693786.2010.507605. [DOI] [PubMed] [Google Scholar]
- 20.Ghahri M, Mirhendi SH, Yadegari MH, Hajizadeh E, Shidfar MR. Identification of pathogenic yeasts isolated from onychomycosis in Tehran, using polymerase chain reaction and enzymatic digestion. Mod J Med Sci Pathol. 2010;13(1):79–91. [Google Scholar]
- 21.Shokohi T, Hajheidari Z, Haghani I, Khalilian A, Aghili SR, Miahi S. The study of 101 cases of onychomycosis and associate factors in patients referred to Boali Sina Hospital and Toba dermatology outpatient clinics in Sari. J Mazandaran Univ Med Sci. 2009;71(18):33–43. [Google Scholar]
- 22.Ghasemi Z, Falahati M, Farahyar S, Nami S, Nozari S, Ahmadi F, et al. Investigation of prevalence of onychomycosis due to yeast fungi in dystrophic nails of patients who referred to Razi hospital (2010-2011) Razi J Med Sci. 2012;19(96):26–33. [Google Scholar]
- 23.Segal R, Kimchi A, Kritzmar A, Inbar R. The frequency of Candida parapsilosis in onychomycosis. An epidemiological study survey in Israel. Mycoses. 2000;43(9-10):349–53. doi: 10.1046/j.1439-0507.2000.00582.x. [DOI] [PubMed] [Google Scholar]
- 24.Vélez A, Linares MJ, Fenández-Roldán JC, Casal M. Study of onychomycosis in Córdoba, Spain: prevailing fungi and pattern of infection. Mycopathologia. 1997;137(1):1–8. doi: 10.1023/a:1006874303991. [DOI] [PubMed] [Google Scholar]
- 25.Kam KM, Au WF, Wong PY, Cheung MM. Onychomycosis in Hong Kong. Int J Dermatol. 1997;36(10):757–61. doi: 10.1046/j.1365-4362.1997.00048.x. [DOI] [PubMed] [Google Scholar]
- 26.Pfaller MA, Meser SA, Boyken L, Rice C, Tendolkar S, Hollis RJ, et al. Use of fluconazole as a surrogate marker to predict susceptibility and resistance to voriconazole among 13,338 clinical isolates of Candida Spp. tested by clinical and laboratory standards institute-recommended broth microdilution methods. J Clin Microbiol. 2007;45(1):70–5. doi: 10.1128/JCM.01551-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bueno JG, Martinez C, Zapata B, Sanclemente G, Gallago M, Mesa AC. In vitro activity of fluconazole, itraconazol, voriconazol and terbinafine against fungi causing onychomycosis. Clin Exp Dermatol. 2010;35(6):658–63. doi: 10.1111/j.1365-2230.2009.03698.x. [DOI] [PubMed] [Google Scholar]
- 28.Fleck R, Dietz A, Hof H. In vitro susceptibility of Candida species to five antifungal agents in a German university hospital assessed by the reference broth microdilution method and Etest. J Antimicrob Chemother. 2007;59(4):767–71. doi: 10.1093/jac/dkl555. [DOI] [PubMed] [Google Scholar]
- 29.Ozçelik B, Kaynak F, Cesur S, Sipahi B, Sultan N. In vitro activities of voriconazole as a triazole derivative and caspofungin as an echinocandin were compared with those of some antifungal agents against Candida species isolated from clinical specimens. Jpn J Infect Dis. 2007;60(5):302–4. [PubMed] [Google Scholar]
- 30.Swinne D, Watelle M, Nolard N. In vitro activities of voriconazole, fluconazole, itraconazole and amphotericin B against non Candida albicans yeast isolates. Rev Iberoam Micol. 2005;22(1):24–8. doi: 10.1016/s1130-1406(05)70002-4. [DOI] [PubMed] [Google Scholar]
- 31.Rubio MC, de Ocáriz IR, Gil J, Benito R, Rezusta A. Potential fungicidal effect of voriconazole against Candida spp. Int J Antimicrob Agents. 2005;25(3):264–7. doi: 10.1016/j.ijantimicag.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Otašević S, Barac A, Pekmezovic M, Tasic S, Ignjatović A, Momčilović S, et al. The prevalence of Candida onychomycosis in Southeastern Serbia from 2011 to 2015. Mycoses. 2016;59(3):167–72. doi: 10.1111/myc.12448. [DOI] [PubMed] [Google Scholar]
- 33.Katiraee F, Teifoori F, Soltani M. Emergence of azole-resistant Candida species in AIDS patients with oropharyngeal candidiasis. Curr Med Mycol. 2015;1(3):11–6. doi: 10.18869/acadpub.cmm.1.3.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yenisehirli G, Bulut N, Yenisehirli A, Bulut Y. In vitro susceptibilities of Candida albicans Isolates to Antifungal Agents in Tokat, Turkey. Jundishapur J Microbiol. 2015;8(9):e28057. doi: 10.5812/jjm.28057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rex JH, Pfaller MA. Has antifungal susceptibility testing come of age? Clin Infect Dis. 2002;35(8):982–9. doi: 10.1086/342384. [DOI] [PubMed] [Google Scholar]