Abstract
Cycloclasticus bacteria are ubiquitous in oil-rich regions of the ocean and are known for their ability to degrade polycyclic aromatic hydrocarbons (PAHs). In this study, we describe Cycloclasticus that have established a symbiosis with Bathymodiolus heckerae mussels and poecilosclerid sponges from asphalt-rich, deep-sea oil seeps at Campeche Knolls in the southern Gulf of Mexico. Genomic and transcriptomic analyses revealed that in contrast to all known Cycloclasticus, the symbiotic Cycloclasticus appeared to lack the genes needed for PAH degradation. Instead, these symbionts use propane and other short-chain alkanes such as ethane and butane as carbon and energy sources, thus expanding the limited range of substrates known to power chemosynthetic symbioses. Analyses of short-chain alkanes in the environment of the Campeche Knolls symbioses revealed that these are present at high concentrations (in the µM to mM range). Comparative genomic analyses revealed high similarities between the genes used by the symbiotic Cycloclasticus to degrade short-chain alkanes and those of free-living Cycloclasticus that bloomed during the Deepwater Horizon (DWH) oil spill. Our results indicate that the metabolic versatility of bacteria within the Cycloclasticus clade is higher than previously assumed, and highlight the expanded role of these keystone species in the degradation of marine hydrocarbons.
Keywords: Cycloclasticus, Bathymodiolus, sponge, symbiosis, hydrocarbon, short-chain alkane, pHMO, pMMO, polycyclic aromatic hydrocarbon, PAH, Gulf of Mexico, deep sea, Deepwater Horizon
Fossil hydrocarbons, abundant in seafloor reservoirs as petroleum and natural gas deposits, fuel the global economy and play an important role in biogeochemical cycles1. Natural seepage of fossil hydrocarbon has a pronounced effect on marine ecosystems2 and the atmosphere3–5, while anthropogenic oil discharge has far-reaching ramifications for the environment6–8. The presence of oil and natural gas in the water column affects the microbial community, which responds rapidly to hydrocarbon infusions 9–12. These intrinsic microbes play an important role in the bioremediation of hydrocarbon pollution13. For example, oil plume bacteria have the potential to mitigate persistent and toxic pollutants, such as polycyclic aromatic hydrocarbons (PAHs)14–16. One group of ubiquitous marine PAH degraders are Cycloclasticus17, so named for their metabolic capability18. To date, all cultured Cycloclasticus are able to degrade PAHs and there is considerable physiological and genomic evidence that uncultivated Cycloclasticus are also able to derive carbon and energy from the oxidation of PAH12,19–23.
Natural hydrocarbon seepage from the subsurface reservoirs at Campeche Knolls, in the southern Gulf of Mexico, provides an opportunity to study microbial hydrocarbon degradation in the deep sea. At Campeche Knolls, asphalt volcanoes at about 3000 m water depth such as Chapopote and Mictlan, are characterized by flows of heavy oil with hotspots of fresh oil and gas emission24,25. Despite the hostile conditions, Chapopote and Mictlan harbor rich invertebrate communities typical of many other hydrocarbon seeps in the Gulf of Mexico (Fig. 1). These include vestimentiferan tubeworms, bathymodiolin mussels, vesicomyid clams and sponges24–26. These invertebrates live in symbiosis with chemosynthetic bacteria that provide them with nutrition from carbon and energy sources that cannot be used by animals, such as hydrogen sulfide and methane24,25.
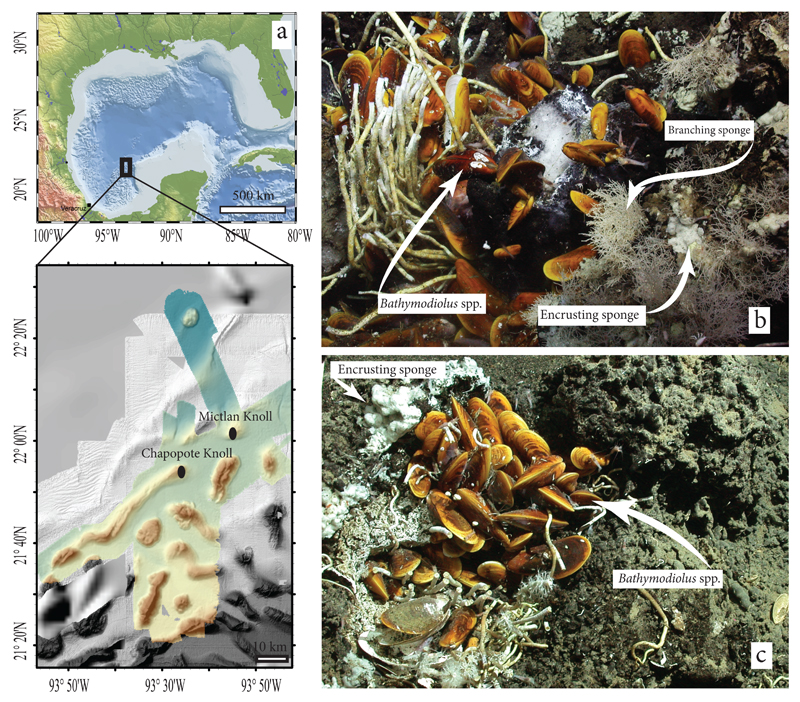
Figure 1. Invertebrate collection sites at Campeche Knolls.
a, Geographic location of Campeche Knolls and the invertebrate collection sites, Chapopote and Mictlan. b, MARUM-Quest ROV image of symbiont-bearing fauna at Chapopote seepage hotspot. Bathymodiolus heckerae, B. brooksi (undistinguishable from B. heckerae in the image), encrusting and branching sponges were collected. c, MARUM-Quest ROV image of symbiont-bearing fauna at Mictlan seepage hotspot. We found Bathymodiolus spp. and encrusting sponges at this site.
Two bathymodiolin mussel species co-occur at Campeche Knolls, Bathymodiolus heckerae and B. brooksi. The gill tissues of these mussels are densely colonized by two types of endosymbiotic gammaproteobacteria that use reduced sulfur compounds and methane as energy sources27,28. In addition to these thiotrophic and methanotrophic symbionts, Cycloclasticus were described from gill cells of the only two B. heckerae individuals collected at Chapopote in 200628. The authors hypothesized that similar to cultivated Cycloclasticus, the B. heckerae Cycloclasticus might use PAHs as a carbon and energy source. This assumption was supported by PCR amplification of two key genes for the use of aromatic compounds and stable carbon isotope signatures consistent with the use of PAHs from the Chapopote environment28.
In March of 2015, we revisited the Campeche Knolls and investigated the association of Cycloclasticus with B. heckerae in more detail. In addition, we examined sponges from the Campeche Knolls, as Cycloclasticus were recently found in the poecilosclerid sponge Myxilla methanophila from hydrocarbon seeps on the upper Louisiana slope of the Gulf of Mexico29. We were able to extract and assemble high quality Cycloclasticus draft genomes from the metagenomes of four B. heckerae mussels, one sponge with an encrusting morphotype (two individuals), and one sponge with a branching morphotype (one individual). Metabolic reconstruction of the Cycloclasticus genomes, complemented by transcriptomic and proteomic analyses, provided insights into the carbon and energy sources that fuel these symbionts. Analyses of gas and oil drops as well as asphalt and gas hydrates from the collection sites confirmed the presence of substrates predicted to play an important role in the metabolism of the Cycloclasticus symbionts. By identifying homologous key genes and transcripts in environmental datasets from the Deepwater Horizon (DWH) oil spill, we show that the carbon and energy pathways we discovered in the symbiotic Cycloclasticus may have also played a central role in the metabolism of free-living Cycloclasticus that bloomed en masse in the subsurface DHW hydrocarbon plume.
Results and Discussion
Bathymodiolus heckerae mussels and two sponge species have host-specific Cycloclasticus 16S rRNA phylotypes
Eight B. heckerae individuals collected at Chapopote in 2015 were examined in this study. All of them had 16S rRNA sequences that were highly similar (> 99.9%) to those of Cycloclasticus symbionts found in the two B. heckerae individuals sampled from Chapopote in 200628 based on sequence analyses of PCR products (Supplementary Methods). Fluorescence in situ hybridization (FISH) with a probe targeting Cycloclasticus showed the intracellular location of the symbionts in B. heckerae gill bacteriocytes, where they co-occurred with previously described thiotrophic and methanotrophic symbionts28 (Fig. 2a, b). The relative abundance of Cycloclasticus in five B. heckerae individuals, based on estimates from Illumina metagenomic reads that mapped to the Cycloclasticus 16S rRNA gene, ranged from 5.1 – 10.7% (Supplementary Table 1). We also examined four B. brooksi individuals co-occurring with B. heckerae in Chapopote using sequencing (15 - 49 million Illumina reads), FISH and PCR (see Supplementary Methods). As in the two B. brooksi individuals collected at Chapopote in 2006, no Cycloclasticus were found in B. brooksi individuals collected in 2015.
