SUMMARY
Mitochondrial network connectivity enables rapid communication and distribution of potential energy throughout the cell. However, this connectivity puts the energy conversion system at risk since damaged elements could jeopardize the entire network. Here we demonstrate the mechanisms for mitochondrial network protection in heart and skeletal muscle (SKM). We find that the cardiac mitochondrial reticulum is segmented into subnetworks comprised of many mitochondria linked through abundant contact sites at highly specific intermitochondrial junctions (IMJs). In both cardiac and SKM subnetworks, a rapid electrical and physical separation of malfunctioning mitochondria occurs consistent with detachment of IMJs and retraction of elongated mitochondria into condensed structures. Regional mitochondrial subnetworks limit the cellular impact of local dysfunction while the dynamic disconnection of damaged mitochondria allows the remaining mitochondria to resume normal function within seconds. Thus, mitochondrial network security is comprised of both proactive and reactive mechanisms in striated muscle cells.
Keywords: Energy distribution, muscle energetics, oxidative phosphorylation, 3D electron microscopy, mitochondrial retraction, mitochondrial dynamics
INTRODUCTION
Cellular mitochondrial networks allow for sharing of metabolites and proteins as well as mitochondrial DNA (Bleazard et al., 1999; Molina et al., 2009; Szabadkai et al., 2004). Coupled mitochondria also provide a rapid conductive path for the distribution of potential energy (Glancy et al., 2015). However, this extensive coupling presents a major risk as local failures can also spread quickly over the entire network and compromise cellular energy conversion. Like many power networks that physically segment elements with circuit breakers, similar strategies may be in place to protect cells with coupled mitochondrial networks from propagating local failures.
The heart has a high constant energy demand with a putative physically connected mitochondrial network (Bakeeva et al., 1983). However, there are contradictory reports of the functional connectivity of the cardiac mitochondrial reticulum. (Amchenkova et al., 1988; Aon et al., 2003; Duchen et al., 1998; Romashko et al., 1998; Wang et al., 2008). We hypothesized that these discrepancies could be explained, at least in part, by the induction of mitochondrial network protection mechanisms in place to mitigate the risks associated with connectivity in the heart. Here, we demonstrate a physically and electrically connected mitochondrial reticulum arranged into longitudinal subnetworks within the cardiac cell. Additionally, we show that both physical and electrical connectivity can be dynamically modulated in response to localized damage in heart and SKM. These rapid alterations in mitochondrial connectivity allow muscle cells to respond to local dysfunction within seconds and restore the energy distribution systems to the remainder of the cell.
RESULTS
The Cardiac Mitochondrial Reticulum is Physically Coupled through Abundant Intermitochondrial Junctions
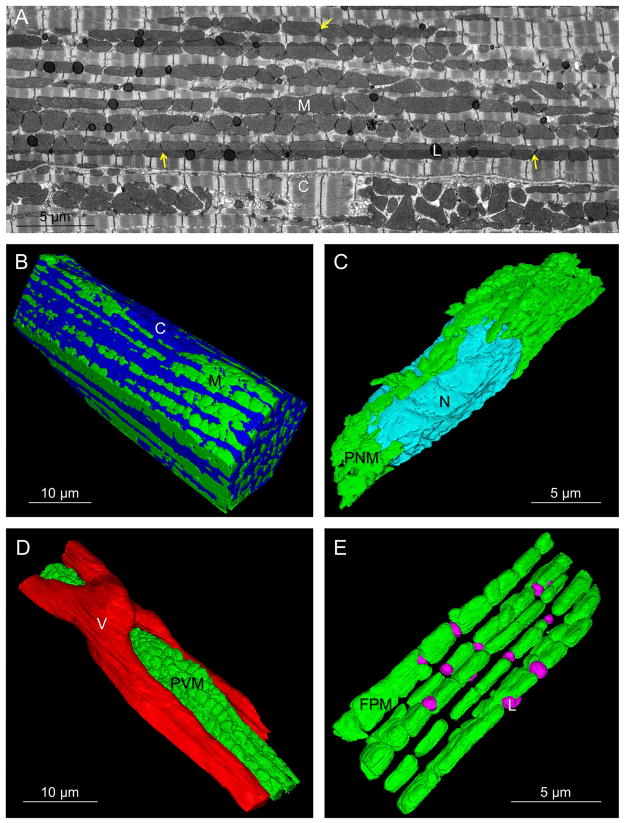
To analyze the structural connectivity of the cardiac mitochondrial reticulum in 3D, focused ion beam scanning electron microscopy (FIB-SEM) volumes of murine left ventricles were collected (Figure 1, Movie S1). Cardiac mitochondrial network structure was significantly different from the SKM described previously (Glancy et al., 2015). Paravascular mitochondria (PVM, Figure 1D) were much less frequent than in SKM but still associated with shallow capillary grooves (Glancy et al., 2014). Paranuclear mitochondria (PNM, Figure 1C) were primarily located on opposite sides of the long axis of the nuclei. The most extensive mitochondrial pool ran parallel to the fiber axis between myofibrils in long, closely packed rows of mitochondria. These fiber parallel mitochondria (FPM, Figure 1E) were larger but morphologically similar to the fiber parallel mitochondrial segments detected in SKM. Notably, the network of thin, tubular I-band mitochondrial segments that ran perpendicular to the long axis of the SKM fiber was not observed in the heart.
Figure 1. Heart Mitochondria Form Networks.
A) Single FIB-SEM frame. M – mitochondria, C – contractile proteins, L – lipid droplets, yellow arrows - intermitochondrial junctions (IMJ). B) 3D rendering of cardiomyocyte (upper cell in A). C) 3D rendering of paranuclear mitochondria (PNM) and nucleus (N). D) 3D rendering of paravascular mitochondria (PVM) and a capillary (V). E) 3D rendering of fiber parallel mitochondria (FPM) and lipid droplets (L). Represents 5 volumes, 4 animals. See Movie S1 and Dataset S1.
Intermitochondrial junctions (IMJs) are defined by the close apposition of both the inner and outer membranes of two adjacent mitochondria with high electron density (Figure 2A,B). While IMJ function and molecular composition have yet to be explicitly defined, IMJs have been suggested to permit communication and/or energy distribution (Bakeeva et al., 1983; Duvert et al., 1985; Glancy et al., 2015; Picard et al., 2015). Indeed, the PNM, PVM and FPM pools were all highly coupled by many IMJs (see Figure 1A). These IMJs are not simply due to close packing of mitochondria or artifacts associated with fixation as IMJs are not observed in EM images of mouse kidney or liver (Xu et al., 2008) despite high mitochondrial content. However, the precise physical nature as well as the frequency and conductivity of these specialized IMJs remain unclear.
