Abstract
While antibody titers and neutralization are considered the gold standard for the selection of successful vaccines, these parameters are often inadequate predictors of protective immunity. As antibodies mediate an array of extra-neutralizing Fc-functions, when neutralization fails to predict protection, investigating Fc-mediated activity may help identify immunological correlates and mechanism(s) of humoral protection. Here, we used an integrative approach termed Systems Serology to analyze relationships among humoral responses elicited in four HIV vaccine-trials. Each vaccine regimen induced a unique humoral “Fc-fingerprint”. Moreover, analysis of case:control data from the first moderately protective HIV vaccine trial, RV144, pointed to mechanistic insights into immune complex composition that may underlie protective immunity to HIV. Thus, multi-dimensional relational comparisons of vaccine humoral fingerprints offer a unique approach for the evaluation and design of novel vaccines against pathogens for which correlates of protection remain elusive.
Introduction
Although over 80 vaccines, covering more than 20 diseases, have been licensed in the US, vaccine design efforts against persisting infections, including malaria, tuberculosis, and HIV continue to fail. These setbacks have driven a shift from empirical vaccine design approaches to rational vaccine development strategies that consider pathogen life cycle, pathogen structural information, and immunological correlates of protection. Yet, the immune correlates for most globally lethal pathogens have yet to be defined, complicating vaccine design efforts. Prospective immunogens are frequently chosen based on measures of antibody (Ab) titer and neutralization, irrespective of their mechanistic effects in immunity. However, for most clinically approved vaccines, titer and neutralization activity alone do not account for protective immunity (Pulendran and Ahmed, 2011). Instead, protective immunity is often observable in absence of neutralization, and accumulating evidence across a spectrum of vaccines has suggested a critical role for extra-neutralizing Ab functions such as Ab-dependent cellular cytotoxicity (ADCC), Ab-dependent cellular phagocytosis (ADCP), Ab-dependent complement deposition (ADCD), and Ab-dependent respiratory burst (ADRB) in both protection from and post-infection control of HIV (Barouch et al., 2015; Bournazos et al., 2014; Hessell et al., 2007), influenza (DiLillo et al., 2014; Jegerlehner et al., 2004), HSV (Kohl and Loo, 1982; Kohl et al., 1981), Ebola virus (Warfield et al., 2007), and malaria (Joos et al., 2010; Osier et al., 2014).
Following vaccination, Abs targeting an extensive array of epitopes with different affinities and Fc-effector profiles collectively contribute to the formation of immune complexes that direct antimicrobial functions via their constant domains (Fc). In addition to the rapid diversification of the antigen (Ag)-binding domain (Fab), the Fc domain is also rapidly tuned during an immune response, altering the affinity of Ab interactions with innate immune receptors (e.g. Fc receptors and complement) expressed on all innate immune cells (Ackerman and Alter, 2013; Chung and Alter, 2014). The diversity of Fc profiles, potential Fab variants, and tissue-specific Fc receptor expression results in a flexible humoral immune response poised for the elimination of pathogens via mechanisms beyond simple neutralization. Hence, analytical approaches able to integrate diverse facets of the humoral immune response will be critical to: define unexpected correlates of protection from infection in protection studies or studies of natural disease resistance; guide the selection of promising vaccines/immunogens through principled analysis of humoral immune profiles; and define the relationships between Ab populations and functions that point to mechanisms of protective immunity.
As a prominent example, the ability to select HIV vaccine candidates has been hindered by an inadequate understanding of the immunological correlates of protection from HIV. However, several phase III trials have been conducted, one of which (RV144) demonstrated a modest level of protection (31.2% reduction in the risk of infection) (Rerks-Ngarm et al., 2009), potentially harboring clues that may guide future vaccine development. This protection was observed in the absence of neutralizing Abs, cytotoxic T cell responses, and high Ab titers. Univariate and multivariate logistic regression analyses linked the reduced risk of infection with non-IgA Ab responses targeting the V1V2 region of the HIV envelope and ADCC activity (Haynes et al., 2012; Zolla-Pazner et al., 2014). Follow-up analyses identified additional features of the humoral immune response associated with protection, including the preferential induction of IgG3 responses, which coordinated multiple Ab effector functions including ADCC and ADCP (Chung et al., 2014b; Yates et al., 2014). However, in the correlates analysis, although many Ab assays were initially considered, the identification of immune correlates in RV144 was constrained by the selected assays that deeply interrogated neutralization and Ab specificity, but profiled only a limited set of Fc features, including only a few Ab subclasses/isotypes (IgG, IgG3, IgA) and a single function, ADCC.
Here, we aimed to consider more integrative and network-oriented relationships between a broader array of polyclonal Ab features and functional properties associated with vaccine regimens and outcomes. As an initial test of this approach, termed “Systems Serology”, we examined recent HIV vaccine trials including the moderately protective RV144 vaccine (ALVAC/AIDSVAX B/E (Rerks-Ngarm et al., 2009)), two trials that did not demonstrate efficacy in phase 2b trials (VAX003; AIDSVAX B/E (Pitisuttithum et al., 2006); and HVTN204; DNA/rAD5 (Churchyard et al., 2011)), and one experimental phase 1 study designed to evaluate the prototype vaccine Ad26 vector (IPCAVD001; Ad26.ENVA.01 (Barouch et al., 2013a)). A battery of modeling techniques that emphasize co-variation among measurements was applied to these data, revealing features of vaccine–induced “fingerprints” offering new insights concerning polyclonal Ab immune responses elicited by vaccines, toward improved evaluation and development of future vaccines.
Results
Systems Serology
Beyond their role in neutralization, Abs mediate a vast array of additional functions via their Fc domains. Thus, a “Systems Serology” approach was developed to broadly profile the extra-neutralizing Ab activity of vaccine-induced polyclonal Abs (Fig 1). The initial platform interrogated six Fc effector functions (ADCC, ADCP, ADCD, and three Ab-dependent NK cell activities (Fig 1). Linked to these 6 functions, 58 biophysical measurements were simultaneously captured, including binding to Fcγ receptors (FCGRs) and the relative abundances of an array of antigenic-specific antibodies (Table S1) in 120 samples from four HIV vaccine trials.
Figure 1. System Serology Analysis.
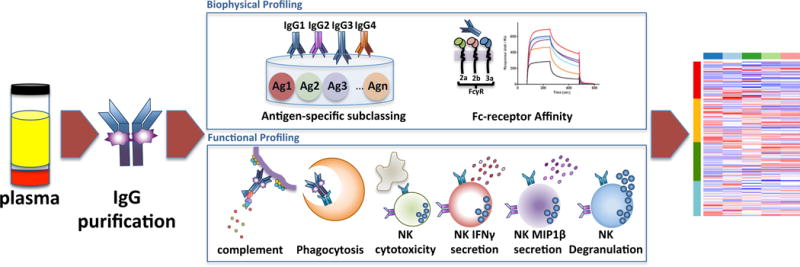
This Systems Serology platform allows for the broad characterization of the polyclonal extra-neutralizing IgG immune profile induced by vaccination. IgG was purified from subjects enrolled in four different HIV vaccine trials (RV144, VAX003, HVTN204 and IPCAVD001). Six Fc effector functions and 58 biophysical measurements were assayed (complete list described in Table S1). All 64 parameters were collected to create an extra-neutralizing serological signature for the four vaccine trials using an array of unsupervised and supervised machine learning algorithms. See also Tables S1 and S2.