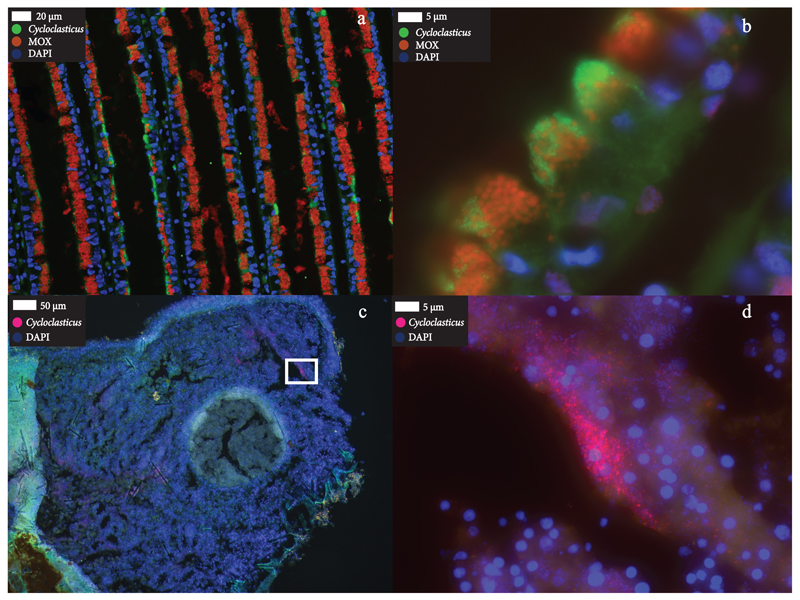
Figure 2. FISH images of Cycloclasticus endosymbionts.
a, Patches of Cycloclasticus symbionts (green) in B. heckerae gill filaments that also host methanotrophic symbionts (MOX, red). b, Intracellular Cycloclasticus symbionts (green) co-occur with the larger methanotrophic symbionts (red) intracellularly in bacteriocytes of B. heckerae gill tissues. c, Cycloclasticus (pink) in the encrusting sponge. d, High-resolution image of sponge tissue framed with a white box in (c), showing Cycloclasticus within the sponge tissues (For a 3D reconstruction from 2D z-stacks, see Supplement). Symbiotic Cycloclasticus were detected with FISH in gill tissues from four B. heckerae and one encrusting sponge individual.
Cycloclasticus were found in all three sponge individuals collected at Chapopote and Mictlan (Fig. 2c and d, Supplementary Table 1). Their abundances ranged between 4.1% and 8.0% based on the number of reads that mapped to the 16S rRNA gene of the sponge Cycloclasticus (Supplementary Table 1). The two encrusting sponges collected from Chapopote and Mictlan belonged to the same species based on 100% identical cytochrome c oxidase subunit I (COI) gene sequences, while the branching sponge individual belonged to a different species based on COI comparisons (10% nucleotide difference to COI of the encrusting sponge) and phylogenetic analyses (Supplementary Fig. 1). Both the encrusting and the branching sponge belong to the order Poecilosclerida based on their COI phylogeny (Suppplementary Fig. 1).
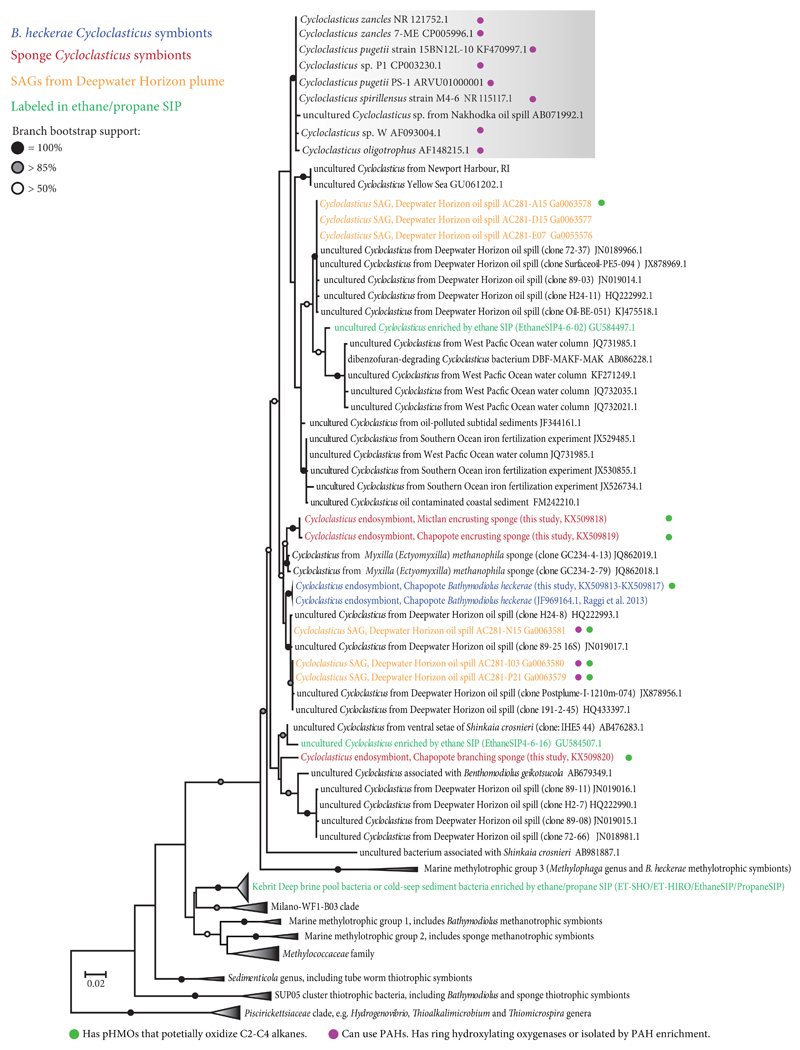
Phylogenetic analyses of the Cycloclasticus 16S rRNA gene sequences from the Campeche Knoll B. heckerae mussels and sponges revealed that they belonged to a closely-related clade (98% similarity) of cultivated and environmental Cycloclasticus often found in oil-contaminated environments (Fig. 3). The mussel and sponge Cycloclasticus were phylogenetically distinct from cultivated Cycloclasticus, such as C. pugetii, which have all been shown to grow on PAH (Fig. 3). In all three Campeche Knoll invertebrate species, the Cycloclasticus sequences were specific to the host species from which they originated and phylogenetically distinct from each other. The closest relatives of the Cycloclasticus from the encrusting sponge were Cycloclasticus from the hydrocarbon seep sponge Myxilla methanophila (Gulf of Mexico), while the Cycloclasticus of B. heckerae was most closely related to uncultured Cycloclasticus in the oil and gas plume from the DWH well blow-out (Fig. 3). The branching sponge Cycloclasticus were also related to bacteria from uncultured DWH Cycloclasticus, as well as bacteria from an ethane enrichment and from invertebrate hosts from chemosynthetic environments (Fig. 3).
Figure 3. Phylogeny of Cycloclasticus 16S rRNA genes.
The dataset included metagenomic 16S rRNA gene sequences from this study, sequences from the NCBI and Silva nucleotide database, and the JGI/IMG (158 sequences total). Bootstrap values below 50% are not shown. The tree is drawn to scale, with branch lengths representing the number of substitutions per site. The analysis included 1257 nucleotide positions.
Given that B. heckerae and the two sponge species each harbored a specific Cycloclasticus 16S rRNA phylotype that differed between these three host species, but was nearly identical between individuals of the same host species (including host individuals from the two collection sites Chapopote and Mictlan, separated by 25 km), we use the term 'symbiotic' for these host-specific bacteria.
Symbiotic Cycloclasticus genotypes are specific to their host species
We assembled draft Cycloclasticus genomes from the metagenomic sequencing of four B. heckerae individuals collected at Chapopote in 2015. The B. heckerae Cycloclasticus genomes had estimated sizes of 2.1-2.2 Mb, GC contents of 42% and were 93-97% complete (Supplementary Table 3). The Cycloclasticus genomes from the four B. heckerae individuals were highly similar based on their average nucleotide identity (ANI) (≥ 99.95%, Supplementary Table 4) and phylogenomic analyses based on eleven single copy genes (Supplementary Figure 2).
