Abstract
The commercial success of tissue engineering products requires efficacy, cost effectiveness and the possibility of scale-up. Advances in tissue engineering require increased sophistication in the design of biomaterials, often challenging our current manufacturing techniques. Interestingly, several of the properties that are desirable for biomaterials design are embodied in the structure and function of plants. This study demonstrates that decellularized plant tissues can be used as adaptable scaffolds for culture of human cells. With simple biofunctionalization technique it’s possible to enable adhesion of human cells on a diverse set of plant tissues. The elevated hydrophilicity and excellent water transport abilities of plant tissues allows cell expansion over prolongued periods of culture. Moreover, cells are able to conform to the microstructure of the plant frameworks, resulting in cell alignment and pattern registration. In conclusion, the current study shows that it is feasible to use plant tissues as an alternative feedstock of scaffolds for mammalian cells.
Keywords: biomaterials, decellularized scaffolds, plants, biofunctionalization, cell expansion
TOC Image
Biomaterials are commonly used to provide “scaffolding” for biomedical applications, such as tissue engineering and cellular biomanufacturing. These scaffolds provide a critical framework for 3-dimensional cell growth and neo-tissue formation. The most desirable scaffolds include interconnected porosity for fluid transport, biochemical properties that support cell function, and a diversity of physical and mechanical properties that can be customized for specific biological or medical needs. Recent advanced manufacturing approaches, such as 3-D printing or decellularization of animal tissues, have produced scaffolds with biomimetic [1] or unique physical properties [2]. Despite these advances, the ability to manufacture biomaterials with a diverse range of achievable physical and biological properties remains a challenge and an active area of inquiry[3].
Interestingly, several of the properties that are desirable for design of biomaterials are embodied in the structure and function of plants. Plant tissues are composite materials, which comprise hard and soft components arranged in complex hierarchical structures[4]. For example, in plant cell walls the cellulose fibers reinforce a pliant and highly hydrophilic matrix of hemicelluloses and either pectin or lignin[5]. The cellulose itself is being synthesized in the form of microfibrils, and is deposited in orderly structures. The hemicelluloses, which are polysaccharides such as arabinoxylans and xylans, have little strength on their own, but by forming hydrogen bonds with the cellulose microfibrils they act as a coupling agent between the matrix and the stiff cellulose microfibrils [6]. The pectins are a group of polysaccharides rich in galacturonic acid units [7], they crosslink together hemicellulose molecules in adjacent microfibrils [5]. Overall, these cellulose polymers are characterized by strong intra-molecular hydrogen bonds and low inter-molecular attractive forces, thus combining strength with flexibility [8].Moreover, the resulting materials offer mechanical properties that often surpass those of their single components by orders of magnitude[4a]. Furthermore, over 700 million years of evolution[9] have developed plants with considerable diversity of properties that are often in defiance of the conundrum of materials that are both strong and tough [4a]. Plant materials also offer unique transport properties, as hydraulic conductance in plants is achieved by the branching of wide conduits into smaller vessels [10]. Transpiration of water from the leaves creates a negative pressure that generates the motive force for the ascent of sap [11]. This energy-efficient mechanism is possible because plant structures are designed to include highly interconnected pores and vessels to maintain hydraulic continuity and allow the propagation of the negative pressure generated in the leaves [9]. Here we hypothesized that plants would provide a diverse feedstock of biomaterials for mammalian cell culture. We reasoned that decellularized plant tissues would include the needed structural sophistication and ease of production to serve as scaffolds, and that they could be biofunctionalized to support human cell growth. We used a broad and diverse array of plant species to demonstrate that decellularized and biofunctionalized plant tissues can support human cell attachment, expansion, alignment, and patterning.
Portions of desired sizes were cut from fresh plants and decellularized using a series of detergents and bleach to remove the plant’s proteins and DNA, respectively (Figure 1a). The use of detergents in decellularization protocols is common practice and they are widely used to decellularize tissue-derived ECM [12] [1] or even plant tissues [13]. Decellularized Petroselinum crispum (parsley) stems showed a more translucent appearance due to the loss of the plant’s pigments and waxy cuticle, and also had a markedly decreased DNA content when compared to non-decellularized parsley stems (Figure 1b–c, Supplementary Figure 1). Electron micrographs of plants before and after decellularization (Figure 2) showed a highly porous ultrastructure, with pore sizes below 100 μm (Figure 2n). As expected, stems from monocot plants such as Vanilla planifolia (Vanilla) had vascular bundles scattered throughout their cross-section (Figure 2k), while dicot plants like parsley (Figure 2i, Supplementary Figure 3) had their vascular bundles arranged in a ring surrounding the pit. Stems maintained their porosity after decellularization (Figure 2), and the ultrastructure was unchanged in the Laelia ancepts (orchid) pseudobulbs, Anthurium waroqueanum (Anthurium) and Bambusoideae (bamboo) stems (Figure 2). However, the size of pores was significantly enlarged in Calathea zebrina (Calathea), parsley and vanilla stems, with parsley stems showing the largest increase in pore size (Figure 2n). Decellularized stems were also able to retain substantial amounts of water, consistent with the mechanism for hydraulic continuity in plants[9]. Stiffer stems like bamboo were able to retain almost 4 times their weight in water, while softer stems from parsley and Solenostemon scutellarioides ‘wasabi’ (Solenostemon) were able to retain more substantial amounts of water (20 and 40 times respectively, Figure 2m). The ability of decellularized plants to retain their hierarchical, hydrophilic, and interconnected ultrastructure led us to explore the ability of plants to serve as scaffolding for mammalian cells.