Identification of vaccine-specific signatures
Unsupervised hierarchical clustering grouped vaccine regimens primarily by immunogen type (Fig 2 and Table S2), including an adenovirus (Ad) vector cluster, composed of mixed HVTN204 (DNA/Ad5) and IPCAVD001 (Ad26) samples (cluster 1: green and yellow, respectively) and a protein immunogen cluster containing largely mixed VAX003 (protein alone) and RV144 (poxvirus prime/protein boost) samples (cluster 2: blue and red, respectively). While this clustering highlights the dominant influence of immunogen type in directing distinct humoral profiles, specific features driving this separation cannot be clearly discerned.
Figure 2. Hierarchical clustering of vaccine trial profiles by biophysical properties and functional responses.
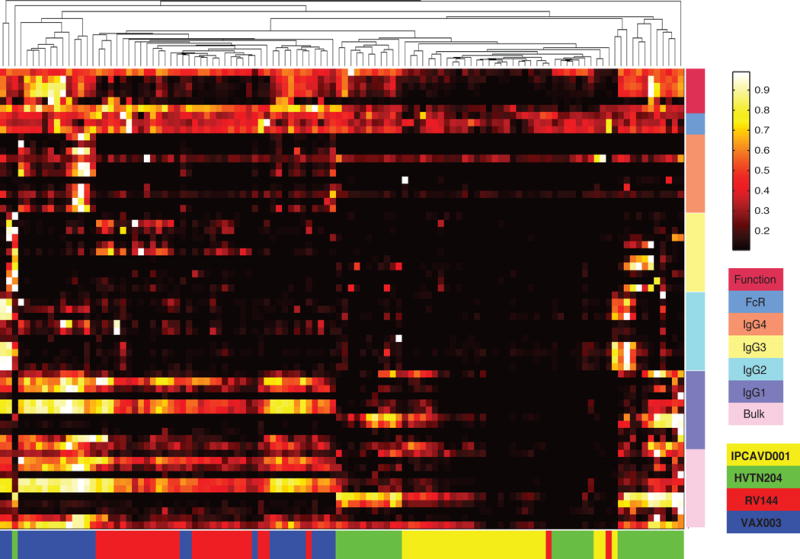
Data was compiled for the four different vaccine trials. Each column represents the full Ab profile of an individual subject. Colored bars along the bottom correspond to the vaccine trial for each subject. Ab properties are grouped by generalized features (Function, FcR affinity, Bulk IgG, IgG1, IgG2, IgG3, IgG4) indicated by the colored bars on the right. Specific features are listed in Table S2. See also Table S1.
To gain enhanced resolution on the key features contributing to profile differences, a multidimensional combined feature selection method (LASSO) (Tibshirani, 1997) and partial least squares discriminant analysis (PLSDA) (Arnold et al., 2015; Lau et al., 2011) was used. Focusing initially on RV144 and VAX003, which shared the same protein immunogen but provided different efficacies, as few as 7 of the 64 features accounted for 76% of the variance across the two trials, driving nearly complete resolution of the vaccine profiles (Fig 3A, B). Separation of the Ab profiles was observed in the scores plot, with points representing individual RV144 (red) or VAX003 (blue) vaccinees (Fig 3A). Differences between vaccine-elicited Ab profiles were largely captured along the first dimension (LV1), which accounted for the majority of the variance between the two trials (61%). The corresponding loadings plot (Fig 3B) illustrates the contribution of the 7 LASSO features, where the relative location of an individual feature is associated with the corresponding vaccine subpopulation in the scores plot (Fig 3A). Elevated gp120-specific IgG3 levels relative to other features (Fig 3B) uniquely marked the RV144 vaccine profile (Fig 3A), as previously described (Chung et al., 2014b; Yates et al., 2014). By contrast, the VAX003 Ab profile was associated with known induction of higher total HIV gp140-specific Ab titers, dominated by IgG4 (Chung et al., 2014b). Yet, additional novel features were identified that associated with the non-protective VAX003 profile, including elevated total gp140-specific responses, higher Ab-driven NK cell degranulation, and chemokine secretion. This result suggests that differences in relationships between Ab features rather than the total Ab amount may be essential for resolving “protective” from “non-protective” vaccine profiles. Moreover, the scores plot highlights an unappreciated level of heterogeneity among the RV144 vaccinees with respect to the magnitude of the IgG3 response, where 26% of the RV144 vaccinees exhibited a more highly skewed IgG3 response specifically across LV2 (Fig 3A).
Figure 3. PLSDA and LASSO identify unique combinations of features that differentiate vaccine trial Ab profiles.
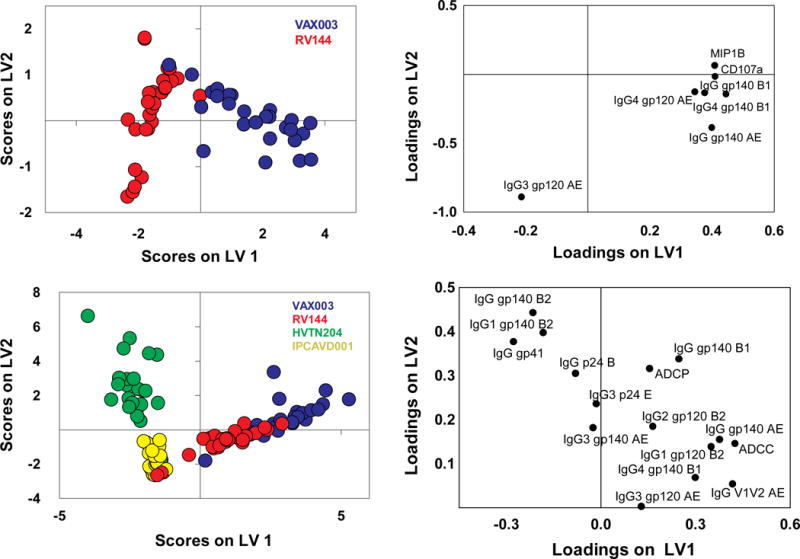
(A) The scores plot represents the RV144 (red) and VAX003 (blue) vaccine profile distribution for each vaccinee tested (dots) from the LASSO and PLSDA. Remarkably, as few as, 7 Ab features, listed on the loadings plot (B), separated the vaccine profiles with 100% calibration and 97% cross-validation accuracy. LV1 captured 61% of X variance and 72% of the Y variance. (C) LASSO and PLSDA of all 4 vaccine profiles identified 15 Ab features (D) able to discriminate between the distinct vaccine regimens (red, RV144; blue, VAX003; green, HVTN204; and yellow, IPCAVD001) with 84% cross validation accuracy. Together LV1 and LV2 captured 57% of the X variance and 45% of the Y variance, respectively. See also Tables S1 and S2.