Draft Cycloclasticus genomes from the two sponge species had estimated sizes of 1.6-2.3 Mb, GC contents of 43-44%, and were 90-95% complete (Supplementary Table 3). The Cycloclasticus genomes from the two encrusting sponge individuals were highly similar to each other (ANI = 99.8%), despite the fact that these hosts were collected at two different sites separated by 20 km (Chapopote and Mictlan, Figure 1). Their genomes differed considerably from the Cycloclasticus genome of the branching sponge (ANI = 79.8%) and the B. heckerae mussels (ANI values below 80%, Supplementary Table 4). Correspondingly, phylogenomic analyses confirmed that the sponge Cycloclasticus symbionts differ from each other and those of B. heckerae. Taken together, ANI values, phylogenetic 16S rRNA and phylogenomic analyses, all provide support for the conclusion that the three invertebrate species examined here, B. heckerae and two sponge species, harbor highly host-specific Cycloclasticus symbionts.
Genes for PAH degradation could not be detected in symbiotic Cycloclasticus
One of the key characteristics of Cycloclasticus is their ability to degrade PAHs, which they can use as their sole energy and carbon source. Correspondingly, the genomes of cultivated Cycloclasticus contain multiple clusters of genes involved in the degradation of PAHs (Supplementary Fig. 3, Supplementary Table 5). Remarkably, none of the symbiotic Cycloclasticus genomes examined in this study, neither from the mussels nor the two sponge species, contained genes or transcripts attributable to PAH degradation (Supplementary Note 1-2, Supplementary Fig. 3). These genomic and transcriptomic results were supported by incubation experiments on board with Cycloclasticus-bearing B. heckerae gill tissues: Despite numerous attempts, we never observed oxidation of 14C-labelled naphthalene to 14CO2 by gill tissues, while control experiments with Cycloclasticus pugetii ATCC 51542 always showed naphthalene oxidation (see Supplementary Note 1 and Supplementary Methods). These results contradict an earlier study on two B. heckerae individuals collected at Chapopote in 2006, in which ring-hydroxylating oxygenase genes known to play a role in the degradation of aromatic compounds were amplified from mussel gill tissues28. It is possible that these PCR amplified sequences originated from environmental contaminants rather than gill-associated symbionts.
Symbiotic Cycloclasticus use short-chain alkanes as an energy and carbon source
We found genomic, transcriptomic and proteomic evidence for the use of short-chain alkanes as an energy and carbon source in the Cycloclasticus symbionts examined in this study (Fig. 4). The first step in the oxidation of short-chain alkanes is their oxidation to an alcohol, and we hypothesize that this step is catalyzed by copper-containing membrane-associated monooxygenases (CuMMO) in the symbiotic Cycloclasticus (Fig. 4 and 5). CuMMOs that oxidize hydrocarbon compounds are called particulate hydrocarbon monooxygenases (pHMOs), and the most well characterized of these is particulate methane monooxygenase (pMMO), which oxidizes methane to methanol. It is now known that the phylogenetic and functional diversity of pHMOs is much greater than previously recognized, and pHMOs that are capable of oxidizing short-chain hydrocarbons have been recently described30.
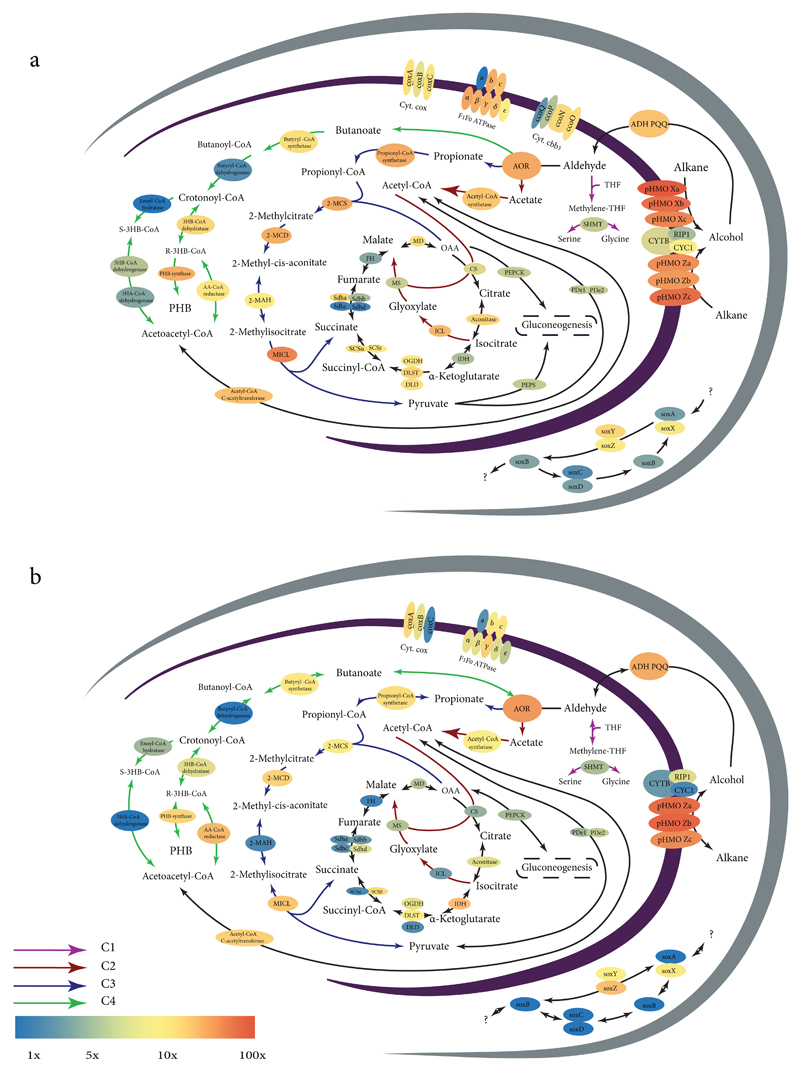
Figure 4. Reconstruction of central carbon and energy metabolic pathways in symbiotic Cycloclasticus.
a, B. heckerae Cycloclasticus symbiont. b, Encrusting sponge Cycloclasticus symbiont. Pathways used for the assimilation of C1-C4 alkanes, as well as for respiration and sulfur oxidation (see Supplementary Notes 7, 10) are shown. Colored arrows indicate C1-C4 assimilation pathways, black arrows indicate general metabolic pathways. Enzyme background colors represent mean expression values in the B. heckerae transcriptomes (BHT1-4) or sponge transcriptomes (ST1-2), normalized to DNA gyrase subunit B (gyrB) expression values in respective samples. These values are described in detail in Supplementary Tables 5 and 6.
The following enzymes are abbreviated: particulate hydrocarbon monooxygenase (pHMO, subunits a-c); PQQ-dependent alcohol dehydrogenase (ADH PQQ); aldehyde oxidoreductase (AOR); cytochrome C oxidase (Cyt. Cox, coxA-C subunits); cytochrome C oxidase cbb3 type (Cyt. Cbb3, ccoN-Q subunits); cytochrome C reductase (CYTB, RIP1, CYC1 subunits); citrate synthase (CS); isocitrate dehydrogenase (IDH); 2-oxoglutarate dehydrogenase complex (DLST – dihydrolipoamide succinyltransferase component E2, DLD – dihydrolipoamide dehydrogenase, OGDH – 2-oxoglutarate dehydrogenase E1); succinyl-CoA ligase (SCS, alpha and beta chains); succinate dehydrogenase (Sdha – iron-sulfur protein, Sdhb – flavoprotein subunit, Sdhc – succinate dehydrogenase cytochrome b-556 subunit and Sdhd – succinate dehydrogenase hydrophobic membrane anchor protein); fumarate hydratase (FH); malate dehydrogenase (MD); isocitrate lyase (ICL); malate synthase (MS); 2-methylcitrate synthase (2-MCS); 2-methylcitrate dehydratase (2-MCD); 2-methylaconitate isomerase (2-MAH); methylisocitrate lyase (MICL); phosphoenolpyruvate carboxykinase (PEPCK); phosphoenolpyruvate synthase (PEPS); serine hydroxymethyltransferase (SHMT); pyruvate dehydrogenase (PD, e1 and e2 subunits).
