Abstract
Autosomal dominant polycystic kidney disease (ADPKD) is driven by mutations in PKD1 and PKD2 genes. Recent work suggests that epigenetic modulation of gene expression and protein function may play a role in ADPKD pathogenesis. In this study, we identified SMYD2, a SET and MYND domain protein with lysine methyltransferase activity, as a regulator of renal cyst growth. SMYD2 was upregulated in renal epithelial cells and tissues from Pkd1-knockout mice as well as in ADPKD patients. SMYD2 deficiency delayed renal cyst growth in postnatal kidneys from Pkd1 mutant mice. Pkd1 and Smyd2 double-knockout mice lived longer than Pkd1-knockout mice. Targeting SMYD2 with its specific inhibitor, AZ505, delayed cyst growth in both early- and later-stage Pkd1 conditional knockout mouse models. SMYD2 carried out its function via methylation and activation of STAT3 and the p65 subunit of NF-κB, leading to increased cystic renal epithelial cell proliferation and survival. We further identified two positive feedback loops that integrate epigenetic regulation and renal inflammation in cyst development: SMYD2/IL-6/STAT3/SMYD2 and SMYD2/TNF-α/NF-κB/SMYD2. These pathways provide mechanisms by which SMYD2 might be induced by cyst fluid IL-6 and TNF-α in ADPKD kidneys. The SMYD2 transcriptional target gene Ptpn13 also linked SMYD2 to other PKD-associated signaling pathways, including ERK, mTOR, and Akt signaling, via PTPN13-mediated phosphorylation.
Keywords: Genetics, Nephrology
Introduction
Autosomal dominant polycystic kidney disease (ADPKD) is caused by mutations in one of two genes, PKD1 or PKD2, which encode the proteins polycystin-1 (PC1) and polycystin-2 (PC2), respectively (1, 2). A number of PKD-associated signaling pathways have been identified, and potential therapeutic targets have been tested in ADPKD animal models (3, 4). The role of epigenetic modulation of gene expression and protein function in ADPKD has recently become the focus of scientific investigation (5, 6). Our recent study showed that targeting SIRT1, a histone deacetylase (HDAC), with nicotinamide (vitamin B3) delayed cyst growth in Pkd1-knockout mouse models through the deacetylation of Rb and p53 to regulate cystic renal epithelial cell proliferation and apoptosis (6). This suggested that epigenetic regulators can mediate the modification of non-histone substrates to mechanistically regulate cyst formation. SMYD2, as a SET domain–containing histone methyltransferase, methylates both histone and non-histone proteins, such as the tumor suppressors Rb and p53 and the molecular chaperone Hsp90, which have all been identified in the pathogenesis of ADPKD (6–11). However, the functional roles of SMYD2 in cyst formation in ADPKD have not to our knowledge been explored.
SMYD2 belongs to the 5-member SET and MYND domain (SMYD) family of proteins, including SMYD1–5, which possess SET-dependent methyltransferase (12, 13). All members of the SMYD family show high identity and similarity in the SET and MYND domains (14). The SMYD proteins have distinct substrate specificities and play diverse roles in muscle development and/or cancer (15, 16). Only SMYD2 and SMYD3 are expressed in kidneys, and SMYD2 has been reported to methylate ADPKD-associated non-histone substrates Rb, p53, and Hsp90 (7–9, 14). SMYD2 methylates Rb on lysines, resulting in either the repression of specific Rb/E2F genes or an increase in Rb phosphorylation on serines to regulate cell cycle progression. SMYD2 methylates p53 on lysine to impede binding of p53 to its target gene promoters, and knockdown of Smyd2 enhances DNA damage–induced, p53-dependent apoptosis (8). In addition, SMYD2 can methylate histone H3 at lysine 4 (H3K4) to induce genes involved in cell cycle and transcription regulation, and this process can be enhanced by its interaction with Hsp90α (14, 17). SMYD2 is also able to methylate histone H3 lysine 36 (H3K36) to repress transcriptional activity via its association with the HDAC repressor complex (14). Overexpression of SMYD2 has been reported in primary tumor samples of esophageal squamous cell carcinoma (ESCC). Genetic knockdown of SMYD2 leads to decreased ESCC cell proliferation via cell cycle regulation and apoptosis (18).
In this study, we found that SMYD2 was upregulated in Pkd1 mutant renal epithelial cells and tissues, and that double conditional knockout of Pkd1 and Smyd2 delayed renal cyst growth and preserved renal function. We further found that targeting SMYD2 with its specific inhibitor, AZ505, delayed cyst growth, possibly unveiling a novel therapeutic agent for the treatment of ADPKD. In addition, the key regulatory components identified by ChIP-sequencing (ChIP-seq) analysis may also serve as effective targets to slow disease progression. Thus, the results of this study should prove to be therapeutically relevant, with the potential for translation into the clinic.
Results
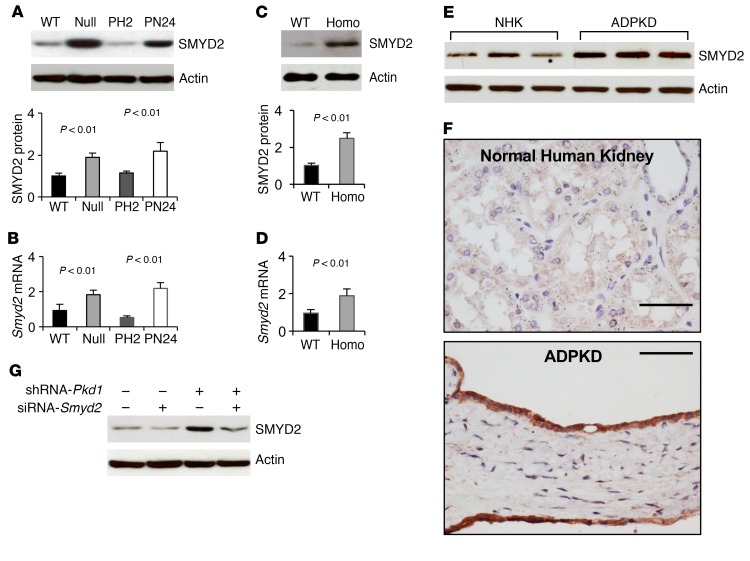
The expression of SMYD2 is upregulated in Pkd1 mutant renal epithelial cells and ADPKD tissues. We found that SMYD2 was upregulated in Pkd1-null mouse embryonic kidney (MEK) collecting duct cells and postnatal Pkd1-homozygous mutant proximal tubule PN24 cells compared with control Pkd1 WT MEK cells and postnatal Pkd1-heterozygous PH2 cells as examined by Western blot analysis (Figure 1A) and quantitative RT-PCR (qRT-PCR) (Figure 1B). We also found that the expression of SMYD2 protein and mRNA was upregulated in kidneys from Pkd1fl/fl:Ksp-Cre mice, a well-characterized animal model for ADPKD, as compared with age-matched Pkd1 WT kidneys at P7 (Figure 1, C and D). The expression of SMYD2 was also increased in human ADPKD cells compared with normal human kidney (NHK) cells (Figure 1E). Our immunohistochemistry analysis indicated that elevated SMYD2 expression was localized to cyst-lining epithelial cells in human ADPKD kidneys (Figure 1F) but was absent in normal human kidneys. In addition, we found that knockdown of Pkd1 with shRNA increased the expression of SMYD2 in mouse inner medullary collecting duct (mIMCD3) cells (Figure 1G).