When all four vaccine trials were analyzed simultaneously, 15 of the 64 features separated the vaccine profiles, accounting for 57% of the variance. The first dimension (LV1) revealed a similar separation as the hierarchical clustering analysis, separating based on protein- (right, Fig 3C) versus Ad-based vectored immunization (33% variance), confirming the dominant effect of immunogen type in directing humoral profiles. LV1 separation was strongly driven by gp41-skewed immunity, due to the inclusion of gp41 in the Ad regimens but not included in VAX003 and only partially included in RV144. Furthermore, Abs targeting clade AE Ags (gp120 and V1V2) uniquely marked RV144 and VAX003 profiles, as subjects were immunized with clade AE–derived immunogens. Thus the antigen itself, rather than the vector/immunization regimen alone, was a critical determinant influencing vaccine-induced humoral profiles.
The second dimension identified additional features that further split the vaccine profiles, accounting for an additional 24% of the variance, contributing to an unexpected grouping and separation of RV144/Ad26 and VAX003/Ad5 profiles. This separation was primarily related to differences in IgG3 subclass and V1V2 levels, which scattered in multidimensional space more closely with RV144 and Ad26 profiles (Fig 3D). Therefore, markers previously associated with reduced risk of infection in RV144 co-segregated with the experimental Ad26 vaccine trial, that used a vector similar to the ones used in regimens recently shown to protect non-human primates from infection through non-neutralizing polyfunctional Abs (Barouch et al., 2015).
Thus, use of the LASSO and PLSDA, incorporating co-variation between features, identified key variables involved in classifying vaccine regimens and provided enhanced resolution of the specific Ab features associated with differentiating vaccine profiles, objectively identifying novel correlates of Ab-mediated protection.
Correlation networks highlight distinct humoral relationships
We next aimed to gain insights into relationships between features contributing to differences among vaccine-induced polyclonal profiles, adapting correlation network analysis tools commonly used in the transcriptomics field. The resulting network models revealed remarkably different Ab co-regulation interactions among the vaccine regimens, providing novel insights into the specific Ab features that may contribute to unique vaccine effector profiles.
VAX003 exhibited the most interconnected network, comprised of four dense subnetworks (Fig 4A). The most prominent subnetworks included an unusual tightly tethered mixture of IgG2 and IgG3 responses that are rarely co-selected (Chaudhuri and Alt, 2004; Chaudhuri et al., 2007), pointing to the induction of a non-coherent poorly coordinated functional response (Chung et al., 2014b). Interestingly, all Fc effector functions were connected to a third subnetwork, consisting largely of IgG1 and bulk IgG responses specific for a broad array of Ags that was unexpectedly connected to the fourth, IgG4 subnetwork, which have been shown to actively compete for immune complex occupancy and dampen Ab function (Chung et al., 2014b). Thus, the VAX003 network exhibited linked IgG1/IgG4 responses staggered next to a dense IgG2/IgG3 cluster, highlighting the peculiar subclass co-selection profiles driven by the non-protective VAX003 strategy.
Figure 4. Correlation networks of vaccine trial-elicited humoral immune responses probe immune complex dynamics.
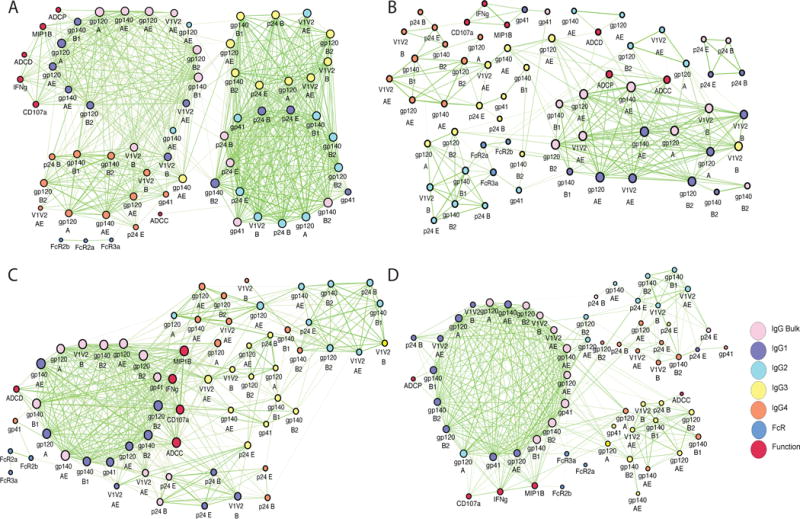
Correlation networks were generated for VAX003 (A), RV144 (B), HVTN204 (C), and IPCAVD001 (D). Each node (circle) represents either a biophysical feature or an effector function. Nodes are connected with an edge (line) if they are significantly correlated. The different Ab isotypes are identified by different colors as indicated. Edge thickness and color intensity of the connecting lines are directly proportional to statistical significance and edge weight, respectively (thicker and brighter network interactions represent a stronger correlation). The size of each node is directly proportional to its degree of connected-ness (ie. the number of features to which that node is connected). See also Figure S1 and Tables S1 and S2.
While less prominent clusters emerged in the RV144 network model (Fig 4B), ADCP, ADCD, and ADCC were largely tethered to a network of gp120-, gp140- or V1V2-specific IgG1 and/or IgG3 responses. The V1V2B–specific IgG3 response was highly associated with the large IgG1 network, suggesting that high IgG3 V1V2B-specific responses acts as a critical surrogate of a coordinated IgG3 and IgG1 response. The total IgG V1V2AE response was directly tethered to both ADCP and ADCC, suggesting that this specific V1V2-response may play an influential role in driving Ab functionality. IgG3 V1V2B and IgG3 V1V2AE responses were not directly correlated, suggesting that these V1V2 responses may represent disparate humoral immune responses rather than a single cross-reactive response. Moreover, because depletion of IgG3 only results in 30% reduction in Ab functionality (Chung et al., 2014b) linked to the networks, it is likely that IgG3 responses may serve as a surrogate for a subpopulation of vaccine-induced IgG1 Abs that direct the polyfunctional Ab responses observed in RV144.
The HVTN204 (DNA/Ad5) network (Fig 4C) contained a highly connected subnetwork, with multiple tethers to less well-connected subnetworks of additional Ab subclasses. The dominant subnetwork consisted of IgG1 Env- and V1V2-specific responses with ADCC, ADCD, and NK cell responses tightly intercalated within the subnetwork, sandwiched between IgG1 and IgG3 responses. However, ADCP did not appear in the network. This exclusion of ADCP suggests that Ad5 and/or DNA may preclude the induction of phagocytic Ab responses, which have been linked to protection from SIV acquisition (Barouch et al., 2013b).
Conversely, the vaccine profile induced by the experimental IPCAVD001 (Ad26) exhibited a nearly single densely connected network tethered to Ab functions and Fc-receptor binding activity (Fig 4D). The large network consisted of a tight grid of related bulk IgG/IgG1 responses, while IgG2, IgG3, and IgG4 formed sparse external clusters, including a less functional, interconnected IgG2/IgG4 cluster (top right). The clear linkages between Ab functions and IgG1 features, including an IgG V1V2–driven ADCP response, further supports the potential role of IgG3 as a surrogate of a highly effective, polyfunctional IgG1 response.
