Abstract
Searching of new enantiomerically pure chiral derivatives of xanthones (CDXs) with potential pharmacological properties, particularly those with anti-inflammatory activity, has remained an area of interest of our group. Herein, we describe in silico studies and in vitro inhibitory assays of cyclooxygenases (COX-1 and COX-2) for different enantiomeric pairs of CDXs. The evaluation of the inhibitory activities was performed by using the COX Inhibitor Screening Assay Kit. Docking simulations between the small molecules (CDXs; known ligands and decoys) and the enzyme targets were undertaken with AutoDock Vina embedded in PyRx—Virtual Screening Tool software. All the CDXs evaluated exhibited COX-1 and COX-2 inhibition potential as predicted. Considering that the (S)-(−)-enantiomer of the nonsteroidal anti-inflammatory drug ketoprofen preferentially binds to albumin, resulting in lower free plasma concentration than (R)-(+)-enantiomer, protein binding affinity for CDXs was also evaluated by spectrofluorimetry as well as in in silico. For some CDXs enantioselectivity was observed.
Keywords: chiral derivatives of xanthones, cyclooxygenase, albumin, enantioselectivity, docking
1. Introduction
A key role of chirality in drug design and development is associated with significant effects on the behavior of this kind of compounds in vivo, with enantiomers being able to interact differently with proteins, and other chiral biomolecules [1,2]. These events can be translated into implications in pharmacokinetics (PK) [3], pharmacodynamics (PD) [4] as well as in toxicity [5]. Frequently only one of the two enantiomers exerts the desired effect while the other might be inactive, less potent or even toxic [6,7]. Consequently, enantioselectivity can be considered an essential issue to take into consideration when studying chiral compounds.
There are several classes of compounds illustrating the importance of chirality on both PK and PD events such as non-steroidal anti-inflammatory drugs (NSAIDs) [8]. Considering ketoprofen, for example, the (S)-(+)-enantiomer preferentially binds to human serum albumin (HSA), resulting in lower free plasma concentration than (R)-(−)-enantiomer [9]. Moreover, the (S)-(+)-ketoprofen is several times more potent than the racemate and etodolac, in which the (S)-enantiomer has cyclooxygenase (COX) inhibitory activity whereas its antipode does not [10]. Furthermore, some NSAIDs have also demonstrated enantioselectivity in antitumor activity, as for example (R)-enantiomer of etodolac binds retinoid X receptor and induces tumor-selective apoptosis in malignant cells [11].
The binding of drugs to plasma proteins is an important parameter since it has implications on drug action in vivo by affecting free concentration in serum [12,13], which has direct implications in pharmacological effects and metabolizing processing [14]. HSA, the most abundant protein in plasma (Mr 66 kDa, concentration 0.53 to 0.75 mM), interacts reversibly with a broad spectrum of drugs, especially neutral and negatively charged hydrophobic compounds [15,16]. According to current point of views in the drug discovery pipeline, the binding of new compounds with HSA at an early stage is of crucial importance, insofar as it affects not only distribution and elimination but also duration and intensity of the pharmacological action of drugs [12,13,17]. Moreover, enantioselectivity for HSA binding has been reported for several drugs such as verapamil and ibuprofen [18], being also predicted by docking studies [19,20]. One group of compounds described with antitumor [21,22,23], anti-inflammatory [24,25,26], among others activities [27,28], concerns xanthone derivatives. Indeed, the xanthone scaffold can be considered a privileged structure [28]. This group of oxygenated heterocyclic compounds can be isolated from natural sources [29,30], including products from marine origin [31], or obtained by synthesis [32,33]. Among them, synthetic chiral derivatives of xanthones (CDXs) have also revealed interesting biological activities [34,35,36,37] and, in some cases, the activity demonstrated to be depending on the stereochemistry of the respective molecules [36,38,39].
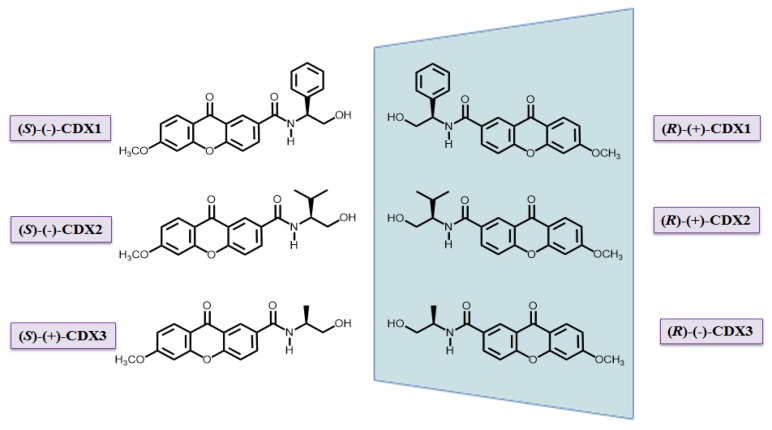
Search of new bioactive CDXs and investigation of enantioselectivity on their biological activity have remained an area of interest of our group [40,41]. Recently, we described the synthesis of new CDXs in enantiomerically pure form, and some of them exhibited growth inhibitory effects on different tumor cell lines as well as enantioselectivity [40]. In this context, herein we described the evaluation of COX inhibition activity and protein binding affinity for three enantiomeric pairs of CDXs (Figure 1).
Figure 1.
Structures of the enantiomers of CDXs 1–3.
Molecular modeling studies by docking technique [42,43,44,45] were also carried out in order to understand the interactions of the CDXs with the active site of the referred biological targets and the structural features associated with the chiral recognition.
2. Results and Discussion
2.1. Cyclooxygenase Inhibition Studies
The effect of CDXs previously obtained in our group [40,41] as inhibitors of membrane located enzymes that might be involved in inflammatory processes, namely COX-1 and COX-2, was evaluated. Three enantiomeric pairs of CDXs were chosen to study the inhibitory effect of both enantiomers of each pair face to biological targets in order to evaluate potency and enantioselectivity. The effect on COXs activity was studied by spectrofluorimetry using a commercial kit measuring the peroxidation activity of COXs. The kit includes isoenzyme-specific inhibitors for distinguishing COX-1 from COX-2 activities.
The results, given as percentage (%) of inhibition and expressed as mean ± standard deviation of two independent experiments, are summarized in Table 1. The overall results indicate that all the CDXs evaluated exhibited COX-1 and COX-2 inhibition potential, although about 20 times less active than indomethacin (positive control). Paired t-test was also performed to compare inhibitory effects within each enantiomeric pair to verify the occurrence of enzyme-type and enantioselectivity. Among all the compounds, (R)-(+)-CDX2 was the only compound presenting statistically significant enzyme-type selectivity (p < 0.05). This enantiomer was more active at inhibiting COX-2 than COX-1. All pairs demonstrated enantioselectivity for COX-1 and |t|calculated values were 3.613, 7.249 and 2.891 for CDX1, CDX2 and CDX3 pairs, respectively (ttab(p = 0.05; d.f. = 10) = 2.228). (S)-(−)-CDX1, (S)-(−)-CDX2 and (S)-(+)-CDX3 were more active than their antipode.