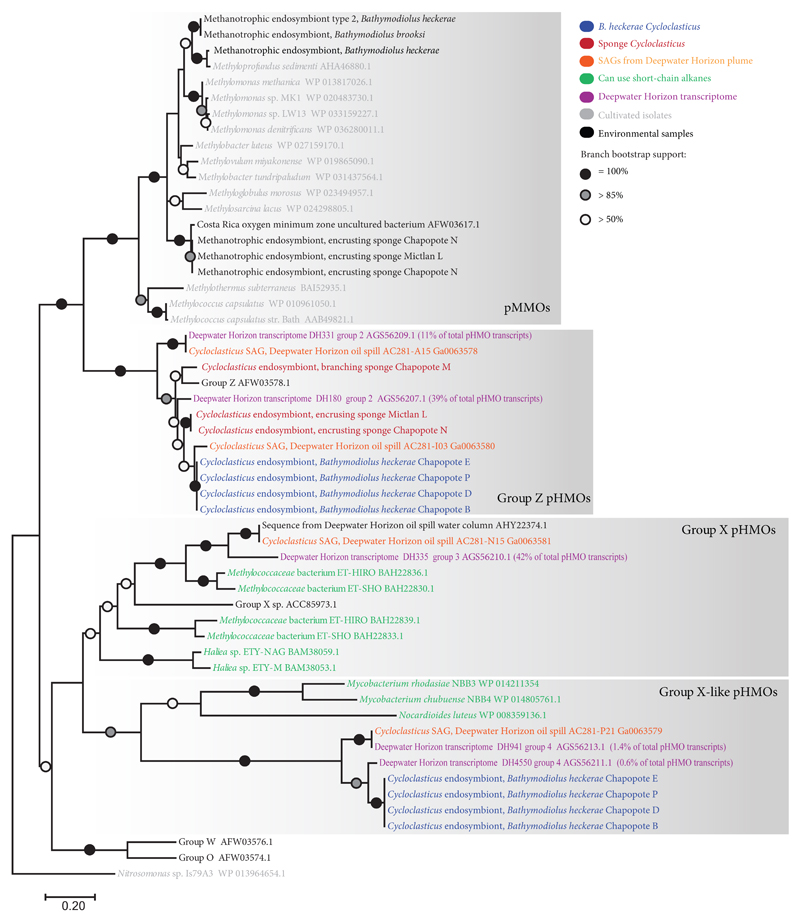
Figure 5. Phylogeny of particulate hydrocarbon monooxygenase (pHMO) subunit A protein sequences.
The tree is drawn to scale, with branch lengths representing the number of substitutions per site. Bootstrap values below 50% are not shown. The analysis was based on 55 protein sequences and included 219 amino acid positions. pMMO = particulate methane monooxygenase, pHMO = particulate hydrocarbon monooxygenase.
We found genes encoding pHMOs (pmoCAB) in the Cycloclasticus symbionts of all three host species. These genes were genetically distant and phylogenetically distinct from the pMMO genes of the methane-oxidizing symbionts of these hosts and were most closely related to bacteria found in the DWH oil plume (Fig. 5). The Cycloclasticus symbiont pmoA sequences belonged to two groups: Group Z31 and a clade of pHMOs related to those of short-chain alkane degrading Nocardioides32 and Mycobacterium33. The latter clade formed a sister group to Group X pHMOs, which are suggested to have a high affinity to ethane34 or ethylene35 (Fig. 5). Hereafter we refer to this clade as Group X-like pHMOs. Transcriptomic analyses of the symbiotic Cycloclasticus from four B. heckerae individuals and the two encrusting sponge individuals revealed that their pHMO genes were the most highly expressed genes compared to all other genes involved in carbon metabolism (Fig. 4, Supplementary Tables 6, 7) (transcriptomic sequencing of the branching sponge was not possible, as insufficient RNA was recovered). Proteomics revealed that the subunit A of Group Z pHMO was among the most highly expressed Cycloclasticus proteins in three B. heckerae individuals (Supplementary Table 8).
The second step in the oxidation of short-chain alkanes, after these have been oxidized to alcohol, is the formation of an aldehyde. In many bacteria that oxidize alcohols, this reaction is catalyzed by pyrroloquinoline quinone-dependent alcohol dehydrogenases (PQQ-ADH). Indeed, we found genes encoding PQQ-ADHs in the symbiotic Cycloclasticus of all three host species, and these were also highly expressed in their transcriptomes (Fig. 4, Supplementary Tables 6 and 7). The membrane-bound PQQ-ADH was also found in the Cycloclasticus proteome of one B. heckerae individual (Supplementary Table 8). The symbiotic Cycloclasticus PQQ-ADH sequences were highly similar (91%) to PQQ-ADHs assumed to oxidize ethanol and higher alcohols, rather than methanol36 (Supplementary Fig. 4).
In the third step of short-chain alkane degradation, the aldehydes are oxidized to carboxylic acids. This step could be carried out by the PQQ-ADHs of the symbiotic Cycloclasticus, as these enzymes have been shown to oxidize aldehydes to carboxylic acids in butane-degrading Pseudomonas spp.37. This third step could also be achieved via tungsten-containing aldehyde ferredoxin oxidoreductases (AORs), which are known to use short-chain alkane derived aldehydes as their substrate38. AORs were highly expressed in the transcriptomes of Cycloclasticus symbionts of all three host species, and also present in the proteomes of Cycloclasticus from two B. heckerae individuals (Supplementary Table 8, 9).
The genomes of the Cyclolcasticus symbionts from all three host species encoded genes that enabled them to fix carbon derived from C2-C4 alkanes (Fig. 4), and many of these genes were highly expressed. These pathways are discussed in detail in the Supplementary Notes 7 - 8. While carbon derived from C2 – C4 alkanes appears to play a central role in the metabolism of the symbiotic Cycloclasticus, it is unlikely that they can use methane or other C1 compounds. While pMMOs and PQQ-ADHs are also highly expressed in metabolically active methanotrophs39,40, their genes are phylogenetically distant from those of the symbiotic Cycloclasticus, suggesting functional divergence. Moreover, genes encoding key enzymes in C1 assimilation pathways such as the ribulose monophosphate (RuMP) pathway, the serine cycle, and the Calvin-Benson-Bassham (CBB) cycle were not present in the Cycloclasticus symbiont genomes.
In summary, given that genes involved in PAH degradation were not detected in our metagenomic, metatranscriptomic and metaproteomic data, and the high expression levels of genes involved in the use of short-chain alkanes such as pHMOs, PQQ-ADHs, and AORs, the symbiotic Cycloclasticus investigated in this study most likely use gaseous non-aromatic short-chain hydrocarbons as energy and carbon sources. This is surprising, as all cultivated Cycloclasticus are able to degrade PAH, but do not appear to use short-chain alkanes. The genomes of cultivated Cycloclasticus lack genes coding for the first two enzymes needed for their oxidation, pHMOs and PQQ-ADHs. AORs, the third group of enzymes, are present in the genomes of cultivated Cycloclasticus but their protein sequences are only 70% similar to those of the symbiotic Cycloclasticus (Supplementary Fig. 5).
Deepwater Horizon Cycloclasticus may also be able to degrade short-chain alkanes
We found genes encoding the first three enzymes central to the oxidation of short-chain alkanes, pHMO, PQQ-ADH and AOR, in genomes and transcriptomes from the DWH hydrocarbon plume that were highly similar to those of the symbiotic Cycloclasticus from Campeche Knoll hosts (Fig. 5, Supplementary Fig. 4 and 5). The Group X-like pHMOs, which were highly expressed in the mussel Cycloclasticus, were also present in a single amplified genome, SAG AC281-P21, and transcripts from the DWH oil plume36. These sequences formed a highly supported monophyletic clade with the Cycloclasticus symbionts (Fig. 5). Correspondingly, the 16S rRNA sequence from the DWH SAG AC281-P21 belonged to the same clade as those from the B. heckerae Cycloclasticus symbionts (Fig. 3). The Group Z pHMOs from the Campeche Knoll mussel and sponge Cycloclasticus belonged to a monophyletic clade that included two DWH SAGs, as well as transcripts that accounted for 50% of all pmoA transcripts in a DWH plume transcriptome36 (Fig. 5). This close phylogenetic relationship was supported by similarly close 16S rRNA gene sequences of one of the two SAGs (AC281-I03) and those of the Cycloclasticus symbionts from B. heckerae and the encrusting sponge (Fig. 3). (The second SAG, AC281-N15, also belonged to the genus Cycloclasticus based on its 16S rRNA gene sequence, but was more distantly related to the symbiotic Cycloclasticus.)
As with the pHMOs, the PQQ-ADH sequences of the Cycloclasticus symbionts were most closely related to PQQ-ADH sequences from DWH bacteria, and formed a highly supported clade with SAG AC281-P21, as well as transcripts from the hydrocarbon plume, with the latter constituting 38% of the total PQQ-ADH transcripts in the plume36 (Supplementary Fig. 4). Symbiotic, free-living and cultivated Cycloclasticus contain AORs (Supplementary Fig. 5). AOR sequences vary considerably within the Cycloclasticus genus and form several distinct phylogenetic clades. Interestingly, SAG AC281-P21, which contained both pHMOs and PQQ-ADH, also had two copies of AOR, one that was similar to those of cultivated Cycloclasticus and the other that was similar to those of the symbiotic Cycloclasticus. SAG AC281-P21, AC281-I03 and AC281-N15, also contained PAH-degradation genes such as dioxygenases similar to those of cultivated Cycloclasticus (Fig.3). Hence, unlike the cultivated and the symbiotic Cycloclasticus, some free-living Cycloclasticus may have the ability to use both PAH and short-chain alkanes.