Overall, these network analyses point to unique relationships between all features and functions among the four vaccine trials, which both may aide in the identification of “desirable” Ab networks delineating specific biophysical Ab feature/function relationships that are associated with protective immunity and may help identify mechanisms underlying correlates, such as the association of IgG3 and V1V2 features with reduced risk of HIV infection by RV144.
System serology analysis of interactions between RV144 surrogates of reduced risk of infection
Systems Serology approaches can complement existing methods for identifying predictive mechanism(s) of protective immunity. While logistic regression involves stepwise evaluation of strongly correlated individual variables, Systems Serology approaches can additionally identify relationships between Ab features that are predictive of protection. Toward this purpose, we next examined RV144 profiles segregating with known correlates of reduced risk of infection. Thus, we dissected two Ab features (IgG/IgG3 V1V2), that were previously positively associated with reduced acquisition in the RV144 case:control analysis, in our cohort of uninfected vaccinees. Importantly, IgA levels were also included in this analysis, due their implicated role as correlates of risk (Haynes et al., 2012). Profiles were then compared between “responders” (top 33% for each correlate of reduced risk (Rerks-Ngarm et al., 2009; Zolla-Pazner et al., 2014)) or “non-responders”. The IgG V1V2 responder profile (Zolla-Pazner et al., 2014) was driven by 16 features (Fig 5A, 5B), including elevated V1V2-responses and a polyfunctional Fc effector profile linked to higher Ab-dependent NK cell degranulation (i.e., CD107a and IFNγ expression), ADCP, and ADCC. Conversely, the non-responders exhibited elevated gp120-specific IgA and increased binding to FCGR2B, the sole inhibitory Fcγ receptor, both features that have been previously associated with antagonism of Fc effector activity (Tomaras et al., 2013; White et al., 2014).
Figure 5. Identification of V1V2high-associated signatures within RV144 vaccine responses.
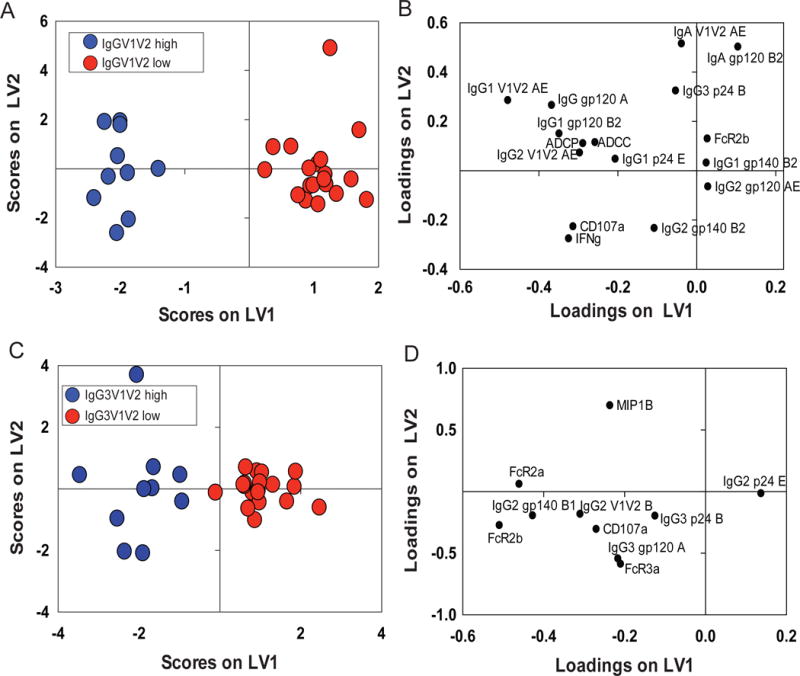
(A) RV144 vaccinees were classified within the IgG V1V2AEhigh (red) (top 30%) or IgG V1V2AElow (blue) groups. (A) LASSO identified a profile of 16 features that differentiated the two groups with 100% calibration and 80% cross-validation accuracy. The loadings plot (right panel) illustrates the features that separated IgG V1V2AEhigh or IgGV1V2AElow responders. Together LV1 and LV2 captured 33% of the X variance and 94% of the Y variance, respectively. (B) The same analysis was repeated for RV144 vaccinees classified as IgG3 V1V2high/IgG3V1V2low (B) with 92% cross-validation and 100% calibration accuracy. (D) LASSO identified a signature of 10 features that best separated these two groups. Together LV1 and LV2 captured 39% of the variance in X and 84% of the variance in Y, respectively. See also Tables S1 and S2.
Similar analysis of the IgG3 V1V2 correlate of reduced risk pointed to 10 Ab features that distinguished responder/non-responder profiles (Fig 5C, 5D), marked by increased broad Fcγ receptor binding among responders, particularly to activating FCGR2A, involved in ADCP, and to FCGR3a, critical for NK cell degranulation and chemokine secretion. Surprisingly, IgA was not selected as a negative predictor of the IgG3 V1V2 responder profile. These findings confirm that IgG V1V2 (Fig 5A) responders exhibit a balanced polyfunctional profile, while IgG3 V1V2 responders (Fig 5B) possessed Abs selectively enhanced for binding to FCGR2A associated with ADCP that has been linked to protection in NHP (Barouch et al., 2013b).
Defining integrative signatures of protective humoral immune profiles in RV144
Finally, to assess whether our approach could provide enhanced resolution of mechanism(s) of potential reduced risk of infection in the RV144 trial, we next analyzed data from the case:control study (Haynes et al., 2012). Specifically, data characterizing distinct Ab subclass levels targeting multiple vaccine Ags and functions comparable to those included in our original profiling data were included in the analysis. PLSDA using data from all cases and controls separated placebos from vaccinees, as expected, along LV1 (Fig 6A). In contrast, PLSDA of vaccinees alone was unable to separate the 40 infected from 201 uninfected vaccinees included in the case:control analysis (Fig S1A, B). Similarly, network analyses showed only modest differences between vaccinated cases and controls (Fig S1C, D), likely related to the fact that it is unclear which uninfected vaccinees were actually exposed and protected.
Figure 6. Defining novel signatures of protection in the RV144 case:control data.
