Abstract
Huntington’s disease is a neurodegenerative disorder caused by a polyglutamine repeat in the Huntingtin gene (HTT). Although suppressing the expression of mutant HTT (mHTT) has been explored as a therapeutic strategy to treat Huntington’s disease, considerable efforts have gone into developing allele-specific suppression of mHTT expression, given that loss of Htt in mice can lead to embryonic lethality. It remains unknown whether depletion of HTT in the adult brain, regardless of its allele, could be a safe therapy. Here, we report that permanent suppression of endogenous mHTT expression in the striatum of mHTT-expressing mice (HD140Q-knockin mice) using CRISPR/Cas9-mediated inactivation effectively depleted HTT aggregates and attenuated early neuropathology. The reduction of mHTT expression in striatal neuronal cells in adult HD140Q-knockin mice did not affect viability, but alleviated motor deficits. Our studies suggest that non–allele-specific CRISPR/Cas9-mediated gene editing could be used to efficiently and permanently eliminate polyglutamine expansion–mediated neuronal toxicity in the adult brain.
Keywords: Neuroscience
Introduction
Expansion of a CAG/glutamine repeat in various genes causes at least 9 different neurodegenerative diseases, including Huntington’s disease (HD). In HD, the expanded CAG repeat encodes a polyglutamine (polyQ) tract in the N-terminal region of huntingtin (HTT) and leads to a wide range of cellular dysfunctions (1). The gain of toxic function of mutant huntingtin (mHTT) has led to considerable efforts to use siRNA, antisense oligonucleotides, or CRISPR/Cas9 to selectively suppress the expression of mHTT (2–4). Indeed, siRNA and antisense oligonucleotides have shown promising therapeutic effects in HD mice that express transgenic mHTT (2, 5). However, this relies on SNPs that are specific to the mutant allele. Also, whether this strategy can be successfully used in HD mice that express mHTT at the endogenous level remains unknown, since normal and expanded alleles of the endogenous Htt gene are not readily distinguished by siRNA and antisense oligonucleotides (6).
Our recent studies using conditional Htt-KO mice revealed that depletion of normal HTT in adult mouse brains does not affect animal survival, growth, or neuronal viability (7). In addition, knockin (KI) mice that express N-terminal mHTT have shown that the N-terminal region of HTT is not essential for early embryonic development (8). These findings suggest that removal of N-terminal HTT containing the polyQ domain, regardless of its allele, could be a potential therapeutic strategy to treat HD. Here, we report that permanent suppression of the endogenous expression of mHTT via CRISPR/Cas9 in the striatum of HD140Q-KI mice, which express a human HD HTT, can effectively deplete HTT aggregates and early neuropathology, even after the formation of abundant HTT aggregates. Reducing HTT expression in striatal neuronal cells does not affect the viability of the adult HD140Q-KI mice, but alleviates their motor deficits and neurological symptoms. Our findings suggest that depletion of HTT via CRISPR/Cas9 in a non–allele-specific manner can efficiently and permanently eliminate polyQ expansion–mediated neuronal toxicity in the adult brain. This also opens up a new avenue for treating other neurodegenerative diseases caused by the gain-of-function mechanism.
Results and Discussion
To delete the polyQ domain of mHTT using CRISPR/Cas9, we designed 4 guide RNAs (gRNAs) to target the DNA regions (T1, T2, T3, and T4) flanking the CAG repeat in exon 1 of human HTT (Figure 1A and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI92087DS1). We transfected HEK293 cells stably expressing exon 1 of human HTT containing 120 CAG repeats with each of the 4 gRNAs and Cas9. Western blotting showed a reduction of mHTT in the transfected cells (Supplemental Figure 1, B and C). We also tested the activities of combining 2 HTT-gRNAs in the stable HEK293 cells. A combination of T1 and T3 HTT-gRNAs led to the greatest reduction in mHTT (Supplemental Figure 1, D and E) and was used for our subsequent studies.
