Abstract
Copy number variations (CNVs) play a crucial role in the intricate genetics of autism spectrum disorders. A region on chromosome 15q24 vulnerable to both deletions and duplications has been previously implicated in a range of phenotypes including autism, Asperger’s syndrome, delayed development, and mild to severe mental retardation. Prior studies have delineated a minimal critical region of approximately 1.33 Mb. In this study, a multiplex autism family was evaluated for CNVs using genotyping data from the Illumina 1M BeadChip and analyzed with the PennCNV algorithm. Variants were then identified that co-segregate with autism features in this family. Here, we report autistic first cousins who carry two microduplications concordant with disease. Both duplications were inherited maternally and found to be identical by descent. The first is an approximately 10,000 base pair microduplication within the minimal region on 15q24 that falls across a single gene, ubiquitin-like 7. This is the smallest duplication in the region to result in a neuropsychiatric disorder, potentially narrowing the critical region for susceptibility to developmental and autism spectrum disorders. The second is a novel, 352 kb tandem duplication on 7p21 that replicates part of the neurexophilin 1 and islet cell autoantigen 1 genes. The breakpoint junction falls within the intronic regions of these genes and demonstrates a microhomology of four base pairs. Each of these microduplications may contribute to the complex etiology of autism spectrum disorders.
Keywords: autism spectrum disorder (ASD), copy number variation (CNV), low-copy repeat (LCR), non-allelic homologous recombination
INTRODUCTION
Genetic discoveries have highlighted the contribution of dosage sensitive genes to human disease Stankiewicz and Lupski, 2010]. The availability of dense single nucleotide polymorphism (SNP) arrays and comparative genomic hybridization arrays has further increased the identification of genomic structural aberrations. Within the chromosome 15q24 region, both deletions and duplications have been identified that result in a range of clinical phenotypes including autism, mental retardation, developmental delay, and Asperger’s syndrome [Smith et al., 2000; Cushman et al., 2005; Sharp et al., 2006, 2007; Kiholm Lund et al., 2008; Klopocki et al., 2008; Marshall et al., 2008; Andrieux et al., 2009; El-Hattab et al., 2009, 2010; Masurel-Paulet et al., 2009; McInnes et al., 2010; Pinto et al., 2010]. A majority of these copy number variations (CNVs) appear to be recurrent rearrangements, suggesting that they stem from non-allelic homologous recombination events between the five low-copy repeats (LCRs or segmental duplications) in the region, designated 15q24A, 15q24B, 15q24C, 15q24D, and 15q24E (Fig. 1). These five LCRs are complex in nature and share 15 homologous subunits in both forward and inverted orientations [El-Hattab et al., 2009]. Thus far, 20 deletions and 6 duplications have been reported to define a minimal critical region of approximately 1.33Mb [Smith et al., 2000; Cushman et al., 2005; Sharp et al., 2006, 2007; Kiholm Lund et al., 2008; Klopocki et al., 2008; Marshall et al., 2008; Andrieux et al., 2009; El-Hattab et al., 2009, 2010; Masurel-Paulet et al., 2009; McInnes et al., 2010; Pinto et al., 2010].
FIG. 1.
Copy number variations (CNVs) identified in the 15q24 region. Green lines denote duplications and red lines signify deletions. Each line represents all individuals carrying a CNV from one family. The purple bars correspond to the low-copy repeats in the region. The microduplication found in family 17122 falls between 15q24B and 15q24C, within the 1.33 Mb critical region.
In this report, we identify two distinct microduplications in a multiplex autism family that co-segregate with disease. The first is a duplication in the minimal 15q24 region, potentially reducing the critical region to approximately 10,000 nucleotides. To our knowledge, this duplication is the smallest reported in the region, encompassing a single known gene and a hypothetical gene. Data also suggest that, unlike a majority of the previously reported CNVs in the 15q24 region, the duplication described here does not appear to be caused by non-allelic homologous recombination between the currently defined LCRs. The second CNV that we found concordant with disease is a novel 352 kb duplication located on 7p21.3, whose breakpoints were mapped within the neurexophilin 1 and islet cell autoantigen 1 genes. From what is known about these genes on chromosome 7 as well as the previously reported patients on chromosome 15, it is possible that each of these microduplications may contribute to this family’s autistic features.
MATERIALS AND METHODS
Patient Ascertainment
The family of interest (17122) was ascertained as part of a larger study on autism genetics whose participants were enrolled under protocols previously described [Ma et al., 2009]. All protocols were approved by the Institutional Review Boards at the University of Miami, the University of South Carolina, and Vanderbilt University. Briefly, participants were ascertained on the basis of a previously existing clinical diagnosis of autism spectrum disorder and enrolled if they met the following minimal inclusion criteria: (1) age between 3 and 21 years, (2) expert clinical determination of diagnosis based on the DSM-IV criteria for an autism spectrum disorder, (3) absence of severe sensory or significant motor impairments, and (4) absence of identified metabolic, genetic, or progressive neurological disorders. Research diagnoses were assigned using DSM-IV criteria [American Psychiatric Association, 1994] supported by structured diagnostic instruments including the autism diagnostic interview-revised (ADI-R) and the autism diagnostic observation schedule (ADOS) [Lord et al., 1999; Rutter et al., 2003b]. In addition to diagnostic and psychometric assessments, whole blood was collected from participants by venipuncture.