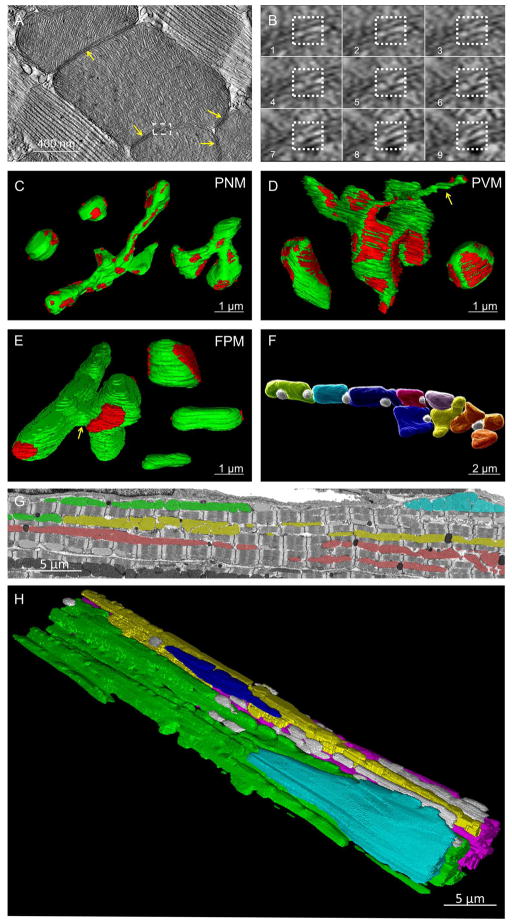
Figure 2. Heart Mitochondrial Morphology and Connectivity.
A) Single IMJ (yellow arrows) tomogram slice. Dashed box shown in B. B) Sequential tomograms of membrane to membrane contact site (dotted box). C–E) Mitochondrial morphologies (green) and locations of IMJ (red). C) Compact (upper left) and Elongated (middle and right) PNM. D) Elongated, Nanotube (yellow arrow), and Compact PVM (left to right). E) Connector (left), Compact (right upper), Elongated (right middle), and Non-connected (right lower) FPM. Yellow arrow – sheet-like connecting structure. F) Adjacent FPM (assorted colors) and lipid droplets (white). G) FIB-SEM frame. Mitochondria in the same color are IMJ-coupled. H) 3D rendering of mitochondrial subnetworks. Each color represents a different IMJ-coupled subnetwork. Grey - not network connected. Represents 5 volumes, 4 animals. See Table S1 and Movie S1.
To provide insight into how IMJs may participate in the connectivity of a cardiac mitochondrial reticulum, we determined the morphology and IMJ linkage sites of the different types of mitochondria in each region of the heart cell (Figures 2C–E). The PNM pool consisted of two basic mitochondrial morphologies: “Compact” PNM were smaller, somewhat spherical mitochondria while “Elongated” PNM were larger and often had several branches (Table S1, Figure 2C). Though there was no difference in surface/volume ratio, Elongated PNM had slightly more surface area participating in an IMJ compared to Compact PNM. PVM also consisted primarily of Compact and Elongated morphologies, although they were generally larger and had lower surface/volume ratios than PNM. Large mitochondria with nanotube (Huang et al., 2013) projections (see yellow arrow in Figure 2D) were also present in the PVM pool, though they made up less than 10% of all PVM. The ~1–4 μm long, thin nanotubes were observed as a connecting structure within an individual mitochondrion or to form an IMJ with one or more adjacent mitochondria. FPM were made up of four basic structures (Figure 2E). Mitochondria with no IMJ coupling (“Non-connected”) were generally smaller and classical bean shaped structures. These may be newly formed mitochondria soon to be connected with the rest of the network or recently disconnected mitochondria ready for degradation. The most prevalent FPM morphology was an Elongated structure often spanning the length of two or more sarcomeres with IMJs at each end. Another common FPM morphology was a more Compact structure with IMJ coupling to several adjacent mitochondria similar to that observed in the PNM and PVM. The fourth FPM morphology was a large mitochondrion containing thin, sheet-like portions (see yellow arrow in Figure 2E) wrapping around the width of one or more sarcomeres and “Connecting” multiple rows of mitochondria through IMJs. Another distinguishing characteristic of the heart mitochondria was the presence of highly embedded, spherical lipid droplets nearly surrounded by the mitochondria (Figure 2F) suggesting direct, local access of these mitochondria to fatty acids. Consistent with this notion, at the lipid-mitochondrial interface, their respective membranes are difficult to resolve on the EM scale suggesting a highly specialized interaction potentially supporting lipid transport to the mitochondria.
Mitochondrial Subnetworks Proactively Limit the Spread of Dysfunction within the Cell
The extent of a potential IMJ-facilitated conductive pathway through mitochondrial networks in the heart was examined by manually tracing mitochondrial pools coupled through IMJs as defined by their increased electron density at the point where the membranes of two adjacent mitochondria were indistinguishable from each other (see yellow arrows in Figure 1A). Mitochondria in the heart do not form a single network coupled via IMJs, but rather several subnetworks of tens to hundreds of mitochondria each (Figure 2G–H, Movie S1). In general, all of the PVM or PNM in a given region of the cell were completely coupled through IMJ structures. In addition, all PVM and PNM pools observed were coupled by IMJs to nearby FPM pools providing a link to different regions of the cell. Most of the mitochondrial subnetworks ran in rows along the long axis of the cardiac cell with multiple rows often joined by the FPM Connector mitochondria described above (Figure 2E, Table S1). However, in many cases, two rows of coupled mitochondria would approach within 100 nm of each other but not form an IMJ. Thus, mitochondria in the heart cell form several regional subnetworks along the long axis of the cell comprised of one or more rows of mitochondria with few perpendicular connections.