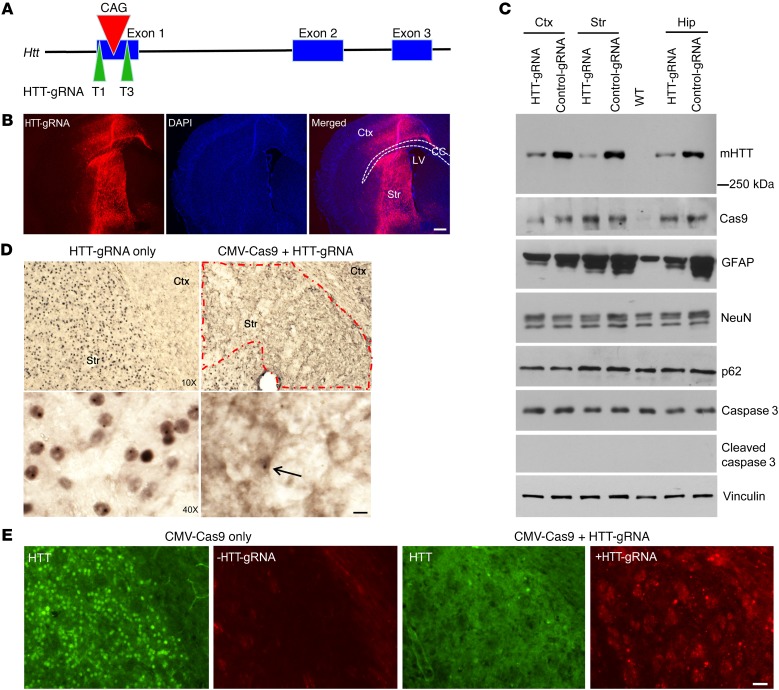
Figure 1. CRISPR/Cas9 depletes the ubiquitous expression of mHTT in homozygous HD140Q-KI mouse striatum.
(A) Schematics of the designed HTT-gRNA (T1 and T3). (B) Immunofluorescence showing the transduction of AAV-HTT-gRNA in the striatum and part of the cortex. Ctx, cortex; Str, striatum; CC, corpus callosum; LV, lateral ventricle. Scale bar: 100 μm. (C) Different brain regions from 9-month-old homozygous HD140Q-KI mice injected with AAV-CMV-Cas9 and AAV-HTT-gRNA (T1 and T3) or control-gRNA were analyzed by Western blotting with 1C2 for mHTT and antibodies against Cas9, GFAP, NeuN, p62, caspase 3, and cleaved caspase 3. Vinculin was used as a loading control. Hip, hippocampus. (D) Low- and high-magnification images show the reduction of nuclear HTT and HTT aggregates in the AAV-HTT-gRNA/AAV-CMV-Cas9–injected area in 9-month-old homozygous HD140Q-KI mice compared with the contralateral striatum injected with AAV-HTT-gRNA only. Arrow indicates a remaining cell with nuclear HTT inclusion. Scale bar: 10 μm. The red dashed outline indicates the injected region where mHTT aggregates are markedly reduced. (E) Double immunostaining confirmed the depletion of mHTT in the area expressing HTT-gRNA in the injected striatum of 9-month-old homozygous HD140Q-KI mice. The striatum of a HD140Q-KI mouse injected with AAV-CMV-Cas9 only was used as a control. Scale bar: 20 μm.