The control group consisted of 727 apparently healthy Caucasian individuals with no known indicators of behavioral, developmental, or cognitive abnormalities (357 males and 370 females). Three hundred eighteen of the control samples came from Caucasian singleton live births at Centennial Women’s Hospital and The Perinatal Research Center, in Nashville, Tennessee. Inclusion criteria consisted of full term births (>37 weeks of gestation) to mothers between ages 18 and 40 years on admission to the hospital with no medical or obstetrical complications during pregnancy or delivery [Velez et al., 2009]. Umbilical cord blood samples were collected after placental delivery. Gender information was recorded when known. The remaining 409 DNA samples were purified from the saliva of children ages 3–21 years recruited via word of mouth, health fairs, daycares, and other child organizations. The social communication questionnaire was collected to rule out developmental problems along with a brief medical history and information on the family’s ethnicity [Rutter et al., 2003a].
SNP Array to Identify Copy Number Variations
Family 17122 was genotyped with the Illumina Human 1M Beadchip, assaying over one million SNPs. Samples were processed and quality control checks were performed as previously described [Ma et al., 2009]. The PennCNV algorithm using the Log R ratio and B allele frequency was used to identify CNVs that spanned across multiple SNPs [Wang et al., 2007]. Individual CNV calling was modified according to default parameters to correct for intensity wave artifacts (GC-waves) across the genome. CNVs were then sorted to identify deletions and duplications concordant with autism in all affected members within a family.
Real-Time PCR Copy Number Assays
TaqMan real-time polymerase chain reaction (PCR) copy number assays predesigned by Applied Biosystems (Foster City, CA) were used for validation and delineation of the regions identified by the SNP array. Assays were chosen within each region using the GeneAssist™ Copy Number Assay Workflow Builder. Each sample was tested in quadruplicate using 10 µl reactions according to the manufacturer’s protocol. Analysis of the data was performed in the CopyCaller Software v1.0 (Applied Biosystems).
Long Range PCR With Outward Facing Primer Sets
To determine whether the duplications occurred in tandem, we designed outward facing primers for each duplication. Individuals carrying the CNVs were assayed with the Roche Expand Long Range dNTPack. In a 25 µl master mix, reagents were combined as follows: 5 µl of 5× buffer with MgCl2, 1 µl of 10mM dNTP mix, 4 ng/µl of each primer, 0.28 µl of the expand enzyme mix, and 50 ng of DNA and water. Long range PCR was performed under the following conditions: 2 min at 92° C, thirty-five cycles through 10 sec at 92° C, 30 sec at 58° C, and 4 min at 68° C, followed by 10 min at 68° C, and a 4° C hold. PCR products were run on a 1% agarose gel, bands were extracted, purified using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA), and sequenced using the Applied Biosystems Big Dye Terminator v3.1 Cycle Sequencing Kit. For the chromosome 7 region, the following primers were used: upstream in the islet cell autoantigen 1 gene, CTGAAGCCGTGCATTTACCT, and downstream in the neurexophilin 1 gene, GCTCAAGATTCCTGCTTTGC.
In Silico Sequence Analysis
The NCBI BLAST 2 program was used to evaluate the breakpoint regions and determine whether there were shared regions of homology (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
RESULTS
Clinical Findings
This family is a multiplex family of European descent (Fig. 2). Individual 0001 met DSM-IV and ADI-R diagnostic criteria for autism. Pregnancy was unremarkable and the patient was born with a bilateral congenital hip dislocation. Initial parental concerns (~20 months of age) included language delay, social disinterest, and the appearance of odd mannerisms. Individual 0001 first walked at 18 months and acquired first words at 24 months. There is a history of multiple abnormal EEGs, but no evidence of frank seizures. A brain MRI was described as normal and fragile X-testing was negative. Individual 0001 did not display dysmorphic features but has macrocephaly. Additional clinical findings from his medical records include hypotonia, allergies, fine and gross motor delay, and minimal language regression. Cognitive assessment reveals moderate intellectual disability (full scale IQ = 46). Adaptive skills fell in the low range (Vineland adaptive behavior composite = 61).
FIG. 2.
Pedigree of the multiplex family 17122. Individuals 0001, 0100, and 0101 were clinically evaluated and diagnosed with autism, while individual 1004 was reported to have mental retardation, repetitive behaviors, and be non-verbal.