Figure 1. Decellularization of plant tissues.
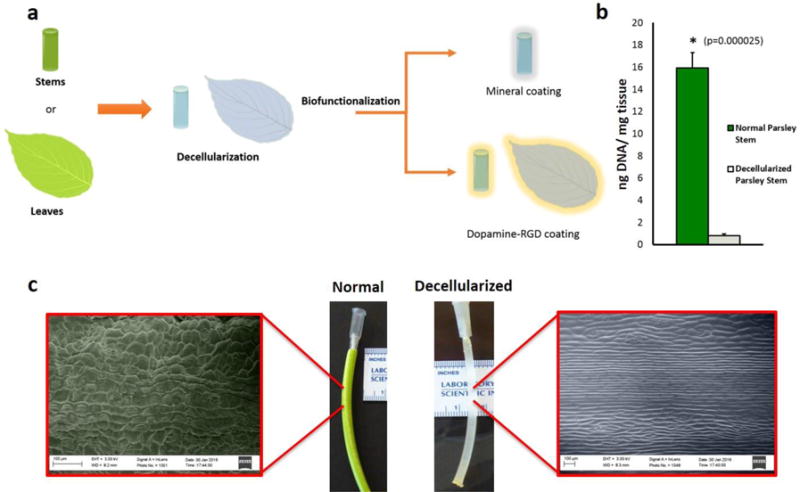
a, Conceptual illustration of plant tissue processing. The cellular component of plant stems or leaves was eliminated by immersion in a series of detergents and bleach. Plant tissues were then biofunctionalized to provide a substrate for adhesion of human cells, using either biomineralization or coating with dopamine-conjugated RGD peptides (RGDOPA). b, DNA quantification in parsley stems, measured using the CyQuant® assay, showing a marked decrease in DNA content after decellularization. (*) represent statistically significant differences using paired student’s t-test n=3, p<0.05. c, Images displaying different appearance between normal and decellularized parsley stems. Color-enhanced SEM micrographs highlight the presence of a waxy cuticle on the surface of normal parsley stems, while the immersion in a hexane bath during the decellularization process dissolved the hydrophobic waxy layer and left a grooved surface structure.
Figure 2. Plant’s stems maintain high porosity after decellularization.
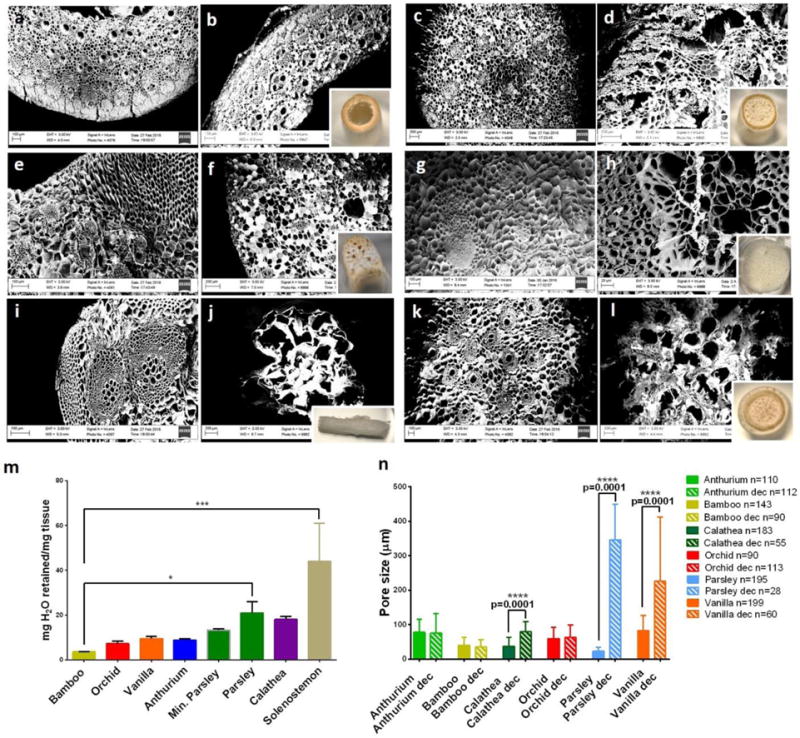
a–l, Paired SEM micrographs of plant stems before (left) and after (right) decellularization: (a–b) Bamboo, (c–d) Anthurium waroqueanum, (e–f) Calathea zebrina, (g–h) Orchid’s pseudobulb, (i–j) Parsley, (k–l) Vanilla. The inset photographs show the visual appearance of decellularized plants after decellularization. m, Mass of water retained normalized to each stem’s mass. (*) represent statistically significant differences using one-way ANOVA followed by Tukey’s multiple comparisons test, n=4, *p<0.05 and *** p<0.001. n, Comparison of average pore size before and after decellularization of stems. Differences were assessed using a paired t-test; p<0.05. The robust regression and outlier removal (ROUT) method was used to identify outliers using the software GraphPad Prism.