Table 1.
Inhibitory effects of CDXs on COX-1 and COX-2.
| CDX | COX-1 | COX-2 |
|---|---|---|
| (S)-(−)-CDX1 | 87.6 ± 2.1 | 80.1 ± 12.8 |
| (R)-(+)-CDX1 | 79.6 ± 5.0 | 84.7 ± 5.7 |
| (S)-(−)-CDX2 | 82.9 ± 5.2 | 85.7 ± 4.5 |
| (R)-(+)-CDX2 | 66.8 ± 1.6 | 73.2 ± 0.4 |
| (S)-(+)-CDX3 | 91.7 ± 10.7 | 93.4 ± 11.4 |
| (R)-(−)-CDX3 | 75.2 ± 9.0 | 75.1 ± 7.2 |
| Indomethacin | 83.2 ± 6.4 | 80.7 ± 9.5 |
Values correspond to percentage of enzyme inhibition (mean ± standard deviation). Each compound was analyzed in triplicate in two independent days. The concentration of CDXs was 20 µmol/L. Indomethacin 1 µmol/L was used as positive control.
Concerning enantioselectivity for COX-2, the results obtained for the pairs of CDX2 and CDX3 should be highlighted. For instance, the % of COX-2 inhibition for (S)-(+)-CDX3 was 93.4 ± 11.4, however the inhibitory effect of (R)-(−)-CDX3 was statistically significantly lower. |t|calculated value was 2.891 for COX-2 (ttab(p = 0.05; d.f. = 10) = 2.228). Similarly, (S)-(−)-CDX2 was more active than (R)-(+)-CDX2 at inhibiting COX-2 (|t|calculated = 6.777; ttab(p = 0.05; d.f. = 10) = 2.228). Accordingly, for these two pairs, weak enantioselectivity was observed. It is important to stress that even though enantioselectivity was observed, its extent was not as evident as that obtained for ketoprofen for instance [10].
Molecular docking studies were also performed in order to predict the potential anti-inflammatory activity and to postulate a hypothetical binding model of the tested compounds. The binding affinity between the target and the small molecule was evaluated by the binding free energy approximation (ΔGb, kcal/mol) using AutoDock Vina. The best scored conformation of each compound predicted by AutoDock Vina was selected and further evaluated. The docking score was used to predict the strength of the non-covalent interactions between two molecules after they have been docked (also referred to as binding energy). The docking score is a mathematical approximation of the binding free energy between the ligand and its target.
Diclofenac, indomethacin, naproxen, and piroxicam [46] were used as positive controls and showed negative binding energy values (Table 2). Ligands obtained from the database established more stable complexes with COX-1, with an average binding free energy of −7.8 kcal/mol. Moreover, the docking scores predicted for decoys into the COX-1 was surprisingly low (−7.3 kcal/mol). From the tested compounds, only (S)- and (R)-CDX3 and (S)-CDX2 presented docking scores more negative than the known COX-1 inhibitors indomethacin and piroxicam. However, as only a very small difference was observed between known ligands and decoys scores, this model cannot be used to predict COX-1 inhibition. Hence, more detailed analysis of docking poses and binding mechanisms was performed for the other studied COX isoform: COX-2. Diclofenac, indomethacin, celecoxib, and valdecoxib [46,47] were used as positive controls for COX-2 inhibition. The average binding energy predicted for decoys and known ligands into the COX-2 was −7.6 and −9.3 kcal/mol, respectively (Table 2). Both diclofenac and indomethacin presented −7.9 kcal/mol, whereas celecoxib and valdecoxib exhibited −11.5 and −9.5 kcal/mol, respectively. Among the tested CDXs, (R)- and (S)-CDX1 exhibited the highest binding affinities and, therefore, lower binding free energies than negative controls.
Table 2.
Docking scores of corresponding test compounds at COX-1 and COX-2 targets.
| Compounds | Docking Score (kcal/mol) | ||
|---|---|---|---|
| COX-1 | COX-2 | ||
| Known ligands | Diclofenac | −6.1 | −7.9 |
| Indomethacin | −5.1 | −7.9 | |
| Naproxen | −7.8 | ||
| Piroxicam | −5.2 | ||
| Celecoxib | −11.5 | ||
| Valdecoxib | −9.5 | ||
| Ligands from database | −7.8 | −9.3 | |
| Decoys from database | −7.3 | −7.6 | |
| (R)-(+)-CDX1 | −4.2 | −7.8 | |
| (S)-(−)-CDX1 | −4.5 | −8.0 | |
| (R)-(+)-CDX2 | −3.4 | −6.5 | |
| (S)-(−)-CDX2 | −5.4 | −7.0 | |
| (R)-(−)-CDX3 | −5.3 | −6.9 | |
| (S)-(+)-CDX3 | −5.6 | −7.5 | |
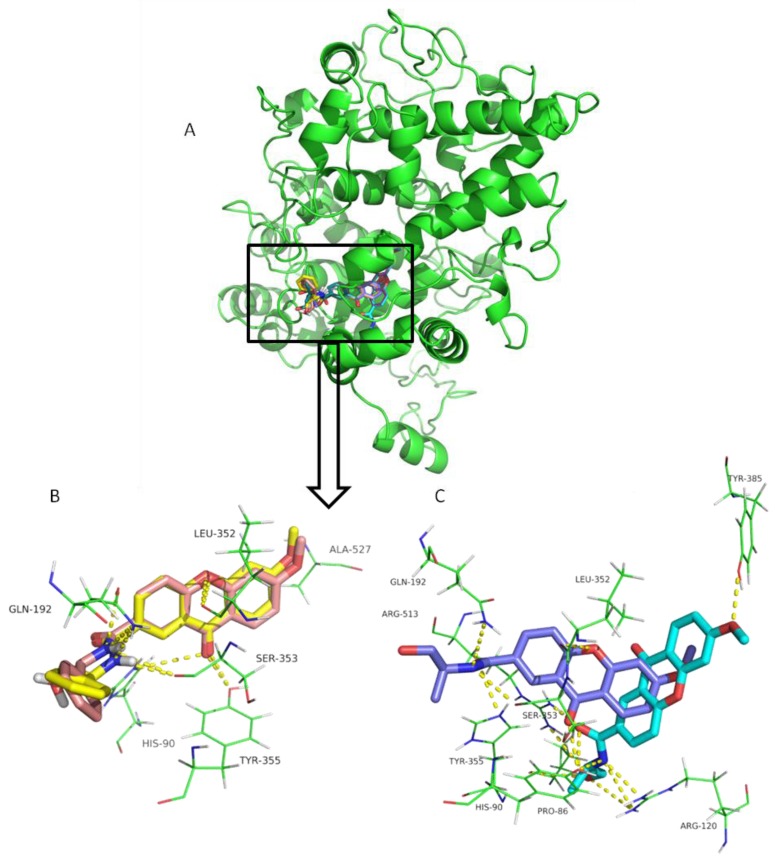
Both positive controls and CDXs could dock into the active site of COX-2 successfully (Figure 2A). The binding mode of celecoxib, valdecoxib, indomethacin, and diclofenac (Supplementary Data, Figure S1) are in accordance to the previously reported binding modes. This is of particular importance considering docking accuracy. (S)-CDX1 presented the highest binding energy (−8.0 kcal/mol) into the COX-2 model, similar to the known ligands value. Docking energies of −7.8, −7.5, and −7.0 kcal/mol were obtained to the (R)-CDX1, (S)-CDX3, and (S)-CDX2, respectively. CDX3 enantiomers presented very different poses within the binding site of COX-2, whereas CDX1 enantiomers showed a minor difference (Figure 2).