We next tested the effect of CRISPR/Cas9-mediated HTT depletion in HD140Q-KI mice. In this KI mouse model, exon 1 of human HTT with 140 CAG repeats replaces exon 1 of endogenous mouse Htt (9), resulting in the expression of full-length mHTT with 140Q under the control of the endogenous mouse Htt promoter. In HD140Q-KI mice, accumulated mHTT in striatal neuronal nuclei is detectable between 4 and 6 months and forms obvious aggregates at 9 to 10 months (8, 10–12). We focused on the striatum to investigate the effect of removing mHTT. Two gRNAs (T1 and T3) are expressed under the U6 promoter in an adeno-associated virus (AAV) vector that also expresses red fluorescent protein (RFP) (AAV-HTT-gRNA), and Cas9 is expressed in another AAV vector under the CMV promoter (AAV-CMV-Cas9) (13). The 2 viruses were mixed at a ratio of 1:4 for stereotaxic injection into mouse striatum (Supplemental Figure 2A). After 3 weeks, Western blotting verified that RFP and Cas9 were predominantly expressed in the injected striatum (Supplemental Figure 2B).
We injected AAV-HTT-gRNA and AAV-CMV-Cas9 into one side of the striatum in homozygous HD140Q-KI mice at the age of 3 or 9 months. The contralateral striatum was injected with AAV-HTT-gRNA or AAV-CMV-Cas9 alone, which allowed us to rigorously examine the efficiency of HTT-gRNA/Cas9–mediated mHTT knockdown. HD140Q-KI mice are known to develop age-dependent motor deficits and nuclear accumulation of mHTT (9, 14). We found that most of the striatum and the needle pathway in the cortex and hippocampus were transduced by AAVs 3 weeks after injection (Figure 1B). Western blotting showed that HTT-gRNA, but not control-gRNA, caused a significant reduction of mHTT in the striatum of 9-month-old HD140Q-KI mice (Figure 1C and Supplemental Figure 2C). Compared with the contralateral striatum injected with HTT-gRNA alone, immunostaining revealed a dramatic decrease in the nuclear accumulation and aggregation of mHTT in the HTT-gRNA/Cas9–injected striatum (Figure 1D). Double immunofluorescence staining further verified that the decrease in mHTT staining is dependent on the expression of HTT-gRNA (Figure 1E). In HD KI mouse brain, a well-known early neuropathology is reactive astrocytes (15, 16). In brain regions transduced by HTT-gRNA/Cas9, attenuation of the increased glial fibrillary acidic protein (GFAP) was associated with knockdown of mHTT compared with brain regions injected with control-gRNA/Cas9 (Figure 1C and Supplemental Figure 2C), indicating that a reduction of mHTT alleviated reactive astrocytes. We also checked several other proteins such as NeuN (a neuronal marker), p62 (an autophagy marker) and caspase 3 (an apoptosis marker), which are frequently studied in neurodegenerative diseases, and found that these proteins remained unchanged (Figure 1C and Supplemental Figure 2C). The results were corroborated by immunohistochemical studies using GFAP and NeuN antibodies (Supplemental Figure 3, A and B).
Most HD patients are heterozygous for the HD gene mutation. Also, neurons are preferentially affected in HD. Thus, we tested the therapeutic potential of CRISPR/Cas9 in heterozygous HD140Q-KI mice, using AAV-HTT-gRNAs (T1 and T3) with AAV-Cas9 that was expressed under the neuronal methyl-CpG–binding protein (Mecp2) promoter (AAV-MECP2-Cas9) (Figure 2A). As a control, AAV-control-gRNA with AAV-MECP2-Cas9 were used. These viruses were mixed at a ratio of 1:4 (gRNA/Cas9) and injected into both sides of the striatum of 9-month-old heterozygous HD140Q-KI mice to maximize the therapeutic effects. Immunostaining of the injected striatum revealed the presence of RFP in dopamine- and cAMP-regulated phosphoprotein as well as dopamine- and cAMP-regulated neuronal phosphoprotein (DARPP-32) (Figure 2B) and NeuN-positive (Supplemental Figure 4) neurons, indicating that medium spiny neurons in the striatum had been transduced by the injected AAVs.
