ABSTRACT
It is now possible to create individual-specific genetic scores, called genome-wide polygenic scores (GPS). We used a GPS for years of education (EduYears) to predict reading performance assessed at UK National Curriculum Key Stages 1 (age 7), 2 (age 12) and 3 (age 14) and on reading tests administered at ages 7 and 12 in a UK sample of 5,825 unrelated individuals. EduYears GPS accounts for up to 5% of the variance in reading performance at age 14. GPS predictions remained significant after accounting for general cognitive ability and family socioeconomic status. Reading performance of children in the lowest and highest 12.5% of the EduYears GPS distribution differed by a mean growth in reading ability of approximately two school years. It seems certain that polygenic scores will be used to predict strengths and weaknesses in education.
Introduction
The origins of most individual differences in diverse reading skills lie with genetic differences (twin heritability estimates > 50%) that help or hinder the process of learning to read (Olson, Keenan, Byrne, & Samuelsson, 2014). Genetic transmission explains entirely the extensive familial resemblance for reading ability (assortative mating r = ~0.40; parent-offspring r = ~0.20; Swagerman et al., 2015). Although family-based designs have contributed immensely to our understanding of individual differences in reading ability, it is of substantial interest to use DNA alone to quantify genetic effects related to reading performance. Over the past two decades, reading ability has also been extensively studied using various molecular genetics methods, which is reviewed in the following paragraphs.
It is the goal of the present study to aid the search for the genetic origins of reading using a new statistical genetic approach, the polygenic score method. This method makes it possible to create individual-specific genetic estimates that aggregate the effects of trait-associated DNA differences. The possibility of quantifying genetic risk and resilience for individuals has far-reaching implications for prediction and intervention to prevent reading difficulties, which have been shown to impact on academic achievement, occupational, and social outcomes, as well as mental health (Maughan, 1995).
Linkage studies
Although dramatic advances have been made recently in the molecular genetics of many behavioral traits (Knopik, Neiderhiser, DeFries, & Plomin, 2017), the field of reading has not as yet profited from these advances. This is surprising because, 20 years ago, reading was the first behavioral trait investigated by quantitative (continuous) trait linkage analysis, which studied many small pedigrees (usually just sibling pairs) in contrast to traditional linkage analyses that studied a few large pedigrees (Cardon et al., 1994). Since then, numerous studies using linkage analyses identified several chromosomal regions linked to reading ability with varying degrees of supportive evidence, including 1p35–1p36, 2p11–2p16, 2q22.3, 3p12–3q13, 6p22, 6q12-q15, 7q32, 11q13.4, 15q15–15q21, 18p11.2, and Xq26-Xq28 (Poelmans, Buitelaar, Pauls, & Franke, 2011; Schumacher, Hoffmann, Schmäl, Schulte-Körne, & Nöthen, 2007). Although subsequent studies identified a range of candidate genes located on various chromosomal regions, it has been difficult to replicate these findings (Bishop, 2015; Scerri & Schulte-Körne, 2010). Currently, only candidate genes DCDC2 and K1AA0319 at the DYX2 locus on chromosome 6p22 have shown some replication of associations with reading ability (Carrion-Castillo, Franke, & Fisher, 2013; Schumacher et al., 2007).
Genetic research on behavioral traits began to move away from linkage approaches, as this method can detect only genetic variants that have a large effect on a trait (Manolio et al., 2009). There are thousands of rare single-gene disorders in which a single genetic variant is necessary and sufficient to cause a disorder (Antonarakis & McKusick, 2000; Hamosh, Scott, Amberger, Bocchini, & McKusick, 2005), and linkage is powerful for identifying the chromosomal neighborhood of the culprit genes. Despite the identification of some rare genetic variants of large effect related to reading disability, it is now commonly accepted that complex traits like reading ability and common disorders like reading disabilities are influenced by many genetic variants each of small effect (Bishop, 2015; Gratten, Wray, Keller, & Visscher, 2014).
Association studies
For this reason, attempts to identify genetic variants responsible for heritability of complex traits and common disorders have switched from linkage to association, which is a more powerful approach for detecting small effects. Association studies test for correlations between alleles and a quantitative trait for unrelated individuals in the population, so that greater power requires only larger numbers of unrelated individuals, not special family pedigrees. As an example of association, a particular allele in a gene called FTO is associated with body weight (Frayling et al., 2007). This variant is a single nucleotide polymorphism (SNP; the most common form of genetic differences between individuals) with two alleles that differ by a single nucleotide base, A or T. More specifically, in comparison to individuals with a TT genotype, individuals with an AT and an AA genotype show an increase in weight of ~1.2 kg and ~2.4 kg on average, respectively. The correlation between the number of A alleles (0, 1, or 2) and body mass index is about 0.10 in multiple studies (e.g., Dina et al., 2007; Frayling et al., 2007). This means that this SNP association accounts for about 1% of the variance of body mass index. Although 1% might seem to be a small effect size, it is one of the largest effect sizes yet detected for the association between a single common genetic variant and a quantitative trait.
Genome-wide association
Because of the expense of genotyping large samples, early association studies had to resort to investigating a few “candidate” genes thought to be involved in pathways that affect the trait. However, in 2007, technological developments made it possible to genotype hundreds of thousands of DNA variants in an individual quickly and inexpensively using DNA arrays. This technological advance enabled genome-wide association (GWA), an atheoretical search for association across the entire genome (Hirschhorn & Daly, 2005). GWA has been successful in identifying thousands of associations for hundreds of continuous traits and disorders (Eicher et al., 2015). The most striking finding from GWA studies of quantitative traits is that most GWA effects account for less than 0.1% of the variance (Visscher, Brown, McCarthy, & Yang, 2012), which implies that many genetic variants of small effect contribute to heritability (polygenicity).
This finding of extreme polygenicity supports a fundamental principle of quantitative genetics that the heritability of complex traits and common disorders is caused by many genetic variants of small effect (Fisher, 1918). From this, it follows that thousands of DNA variants contribute to heritability (Chabris, Lee, Cesarini, Benjamin, & Laibson, 2015). For example, if the effect size of each associated genetic variant was 0.01%, 5,000 DNA variants would be needed to reach a heritability of 50%.
Extremely large sample sizes are required to detect such small effect sizes, especially to reach genome-wide significance that corrects for all the multiple testing involved. As with all null-hypothesis significance testing, there is a trade-off between power and effect size. Based on the number of independent SNPs in the whole genome that might be tested in a GWA study, the criterion for genome-wide significance corrected for multiple testing is a p value of less than .00000005 (Jannot, Ehret, & Perneger, 2015). This stringent threshold captures only the largest effect sizes even with huge sample sizes. The exemplar for GWA of quantitative traits is height, although height is much more heritable than most behavioral traits. A GWA of 250,000 individuals yielded 697 genome-wide significant associations (Wood et al., 2014). For reading, the largest GWA study to date, which had a sample size of ~5,000 individuals, yielded no genome-wide significant associations (Gialluisi et al., 2014; Luciano et al., 2013). Three other GWA studies of reading ability had even smaller samples and were also unsuccessful in identifying genome-wide significant associations (Eicher et al., 2013; Gialluisi et al., 2014; Meaburn, Harlaar, Craig, Schalkwyk, & Plomin, 2008).