The sole sibling of 0001, individual 0100, also met DSM-IV and ADI-R criteria for autism. During pregnancy, individual 0100 was found to be large for his gestational age. Initial parental concerns (~12 months of age) were because of language problems. Individual 0100 first walked at 18 months and acquired his first word at 40 months; he developed phrase speech at 57 months. His medical records include EEG findings of uncertain significance, but no clinical seizures. In addition, he has a history of allergies, sensory problems, and hyperactivity. No dysmorphic features were observed, but individual 0100 had head circumference measurements in the high normal range. Fragile × testing was negative. Cognitive testing revealed a moderate intellectual disability (full scale IQ = 46) and adaptive skills fell in the low range (Vineland adaptive behavior composite = 55). While no formal evaluation was conducted, the mother of individuals 0001 and 0100, individual 1001, was reported to have migraines, mitral valve prolapse, and allergies.
A third individual in this family, 0101, carries a research diagnosis of an autism spectrum disorder based on both DSM-IV and ADI-R criteria. This individual is the first cousin to individuals 0001 and 0100, related through their mothers (individuals 1001 and 1006, Fig. 2). Initial parental concerns (~96 months of age) were not specific to autism, but included attention problems, language difficulties, and learning impairments. Subsequently, he was diagnosed with ADHD, depressive disorder, and a reading disability. A review of his medical records reveals that he showed significant social problems and no inclination to interact with peers. As part of this research study, a standard diagnostic measure (ADI-R) was completed with his parents. In conjunction with medical record and observational data, a best estimate research diagnosis of autism spectrum disorder was assigned. Individual 0101 first walked at 11 months, acquired first words at 12 months, and phrase speech at 24 months. His medical records reveal that he has dental misalignment, articulation and oral motor problems, an awkward gait, and overly acute hearing. Cognitive assessment placed him in the average range of intellectual functioning (full scale IQ = 99). However, adaptive skills fell below the average range (Vineland adaptive behavior composite = 65). His mother, individual 1006, was reported to have a history of allergies, Raynaud’s syndrome, and hearing loss due to high fever with varicella infection as a child. In addition, she reported that she has a “tipped uterus” and that she experienced three first trimester miscarriages. Finally, she described suffering from multiple fears.
Interestingly, there is a fourth individual in the family (1004), the maternal uncle of the other ASD individuals, who has several features of an autism spectrum disorder, although he has never been clinically evaluated or diagnosed. Anecdotally, this individual was reported to be non-verbal, have mental retardation and perform repetitive behaviors such as rocking, humming, and hand flapping. These problems may have been related to birth trauma. While the family has four members who exhibit features along the autism spectrum, individuals 0001 and 1004 were unavailable for genetic testing.
Detection and Validation of the Microduplications
Two duplications were identified as being shared between two autism spectrum disorder individuals (individuals 0100 and 0101) and present in their mothers as well (individuals 1001 and 1006). The first shared duplication was isolated on chromosome 15q24.1 and defined by three SNPs for a size of approximately 5.8 kb (Fig. 1). The second duplication encompassed 162 SNPs on chromosome 7p21.3 for a total size of approximately 348.5 kb. Individual 0101’s unaffected sibling 0102 was negative for both microduplications, as were both fathers (individuals 1000 and 1007). Neither of these duplications was identified in 727 pediatric control individuals nor have they been reported in the Database of Genomic Variants (DGV, http://projects.tcag.ca/variation/). Only a single inversion in DGV was reported to cross the 15q24.1 region, with breakpoints mapping to approximately the 15q24B and 15q24C LCRs (Kidd et al., 2008).
The 7p21.3 and 15q24.1 regions were each evaluated with six TaqMan real-time PCR copy number assays to confirm the duplications and refine the breakpoint regions (Table I). Both duplications were validated by real-time PCR. These experiments established that the duplication on 7p21.3 is between 350,803 and 353,216 base pairs in length (maximum region chr7:8,144,214–8,497,430, NCBI Build 36) with the breakpoints falling within the neurexophilin 1 (NXPH1, OMIM:604639) and islet cell autoantigen 1 (ICA1,OMIM:147625) genes (Fig. 3). In addition, the duplication on 15q24.1 was narrowed to be between 9,689 and 11,808 base pairs in size (maximum region chr15:72,536,068–72,547,876) and disrupt a single annotated gene, ubiquitin-like 7 (UBL7, OMIM:609748). Both of the duplications were carried through two sisters (1001, 1006) to their affected sons (0100, 0101). An unaffected son and both unaffected spouses were negative for the CNVs.
TABLE I.