Figure 2.
(A) COX-2 ribbon representation. (B) Comparison of docking poses of CDX1 enantiomers in COX-2 binding site. (R) and (S) enantiomers are represented in pink and yellow sticks, respectively. (C) Comparison of docking poses of CDX3 enantiomers in COX-2 binding site. (R) and (S) enantiomers are represented in light and dark blue sticks, respectively. Hydrogen interactions are represented as yellow dashes and the residues evolved are represented as green lines and labeled.
CDX1 enantiomers interact through hydrogen bonds with His90, Leu352, Ser353, Tyr355, and Ala527, also involved in the binding of known anti-inflammatory compounds to COX-2 [48,49,50]. CDX1 enantiomers bind similarly to COX-2 binding pocket, with the xanthone scaffold aligned approximately in the same special position, with a slightly different orientation of the aromatic ring and OH group of the chiral moiety (Figure 2B). (R)-CDX1 shows an additional hydrogen interaction between OH and Gln-192, similarly to celecoxib and valdecoxib. On the other hand, CDX3 enantiomers bind in very different poses in COX-2 binding pocket, almost perpendicular to each other (Figure 2C). In fact, (S)-CDX3 (Figure 2C, dark blue sticks) binds in a pose similar to CDX1, establishing hydrogen interactions with residues Gln192, His90, Ser353, Leu352, and Tyr355, documented as being important for COX-2 inhibition [50,51,52]. Concerning (R)-CDX3, the aromatic backbone projects deep COX active site from the hydrophobic channel, with the C3 chain establishing hydrogen interactions with Arg513, Pro86, and Arg120, and the C6-methoxy group establishing hydrogen interactions with Tyr385 (Figure 2C, light blue sticks), which is important in the catalysis or inactivation of the enzyme [53]. (S)-CDX-2 establishes hydrogen interactions with Gln192, Leu352, Ser353, His90, Tyr355, and Arg513; and (R)-CDX-2 establishes hydrogen interactions with Tyr385, Tyr355, Arg513, Pro86, and Arg-120 (not shown).
2.2. Human Serum Albumin Affinity Studies
HSA-CDX binding parameters are compiled in Table 3. The binding process has reached a completion state as indicated by the Ymax that reached about 100%. For all the compounds, HSA binding occurred spontaneously (ΔG values < 0) in a single binding site (n = 1). All compounds presented dissociation constants (Kd) below 100 µM, which accounts for high affinity binding to HSA [54]. Paired t-test was performed to compare Kd obtained for (S)- and (R)-enantiomers of each CDX pair. For all the pairs, there was a statistically significant difference between Kd values. Hence, weak enantioselectivity concerning albumin binding was observed. Among all pairs, the highest difference in binding affinity was observed for CDX1 (ca. 2.6 fold) as demonstrated by the |t|calculated value which was 10.103 (ttab(p = 0.05; d.f. = 4) = 4.303). The (S)-enantiomer presented higher binding affinity compared to the (R)-enantiomer. For CDX3 and CDX2, the difference in HSA binding affinity obtained for (S)- and (R)-enantiomers was less pronounced compared to CDX1. For CDX2, the (S)-enantiomer presented slightly higher affinity than the (R) one; |t|calculated value was 5.484 (ttab(p = 0.05; d.f. = 4) = 2.776). On the contrary, (R)-(−)-CDX3 has shown slightly higher affinity than (S)-(+)-CDX3. In this case, |t|calculated value was 3.713 (ttab(p = 0.05; d.f. = 4) = 2.776).
Table 3.
HSA binding parameters obtained for the CDXs and predicted docking scores between HSA and CDXs.
| Compound | Kd | Ymax | ∆G Binding | Docking Score (kcal/mol) | |
|---|---|---|---|---|---|
| Known ligands | Azaprozone | −5.9 | |||
| Diazepam | −7.1 | ||||
| Fusidic acid | −5.8 | ||||
| (S)-Ibuprofen | −7.3 | ||||
| Iophenoxid acid | −4.4 | ||||
| Naproxen | −7.9 | ||||
| Warfarin | −8.5 | ||||
| (R)-(+)-CDX1 | 61.8 ± 6.5 | 109.6 ± 1.6 | −2.4 ± 0.2 | −7.3 | |
| (S)-(−)-CDX1 | 23.6 ± 0.8 | 105.3 ± 0.4 | −1.9 ± 0.1 | −7.0 | |
| (R)-(+)-CDX2 | 29.2 ± 0.9 | 108.2 ± 0.2 | −2.0 ± 0.1 | −7.2 | |
| (S)-(−)-CDX2 | 24.7 ± 1.1 | 107.4 ± 5.4 | −1.9 ± 0.1 | −7.2 | |
| (R)-(−)-CDX3 | 26.4 ± 1.2 | 113.2 ± 1.4 | −1.9 ± 0.1 | −7.2 | |
| (S)-(+)-CDX3 | 31.4 ± 2.0 | 116.2 ± 0.6 | −2.0 ± 0.2 | −7.0 | |
Values correspond to the mean ± standard deviation of triplicate runs; Kd corresponds to the dissociation constant of CDX-HSA in μM; Ymax corresponds to the maximum percentage of HSA fluorescence quenching; ∆G for binding expressed in kcal/mol.