Genome-wide polygenic scores
One goal for GWA is to identify “hits,” genome-wide significant associations, that can be used in bottom-up research tracing mechanisms from genes to brain to behavior. However, another goal is to predict genetic strengths and weaknesses for individuals. This top-down goal can be achieved by changing the focus of GWA from statistically significant “hits” to the pragmatic issue of prediction in independent samples. Predictive power can be greatly increased by aggregating genotypic effects across SNPs even if they do not individually reach genome-wide statistical significance (Palla & Dudbridge, 2015). Therefore, rather than just aggregating SNPs that reach genome-wide significance, a new development is to add SNPs as long as they add to the prediction of a trait in an independent sample (Dudbridge, 2013). From the hundreds of thousands SNPs measured in GWA studies, often tens of thousands of SNPs throughout the genome are used to create what we call a genome-wide polygenic score (GPS; Plomin & Deary, 2015). This is based on the idea that a GPS will be enriched for positive associations that do not reach genome-wide significance even though the GPS will include false-positive associations (Dudbridge, 2013; Wray et al., 2014). For example, in the GWA for height mentioned earlier, a GPS for height that aggregates all SNPs associated at a p value of < 5 × 10–5 (N SNPs ~1,900) accounts for 17% of the variance of height in independent samples (Wood et al., 2014). Here, we propose to label the variance explained by a GPS as GPS heritability, because heritability estimates derived through different statistical methods vary in their meaning and interpretation. GPS heritability refers to the phenotypic variance that is tagged by the variability of the genetic variants included in the GPS.
There are several labels to describe such polygenic scores, most of which involved the word “risk,” such as polygenic risk scores, but we prefer the term polygenic score because for many normally distributed quantitative traits it is unclear which direction of effect, if any, represents a risk (e.g., height, HDL cholesterol). Using the body weight example mentioned earlier, the A allele can be assigned a score of 1 so that the TT, AT, and AA genotypes are assigned values of 0, 1, and 2, respectively, with higher scores predicting greater body weight. These genotypic scores, scored in the direction of greater body weight, can be added across many SNPs. Similar to adding items on a test weighted by their factor loading, each SNP is weighted by its GWA regression weight.
GPS for years of education
As study sizes are likely to be too small, no GPS for reading has yet been created from the four GWA studies of reading. Very large GWA study sample sizes are required to generate powerful GPS in independent samples (Palla & Dudbridge, 2015). Until an adequately powered GWA study for reading is available as a basis for calculating a GPS, one way to begin is to create a GPS based on a trait genetically related to reading.
The largest GWA study conducted to date, which included ~300,000 individuals, has recently been reported for years of education and produced 74 genome-wide significant hits (Okbay et al., 2016). The large sample size was made possible because years of education is assessed in most GWA studies as a standard demographic variable, thus enabling aggregation across many studies. A GPS (EduYears) derived from this GWA study based on all measured SNPs accounted for up to 4% of the variance in years of education in independent samples (Okbay et al., 2016).
EduYears GPS can also be used as a “proxy” for variables related to educational attainment such as cognitive abilities (Rietveld et al., 2014). An earlier GWA study of years of education with a sample size of ~126,000 (Rietveld et al., 2013) showed the value of this approach for general cognitive ability (Rietveld et al., 2014) and tested educational achievement (Krapohl & Plomin, 2015). A GPS for educational attainment may be particularly useful for predicting reading ability, as reading performance and academic achievement are strongly associated. Reading ability assessed at the beginning of formal schooling is a substantial predictor of academic performance across the school years (Arnold & Doctoroff, 2003; Duncan et al., 2007; Herbers et al., 2012). This evidence suggests that reading performance is an important factor for academic success, and therefore a GPS for educational attainment should be closely related to reading ability. However, so far only one study has been published using EduYears GPS in the reading domain. This study investigated mean differences in reading abilities between ages 7–18 in individuals who were categorized into groups of high versus low polygenic scores, using the 2013 EduYears GPS (Belsky et al., 2016). In this study, students with high EduYears polygenic scores acquired reading skills earlier, developed reading abilities more rapidly, and peaked in their performance at a younger age than students with a low polygenic score (Belsky et al., 2016). These findings indicate that the use of EduYears GPS can be extended to investigate reading.
The use of a GPS provides novel advantages to the scientific study of reading ability. Twin and family studies provide population estimates of genetic and environmental influence on reading ability and have highlighted that genetic influence is substantial on individual differences in various components of reading (Knopik et al., 2017). However, these estimates refer to only average genetic influence in the population. Although they can predict average risk in a family, they cannot predict genetic propensities for individuals. GWA analysis could provide genetic estimates based on an individual’s genotype, but no adequately powered GWA studies of reading have as yet been reported. Rather than trait prediction, it is the aim of GWA studies to identify mechanisms and specific pathways. By remaining agnostic about the mechanisms, the top-down approach of a GPS focuses on individual prediction, which is greatly increased by accumulating the effects of many genetic variants. The availability of individuals’ genetic scores is an excellent research tool and offers new possibilities for prevention and intervention through early prediction of genetic risk.
The aim of the present study
In the current study, we assess the links between the latest EduYears GPS and diverse aspects of reading performance, such as reading efficiency and comprehension, across development at ages 7, 12, and 14 in a UK-representative sample of 5,825 unrelated individuals. The association between genetic effects on educational achievement and reading ability has been extensively studied in quantitative genetics, but here we test this association using polygenic scores.
Therefore, the main aim of this study was to test the extent to which individual-specific EduYears GPS estimates of years of education in adults predict reading performance across the school years, whether this prediction remains after controlling for general cognitive ability and family socioeconomic status (SES), and whether EduYears GPS differentially predicts distinct reading abilities or overall reading ability at different stages of development.
Methods
Sample
This study included the genotyped subsample (N = 5,825) of the multivariate longitudinal Twins Early Development Study (TEDS), which comprises only unrelated individuals (one twin per pair). TEDS recruited more than 15,000 twin pairs born in England and Wales between 1994 and 1996 (Haworth, Davis, & Plomin, 2013). The sample is representative of British families in ethnicity, family SES, and parental occupation (Haworth et al., 2013). The genotyped subsample is representative of UK census data at first contact (Selzam et al., 2016). The Institute of Psychiatry, Psychology and Neuroscience ethics committee (05.Q0706/228) granted project approval, and parental consent was obtained prior to data collection. Additional consent (75% of the genotyped subsample) was obtained from TEDS participants or their parents for access to their National Pupil Database (NPD) data.
Genotyping
AffymetrixGeneChip 6.0 SNP arrays were used to genotype 3,666 individuals of European ancestry at Affymetrix, Santa Clara (California, USA) based on DNA samples extracted from buccal cells. Genotypic data was generated at the Wellcome Trust Sanger Institute (Hinxton, UK) as part of the Wellcome Trust Case Control Consortium 2 (https://www.wtccc.org.uk/ccc2/). In addition, 3,497 other individuals of European ancestry were genotyped on HumanOmniExpressExome-8v1.2 arrays at the Molecular Genetics Laboratories of the Medical Research Council Social, Genetic Developmental Psychiatry Centre, based on DNA that was extracted from saliva samples. After quality control, 652,010 SNPs were available for AffymetrixGeneChip 6.0 genotypes and 524,177 SNPs for HumanOmniExpressExome genotypes. Imputation based on the Haplotype Reference Consortium (S. McCarthy et al., 2015) and Minimac3 1.0.13 (Fuchsberger, Abecasis, & Hinds, 2015; Howie, Fuchsberger, Stephens, Marchini, & Abecasis, 2012) was performed separately on both samples before merging genotype data obtained from both arrays (for full details, see Selzam et al., 2016). A total of 5,825 samples passed quality control, with 3,127 individuals genotyped on Affymetrix and 2,698 individuals genotyped on Illumina. To control for platform (Affymetrix vs. Illumina) and plate effects, we regressed the GPS used in this study on platform and plate data and used z-standardized residuals in subsequent analyses.