Figure 1. Pkd1 mutant renal epithelial cells and tissues demonstrated increased expression of SMYD2.
(A) Western blot analysis of SMYD2 expression from whole cell lysates in Pkd1 WT, Pkd1null/null MEK cells (Null), Pkd1-heterozygous PH2 cells, and Pkd1-homozygous PN24 cells (top panel). Relative SMYD2 expression was quantified from 3 independent immunoblots and standardized to actin (bottom panel). (B) qRT-PCR analysis of relative Smyd2 mRNA expression in WT, Null, PH2, and PN24 cells. (C) Western blot analysis of SMYD2 expression in P7 kidneys from Pkd1+/+:Ksp-Cre (WT) and Pkd1fl/fl:Ksp-Cre (Homo) neonates (top panel). Relative SMYD2 expression in the kidneys (bottom panel) as standardized to actin. (D) qRT-PCR analysis of relative Smyd2 mRNA expression in the kidneys described in C. n = 3. (E) Western blot analysis of SMYD2 expression in primary human ADPKD and NHK cells. Data are representative of 2 independent experiments. (F) Immunohistochemistry analysis indicated that SMYD2 expression was increased in cyst-lining epithelia in human ADPKD kidneys (bottom panel) but not in normal human kidneys (top panel). Scale bars: 50 μm. (G) Western blot analysis of SMYD2 expression in mIMCD3 cells with or without knockdown of Pkd1 with shRNA and/or Smyd2 with siRNA. Representative data from 3 independent experiments are shown.
Smyd2 and Pkd1 double conditional knockout delayed renal cyst growth.
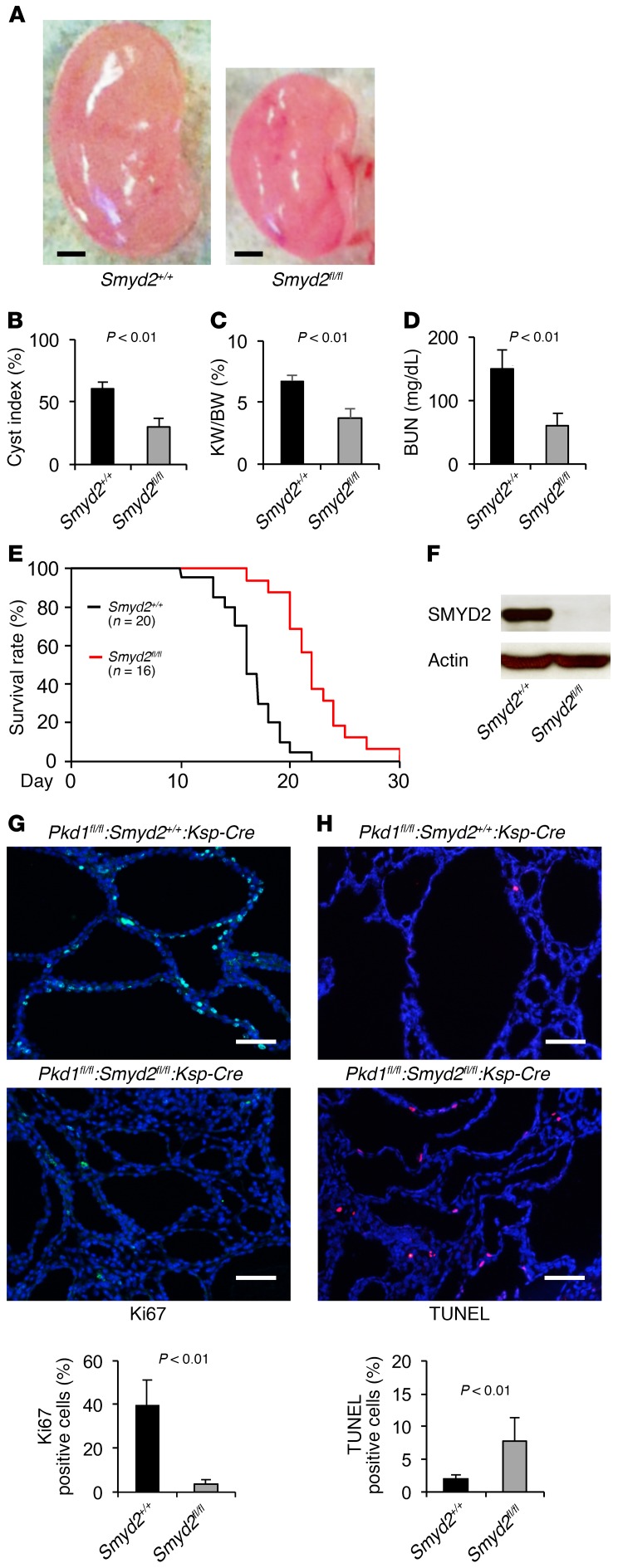
To investigate the functional role of SMYD2 in vivo, we generated Pkd1 and Smyd2 double conditional knockout Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre mice, which had kidney-specific cadherin (Ksp-cadherin) driving Cre expression (19). We found that cyst formation was significantly delayed in the absence of SMYD2 in Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre mice (n = 12) at P7 compared with that in age-matched Pkd1fl/fl:Smyd2+/+:Ksp-Cre mice (n = 14) (P < 0.01) (Figure 2, A and B). The kidney weight to body weight (KW/BW) ratios and blood urea nitrogen (BUN) levels from Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre mice were dramatically reduced compared with those from Pkd1fl/fl:Smyd2+/+:Ksp-Cre mice (P < 0.01) (Figure 2, C and D), which indicated that cyst growth and renal function were normalized. We further found that Pkd1 and Smyd2 double-knockout mice lived to a mean age of 22.2 days, while Pkd1fl/fl:Smyd2+/+:Ksp-Cre mice died of polycystic kidney disease (PKD) at a mean age of 16.3 days (P < 0.01) (Figure 2E). Expression of SMYD2 could not be detected in kidneys from double conditional knockout mice as analyzed by Western blotting (Figure 2F). We found that Ki67-positive cells were significantly decreased in kidneys from Pkd1 and Smyd2 double conditional knockout mice (Figure 2G and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI90921DS1). Unexpectedly, we found that double conditional knockout induced cyst-lining epithelial cell apoptosis, as analyzed by TUNEL assay and H&E staining (Figure 2H and Supplemental Figure 1B). These results suggested that SMYD2 is involved in regulating renal cyst growth in Pkd1-knockout mice.