To provide a mechanism for cell attachment on a heterogeneous set of plant surfaces, RGD peptides were conjugated with dopamine, a unique catechol moiety found in adhesive proteins and capable of strong molecular adhesion in aqueous environments [14] (Supplementary Figures 4–6). Decellularized plants coated with the RGD-dopamine conjugate (RGDOPA) supported adhesion of human dermal fibroblasts (hDF), while non-coated plants did not support cell attachment on parsley stems (Figure 3) and Impatiens capensis stems (Supplementary Figure 7). A further advantage of the RGDOPA coating was its ability to functionalize plant tissues without clogging pores, thereby maintaining their topographical features and only minimally affecting their surface area (Figure 3h–i, Supplementary Table S1). Decellularized plants were also functionalized via biomineralization (Supplementary Figure S8), a method used in a variety of previous studies to coat biomaterials [15]. Biomineralized plants also supported attachment of hDF (Figure 3b). Scanning electron microscopy (SEM) showed that the biomineralization process preserved the structural features of vascular bundles and larger pores in dicot stems (Figure 3g), but also changed the topography of decellularized stems (Figure 3f, Supplementary Figure 9) and occluded some of the smallest pores (Supplementary Table S1).
Figure 3. Biofunctionalized plants as scaffolds for human cells.
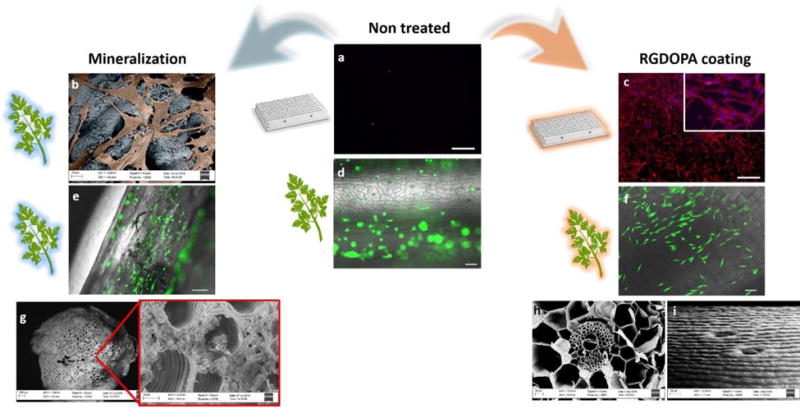
a, Rhodamine-phalloidine staining of actin filaments (red) and dapi staining of nuclei (blue) of hDFs cultured for 2 days in ultralow attachment polystyrene wells (scalebars 500μm). b, Color-enhanced SEM micrographs displaying hDFs (orange) adhering on a biomineralized parsley stem (blue). c, Rhodamine-phalloidin and DAPI staining of hDFs cultured for 2 days on RGDOPA-coated ultralow attachment polystyrene well (scalebars 500μm). d–f, calcein staining of hDFs seeded on decellularized parsley stems untreated (d), biomineralized (e), and coated with RGDOPA (f) (scalebars 100μm). The images show selective cell adhesion on functionalized surfaces. g, a decellularized parsley stem following 7 days incubation in SBF showing growth of a mineral coating with spheroidal morphology within the pores of the stem. The smallest pores appear to be occluded by the mineral, but larger pores and vascular bundles are open and morphological features are maintained after the mineralization. h–i, SEM micrographs of decellularized stems coated with RGDOPA. A cross-section of a Calathea zebrina stem (h) shows that the RGDOPA coating does not occlude even the smallest (~2 μm) pores; a side-view of a vanilla stem (i) shows that the topographical cues of the stems are still evident after the RGDOPA coating.
Biofunctionalized plant tissues provided highly efficient and scalable scaffolds for expansion of primary human cells. Human mesenchymal stem cells (MSCs) and human dermal fibroblasts (hDFs) attached to a variety of RGDOPA-coated stems and biomineralized parsley stems, and the cell populations expanded for a period of 50 days. Cells were viable in all stems (Supplementary Figure 10) and during the first 10 days of culture both cell types showed an increase in metabolic activity in all samples (Figure 4). Over the longer timeframes, the increase in metabolic activity of MSCs was observed only on parsley, mineralized parsley and in standard monolayer culture. This result was confirmed also by DNA quantification, which showed significant MSC expansion in parsley stems, but a significant decrease in cell number in calathea and vanilla stems (Figure 4C). hDFs seeded on parsley and mineralized parsley showed the highest increases in metabolic activity, and hDFs also expanded significantly on orchid pseudobulbs (Figure 4D). Importantly, cell expansion efficiency – the cell expansion normalized to the cell seeding area - was substantially higher on plant stems when compared to standard monolayer cultures (Supplementary Table S2) (Figure 4). Specifically, hDFs underwent a 12.5-fold expansion on mineralized parsley stems and a 14.5-fold expansion on RGDOPA-coated parsley stems, versus only a 2.8-fold expansion in standard monolayer culture. Similarly, MSCs expanded 8.7-fold and 17.5-fold on mineralized and RGDOPA-coated parsley stems, respectively, versus only 1.6-fold in monolayer culture. These data indicate that expansion of human cells on the plant scaffolds was highly efficient relative to standard monolayer culture, likely due to the highly interconnected, porous surface area presented by the decellularized plants. Interestingly, the decellularization of plant tissues is a simple process that can yield large scaffolds. For example, the tropical plant Anthurium magnificum (length=40 cm, width=30 cm) was used to produce several scaffolds (Figure 4i), which could each be functionalized with RGDOPA to support adhesion of primary human cells (human umbilical vein endothelial cells, Figure 4j–l).
Figure 4. Expansion of human cells on decellularized plant stems.