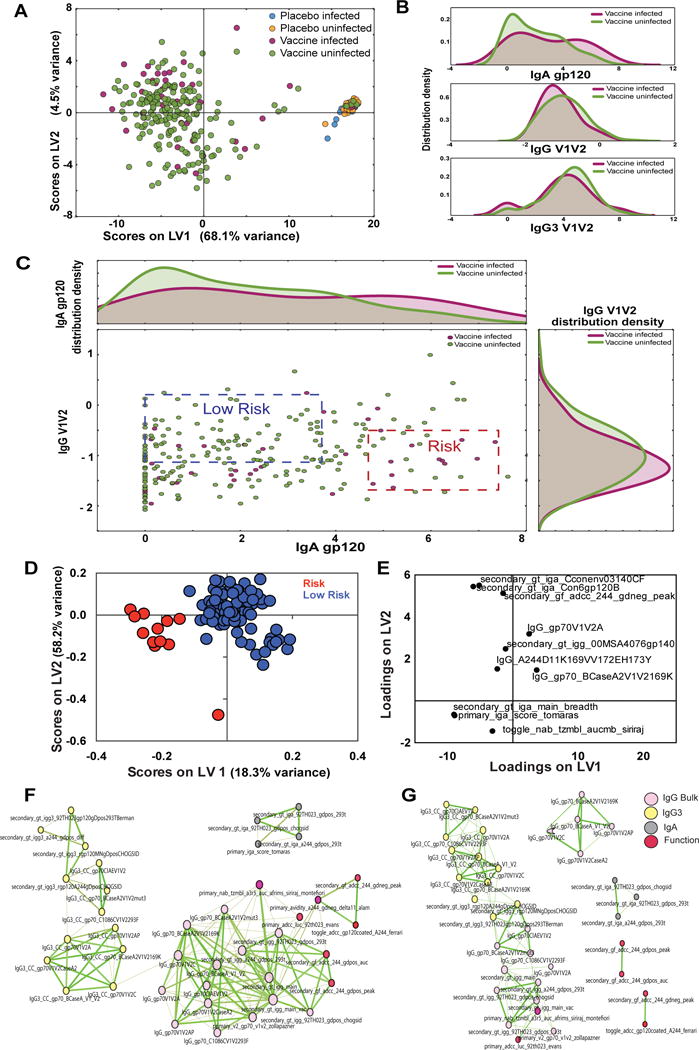
(A) The PLSDA shows the distribution of all case:control data including all infected and uninfected placebos as well as infected and uninfected vaccinees using 101 humoral features (described in Table S3). LV1 accounted for 68.1% of all variance, separating most placebos from the vaccines, while LV2 only contributed to 4.5% of the variance. (B) Further insights into the distribution of IgA gp120, IgG V1V2 and IgG3 V1V2 levels were analyzed using histograms demonstrating unique multi-modal differences in feature distribution among the infected and uninfected vaccinees. (C) The scatter plot, in the central panel, represents the bivariate distribution of IgA gp120 and IgG V1V2 in the vaccines, and is framed by the histogram distributions for uni-dimensional reference. The blue and red dash-lined boxes represent quadrants within the data that comprise of the fewest cases:controls (low risk, blue) or the highest ratio of cases:controls (high risk, red). (D and E) LASSO and PLSDA identified 9 features that split low and high risk profile separation with 97.8% accuracy in cross-validation. Together LV1 and LV2 captured 70.4% of the X variance and 30.1% of the Y variance, respectively. Correlation networks were generated for both the low risk (F) and high risk (G) groups. See also Figures S2 and S3, and table S3.
To address this complication, we defined groups representing extreme profiles based on known correlates of risk (Haynes et al., 2012). Given that the IgG3 and IgG V1V2 levels were highly correlated (Fig S2), we elected to focus on the IgG V1V2 and IgA relationship due to the intriguing relationships found for these two parameters in the non-case:control data (Fig 5A). Two sets of samples were identified: 1) a region containing the greatest ratio of uninfected:infected vaccinees was classified as the “low-risk” group (blue box, P=28%, p-value=0.0088) and 2) the area that contained the lowest ratio of uninfected:infected vaccinees was classified as the “high-risk” group (red box, P=-26%, p-value=0.0003). As expected, the lowest frequency of infections were observed in the IgG V1V2high/IgAlow region of the plot, and the highest frequency of cases were observed among the IgG V1V2low/IgAhigh groups. PLSDA analyses clearly separated these two groups (Fig 6D), with the “low-risk” group largely associated with features (Fig 6D positive loadings) that mark high IgG responses against the V1V2A scaffold as well as the V1V2-169K scaffold, corresponding to Ab responses against the viral variant able to evade the vaccine response among the infected vaccinees (Rolland et al., 2012).
Correlation networks further pointed to distinct profiles between the two groups. Three subnetworks were observed in the “low-risk” case:controls, an independent small network of IgA features and two larger linked clusters including: 1) all IgG3 features and 2) IgG responses tethered to Ab functional features (Fig 6E). These two clusters were linked by a single IgG3 and IgG response directed the same V1V2C scaffold, which was linked to all other V1V2 scaffold responses. This again suggests that the IgG3 response may be a surrogate of a highly functional IgG1 response more directly involved in modulating Ab functionality. Conversely, the “risk” group exhibited five clusters, of which four were small groups that appeared to form relationships independent of all the IgG3 features. One of the small clusters, separate from IgG3 and all functions, included several IgG V1V2 responses, highlighting a unique structure of the humoral response among the “risk” group. By contrast, all IgG3 features were tightly inter-connected and directly tethered to IgG features and the primary ADCC and neutralization results but not to the secondary ADCC features.
These findings indicate a mis-coordinated IgG/IgG3 V1V2 response largely separate from Ab function in the vaccinees that went on to become infected, whereas IgG/IgG3 V1V2 responses were well integrated within the network profile in vaccinees with reduced correlates of risk (i.e., IgG V1V2high/IgAlow). Even though many of the desirable features, in particular poly-functional responses identified in Fig 5, were not available for analysis, these data highlight the IgG V1V2 responses that likely drive protective immunity.
Discussion/Conclusion
Because the humoral immune response consists of waves of B cell responses that progressively induce higher affinity, broadly targeting, and functionally enhanced complexes of Abs poised to eliminate a pathogen, we aimed to develop a multivariate approach that could capture the complexity of interactions between antibodies at unprecedented depths. The Systems Serology approach described here, not only identified features reported in previous correlates analyses, including elevated IgG3 responses in RV144 (Chung et al., 2014b; Yates et al., 2014) and Ab binding to V1V2 (Zolla-Pazner et al., 2014), it also pointed to largely indirect connections between V1V2 IgG or IgG3 responses and Ab function (ADCC, ADCP, and ADCD) in vaccinees (Fig 4B) and the low-risk RV144 case:control samples (Fig 6F). Instead, vaccine-specific IgG1 responses were largely directly tethered to Ab function (Fig 4B). This suggests that the IgG3 “protective” signatures may either represent a surrogate of an effective Ab response or only contribute in combination with multiple other Ab features (eg. IgG1) to induce antiviral activity. Along these lines, while depletion of IgG3 Abs from RV144 vaccinees resulted in a significant loss of ADCP and ADCC activity, the activity was not completely depleted with the removal of this subclass of Abs (Chung et al., 2014b), suggesting that IgG3 Abs alone do not mediate the activity in polyclonal R144 sera and that function was also mediated by Abs remaining in the depleted purified IgGs. Therefore, the induction of IgG3 responses in RV144 may mark the coordinated production of highly functional IgG1 responses that may be functionally enhanced through altered IgG1 glycosylation, known to impact Fc-receptor affinity (Chung et al., 2014a), rather than subclass selection differences alone (Chung et al., 2014a; Hristodorov et al., 2013). Thus together with the IgG3 Abs, these IgG1 Abs may form highly functional immune complexes that are able to rapidly and effectively clear the virus or infected cells.