Flanking SNPs on Illumina 1 M Chip and TaqMan Real-Time PCR Copy Number Assays
| Chr. | SNP | CNV assay ID | Location (bp)a | Gene | Copy number |
|---|---|---|---|---|---|
| 7 | rs17330124 | 8144214 | ICA1 | 2 | |
| Hs02960372 | 8145214 | ICA1 | 3 | ||
| rs3807843 | 8146180 | ICA1 | 3 | ||
| Hs01817569 | 8227474 | ICA1 | 3 | ||
| Hs04963570 | 8443081 | NXPH1 | 3 | ||
| rs10486229 | 8494690 | NXPH1 | 3 | ||
| Hs04978506 | 8496017 | NXPH1 | 3 | ||
| Hs04933524 | 8497430 | NXPH1 | 2 | ||
| rs6959225 | 8499029 | NXPH1 | 2 | ||
| 15 | rs7166701 | 72536068 | UBL7 | 2 | |
| Hs02844041 | 72538187 | UBL7 | 3 | ||
| Hs02194368 | 72540549 | UBL7 | 3 | ||
| rs4887152 | 72543407 | LOC440288 | 3 | ||
| Hs04451741 | 72544757 | LOC440288 | 3 | ||
| Hs03910824 | 72547876 | LOC440288 | 3 | ||
| rs4077703 | 72549215 | LOC440288 | 3 | ||
| Hs03898004 | 72550460 | LOC440288 | 2 | ||
| Hs03905957 | 72554610 | LOC440288 | 2 | ||
| rs8027459 | 72557387 | LOC440288 | 2 |
NCBI Build 36.
FIG. 3.
A,B: Results from the real-time PCR TaqMan assays demonstrating the presence of the each of the microduplications by an increase in the copy number size from two to three. Arrow bars show the range of the raw results from each sample that was performed in quadruplicate. C: The genomic region encompassed by the 7p21.3 duplication, chr7:8,144,894–8,497,305, contains two known genes, ICA1 and NXPH1. D: The duplication on 15q24.1 contains a single annotated gene, UBL7 (maximum region chr15:72,536,068–72,547,876).
Identification of the 7p21.3 Breakpoints
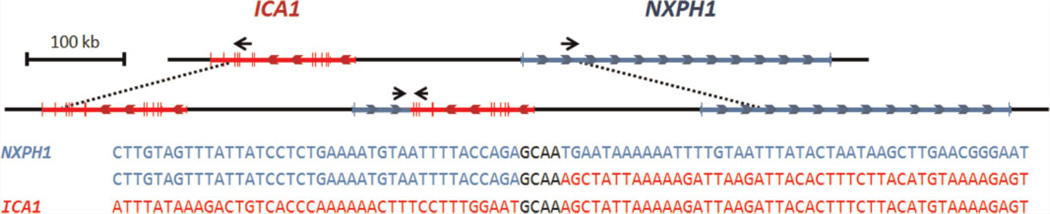
We hypothesized that the CNVs may result in tandem duplications and therefore attempted to sequence across these regions by designing outward facing primers from each of the breakpoints and amplifying by long range PCR. Primers were constructed to test whether the duplications might be either in direct or inverted positions to their original location. Using this method, we successfully identified the chromosome 7 CNV as a tandem duplication of the chr7:8,144,894–8,497,305 region, with the breakpoints falling within the intronic regions of the NXPH1 and ICA1 genes (Figs. 3C and 4). This generates a new, fused gene composed of the first 2 exons of NXPH1 and the first 12 exons of ICA1 oriented to face each other. The breakpoint junction was identified by Sanger sequencing of individual 1006 and confirmed to be identical throughout the family by also sequencing individuals 0100, 0101, and 1001. When evaluating the sequence at the breakpoint junction, a microhomology of the four base pairs “GCAA” was identified (Fig. 4).
FIG. 4.
The tandem duplication of the 7p21.3 region (chr7:8,144,894–8,497,305). Exons are marked with vertical lines and colored arrows show the direction of gene transcription. Outward facing primers, denoted with black arrows, were designed to amplify a product if the upstream and downstream breakpoint regions were brought into proximity of each other. The middle line of sequence shows the nucleotides at the joined breakpoints. The four base pairs in black, “GCAA,” demonstrate a region of microhomology between the ICA1 and NXPH1 genes at the breakpoint junction.
Evaluation of Genomic Architectural Elements on 15q24.1
A previous investigation of the large LCRs in the chromosome 15 region identified five complex segmental duplications (Fig. 1) that appear to facilitate the generation of most CNVs in the region through non-allelic homologous recombination (El-Hattab et al., 2009). Since the 15q24 microduplication we describe here does not align with the LCRs in the region, we appraised the area for other genomic elements that may have contributed to the generation of the CNV. We evaluated the proximal (chr15:72,536,068–72,538,187) and distal (chr15:72,549,215–72,550,460) breakpoint regions with the BLAST 2 program for areas of homology. The proximal breakpoint falls within UBL7’s fourth intron while the distal breakpoint is located in the second intron of the hypothetical gene LOC440288. Four Alu transposable elements were identified in the proximal breakpoint region that had 73–78% homology to a single AluJb segment located in the distal breakpoint region (Table II).
TABLE II.
Areas of Homology on Chromosome 15 Across the Breakpoint Regions
| Proximal regiona | Elements in region |
Distal regiona | Elements in region |
Homology (%) | Length in nucleotides |
|---|---|---|---|---|---|
| 72536210–72536507 | AluJb | 72549217–72549512 | AluJb | 78 | 298 |
| 72536656–72536945 | AluSx | 72549219–72549510 | AluJb | 76 | 292 |
| 72537541–72537820 | AluSg | 72549217–72549492 | AluJb | 75 | 285 |
| 72536977–72537141 | AluSg | 72549347–72549512 | AluJb | 73 | 166 |
NCBI Build.