Regarding computational studies, drugs that are described as having high affinity to HSA lead to docking scores between −4.4 kcal/mol (Iophenoxid acid) and −8.5 kcal/mol (warfarin) (Table 3). CDXs presented scores from −7.0 to −7.3 kcal/mol, and therefore it is hypothesized that they will have high affinity to albumin target. There is an offset between the ∆G binding and the docking scores. This relates to the ability of the docking algorithm to predict the strength of ligand binding to the protein target, and therefore, other scoring functions will be used in the future to increase the accuracy.
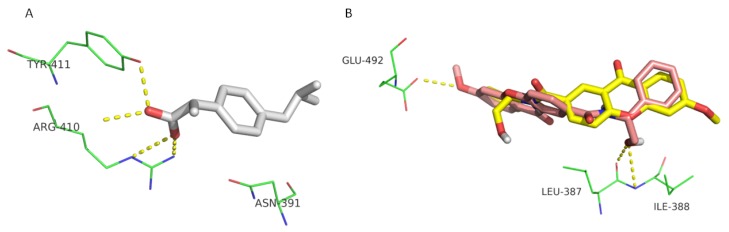
(S)-Ibuprofen binds albumin through hydrogen interactions with Arg-140, Tyr-411, and Lys-414 (Figure 3A), residues described as being involved in binding of substrates to HSA [55]. The present study indicates that CDXs fit within the hydrophobic pocket of subdomain IIIA, presenting low negative docking scores. This groove was selected for the docking studies as it was described as being the binding pocket for (S)-ibuprofen and most ligands [56]. CDXs bind in a similar position in the binding groove, with the central xanthone ring aligned with ibuprofen ring. The binding of CDX enantiomers in HSA subdomain IIIA present differences concerning the number of hydrogen interactions. For example, the complex (R)-CDX1-HSA is stabilized by three hydrogen-bond interactions with residues Leu-430, Ser-489, and Asn-391, already described as being involved in the binding of drugs to HSA [57,58,59], whereas (S)-CDX1 lacks those interactions (Figure 3B). Therefore, there is concordance between in silico and in vitro studies, as enantiosselectivity can be found on the binding of CDX1 to HSA.
Figure 3.
Interactions of crystallographic (S)-ibuprofen (yellow sticks) (A) and docked CDX1 enantiomers (B) with HSA. (R)- and (S)-enantiomers are represented in pink and yellow sticks, respectively. Hydrogen interactions are represented as yellow dashes and the residues evolved are represented as green lines and labeled.
3. Materials and Methods
3.1. Compounds
CDXs (Figure 1) were synthesized in enantiomerically pure form [40,41]. Briefly, a carboxyxanthone derivative, with the structure based on some bioactive xanthones from marine origin [31] was coupled with both enantiomers of commercially available chiral building blocks using O-(benzotriazol-1-yl)-N-N-N’-N’-tetramethyluronium tetrafluoroborate as coupling reagent and a catalytic amount of triethylamine in tetrahydrofuran, at room temperature. Liquid chromatography using different types of chiral stationary phases was used to determine the enantiomeric purity of the synthesized compounds [60,61], achieving enantiomeric excess values higher than 99%.
3.2. In vitro Cyclooxygenase Inhibition Studies
The evaluation of the inhibition of COX-1 and COX-2 by CDXs was conducted using the commercially available COX (ovine/human) Inhibitor Screening Assay Kit (Cayman Chemical, Michigan, MI, USA). Briefly, the assay implies an enzymatic immunoassay based on the competition between prostaglandins (PGs) and a PG-acetylcholinesterase (AChE) conjugate (PG tracer), which is then evaluated by the addition of Ellman’s reagent. All the solutions required for the experiment were prepared according to the manufacturer’s instructions. Each compound was analysed in two independent days, in triplicate. CDX working solutions were prepared in DMSO to a final concentration of 20 µM. Indomethacin (1 µM) was used as positive control. Absorbance measurements at 412 nm were performed in a Synergy HT microplate reader (Bio-Tek Instruments, Winooski, VT, USA) operated with Gen5 software (Bio-Tek Instruments).
3.3. Interaction with Human Serum Albumin by Fluorescence Quenching
Evaluation of the binding of CDXs to HSA was based on the quenching of HSA intrinsic fluorescence. Phosphate buffer consisting of 7.5 mM Na2HPO4, 1.5 mM KH2PO4 and 140 mM NaCl (pH 7.4) was used in the preparation of CDX solutions. Solutions of HSA fraction V (Sigma-Aldrich, St. Louis, MO, USA) were prepared in water. Briefly, 500 µL of HSA (fixed final concentration 2 µM), increasing volumes of drug solution (n = 13, 0–200 µM) and phosphate buffer solution (pH 7.4) were mixed to a final volume of 1500 µL. The corresponding blank solutions were identically prepared and analysed in the absence of the drug. Fluorescence emission spectra were recorded in the range of 300–450 nm upon excitation at 295 nm. Excitation spectra were also recorded between 220 and 310 nm with emission set at 330 nm. For individual measurements, excitation and emission wavelength were set at 295 nm and 330 nm, respectively. For each measurement, fluorescence emission was automatically acquired during 180 s (5 nm bandwidth). For all compounds, UV-vis absorption spectra (200–500 nm) were recorded. These measurements were used to correct the fluorescence intensity values due to inner filter effects at the excitation wavelength [62]. All experiments were performed at room temperature (25 ± 1 °C). Fluorescence and absorbance measurement were performed in a LS-50B spectrometer (Perkin Elmer, Waltham, MA, USA) and a V-660 spectrophotometer (Jasco, Easton, MD, USA) respectively.
Assessment of HSA-CDX Binding Parameters
HSA-CDX binding parameters were calculated using the Origin 8.5.1 software (8.5.1, Northampton, MA, USA). The fitting of the experimental values was made according to the Langmuir binding equation (Equation (1)) to calculate HSA binding parameters:
| (1) |
where [HSA], [CDX] and [HSA-CDX] are given in μM and Kd corresponds to the dissociation constant of HSA-CDX complexes.
In terms of fluorescence quenching mediated by the compound, the previous Langmuir isotherm can be rewritten as follows (Equation (2)):
| (2) |
where ymax corresponds to the highest percentage of quenching induced by a given compound and n accounts for the number of biding sites of the enzyme to the CDX. Finally, free Gibbs energy (ΔG) was also determined for all the interactions.
3.4. Computational Studies
3.4.1. Preparation of CDXs, Controls, Decoys, and Macromolecules
The six CDXs and several known inhibitors (Table 1 and Table 2) were drawn and minimized using Universal Force Field (UFF) of Rappé and coworkers [63] which consists of a molecular mechanics (MM) force field that includes parameterization for the entire periodic table. The calculation is finished when the gradient between any two successive steps in the geometry search is less than 10−1 kcal/mol/Å or the maximum steps are reached, whichever comes first. The line search used is the Broyden-Fletcher-Golfarb-Shanno search which uses an approximate Hessian matrix to guide the search. Charges were calculated using gasteiger method [64] available in Chimera [65].