Interestingly, while vaccine-induced IgA responses were associated with enhanced risk of infection (Haynes et al., 2012), IgA emerged as an antagonist of the IgG V1V2 (Fig 5A, B) response but not the IgG3 response in the PLSDA analyses (Fig 5C, D). Furthermore, IgA responses were not connected to any of the subnetworks containing functional responses identified in the network analyses, suggesting that IgA responses may serve as a marker of a deregulated or less functional humoral immune response rather than a direct antagonist of protective humoral immune responses. Thus, while it is certain that pre-incubation of Ag with IgA monoclonal Abs may prevent IgG1 and IgG3 monoclonal Ab binding (Tomaras et al., 2013), it does not appear that these responses were directly co-induced (Fig 6). Moreover, given that the infected vaccinees exhibited both the lowest and the highest levels of IgA responses (Fig 6B), it is unlikely that IgA responses directly contributed to impaired humoral immune protection. Likewise, monoclonal therapeutics generated as IgAs exhibit potent cytotoxicity and clearance of tumor targets through Fcα receptors expressed on effector cells (e.g., (neutrophils and macrophages) (Black et al., 1996; Dechant and Valerius, 2001) and have been recently linked to protection from SHIV challenge (Watkins et al., 2013). Thus, future studies may aim to define the vaccine strategies that most effectively co-select a highly functional blood IgG response and a highly effective IgA response that may collectively prevent infection at the portal of entry.
Protection from infectious diseases, like HIV, will likely require the targeted containment of viral replication/dissemination at the site of infection. Along these lines, HIV is transmitted across mucosal barriers, where FcγR2-expressing monocytes/macrophages are abundant (Brown and Mattapallil, 2014; Zigmond and Jung, 2013). Moreover, ADCP activity was present in the RV144, VAX003, and IPCAVD001 networks (Fig 4), but was not observed in the HVTN204 network (Fig 4C) that was highly skewed to the elicitation of NK cell–mediated activities. Conversely, ADCP was tightly tethered within the RV144 and IPCAVD001 networks (Fig 4), was enhanced in the high V1V2 IgG3/IgG1 RV144 vaccinees (Fig 5), and was previously associated with protection in NHP (Barouch et al., 2013b). Thus these results raise the possibility that ADCP may represent a critical function, within polyfunctional Ab profiles, required for protection from mucosal transmission.
Beyond HIV, these vaccine-profiling approaches have broad applications and can aid in vaccine design efforts against many of the deadliest global pathogens for which immune correlates of protection have yet to be elucidated. For example, recent clinical evidence suggests that Abs present in Ebola virus–infected convalescent immune sera contribute to improved clinical outcomes in infected patients (Kreil, 2015; Lyon et al., 2014) and recently vesicular stomatitis virus (VSV) vaccination has been shown to drive robust humoral immune responses (Regules et al., 2015) that provide protection from infection (Henao-Restrepo et al., 2015). However, the specific mechanism(s) by which Abs provide protection remains unclear. Yet, a non-neutralizing monoclonal Ab, 13c6, has been shown to provide protection from infection in an Fc-dependent manner (Olinger et al., 2012), suggesting that non-neutralizing Ab functions contribute to antiviral immunity Thus, similar to the application of Systems Serology for the evaluation of HIV vaccine responses, the application of a Systems Serology–guided dissection of natural humoral immune profiles that emerge in Ebola virus survivors and vaccinees may provide insights into the immunological correlates and mechanisms of protection that may help guide future vaccine efforts.
Thus, in this manuscript, Systems Serological profiling provides a novel approach for the dissection of four HIV vaccine regimen profiles at unprecedented depths and a framework for dissecting the immune profiles that segregate with previously defined correlates of risk in efficacy studies. Thus, while Systems Serology complements traditional multivariate approaches aimed at defining independent predictors of vaccine efficacy, aiding in the identification of Ab function/feature relationships that track with protective humoral immune profiles. Thus these relational tools provide an additional powerful method for comparing immune profiles of different vaccine groups/outcomes to provide greater mechanistic insights underlying the relationships of features that may contribute to immune control. While this analysis included 64 humoral features, many other features can be collected, including measures of neutralization, affinity, Fc-glycosylation, etc. Moreover, these techniques may be expanded to examine the protective/immunopathological role of Abs in non-infectious disease settings including malignancies and/or autoimmunity, as well as how Abs may differ among genders, ethnicity and age. Thus, this study lays the groundwork for the evaluation, deep characterization, and comparison of polyclonal vaccine profiles for many future vaccines for which correlates of protective immunity are still elusive.
Experimental Procedures
Vaccine Samples
RV144 (Rerks-Ngarm et al., 2009): Plasma samples from 30 vaccinated subjects at week 26 (2 weeks post final vaccination) were provided by the MHRP. RV144 case: control study data were provided by the RV144 study team. Serum samples from 30 vaccinated subjects at month 30.5 (two weeks post final vaccination) were provided by GSID. HVTN204 (Churchyard et al., 2011): Serum samples from 30 vaccinated subjects taken at 2 weeks post final vaccination were provided by the HVTN. IPCAVD 001 (Barouch et al., 2013a): Serum samples from 30 vaccine subjects taken at 2 weeks post final vaccination were provided by Dan Barouch. Detailed descriptions of each vaccine are included in Supplementary Materials.
Purifying bulk IgG
IgG was purified from all vaccine plasma and serum samples using Melon Gel columns according to the manufacturer’s instructions (Thermo Scientific), and the concentration was calculated using a human IgG ELISA kit (MABTECH).
Antibody-Functional Profiling
The following assays were performed to functionally profile the Fc-effector functions of all vaccine Abs. In order to assess Ab-dependent cellular phagocytosis (ADCP), a THP-1 based ADCP assay was performed as previously described (Ackerman et al., 2011). Ab-dependent cellular cytotoxicity (ADCC) was assayed using a modified rapid fluorescent ADCC (RFADCC) as previously described (Gomez-Roman et al., 2006) (Chung et al., 2014b). Ab-dependent complement deposition (ADCD) was assessed via the measurement of complement component C3b deposition on the surface of target cells. Ab-dependent NK cell degranulation and cytokine/chemokine secretion was measured using the CEM-NKr CCR5+ T lymphoblast cell line pulsed with vaccine-specific gp120 (60 mg/ml), as previously described (Chung et al., 2014b). Detailed methods of each functional assay is described in Supplementary Materials.
Antibody Biophysical profiling
The following assays were performed to assess the biophysical profile of each of the vaccine Ab samples. Ab affinity for FCGRs was determined using surface plasmon resonance as previously described (Chung et al., 2014a). While a customized Luminex isotype assay was used to quantify the relative concentration of each Ab isotype to a panel of HIV-specific Abs. Detailed methods of each of these profiling tools are included in the supplementary Materials
Identification of vaccine-specific signatures with LASSO and PLSDA
The minimum signature of Ab features and functional parameters useful for differentiating vaccine groups was identified using the Least Absolute Shrinkage and Selection Operator (LASSO) method (Tibshirani, 1997) and implemented using Matlab software (aversion 2014a, Mathworks, Natick, MA). Partial least square discriminant analysis (PLSDA) (Arnold et al., 2015; Lau et al., 2011) assessed the predictive ability of LASSO-selected biomarkers for classifying vaccine groups. Detailed description of validation and quality control for this analysis are included in the Supplementary Materials.