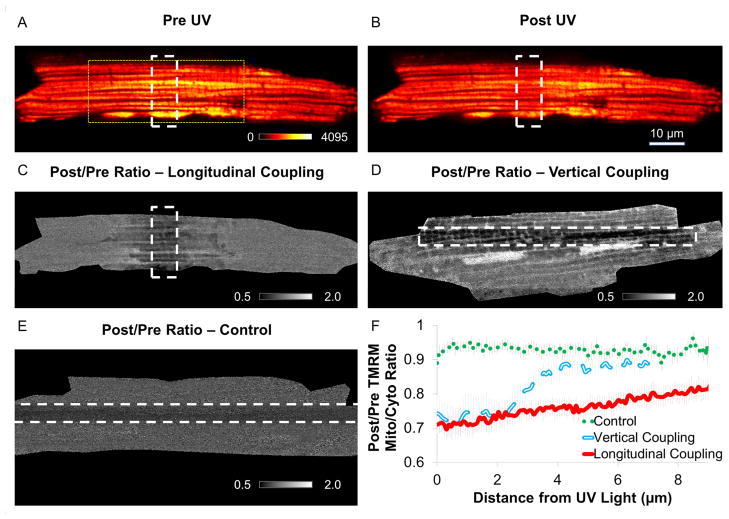
To assess electrical connectivity, we used a similar strategy as previously published in SKM (Glancy et al., 2015) utilizing a photoactivated mitochondrial uncoupler localized to the mitochondrial matrix (MitoPhotoDNP (MPD) (Chalmers et al., 2012)). In these studies, isolated cardiomyocytes (CM) are equilibrated with MPD in the presence of TMRM, a fluorescent dye whose concentration gradient from the cytosol to the mitochondrial matrix is proportional to Δψm. MPD was regionally irradiated to increase the proton permeability of the mitochondrial inner membrane and the redistribution of TMRM associated with any depolarization of Δψm was followed. Within the first frame (~400msec) after irradiation, significant redistribution of TMRM from the mitochondria to the cytosol was detected outside the irradiation region consistent with functionally coupled mitochondrial subnetworks (Figure 3). To examine coupling down the longitudinal axis of the cell, we employed an irradiation zone that was perpendicular to the cell (Longitudinal Coupling, see Figure 3A–C and Movie S2) and to examine vertical coupling, we employed an irradiation zone parallel to the cell (Vertical Coupling, see Figure 4D, Figure S1, and Movie S2). Small, “single-mitochondrion” sized irradiation zones did not result in significant depolarization outside the irradiation region (Figure S1), thus, larger irradiation regions were used to overcome the current capacity of the network. Consistent with the structural data, mitochondrial depolarization extended further outside the irradiation region down the longitudinal axis (Figure 3F). This arrangement of mitochondria into several regional subnetworks as opposed to a single, cell-wide network limits the spread of localized mitochondrial dysfunction to within defined volumes while still providing a uniform potential energy source within each subnetwork.
Figure 3. Membrane Potential Conduction.
TMRM and MPD loaded cardiomyocyte prior to (A) and after (B) UV irradiation. White dashed lines - UV irradiation zone. Yellow dotted lines - size of FIB-SEM image in Figure 1A. C) Ratio TMRM image before and after irradiation. Black pixels – decreasing signal. White pixels – increasing signal. White dashed lines – irradiation zone for Longitudinal coupling. Represents 9 experiments, 6 mice. D) Ratio TMRM image for Vertical coupling. Represents 8 experiments, 4 mice. E) Ratio TMRM image without MPD (Control). Represents 9 experiments, 5 mice. F) Mitochondrial depolarization extends further along longitudinal axis. See Movie S2 and Figures S1,3.
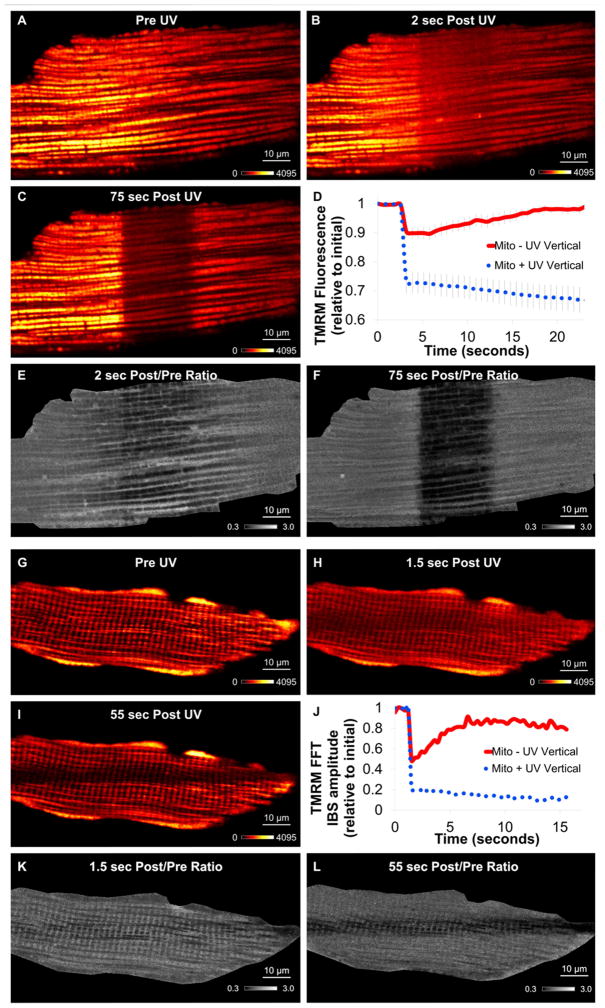
Figure 4. Mitochondrial Circuit Breaker.
TMRM fluorescence before (A), immediately after (B), and 75 seconds after (C) cardiomyocyte regional uncoupling. D) Mitochondrial TMRM timecourse of irradiated (dotted blue) and non-irradiated, depolarized (solid red) regions. Data = mean ± SE. E) Ratio TMRM image before and immediately after irradiation. F) Ratio TMRM image before and 75 sec after UV. Represent 7 experiments, 3 mice. G–I) TMRM fluorescence during muscle fiber regional uncoupling. G) Pre UV. H) Immediately after UV. I) 55 seconds after UV. J) Representative TMRM fast fourier transform (FFT) 0.5 μm−1 frequency amplitude timecourse of irradiated (dotted blue) and non-irradiated (solid red) regions. K) Ratio TMRM image before and immediately after irradiation. L) Ratio TMRM image before and 55 sec after UV. Represents 5 experiments, 5 mice. See Movies S3.