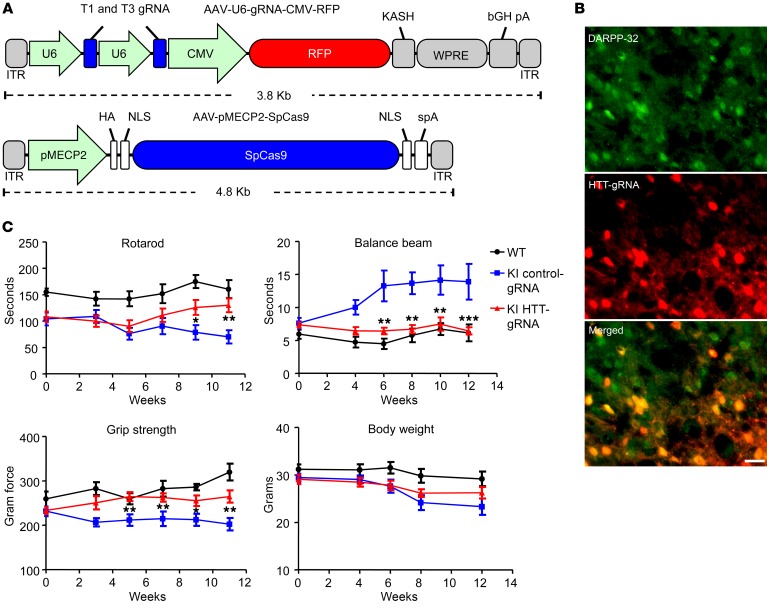
Figure 2. Behavioral analysis of heterozygous HD140Q-KI mice with depletion of neuronal HTT in the striatum by AAV-HTT-gRNA/AAV-MECP2-Cas9 injection.
(A) Schematics showing the viral vectors used. HA, human influenza hemagglutinin; ITR, inverted terminal repeat; KASH, Klarsicht, ANC-1, Syne Homology; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element. (B) Double immunostaining with anti–DARRP-32 indicated that medium spiny neurons expressed AAV-HTT-gRNA. Scale bar: 10 μm. (C) Motor functions of heterozygous HD140Q-KI mice injected with AAV-HTT-gRNA/AAV-MECP2-Cas9 (KI HTT-gRNA) or AAV-control-gRNA/AAV-MECP2-Cas9 (KI control-gRNA) and WT mice injected with AAV-control-gRNA/AAV-MECP2-Cas9 (WT) were evaluated using rotarod, balance beam, and grip strength tests at different time points after injection (n = 12 for each group; *P < 0.05, **P < 0.012, and ***P < 0.001, by 2-way ANOVA with Bonferroni’s test, comparing the KI HTT-gRNA group with the KI control-gRNA group).Data represent the mean ± SEM.
We examined whether CRISPR/Cas9-mediated neuronal mHTT depletion had any impact on the motor function of HD140Q-KI mice. We were able to obtain 24 nine-month-old heterozygous KI mice for examination, at which age the mice show abundant nuclear accumulation of mHTT in striatal neurons and also develop obvious motor dysfunction. These KI mice were injected with either AAV-HTT-gRNA/AAV-MECP2-Cas9 or AAV-control-gRNA/AAV-MECP2-Cas9, and their behaviors were monitored for 3 months. In HD140Q-KI mice, motor dysfunction has been well documented using rotarod, balance beam, and grip strength tests (9, 17). We found that HTT-gRNA/Cas9 could significantly improve performance in these tests and alleviate the motor deficits of HD140Q-KI mice compared with control-gRNA/Cas9–injected KI and WT mice (Figure 2C). In addition, knocking down HTT expression also attenuated body weight reductions (Figure 2C). The efficiency of mHTT reduction in the striatum varied among the individual mice tested (Supplemental Figure 5A). We found that the percentage of mHTT reduction in each mouse correlated with its rotarod and balance beam performance (Supplemental Figure 5, B and C).