COX-1 and COX-2 decoy and ligand sets were downloaded from A Directory of Useful Decoys (DUD) [66], a database from the University of California, San Francisco. A hundred decoys and a hundred ligands were used for docking simulations with COX-2. Fifty decoys and the twenty three available ligands were used for docking simulations with COX-1. These molecules were used with no further manipulation.
The X-ray crystal structures of COX-1 (PDB code: 3n8x) and COX-2 (PDB code: 1cx2) were downloaded from the Protein Data Bank of Brookhaven [67]. An additional docking study was performed using HSA as target (PDB code: 2bxg), and azaprozone, diazepam, fusidic acid, ibuprofen, iophenoxid acid, naproxen, and warfarin as positive controls.
3.4.2. Docking
Docking simulations between the CDXs, the anti-inflammatory targets, and HSA were undertaken in AutoDock Vina (Molecular Graphics Lab, La Jolla, CA, USA) [68]. AutoDock Vina considered the target conformation (biomacromolecule) as a rigid unit while the ligands were allowed to be flexible and adaptable to the target. Vina searched for the lowest binding affinity conformations and returned nine different conformations for each CDX. The lowest binding energy docking poses of each compound were chosen. AutoDock Vina was run using an exhaustiveness of 8 and a grid box with the dimensions of X: 21.6, Y: 22.3, Z: 20.2 for COX-1; X: 22.9, Y: 23.7, Z: 21.7 for COX-2; and X: 18.0, Y: 19.0, Z: 19.0 for HSA. PyMol v1.3 (Schrödinger, New York, NY, USA) [69] and Chimera (UCSF, San Francisco, CA, USA) [65] were used for visual inspection of results and graphical representations.
4. Conclusions
All CDXs evaluated exhibited COX-1 and COX-2 inhibition in in vitro assays. Generally, the inhibitory effects were very similar for both COXs, with exception of CDX2 enantiomeric pair. Among all the compounds, (R)-(+)-CDX2 was more active at inhibiting COX-2 than COX-1. Interestingly, all pairs demonstrated enantioselectivity for COX-1. Concerning COX-2, the percentage of inhibition for (S)-(−)-CDX2 and (S)-(+)-CDX3 was higher. Accordingly, for CDX2 and CDX3 pairs enantioselectivity for COX-2 was also observed.
Regarding in in silico studies, no significant difference was found between known ligands and decoys docking scores on COX-1, therefore, no reliable conclusions can be taken from test ligands binding affinity to COX-1. However, regarding docking studies with COX-2, CDX-3 enantiomers as they presented very different poses within the binding site of the enzyme, it is reasonable to predict enantioselectivity been in accordance to in vitro studies.
Additionally, all CDXs demonstrated to bind with high affinity to HSA in in vitro assays and weak enantioselectivity was observed for all enantiomeric pairs. This effect was particularly evident for CDX-1 pair. The in silico studies also confirmed that CDXs bind to HSA, as they have docking scores similar to positive controls such as ibuprofen and diazepam; and CDX1 enantiomeric pair exhibited enantioselectivity. Regarding HSA affinity studies, agreement between in silico and in vitro data (activity and enantioselectivity) was achieved. Moreover, useful information about the mechanism of molecular recognition for both COX and HSA was obtained. Even though statistical analysis demonstrated only a trend for enantioselectivity, we envision that the introduction of molecular modifications supported by docking studies might enhance those differences.
Taking into account the results of this study, it can be concluded that new knowledge was added in the field of CDXs as potential anti-inflammatory agents, paving a very interesting way to understand the enantioselectivity of this family of compounds facing to COX and HSA.
Acknowledgments
This work was partially supported through national funds provided by FCT/MCTES—Foundation for Science and Technology from the Minister of Science, Technology and Higher Education (PIDDAC) and European Regional Development Fund (ERDF) through the COMPETE—Programa Operacional Factores de Competitividade (POFC) programme, under the Strategic Funding UID/Multi/04423/2013, the project PTDC/MAR-BIO/4694/2014 (reference POCI-01-0145-FEDER-016790; Project 3599—Promover a Produção Científica e Desenvolvimento Tecnológico e a Constituição de Redes Temáticas (3599-PPCDT)) in the framework of the programme PT2020 as well as by the project INNOVMAR - Innovation and Sustainability in the Management and Exploitation of Marine Resources (reference NORTE-01-0145-FEDER-000035, within Research Line NOVELMAR), supported by North Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF) and COXANT-CESPU-2016.
Supplementary Materials
The following are available online at http://www.mdpi.com/1424-8247/10/2/50/s1.
Author Contributions
Carla Fernandes contributed in writing of the manuscript and data interpretation and analysis. Maria Elizabeth Tiritan participated in discussion of the results. Carlos Carneiro performed docking studies with COXs. Andreia Palmeira performed docking studies with HSA, contributed in the data analysis carried out in both in silico studies, and contributed in writing of the manuscript. Honorina Cidade participated in vitro COXs inhibition experiments. Inês I. Ramos performed both in vitro experiments and contributed in writing of the manuscript. Paula C.A.G. Pinto contributed in the data analysis carried out in vitro COXs inhibition studies. M. Lúcia M.F.S. Saraiva and Carlos Afonso gave scientific advice for the development of in vitro HSA affinity studies. Salette Reis coordinated and supervised the in vitro experiments and reviewed the data generated. Corresponding author Madalena M.M. Pinto conceived and designed the experiments, supervised the project, discussed results, and led the preparation of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest
References
- 1.Tiritan M.E., Ribeiro A.R., Fernandes C., Pinto M. Kirk-Othmer Encyclopedia of Chemical Technology. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2016. Chiral pharmaceuticals; pp. 1–28. [Google Scholar]
- 2.Blaser H.U. Chirality and its implications for the pharmaceutical industry. Rend. Lincei. 2013;24:213–216. doi: 10.1007/s12210-012-0220-2. [DOI] [Google Scholar]
- 3.Brocks D.R. Drug disposition in three dimensions: An update on stereoselectivity in pharmacokinetics. Biopharm. Drug Dispos. 2006;27:387–406. doi: 10.1002/bdd.517. [DOI] [PubMed] [Google Scholar]
- 4.Mannschreck A., Kiesswetter R., von Angerer E. Unequal activities of enantiomers via biological receptors: Examples of chiral drug, pesticide, and fragrance molecules. J. Chem. Educ. 2007;84:2012–2017. doi: 10.1021/ed084p2012. [DOI] [Google Scholar]
- 5.Smith S.W. Chiral toxicology: It's the same thing only different. Toxicol. Sci. 2009;110:4–30. doi: 10.1093/toxsci/kfp097. [DOI] [PubMed] [Google Scholar]
- 6.Sekhon B.S. Exploiting the power of stereochemistry in drugs: An overview of racemic and enantiopure drugs. J. Mod. Med. Chem. 2013;1:10–36. doi: 10.12970/2308-8044.2013.01.01.2. [DOI] [Google Scholar]
- 7.Triggle D.J. Stereoselectivity of drug action. Drug Discov. Today. 1997;2:138–147. doi: 10.1016/S1359-6446(97)01010-6. [DOI] [Google Scholar]
- 8.Kasprzyk-Hordern B. Pharmacologically active compounds in the environment and their chirality. Chem. Soc. Rev. 2010;39:4466–4503. doi: 10.1039/c000408c. [DOI] [PubMed] [Google Scholar]
- 9.Moreno J.J., Calvo L., Fernandez F., Carganico G., Bastida E., Bujons J., Messeguer A. Biological activity of ketoprofen and its optical isomers. Eur. J. Pharmacol. 1990;183:2263–2264. doi: 10.1016/0014-2999(90)93806-2. [DOI] [Google Scholar]
- 10.Sánchez T., Moreno J.J. ketoprofen S(+)-enantiomer inhibits prostaglandin production and cell growth in 3T6 fibroblast cultures. Eur. J. Pharmacol. 1999;370:63–67. doi: 10.1016/S0014-2999(99)00108-9. [DOI] [PubMed] [Google Scholar]
- 11.Kolluri S.K., Corr M., James S.Y., Bernasconi M., Lu D., Liu W., Cottam H.B., Leoni L.M., Carson D.A., Zhang X.K. The R-enantiomer of the nonsteroidal antiinflammatory drug etodolac binds retinoid x receptor and induces tumor-selective apoptosis. Proc. Natl. Acad. Sci. USA. 2005;102:2525–2530. doi: 10.1073/pnas.0409721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trainor G.L. The importance of plasma protein binding in drug discovery. Expert Opin. Drug Discov. 2007;2:51–64. doi: 10.1517/17460441.2.1.51. [DOI] [PubMed] [Google Scholar]
- 13.Kerns E.H., Di L. Drug-Like Properties: Concepts, Structure Design and Methods. Elsevier Inc.; Burlington, MA, USA: 2008. [Google Scholar]
- 14.Fasano M., Curry S., Terreno E., Galliano M., Fanali G., Narciso P., Notari S., Ascenzi P. The extraordinary ligand binding properties of human serum albumin. IUBMB Life. 2005;57:787–796. doi: 10.1080/15216540500404093. [DOI] [PubMed] [Google Scholar]
- 15.Shen Q., Wang L., Zhou H., Jiang H.D., Yu L.S., Zeng S. Stereoselective binding of chiral drugs to plasma proteins. Acta Pharmacol. Sin. 2013;34:998–1006. doi: 10.1038/aps.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodman G., Gilman . The Pharmacological Basis of Therapeutics. 9th ed. McGraw-Hill; New York, NY, USA: 1996. [Google Scholar]
- 17.Kratochwil N.A., Huber W., Müller F., Kansy M., Gerber P.R. Predicting plasma protein binding of drugs: A new approach. Biochem. Pharmacol. 2002;64:1355–1374. doi: 10.1016/S0006-2952(02)01074-2. [DOI] [PubMed] [Google Scholar]
- 18.Oravcova J., Bohs B., Lindner W. Drug-protein binding studies—new trends in analytical and experimental methodology. J. Chromatogr. B Biomed. Appl. 1996;677:1–28. doi: 10.1016/0378-4347(95)00425-4. [DOI] [PubMed] [Google Scholar]
- 19.Paal K., Shkarupin A. Paclitaxel binding to the fatty acid-induced conformation of human serum albumin—automated docking studies. Bioorg. Med. Chem. 2007;15:7568–7575. doi: 10.1016/j.bmc.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Tang B., Huang Y.M., Ma X.L., Liao X.X., Wang Q., Xiong X.N., Li H. Multispectroscopic and docking studies on the binding of chlorogenic acid isomers to human serum albumin: Effects of esteryl position on affinity. Food Chem. 2016;212:434–442. doi: 10.1016/j.foodchem.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Pouli N., Marakos P. Fused xanthone derivatives as antiproliferative agents. Anticancer Agents Med. Chem. 2009;9:77–98. doi: 10.2174/187152009787047699. [DOI] [PubMed] [Google Scholar]
- 22.Palmeira A., Paiva A., Sousa E., Seca H., Almeida G.M., Lima R.T., Fernandes M.X., Pinto M., Vasconcelos M.H. Insights into the in vitro antitumor mechanism of action of a new pyranoxanthone. Chem. Biol. Drug Des. 2010;76:43–58. doi: 10.1111/j.1747-0285.2010.00978.x. [DOI] [PubMed] [Google Scholar]
- 23.Sousa E., Paiva A., Nazareth N., Gales L., Damas A.M., Nascimento M.S.J., Pinto M. Bromoalkoxyxanthones as promising antitumor agents: Synthesis, crystal structure and effect on human tumor cell lines. Eur. J. Med. Chem. 2009;44:3830–3835. doi: 10.1016/j.ejmech.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Panthong K., Hutadilok-Towatana N., Panthong A. Cowaxanthone f, a new tetraoxygenated xanthone, and other anti-inflammatory and antioxidant compounds from garcinia cowa. Can. J. Chem. 2009;87:1636–1640. doi: 10.1139/V09-123. [DOI] [Google Scholar]
- 25.Nakatani K., Nakahata N., Arakawa T., Yasuda H., Ohizumi Y. Inhibition of cyclooxygenase and prostaglandin E2 synthesis by γ-mangostin, a xanthone derivative in mangosteen, in C6 rat glioma cells. Biochem. Pharmacol. 2002;63:73–79. doi: 10.1016/S0006-2952(01)00810-3. [DOI] [PubMed] [Google Scholar]