Network interactions
Networks were constructed based on the pairwise correlation coefficients between all biophysical features and functional responses. Edges between nodes are weighted using significant correlation coefficients, ρij, after correcting for multiple comparisons (Benjamini-Hochberg q-value < 0.05, testing the hypothesis of zero correlation) as follows:
with α = 6.
To assess the significance of the variable groupings observed in the network, we calculate the network clustering coefficient for the original network and for 100 randomized networks. Random networks are generated by randomly swapping edges while preserving the degree of all nodes (degree preserving edge shuffle) (Fig S2).
RV144 case: control study data processing
RV144 case: control study data included results from 281 of patients, including 101 Ab features and functional parameters. Specific features used within this analysis are documented in Table S3. Subjects were categorized into four groups including: placebo infected, placebo uninfected, vaccine infected, and vaccine uninfected for all analyses. Because IgG3 and IgG V1V2 levels were highly correlated (Fig S3), vaccinees were classified based on their IgG V1V2 and IgA levels. A “high” and “low“ risk group was defined as the region of the IgG V1V2 vs IgA plot that contained the fewest cases or the fewest controls, respectively, in a mutually exclusive manner. The percentage difference between infected versus uninfected vaccinees was defined as,
where for any given region r the percentage of infected people Ir over the infected population It was calculated as well as for uninfected individuals. The enriched region, with the highest P was defined as the “risk” group, whereas the region with the lowest P was defined as the “low risk” group. Fisher’s exact test was used to estimate the significance of the enriched region from a null hypothesis.
Supplementary Material
Acknowledgments
The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: CEM.NKR-CCR5. We would like to thank the: 1) NIH National Institute of Allergy and Infectious Diseases (NIAID) and the NIAID-funded HIV Vaccine Trials Network for providing specimens for the HVTN204 vaccine trial; 2) the Military HIV Research Program (MHRP) Diseases for specimens from the RV144 vaccine trial; 3) Global Solutions for Infectious Disease (GSID) for samples from the VAX003 vaccine trial; and 4) Dan Barouch for specimens from the experimental Ad26 vaccine trial. We would like to thank the RV144 study team for permission to include the case: control data in our manuscript, and would like to specifically thank Drs. Peter Gilbert and Allan DeCamp and Elizabeth Heger for their assistance in RV144 case:control data collection. The opinions herein are those of the authors and should not be construed as official or representing the views of the U.S. Department of Defense or the Department of the Army. This work was supported by the National Institute of Health (R01 AI080289), the Bill and Melinda Gates Foundation CAVD (OPP1032817: Leveraging Antibody Effector Function) and the Ragon Institute of MGH, MIT and Harvard. Hanneke Schuitemaker and Maria G. Pau are employees of Crucell Holland B.V., a Janssen pharmaceutical company of Johnson & Johnson and shareholders of Johnson & Johnson.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
Project planning was performed by AWC, MPK, KBA, AEM, DL, WHY, GA. Experimental work was performed by AWC, MKS, CL, AEM, MH, Data analysis was performed by AWC, MPK, KBA, WHY. Vaccine trials were designed and conducted by HS, MGP, LRB, JHK, NLM, DHB, Manuscript composition was performed by AWC, MPK, KBA, WHY, TJS, NF, HS, MEA, JM, HS, DL, GA
References
- Ackerman ME, Alter G. Opportunities to exploit non-neutralizing HIV-specific antibody activity. Current HIV research. 2013;11:365–377. doi: 10.2174/1570162x113116660058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman ME, Moldt B, Wyatt RT, Dugast AS, McAndrew E, Tsoukas S, Jost S, Berger CT, Sciaranghella G, Liu Q, et al. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. Journal of immunological methods. 2011;366:8–19. doi: 10.1016/j.jim.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold KB, Burgener A, Birse K, Romas L, Dunphy LJ, Shahabi K, Abou M, Westmacott GR, McCorrister S, Kwatampora J, et al. Increased levels of inflammatory cytokines in the female reproductive tract are associated with altered expression of proteases, mucosal barrier proteins, and an influx of HIV-susceptible target cells. Mucosal immunology. 2015 doi: 10.1038/mi.2015.51. [DOI] [PubMed] [Google Scholar]
- Barouch DH, Alter G, Broge T, Linde C, Ackerman ME, Brown EP, Borducchi EN, Smith KM, Nkolola JP, Liu J, et al. Protective efficacy of adenovirus-protein vaccines against SIV challenges in rhesus monkeys. Science. 2015 doi: 10.1126/science.aab3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Liu J, Peter L, Abbink P, Iampietro MJ, Cheung A, Alter G, Chung A, Dugast AS, Frahm N, et al. Characterization of humoral and cellular immune responses elicited by a recombinant adenovirus serotype 26 HIV-1 Env vaccine in healthy adults (IPCAVD 001) The Journal of infectious diseases. 2013a;207:248–256. doi: 10.1093/infdis/jis671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Stephenson KE, Borducchi EN, Smith K, Stanley K, McNally AG, Liu J, Abbink P, Maxfield LF, Seaman MS, et al. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell. 2013b;155:531–539. doi: 10.1016/j.cell.2013.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black KP, Cummins JE, Jr, Jackson S. Serum and secretory IgA from HIV-infected individuals mediate antibody-dependent cellular cytotoxicity. Clinical immunology and immunopathology. 1996;81:182–190. doi: 10.1006/clin.1996.0175. [DOI] [PubMed] [Google Scholar]
- Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, Ravetch JV. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell. 2014;158:1243–1253. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D, Mattapallil JJ. Gastrointestinal tract and the mucosal macrophage reservoir in HIV infection. Clinical and vaccine immunology: CVI. 2014;21:1469–1473. doi: 10.1128/CVI.00518-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nature reviews Immunology. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Advances in immunology. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- Chung AW, Alter G. Dissecting the Antibody Constant Region Protective Immune Parameters in HIV Infection. Future Virol. 2014;9:397–414. [Google Scholar]
- Chung AW, Crispin M, Pritchard L, Robinson H, Gorny MK, Yu X, Bailey-Kellogg C, Ackerman ME, Scanlan C, Zolla-Pazner S, et al. Identification of antibody glycosylation structures that predict monoclonal antibody Fc-effector function. Aids. 2014a;28:2523–2530. doi: 10.1097/QAD.0000000000000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung AW, Ghebremichael M, Robinson H, Brown E, Choi I, Lane S, Dugast AS, Schoen MK, Rolland M, Suscovich TJ, et al. Polyfunctional Fc-effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines. Science translational medicine. 2014b;6:228–238. doi: 10.1126/scitranslmed.3007736. [DOI] [PubMed] [Google Scholar]