Short-chain alkanes are abundant in natural gas and oil seeps
Given the strong genomic and transcriptomic evidence for the use of short-chain alkanes by symbiotic Cycloclasticus from Campeche Knolls, we asked if these alkanes are present in their environment. In previous studies at Chapopote, high concentrations of C2 – C4 alkanes (in the low mM range) were measured at 2 – 10 cm depth in surface asphalts and the fractions of these compounds ranged between 3 – 33% of total light hydrocabons41. In this study, we measured the concentrations and relative proportions of short-chain alkanes in the environment of the symbiont-bearing invertebrates at Campeche Knolls by sampling and analyzing gas and oil bubbles a few centimeters above the invertebrate collection sites at Chapopote and Mictlan, and a piece of surface asphalt with gas hydrate from the immediate vicinity of mussels and sponges at Chapopote. Concentrations of ethane, propane and butane in the asphalt were in the µM to low mM range, and their relative fractions ranged between 7-8% for C2, 1-4% for C3 and 0.5 – 1% for C4 isomers (Table 1a). Gas and oil bubbles from Chapopote and Mictlan contained considerable fractions of C2 (1.5% and 16%), C3 (0.4% and 14.5%), and C4 (0.06 – 3.0%) (Table 1b). For comparison, in the DWH plume, which was dominated by propane and ethane degrading bacteria in the late stages of the plume10, the dissolved gas in the plume contained similar concentrations and proportions of ethane and propane (for C2: 0.01-0.03 mM, 8-9%, for C3: ca. 0.01-0.02 mM, 4-6%, for C4: 0.01-0.03 mM, 2-3%)42. These results show that in both anthropogenically-induced and natural seafloor hydrocarbon seepage in the Gulf of Mexico, short-chain alkanes are present in considerable concentrations that are likely sufficient to support the growth of alkane-degrading bacteria.
Table 1. Alkanes in fresh surface asphalts.
a, Concentrations of low-molecular-weight alkanes [in µmol L-1 bulk asphalt] and resulting C1/(C2-C5) ratios in gas samples prepared from an asphalt/gas hydrate piece from one of the Chapopote invertebrate collection sites (GeoB19325-9). b, The relative composition of light hydrocarbons [in mol% Σ(C1-C6)] and resulting C1/(C2-C6) ratios in gas (Chapopote) and oil (Mictlan) bubbles above invertebrate collection sites (see Fig. 1).
| a) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample location on asphalt / hydrate piece | C2 | C3 | i-C4 | n-C4 | n-C5 | C1/(C2-C5) | C1/C2 | C1/C3 | |
| outside of microbial mat | 1,026.5 | 8.8 | n.d. | 9.7 | 4.4 | 20.5 | 20.9 | 2440.1 | |
| inside microbial mat | 396.9 | 67.9 | 23.2 | 73.9 | 28.6 | 8.9 | 13.2 | 77.2 | |
| below microbial mat | 839.4 | 376.7 | 57.1 | 411.6 | 128.7 | 4.7 | 10.2 | 22.8 | |
| b) | |||||||||
| Collection site | Station no. GeoB | C1 | C2 | C3 | i-C4 | n-C4 | Σ C5-isomers | Σ C6-isomers | C1/(C2-C6) |
| Chapopote | 19325-13 | 97.6 | 1.5 | 0.4 | 0.1 | 0.2 | 0.1 | 0.1 | 41.4 |
| Mictlan | 19336-5 | 64.1 | 16.0 | 14.5 | 0.1 | 3.0 | 1.2 | 0.4 | 1.8 |
Conclusions
Our study provides genomic and transcriptomic evidence for the use of short-chain alkanes by symbiotic and free-living Cycloclasticus. We thus provide a direct link between the phylogenetic identity of these bacteria and their metabolic function. Our results confirm earlier studies that found indirect evidence for the use of short-chain alkanes by Cycloclasticus based on their dominance in DWH plumes with high rates of propane and ethane consumption, and stable isotope probing with propane and ethane at natural hydrocarbon seeps10,34. These results show that metabolic versatility within the clade of closely related Cycloclasticus (>98% 16S rRNA identity) is higher than previously assumed and not limited to PAH degradation.
Within the very closely related clade of Cycloclasticus that have been cultivated (Fig. 3, top clade), we found no evidence for genes involved in the use of short-chain alkanes. These bacteria appear to rely solely on PAH degradation. In contrast, SAGs from the DWH contained genes for the use of both PAH and short-chain alkanes. Such a versatile metabolism could be advantageous for Cycloclasticus in their ephemeral environment. During the early development of hydrocarbon plumes, short-chain alkanes are abundant, and their use costs less energy than that of PAHs. However, as plumes mature, the concentrations of short-chain alkane can decrease, and the plume Cycloclasticus can then switch to using PAHs10,43,44.
It is intriguing that the symbiotic Cycloclasticus are the only bacteria within this genus that lack the ability to degrade PAHs, despite the high concentrations of PAHs in Campeche Knoll asphalts45. (Some SAGs from DWH do not have genes for PAH degradation (Fig. 3), but these genomes are only 25 – 50 % complete.). Unlike DWH oil plume Cycloclasticus, symbionts experience a consistent supply of short-chain alkanes, determined by the host’s location above a hydrocarbon source. The host can enhance the metabolic capacity of symbionts by providing a continuous supply of dissolved gases via active pumping. Moreover, given their protected environment within host tissues, the Cycloclasticus symbionts would not experience competition with free-living short-chain alkane degraders. Furthermore, the low aqueous solubility of PAH compared to short-chain alkanes, as well as diffusion barriers across the host cell membrane could limit the access of these symbionts to PAH. The selective advantage in maintaining the large suite of genes needed for PAH degradation appears to no longer exist for symbiotic Cycloclasticus. Similarly, we could envision a similar selection-driven loss in free-living Cycloclasticus from environments with a continuous supply of short-chain alkanes, such as surface asphalt and sediments at hydrocarbon-rich seeps. Future molecular and physiological studies on both symbiotic and free-living Cycloclasticus are needed to test these hypotheses and gain a better understanding how this group of bacteria contributes to the consumption of hydrocarbons.
Methods
Sample collection
Mussels and sponges were collected with the remotely-operated vehicle (ROV) MARUM-Quest 4000 m during the RV Meteor M114-2 cruise to the Campeche Knolls in March 2015 (Fig. 1a). B. heckerae and B. brooksi mussels were collected during three dives from the Chapopote “bubble” site (21°54' N; 93°26' W) at a water depth of 2925 m. This site is characterized by the presence of fresh asphalts, exposed gas hydrates and flourishing faunal communities (Fig. 1b). An asphalt piece inhabited by both branching and encrusting sponges was also recovered from the “bubble” site. Another encrusting sponge was collected at Mictlan Knolls from a site at 3106 m water depth that was characterized by the presence of fresh asphalts and considerable gas and oil bubbling (22°1' N; 93°14' W, Fig. 1c). A detailed description of the collection sites is available elsewhere24,25.
Mussels were identified on board based on their morphology46. Their correct morphological identification was later confirmed based on 100% similarity of their cytochrome c oxidase subunit I (COI) genes to those collected from Chapopote in 200628 and the published COI sequences for these species47 (see 'Genome analysis' below). The symbiont-bearing gills of the mussels were dissected and fixed immediately after retrieval as described below. The sponges appeared intact immediately after collection, with no visible evidence of tissue damage. Encrusting sponge tissue was carefully removed from the underlying asphalt with a scalpel. The Mictlan sponge individual was fixed immediately upon retrieval. The Chapopote sponges were kept for several hours on the asphalt piece to which they were attached in a bucket containing water collected together with the sample at 4 °C until processed. Samples for transcriptomic and metagenomic analyses were fixed in RNAlater (Sigma, Steinheim, Germany) according to the manufacturer’s instructions and stored at -80 °C. Samples for microscopy were fixed in 2% paraformaldehyde in 1X phosphate-buffered saline (PBS) for at most 12 h at 4 °C, rinsed three times in 1X PBS, and stored at 4 °C in 0.5X PBS/50% ethanol.
Gas sampling and analysis
Gas bubbles (sample GeoB19325-13 from Chapopote, dive 354) and oil drops (GeoB19336-5 from Mictlan, dive 357) were collected with the MARUM-Quest ROV several centimeters above mussel beds with the pressure-tight Gas Bubble Sampler48. Additionally, three gas samples were prepared on board from different parts of an asphalt/gas hydrate piece (GeoB19325-9) recovered from Chapopote Knoll (this piece was not the same as the asphalt sample described above from which the sponges were collected). Molecular compositions of C1-C6-alkanes in gas and oil samples were determined by gas chromatography48.