Figure 2. Double conditional knockout of Smyd2 and Pkd1 delayed renal cyst formation.
(A) Representative kidneys from Pkd1fl/fl:Smyd2+/+:Ksp-Cre (Smyd2+/+) and Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre (Smydfl/fl) neonates. Scale bars: 2 mm. (B) Percent cystic area relative to total kidney section area was significantly decreased in P7 kidneys from Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre versus Pkd1fl/fl:Smyd2+/+:Ksp-Cre neonates. Data reflect all sections quantified for each condition (n = 12 in Smyd2+/+ group and n = 14 in Smyd2fl/fl group). (C) KW/BW ratios were reduced in P7 Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre versus Pkd1fl/fl:Smyd2+/+:Ksp-Cre neonates. (D) BUN levels were significantly decreased in P7 serum from Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre neonates compared with Pkd1fl/fl:Smyd2+/+:Ksp-Cre neonates. (E) Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre mice lived to a mean age of 22.2 days (n = 16), whereas Pkd1fl/fl:Smyd2+/+:Ksp-Cre mice died of PKD at a mean age of 16.3 days (n = 20). P < 0.01. (F) Western blot analysis of SMYD2 expression in P7 kidneys from Pkd1fl/fl:Smyd2+/+:Ksp-Cre (Smyd2+/+) and Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre (Smydfl/fl) neonates. (G) Cell proliferation was decreased in P7 kidneys from Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre neonates versus those from Pkd1fl/fl:Smyd2+/+:Ksp-Cre neonates, as detected with Ki67 staining. The percentage of Ki67-positive nuclei in cyst-lining epithelial cells was calculated from an average of 1,000 nuclei per mouse kidney section. Scale bars: 100 μm. (H) Knockout of Smyd2 induced cyst-lining epithelial cell death in kidneys from P7 Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre neonates, while apoptosis was rare in kidneys from Pkd1fl/fl:Smyd2+/+:Ksp-Cre neonates, as detected by TUNEL assay. Scale bars: 100 μm. The images in G and H are also shown in Supplemental Figure 1, A and B.
A specific SMYD2 inhibitor, AZ505, delays cyst growth in Pkd1 mutant kidneys.
AZ505 is a specific SMYD2 inhibitor as identified by high-throughput chemical screen (20). We found that knockdown of Smyd2 inhibited Pkd1 mutant renal epithelial cell growth as examined by MTT assay (Supplemental Figure 2, A and B) and decreased S-phase entry as determined by flow cytometry (FACS) analysis (Supplemental Figure 2C). Treatment with AZ505 further decreased S-phase entry in Smyd2-knockdown Pkd1 mutant renal epithelial cells compared with that in DMSO-treated Smyd2-knockdown cells (Supplemental Figure 2C), which might be caused by the inhibition of AZ505 on the remaining SMYD2 in the Smyd2-knockdown Pkd1 mutant renal epithelial cells. These results support the specificity of AZ505 for the inhibition of SMYD2.
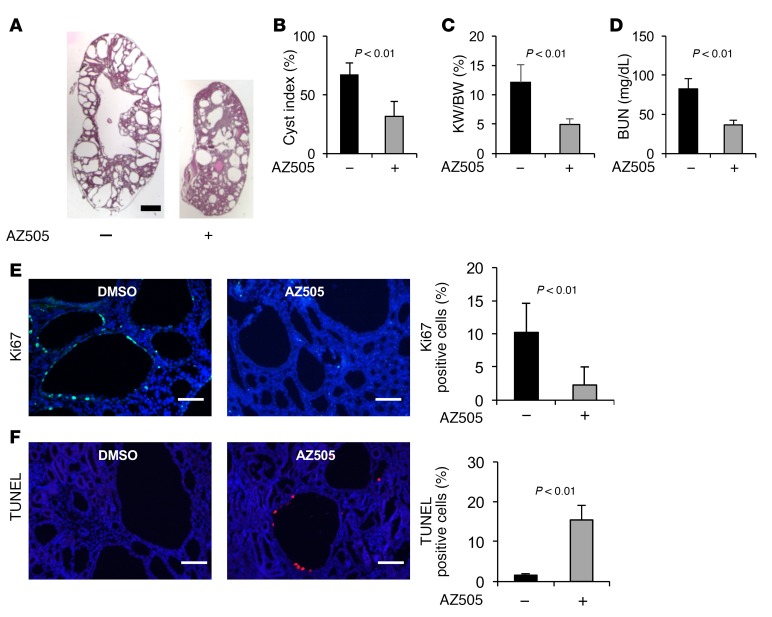
To test whether inhibiting the activity of SMYD2 would suppress cyst formation in vivo, we examined whether AZ505 could delay cyst growth in a progressive hypomorphic Pkd1nl/nl mouse model (21). We found that administration of AZ505 (5 mg/kg) (n = 12) delayed cyst growth, as indicated by decreased cyst index, KW/BW ratio, and BUN level (Figure 3, A–D); inhibited cystic epithelial cell proliferation (Figure 3E and Supplemental Figure 3A); and induced cystic epithelial cell apoptosis (Figure 3F and Supplemental Figure 3B) in P28 kidneys from Pkd1nl/nl mice compared with those in age-matched kidneys from DMSO-injected Pkd1nl/nl mice (n = 12). No sex difference was observed for the effect of AZ505 on delaying cyst growth in Pkd1nl/nl mice when 6 male mice and 6 female mice were compared in each group (data not shown).
Figure 3. AZ505 treatment delayed cyst formation in Pkd1-knockout mice.
(A) Histological examination of kidneys from Pkd1nl/nl mice injected daily with AZ505 or DMSO vehicle control from P7 to P27. Scale bar: 2 mm. (B) Percent cystic area relative to total kidney area of kidneys from Pkd1nl/nl mice treated with AZ505 (n = 12) or DMSO (n = 12). (C and D) Treatment with AZ505 compared with DMSO decreased KW/BW ratios (C) and BUN levels (D) in Pkd1nl/nl mice. (E) AZ505 treatment reduced cyst-lining epithelial cell proliferation in kidneys from Pkd1nl/nl mice, as detected with Ki67 staining. The percentage of Ki67-positive nuclei in cystic epithelial cells was calculated from an average of 1,000 nuclei per mouse kidney section. Scale bars: 100 μm. (F) AZ505 treatment induced cyst-lining epithelial cell death in kidneys from Pkd1nl/nl mice as detected by TUNEL assay. Scale bars: 100 μm. The images in E and F are also shown in Supplemental Figure 3, A and B.