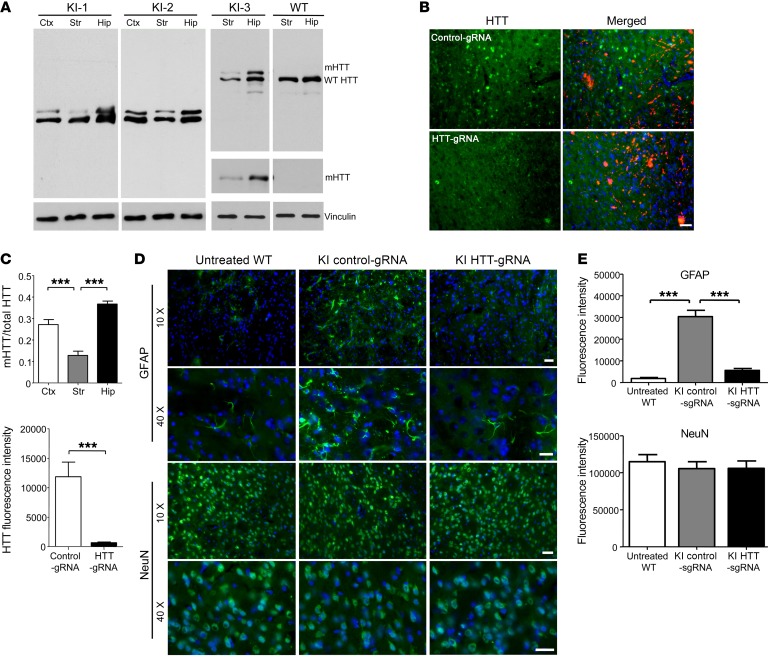
Many studies have shown rare off-targets when specific gRNAs are used (18, 19). Whole-genome sequencing analysis using genomic DNA extracted from HTT-gRNA/Cas9–injected striatum verified that genomic mutations predominantly occurred around the HTT-gRNA targeted sequences in the Htt gene, but not in potential off-target loci (Supplemental Figure 6). This result was further corroborated by a T7E1 assay showing a lack of DNA mutations in selected potential off-target loci (Supplemental Figure 7A). In addition, DNA sequencing confirmed the presence of frameshift mutations around the targeted region of the HTT-gRNA (Supplemental Figure 7B). Western blotting analysis of individual injected mice showed an obvious reduction of mHTT in the striatum and part of the cortex containing the injection pathway compared with mHTT levels in the hippocampus (Figure 3, A and C). Double immunostaining confirmed a specific reduction of mHTT by HTT-gRNA, but not control-gRNA, in the injected striatal area (Figure 3, B and C). Furthermore, AAV-MCEP2-Cas9, which selectively depleted HTT in neuronal cells, could also reduce reactive astrocytes but did not alter expression of the neuronal marker NeuN (Figure 3, D and E). We also examined striatal volume and brain weight and did not find significant differences between HTT-gRNA– and control-gRNA–injected HD140Q-KI mice (Supplemental Figure 8). These results suggest that neuronal mHTT causes early neuropathology to increase reactive astrocytes in HD140Q-KI mouse brains, which could be diminished by eliminating the expression of HTT. Taken together, removal of endogenous HTT in neuronal cells via CRISPR/Cas9 can efficiently alleviate mHTT-mediated neuropathology in HD140Q-KI mice.
Figure 3. Removal of mHTT in neuronal cells alleviates neuropathology in 13-month-old heterozygous HD140Q-KI mouse striatum.