Depolarized Mitochondria are Electrically Separated from the Mitochondrial Reticulum in Seconds
While the initial (<5 seconds) response to regional mitochondrial depolarization permitted assessment of the functional connectivity of the mitochondrial reticulum as described above, observation over longer periods of time allowed us to evaluate how the mitochondrial network responded to this locally induced dysfunction. Shortly after the immediate, network-wide Δψm depolarization (Figure 4A,B), the center of the cell where the uncoupler was activated continued to depolarize as evident by the loss of TMRM signal in this region (Figure 4C). In contrast, the TMRM signal outside of the irradiated region quickly redistributed back from the cytosol into the mitochondria consistent with a repolarization of Δψm with a recovery time constant of 7.6±1.0 seconds (n=7) (Figure 4D,F, Movie S3). Thus, after the initial network-wide depolarization, there was an apparent electrical separation of the uncoupled mitochondria from the healthy mitochondria. This resulted in the continued loss of TMRM from the damaged region while the heathy region, no longer burdened by the current drain of the uncoupled region, recovered the Δψm.
An electrical separation mechanism is also present in isolated mouse soleus fibers (Figure 4G–L, Movie S3) which form a large, grid-like reticulum electrically coupled by many long, branched mitochondria linked together through IMJs (Glancy et al., 2015). Due to the larger network configuration in the SKM cells, the initial response to local uncoupling was a more homogeneous depolarization of the mitochondrial reticulum than in the cardiac cells (Figure 4K), but the secondary electrical separation of the uncoupled and control regions of the cell was still observed (Figure 4J,L) and with a similar recovery time constant (7.9±0.3 seconds, n=5). These results were not consistent with the electrical response of mitochondria to photodamage (Movie S3).
Depolarized Mitochondria are Rapidly Physically Removed from the Mitochondrial Reticulum
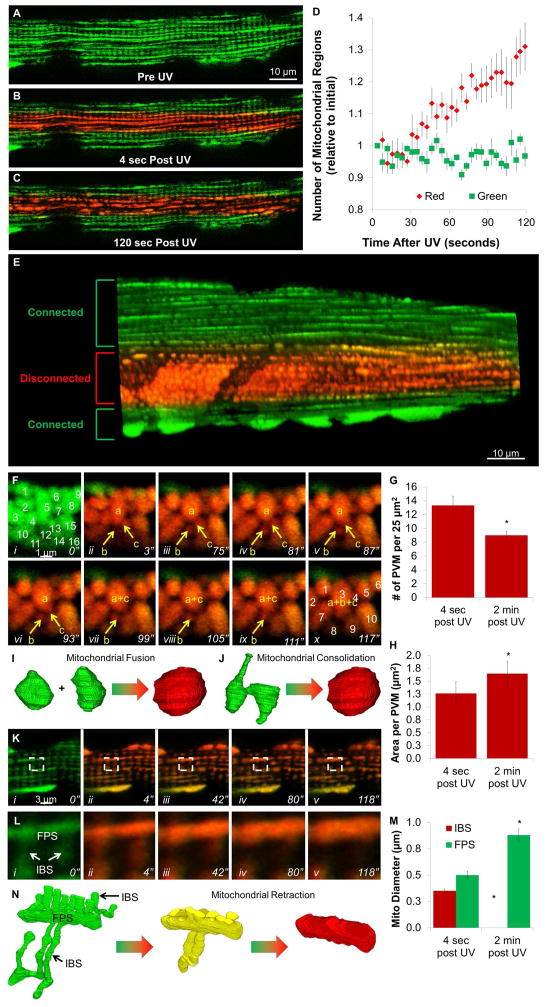
We hypothesized that the electrical separation of the mitochondrial network observed after localized damage involved a rapid, physical separation of the mitochondria via fission or other mechanical event. In order to observe mitochondrial structural changes, we used mice that express a green-to-red photo-switchable fluorescent protein localized to the mitochondrial matrix (MitoDendra2) (Pham et al., 2012). By combining MPD with MitoDendra2, we were able to simultaneously depolarize and photoconvert mitochondria in the center of the cell allowing for separate structural observations of the uncoupled (red) and normal (green) regions of the mitochondrial network. We focused primarily on SKM fibers for these studies as the lower mitochondrial content allowed for better visualization of individual mitochondrial structures. In the absence of MPD, mitochondrial dynamics were negligible in the two minutes after photoactivation (Movie S4) consistent with previous reports in muscle (Eisner et al., 2014). Prior to and immediately after activating MPD in the center of the cell, both the uncoupled and normal regions of the cell retained their normal, grid-like mitochondrial network structure (Figure 5A,B). However, quickly after photoconverting MPD, the red, uncoupled mitochondria became highly dynamic while the green, control regions of the cell maintained its static mitochondrial structure (Figure 5C, Movie S4). Over time, the uncoupled region became progressively more disconnected as observed by the increasing number of separate mitochondrial regions in these 2D images (Figure 5D). Within a few minutes, the irradiated regions became filled with apparently separated, spherical mitochondrial structures, while the control regions of the cell continued to maintain the normal, grid-like mitochondrial network (Figure 5E).
Figure 5. Mitochondrial Network Disconnection.
MitoDendra2 fluorescence prior to (A), immediately after (B), and 120 seconds after (C) UV irradiation of muscle fiber w/MPD. D) Timecourse of network disconnectivity. Mean±SE, n=6 fibers, 3 mice. E) 3D rendering after regional uncoupling. F) Timeseries of muscle fiber PVM MitoDendra2 fluorescence after regional uncoupling. G) Number of PVM immediately and 2 minutes after regional uncoupling (n=3). H) Area per PVM immediately and 2 minutes after regional uncoupling (n=3). I,J) 3D schematic of PVM fusion (I) or consolidation (J). K) Timeseries of intrafibrillar MitoDendra2 fluorescence after regional uncoupling. L) Boxed region from K shows retraction of I-band mitochondrial segments (IBS) into fiber parallel segments (FPS). M) Mitochondrial diameters immediately after and 2 minutes after irradiation (n= 16 mitochondria, 3 experiments). N) 3D schematic of mitochondrial retraction. Structures from (Glancy et al., 2015). Represents 6 fibers, 3 mice. *significantly different from 4 sec after UV. See Figure S2 and Movies S4,5.