Measures
Twins Early Development Study (TEDS) measures
TEDS reading measures are described in greater detail elsewhere (Davis et al., 2014; Davis, Haworth, & Plomin, 2009; Harlaar, Spinath, Dale, & Plomin, 2005).
Reading efficiency
The Test of Word Reading Efficiency (Torgesen, Wagner, & Rashotte, 1999) was administered to TEDS participants via telephone at age 7 (M = 7.12, SD = 0.24) and 12 (M = 11.87, SD = 0.60). Test data and genotype data were available for 3,076 individuals at age 7 and 3,240 individuals at age 12. This test includes two subtests that assess accuracy and fluency in the pronunciation of written words (Sight Word Efficiency) and in the phonemic regulation of nonwords (Phonemic Decoding Efficiency). Because they correlate highly (r = .82 at age 7; r = .71 at age 12), we created a mean composite Test of Word Reading Efficiency score from the z-standardized subtests Sight Word Efficiency and Phonemic Decoding Efficiency at age 7 as well as age 12.
Reading efficiency was also assessed using an online version of the Woodcock-Johnson III Reading Fluency subtest (Woodcock, McGrew, & Mather, 2001), which was administered in a large web-based battery of cognitive tests at age 12 (M = 11.57, SD = 0.69; Haworth et al., 2007). A total of 3,825 individuals were genotyped and had test data available. This test consisted of a sequence of 98 simple, short written statements (e.g., “Most dogs can fly over the tops of mountains”). The participant indicated whether each statement was correct or incorrect.
Reading comprehension
We used a web-based reading subtest of the Peabody Individual Achievement Test (Markwardt, 1997) to measure children’s ability to understand the meaning of written text at age 12 (M = 11.57, SD = 0.69). Test data and genotypes were available for 3,851 individuals. In this test, the participant was presented with a sentence on the screen (e.g., “It is a red duck”), which was replaced by a set of four images after 20s. The participant was asked to choose the picture that matched the sentence most accurately within a time limit of 20s. This test included three practice items and 82 main items that increased in the level of difficulty throughout the assessment.
Reading comprehension was also tested at age 12 using a web version of the GOAL Formative Assessment in Literacy (Key Stage 3) (GOAL plc, 2002). Genotype data and GOAL test data (M = 11.57, SD = 0.69) were available for 3,810 individuals. This web test included 36 reading comprehension questions of increasing difficulty. For each item (e.g., “Which word is the odd one out?”), the participant had four possible responses to choose one correct answer (e.g., “cheerful,” “glad,” “happy,” “sleepy”).
To inform the calculation of reading composites, we performed confirmatory factor analysis based on structural equation modeling implemented in the R package lavaan (Rosseel, 2012). Results indicated that our data were best explained by a two-factor solution comprising a “reading efficiency” factor and a “reading comprehension” factor (N = 4,007; imputation of missing values was based on maximum likelihood; Supplementary Table S1). We extracted and used factor scores for all further analyses. Because the two reading factors correlated substantially (Supplementary Table S2), we also calculated an overall reading ability composite by taking the mean of both reading efficiency and reading comprehension factor scores.
Correlations across all individual tests of reading ability can be found in Supplementary Table S3.
General cognitive ability
The participants’ general cognitive ability was assessed using web-based tests of verbal and nonverbal abilities at ages 7, 12, and 16. A mean score composite was obtained from four tests at age 7 (Similarities: Wechsler, 1992; Vocabulary: Wechsler, Golombok, & Rust, 1992; Conceptual Grouping: McCarthy, 1972; Picture Completion: Wechsler et al., 1992), three tests at age 12 (Raven’s Progressive Matrices: Raven, Court, & Raven, 1996; Picture Completion: Wechsler et al., 1992; General Knowledge: Kaplan, Fein, Kramer, Delis, & Morris, 1999), and two tests at age 16 (Raven’s Progressive Matrices: Raven et al., 1996; Mill Hill Vocabulary test: Raven, Raven, & Court, 1998). We performed imputation for any missing measures. Behavioral and genotypic data were available for 3,559 individuals at age 7 (M = 7.12, SD = 0.24), 3,248 individuals at age 12 (M = 11.46, SD = 0.64), and 1,598 individuals at age 16 (M = 16.52, SD = 0.30). On average, general cognitive ability composites at the different ages correlated 0.48. To maximize sample size, we calculated a general cognitive ability mean composite based on data available at ages 7, 12, and 16. Inclusion criteria required the availability of at least one general cognitive ability composite at any one of the ages, which resulted in a total sample size of 4,852 participants.
Family SES
Family SES was assessed at first contact at age 2 and is a mean composite of measures of parental education and occupation, as well as maternal age at birth of the first child. Data were available for 5,379 genotyped individuals.
NPD measures
The following reading measures were obtained from NPD, a rich database containing information about students’ school-related characteristics and achievements (https://www.gov.uk/government/collections/national-pupil-database) that are generally assessed at the end of each National Curriculum (NC) Key Stage in the United Kingdom. Reading-related measures were available at NC Key Stage 1 (KS 1), which corresponds to School Year 1 and 2 (in the present sample, M = 7.29 years, SD = 0.29), NC Key Stage 2 (KS 2), which corresponds to School Year 3 to 6 (in the present sample, M = 11.29 years, SD = 0.29), and NC Key Stage 3 (KS 3), which corresponds to School Year 7 to 9 (in the present sample, M = 14.26 years; SD = 0.34; https://www.gov.uk/national-curriculum/overview).
NC teacher reading ratings
Children in the United Kingdom are evaluated by their teachers on core areas of ability such as English (reading, writing, and speaking and listening), mathematics (numbers, shapes, space, using and applying mathematics, and measures), and science (life process, scientific enquiry, and physical process) throughout the school years. These assessments represent the requirements related to key achievements set by the core NC, which was established by the Qualifications and Curriculum Authority (www.qca.org.uk), and the National Foundation for Educational Research (www.nfer.ac.uk/index.cfm).
In the current study, we used NC teacher ratings of reading ability. Children’s reading performance was rated on a scale of nine levels of achievement, with Level 2 representing ability levels common for 7 year-olds, Level 4 representing skills common for 11-year-olds, and Levels 5–6 representing performance that is expected for most 14-year-olds. Level 9 is assigned only to students with “exceptional performance.” Genotypes and teacher ratings on reading performance were available for 3,910 individuals at KS 1, 3,893 individuals at KS 2 and 922 individuals at KS 3.
NC reading tests: Knowledge, skills, and understanding
As part of the NC, students were tested on their reading knowledge, skills, and understanding using UK standardized tests that were administered in the classroom. At all Key Stages, these untimed assessments required the students to read text and to respond to questions using multiple-choice answers. These tests require skills including reading fluency, accuracy, and comprehension. Reading test and genotype data were available for 3,774 pupils at KS 1, 3,864 pupils at KS 2, and 909 students at KS 3.