- 26.Nakatani K., Yamakuni T., Kondo N., Arakawa T., Oosawa K., Shimura S., Inoue H., Ohizumi Y. Γ-mangostin inhibits inhibitor-κB kinase activity and decreases lipopolysaccharide-lnduced cyclooxygenase-2 gene expression in C6 rat glioma cells. Mol. Pharmacol. 2004;66:667–674. doi: 10.1124/mol.104.002626. [DOI] [PubMed] [Google Scholar]
- 27.Shagufta, Ahmad I. Recent insight into the biological activities of synthetic xanthone derivatives. Eur. J. Med. Chem. 2016;116:267–280. doi: 10.1016/j.ejmech.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 28.Pinto M.M.M., Sousa M.E., Nascimento M.S.J. Xanthone derivatives: New insights in biological activities. Curr. Med. Chem. 2005;12:2517–2538. doi: 10.2174/092986705774370691. [DOI] [PubMed] [Google Scholar]
- 29.Vieira L.M.M., Kijjoa A. Naturally-occurring xanthones: Recent developments. Curr. Med. Chem. 2005;12:2413–2446. doi: 10.2174/092986705774370682. [DOI] [PubMed] [Google Scholar]
- 30.Masters K.S., Bräse S. Xanthones from fungi, lichens, and bacteria: The natural products and their synthesis. Chem. Rev. 2012;112:3717–3776. doi: 10.1021/cr100446h. [DOI] [PubMed] [Google Scholar]
- 31.Pinto M.M.M., Castanheiro R.A.P., Kijjoa A. Encyclopedia of Analytical Chemistry. Vol. 27. John Wiley & Sons, Ltd.; Hoboken, NJ, USA: 2014. Xanthones from marine-derived microorganisms: Isolation, structure elucidation, and biological activities; pp. 1–21. [Google Scholar]
- 32.Azevedo C.M.G., Afonso C.M.M., Sousa D., Lima R.T., Helena Vasconcelos M., Pedro M., Barbosa J., Corrêa A.G., Reis S., Pinto M.M.M. Multidimensional optimization of promising antitumor xanthone derivatives. Bioorg. Med. Chem. 2013;21:2941–2959. doi: 10.1016/j.bmc.2013.03.079. [DOI] [PubMed] [Google Scholar]
- 33.Sousa M.E., Pinto M.M.M. Synthesis of xanthones: An overview. Curr. Med. Chem. 2005;12:2447–2479. doi: 10.2174/092986705774370736. [DOI] [PubMed] [Google Scholar]
- 34.Waszkielewicz A.M., Słoczyńska K., Pękala E., Żmudzki P., Siwek A., Gryboś A., Marona H. Design, synthesis, and anticonvulsant activity of some derivatives of xanthone with aminoalkanol moieties. Chem. Biol. Drug Des. 2017;89:339–352. doi: 10.1111/cbdd.12842. [DOI] [PubMed] [Google Scholar]
- 35.Szkaradek N., Rapacz A., Pytka K., Filipek B., Siwek A., Cegła M., Marona H. Synthesis and preliminary evaluation of pharmacological properties of some piperazine derivatives of xanthone. Bioorg. Med. Chem. 2013;21:514–522. doi: 10.1016/j.bmc.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 36.Marona H., Pekala E., Antkiewicz-Michaluk L., Walczak M., Szneler E. Anticonvulsant activity of some xanthone derivatives. Bioorg. Med. Chem. 2008;16:7234–7244. doi: 10.1016/j.bmc.2008.06.039. [DOI] [PubMed] [Google Scholar]
- 37.Sousa E.P., Silva A.M.S., Pinto M.M.M., Pedro M.M., Cerqueira F.A.M., Nascimento M.S.J. Isomeric kielcorins and dihydroxyxanthones: Synthesis, structure elucidation, and inhibitory activities of growth of human cancer cell lines and on the proliferation of human lymphocytes in vitro. Hel. Chim. Acta. 2002;85:2862–2876. doi: 10.1002/1522-2675(200209)85:9<2862::AID-HLCA2862>3.0.CO;2-R. [DOI] [Google Scholar]
- 38.Rewcastle G.W., Atwell G.J., Baguley B.C., Boyd M., Thomsen L.L., Zhuang L., Denny W.A. Potential antitumor agents. 63. Structure-activity relationships for side-chain analogues of the colon 38 active agent 9-oxo-9h-xanthene-4-acetic acid. J. Med. Chem. 1991;34:2864–2870. doi: 10.1021/jm00113a027. [DOI] [PubMed] [Google Scholar]
- 39.Sousa M.E., Tiritan M.E., Belaz K.R.A., Pedro M., Nascimento M.S.J., Cass Q.B., Pinto M.M.M. Multimilligram enantioresolution of low-solubility xanthonolignoids on polysaccharide chiral stationary phases using a solid-phase injection system. J. Chromatogr. A. 2006;1120:75–81. doi: 10.1016/j.chroma.2005.12.071. [DOI] [PubMed] [Google Scholar]
- 40.Fernandes C., Masawang K., Tiritan M.E., Sousa E., De Lima V., Afonso C., Bousbaa H., Sudprasert W., Pedro M., Pinto M.M. New chiral derivatives of xanthones: Synthesis and investigation of enantioselectivity as inhibitors of growth of human tumor cell lines. Bioorg. Med. Chem. 2014;22:1049–1062. doi: 10.1016/j.bmc.2013.12.042. [DOI] [PubMed] [Google Scholar]
- 41.Fernandes C., Oliveira L., Tiritan M.E., Leitao L., Pozzi A., Noronha-Matos J.B., Correia-De-Sá P., Pinto M.M. Synthesis of new chiral xanthone derivatives acting as nerve conduction blockers in the rat sciatic nerve. Eur. J. Med. Chem. 2012;55:1–11. doi: 10.1016/j.ejmech.2012.06.049. [DOI] [PubMed] [Google Scholar]
- 42.Cidade H., Rocha V., Palmeira A., Marques C., Tiritan M.E., Ferreira H., Lobo J.S., Almeida I.F., Sousa M.E., Pinto M. In silico and in vitro antioxidant and cytotoxicity evaluation of oxygenated xanthone derivatives. Arab. J. Chem. 2017 doi: 10.1016/j.arabjc.2017.01.006. in press. [DOI] [Google Scholar]
- 43.Pereira D., Lima R.T., Palmeira A., Seca H., Soares J., Gomes S., Raimundo L., Maciel C., Pinto M., Sousa E., et al. Design and synthesis of new inhibitors of p53-MDM2 interaction with a chalcone scaffold. Arab. J. Chem. 2016 doi: 10.1016/j.arabjc.2016.04.015. in press. [DOI] [Google Scholar]
- 44.Palmeira A., Rodrigues F., Sousa E., Pinto M., Vasconcelos M.H., Fernandes M.X. New uses for old drugs: Pharmacophore-based screening for the discovery of p-glycoprotein inhibitors. Chem. Biol. Drug Des. 2011;78:57–72. doi: 10.1111/j.1747-0285.2011.01089.x. [DOI] [PubMed] [Google Scholar]
- 45.Lima R.T., Seca H., Palmeira A., Fernandes M.X., Castro F., Correia-da-Silva M., Nascimento M.S.J., Sousa E., Pinto M., Vasconcelos M.H. Sulfated small molecules targeting EBV in Burkitt lymphoma: From in silico screening to the evidence of in vitro effect on viral episomal DNA. Chem. Biol. Drug Des. 2013;81:631–644. doi: 10.1111/cbdd.12109. [DOI] [PubMed] [Google Scholar]