Based on our initial results, we hypothesized that this physical disconnection of damaged mitochondria involved breaking the IMJs between adjacent mitochondria. One area to examine this is the PVM pool of SKM that does not compete for space with the contractile elements (Glancy et al., 2014), and as such, are tightly packed together through IMJs but with highly irregular shapes (Glancy et al., 2015). Both before and immediately after depolarization of Δψm, PVM remained closely associated and with varying size and shape (Figure 5Fi,ii). However, within two minutes after regional uncoupling, 32 ± 4% fewer PVM were detected within a given volume (Figure 5G) as they became 33 ± 8% larger (Figure 5H) as well as more round in shape. A time series of these changes (Figure 5F, Movie S5) shows what appear to be three individual mitochondria (a–c) prior to depolarization eventually merging into a single mitochondrion through two separate events. Due to the 2D resolution of these images, it is difficult to discern whether the observed merging of PVM is caused by fusion of multiple mitochondria (Figure 5I) or consolidation of a single, non-uniform 3D mitochondrion which may appear as multiple structures in a 2D image (Figure 5J).
In the intrafibrillar regions of the cell, network disconnections also appeared to occur at locations consistent with IMJs between two adjacent mitochondria (Figure 5K, Movie S5). This physical separation of IMJs was particularly evident in the uncoupled regions of MitoDendra2 isolated CM (Figure S2). Upon network disconnection in the SKM, the thin, tubular I-band mitochondrial segments (IBS) were observed to retract into the thicker, fiber parallel mitochondrial segments (FPS) until the IBS were no longer distinct from the now 82 ± 10% wider FPS (Figure 5K–N, Movie S5). Thus, similar to the tightly packed PVM, mitochondria in the intrafibrillar space of the SKM fiber also appear to disconnect at IMJs and consolidate into more compact structures.
DISCUSSION
The large differences in mitochondrial reticulum structure, conductivity, and protein programming (Figure S3) between heart and SKM suggest that mitochondrial network configuration is tuned to match the functional requirements of the cell. The SKM high mechanical power, and associated myofibril content, limits the mitochondrial volume to support energy homeostasis. Thus, a fine, grid-like network (Glancy et al., 2015) provides a mechanism allowing a minimal mitochondrial volume to rapidly and efficiently distribute potential energy. Conversely, the constantly active heart prioritizes energy homeostasis over mechanical power resulting in a much greater mitochondrial content. However, even with ATP producing and utilizing enzymes in such close proximity, simple diffusion of metabolites is inadequate to match ATP production and utilization in the heart (Birkedal et al., 2014). Thus, the heart requires specialized energy distribution systems to prevent potential energy gradients that could compromise steady state function. While creatine kinase facilitates cytosolic energy distribution, an electrically conductive mitochondrial reticulum would provide a rapid summation of ATP production capacity and ensure cellular energy homogeneity across large regions of the cell.
While relatively greater Δψm conductivity down the cardiomyocyte longitudinal axis (Figure 3) was consistent with structural connectivity (Figures 1,2), it has also been shown that cytosolic diffusion rates are faster longitudinally compared to vertically in CM (Vendelin and Birkedal, 2008) and might channel TMRM diffusion confounding our interpretation. However, the increase in cytosolic TMRM remote from the irradiation region was associated with a loss of mitochondrial TMRM from the same region consistent the local release of TMRM and not long range diffusion from the irradiated zone.
Though our irradiation scheme using 355 nm light to photoactivate MPD only has a small photobleaching effect on TMRM alone (Figure 3E), it is possible that the presence of MPD increases the sensitivity of the SKM and cardiac cells to light induced damage. We tested this by irradiating MPD-loaded MitoDendra2 CM simultaneously with 355 nm light in one region of the cell and 405 nm light in another region of the cell (Figure S2). Each light source resulted in similar red-to-green photoconversion of MitoDendra2, however, only the 355 nm light resulted the remodeling of mitochondria into more consolidated structures. Since 405 nm light does not activate MPD (Chalmers et al., 2012), these results suggest mitochondrial depolarization, not photodamage, causes the consolidation of mitochondrial structures in MitoDendra2 cardiac and SKM cells.
It is tempting to predict that irradiating a single MPD-loaded mitochondrion within the mitochondrial reticulum would depolarize the connecting mitochondria and reveal the extent of mitochondrial subnetworks. However, we do not observe any measurable depolarization outside the irradiated region when using small irradiation schemes in both cell types (Figure S1). This is likely due to several aspects of a coupled, high current capacity network. The rate of oxidative phosphorylation (i.e. the current) in mitochondria of non-contracting CM can be increased at least ten-fold (Monge et al., 2009) and higher in SKM. Thus, when only 1 or 2 out of several hundred mitochondria in a network are uncoupled, the remainder of the network is able to easily increase its current to maintain the voltage. This is also likely a safety feature of the network in the short term. Due to the coupling and reserve capacity of the network, we found it necessary to photoactivate regions large enough to overcome the current capacity of all connected mitochondria. Small regions of photoactivated uncoupling cannot pull this high capacity network down.
Depolarization of Δψm is known to cause mitochondrial fission (Narendra et al., 2008), and fission has even been suggested play a role preventing damage in mitochondrial networks (Skulachev et al., 2004). However, the depolarization-induced mitochondrial dynamics shown in Movie S5 (Figure 5) are not consistent with the simple division of one mitochondria in two (Nunnari et al., 1997). In the SKM PVM, localized depolarization resulted in fewer and larger mitochondria (Figure 5F–H) whereas mitochondrial fission would produce many smaller mitochondria. Moreover, several consolidation events between mitochondria were observed in the depolarized PVM as opposed to the splitting events mitochondrial fission would cause. In the intrafibrillar regions, localized depolarization induced MitoDendra2 mitochondria to retract such that all perpendicular IBS were no longer present and FPS became more condensed and with larger diameters (Figure 5K–M) which is not consistent with fission. The rate of mitochondrial fission in adult SKM is much lower than the already low fusion rate (Eisner et al., 2014). Moreover, depolarization-induced fission is often measured over hours or even days (Narendra et al., 2008; Skulachev et al., 2004). Thus, retracting mitochondria into consolidated structures may be a more efficient protection mechanism to quickly prevent the spread of localized dysfunction throughout the muscle mitochondrial reticulum. These data also suggest that the complex mitochondrial structures observed in both muscles are under tension by cytoskeletal tethering that once released results in the condensed structures observed after uncoupling.