For assessment at the end of KS 1, students were tested on reading material in accordance with NC Level 2 and NC Level 3 attainment targets. For NC Level 2 tests, reading performance was categorized as Level 2 not achieved (0–6 points), Level 2C achieved (7–14 points), Level 2B achieved (15–21 points), or Level 2A achieved (22–30 points). For NC Level 3, reading performance was scored as Level 3 not achieved (0–14 points) and Level 3 achieved (15–26 points).
To test reading ability at the end of KS 2, students were assessed on reading material in accordance to NC Levels 3–5 and on material in line with NC Levels 4–7. At the end of KS 3, students were tested based on standards of NC Levels 4–7. At both key stages, reading performance was scored from 0 to 50.
NPD NC teacher ratings and reading tests were highly correlated at all three key stages (average r = .85). Therefore, reading performance composites were created based on the mean of z-standardized NC teacher ratings and reading tests at KS 1, KS 2, and KS 3.
For most measures, small but significant effects of gender and age were found (Supplementary Table S4). Therefore, all measures were corrected for gender and age effects using the regression method. Z-standardized residuals of these measures were used in further analyses. All data points 3 or more standard deviations from the mean were removed to control for outliers.
Statistical analyses
Genome-wide polygenic scores
In the current study, we used the software PRSice (Euesden, Lewis, & O’Reilly, 2015) to create GPS for 5,825 unrelated individuals based on p-values and β-weights obtained from the 2016 GWA study on educational attainment (Okbay et al., 2016). Those SNPs that passed quality control were clumped for linkage disequilibrium by applying an R 2 = 0.1 cutoff within a 250-kb window, leaving 108,737 SNPs for further analyses. To calculate GPS in our sample, all trait-associated alleles (0, 1, or 2) were counted using the sign of the beta weights to determine which allele is the “increasing” allele, and multiplied by their effect size, as identified from GWA analysis, to account for their relative importance. These weighted counts were then summed to form a personal score for each individual. Informed by the p value for SNP–trait association supplied by GWA summary statistics, we calculated several GPS for different significant thresholds (0.001, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5), including increasing numbers of SNPs (see Supplementary Table S5 for details). After z-standardization (M = 0, SD = 1), a value that is greater than zero indicates that the individual carries more education-associated alleles than average in the population, and a value that is lower than the mean zero implies that the individual carries fewer than average education-associated alleles. For analyses in the current study, we used the GPS based on a p value threshold of .05, which included 19,415 SNPs (see Supplementary Table S3 for results using GPS based on all other thresholds). This GPS was perfectly normal distributed (Supplementary Figure S1). To control for population stratification, we regressed the GPS on the first 10 principal components and used the residuals in all subsequent analyses.
To test the association between EduYears GPS and the various reading measures, we performed regression analyses. To test differences between EduYears GPS predictions of the various reading measures, we used the Williams modification of the Hotelling test (Williams, 1959), which corrects for correlations between outcome variables. We used t-tests to compare the differences between the reading ability of students in the lowest and highest 12.5% of GPS distribution. To test nonlinear effects between reading ability and EduYears GPS, we added cubic and quadratic polynomials to the regression equation. All significance thresholds were corrected for multiple testing using the Nyholt- Šidák correction, which adjusts the p-value threshold (.05) for the number of independent tests (6.71). This resulted in a corrected p-value threshold of .008, which was applied to all statistical analyses (see Supplementary Methods S2).
Power was calculated using the AVENGEME software (Palla & Dudbridge, 2015). We estimated that with a GWA discovery sample size of 328,918 and a mean target sample size of 3,490, our EduYears GPS has more than 80% power to detect 0.3% of the phenotypic variance (p < .008), which means that power is substantial in the present study to detect associations where we expect EduYears GPS to account for at least 1.0% of the phenotypic variance of reading traits. (See Supplementary Methods S1a for power calculation parameters and Supplementary Methods S1b and S1c for additional power analyses based on varying parameters.)
Results
Polygenic score analyses
EduYears GPS accounted for 2.1% to 5.1% (p < .001) of the variance in the reading measures (Figure 1). The EduYears GPS correlated as much with the efficiency factor as with the comprehension factor. There was a trend toward increasing GPS heritability with age for Key Stage reading performance—2.1% at age 7, 3.5% at age 12, and 5.1% at age 14—but these differences were not significant as indicated by the Williams modification of the Hotelling test (Supplementary Table S6). As predicted, the variance explained in reading ability was considerably larger for EduYears GPS than for other GPS, such as childhood general cognitive ability, adult verbal and numerical reasoning, and schizophrenia (Supplementary Methods S3), which explained between 0% and 1.1% of the variance in reading (Supplementary Table S7).
Figure 1.
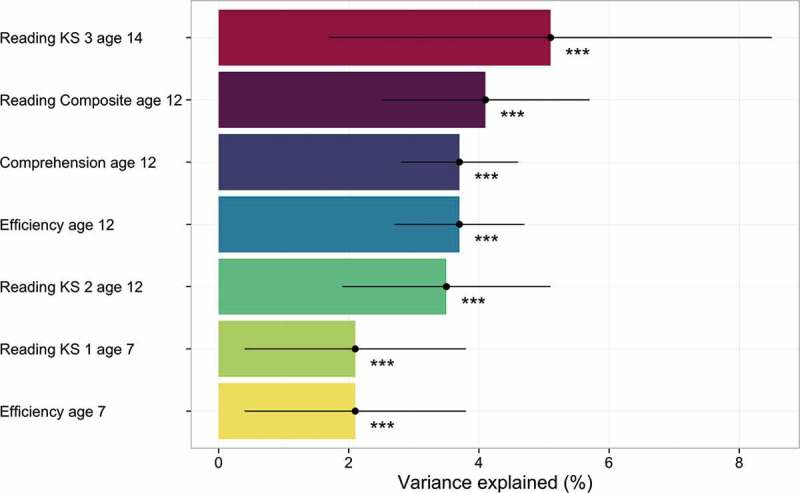
EduYears GPS heritability estimates for reading measures. Error bars represent standard errors. KS = Key Stage; Efficiency = reading efficiency factor based on TOWRE and Woodcock-Johnson test assessed at age 12; Comprehension = reading comprehension factor based on GOAL and PIAT test assessed at age 12; Reading Composite = mean score of efficiency factor and comprehension factor; ***p < 0.001. Note. KS = Key Stage.
EduYears GPS accounted for 2.7% (p < .001) of the variance in general cognitive ability. Although general cognitive ability and reading performance are related (r = .31–.56; Supplementary Table S3), EduYears GPS remained a significant predictor of all reading measures after controlling for general cognitive ability, accounting for 0.6% to 2.4% of the variance (Supplementary Table S8). We repeated these analyses by correcting reading measures at various ages for general cognitive ability assessed at each respective age, obtaining similar results to using the general cognitive ability composite (Supplementary Table S8).
Results also showed strong correlations between family SES and measures of reading ability (r = .25–.39; Supplementary Table S3). Even though family SES is highly correlated with educational attainment, which was the target trait for the EduYears GWA study, we found that EduYears GPS was still a significant predictor of reading ability after accounting for family SES, explaining 0.8% to 2.5% of the variance (Supplementary Table S8). Even after controlling for both age-specific general cognitive ability and family SES, EduYears GPS significantly predicted between 0.5% and 1% of the variance in reading ability (Supplementary Table S8).