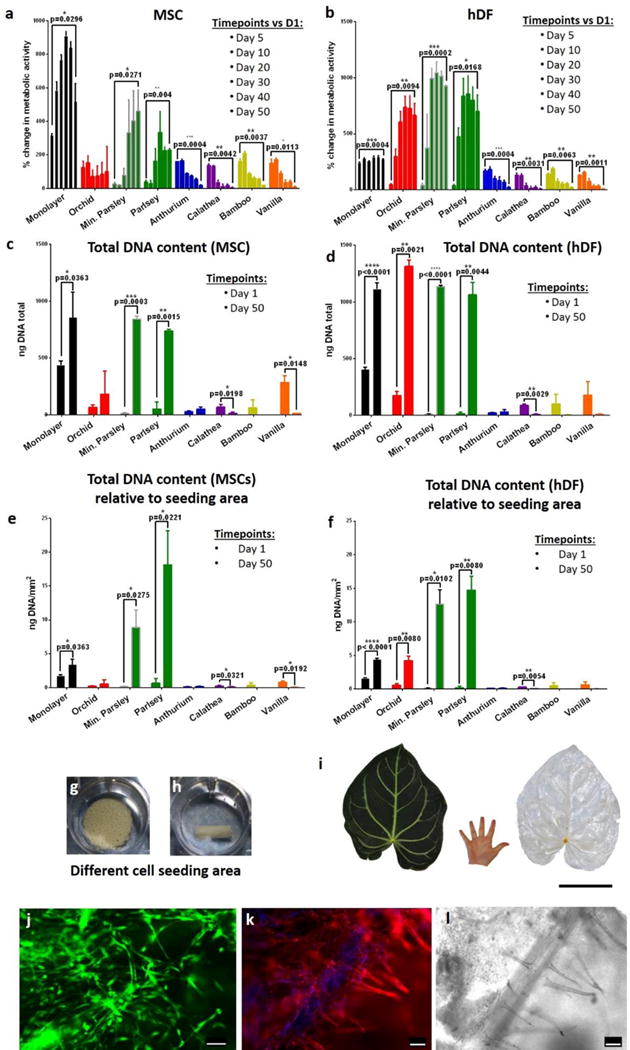
a–b, The metabolic activity of cells was measured using the CellTiter-Blue® assay. MSCs (a) show a steady increase in metabolic activity only on monolayer and on parsley stems, and they decrease in other plant stems. Similar behavior was observed also in hDF (b), however, in this case there was a significant increase in metabolic activity also in orchid’s pseudobulb stems. n=3, p<0.05 paired student’s t-test. c–d, Quantification of total DNA content assessed using the PicoGreen® assay and compared between day 1 and day 50 of culture, n=3, p<0.05 paired student’s t-test. e–f, Total DNA content (ng/mm2) was normalized by seeding area; n=3, p<0.05 paired student’s t-test. g–h, Orchid’s pseudobulb and a mineralized parsley stem respectively in ultralow attachment polystyrene wells. They clearly have different volumes and offer different cell seeding areas. i, Tropical Anthurium magnificum leaf before and after decellularization. The leaves of Anthurium magnificum are on average 30 cm wide and 40 cm long, in the images their size is directly compared to that of a human hand. Scalebar 15cm. j, A decellularized Anthurium magnificum leaf was cut using 8 mm biopsy punch and was used as scaffold for culture of HUVEC cells. After 5 days of culture, live cells were stained using calcein (green). K–l, Rhodamine-phalloidin staining (k) of actin filaments (red) and DAPI staining of nuclei (blue) and relative brightfield image (l) of HUVEC cells cultured on the decellularized Anthurium magnificum leaf for 5 days. The brightfield image (l) dislays the presence of vascular structures and cells appear to register the shape of the vessels (k). From a single leaf of Anthurium magnificum it was possible to obtain numerous pre-vascularized scaffolds. Scalebars 100 μm.
The differences in cell expansion among the plants probed in this study may be attributable to differences in plant stiffness, hydrophilicity, pore sizes and overall size. Parsley stems were among the stems with the highest level of hydrophilicity and the largest pore sizes after decellularization (Figure 2), which may have enabled more efficient cell attachment and expansion. Interestingly, orchid pseudobulbs did not display high hydrophilicity or large pores, yet they supported considerable hDF expansion.