It is important to note that the physical sequestration mechanism could not be detected until ~30 seconds after the depolarization event (Figure 5D) whereas electrical separation begins within 5 seconds after depolarization (Figure 4). Thus, electrical and physical separation appear to be sequential steps in the mitochondrial network protection system. While the structural sequestration and consolidation step is not likely to be reversible and would appear to facilitate the lysosomal removal of these damaged elements via mitophagy (Gomes et al., 2011; Lemasters, 2005), electrical network separation may be reversible if the IMJs are the conductive elements between adjacent mitochondria as the data here suggests. Indeed, the capacity to rapidly modulate electrical network connectivity may also explain why transient “flickers” (Duchen et al., 1998) or “flashes” (Wang et al., 2008) of the Δψm of individual mitochondria can occur without depolarizing the entire network. In fact, we can induce localized Δψm flickering, after a short delay, by increasing our UV irradiation power to cause photodamage (Movie S3). We propose that the electrical separation mechanism is designed to quickly prevent the spread of transient local dysfunction while the physical separation mechanism targets unrecoverable mitochondria for repair or replacement (Lemasters, 2005).
Currently, IMJs are the only structural element that would permit rapid coupling of Δψm across adjacent mitochondria within the muscle mitochondrial reticulum. However, the specific molecular composition of IMJs remains unknown thereby precluding targeted manipulation and direct observation of changes to the connectivity and protection system of the muscle mitochondrial reticulum to test this hypothesis. Though the decades long search and eventual discovery of the mitochondrial calcium uniporter protein(s) (Baughman et al., 2011; De Stefani et al., 2011; Deluca and Engstrom, 1961) can provide some guidance in the search for IMJ proteins, development of IMJ abundant cell systems amenable to rapid screening will be critical in this pursuit. However, regardless of the degree to which IMJs are involved (or not), we demonstrate here that a multi-tiered network security scheme is in place to protect the electrically connected muscle mitochondrial reticulum. This mitochondrial network protection system limits the propagation of local failures and allows for the quick recovery of undamaged mitochondria in order to sustain cellular function.
EXPERIMENTAL PROCEDURES
Mice
2–4 month old male C57BL/6N (Taconic) and male MitoDendra2 (JAX # 018397) mice were used. Procedures were approved by the NHLBI Animal Care and Use Committee and performed in accordance with the Animal Care and Welfare Act (7 USC 2142 § 13).
Focused Ion Beam Scanning Electron Microscopy (FIB-SEM)
Mice prepared as described previously (Glancy et al., 2014). Mouse hearts were perfusion fixed, stained, and embedded as in (Sun et al., 2012). Five left ventricular myocyte data sets from three mice were collected were imaged on a Zeiss Nvision 40 as described previously (Glancy et al., 2015). A raw, 2.5 GB TIFF stack for the dataset shown in Movie S1 can be downloaded at https://labalaban.nhlbi.nih.gov/files/HeartDatasetS1.tif.
FIB-SEM Analysis
Whole image segmentation was performed with a 3D, two pixel median filter. The intensity threshold was manually adjusted to create binary images including primarily mitochondria and followed by the Remove Outliers tool. Manual tracing was done by three different observers using TrakEM (ImageJ).
Electron Tomography
Resin-embedded heart was cut into 200–230nm sections, placed on carbon-coated formvar grids with gold fiducial markers, stained with uranyl acetate and lead citrate and imaged on a FEI Tecnai T12 microscope at a tilt interval of 2° over a ± 60° range, using a pixel size of 0.56 nm. Reconstructions were generated in IMOD using the weighted backprojection method (Kremer et al., 1996).
Heart Immunostaining
Excised hearts were frozen and embedded in optimal cutting temperature compound then cryosectioned and air dried for 5 min prior to fixation in 10% formalin for 7 min at 25C°. Sections were washed 3X in PBS for 5 min, permeabilized for 5 min with 0.01% Triton-X 100 in PBS, blocked for 20 min with 10% goat serum, and incubated with primary antibodies. Complex IV and V distribution was imaged and assessed by the mean Complex IV/V ratio as described previously (Glancy et al., 2015) in 11 heart cells from 3 mice.
Myocyte Isolation
Adult mouse CM were isolated as described previously (Sun and Murphy, 2010) and suspended in an incubation medium (IM) composed of: (mM): NaHEPES (10), NaCl (137), KCl (5.4), CaCl2 (1.8), MgCl2 (0.5), NaH2PO4 (0.5), Glucose (10), NaPyruvate (1) and Butanedione monoxomine (BDM, 20), pH 7.4. CM were incubated in IM containing 5 nM TMRM and/or 20 μM MPD for >20 minutes. Mouse soleus fibers were prepared as described previously (Glancy et al., 2015).
Light Microscopy
Confocal microscopy and MPD photoactivation of isolated CM was conducted as described previously (Glancy et al., 2015). MitoDendra2 muscle cells were excited with a 488 nm laser at 0.15% power and emission collected from 490 nm to 553 nm (green) and excited with a 561 nm laser at 2% power and emission collected from 570 nm to 695 nm (red). Green to red photoconversion was performed with the same 355 nm laser pulse used to photoactivate MPD.
Confocal Image Analysis
Mitochondrial functional connectivity was analyzed from averages of five images taken before (Pre) and immediately after UV exposure (Post). Mitochondrial and cytosolic/nuclei TMRM signals were separated for analysis by intensity thresholding each image. Ratiometric images were created by dividing the Post images by the Pre image.
FFT analysis of IBS-specific TMRM fluorescence was performed as described in (Glancy et al., 2015). The IBS recovery time constant (τ) was fit using a linear least squares approach according to IBSt = IBSi + ΔIBS(1−e−t/τ) where IBSt is the IBS amplitude at any time point, t, IBSi is the initial IBS amplitude, and ΔIBS is the difference in IBS amplitude between the first point after UV irradiation and the initial state.
Cardiomyocyte TMRM response to uncoupling was assessed using ROIs in irradiated and adjacent regions. The cardiomyocyte TMRM recovery time constant (τ) was fit using a linear least squares TMRMt = TMRMi + ΔTMRM(1−e−(t−TD)/τ) where TMRMt is the TMRM fluorescence at any time point, t, TMRMi is the initial TMRM fluorescence, ΔTMRM is the difference in TMRM fluorescence between the first point after UV irradiation and the initial state, and TD is the time delay between UV irradiation and the recovery response.