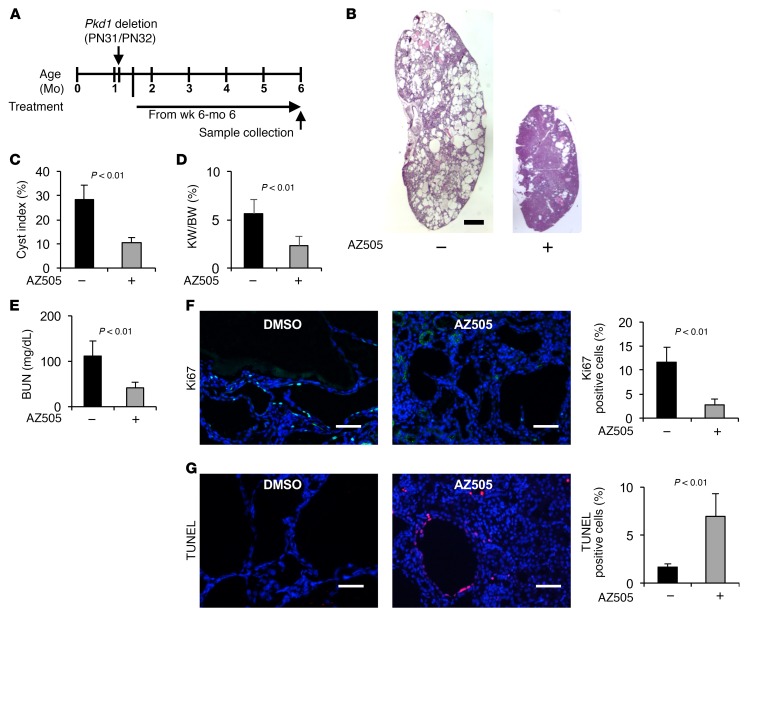
To extend the translational significance of our in vivo findings in early-stage animal models, we also tested the long-term effects of AZ505 on renal cyst formation in Pkd1fl/fl:tamoxifen-Cre mice, in which the Cre is driven by the Esr1+ promoter. We induced Pkd1 deletion in Pkd1fl/fl:tamoxifen-Cre mice via i.p. injection of tamoxifen (125 mg/kg body weight, formulated in corn oil) on two sequential postnatal days (P31 and P32), which resulted in renal cyst formation in the test group after 4 months (10, 22). We administered AZ505 (10 mg/kg) to these mice through i.p. injection up to 6 months (see Methods for details). The Pkd1fl/fl:tamoxifen-Cre mice were treated 3 times per week from P42 (10 days after Pkd1 deletion) to 3.5 months and only one time per week from 3.5 months to 6 months (Figure 4A). We found that AZ505 treatment delayed cyst growth as indicated by decreased cyst index, KW/BW ratio, BUN level, and proliferation of cyst-lining epithelial cells; and by increased apoptosis of cyst-lining epithelial cells (14 mice for each group, including 7 male and 7 female mice) (Figure 4, B–G, and Supplemental Figure 3, C and D). Treatment with AZ505 compared with the control had no effects on body weights in these mouse models (Supplemental Figure 4, A and B). These results suggest that targeting SMYD2 with pharmacological inhibition might delay cyst growth in ADPKD patients.
Figure 4. Treatment with AZ505 delayed cyst growth in conditional Pkd1-knockout mice.
(A) Schedule of the induction and treatment of Pkd1 inducible knockout mice. (B) Histological examination of kidneys from Pkd1fl/fl:tamoxifen-Cre mice injected daily with AZ505 or DMSO vehicle control from P42 to 6 months. Scale bar: 2 mm. (C) Percent cystic area relative to total kidney section area of kidneys from Pkd1fl/fl:tamoxifen-Cre mice treated with AZ505 (n = 14) or DMSO (n = 14). (D and E) Treatment with AZ505 compared with DMSO decreased KW/BW ratios (D) and BUN levels (E) in Pkd1fl/fl:tamoxifen-Cre mice. (F) AZ505 treatment reduced cyst-lining epithelial cell proliferation in kidneys from Pkd1fl/fl:tamoxifen-Cre mice as detected with Ki67 staining. Scale bars: 100 μm. (G) AZ505 treatment induced cyst-lining epithelial cell death in kidneys from Pkd1fl/fl:tamoxifen-Cre mice as detected by TUNEL assay. Scale bars: 100 μm. The images in F and G are also shown in Supplemental Figure 3, C and D.
Silencing SMYD2 decreased phosphorylation and activation of STAT3 and NF-κB in Pkd1 mutant renal epithelial cells and tissues.
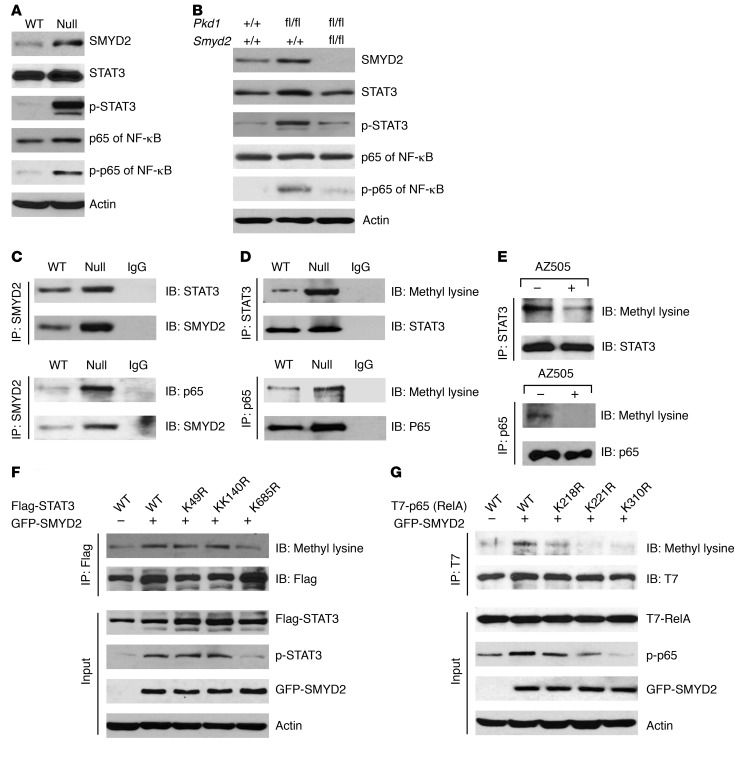
Given the evidence that SMYD2 promotes renal cyst growth in vivo, we investigated the mechanisms involved in this process. It has been reported that the transcription factor STAT3 has sustained activation in Pkd1-knockout polycystic kidneys and the STAT3 inhibitor S3I-201 reduces cyst growth in a neonatal PKD mouse model (23). Recently, we reported that TNF-α mediates NF-κB–regulated cystic renal epithelial cell survival and death (24). We found that the phosphorylation of STAT3 (p-STAT3) and the p65 subunit of NF-κB (p-p65) was increased in Pkd1 mutant cells (Figure 5A) and tissues (Figure 5B). We further found that overexpression of exogenous GFP–tagged SMYD2 increased the phosphorylation of STAT3 and p65 in mIMCD3 cells (Supplemental Figure 5A). Targeting Smyd2 with small siRNA and AZ505 and knockout of Smyd2 decreased the levels of tyrosine phosphorylated active STAT3 and p65, but had no effect on the levels of STAT3 and p65 in Pkd1 mutant cells (Supplemental Figure 5, B and C) and kidneys (Figure 5B).