Interestingly, human cells conformed to the microstructure of the plant frameworks, resulting in cell alignment and registration between the cell patterns and the plant microstructures. Previous studies have found that cells are sensitive not only to the chemical milieu of their surroundings but they are also responsive to topographical cues [16]. This phenomenon is known as contact guidance, and it is characterized by the cell’s response to structures on the micron and sub-micron scale [17]. Contact guidance can direct cell migration and influence a number of cell behaviors regardless of the underlying material chemistry [16]. Previous studies have shown that human cells such as fibroblasts are able to sense topographical features as thin as 70 nm [18]. Each leaf or stem used in the current study had unique topographies as a result of the patterning of micro-grooves on its surface, and hDFs responded to plant topographical cues by aligning with the characteristic structural patterns of plants (Figure 5 and Supplementary Figure 11). For example, hDFs adhered and grew preferably in proximity to the plant’s stomata or within grooves (Figure 5 and Supplementary Figure 12). To better understand the level of cell alignment to these plant topographies, we measured an “orientation angle” (OA), defined as the angle between the orientation of the plant topography and the orientation of the attached cells (OA=0° would be perfect alignment). The greatest alignment to plant’s topography was observed on Solenostemon stems (Figure 5, Supplementary Figures 13–14), where 44.09 ± 7.16% of the cells had an OA less than 20°. In contrast, 28.64 ± 3.39% of the cells on parsley stems had an OA less than 20°. Further, hDFs seeded onto the Buddleja davidii (summer lilac) leaf were able to populate the entire leaf and grow around the patterned vascular structure (Figure 5n–o). The cell patterning and alignment observed here on plant scaffolds could be important in future studies, as topographical cues have been used in other contexts to direct cell differentiation [17b], and spatial patterning can facilitate development of complex mammalian tissues. Another aspect that deserves some consideration is the biocompatibility of the plant tissues. With thousands of different plant species, it is difficult to make general assertions regarding biocompatibility. Some plant species will be more suitable than others for regenerative medicine applications. Particular attention should be devoted in avoiding the use of plants that secrete toxic compounds. In addition, it was found that plants can accumulate heavy metals and other trace elements (TE) from their growth environment with different rates[19]. TE accumulation varies in different categories of plants, for example, it was found to be low in legumes, moderate in root vegetables, and high in leafy vegetables[19]. However, it was found that TE concentrations in plants are highly related to the chemical composition of the growth media [19]. Therefore, the TE accumulation in plants can be prevented simply by growing plants in controlled environments devoid of heavy metals and other elements that can cause complications. Moreover, eventual TE will most likely be removed by the numerous washes that plant-derived scaffolds undergo during the decellularization process. In fact, the concentration of TE in the decellularized stems was so low that we were not able to detect them during the EDS analysis (Supplementary Figure S8). To date there is only limited knowledge about the tolerance of mammalian tissues to plant tissues in vivo, however, a recent study revealed that subcutaneous implantation of plant-derived cellulose materials evoked only a mild immune response that disappeared 8 weeks post-implantation [13]. This is in line with what has been found following implantation of other cellulosic biomaterials [20]. Interestingly, it was shown that the highly crystalline celluloses evoked no immulogical response [20d]. Considering that the degree of crystallinity in many plants is estimated to be around 50% [21], we can speculate that limited immunological reactions may be expected after implantation of decellularized plants. Another issue that should be considered is scaffold degradation. In nature, cellulose (especially in the amorphous form) can be degraded by a family of hydrolytic enzymes called cellulases [9]. However, the tightly packed and orderly structure of crystalline chains of cellulose is impervious to enzymatic degradation. For this reason, cellulosic materials are durable and show limited degradation over time [9, 22]. The recalcitrance of plant tissues could also limit their adoption as scaffolds for regenerative medicine. However, the increase of cellulose-based biomaterials is pushing towards designing novel strategies for controlled degradation in vivo. Some studies have already shown that following hydrolysis pre-treatment and co-delivery of cellulases it is possible to obtain cellulose scaffolds that are resorbable to differing degrees [20c, d, 23].
Figure 5. Cell orientation follows the plant’s topographical cues.
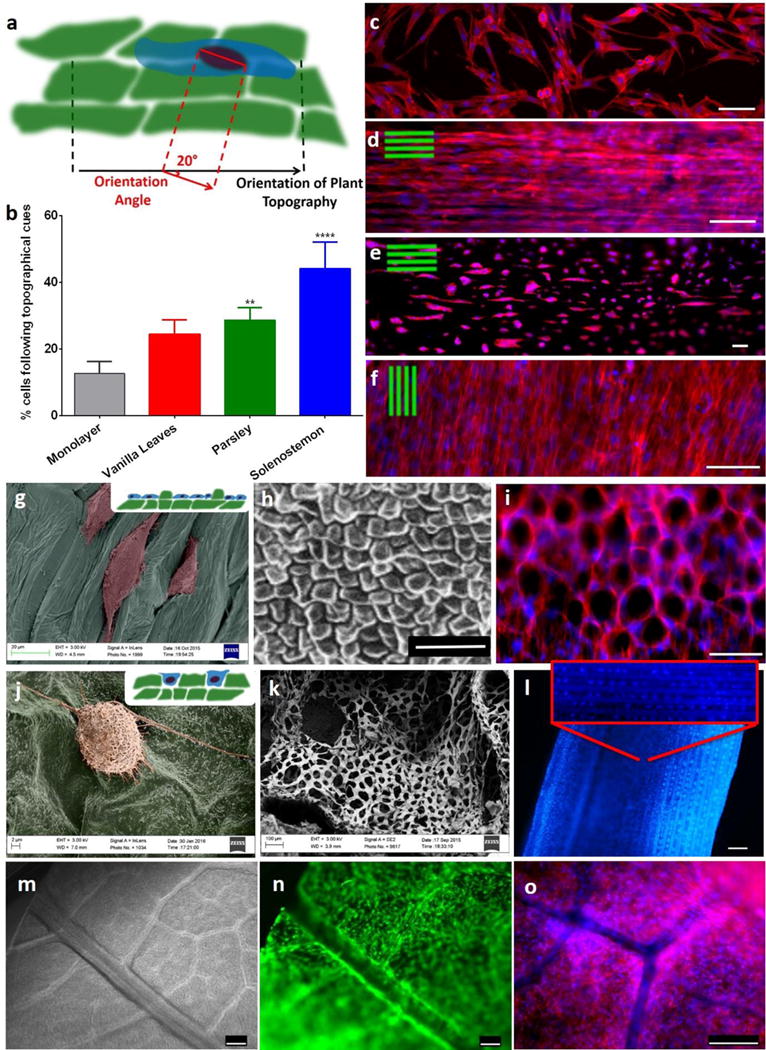
a, Conceptual illustration of the method used for the quantification of orientation angle (OA) between the mammalian cell and the underlying plant structure. b, Only 15% of the cells seeded on monolayer had an OA < 20°, whereas more than 40% of the cells seeded on Solenostemon were aligned with the stem’s topography (OA<20°). n=3, p<0.05, One-way ANOVA followed by Dunnett’s multiple comparisons test. c–f, Rhodamine-phalloidin staining of actin filaments (red) and DAPI staining of nuclei (blue) of hDFs cultured on monolayer (c), Solenostemon (d), Parsley (e), and Vanilla leaves (f), scalebars 100 μm. Cells appear to follow the topographical cues of each of the plant tissues. g, Color-enhanced SEM micrograph of hDFs seeded on parsley stems shows that cells grow preferably in the concave areas of the plant tissues. h, SEM micrograph displaying the surface topography of an Anthurium waroqueanum stem. i, hDF seeded on Anthurium stems conform into a pattern reminiscent of the plant topography. Actin filaments were stained red with Rhodamine-phalloidine nuclei stained blue with dapi. j, Color-enhanced SEM micrograph displaying a hDF cell adhering on the surface of a summer lilac leaf. k, SEM micrograph showing a cross-section of a Schoenoplectus tabernaemontani stem. l, DAPI staining of hDF cultured on a Schoenoplectus tabernaemontani stem for 30 days. Scalebar 250 μm. m, Brightfield image of a summer lilac leaf after decellularization. Our decellularization protocol allowed for maintenance of the vasculature of the leaf. n–o, hDF seeded on a summer lilac leaf and live-stained with calcein (n) or stained using rhodamine-phalloidine, DAPI (o). Cells were expanded on the leaves for 4 days, and during this period they populated the entire leaf and grew around the vasculature, thereby using the leaf’s structure as a template. Scalebars 250 μm.