Mitochondrial network connectivity was assessed with MitoDendra2 as the number of discrete mitochondria segments within an image frame over time. Dendra2 fluorescence was amplitude thresholded to remove background pixels and the Find Connected Regions plugin in ImageJ was used to count the number of regions containing at least 10 pixels.
Statistical Analyses
Differences in mitochondrial morphology were assessed by ANOVA with a Tukey’s HSD post hoc test and a p-value of 0.05. Differences in the relative distribution of Complex IV and Complex V between the peripheral and interior mitochondrial pools were determined using paired t-tests and a p-value of 0.05.
Supplementary Material
Upper: A 3D FIB-SEM image stack of a mouse left ventricle. Longitudinal images are shown and time represents sequential images moving deeper into the fiber. Data shown has been binned by 3 in x and y and by 10 in z to reduce file size. A 2.5 Gb raw image stack is available as Supplementary Dataset 1. Lower: 360° rotation of mitochondrial subnetworks within part of a single heart cell. Each color represents a different subnetwork of tens to hundreds of mitochondria. White mitochondria are not network connected.
Top) Timecourse loop of confocal microscopy images of the TMRM signal in a live, isolated cardiomyocyte before and after UV light induced release of MitoPhotoDNP in the cell interior to assess Longitudinal coupling. Bottom) Timecourse loop of confocal microscopy images of the TMRM signal in a live, isolated cardiomyocyte before and after UV light induced release of MitoPhotoDNP in the cell interior to assess Vertical coupling.
Upper) TMRM fluorescence in an isolated mouse left ventricular cardiomyocyte before and after UV activation of MitoPhotoDNP. Middle) TMRM fluorescence in an isolated mouse soleus fiber before and after UV activation of MitoPhotoDNP. Lower: TMRM fluorescence in an isolated mouse soleus fiber before and two minutes after UV photodamage (~8-fold higher 355 nm laser power per μm2 than used to activate MitoPhotoDNP).
Upper three panels: MitoDendra2 fluorescence in an isolated mouse soleus fiber before and two minutes after green to red photoconversion of MitoDendra2. Lower panel: MitoDendra2 fluorescence in an isolated mouse soleus fiber before and two minutes after simultaneous green to red photoconversion of MitoDendra2 and photoactivation of MitoPhotoDNP.
Upper: MitoDendra2 fluorescence in the paravascular region of an isolated mouse soleus fiber before and two minutes after simultaneous green to red photoconversion of MitoDendra2 and photoactivation of MitoPhotoDNP. Lower: MitoDendra2 fluorescence in the intrafibrillar region of an isolated mouse soleus fiber before and two minutes after simultaneous green to red photoconversion of MitoDendra2 and photoactivation of MitoPhotoDNP.
Acknowledgments
We thank R Swenson and H Wu of the NIH IPDC for MPD, ZX Yu of the NHLBI Pathology Core for immunofluorescence, PS Connelly of the NHLBI EM Core for heart blocks, and N Porat-Shliom (NCI) for MitoDendra2 mice. Supported by DIR NHLBI and the CCR, NCI. The authors declare no conflicts of interest.
Footnotes
AUTHOR CONTRIBUTIONS
R.S.B., S.S., B.G., and L.M.H. designed and L.M.H performed the FIB-SEM and electron tomographic experiments. R.S.B., A.F., and B.G. performed the structural segmentations and analyzed the data. R.S.B. and B.G. designed and B.G. performed the dual immunolabeling experiments. R.S.B., E.M., C.A.C., and B.G. designed and J.S., C.A.C., and B.G. performed the isolated cardiomyocyte experiments. R.S.B., C.A.C., and B.G. designed, performed and analyzed the isolated muscle fiber experiments.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amchenkova AA, Bakeeva LE, Chentsov YS, Skulachev VP, Zorov DB. Coupling membranes as energy-transmitting cables. I. Filamentous mitochondria in fibroblasts and mitochondrial clusters in cardiomyocytes. J Cell Biol. 1988;107:481–495. doi: 10.1083/jcb.107.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aon MA, Cortassa S, Marban E, O’Rourke B. Synchronized whole cell oscillations in mitochondrial metabolism triggered by a local release of reactive oxygen species in cardiac myocytes. J Biol Chem. 2003;278:44735–44744. doi: 10.1074/jbc.M302673200. [DOI] [PubMed] [Google Scholar]
- Bakeeva LE, Chentsov Yu S, Skulachev VP. Intermitochondrial contacts in myocardiocytes. J Mol Cell Cardiol. 1983;15:413–420. doi: 10.1016/0022-2828(83)90261-4. [DOI] [PubMed] [Google Scholar]
- Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkedal R, Laasmaa M, Vendelin M. The location of energetic compartments affects energetic communication in cardiomyocytes. Frontiers in physiology. 2014;5:376. doi: 10.3389/fphys.2014.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers S, Caldwell ST, Quin C, Prime TA, James AM, Cairns AG, Murphy MP, McCarron JG, Hartley RC. Selective uncoupling of individual mitochondria within a cell using a mitochondria-targeted photoactivated protonophore. J Am Chem Soc. 2012;134:758–761. doi: 10.1021/ja2077922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca HF, Engstrom GW. Calcium uptake by rat kidney mitochondria. Proc Natl Acad Sci U S A. 1961;47:1744–1750. doi: 10.1073/pnas.47.11.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen MR, Leyssens A, Crompton M. Transient mitochondrial depolarizations reflect focal sarcoplasmic reticular calcium release in single rat cardiomyocytes. J Cell Biol. 1998;142:975–988. doi: 10.1083/jcb.142.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvert M, Mazat JP, Barets AL. Intermitochondrial junctions in the heart of the frog, Rana esculenta. A thin-section and freeze-fracture study. Cell Tissue Res. 1985;241:129–137. doi: 10.1007/BF00214634. [DOI] [PubMed] [Google Scholar]