Fluorescence in situ hybridization (FISH)
FISH with mono-labeled probes was performed on 8 μm sections of both sponge species and gill tissues from four B. heckerae individuals as previously described27. The Cycloclasticus probe CYPU-82949 was used at 10% formamide as previously described28 for the symbiotic Cycloclasticus together with probes targeting the B. heckerae methanotrophic and thiotrophic symbionts27. Images were acquired with Zeiss Axioplan 2 epifluorescence microscope (Zeiss, Jena, Germany).
DNA and RNA extraction and sequencing
To account for a potentially uneven spatial distribution of Cycloclasticus, we homogenized whole gills from mussels with shell lengths < 10 cm, with innuSPEED lysis tubes E (Analytik Jena, Jena, Germany). For larger mussels (> 15 cm), a 5 cm long gill subsample was homogenized. Following homogenization, DNA was extracted from six B. heckerae individuals (Supplementary Table 1), according to the standard protocol50, with the following adaptations: 25 μl homogenate aliquots were treated with 40 μl proteinase k and with 20 μl of 20 mg μl-1 RNase for 5 min at 37 °C. RNA was extracted using the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) following manufacturer’s instructions for both B. heckerae gill and sponge samples (Supplementary Table 1).
For the sponges, DNA and RNA were extracted in parallel with the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany). DNA/RNA quality was assessed with the Agilent 2100 Bioanalyzer. (Agilent, Santa Clara, USA). For the branching sponge individual only the amount of DNA, but not of RNA was sufficient for sequencing. Ovation RNA-seq System V2 (NuGen, San Carlos, CA, USA) was used to synthesize cDNA. Genomic DNA and cDNA libraries were generated with the DNA library prep kit for Illumina (BioLABS, Frankfurt am Main, Germany). Genomic DNA was sequenced as follows: 12.5 million 250 bp paired-end reads were generated, while further sequencing was done as 150 bp paired-end reads (Supplementary Table 1). cDNA libraries were sequenced as 100 bp paired-end reads. All samples were sequenced on the Illumina HiSeq 2500 platform at the Max Planck Genome Centre (Cologne).
Genome analysis
Individual metagenomes were assembled with IDBA-UD51, following decontamination, quality filtering (QV=2) and adapter-trimming of the reads with the BBDuk tool from the BBMap suite (Bushnell B, http://sourceforge.net/projects/bbmap/). To determine the relative abundance of Cycloclasticus 16S rRNA gene sequences within the metagenomes, we used the phyloFlash suite (https://github.com/HRGV/phyloFlash). Host identity was verified by aligning the COI gene sequences from the metagenomes with the respective sequences in the NCBI database.
The Cycloclasticus symbiont genomes were binned based on genome coverage, GC content and taxonomic affiliation using gbtools52, and the differential coverage approach53 when applicable (Supplementary Figure 3). To reassemble the genomes, we re-mapped Illumina reads to the bins using BBMap with 0.95 minimum identity. Paired read overlap was merged with BBMerge (BBMap suite) for the 250 bp reads. The reads were reassembled with Spades V3.754,55, using a maximum k-mer length of 127 and by supplying the merged reads as long single reads. Following manual removal of contigs shorter than 800 bp and contamination screening, quality metrics were calculated with Quast56 and CheckM57 (Supplementary Table 3). The genomes were annotated with RAST58 and the DOE-JGI Microbial Genome Annotation Pipeline59. The annotations were manually cross-checked and the annotations for the main genes discussed here were verified using NCBI’s BLAST60.
Average Nucleotide Identity61 (ANI) were calculated with the ANI calculator (http://enve-omics.ce.gatech.edu/ani/). To identify the presence/absence of key metabolic genes in symbiotic and cultivated Cycloclasticus (Cycloclasticus sp. P1), we blasted their genomes against each other using tblastx (minimum identity of 0.4 and 0.00001 P value cutoff). We used GView62 to explore the graphical representation of blast hits. We also mapped our metagenomic reads with BBMap using a minimum identity of 0.6 to the Cycloclasticus pugetii genome (GenBank accession number: NZ_ARVU01000001), to verify the results of the BLAST analyses (Supplementary Table 5).
Transcriptome analysis
Adapters and symbiont ribosomal RNA genes were removed from transcriptome reads with BBDuk. Transcriptome reads from each individual were mapped to the assembled Cycloclasticus genomes from the respective host with BBMap using a minimum identity value of 0.98. Mapped reads were assigned to genomic features with featureCounts63. To compare the transcriptome libraries of each individual, a normalization factor was estimated with calcNormFactors based on the trimmed mean of M-values (TMM) implemented in the edgeR package64. The TMM normalized read counts were converted to reads per kilobases of exon per million reads mapped (RPKM) with Rsubread R package (http://www.bioconductor.org). The expression values were normalized to expression of housekeeping genes with high general expression stability, such as those encoding the DNA gyrase subunit A and B, and RecA65. Sponge and mussel metatranscriptomes were assembled with Trinity66 to verify the absence of transcripts involved in PAH degradation pathways (two individual sponge assemblies and one mussel assembly combined from four B. heckerae transcriptomic libraries). Blast+67 was used to search for candidate genes, such as aromatic hydrocarbon mono- and dioxygenases.
Proteome analysis
Protein extraction and peptide preparation
We prepared tryptic digests of gill samples from three B. heckerae individuals also used for transcriptomic analyses, following the filter-aided sample preparation (FASP) protocol68 with some small modifications69. Peptides were not desalted. Approximate peptide concentrations were determined using the Pierce Micro BCA assay (Thermo Scientific Pierce, Rockford, IL, USA) following the manufacturer’s instructions.
1D-LC-MS/MS
Samples were analyzed by 1D-LC-MS/MS. For each mussel sample, three technical replicates were run. Two wash runs and one blank run were done between samples to reduce carry over. For each run, 2 to 7 µg of peptide were loaded onto a 5 mm, 300 µm ID C18 Acclaim® PepMap100 pre-column (Thermo Fisher Scientific) using an UltiMateTM 3000 RSLCnano Liquid Chromatograph (Thermo Fisher Scientific), and desalted on the pre-column. After desalting the peptides, the pre-column was switched in line with a 50 cm x 75 µm analytical EASY-Spray column packed with PepMap RSLC C18, 2µm material (Thermo Fisher Scientific), which was heated to 45° C. The analytical column was connected via an Easy-Spray source to a Q Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Peptides were separated on the analytical column and mass spectra acquired in the Orbitrap as described previously70. Roughly 650,000 MS/MS spectra were acquired per sample (three technical replicates combined).
Protein identification and quantification
For protein identification, a database was created using all protein sequences predicted from the B. heckerae Cycloclasticus symbiont genome published in this study (PRJNA318571), from preliminary genomes of the other B. heckerae symbionts, and from an EST library of the related host species B. azoricus71. CD-HIT was used to remove redundant sequences from the database72. The cRAP protein sequence database (http://www.thegpm.org/crap/) containing protein sequences of common laboratory contaminants was appended to the database. The final database contained 52,997 protein sequences. For protein identification, MS/MS spectra were searched against the database using the Sequest HT node in Proteome Discoverer version 2.0.0.802 (Thermo Fisher Scientific) as described previously70. Only proteins identified with medium or high confidence were retained, resulting in an overall false discovery rate of <5%. Between 4960 and 7742 proteins were identified per sample. The identified Cycloclasticus proteins are summarized in Supplementary Table 8.
For protein quantification, normalized spectral abundance factors (NSAFs) were calculated for the Cycloclasticus proteins based on the number of PSMs per protein73 and multiplied by 100. The NSAF% gives the relative abundance of a protein in a sample in % as compared to all other Cycloclasticus proteins (Supplementary Table 9).
Cycloclasticus SAG generation and analysis
Samples were collected from the deep sea hydrocarbon plume created by the Deepwater Horizon on June 15, 201010. The water sample used for SAG generation was collected from site 15, 12 km from the leaking wellhead at a depth of 1120 m. 1 ml seawater was cryopreserved with 6% glycine betaine and stored at -80°C. Single cell sorting, whole genome amplification, and 16S rRNA gene PCR screening were performed at the Bigelow Single Cell Genomics Center, as described by Swan and colleagues74. High-quality 16S rRNA gene sequences were obtained from 170 SAGs; 54 of these were identified as Cycloclasticus and 6 were selected for sequencing. SAG sequencing, assembly, contamination screening, and annotation were performed by the DOE Joint Genome Institute (JGI) following standard JGI protocols for microbial single cell genomes.
Phylogenetic analyses
The evolutionary history of protein and 16S rRNA gene sequences was inferred by using the Maximum Likelihood method based on the Le Gascuel 2008 model75 and Kimura 2-parameter model76 for amino acid and nucleotide sequences, respectively. The trees with the highest log likelihood were chosen. The percentage of trees in which the associated taxa clustered together was determined based on 200 and 1000 bootstrap resamples for amino acid and nucleotide sequences, respectively. Phylogenetic analyses were conducted in MEGA777.