(A) Western blotting shows the reduction of mHTT in brain tissues from 3 heterozygous HD140Q-KI (KI-1, KI-2, and KI-3) and WT mice. 2166 Antibody was used to show both mHTT and WT HTT. 1C2 Antibody was used to show only mHTT. Replicate samples run on separated blots are presented. (B) Double immunostaining with 1C2 antibody confirmed the depletion of mHTT by AAV-HTT-gRNA. A heterozygous HD140Q-KI mouse injected with AAV-control-gRNA served as a control. Scale bar: 20 μm. (C) Quantitative assessments of the relative ratio of mHTT to total HTT in A (left; n = 8; ***P < 0.001, by 1-way ANOVA with Tukey’s test) and relative levels of mHTT staining in B (right; n = 8; ***P < 0.001, by Student’s t test). (D) Double immunostaining of striatum (from 9-month-old injected mice examined at 13 months of age) shows decreased GFAP levels by HTT-gRNA compared with control-gRNA. There was no difference in NeuN staining. Scale bars: 20 μm. (E) Quantitative assessment of the relative levels of GFAP and NeuN staining (n = 8). The staining intensity for each mouse was the average from three ×10 images. ***P < 0.001, by 1-way ANOVA with Tukey’s test. Data represent the mean ± SEM.
Although shutting off the expression of transgenic mHTT can alleviate neurological symptoms in HD mice (20, 21), whether reducing the expression of endogenous HTT can be used to treat HD without deleterious effects remains unknown. When we used homozygous HD140Q-KI mice in which 2 alleles of the mHTT gene could be disrupted by CRISPR/Cas9, we found that removal of HTT did not affect the expression of NeuN or caspase 3. Instead, mHTT depletion significantly reduced reactive astrocytes, an early pathological event in HD KI mouse brains (15, 16). The results also support our recent findings that depletion of endogenous mouse HTT in adult neurons is nondeleterious and that the function of HTT is cell type and age dependent (7). Using heterozygous HD140Q-KI mice to further analyze their pathology and behaviors, we also verified that CRISPR/Cas9 could effectively alleviate HD-related phenotypes. At the age of 9 months, HD140Q-KI mice show abundant nuclear mHTT accumulation in the striatum and obvious motor deficit phenotypes (8, 10–12). Our findings revealed that CRISPR/Cas9-mediated gene inactivation could reverse the neuropathology and behavioral phenotypes, even when the mice were 9 months old, suggesting that old neuronal cells still have the ability to clear the accumulated mutant proteins and repair early injury once the expression of mutant proteins is blocked. Thus, reducing mHTT expression in the brains of elder HD patients might still be effective in alleviating neurological symptoms.
Given that CRISPR/Cas9 can permanently eliminate the expression of targeted genes, using CRISPR/Cas9 should more efficiently deplete the expression of mHTT than has been possible with previous therapeutic approaches, which require continuous administration. Also, the severe neurological symptoms of many neurodegenerative diseases are often associated with the preferential vulnerability of selective neuronal populations. The use of specific promoters allows CRISPR/Cas9 to target specific types of neurons. Thus, using CRISPR/Cas9 to inhibit mutant protein expression in specific brain regions opens up a new avenue for treating HD as well as other neurodegenerative diseases that are caused by a toxic gain of function of mutant genes.
Methods
Study approval.
All procedures were performed in accordance with NIH guidelines and the US Public Health Service’s Guide for the Care and Use of Laboratory Animals and were approved by the IACUC of Emory University, which is accredited by the American Association for Accreditation of Laboratory Care (AAALC).
Whole-genome sequencing.
Whole-genome sequencing data have been deposited in the NCBI’s Sequence Read Archive (SRA accession number SRP105422).
Statistics.
Statistical significance was determined by 2-tailed Student’s t test, 1-way ANOVA, or 2-way ANOVA using GraphPad Prism 5.0 (GraphPad Software). A P value of less than 0.05 was considered statistically significant.
Author contributions
SY, RC, SL, and XJL designed the study. SY, RC, HY, TZ, and YH, performed experiments and collected the data, HEK, XS, ZQ, and PJ performed whole-genome sequencing–related work. SY, RC, SL, and XJL analyzed the data. SY, RC, and XJL wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (NS036232 and NS101701, to XJL, and NS095279, to SHL) and the National Natural Science Foundation of China (grant 91332206).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(7):2719–2724.https://doi.org/10.1172/JCI92087.