Moreover, a new set of enzymes classified as CBM33 and GH61 were recently found to catalyze the oxidative cleavage of polysaccharides [24]. These enzymes are abundant in genomes of biomass-converting microorganisms and are capable of binding effectively to crystalline chains of cellulose and disrupting their structure, thus increasing their accessibility for hydrolytic enzymes such as cellulases [24]. Therefore, degradation of plant-derived scaffolds could be achieved by designing hydrolysis pre-treatments and by the administration of cocktails of cellulases and CBM33 or GH61 enzymes. As tissue engineering approaches more sophisticated designs, some of the current limitations become more noticeable. For example, design of smaller scaffolds is not always achievable with the literal downsizing of conventional engineering techniques [25]. The resulting scaffolds have limited resolution, high costs and often require extensive adjustments that stretch the time between design and implementation [4a]. Nature’s creative use of biopolymer building blocks provides an alternative feedstock of manufactured scaffolds for tissue engineering applications. Plant development results in complex hierarchical structures in layers up to 1 μm in thickness, a resolution that is out of reach for most conventional manufacturing techniques [5]. Herein we show that it is possible to maintain the structural complexity of plant tissues after decellularization, and with simple biofunctionalization these surfaces can support adhesion of human cells. The highly hydrophilic nature of plant tissues and their efficacy in transport of fluids also enabled efficient expansion of human cells over extended periods of time. Human cells also sensed the topographical features of plant tissues and conformed to the structural motifs, resulting in cell patterning into concave areas, alignment along plant micropatterns, or growth around the plant vasculature. Decellularized plant tissues may provide a diverse array of complex biomaterials with limited costs. In addition, the ability to borrow scaffold structures from the plant kingdom offers the potential to shorten developmental time, while allowing mass production of complex biomaterials with low costs.
Experimental Section
Decellularization of plant tissues
Most of the plants used in this study (Calathea zebrina, Anthurium waroquaenum, Anthurium magnificum, Solenostemon, Vanilla, Laelia ancepts, Bamboo) were obtained from the Olbrich Botanical Gardens in Madison, Here, among the available tropical plants we selected plants with stems of high porosity and fast growth rate to ensure continuity of supply for the study and for potential applications. Parsley was purchased from a local market and Schoenoplectus tabernaemontani plants were collected at the UW Arboretum. Leaves and stems were collected from fresh plant tissues to minimize disruptions to the tissues structure. Leaves were cut into discs using 8 mm punches, while stems were manually cut at about 8 mm length. Plant tissues were then immersed in a solution of 10X sodium dodecyl sulfate (SDS) in water for 5 days in gentle agitation. Successively, the tissues were incubated in 0.1% Triton-X-100 in a 10% solution of bleach for 48 hours. After which the stems and leaves lost all their pigments and became translucent. The waxy cuticle was dissolved by 1 minute incubation in hexane followed by 1 minute wash in 1X Phosphate buffered saline (PBS), the process was repeated at least twice. Deionized water was used to remove eventual residues of detergents and bleach, the tissues were incubated in H2O for at least 2 days, after which were lyophilized and stored dry.
RGDOPA synthesis
A custom peptide with the sequence RGDGGG was purchased from Genscript®. The peptide was reconstituted at a concentration of 16 mM in a buffer solution of 10mM 2-(N-morpholino)ethanesulfonic acid (MES) at pH 6. While stirring, dopamine hydrochloride (Sigma-Aldrich®, H8502) was also dissolved into this solution at a final concentration of 200 mM. The conjugation of dopamine to the custom peptide was obtained by using the zero-length crosslinking agent 1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) and N-hydroxysuccinimide (NHS). NHS (ThermoFisher Scientific™, 24500) was first added to the reaction mix at a concentration of 5 mM and successively EDC (Sigma-Aldrich®, E6383) at a concentration of 0.1M. The reaction mix was stirred for 2 hours at room temperature and then dialyzed using a 100–500 Da dialysis membrane (Spectrum®Labs, 131060) in deionized water for 5 days. The purity of the reaction product was assessed by High Performance Liquid Chromatography (HPLC). RGDOPA was then lyophilized and stored dry at −20°C.