DISCUSSION
This extended family presents an intriguing case in which two potentially significant microduplications are concordant with disease. The region on chromosome 15 is especially interesting due to reported cases of autism, mental retardation, developmental delay, and Asperger’s syndrome caused by CNVs in this area [Smith et al., 2000; Cushman et al., 2005; Sharp et al., 2006, 2007; Kiholm Lund et al., 2008; Klopocki et al., 2008; Marshall et al., 2008; Andrieux et al., 2009; El-Hattab et al., 2009; Masurel-Paulet et al., 2009; McInnes et al., 2010; Pinto et al., 2010]. Previous cases of duplications and deletions within 15q24 defined a minimal critical region of approximately 1.33 Mb. This region contains 26 genes:GOLGA6, ISLR2, ISLR, STRA6, CCDC33, CYP11A1, SEMA7A, UBL7, ARID3B, CLK3, EDC3, CYP1A1, CYP1A2, CSK, LMAN1L, CPLX3, ULK3, SCAMP2, MP1, C15orf17, COX5A, RPP25, SCAMP5, PPCDC, C15orf39, and COMMD4. From the microduplication identified in this family, the maximum region of the CNV has now been reduced to approximately 10,000 base pairs (chr15:72,536,068–72,547,876) and only includes one known gene, the first four exons of the UBL7 gene and two predicted exons of the hypothetical gene LOC440288 (Fig. 3D). Expression of UBL7 (also known as BMSC-UbP) is widespread, including in the brain and spinal cord, though its function is unknown [Liu et al., 2003]. It is also possible that the CNV contains a regulatory element and is causing misregulation of a nearby gene or genes. One such candidate is the gene directly upstream of UBL7, semaphorin 7A (SEMA7A, OMIM:607961). Previous data has demonstrated that SEMA7A acts to promote axonal growth during embryonic development [Pasterkamp et al., 2003]. Significantly, the closely relatedSEMA5A gene on chromosome 5 was identified in an autism genome wide association study and there is evidence of decreased expression of this gene in patients with autism [Melin et al., 2006; Weiss et al., 2009].
There are also five reported CNVs identified in individuals with either autism or developmental delay that fail to overlap with our newly defined minimal region on 15q24 (Fig. 1) [El-Hattab et al., 2009, 2010; McInnes et al., 2010; Pinto et al., 2010]. Two deletions and a single duplication fall completely outside the previously delineated 1.33Mb critical region and, therefore, the pathogenic implications are unclear [El-Hattab et al., 2009; McInnes et al., 2010; Pinto et al., 2010]. The fourth CNV is a duplication located at 72.67–73.09Mb in an individual reported to have delays in development, speech, and motor skills, approximately 117.8 kb away from our newly recognized CNV [El-Hattab et al., 2010]. Perhaps these CNVs are close enough to alter distinct regulatory regions for the same gene. The final, non-overlapping CNV is an 82 kb duplication in individual 3016_003 located at 72.20–72.28Mb [Pinto et al., 2010]. However, the presence of small CNVs previously identified in control individuals across this area as well as the duplication’s proximity to the 15q24B LCR (72.199–72.281 Mb) led us to hypothesize that the causative gene is not located within this region [Locke et al., 2006; Redon et al., 2006].
While evidence of duplications and deletions in the chromosome 15 region demonstrate that the 15q24 duplication could contribute strongly to the clinical phenotypes in this family, the tandem duplication identified on chromosome 7 may also play a role. Two genes fall within the duplicated 7p21.3 region (Fig. 3), NXPH1 and ICA1.NXPH1 is an appealing candidate due to previous evidence of the protein NXPH1 complexing with neurexin 1α, which has already been implicated in autism [Kim et al., 2008; Yan et al., 2008]. The neurexophilin family of proteins is preferentially expressed in the brain, undergo glycosylation, and act as ligands forα-neurexins [Missler and Sudhof, 1998]. ICA1 is also an intriguing candidate due to its role in AMPA regulation [Cao et al., 2007]. It is possible that the 7p21 and 15q24 microduplications are acting in concert within family 17122. This synergistic effect was also suggested for three other families carrying 15q24CNVs where a second, potentially deleterious CNV was identified [El-Hattab et al., 2010].