Sequence data availability
COI gene sequences were submitted to GenBank under accession numbers KU659139 (B. heckerae) and KU659136-8 (sponges). Genomes with curated metadata are available through the Integrated Microbial Genome (IMG) database78 (analysis project GOLD IDs are summarized in Supplementary Table 2). Genomes were also submitted to NCBI in BioProjects PRJNA318571-3. Transcriptome reads that mapped to the Cycloclasticus symbionts were deposited in the European Nucleotide Archive under the accession number PRJEB12576. SAG assembly statistics and annotations are available through the Integrated Microbial Genome (IMG) database under IMG taxon IDs 2599185276, 2599185294, 2599185283, 2602042074, 2599185280, and 2599185270. The mass spectrometry proteomics data and the protein sequence database were deposited in the ProteomeXchange Consortium79 via the PRIDE partner repository with the dataset identifier PXD005351.
Supplementary Material
Acknowledgements
The authors thank all who helped during the R/V Meteor research cruise M114, including onboard technical and scientific personnel, the captain and crew, and the ROV MARUM-Quest 4000 m team. Dr. Andrew Crombie, University of East Anglia, UK, is thanked for useful discussions on propane metabolism. We thank Dr. Adam Rivers, DOE Joint Genome Institute, for sharing DWH plume PQQ-ADH transcript sequences. We thank Marc Strous for access to proteomics equipment and Erin Thorson for technical assistance with the determination of peptide concentrations. The purchase of the proteomics equipment was supported by a grant of the Canadian Foundation for Innovation to Marc Strous. We thank the Max Planck-Genome-Centre Cologne (http://mpgc.mpipz.mpg.de/home/) for generating the metagenomic and the metatranscriptomic data used in this study. Cycloclasticus SAG sequencing was supported by the Joint Genome Institute’s Community Science Program. The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science, Biological and Environmental Research Program of the U.S. Department of Energy, and by the University of California, Lawrence Berkeley National Laboratory under Contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under Contract No. DE-AC02-06NA25396. The Campeche Knoll cruise was funded by the German Research Foundation (DFG – Deutsche Forschungsgemeinschaft). Additional support was provided through the MARUM DFG-Research Center / Excellence Cluster “The Ocean in the Earth System” at the University of Bremen. We are grateful to the Mexican authorities for granting permission to conduct this research in the southern Gulf of Mexico (permission of DGOPA: 02540/14 from 5 November 2014). This study was funded by the Max Planck Society and the MARUM DFG-Research Center / Excellence Cluster “The Ocean in the Earth System” at the University of Bremen. Further support was provided by an ERC Advanced Grant (BathyBiome, 340535), and a Gordon and Betty Moore Foundation Marine Microbial Initiative Investigator Award to ND (Grant GBMF3811). DLV and MCR received funding from the US National Science Foundation, grants OCE-1155855 and OCE-1046144. CPA was supported by a postdoctoral fellowship from the Humboldt Foundation. MK was supported by a NSERC Banting Postdoctoral Fellowship.
Footnotes
Author contributions
MRB, CB, CPA and ND conceived the study. HS and GB provided the framework for deep sea sample collections. MRB and CB processed the samples onboard. MRB, CPA and LS analyzed the biological samples. MK prepared samples for proteomics, generated, processed and analyzed proteomic data. TP provided the short-chain alkane analyses. MCR and DLV collected, prepared and sequenced the samples for the Deepwater Horizon Cycloclasticus SAGs and MCR; DLV, MRB and CPA analyzed their genomes. MRB and ND wrote the manuscript with contributions from all co-authors.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Bolin B. Changing global biogeochemistry. In: Brewer PG, editor. Oceanography. Springer; US, New York: 1983. pp. 305–326. [Google Scholar]
- 2.Dando PR, Hovland M. Environmental effects of submarine seeping natural gas. Cont Shelf Res. 1992;12:1197–1207. [Google Scholar]
- 3.Cicerone RJ, Oremland R. Biogeological aspects of atmospheric methane. Global Biogeochem Cycles. 1988;2:229–327. [Google Scholar]
- 4.Collins WJ, Derwent RG, Johnson CE, Stevenson DS. The oxidation of organic compounds in the troposphere and their global warming potentials. Clim Change. 2002;52:453–479. [Google Scholar]
- 5.Katzenstein AS, Doezema LA, Simpson IJ, Blake DR, Rowland FS. Extensive regional atmospheric hydrocarbon pollution in the southwestern United States. Proc Natl Acad Sci USA. 2003;100:11975–11979. doi: 10.1073/pnas.1635258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterson C, et al. Long-term ecosystem response to the Exxon Valdez oil spill. Science. 2003;302:2082–2086. doi: 10.1126/science.1084282. [DOI] [PubMed] [Google Scholar]
- 7.Head IM, Swannell RP. Bioremediation of petroleum hydrocarbon contaminants in marine habitats. Curr Opin Biotechnol. 1999;10:234–239. doi: 10.1016/S0958-1669(99)80041-X. [DOI] [PubMed] [Google Scholar]
- 8.Joye SB, et al. The Gulf of Mexico ecosystem, six years after the Macondo oil well blowout. Deep Sea Res Part II. 2016;129:4–19. [Google Scholar]
- 9.Redmond MC, Valentine DL. Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc Natl Acad Sci USA. 2012;109:20292–20297. doi: 10.1073/pnas.1108756108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valentine DL, et al. Propane respiration jump-starts microbial response to a deep oil spill. Science. 2010;330:208–211. doi: 10.1126/science.1196830. [DOI] [PubMed] [Google Scholar]
- 11.Kleindienst S, et al. Diverse, rare microbial taxa responded to the Deepwater Horizon deep-sea hydrocarbon plume. ISME J. 2015;10:400–415. doi: 10.1038/ismej.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dombrowski N, et al. Reconstructing metabolic pathways of hydrocarbon-degrading bacteria from the Deepwater Horizon oil spill. Nat Microbiol. 2016;1:16057. doi: 10.1038/nmicrobiol.2016.57. [DOI] [PubMed] [Google Scholar]
- 13.Hazen TC, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 14.Seo J-S, Keum Y-S, Li QX. Bacterial degradation of aromatic compounds. Int J Environ Res Public Health. 2009;6:278–309. doi: 10.3390/ijerph6010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez T, et al. Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP. ISME J. 2013;7:2091–2104. doi: 10.1038/ismej.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kappell AD, et al. The polycyclic aromatic hydrocarbon degradation potential of Gulf of Mexico native coastal microbial communities after the Deepwater Horizon oil spill. Front Microbiol. 2014;5:00205. doi: 10.3389/fmicb.2014.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasai Y, Kishira H, Harayama S. Bacteria belonging to the genus Cycloclasticus play a primary role in the degradation of aromatic hydrocarbons released in a marine environment. Appl Environ Microbiol. 2002;68:5625–5633. doi: 10.1128/AEM.68.11.5625-5633.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yakimov MM, Timmis KN, Golyshin PN. Obligate oil-degrading marine bacteria. Curr Opin Biotechnol. 2007;18:257–266. doi: 10.1016/j.copbio.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Kasai Y, Shindo K, Harayama S, Misawa N. Molecular characterization and substrate preference of a polycyclic aromatic hydrocarbon dioxygenase from Cycloclasticus sp. strain A5. Appl Environ Microbiol. 2003;69:6688–6697. doi: 10.1128/AEM.69.11.6688-6697.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyksterhouse SE, Gray JP, Herwig RP, Lara JC, Staley JT. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int J Syst Bacteriol. 1995;45:116–123. doi: 10.1099/00207713-45-1-116. [DOI] [PubMed] [Google Scholar]
- 21.Chung WK, King GM. Isolation, characterization, and polyaromatic hydrocarbon degradation potential of aerobic bacteria from marine macrofaunal burrow sediments and description of Lutibacterium anuloederans gen. nov., sp. nov., and Cycloclasticus spirillensus sp. nov. Appl Environ Microbiol. 2001;67:5585–5592. doi: 10.1128/AEM.67.12.5585-5592.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui Z, Xu G, Li Q, Gao W, Zheng L. Genome sequence of the pyrene- and fluoranthene-degrading bacterium Cycloclasticus sp. strain PY97M. Genome Announc. 2013;1:e00536–13. doi: 10.1128/genomeA.00536-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Messina E, et al. Genome sequence of obligate marine polycyclic aromatic hydrocarbons-degrading bacterium Cycloclasticus sp. 78-ME, isolated from petroleum deposits of the sunken tanker Amoco Milford Haven, Mediterranean Sea. Mar Genomics. 2015;25:11–13. doi: 10.1016/j.margen.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 24.MacDonald IR, et al. Asphalt volcanism and chemosynthetic life in the Campeche Knolls, Gulf of Mexico. Science. 2004;304:999–1002. doi: 10.1126/science.1097154. [DOI] [PubMed] [Google Scholar]