Figure 5. SMYD2 regulates the phosphorylation of STAT3 and the p65 subunit of NF-κB.
(A) Western blot analysis of p-STAT3 and p-p65 expression from whole cell lysates of Pkd1 WT MEK and Pkd1-null MEK cells. The phosphorylation of STAT3 and p65 was increased in Pkd1-null MEK versus WT MEK cells, whereas the expression of STAT3 and p65 was almost at the same level in these cells. (B) Western blot analysis of the expression of SMYD2, p-STAT3, STAT3, p-p65, and p65 in kidneys from Pkd1+/+:Smyd2+/+:Ksp-Cre, Pkd1fl/fl:Smyd2+/+:Ksp-Cre, and Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre neonates at P7. The expression of SMYD2 and the phosphorylation of STAT3 and p65 were increased in kidneys from Pkd1fl/fl:Smyd2+/+:Ksp-Cre compared with Pkd1+/+:Smyd2+/+:Ksp-Cre mice, whereas the expression of these 3 proteins was decreased in kidneys from Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre compared with Pkd1fl/fl:Smyd2+/+:Ksp-Cre neonates. (C) Interactions between SMYD2 and STAT3 (top panel) as well as SMYD2 and p65 (bottom panel) in Pkd1 WT and Pkd1-null MEK cells were detected with anti-SMYD2 antibody and then blotted with STAT3 antibody (top panel) and p65 (bottom panel) antibody, respectively. IgG was used as a negative control. (D) Methylation of STAT3 and p65 was increased in Pkd1 WT MEK cells compared with Pkd1-null MEK cells. These cells were immunoprecipitated with anti-STAT3 antibody (top panel) and anti-p65 antibody (bottom panel), and then blotted with STAT3, p65, and pan–methyl lysine antibody, respectively. IgG was used as a negative control. (E) The level of methylated STAT3 and p65 was decreased in Pkd1-null MEK cells treated with AZ505 (2 hours) compared with that in cells treated with DMSO as examined with anti–pan–methyl lysine antibody after these proteins were pulled down with STAT3 and p65 antibodies. (F) GFP-tagged SMYD2 together with Flag-tagged WT STAT3 or mutant STAT3 with lysine-to-arginine substitution were transfected into HEK293T cells. The effect of overexpression of SMYD2 on the methylation and phosphorylation of Flag-tagged STAT3 was examined by Western blot analysis. (G) GFP-tagged SMYD2 together with T7-tagged WT p65 (RelA) or mutant p65 (RelA) with lysine-to-arginine substitution were transfected into HEK293T cells. The effect of overexpression of SMYD2 on the methylation and phosphorylation of T7-tagged p65 (RelA) was examined by Western blot analysis.
SMYD2 interacts with STAT3 and p65 and regulates STAT3 and p65 methylation and activation in Pkd1 mutant renal epithelial cells.
To further investigate the mechanisms mediated by SMYD2, we determined that endogenous SMYD2 could immunoprecipitate STAT3 and p65 in Pkd1 WT and mutant MEK cells, and that SMYD2 in Pkd1-null MEK cells could pull down more STAT3 and p65 than in Pkd1 WT MEK cells (Figure 5C). To investigate whether the interactions between SMYD2 and STAT3 as well as SMYD2 and p65 affect their methylation, we compared the methylation levels of STAT3 and p65 in Pkd1 WT and mutant MEK cells. These experiments used anti-STAT3 antibody and anti-p65 antibody for immunoprecipitation, followed by blotting with anti–methyl lysine antibody. We found that methylation of STAT3 and p65 was increased in Pkd1-null MEK cells compared with Pkd1 WT MEK cells (Figure 5D). Inhibition of SMYD2 with AZ505 decreased the methylation of STAT3 and p65 in Pkd1-null MEK cells (Figure 5E). These results together with those for knockdown of Smyd2 or inhibition with AZ505, showing decreased phosphorylation of STAT3 and p65 (Supplemental Figure 5), suggested that SMYD2 may regulate STAT3 and p65 phosphorylation and activation in cystic renal epithelial cells through methylation of these substrates.
To determine the potential methylation site(s) on STAT3 mediated by SMYD2, we expressed Flag-tagged WT STAT3 and its lysine-to-arginine substitutive mutants, including K49R, K140R, and K685R, with or without GFP-tagged SMYD2 in HEK293T cells. The methylation of Flag-tagged STAT3 was examined with an anti–pan–methyl lysine antibody after the Flag-tagged proteins were pulled down with anti-Flag antibody. We found that the methylation of STAT3 was only decreased when STAT3 was mutated at K685R (Figure 5F). We also found that overexpression of SMYD2 increased the phosphorylation of Flag-tagged WT STAT3 and mutant STAT3 only at K49R and K140R, but had no effect on the phosphorylation of Flag-tagged mutant STAT3 at K685R (Figure 5F). Our results suggested that lysine 685 of STAT3 is critical for SMYD2-mediated STAT3 methylation and phosphorylation.
We also investigated the potential methylation site(s) on p65 mediated by SMYD2 with a similar strategy by expressing T7-tagged WT-p65 (RelA) and its lysine-to-arginine substitutive mutants, K218R, K221R, and K310R, with or without GFP-tagged SMYD2 in HEK293T cells. We found that the methylation of p65 was strikingly decreased when p65 was mutated at K221R and K310R and slightly decreased when p65 was mutated at K218 compared with the WT p65 as examined by immunoprecipitation with anti-T7 antibodies followed by immunoblotting with anti–pan–methyl lysine antibodies (Figure 5G). We further found that overexpression of GFP-tagged SMYD2 could markedly increase the phosphorylation of WT p65, only mildly increased the phosphorylation of p65 with K-to-R mutants at K218 and K221, and had no obvious effect on the phosphorylation of p65 with a K-to-R mutant at K310 (Figure 5G). These results suggest that K310 of p65 is the site that most likely contributes to the SMYD2-mediated methylation and phosphorylation of p65, and the lysine K221 of p65 may partially contribute to this process.
Because STAT3 and NF-κB both function as transcription factors, we determined the functional effects of SMYD2 on STAT3 and NF-κB activity by measuring the expression of validated STAT3 and NF-κB target genes, including those encoding BCL2, cyclin D1, TNF-α, c-Myc, and TNFAIP3. Cyclin D1, TNF-α, and c-Myc have been reported to be involved in cyst formation (25–27). First, we found that the levels of these STAT3 and NF-κB target genes were upregulated in Pkd1-null MEK cells compared with Pkd1 WT MEK cells (Supplemental Figure 6A). Next, we found that knockdown of Smyd2 and inhibition of SMYD2 with AZ505 significantly decreased the mRNA levels of these STAT3 and NF-κB targeting genes in cystic renal epithelial cells (Supplemental Figure 6, B and C) and in kidneys from Smyd2 and Pkd1 double-knockout mice compared with Pkd1 single-knockout mice (Supplemental Figure 6D). These results suggested that SMYD2 regulates STAT3 and NF-κB transcriptional activity in Pkd1 mutant renal epithelial cells and tissues.