Plant tissue functionalization with RGDOPA
Plant’s stems or leaves were washed in a solution of 10 mM Tris-HCl at a pH of 8.5 and incubated at room temperature for 30 minutes under gentle agitation. In the meantime, RGDOPA powder was reconstituted in 10 mM Tris-HCl (pH 8.5) at a final concentration of 1mg/mL. Plant tissues to be functionalized were then immersed in the RGDOPA solution and incubated for 24 hours under gentle agitation at room temperature. After functionalization, plant tissues acquired a slightly grey/black color. To remove unbound RGDOPA the tissues are then washed twice in 10 mM Tris-HCl (pH 8.5) and once in PBS (1X) prior to be used for cell culture.
Mineralization of plant stems
To form a mineralized coating, plant’s stems were incubated in modified simulated body fluid (mSBF) for 7 days under gentle agitation. The mSBF was prepared adding the following reagents into deionized water in the following order: 141mM NaCl, 4 mM KCl, 0.5 mM MgSO4, 1 mM MgCl2, 150 mM NaHCO3, 20 mM HEPES, 5 mM CaCl2 and 2 mM KH2PO4. The pH of the mSBF was then adjusted to 6.8 and throughout the 7 days of coating, the mSBF solution was changed daily. After the 7 days of coating, plant stems were rinsed in deionized water and lyophilized.
Supplementary Material
Acknowledgments
We thank Bradley Herrick and the UW Arboretum (Madison, Wisconsin) for allowing us to collect plant samples; and all the staff in Olbrich Botanical Gardens (Madison, Wisconsin) for their assistance regarding collection of tropical plants; and Ellen Leiferman (University of Wisconsin-Madison) for her help with the faxitron images. This work is supported by the Environmental Protection Agency (STAR grant no. 83573701), the National Institutes of Health (R01HL093282-01A1 and UH3TR000506) and the National Science Foundation (IGERT DGE1144804).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Gianluca Fontana, Department of Orthopedics and Rehabilitation, University of Wisconsin School of Medicine and Public Health, Madison, WI 53705 USA
Joshua Gershlak, Biomedical Engineering, Worcester Polytechnic Institute, Worcester, MA 01609 USA.
Michal Adamski, Department of Surgery, University of Wisconsin School of Medicine and Public Health, Madison, WI 53705 USA.
Dr. Jae-Sung Lee, Department of Orthopedics and Rehabilitation, University of Wisconsin School of Medicine and Public Health, Madison, WI 53705 USA
Shion Matsumoto, Biomedical Engineering, Worcester Polytechnic Institute, Worcester, MA 01609 USA.
Prof. Hau D. Le, Department of Surgery, University of Wisconsin School of Medicine and Public Health, Madison, WI 53705 USA
Dr. Bernard Binder, Department of Surgery, University of Wisconsin School of Medicine and Public Health, Madison, WI 53705 USA
John Wirth, Olbrich Botanical Gardens, Madison, WI 53704 USA.
Prof. Glenn Gaudette, Biomedical Engineering, Worcester Polytechnic Institute, Worcester, MA 01609 USA
Prof. William L. Murphy, Biomedical Engineering, Material Sciences and Engineering, Department of Orthopedics and Rehabilitation, Department of Surgery, University of Wisconsin-Madison, Madison, WI 53705 USA
References
- 1.Hoshiba T, Chen G, Endo C, Maruyama H, Wakui M, Nemoto E, Kawazoe N, Tanaka M. Stem cells international. 2016;2015 doi: 10.1155/2016/6397820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Paulsen SJ, Miller JS. Dev Dyn. 2015;244:629–640. doi: 10.1002/dvdy.24254. [DOI] [PubMed] [Google Scholar]; b) Gu BK, Choi DJ, Park SJ, Kim MS, Kang CM, Kim CH. Biomater Res. 2016;20:12. doi: 10.1186/s40824-016-0058-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Place ES, Evans ND, Stevens MM. Nature materials. 2009;8:457–470. doi: 10.1038/nmat2441. [DOI] [PubMed] [Google Scholar]
- 4.a) Wegst UG, Bai H, Saiz E, Tomsia AP, Ritchie RO. Nature materials. 2015;14:23–36. doi: 10.1038/nmat4089. [DOI] [PubMed] [Google Scholar]; b) Burgert I, Fratzl P. Philos Trans A Math Phys Eng Sci. 2009;367:1541–1557. doi: 10.1098/rsta.2009.0003. [DOI] [PubMed] [Google Scholar]
- 5.Gibson LJ. J R Soc Interface. 2012;9:2749–2766. doi: 10.1098/rsif.2012.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgert I. Am J Bot. 2006;93:1391–1401. doi: 10.3732/ajb.93.10.1391. [DOI] [PubMed] [Google Scholar]
- 7.Ridley BL, O’Neill MA, Mohnen D. Phytochemistry. 2001;57:929–967. doi: 10.1016/s0031-9422(01)00113-3. [DOI] [PubMed] [Google Scholar]
- 8.Lenting HB, Warmoeskerken MM. J Biotechnol. 2001;89:217–226. doi: 10.1016/s0168-1656(01)00300-5. [DOI] [PubMed] [Google Scholar]
- 9.Raven PH, Evert RF, Eichhorn SE. Biology of plants. Macmillan; 2005. [Google Scholar]