Quantile GPS analyses
To test the magnitude of the differences in reading performance between the extreme EduYears GPS groups, we performed t-tests between mean reading scores of individuals in the highest and lowest EduYears GPS octiles. We observed substantial mean differences in reading ability across the school years (Cohen’s d 0.49–0.67; see Supplementary Table S9). Figure 2 depicts the relationship between EduYears GPS octiles and reading efficiency and reading comprehension factors at age 12. Furthermore, we found that for all reading measures, the association with EduYears GPS was linear (Supplementary Table S10).
Figure 2.
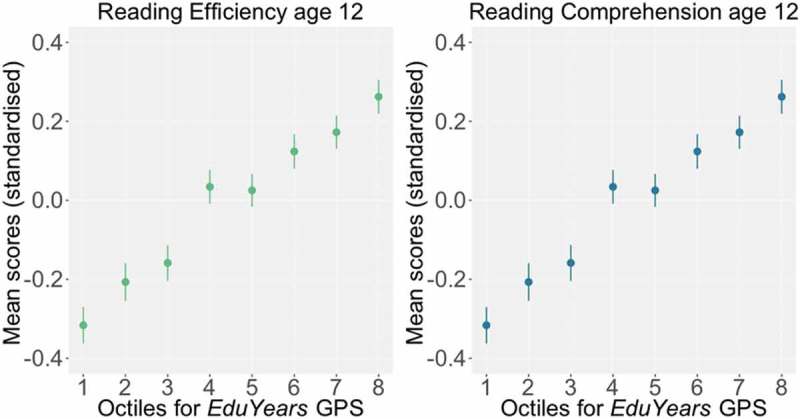
Reading scores as a function of EduYears GPS divided into octiles (1 = lowest, 8 = highest). Z-Standardized means and standard errors for the reading efficiency factor (based on TOWRE and GOAL test) and reading comprehension factor (based on PIAT and Woodcock-Johnson test). All tests were assessed at age 12. Note. GPS = genome-wide polygenic scores.
Discussion
The present study provides the first GPS for reading ability and disability. Although a GPS heritability of 5% at age 14 may not seem like much, it is an impressive result for five reasons. First, predicting 5% of the variance of reading is substantial in comparison to most other findings in reading. As one of countless examples, much is made of sex differences in reading, but meta-analytic studies show that sex differences account for less than 1% of the variance (Hyde, 2014).
Second, 5% compares very favorably with the GPS heritability estimate of 1% from a GPS for general cognitive ability. This difference in prediction further demonstrates that the utility of EduYears GPS in predicting specific skills like reading beyond general cognitive ability. EduYears GPS also predicts more of the variance of general cognitive ability than do GPS based on GWA studies of general cognitive ability, but, even so, the EduYears GPS heritability accounts for only 2.9% of the variance of general cognitive ability, as compared to 5% for reading ability (Supplementary Table S3).
A third reason is that the bottom and top octiles of the GPS distribution differ significantly across the school years, with a difference of ~0.50 of a standard deviation in their reading performance at age 7, ~0.60 of a standard deviation at age 12, and ~0.70 of a standard deviation at age 14. For example, a difference of 0.60 of a standard deviation at age 12 corresponds to the mean growth of reading ability between Grades 5 and 7 (d = ~0.55; Lipsey et al., 2012). These comparisons suggest that reading performance of children in the lowest GPS octile lags two entire school years behind the reading performance of children within the highest GPS octile.
Fourth, the EduYears GPS is based on years of education. For this reason, a comparably large GWA study of reading would be expected to predict much more variance. The expectation for the predictive power of a GPS based on a GWA of reading depends on the genetic correlation between years of education and reading, a genetic correlation that is not known. Unless the correlation between EduYears GPS and a reading GPS is 1, variance in reading ability explained by EduYears GPS would underestimate the variance in reading ability that a reading GPS could explain. In Supplementary Figure S2, we show algebraically that it is possible to estimate the increased predictive power of a GPS based on a GWA study of reading. This estimate depends on the EduYears GPS prediction of reading ability and the correlation between EduYears GPS and a reading GPS. As illustrated in Supplementary Figure S2, the expected correlation between EduYears GPS and reading is equal to the correlation between the two GPS times the correlation between GPSreading and reading. Therefore, the amount of variance in reading that could be explained by a reading GPS can be estimated from the squared correlation between GPSEduYears and reading ability (the variance in reading ability explained by GPSEduYears)—0.05 in the present study (i.e., 0.222)—divided by the squared correlation between the two GPS measures (see caption of Supplementary Figure 2). Because years of education is such a broad measure that goes well beyond reading to include other cognitive abilities, personality, mental health and illness, motivation and socioeconomic background, we would be surprised if the genetic correlation between years of education and reading ability during the school years were greater than 0.50. Although a correlation between EduYears GPS and a GPS for reading cannot be interpreted like a genetic correlation estimated through twin studies, it would reflect the genetic relationship between the two traits. Following the equation derived from Supplementary Figure S2, if the correlation between EduYears GPS and reading performance GPS were 0.50, then a reading GPS could explain 20% (i.e., 0.05/0.502) of the observed variance in reading performance.
Fifth, GWA studies and GPS are relatively new, so optimism seems warranted in this fast-moving area of research. For example, GWA studies have been limited to the additive effects of common SNPs (i.e., with minor allele frequencies > 1%) on commercially available DNA arrays. As the vast majority of SNPs are much less common, a GPS heritability estimate of reading ability of up to 5% based only on common variants is impressive. Whole-genome sequencing—sequencing all 3 billion base pairs of DNA—will progress our understanding even further because it can capture DNA variation of any kind, rare as well as common variants, and DNA variants of any kind, not just SNPs. A related issue is missing heritability, which refers to the gap between the variance explained by GPS and twin study estimates of heritability. Heritability of reading ability has been estimated at ~70% in twin studies. Our GPS heritability estimate that explains 5% of the total variance of reading ability accounts for only ~7% (5%/70%) of this twin-study estimate of heritability. However, as the use of GPS is limited to the additive effects of common genetic variants, it is more informative to compare GPS heritability to SNP-heritability estimates derived from Genome-wide Complex Trait Analysis (Yang, Lee, Goddard, & Visscher, 2011). Genome-wide Complex Trait Analysis estimates genetic influence of common variants by comparing genetic relatedness to phenotypic similarity in thousands of unrelated individuals. SNP-heritability of reading performance has been estimated at 22% (Harlaar, Trzaskowski, Dale, & Plomin, 2014), which represents a ceiling for GPS heritability. This means that the current EduYears GPS accounts for more than 20% of the heritable variance derived from common SNPs.
Our results highlight the ability to predict specific learning abilities using individuals’ DNA, even at the beginning of formal schooling. It is remarkable that EduYears GPS significantly predicted reading even at age 7, even though EduYears GPS is based on years of education in adults. It is also noteworthy that this association remained significant even after controlling for general cognitive ability and family SES. Prediction from adult years of education to reading performance at age 7 reflects the high genetic stability of reading ability (average age-to-age genetic correlation = 0.97), as shown in longitudinal twin analyses (Hulslander, Olson, Willcutt, & Wadsworth, 2010). Although GPS heritability was substantially higher at KS 3 (5.1%) in comparison to KS 1 (2.1%), this was not statistically significant, which is likely due to the differences in sample sizes and the corresponding magnitude of standard errors of the mean.