Synergistic effects exist among SMYD2, STAT3, and NF-κB p65 in cystic renal epithelial cells.
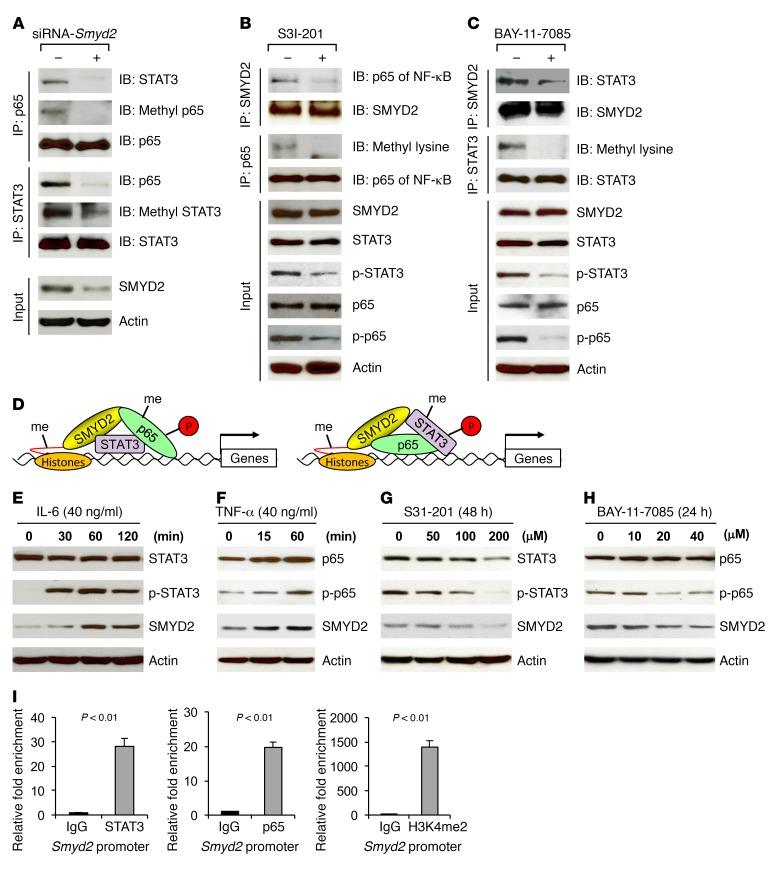
STAT3 has been reported to modify p65 post-translationally by recruitment of the acetyltransferase p300, mediating the acetylation of NF-κB and prolonging its nuclear retention (28). To address whether SMYD2 contributes to the interaction between STAT3and NF-κB, we knocked down Smyd2 in cystic renal epithelial cells, and found that anti-STAT3 antibody pulled down less p65 and anti-p65 antibody also pulled down less STAT3 (Figure 6A). We further found that the methylation of STAT3 and p65 was decreased in the Smyd2-knockdown cells (Figure 6A). Inhibition of STAT3 with its specific inhibitor S3I-201 disrupted the interaction between SMYD2 and p65, leading to a decrease in the methylation and phosphorylation of p65 (Figure 6B), while inhibition of NF-κB with its specific inhibitor BAY-11-7085 disrupted the interaction between SMYD2 and STAT3, leading to a decrease in the methylation and phosphorylation of STAT3 (Figure 6C). These results together with the results in Figure 5 suggested that the interaction between STAT3 and p65 is SMYD2 dependent and STAT3 modifies p65 post-translationally by recruitment of SMYD2, mediating the methylation and activation of the NF-κB p65 subunit in cystic renal epithelial cells (Figure 6D, left panel), and p65 modifies STAT3 methylation and activation by recruitment of SMYD2 (Figure 6D, right panel).
Figure 6. Synergistic effects exist among SMYD2, STAT3, and the p65 subunit of NF-κB in Pkd1 mutant renal epithelial cells.
(A) The interaction between STAT3 and p65 was disrupted in Smyd2-knockdown Pkd1-homozygous PN24 cells as examined by co-IP with either anti-STAT3 or anti-p65 antibody, and the methylation of STAT3 and p65 was also decreased in these cells. (B) Inhibition of STAT3 with its inhibitor S3I-201 disrupted the interaction between SMYD2 and NF-κB p65, and decreased p65 methylation and phosphorylation in Pkd1-homozygous PN24 cells. (C) Inhibition of NF-κB with its inhibitor BAY-11-7085 disrupted the interaction between SMYD2 and STAT3, and decreased STAT3 methylation and phosphorylation in Pkd1-homozygous PN24 cells. (D) Potential working models of the synergistic effect among SMYD2, STAT3, and p65. The interaction between SMYD2 and STAT3 may facilitate the recruitment of NF-κB to this complex, leading to the methylation (me) and phosphorylation of NF-κB (left panel); and the interaction between SMYD2 and NF-κB may facilitate the recruitment of STAT3 to this complex, leading to the methylation and phosphorylation of STAT3 (right panel), in Pkd1 mutant renal epithelial cells. (E and F) Stimulation with cytokines IL-6 (E) and TNF-α (F) induced SMYD2 expression in a time-dependent manner in mIMCD3 cells. (G and H) Western blot analysis of SMYD2 expression in the presence of the STAT3 inhibitor S3I-201 (G) or NF-κB inhibitor BAY-11-7085 (H) in Pkd1-null MEK cells. SMYD2 expression was decreased in Pkd1-null MEK cells treated with these inhibitors in a dose-dependent manner. (I) STAT3 and p65 bound to the promoter of Smyd2. ChIP-qPCR assay was performed with anti-STAT3 antibody, anti-p65 antibody, or normal rabbit IgG in Pkd1-null MEK cells. Anti-H3K4me2 antibody was used as a positive control.
Feedback loops among SMYD2, STAT3, NF-κB, and cytokines in cystic renal epithelial cells.
Both STAT3 and NF-κB p65 regulate gene expression involved in cell proliferation and apoptosis through direct promoter binding. These two proteins can also affect signaling pathways indirectly through regulation of downstream cytokines. IL-6 and TNF-α play particularly important roles, because they can form feedback loops with STAT3 and NF-κB, respectively (29, 30). We found that IL-6 treatment not only increased the phosphorylation of STAT3 but also increased the expression of SMYD2 (Figure 6E), and that treatment with TNF-α also increased the phosphorylation of p65 and the expression of SMYD2 (Figure 6F). We further found that treatment with the STAT3 inhibitor S3I-201 (Figure 6G) and NF-κB inhibitor BAY-11-7085 (Figure 6H) decreased the expression of SMYD2 in Pkd1-null MEK cells in a dose-dependent manner. To further support that STAT3 and NF-κB could regulate the expression of SMYD2 in Pkd1-null cells, we found that STAT3 and p65 bound to the Smyd2 promoter in Pkd1-null MEK cells by ChIP coupled with quantitative PCR (ChIP-qPCR) assay (Figure 6I). Our results suggest that there are two positive feedback loops, SMYD2/IL-6/STAT3/SMYD2 and SMYD2/TNF-α/NF-κB/SMYD2, in cystic renal epithelial cells.