- 10.McCulloh KA, Sperry JS, Adler FR. Nature. 2003;421:939–942. doi: 10.1038/nature01444. [DOI] [PubMed] [Google Scholar]
- 11.a) Kim HK, Park J, Hwang I. Journal of experimental botany. 2014:eru075. doi: 10.1093/jxb/eru075. [DOI] [PubMed] [Google Scholar]; b) Tyree MT, Zimmermann MH. Xylem structure and the ascent of sap. Springer Science & Business Media; 2013. [Google Scholar]; c) Petit G, Anfodillo T. Journal of Theoretical Biology. 2009;259:1–4. doi: 10.1016/j.jtbi.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Guyette JP, Gilpin SE, Charest JM, Tapias LF, Ren X, Ott HC. Nat Protocols. 2014;9:1451–1468. doi: 10.1038/nprot.2014.097. [DOI] [PubMed] [Google Scholar]
- 13.Modulevsky DJ, Cuerrier CM, Pelling AE. PLoS One. 2016;11:e0157894. doi: 10.1371/journal.pone.0157894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Mehdizadeh M, Yang J. Macromolecular bioscience. 2013;13:271–288. doi: 10.1002/mabi.201200332. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wilker JJ. Nat Mater. 2014;13:849–850. doi: 10.1038/nmat4070. [DOI] [PubMed] [Google Scholar]
- 15.a) Suarez-Gonzalez D, Barnhart K, Migneco F, Flanagan C, Hollister SJ, Murphy WL. Biomaterials. 2012;33:713–721. doi: 10.1016/j.biomaterials.2011.09.095. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Choi S, Murphy WL. Acta Biomater. 2010;6:3426–3435. doi: 10.1016/j.actbio.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Murphy WL, Mooney DJ. J Am Chem Soc. 2002;124:1910–1917. doi: 10.1021/ja012433n. [DOI] [PubMed] [Google Scholar]
- 16.Stevens MM, George JH. Science. 2005;310:1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 17.a) Wilkinson C, Riehle M, Wood M, Gallagher J, Curtis A. Materials Science and Engineering: C. 2002;19:263–269. [Google Scholar]; b) Bettinger CJ, Langer R, Borenstein JT. Angewandte Chemie International Edition. 2009;48:5406–5415. doi: 10.1002/anie.200805179. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wolf K, Müller R, Borgmann S, Bröcker E-B, Friedl P. Blood. 2003;102:3262–3269. doi: 10.1182/blood-2002-12-3791. [DOI] [PubMed] [Google Scholar]
- 18.a) Teixeira AI, Nealey PF, Murphy CJ. Journal of Biomedical Materials Research Part A. 2004;71:369–376. doi: 10.1002/jbm.a.30089. [DOI] [PubMed] [Google Scholar]; b) Price R, Haberstroh K, Webster T. Medical and Biological Engineering and Computing. 2003;41:372–375. doi: 10.1007/BF02348445. [DOI] [PubMed] [Google Scholar]
- 19.Kabata-Pendias A, Pendias H. Trace elements in soils and plants - Fourth Edition. CRC press Boca Raton; 2010. [Google Scholar]
- 20.a) Hart J, Silcock D, Gunnigle S, Cullen B, Light ND, Watt PW. The international journal of biochemistry & cell biology. 2002;34:1557–1570. doi: 10.1016/s1357-2725(02)00062-6. [DOI] [PubMed] [Google Scholar]; b) Helenius G, Bäckdahl H, Bodin A, Nannmark U, Gatenholm P, Risberg B. Journal of Biomedical Materials Research Part A. 2006;76:431–438. doi: 10.1002/jbm.a.30570. [DOI] [PubMed] [Google Scholar]; c) Märtson M, Viljanto J, Hurme T, Laippala P, Saukko P. Biomaterials. 1999;20:1989–1995. doi: 10.1016/s0142-9612(99)00094-0. [DOI] [PubMed] [Google Scholar]; d) Miyamoto T, Takahashi Si, Ito H, Inagaki H, Noishiki Y. Journal of biomedical materials research. 1989;23:125–133. doi: 10.1002/jbm.820230110. [DOI] [PubMed] [Google Scholar]; e) Svensson A, Nicklasson E, Harrah T, Panilaitis B, Kaplan D, Brittberg M, Gatenholm P. Biomaterials. 2005;26:419–431. doi: 10.1016/j.biomaterials.2004.02.049. [DOI] [PubMed] [Google Scholar]; f) Wippermann J, Schumann D, Klemm D, Kosmehl H, Salehi-Gelani S, Wahlers T. European Journal of Vascular and Endovascular Surgery. 2009;37:592–596. doi: 10.1016/j.ejvs.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 21.a) Klemm D, Heublein B, Fink HP, Bohn A. Angewandte Chemie International Edition. 2005;44:3358–3393. doi: 10.1002/anie.200460587. [DOI] [PubMed] [Google Scholar]; b) Wertz J-L, Mercier JP, Bédué O. Cellulose science and technology. CRC Press; 2010. [Google Scholar]
- 22.Reese ET. Applied microbiology. 1956;4:39. doi: 10.1128/am.4.1.39-45.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Entcheva E, Bien H, Yin L, Chung CY, Farrell M, Kostov Y. Biomaterials. 2004;25:5753–5762. doi: 10.1016/j.biomaterials.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 24.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink V. Biotechnology for biofuels. 2012;5:45. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ball P. Nanotechnology. 2002;13:R15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