- 46.Hawkey C.J. Cox-1 and cox-2 inhibitors. Best Pract. Res. Clin. Gastroenterol. 2001;15:801–820. doi: 10.1053/bega.2001.0236. [DOI] [PubMed] [Google Scholar]
- 47.Atukorala I., Hunter D.J. Valdecoxib: The rise and fall of a cox-2 inhibitor. Expert Opin. Pharmacother. 2013;14:1077–1086. doi: 10.1517/14656566.2013.783568. [DOI] [PubMed] [Google Scholar]
- 48.Bali A., Ohri R., Deb P.K. Synthesis, evaluation and docking studies on 3-alkoxy-4-methanesulfonamido acetophenone derivatives as non-ulcerogenic anti-inflammatory agents. Eur. J. Med. Chem. 2012;49:397–405. doi: 10.1016/j.ejmech.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 49.Hegazy G.H., Ali H.I. Design, synthesis, biological evaluation, and comparative cox1 and cox2 docking of p-substituted benzylidenamino phenyl esters of ibuprofenic and mefenamic acids. Bioorg. Med. Chem. 2012;20:1259–1270. doi: 10.1016/j.bmc.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 50.El-Sayed M.A., Abdel-Aziz N.I., Abdel-Aziz A.A., El-Azab A.S., ElTahir K.E. Synthesis, biological evaluation and molecular modeling study of pyrazole and pyrazoline derivatives as selective cox-2 inhibitors and anti-inflammatory agents. Part 2. Bioorg. Med. Chem. 2012;20:3306–3316. doi: 10.1016/j.bmc.2012.03.044. [DOI] [PubMed] [Google Scholar]
- 51.Ryn J., Trummlitz G., Pairet M. Cox-2 selectivity and inflammatory processes. Curr. Med. Chem. 2000;7:1145–1161. doi: 10.2174/0929867003374255. [DOI] [PubMed] [Google Scholar]
- 52.Abdel-Aziz A.A., ElTahir K.E., Asiri Y.A. Synthesis, anti-inflammatory activity and cox-1/cox-2 inhibition of novel substituted cyclic imides. Part 1: Molecular docking study. Eur. J. Med. Chem. 2011;46:1648–1655. doi: 10.1016/j.ejmech.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 53.Rowlinson S.W., Kiefer J.R., Prusakiewicz J.J., Pawlitz J.L., Kozak K.R., Kalgutkar A.S., Stallings W.C., Kurumbail R.G., Marnett L.J. A novel mechanism of cyclooxygenase-2 inhibition involving interactions with ser-530 and tyr-385. J. Biol. Chem. 2003;278:45763–45769. doi: 10.1074/jbc.M305481200. [DOI] [PubMed] [Google Scholar]
- 54.Parikh H.H., McElwain K., Balasubramanian V., Leung W., Wong D., Morris M.E., Ramanathan M. A rapid spectrofluorimetric technique for determining drug-serum protein binding suitable for high-throughput screening. Pharm. Res. 2000;17:632–637. doi: 10.1023/A:1007537520620. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe H., Tanase S., Nakajou K., Maruyama T., Kragh-Hansen U., Otagiri M. Role of arg-410 and tyr-411 in human serum albumin for ligand binding and esterase-like activity. Biochem. J. 2000;349(Pt 3):813–819. doi: 10.1042/bj3490813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghuman J., Zunszain P.A., Petitpas I., Bhattacharya A.A., Otagiri M., Curry S. Structural basis of the drug-binding specificity of human serum albumin. J. Mol. Biol. 2005;353:38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 57.Zaidi N., Ahmad E., Rehan M., Rabbani G., Ajmal M.R., Zaidi Y., Subbarao N., Khan R.H. Biophysical insight into furosemide binding to human serum albumin: A study to unveil its impaired albumin binding in uremia. J. Phys. Chem. B. 2013;117:2595–2604. doi: 10.1021/jp3069877. [DOI] [PubMed] [Google Scholar]
- 58.Karthikeyan S., Bharanidharan G., Mani K.A., Srinivasan N., Kesherwani M., Velmurugan D., Aruna P., Ganesan S. Determination on the binding of thiadiazole derivative to human serum albumin: A spectroscopy and computational approach. J. Biomol. Struct. Dyn. 2016:1–12. doi: 10.1080/07391102.2016.1162751. [DOI] [PubMed] [Google Scholar]
- 59.Singh D.V., Bharti S.K., Agarwal S., Roy R., Misra K. Study of interaction of human serum albumin with curcumin by nmr and docking. J. Mol. Model. 2014;20:2365. doi: 10.1007/s00894-014-2365-7. [DOI] [PubMed] [Google Scholar]
- 60.Fernandes C., Brandão P., Santos A., Tiritan M.E., Afonso C., Cass Q.B., Pinto M.M. Resolution and determination of enantiomeric purity of new chiral derivatives of xanthones using polysaccharide-based stationary phases. J. Chromatogr. A. 2012;1269:143–153. doi: 10.1016/j.chroma.2012.07.058. [DOI] [PubMed] [Google Scholar]
- 61.Fernandes C., Tiritan M.E., Cass Q., Kairys V., Fernandes M.X., Pinto M. Enantioseparation and chiral recognition mechanism of new chiral derivatives of xanthones on macrocyclic antibiotic stationary phases. J. Chromatogr. A. 2012;1241:60–68. doi: 10.1016/j.chroma.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 62.Coutinho A., Prieto M. Ribonuclease t1 and alcohol dehydrogenase fluorescence quenching by acrylamide—A laboratory experiment for undergraduate students. J. Chem. Edu. 1993;70:425–428. doi: 10.1021/ed070p425. [DOI] [Google Scholar]
- 63.Casewit C.J., Colwell K.S., Rappe A.K. Application of a universal force field to main group compounds. J. Am. Chem. Soc. 1992;114:10046–10053. doi: 10.1021/ja00051a042. [DOI] [Google Scholar]
- 64.Gasteiger J., Marsili M. Iterative partial equalization of orbital electronegativity—A rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. doi: 10.1016/0040-4020(80)80168-2. [DOI] [Google Scholar]
- 65.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 66.Mysinger M.M., Carchia M., Irwin J.J., Shoichet B.K. Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rose P.W., Prlic A., Bi C., Bluhm W.F., Christie C.H., Dutta S., Green R.K., Goodsell D.S., Westbrook J.D., Woo J., et al. The RCSB protein data bank: Views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015;43:D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trott O., Olson A.J. Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seeliger D., de Groot B.L. Ligand docking and binding site analysis with pymol and autodock/vina. J. Comput. Aided Mol. Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.