SMYD2 regulates PKD-associated signaling via histone methylation.
SMYD2 has been identified as an H3K36 methyltransferase, and an H3K4 methyltransferase when bound to Hsp90 (14, 17). However, the full spectrum of the impact of SMYD2 on transcriptional regulation is still largely unknown. We found that inhibition of SMYD2 with AZ505 decreased the mono-, di-, and tri-methylation of H3K4 and H3K36 (Supplemental Figure 7, A and B). We further tested the effect of AZ505 on methylations in Smyd2-knockdown Pkd1-null MEK cells and found that knockdown of Smyd2 with siRNA decreased the mono-, di-, and tri-methylations of H3K4 and H3K36 in the nuclear fraction of these cells; and that treatment with AZ505 just slightly decreased the methylations of histone H3K4 and H3K36 in the nuclear fraction of Smyd2-knockdown cells compared with those in the Smyd2-knockdown cells treated with DMSO (Supplemental Figure 7C). The slight decrease in the methylation of histones by AZ505 in Smyd2-knockdown cystic renal epithelial cells might be caused by the inhibition of AZ505 on the remaining SMYD2 in these cells, which further supported the specificity of AZ505 on inhibition of SMYD2 in these cells (Supplemental Figure 7C). To identify potential transcriptional targets of SMYD2, we performed ChIP-seq analysis in Pkd1 WT and Pkd1-null MEK cells with an anti-SMYD2 antibody. We identified 91 potential SMYD2 target genes in Pkd1 WT MEK cells and 116 potential SMYD2 target genes in Pkd1-null MEK cells. There were 14 genes identified in both Pkd1 WT and -null MEK cells. Thus, the number of potential SMYD2 target genes in Pkd1-null MEK cells was 116–14 = 102 genes (Supplemental Figure 8A).
Our ChIP assay indicated that SMYD2 bound to the promoter of Ptpn13 (which encodes a protein of the protein tyrosine phosphatase [PTP] family) (Supplemental Figure 9, A and B). The mRNA and protein expression levels of PTPN13 were downregulated in Pkd1-homozygous PN24 cells compared with Pkd1-heterozygous PH2 control cells (Supplemental Figure 9, C and D). In contrast, knockdown of Smyd2 with siRNA increased the mRNA and protein levels of PTPN13 in PN24 cells (Supplemental Figure 9, E and F).
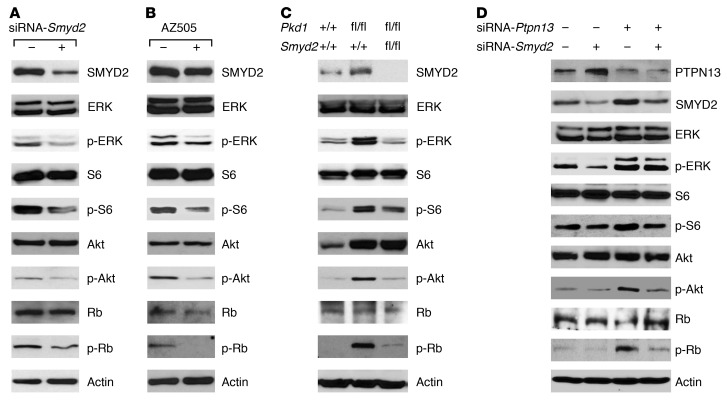
PTPN13 regulates a variety of cellular processes and oncogenic transformation by removing phosphate groups from phosphorylated tyrosine residues on proteins, including ERK and STAT (31). Since knockdown of Smyd2 and inhibition of SMYD2 with AZ505 decreased the phosphorylation of ERK, S6, and Akt in cystic renal epithelial cells (Figure 7, A and B), and knockout of Smyd2 led to similar results (Figure 7C), we suspected that PTPN13 might be involved in regulating these processes. To test this possibility, we knocked down Ptpn13 with siRNA in mIMCD3 cells. We found that knockdown of Ptpn13 increased the phosphorylation of ERK, S6, and Akt in these cells (Figure 7D), whereas knockdown of Smyd2 blocked the increases in the phosphorylation of ERK, S6, and Akt in Ptpn13-knockdown cells (Figure 7D).
Figure 7. PKD-associated signaling pathways could be affected by SMYD2 in Pkd1 mutant renal epithelial cells and cystic tissues.
(A and B) Western blot analysis of the phosphorylation of ERK, S6, AKT, and Rb as well as the total protein levels of these proteins in Pkd1-null MEK cells with or without knockdown of Smyd2 with siRNA for 24 hours (A) and with or without AZ505 treatment for 2 hours (B). Representative data are shown from 3 independent experiments. (C) Western blot analysis of the phosphorylation of ERK, S6, AKT, and Rb in kidneys from Pkd1 conditional knockout mice. Data are representative of 2 independent experiments. (D) Western blot analysis of the phosphorylation of ERK, S6, AKT, and Rb, as well as the expression of SMYD2 and PTPN13 in mIMCD3 cells transfected with or without Smyd2 siRNA and/or Ptpn13 siRNA. Representative data from 3 independent experiments are shown.
SMYD2 regulates p53-dependent cystic renal epithelial cell apoptosis.
We recently reported that p53 signaling is involved in regulating cystic renal epithelial cell apoptosis (6). Previous studies demonstrated that SMYD2 inhibits p53 activity in H1299 cells and that reducing SMYD2 by siRNA enhances p53-mediated apoptosis in DNA damage conditions but not in non–DNA damage conditions in U2OS cells (8). We found that knockdown of Smyd2 with siRNA significantly induced Pkd1-null MEK cell apoptosis but had no effect on the apoptosis of Pkd1 WT MEK cells (Supplemental Figure 10, A and B) as analyzed by TUNEL assay. However, knockdown of P53 with siRNA significantly decreased apoptosis induced by knockdown of Smyd2 in Pkd1-null MEK cells, while knockdown of P53 alone had no effect on cell survival and death (Supplemental Figure 10, C and D). To investigate the potential role of SMYD2 in regulating p53-dependent cystic renal epithelial cell death, we found by co-IP analysis that SMYD2 interacted with p53 and that this interaction increased the methylation of p53 in Pkd1-null MEK cells compared with Pkd1 WT MEK cells (Supplemental Figure 10E). Knockdown of Smyd2 decreased the methylation of p53 but unexpectedly increased the expression of p53 in Pkd1-null MEK cells as analyzed by Western blot and qRT-PCR (Supplemental Figure 10, F and G). We further found that SMYD2 bound to the promoter of P53 in Pkd1-null MEK cells as analyzed by ChIP assay (Supplemental Figure 10H). These results suggested that SMYD2 regulates p53-dependent cystic renal epithelial cell apoptosis through regulation of the methylation and expression of p53 in cystic renal epithelial cells. In addition to SMYD2-mediated p53 signaling, we also found that NF-κB signaling contributed to SMYD2-mediated apoptosis, since inhibition of NF-κB with its inhibitor induced more Pkd1-null MEK cell death (Supplemental Figure 10, I and J).