- 25.Sahling H, et al. Massive asphalt deposits, oil seepage, and gas venting support abundant chemosynthetic communities at the Campeche Knolls, southern Gulf of Mexico. Biogeosciences. 2016;13:4491–4512. [Google Scholar]
- 26.Petersen JM, Wentrup C, Verna C, Knittel K, Dubilier N. Origins and evolutionary flexibility of chemosynthetic symbionts from deep-sea animals. Biol Bull. 2012;223:123–137. doi: 10.1086/BBLv223n1p123. [DOI] [PubMed] [Google Scholar]
- 27.Duperron S, et al. Diversity, relative abundance and metabolic potential of bacterial endosymbionts in three Bathymodiolus mussel species from cold seeps in the Gulf of Mexico. Environ Microbiol. 2007;9:1423–1438. doi: 10.1111/j.1462-2920.2007.01259.x. [DOI] [PubMed] [Google Scholar]
- 28.Raggi L, Schubotz F, Hinrichs K-U, Dubilier N, Petersen JM. Bacterial symbionts of Bathymodiolus mussels and Escarpia tubeworms from Chapopote, an asphalt seep in the Southern Gulf of Mexico. Environ Microbiol. 2013;15:1969–1987. doi: 10.1111/1462-2920.12051. [DOI] [PubMed] [Google Scholar]
- 29.Arellano SM, et al. Deep sequencing of Myxilla (Ectyomyxilla) methanophila, an epibiotic sponge on cold-seep tubeworms, reveals methylotrophic, thiotrophic, and putative hydrocarbon-degrading microbial associations. Microb Ecol. 2013;65:450–461. doi: 10.1007/s00248-012-0130-y. [DOI] [PubMed] [Google Scholar]
- 30.Li M, Jain S, Baker BJ, Taylor C, Dick GJ. Novel hydrocarbon monooxygenase genes in the metatranscriptome of a natural deep-sea hydrocarbon plume. Environ Microbiol. 2014;16:60–71. doi: 10.1111/1462-2920.12182. [DOI] [PubMed] [Google Scholar]
- 31.Tavormina PL, Ussler W, Orphan VJ. Planktonic and sediment-associated aerobic methanotrophs in two seep systems along the North American Margin. Appl Environ Microbiol. 2008;74:3985–3995. doi: 10.1128/AEM.00069-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamamura N, Yeager CM, Arp DJ. Two distinct monooxygenases for alkane oxidation in Nocardioides sp. Strain CF8. Appl Environ Microbiol. 2001;67:4992–4998. doi: 10.1128/AEM.67.11.4992-4998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coleman NV, et al. Hydrocarbon monooxygenase in Mycobacterium: recombinant expression of a member of the ammonia monooxygenase superfamily. ISME J. 2012;6:171–182. doi: 10.1038/ismej.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redmond MC, Valentine DL, Sessions AL. Identification of novel methane-, ethane-, and propane-oxidizing bacteria at marine hydrocarbon seeps by stable isotope probing. Appl Environ Microbiol. 2010;76:6412–6422. doi: 10.1128/AEM.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki T, Nakamura T, Fuse H. Isolation of two novel marine ethylene-assimilating bacteria, Haliea species ETY-M and ETY-NAG, containing particulate methane monooxygenase-like genes. Microbes Environ. 2012;27:54–60. doi: 10.1264/jsme2.ME11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivers AR, et al. Transcriptional response of bathypelagic marine bacterioplankton to the Deepwater Horizon oil spill. ISME J. 2013;7:2315–2329. doi: 10.1038/ismej.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vangnai AS, Arp DJ. An inducible 1-butanol dehydrogenase, a quinohaemoprotein, is involved in the oxidation of butane by ‘Pseudomonas butanovora’. Microbiology. 2001;147:745–756. doi: 10.1099/00221287-147-3-745. [DOI] [PubMed] [Google Scholar]
- 38.White H, Huber C, Feicht R, Simon H. On a reversible molybdenum-containing aldehyde oxidoreductase from Clostridium formicoaceticum. Arch Microbiol. 1993;159:244–249. [Google Scholar]
- 39.Vorobev A, et al. Genomic and transcriptomic analyses of the facultative methanotroph Methylocystis sp. strain SB2 grown on methane or ethanol. Appl Environ Microbiol. 2014;80:3044–3052. doi: 10.1128/AEM.00218-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsen JB, Yang S, Stein LY, Beck D, Kalyuzhnaya MG. Global molecular analyses of methane metabolism in methanotrophic alphaproteobacterium, Methylosinus trichosporium OB3b. Part I: transcriptomic study. Front Microbiol. 2013;4:00040. doi: 10.3389/fmicb.2013.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schubotz F, et al. Petroleum degradation and associated microbial signatures at the Chapopote asphalt volcano, Southern Gulf of Mexico. Geochim Cosmochim Acta. 2011;75:4377–4398. [Google Scholar]
- 42.Joye SB, MacDonald IR, Leifer I, Asper V. Magnitude and oxidation potential of hydrocarbon gases released from the BP oil well blowout. Nat Geosci. 2011;4:160–164. [Google Scholar]
- 43.Head IM, Jones DM, Röling WFM. Marine microorganisms make a meal of oil. Nat Rev Microbiol. 2006;4:173–182. doi: 10.1038/nrmicro1348. [DOI] [PubMed] [Google Scholar]
- 44.Mendes SD, et al. Marine microbes rapidly adapt to consume ethane, propane, and butane within the dissolved hydrocarbon plume of a natural seep. J Geophys Res Ocean. 2015;120:1937–1953. [Google Scholar]
- 45.Schubotz F. Microbial community characterization and carbon turnover in methane-rich marine environments - case studies in the Gulf of Mexico and the Black Sea. PhD dissertation, University of Bremen; 2009. [Google Scholar]
- 46.Gustafson RG, Turner RD, Lutz RA, Vrijenhoek RC. A new genus and five new species of mussels (Bivalvia, Mytilidae) from deep-sea sulfide/hydrocarbon seeps in the Gulf of Mexico. Malacologia. 1998;40:63–112. [Google Scholar]
- 47.Faure B, Schaeffer SW, Fisher CR. Species distribution and population connectivity of deep-sea mussels at hydrocarbon seeps in the Gulf of Mexico. PLoS One. 2015;10:e0118460. doi: 10.1371/journal.pone.0118460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pape T, et al. Molecular and isotopic partitioning of low-molecular-weight hydrocarbons during migration and gas hydrate precipitation in deposits of a high-flux seepage site. Chem Geol. 2010;269:350–363. [Google Scholar]
- 49.Maruyama A, et al. Dynamics of microbial populations and strong selection for Cycloclasticus pugetii following the Nakhodka Oil Spill. Microb Ecol. 2003;46:442–453. doi: 10.1007/s00248-002-3010-z. [DOI] [PubMed] [Google Scholar]
- 50.Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–8. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 52.Seah BKB, Gruber-Vodicka HR. gbtools: Interactive visualization of metagenome Bins in R. Front Microbiol. 2015;6:1451. doi: 10.3389/fmicb.2015.01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albertsen M, et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 54.Nurk S, Bankevich A, Antipov D. Assembling genomes and mini-metagenomes from highly chimeric reads. Res Comput Mol Biol. 2013;10:158–170. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells , and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Overbeek R, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huntemann M, et al. The standard operating procedure of the DOE-JGI Microbial Genome Annotation Pipeline (MGAP v.4) Stand Genomic Sci. 2015;10:86. doi: 10.1186/s40793-015-0077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson M, et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goris J, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 62.Petkau A, Stuart-Edwards M, Stothard P, van Domselaar G. Interactive microbial genome visualization with GView. Bioinformatics. 2010;26:3125–3126. doi: 10.1093/bioinformatics/btq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 64.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rocha DJP, Santos CS, Pacheco LGC. Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie Van Leeuwenhoek. 2015;108:685–693. doi: 10.1007/s10482-015-0524-1. [DOI] [PubMed] [Google Scholar]
- 66.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 69.Hamann E, et al. Environmental Breviatea harbour mutualistic Arcobacter epibionts. Nature. 2016;534:254–258. doi: 10.1038/nature18297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petersen JM, et al. Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation. Nat Microbiol. 2016;2:16195. doi: 10.1038/nmicrobiol.2016.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bettencourt R, et al. High-throughput sequencing and analysis of the gill tissue transcriptome from the deep-sea hydrothermal vent mussel Bathymodiolus azoricus. BMC Genomics. 2010;11:559. doi: 10.1186/1471-2164-11-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 73.Florens L, et al. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swan BK, et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science. 2011;333:1296–300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- 75.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 76.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 77.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Markowitz VM, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42:560–567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vizcaíno J, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotech. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.