- Eisner V, Lenaers G, Hajnoczky G. Mitochondrial fusion is frequent in skeletal muscle and supports excitation-contraction coupling. J Cell Biol. 2014;205:179–195. doi: 10.1083/jcb.201312066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glancy B, Hartnell LM, Malide D, Yu ZX, Combs CA, Connelly PS, Subramaniam S, Balaban RS. Mitochondrial reticulum for cellular energy distribution in muscle. Nature. 2015;523:617–620. doi: 10.1038/nature14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glancy B, Hsu LY, Dao L, Bakalar M, French S, Chess DJ, Taylor JL, Picard M, Aponte A, Daniels MP, et al. In vivo microscopy reveals extensive embedding of capillaries within the sarcolemma of skeletal muscle fibers. Microcirculation. 2014;21:131–147. doi: 10.1111/micc.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Sun L, Ji S, Zhao T, Zhang W, Xu J, Zhang J, Wang Y, Wang X, Franzini-Armstrong C, et al. Kissing and nanotunneling mediate intermitochondrial communication in the heart. Proc Natl Acad Sci U S A. 2013;110:2846–2851. doi: 10.1073/pnas.1300741110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation research. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- Molina AJ, Wikstrom JD, Stiles L, Las G, Mohamed H, Elorza A, Walzer G, Twig G, Katz S, Corkey BE, et al. Mitochondrial networking protects beta-cells from nutrient-induced apoptosis. Diabetes. 2009;58:2303–2315. doi: 10.2337/db07-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monge C, Beraud N, Tepp K, Pelloux S, Chahboun S, Kaambre T, Kadaja L, Roosimaa M, Piirsoo A, Tourneur Y, et al. Comparative analysis of the bioenergetics of adult cardiomyocytes and nonbeating HL-1 cells: respiratory chain activities, glycolytic enzyme profiles, and metabolic fluxes. Can J Physiol Pharmacol. 2009;87:318–326. doi: 10.1139/Y09-018. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Marshall WF, Straight A, Murray A, Sedat JW, Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Molecular biology of the cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham AH, McCaffery JM, Chan DC. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis. 2012;50:833–843. doi: 10.1002/dvg.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, McManus MJ, Csordas G, Varnai P, Dorn GW, 2nd, Williams D, Hajnoczky G, Wallace DC. Trans-mitochondrial coordination of cristae at regulated membrane junctions. Nature communications. 2015;6:6259. doi: 10.1038/ncomms7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romashko DN, Marban E, O’Rourke B. Subcellular metabolic transients and mitochondrial redox waves in heart cells. Proc Natl Acad Sci U S A. 1998;95:1618–1623. doi: 10.1073/pnas.95.4.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skulachev VP, Bakeeva LE, Chernyak BV, Domnina LV, Minin AA, Pletjushkina OY, Saprunova VB, Skulachev IV, Tsyplenkova VG, Vasiliev JM, et al. Thread-grain transition of mitochondrial reticulum as a step of mitoptosis and apoptosis. Mol Cell Biochem. 2004;256–257:341–358. doi: 10.1023/b:mcbi.0000009880.94044.49. [DOI] [PubMed] [Google Scholar]
- Sun J, Kohr MJ, Nguyen T, Aponte AM, Connelly PS, Esfahani SG, Gucek M, Daniels MP, Steenbergen C, Murphy E. Disruption of caveolae blocks ischemic preconditioning-mediated S-nitrosylation of mitochondrial proteins. Antioxidants & redox signaling. 2012;16:45–56. doi: 10.1089/ars.2010.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Murphy E. Calcium-sensing receptor: a sensor and mediator of ischemic preconditioning in the heart. Am J Physiol Heart Circ Physiol. 2010;299:H1309–1317. doi: 10.1152/ajpheart.00373.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabadkai G, Simoni AM, Chami M, Wieckowski MR, Youle RJ, Rizzuto R. Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Molecular cell. 2004;16:59–68. doi: 10.1016/j.molcel.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Vendelin M, Birkedal R. Anisotropic diffusion of fluorescently labeled ATP in rat cardiomyocytes determined by raster image correlation spectroscopy. Am J Physiol Cell Physiol. 2008;295:C1302–1315. doi: 10.1152/ajpcell.00313.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Fang H, Groom L, Cheng A, Zhang W, Liu J, Wang X, Li K, Han P, Zheng M, et al. Superoxide flashes in single mitochondria. Cell. 2008;134:279–290. doi: 10.1016/j.cell.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Ackerley C, Maj MC, Addis JB, Levandovskiy V, Lee J, Mackay N, Cameron JM, Robinson BH. Disruption of a mitochondrial RNA-binding protein gene results in decreased cytochrome b expression and a marked reduction in ubiquinol-cytochrome c reductase activity in mouse heart mitochondria. Biochem J. 2008;416:15–26. doi: 10.1042/BJ20080847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Upper: A 3D FIB-SEM image stack of a mouse left ventricle. Longitudinal images are shown and time represents sequential images moving deeper into the fiber. Data shown has been binned by 3 in x and y and by 10 in z to reduce file size. A 2.5 Gb raw image stack is available as Supplementary Dataset 1. Lower: 360° rotation of mitochondrial subnetworks within part of a single heart cell. Each color represents a different subnetwork of tens to hundreds of mitochondria. White mitochondria are not network connected.
Top) Timecourse loop of confocal microscopy images of the TMRM signal in a live, isolated cardiomyocyte before and after UV light induced release of MitoPhotoDNP in the cell interior to assess Longitudinal coupling. Bottom) Timecourse loop of confocal microscopy images of the TMRM signal in a live, isolated cardiomyocyte before and after UV light induced release of MitoPhotoDNP in the cell interior to assess Vertical coupling.
Upper) TMRM fluorescence in an isolated mouse left ventricular cardiomyocyte before and after UV activation of MitoPhotoDNP. Middle) TMRM fluorescence in an isolated mouse soleus fiber before and after UV activation of MitoPhotoDNP. Lower: TMRM fluorescence in an isolated mouse soleus fiber before and two minutes after UV photodamage (~8-fold higher 355 nm laser power per μm2 than used to activate MitoPhotoDNP).
Upper three panels: MitoDendra2 fluorescence in an isolated mouse soleus fiber before and two minutes after green to red photoconversion of MitoDendra2. Lower panel: MitoDendra2 fluorescence in an isolated mouse soleus fiber before and two minutes after simultaneous green to red photoconversion of MitoDendra2 and photoactivation of MitoPhotoDNP.
Upper: MitoDendra2 fluorescence in the paravascular region of an isolated mouse soleus fiber before and two minutes after simultaneous green to red photoconversion of MitoDendra2 and photoactivation of MitoPhotoDNP. Lower: MitoDendra2 fluorescence in the intrafibrillar region of an isolated mouse soleus fiber before and two minutes after simultaneous green to red photoconversion of MitoDendra2 and photoactivation of MitoPhotoDNP.