We also found that EduYears GPS heritability estimates were not significantly different for distinct components of reading ability. For example, EduYears GPS accounted for 3.7% of the variance in both reading efficiency and reading comprehension at age 12. It has been suggested that individual differences in reading comprehension partly depend on reading efficiency (Tunmer & Chapman, 2012), and quantitative genetic research has shown that reading efficiency substantially accounts for the genetic variance in reading comprehension (Keenan, Betjemann, Wadsworth, DeFries, & Olson, 2006). This indicates that both reading efficiency and reading comprehension share much of the same genetic architecture. This finding is further supported by a twin study based on ~5,000 TEDS twins, which showed that reading efficiency and reading comprehension substantially share the same genetic and common environmental factors (Harlaar, Kovas, Dale, Petrill, & Plomin, 2012). Overall, these results may explain why the GPS heritability estimates did not differ significantly for these traits. Our findings show that the strength of EduYears GPS lies in the prediction of overall reading performance, rather than in the differentiation of reading ability at different developmental stages, or in distinguishing different forms of reading ability. However, as noted earlier, years of education is a broad downstream measure and would not be expected to correlate more highly with any single facet of reading. For this reason, there is still much potential for finding DNA variants specific to components of reading, as well as DNA variants general to all components of reading identified in the present research.
We have also shown that EduYears GPS is linearly associated with all measures of reading ability across the reading distribution at all ages. This may be indicative of an underlying genetic continuum that influences both reading ability and reading disability equally, suggesting that both traits are not causally distinct. Although a GPS is additively constructed based on prediction weights derived from a linear model (GWA) and is normally distributed (Supplementary Figure S1), it can identify nonlinear effects in cross-trait analysis, between EduYears and reading in the present study. Additive genetic effects associated with the trait used to calculate the GPS (EduYears) may relate nonlinearly to a different phenotype (reading), despite the additive construction of the polygenic score. As the current GPS may not be sufficiently powerful to disentangle the nature of nonlinear relationships, it is unclear whether our findings represent the true underlying relationship between EduYears GPS and measures of reading ability. However, it is likely that our results would have shown some evidence if there were a strong nonlinear relationship between EduYears GPS and reading performance. When more powerful GPS become available in the future, we intend to reassess linearity of this association.
The findings of this study do not have specific implications for existing theories or applications in themselves. However, they represent a starting point for future research in the scientific study of reading that uses DNA information to further our knowledge of individual differences in reading ability. A key benefit of finding inherited DNA sequence variation associated with individual differences in reading performance is its potential for early prediction of reading problems. Because inherited DNA sequence does not change during development, the prediction of reading ability and disability from the EduYears GPS is just as strong infancy, at birth and even prenatally, long before any cognitive tests can be used to predict reading. The use of GPS in education could facilitate early prediction of reading problems, which could lead to intervention and possible prevention by identifying those at particular genetic risk and tailoring support according to their needs.
For example, GPS could enable research on individual differences in resilience to developing reading difficulties despite high GPS risk. Because GPS predict just as accurately from birth, it facilitates research on the developmental mediators and moderators of risk and resilience. For example, it is likely that a combination of early developmental predictors and a GPS is better in predicting later reading ability than either does individually. This could help to identify risk early, paving the way for interventions to decrease negative outcomes linked to reading difficulties. A final example is that research could address whether children with early reading problems and high GPS risk respond differently to interventions than children with comparable problems without high GPS risk. This may elucidate whether different intervention strategies could be profitably employed depending on the children’s unique genetic characteristics.
A strength of this study is the multivariate longitudinal design of TEDS, which permitted investigation of genetic influences in different reading components across development. Another strength is the large sample size, which enabled excellent power to detect robust effects. Last, the availability of actual academic performance data, as well as teacher ratings through the NPD, made it possible to supplement the cognitive measures in TEDS with measures of reading performance assessed at school as well as to externally validate our measures assessed in TEDS.
The present study has two main limitations. As just discussed, our GWA study statistics are based on years of education in adults and are not specific to reading ability in school children. A second limitation is that the participants in our sample received education prescribed by the NC in the United Kingdom, and these results need to be tested in other samples beyond the United Kingdom.
No policy implications necessarily follow from our finding that DNA can be used to predict individual reading performance. However, we hope that these findings will raise awareness about social and ethical aspects of genetic prediction. We also hope that our findings will contribute to the making of better policy decisions that recognize and respect genetically driven differences between children in their ability to learn to read and in their eventual reading efficiency and comprehension. Polygenic scores may eventually be used as an early screening tool to tailor intervention strategies based on individuals’ needs to prevent or reduce the impact of potential reading difficulties, rather than waiting until children fail at reading in school, with all the collateral damage that entails.
Supplementary Material
Funding Statement
We gratefully acknowledge the ongoing contribution of the participants in the Twins Early Development Study (TEDS) and their families. TEDS is supported by a program grant to RP from the UK Medical Research Council (MR/M021475/1 and previously G0901245), with additional support from the US National Institutes of Health (HD044454; HD059215). SS is supported by the MRC/IoPPN Excellence Award and by the EU Framework Programme 7 (602768). EK is supported by a Medical Research Council studentship. RP is supported by a Medical Research Council Advanced Investigator award (295366). JCDF is supported by grants from HD027802 and AG046938 from the US National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Funding
We gratefully acknowledge the ongoing contribution of the participants in the Twins Early Development Study (TEDS) and their families. TEDS is supported by a program grant to RP from the UK Medical Research Council (MR/M021475/1 and previously G0901245), with additional support from the US National Institutes of Health (HD044454; HD059215). SS is supported by the MRC/IoPPN Excellence Award and by the EU Framework Programme 7 (602768). EK is supported by a Medical Research Council studentship. RP is supported by a Medical Research Council Advanced Investigator award (295366). JCDF is supported by grants from HD027802 and AG046938 from the US National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Supplemental data
Supplemental data for this article can be accessed at www.tandfonline.com/hssr.