It is interesting to note the pattern of inheritance within the family. There are four individuals that appear to fall along the spectrum of autism disorders, all male. Furthermore, both duplications are carried through two sisters, who fail to present with autistic features. For duplications on chromosome 15, this is consistent with earlier studies demonstrating inheritance from individuals that are not autistic and may be the result of incomplete or sex specific penetrance, X-linked inheritance or imprinting [Kiholm Lund et al., 2008; El-Hattab et al., 2009]. In the six previously reported 15q24 duplication families, two duplications were inherited maternally, three duplications were inherited paternally, and the last duplication was undetermined for inheritance status [Kiholm Lund et al., 2008; El-Hattab et al., 2009, 2010; Pinto et al., 2010]. This mirrors our finding of the duplication being inherited from individuals failing to present clinically with autism. In contrast, deletions reported across the 15q24 region all appear to occur de novo, with the exception of a deletion inherited paternally to two autistic sons that falls outside of the critical region [Smith et al., 2000; Cushman et al., 2005; Sharp et al., 2006,; Kiholm Lund et al., 2008; Klopocki et al., 2008; Marshall et al., 2008; Andrieux et al., 2009; El-Hattab et al., 2009, 2010; Masurel-Paulet et al., 2009; McInnes et al., 2010]. This suggests that the deletions falling across the critical region confer a severe and fully penetrant phenotype, while duplications have a milder effect, may be passed through clinically normal individuals, or may need to act in concert with another genetic aberration.
The mechanisms by which each of the duplications arose in this family are still unclear. Nonetheless, we defined the breakpoints in the 7p21.3 region, demonstrating that the tandem duplication is present and there is microhomolgy of four base pairs where the breakpoints join. This microhomology hints at potential mechanisms including fork stalling and template switching (FoSTeS) and microhomology-mediated break-induced replication (MMBIR) [Zhang et al., 2009]. While the chromosome 15 duplication was not delineated to single base pair resolution, an AluJb segment in the distal region was identified that may facilitate non-allelic homologous recombination with one of three Alu elements located in the proximal breakpoint region, a process reviewed by Hedges and Deininger [2007]. It also does not appear that the 15q24 duplication is directly adjacent to the original DNA in either direct or inverted orientations. This leaves open the possibility that some of the clinical phenotypes are not a result of the duplication itself but are perhaps due to a disruption at the insertion site. However, the previous reports of individuals demonstrating autism, mental retardation, developmental delay, and Asperger’s syndrome due to larger duplications and deletions supports the idea that the chromosome 15 duplication itself, and not a secondary effect, is causing a majority of the clinical features [Smith et al., 2000; Cushman et al., 2005; Sharp et al., 2006, 2007; Kiholm Lund et al., 2008; Klopocki et al., 2008; Marshall et al., 2008; Andrieux et al., 2009; El-Hattab et al., 2009, 2010; Masurel-Paulet et al., 2009; Pinto et al., 2010]. If this is true, then we have successfully narrowed the critical region from 1.33Mband 26 genes to approximately 10,000 base pairs and one known gene, UBL7. In conclusion, we have identified two microduplications concordant with disease in a multiplex family which may contribute to the genetics of autism.
ACKNOWLEDGEMENTS
We gratefully acknowledge our patients and their families, without whom this work would be impossible. Funding for this research was provided by the National Institute of Neurological Disorders and Stroke (7P01NS026630 and 9R01MH080647) and the Hussman Foundation.
Footnotes
While this manuscript was in press, Gai and colleagues identified two additional ASD families carrying microduplications on 7p21 [Gai et al., 2011]. Each CNV has breakpoints within the ICA1 and NXPH1 genes, similar in size and position to the duplication that we report here. We feel that this demonstrates an independent validation of the significance of this region to autism spectrum disorders.
The authors declare no competing interests.
REFERENCES
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders (DSM-IV) Washington: American Psychiatric Press, Inc; 1994. [Google Scholar]