- Churchyard GJ, Morgan C, Adams E, Hural J, Graham BS, Moodie Z, Grove D, Gray G, Bekker LG, McElrath MJ, et al. A phase IIA randomized clinical trial of a multiclade HIV-1 DNA prime followed by a multiclade rAd5 HIV-1 vaccine boost in healthy adults (HVTN204) PloS one. 2011;6:e21225. doi: 10.1371/journal.pone.0021225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant M, Valerius T. IgA antibodies for cancer therapy. Critical reviews in oncology/hematology. 2001;39:69–77. doi: 10.1016/s1040-8428(01)00105-6. [DOI] [PubMed] [Google Scholar]
- DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nature medicine. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roman VR, Florese RH, Patterson LJ, Peng B, Venzon D, Aldrich K, Robert-Guroff M. A simplified method for the rapid fluorometric assessment of antibody-dependent cell-mediated cytotoxicity. Journal of immunological methods. 2006;308:53–67. doi: 10.1016/j.jim.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. The New England journal of medicine. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Restrepo AM, Longini IM, Egger M, Dean NE, Edmunds WJ, Camacho A, Carroll MW, Doumbia M, Draguez B, Duraffour S, et al. Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster-randomised trial. Lancet. 2015;386:857–866. doi: 10.1016/S0140-6736(15)61117-5. [DOI] [PubMed] [Google Scholar]
- Hessell AJ, Hangartner L, Hunter M, Havenith CE, Beurskens FJ, Bakker JM, Lanigan CM, Landucci G, Forthal DN, Parren PW, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- Hristodorov D, Fischer R, Linden L. With or without sugar? (A)glycosylation of therapeutic antibodies. Molecular biotechnology. 2013;54:1056–1068. doi: 10.1007/s12033-012-9612-x. [DOI] [PubMed] [Google Scholar]
- Jegerlehner A, Schmitz N, Storni T, Bachmann MF. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J Immunol. 2004;172:5598–5605. doi: 10.4049/jimmunol.172.9.5598. [DOI] [PubMed] [Google Scholar]
- Joos C, Marrama L, Polson HE, Corre S, Diatta AM, Diouf B, Trape JF, Tall A, Longacre S, Perraut R. Clinical protection from falciparum malaria correlates with neutrophil respiratory bursts induced by merozoites opsonized with human serum antibodies. PloS one. 2010;5:e9871. doi: 10.1371/journal.pone.0009871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl S, Loo LS. Protection of neonatal mice against herpes simplex virus infection: probable in vivo antibody-dependent cellular cytotoxicity. J Immunol. 1982;129:370–376. [PubMed] [Google Scholar]
- Kohl S, Loo LS, Pickering LK. Protection of neonatal mice against herpes simplex viral infection by human antibody and leukocytes from adult, but not neonatal humans. Journal of immunology. 1981;127:1273–1275. [PubMed] [Google Scholar]
- Kreil TR. Treatment of ebola virus infection with antibodies from reconvalescent donors. Emerging infectious diseases. 2015;21:521–523. doi: 10.3201/eid2103.141838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau KS, Juchheim AM, Cavaliere KR, Philips SR, Lauffenburger DA, Haigis KM. In vivo systems analysis identifies spatial and temporal aspects of the modulation of TNF-alpha-induced apoptosis and proliferation by MAPKs. Science signaling. 2011;4:ra16. doi: 10.1126/scisignal.2001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon GM, Mehta AK, Varkey JB, Brantly K, Plyler L, McElroy AK, Kraft CS, Towner JS, Spiropoulou C, Stroher U, et al. Clinical care of two patients with Ebola virus disease in the United States. The New England journal of medicine. 2014;371:2402–2409. doi: 10.1056/NEJMoa1409838. [DOI] [PubMed] [Google Scholar]
- Olinger GG, Jr, Pettitt J, Kim D, Working C, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, et al. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18030–18035. doi: 10.1073/pnas.1213709109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osier FH, Feng G, Boyle MJ, Langer C, Zhou J, Richards JS, McCallum FJ, Reiling L, Jaworowski A, Anders RF, et al. Opsonic phagocytosis of Plasmodium falciparum merozoites: mechanism in human immunity and a correlate of protection against malaria. BMC medicine. 2014;12:108. doi: 10.1186/1741-7015-12-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van Griensven F, Hu D, Tappero JW, Choopanya K. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. The Journal of infectious diseases. 2006;194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nature immunology. 2011;12:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regules JA, Beigel JH, Paolino KM, Voell J, Castellano AR, Munoz P, Moon JE, Ruck RC, Bennett JW, Twomey PS, et al. A Recombinant Vesicular Stomatitis Virus Ebola Vaccine - Preliminary Report. The New England journal of medicine. 2015 doi: 10.1056/NEJMoa1414216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. The New England journal of medicine. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, deCamp AC, Carrico C, Menis S, Magaret CA, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. The lasso method for variable selection in the Cox model. Statistics in medicine. 1997;16:385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Tomaras GD, Ferrari G, Shen X, Alam SM, Liao HX, Pollara J, Bonsignori M, Moody MA, Fong Y, Chen X, et al. Vaccine-induced plasma IgA specific for the C1 region of the HIV-1 envelope blocks binding and effector function of IgG. Proceedings of the National Academy of Sciences of the United States of America. 2013 doi: 10.1073/pnas.1301456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield KL, Swenson DL, Olinger GG, Kalina WV, Aman MJ, Bavari S. Ebola virus-like particle-based vaccine protects nonhuman primates against lethal Ebola virus challenge. The Journal of infectious diseases. 2007;196(Suppl 2):S430–437. doi: 10.1086/520583. [DOI] [PubMed] [Google Scholar]
- Watkins JD, Sholukh AM, Mukhtar MM, Siddappa NB, Lakhashe SK, Kim M, Reinherz EL, Gupta S, Forthal DN, Sattentau QJ, et al. Anti-HIV IgA isotypes: differential virion capture and inhibition of transcytosis are linked to prevention of mucosal R5 SHIV transmission. Aids. 2013;27:F13–20. doi: 10.1097/QAD.0b013e328360eac6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AL, Beers SA, Cragg MS. FcgammaRIIB as a key determinant of agonistic antibody efficacy. Current topics in microbiology and immunology. 2014;382:355–372. doi: 10.1007/978-3-319-07911-0_16. [DOI] [PubMed] [Google Scholar]
- Yates NL, Liao HX, Fong Y, deCamp A, Vandergrift NA, Williams WT, Alam SM, Ferrari G, Yang ZY, Seaton KE, et al. Vaccine-induced Env V1-V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Science translational medicine. 2014;6:228–239. doi: 10.1126/scitranslmed.3007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond E, Jung S. Intestinal macrophages: well educated exceptions from the rule. Trends in immunology. 2013;34:162–168. doi: 10.1016/j.it.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Zolla-Pazner S, deCamp A, Gilbert PB, Williams C, Yates NL, Williams WT, Howington R, Fong Y, Morris DE, Soderberg KA, et al. Vaccine-induced IgG antibodies to V1V2 regions of multiple HIV-1 subtypes correlate with decreased risk of HIV-1 infection. PloS one. 2014;9:e87572. doi: 10.1371/journal.pone.0087572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