References
- Antonarakis S. E., & McKusick V. A. (2000). OMIM passes the 1,000-disease-gene mark. Nature Genetics, 25(1), 11. doi: 10.1038/75497 [DOI] [PubMed] [Google Scholar]
- Arnold D. H., & Doctoroff G. L. (2003). The early education of socioeconomically disadvantaged children. Annual Review of Psychology, 54(1), 517–545. doi: 10.1146/annurev.psych.54.111301.145442 [DOI] [PubMed] [Google Scholar]
- Belsky D. W., Moffitt T. E., Corcoran D. L., Domingue B., Harrington H., Hogan S., & Caspi A. (2016). The genetics of success: How single-nucleotide polymorphisms associated with educational attainment relate to life-course development. Psychological Science, 27, 957–972. doi: 10.1177/0956797616643070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D. V. M. (2015). The interface between genetics and psychology: Lessons from developmental dyslexia In Proceedings of the Royal Society B (Vol. 282, pp. 20143139). The Royal Society. doi: 10.1098/rspb.2014.3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon L. R., Smith S. D., Fulker D. W., Kimberling W. J., Pennington B. F., & DeFries J. C. (1994). Quantitative trait locus for reading disability on chromosome 6. Science, 266(5183), 276–279. doi: 10.1126/science.7939663 [DOI] [PubMed] [Google Scholar]
- Carrion-Castillo A., Franke B., & Fisher S. E. (2013). Molecular genetics of dyslexia: An overview. Dyslexia, 19(4), 214–240. doi: 10.1002/dys.1464 [DOI] [PubMed] [Google Scholar]
- Chabris C. F., Lee J. J., Cesarini D., Benjamin D. J., & Laibson D. I. (2015). The fourth law of behavior genetics. Current Directions in Psychological Science, 24(4), 304–312. doi: 10.1177/0963721415580430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis O. S. P., Band G., Pirinen M., Haworth C. M. A., Meaburn E. L., Kovas Y., … Spencer C. C. A. (2014). The correlation between reading and mathematics ability at age twelve has a substantial genetic component. Nature Communications, 5, 4204. doi: 10.1038/ncomms5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis O. S. P., Haworth C. M. A., & Plomin R. (2009). Learning abilities and disabilities: Generalist genes in early adolescence. Cognitive Neuropsychiatry, 14(4–5), 312–331. doi: 10.1080/13546800902797106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina C., Meyre D., Gallina S., Durand E., Körner A., Jacobson P., … Froguel P. (2007). Variation in FTO contributes to childhood obesity and severe adult obesity. Nature Genetics, 39(6), 724–726. doi: 10.1038/ng2048 [DOI] [PubMed] [Google Scholar]
- Dudbridge F. (2013). Power and predictive accuracy of polygenic risk scores. Plos Genet, 9(3), e1003348. doi: 10.1371/journal.pgen.1003348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan G. J., Dowsett C. J., Claessens A., Magnuson K., Huston A. C., Klebanov P., … Japel C. (2007). School readiness and later achievement. Developmental Psychology, 43(6), 1428–1446. doi: 10.1037/0012-1649.43.6.1428 [DOI] [PubMed] [Google Scholar]
- Eicher J. D., Landowski C., Stackhouse B., Sloan A., Chen W., Jensen N., … Johnson A. D. (2015). Grasp v2.0: An update on the genome-wide repository of associations between snps and phenotypes. Nucleic Acids Research, 43, D799–804. doi: 10.1093/nar/gku1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher J. D., Powers N. R., Miller L. L., Akshoomoff N., Amaral D. G., Bloss C. S., … Chang L. (2013). Genome-wide association study of shared components of reading disability and language impairment. Genes, Brain and Behavior, 12(8), 792–801. doi: 10.1111/gbb.12085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euesden J., Lewis C. M., & O’Reilly P. F. (2015). PRSice: Polygenic risk score software. Bioinformatics, 31(9), 1466–1468. doi: 10.1093/bioinformatics/btu848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A. (1918). The correlation between relatives on the supposition of Mendelian inheritance. American Journal of Human Genetics, 2(52), 399–433. [Google Scholar]
- Frayling T. M., Timpson N. J., Weedon M. N., Zeggini E., Freathy R. M., Lindgren C. M., … McCarthy M. I. (2007). A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science (New York, N.Y.), 316(5826), 889–894. doi: 10.1126/science.1141634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsberger C., Abecasis G. R., & Hinds D. A. (2015). minimac2: Faster genotype imputation. Bioinformatics (Oxford, England), 31(5), 782–784. doi: 10.1093/bioinformatics/btu704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gialluisi A., Newbury D. F., Wilcutt E. G., Olson R. K., DeFries J. C., & Brandler W. M., … Luciano M. (2014). Genome-wide screening for DNA variants associated with reading and language traits. Genes, Brain and Behavior, 13(7), 686–701. doi: 10.1111/gbb.12158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOAL plc (2002). GOAL formative assessment: Key stage 3. London, UK: Hodder & Stoughton. [Google Scholar]
- Gratten J., Wray N. R., Keller M. C., & Visscher P. M. (2014). Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nature Neuroscience, 17(6), 782–790. doi: 10.1038/nn.3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A., Scott A. F., Amberger J. S., Bocchini C. A., & McKusick V. A. (2005). Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Research, 33(Suppl. 1), D514–517. doi: 10.1093/nar/gki033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlaar N., Kovas Y., Dale P. S., Petrill S. A., & Plomin R. (2012). Mathematics is differentially related to reading comprehension and word decoding: Evidence from a genetically-sensitive design. Journal of Educational Psychology, 104, 3. doi: 10.1037/a0027646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlaar N., Spinath F. M., Dale P. S., & Plomin R. (2005). Genetic influences on early word recognition abilities and disabilities: A study of 7-year-old twins. Journal of Child Psychology and Psychiatry, 46(4), 373–384. doi: 10.1111/j.1469-7610.2004.00358.x [DOI] [PubMed] [Google Scholar]
- Harlaar N., Trzaskowski M., Dale P. S., & Plomin R. (2014). Word reading fluency: Role of genome-wide single-nucleotide polymorphisms in developmental stability and correlations with print exposure. Child Development, 85(3), 1190–1205. doi: 10.1111/cdev.12207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haworth C. M. A., Davis O. S., & Plomin R. (2013). Twins Early Development Study (TEDS): A genetically sensitive investigation of cognitive and behavioral development from childhood to young adulthood. Twin Research and Human Genetics, 16(01), 117–125. doi: 10.1017/thg.2012.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haworth C. M. A., Harlaar N., Kovas Y., Davis O. S. P., Oliver B. R., Hayiou-Thomas M. E., … Plomin R. (2007). Internet cognitive testing of large samples needed in genetic research. Twin Research and Human Genetics, 10(04), 554–563. doi: 10.1375/twin.10.4.554 [DOI] [PubMed] [Google Scholar]
- Herbers J. E., Cutuli J. J., Supkoff L. M., Heistad D., Chan C.-K., Hinz E., & Masten A. S. (2012). Early reading skills and academic achievement trajectories of students facing poverty, homelessness, and high residential mobility. Educational Researcher, 41(9), 366–374. doi: 10.3102/0013189X12445320 [DOI] [Google Scholar]
- Hirschhorn J. N., & Daly M. J. (2005). Genome-wide association studies for common diseases and complex traits. Nature Reviews Genetics, 6(2), 95–108. doi: 10.1038/nrg1521 [DOI] [PubMed] [Google Scholar]
- Howie B., Fuchsberger C., Stephens M., Marchini J., & Abecasis G. R. (2012). Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nature Genetics, 44(8), 955–959. doi: 10.1038/ng.2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulslander J., Olson R. K., Willcutt E. G., & Wadsworth S. J. (2010). Longitudinal stability of reading-related skills and their prediction of reading development. Scientific Studies of Reading, 14(2), 111–136. doi: 10.1080/10888431003604058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde J. S. (2014). Gender similarities and differences. Annual Review of Psychology, 65, 373–398. doi: 10.1146/annurev-psych-010213-115057 [DOI] [PubMed] [Google Scholar]