- Andrieux J, Dubourg C, Rio M, Attie-Bitach T, Delaby E, Mathieu M, Journel H, Copin H, Blondeel E, Doco-Fenzy M, Landais E, Delobel B, Odent S, Manouvrier-Hanu S, Holder-Espinasse M. Genotype-phenotype correlation in four 15q24 deleted patients identified by array-CGH. Am J Med Genet Part A. 2009;149A:2813–2819. doi: 10.1002/ajmg.a.33097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Xu J, Shen C, Kam C, Huganir RL, Xia J. PICK1-ICA69 heteromeric BAR domain complex regulates synaptic targeting and surface expression of AMPA receptors. J Neurosci. 2007;27:12945–12956. doi: 10.1523/JNEUROSCI.2040-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman LJ, Torres-Martinez W, Cherry AM, Manning MA, Abdul-Rahman O, Anderson CE, Punnett HH, Thurston VC, Sweeney D, Vance GH. A report of three patients with an interstitial deletion of chromosome 15q24. Am J Med Genet Part A. 2005;137A:65–71. doi: 10.1002/ajmg.a.30836. [DOI] [PubMed] [Google Scholar]
- El-Hattab AW, Smolarek TA, Walker ME, Schorry EK, Immken LL, Patel G, Abbott MA, Lanpher BC, Ou Z, Kang SH, Patel A, Scaglia F, Lupski JR, Cheung SW, Stankiewicz P. Redefined genomic architecture in 15q24 directed by patient deletion/duplication breakpoint mapping. Hum Genet. 2009;126:589–602. doi: 10.1007/s00439-009-0706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hattab AW, Zhang F, Maxim R, Christensen KM, Ward JC, Hines-Dowell S, Scaglia F, Lupski JR, Cheung SW. Deletion and duplication of 15q24: Molecular mechanisms and potential modification by additional copy number variants. Genet Med. 2010;12:573–586. doi: 10.1097/GIM.0b013e3181eb9b4a. [DOI] [PubMed] [Google Scholar]
- Gai X, Xie HM, Perin JC, Takahashi N, Murphy K, Wenocur AS, D’arcy M, O’Hara RJ, Goldmuntz E, Grice DE, Shaikh TH, Hakonarson H, Buxbaum JD, Elia J, White PS. Rare structural variation of synapse and neurotransmission genes in autism. Mol Psych. 2011 doi: 10.1038/mp.2011.10. advance online pub Mar 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges DJ, Deininger PL. Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat Res. 2007;616:46–59. doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, Haugen E, Zerr T, Yamada NA, Tsang P, Newman TL, Tuzun E, Cheng Z, Ebling HM, Tusneem N, David R, Gillett W, Phelps KA, Weaver M, Saranga D, Brand A, Tao W, Gustafson E, McKernan K, Chen L, Malig M, Smith JD, Korn JM, McCarroll SA, Altshuler DA, Peiffer DA, Dorschner M, Stamatoyannopoulos J, Schwartz D, Nickerson DA, Mullikin JC, Wilson RK, Bruhn L, Olson MV, Kaul R, Smith DR, Eichler EE. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiholm Lund AB, Hove HD, Kirchhoff M. A 15q24 microduplication, reciprocal to the recently described 15q24 microdeletion, in a boy sharing clinical features with 15q24 microdeletion syndrome patients. Eur J Med Genet. 2008;51:520–526. doi: 10.1016/j.ejmg.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, Lally E, Weiss LA, Najm J, Kutsche K, Descartes M, Holt L, Braddock S, Troxell R, Kaplan L, Volkmar F, Klin A, Tsatsanis K, Harris DJ, Noens I, Pauls DL, Daly MJ, MacDonald ME, Morton CC, Quade BJ, Gusella JF. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopocki E, Graul-Neumann LM, Grieben U, Tonnies H, Ropers HH, Horn D, Mundlos S, Ullmann R. A further case of the recurrent 15q24 microdeletion syndrome, detected by array CGH. Eur J Pediatr. 2008;167:903–908. doi: 10.1007/s00431-007-0616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yu Y, An H, Li N, Lin N, Wang W, Zhang W, Wan T, Cao X. Cloning and identification of a novel ubiquitin-like protein, BMSC-UbP, from human bone marrow stromal cells. Immunol Lett. 2003;86:169–175. doi: 10.1016/s0165-2478(03)00004-x. [DOI] [PubMed] [Google Scholar]
- Locke DP, Sharp AJ, McCarroll SA, McGrath SD, Newman TL, Cheng Z, Schwartz S, Albertson DG, Pinkel D, Altshuler DM, Eichler EE. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am J Hum Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Rutter M, DiLavore P, Risi S. Autism diagnostic observation schedule-WPS (WPS edition) Los Angeles, CA: Western Psychological Association; 1999. [Google Scholar]
- Ma D, Salyakina D, Jaworski JM, Konidari I, Whitehead PL, Andersen AN, Hoffman JD, Slifer SH, Hedges DJ, Cukier HN, Griswold AJ, McCauley JL, Beecham GW, Wright HH, Abramson RK, Martin ER, Hussman JP, Gilbert JR, Cuccaro ML, Haines JL, Pericak-Vance MA. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Ann Hum Genet. 2009;73:263–273. doi: 10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masurel-Paulet A, Callier P, Thauvin-Robinet C, Chouchane M, Mejean N, Marle N, Mosca AL, Ben Salem D, Giroud M, Guibaud L, Huet F, Mugneret F, Faivre L. Multiple cysts of the corpus callosum and psychomotor delay in a patient with a 3.1 mb 15q24.1q24.2 interstitial deletion identified by array-CGH. Am J Med Genet Part A. 2009;149A:1504–1510. doi: 10.1002/ajmg.a.32904. [DOI] [PubMed] [Google Scholar]