Contributor Information
Su Yang, Email: syang33@emory.edu.
Renbao Chang, Email: rbchang@126.com.
Huiming Yang, Email: huiming.yang@emory.edu.
Ting Zhao, Email: tzhao5@emory.edu.
Yan Hong, Email: yhong31@emory.edu.
Ha Eun Kong, Email: haeun.kong@emory.edu.
Xiaobo Sun, Email: xsun28@emory.edu.
Zhaohui Qin, Email: zhaohui.qin@emory.edu.
Peng Jin, Email: peng.jin@emory.edu.
Shihua Li, Email: sli@emory.edu.
Xiao-Jiang Li, Email: xli2@emory.edu.
References
- 1.Bates GP, et al. Huntington disease. Nat Rev Dis Primers. 2015;1:15005. doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 2.Carroll JB, et al. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene/allele-specific silencing of mutant huntingtin. Mol Ther. 2011;19(12):2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drouet V, et al. Allele-specific silencing of mutant huntingtin in rodent brain and human stem cells. PLoS One. 2014;9(6):e99341. doi: 10.1371/journal.pone.0099341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monteys AM, Ebanks SA, Keiser MS, Davidson BL. CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Mol Ther. 2017;25(1):12–23. doi: 10.1016/j.ymthe.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harper SQ, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc Natl Acad Sci U S A. 2005;102(16):5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez-Lebron E, Paulson HL. Allele-specific RNA interference for neurological disease. Gene Ther. 2006;13(6):576–581. doi: 10.1038/sj.gt.3302702. [DOI] [PubMed] [Google Scholar]
- 7.Wang G, Liu X, Gaertig MA, Li S, Li XJ. Ablation of huntingtin in adult neurons is nondeleterious but its depletion in young mice causes acute pancreatitis. Proc Natl Acad Sci U S A. 2016;113(12):3359–3364. doi: 10.1073/pnas.1524575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, et al. N-terminal huntingtin knock-in mice: implications of removing the N-terminal region of huntingtin for therapy. PLoS Genet. 2016;12(5):e1006083. doi: 10.1371/journal.pgen.1006083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J Comp Neurol. 2003;465(1):11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 10.Wheeler VC, et al. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum Mol Genet. 2000;9(4):503–513. doi: 10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- 11.Lin CH, et al. Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Hum Mol Genet. 2001;10(2):137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 12.Menalled LB, et al. Early motor dysfunction and striosomal distribution of huntingtin microaggregates in Huntington’s disease knock-in mice. J Neurosci. 2002;22(18):8266–8276. doi: 10.1523/JNEUROSCI.22-18-08266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swiech L, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33(1):102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pépin J, et al. In vivo imaging of brain glutamate defects in a knock-in mouse model of Huntington’s disease. Neuroimage. 2016;139:53–64. doi: 10.1016/j.neuroimage.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 15.Palfi S, et al. Expression of mutated huntingtin fragment in the putamen is sufficient to produce abnormal movement in non-human primates. Mol Ther. 2007;15(8):1444–1451. doi: 10.1038/sj.mt.6300185. [DOI] [PubMed] [Google Scholar]
- 16.Yu ZX, Li SH, Evans J, Pillarisetti A, Li H, Li XJ. Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington’s disease. J Neurosci. 2003;23(6):2193–2202. doi: 10.1523/JNEUROSCI.23-06-02193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hickey MA, et al. Extensive early motor and non-motor behavioral deficits are followed by striatal neuronal loss in knock-in Huntington’s disease mice. Neuroscience. 2008;157(1):280–295. doi: 10.1016/j.neuroscience.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veres A, et al. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15(1):27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iyer V, et al. Off-target mutations are rare in Cas9-modified mice. Nat Methods. 2015;12(6):479. doi: 10.1038/nmeth.3408. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell. 2000;101(1):57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 21.DiFiglia M, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci U S A. 2007;104(43):17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.