- Jannot A.-S., Ehret G., & Perneger T. (2015). P < 5 × 10−8 has emerged as a standard of statistical significance for genome-wide association studies. Journal of Clinical Epidemiology, 68(4), 460–465. doi: 10.1016/j.jclinepi.2015.01.001 [DOI] [PubMed] [Google Scholar]
- Kaplan E., Fein D., Kramer J., Delis D., & Morris R. (1999). WISC-III as a process instrument (WISC-III-PI). New York, NY: The Psychological Corporation. [Google Scholar]
- Keenan J. M., Betjemann R. S., Wadsworth S. J., DeFries J. C., & Olson R. K. (2006). Genetic and environmental influences on reading and listening comprehension. Journal of Research in Reading, 29(1), 75–91. doi: 10.1111/j.1467-9817.2006.00293.x [DOI] [Google Scholar]
- Knopik V. S., Neiderhiser J. M., DeFries J. C., & Plomin R. (2017). Behavioral genetics (7th ed.). New York, NY: Worth. [Google Scholar]
- Krapohl E., & Plomin R. (2015). Genetic link between family socioeconomic status and children’s educational achievement estimated from genome-wide SNPs. Molecular Psychiatry, 21, 437–443. doi: 10.1038/mp.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsey M. W., Puzio K., Yun C., Herbert M. A., Steinka-Fry K., Cole M. W., … Busick M. D. (2012). Translating the statistical representation of the effects of education interventions into more readily interpretable forms. New York, NY: National Center for Special Education Reserach. [Google Scholar]
- Luciano M., Evans D. M., Hansell N. K., Medland S. E., Montgomery G. W., Martin N. G., … Bates T. C. (2013). A genome-wide association study for reading and language abilities in two population cohorts. Genes, Brain, and Behavior, 12(6), 645–652. doi: 10.1111/gbb.12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., Hunter D. J., … Cho J. H. (2009). Finding the missing heritability of complex diseases. Nature, 461(7265), 747–753. doi: 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwardt F. C. (1997). Peabody Individual Achievement Test—Revised, circle pines. Bloomington, MN: American Guidance Service. [Google Scholar]
- Maughan B. (1995). Annotation: Long-term outcomes of developmental reading problems. Journal of Child Psychology and Psychiatry, 36(3), 357–371. doi: 10.1111/jcpp.1995.36.issue-3 [DOI] [PubMed] [Google Scholar]
- McCarthy D. (1972). McCarthy Scales of Children’s Abilities. New York, NY: the psychological corporation. [Google Scholar]
- McCarthy S., Das S., Kretzschmar W., Durbin R., Abecasis G., & Marchini J. (2015). A reference panel of 64,976 haplotypes for genotype imputation. BioRxiv, 035170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn E. L., Harlaar N., Craig I. W., Schalkwyk L. C., & Plomin R. (2008). Quantitative trait locus association scan of early reading disability and ability using pooled DNA and 100K SNP microarrays in a sample of 5760 children. Molecular Psychiatry, 13(7), 729–740. doi: 10.1038/sj.mp.4002063 [DOI] [PubMed] [Google Scholar]
- Okbay A., Beauchamp J. P., Fontana M. A., Lee J. J., Pers T. H., Rietveld C. A., … Benjamin D. J. (2016). Genome-wide association study identifies 74 loci associated with educational attainment. Nature, advance online publication. doi: 10.1038/nature17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson R. K., Keenan J. M., Byrne B., & Samuelsson S. (2014). Why do children differ in their development of reading and related skills? Scientific Studies of Reading, 18(1), 38–54. doi: 10.1080/10888438.2013.800521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palla L., & Dudbridge F. (2015). A fast method that uses polygenic scores to estimate the variance explained by genome-wide marker panels and the proportion of variants affecting a trait. American Journal of Human Genetics, 97(2), 250–259. doi: 10.1016/j.ajhg.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R., & Deary I. J. (2015). Genetics and intelligence differences: Five special findings. Molecular Psychiatry, 20(1), 98–108. doi: 10.1038/mp.2014.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelmans G., Buitelaar J. K., Pauls D. L., & Franke B. (2011). A theoretical molecular network for dyslexia: Integrating available genetic findings. Molecular Psychiatry, 16(4), 365–382. doi: 10.1038/mp.2010.105 [DOI] [PubMed] [Google Scholar]
- Raven J., Raven J. C., & Court J. H. (1998). Mill Hill Vocabulary Scale. Oxford, UK: OPP. [Google Scholar]
- Raven J. C., Court J. H., & Raven J. (1996). Manual for Raven’s Progressive Matrices and Vocabulary Scales. Oxford, UK: Oxford University. [Google Scholar]
- Rietveld C. A., Esko T., Davies G., Pers T. H., Turley P., Benyamin B., … Koellinger P. D. (2014). Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. PNAS Proceedings of the National Academy of Sciences of the United States of America, 111(38), 13790–13794. doi: 10.1073/pnas.1404623111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld C. A., Medland S. E., Derringer J., Yang J., Esko T., Martin N. W., … Koellinger P. D. (2013). GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science, 340(6139), 1467–1471. doi: 10.1126/science.1235488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosseel Y. (2012). Lavaan: An R package for structural equation modeling. Journal of Statistical Software, 48(2), 1–36. doi: 10.18637/jss.v048.i02 [DOI] [Google Scholar]
- Scerri T. S., & Schulte-Körne G. (2010). Genetics of developmental dyslexia. European Child & Adolescent Psychiatry, 19(3), 179–197. doi: 10.1007/s00787-009-0081-0 [DOI] [PubMed] [Google Scholar]
- Schumacher J., Hoffmann P., Schmäl C., Schulte-Körne G., & Nöthen M. M. (2007). Genetics of dyslexia: The evolving landscape. Journal of Medical Genetics, 44(5), 289–297. doi: 10.1136/jmg.2006.046516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selzam S., Krapohl E., von Stumm S., O’Reilly P. F., Rimfeld K., Dale P. S., Lee J. J., & Plomin R. (2016). Predicting educational achievement from DNA. Molecular Psychiatry, 22, 267–272. doi: 10.1038/mp.2016.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swagerman S. C., Van Bergen E., Dolan C., De Geus E. J. C., Koenis M. M. G., Hulshoff Pol H. E., & Boomsma D. I. (2015). Genetic transmission of reading ability. Brain and Language. doi: 10.1016/j.bandl.2015.07.008 [DOI] [PubMed] [Google Scholar]
- Torgesen J., Wagner R., & Rashotte C. (1999). Test of Word Reading Efficiency. Austin, TX: Pro-Ed. [Google Scholar]
- Tunmer W. E., & Chapman J. W. (2012). The simple view of reading redux vocabulary knowledge and the independent components hypothesis. Journal of Learning Disabilities, 45(5), 453–466. doi: 10.1177/0022219411432685 [DOI] [PubMed] [Google Scholar]
- Visscher P. M., Brown M. A., McCarthy M. I., & Yang J. (2012). Five years of GWAS discovery. American Journal of Human Genetics, 90(1), 7–24. doi: 10.1016/j.ajhg.2011.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D., Golombok S., & Rust J. (1992). WISC-III UK Wechsler Intelligence Scale for Children: UK manual. Sidcup, UK: The Psychological Corporation. [Google Scholar]
- Williams E. J. (1959). The comparison of regression variables. Journal of the Royal Statistical Society. Series B (Methodological), 396–399. [Google Scholar]
- Wood A. R., Esko T., Yang J., Vedantam S., Pers T. H., Gustafsson S., … Frayling T. M. (2014). Defining the role of common variation in the genomic and biological architecture of adult human height. Nature Genetics, 46(11), 1173–1186. doi: 10.1038/ng.3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock R. W., McGrew K. S., & Mather N. (2001). Woodcock-Johnson III. Itasca, IL: Riverside. [Google Scholar]
- Wray N. R., Lee S. H., Mehta D., Vinkhuyzen A. A. E., Dudbridge F., & Middeldorp C. M. (2014). Research review: Polygenic methods and their application to psychiatric traits. Journal of Child Psychology and Psychiatry, and Allied Disciplines, 55(10), 1068–1087. doi: 10.1111/jcpp.12295 [DOI] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., & Visscher P. M. (2011). GCTA: A tool for Genome-wide Complex Trait Analysis. The American Journal of Human Genetics, 88(1), 76–82. doi: 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