- McInnes LA, Nakamine A, Pilorge M, Brandt T, Jiménez González P, Fallas M, Manghi ER, Edelmann L, Glessner J, Hakonarson HH, Betancur C, Buxbaum JD. A large-scale survey of the novel 15q24 microdeletion syndrome in autism spectrum disorders identifies an atypical deletion that narrows the critical region. Mol Autism. 2010;1:5. doi: 10.1186/2040-2392-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin M, Carlsson B, Anckarsater H, Rastam M, Betancur C, Isaksson A, Gillberg C, Dahl N. Constitutional downregulation of SEMA5A expression in autism. Neuropsychobiology. 2006;54:64–69. doi: 10.1159/000096040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Sudhof TC. Neurexophilins form a conserved family of neuropeptide-like glycoproteins. J Neurosci. 1998;18:3630–3638. doi: 10.1523/JNEUROSCI.18-10-03630.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasterkamp RJ, Peschon JJ, Spriggs MK, Kolodkin AL. Semaphorin 7A promotes axon outgrowth through integrins and MAPKs. Nature. 2003;424:398–405. doi: 10.1038/nature01790. [DOI] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bolte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BH, Cochrane L, Corsello C, Crawford EL, Crossett A, Cytrynbaum C, Dawson G, de Jonge M, Delorme R, Drmic I, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Goldberg J, Green A, Green J, Guter SJ, Hakonarson H, Heron EA, Hill M, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Korvatska O, Kustanovich V, Lajonchere CM, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Lionel AC, Liu XQ, Lord C, Lotspeich L, Lund SC, Maestrini E, Mahoney W, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Merikangas A, Migita O, Minshew NJ, Mirza GK, Munson J, Nelson SF, Noakes C, Noor A, Nygren G, Oliveira G, Papanikolaou K, Parr JR, Parrini B, Paton T, Pickles A, Pilorge M, Piven J, Ponting CP, Posey DJ, Poustka A, Poustka F, Prasad A, Ragoussis J, Renshaw K, Rickaby J, Roberts W, Roeder K, Roge B, Rutter ML, Bierut LJ, Rice JP, Salt J, Sansom K, Sato D, Segurado R, Sequeira AF, Senman L, Shah N, Sheffield VC, Soorya L, Sousa I, Stein O, Sykes N, Stoppioni V, Strawbridge C, Tancredi R, Tansey K, Thiruvahindrapduram B, Thompson AP, Thomson S, Tryfon A, Tsiantis J, Van Engeland H, Vincent JB, Volkmar F, Wallace S, Wang K, Wang Z, Wassink TH, Webber C, Weksberg R, Wing K, Wittemeyer K, Wood S, Wu J, Yaspan BL, Zurawiecki D, Zwaigenbaum L, Buxbaum JD, Cantor RM, Cook EH, Coon H, Cuccaro ML, Devlin Ennis S, Gallagher L, Geschwind DH, Gill M, Haines JL, Hallmayer J, Miller J, Monaco AP, Nurnberger JI, Jr, Paterson AD, Pericak-Vance MA, Schellenberg GD, Szatmari P, Vicente AM, Vieland VJ, Wijsman EM, Scherer SW, Sutcliffe JS, Betancur C. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M, Bailey A, Berument SK, LeCouteur A, Lord C, Pickles A. Social communication questionnaire (SCQ) Los Angeles, CA: Western Psychological Services; 2003a. [Google Scholar]
- Rutter M, LeCouteur A, Lord C. Autism diagnostic interview, revised (ADI-R) Los Angeles, CA: Western Psychological Services; 2003b. [Google Scholar]
- Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA, Stewart H, Price SM, Blair E, Hennekam RC, Fitzpatrick CA, Segraves R, Richmond TA, Guiver C, Albertson DG, Pinkel D, Eis PS, Schwartz S, Knight SJ, Eichler EE. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Selzer RR, Veltman JA, Gimelli S, Gimelli G, Striano P, Coppola A, Regan R, Price SM, Knoers NV, Eis PS, Brunner HG, Hennekam RC, Knight SJ, de Vries BB, Zuffardi O, Eichler EE. Characterization of a recurrent 15q24 microdeletion syndrome. Hum Mol Genet. 2007;16:567–572. doi: 10.1093/hmg/ddm016. [DOI] [PubMed] [Google Scholar]
- Smith M, Filipek PA, Wu C, Bocian M, Hakim S, Modahl C, Spence MA. Analysis of a 1-megabase deletion in 15q22-q23 in an autistic patient: Identification of candidate genes for autism and of homologous DNA segments in 15q22-q23 and 15q11-q13. Am J Med Genet. 2000;96:765–770. doi: 10.1002/1096-8628(20001204)96:6<765::aid-ajmg13>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- Velez DR, Fortunato S, Thorsen P, Lombardi SJ, Williams SM, Menon R. Spontaneous preterm birth in African Americans is associated with infection and inflammatory response gene variants. Am J Obstet Gynecol. 2009;200:209.e1–209.e27. doi: 10.1016/j.ajog.2008.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: An integrated hidden markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Arking DE, Daly MJ, Chakravarti A Gene Discovery Project of Johns Hopkins the Autism Consortium. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Noltner K, Feng J, Li W, Schroer R, Skinner C, Zeng W, Schwartz CE, Sommer SS. Neurexin 1alpha structural variants associated with autism. Neurosci Lett. 2008;438:368–370. doi: 10.1016/j.neulet.2008.04.074. [DOI] [PubMed] [Google Scholar]
- Zhang F, Carvalho CM, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]