Discussion
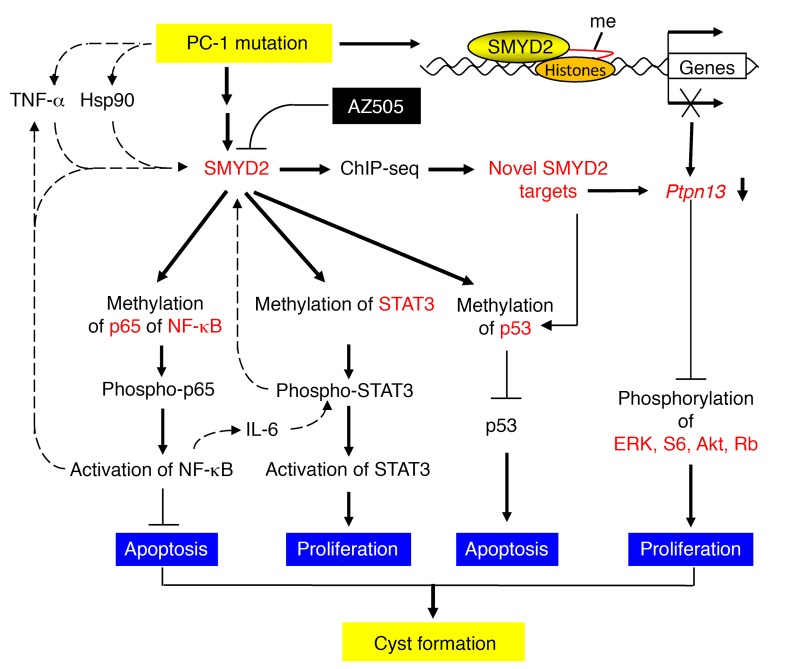
In this study, we found that SMYD2 is an epigenetic regulator of cyst growth in ADPKD. We uncovered the mechanisms underlying SMYD2’s regulation of cyst progression, and identified its non-histone substrates, including STAT3 and NF-κB, and transcriptional target gene, Ptpn13 (Figure 8). Most importantly, we showed that a SMYD2-specific inhibitor, AZ505, delayed cyst growth in the preclinical treatment of ADPKD. Once-weekly treatment with AZ505 during the period from 3.5 to 6 months in the so-called “milder” Pkd1fl/fl:tamoxifen-Cre conditional knockout mice, a mouse model to mimic human ADPKD, proved therapeutic, suggesting that targeting SMYD2 may be a reasonable strategy for treating ADPKD.
Figure 8. Working model of SMYD2 in regulation of cyst growth in ADPKD.
A schematic diagram depicting SMYD2-mediated pathways and processes in Pkd1 mutant renal epithelial cells and kidneys. Pkd1 knockout or mutation results in the upregulation of SMYD2, which may be stabilized via Hsp90, induced by cyst fluid TNF-α or through other unknown mechanisms. Upregulated SMYD2 in Pkd1 mutant renal epithelial cells (a) methylates STAT3, leading to its activation and cystic renal epithelial cell proliferation; (b) methylates the p65 subunit of NF-κB, leading to its activation, which represses cystic renal epithelial cell apoptosis; (c) methylates p53, leading to the repression of p53 and cystic renal epithelial cell apoptosis; and (d) methylates histones to regulate the transcription of the SMYD2 target gene Ptpn13, a protein of the PTP family, which may regulate the phosphorylation and activation of ERK, mTOR, Akt, and Rb signaling. Targeting SMYD2 with its specific inhibitor AZ505 delays cyst growth in Pkd1-knockout mouse kidneys. In addition, two positive feedback loops can be observed: SMYD2/IL-6/STAT3/SMYD2 and SMYD2/TNF-α/NF-κB/SMYD2, which may further increase the levels of SMYD2 in cystic renal epithelial cells.
Protein-lysine methyltransferases represent one of several families of enzymes with critical roles in epigenetic regulation. They catalyze the transfer of methyl groups from S-adenosyl-l-methionine to acceptor lysine residues on histone and/or non-histone protein substrates, impacting a variety of biological processes and disease states (32, 33). SMYD2 methylates H3K4 and H3K36 in cooperation with the HDAC complex, and it methylates key cancer proteins, including p53, Rb, and Hsp90, to alter their functions (7–9, 14). Evidence thus suggests that SMYD2 is involved in cancer via transcriptional activation and inhibition, Rb- or p53-dependent cell-cycle control, and apoptosis. However, the lack of a driver role for p53 in SMYD2-mediated ESCC cell proliferation suggests that the mechanisms by which SMYD2 drives tumor cell proliferation may involve more than one substrate and may differ depending on cell type (18, 34). To date, how SMYD2 specifically recognizes its substrates remains largely unknown. We found that STAT3 and the p65 subunit of NF-κB are SMYD2 non-histone substrates (Figure 5), and we provided evidence that upregulation of SMYD2 led to methylation of STAT3 at lysine 685 and p65 at lysine 310 and partially at lysine 221, followed by their phosphorylation and activation to regulate cystic renal epithelial cell proliferation and apoptosis.
JAK/STAT signaling is activated in ADPKD animal models (35). It is possible that Pkd1 mutation induces the phosphorylation and activation of STAT3, which then serves as a positive regulator of cyst growth (23). Inhibition of STAT3 delayed cyst growth in Pkd1-knockout mice (23). However, the upstream regulator for the activation and phosphorylation of STAT3 was unknown. Our results demonstrated that SMYD2 acts as a mediator between Pkd1 mutation and the activation of STAT3 via lysine methylation at K685, resulting in its phosphorylation and activation. STAT3 can also be methylated by other methyltransferases, including EZH2 and SET9 (36, 37). However, methylation of K140 by SET9 is a negative regulatory event, because its blockade greatly increases the steady-state amount of activated STAT3 and the expression of many (i.e., SOCS3) but not all STAT3 target genes (37). Thus, SET9 may not be involved in the activation of STAT3 in cystic renal epithelial cells. The involvement of EZH2 in PKD will be investigated in the future.
It has been shown that the p65 subunit of NF-κB interacts physically with STAT3, facilitating NF-κB recruitment to Stat3 promoters and vice versa, suggesting that NF-κB and STAT3 may cooperatively regulate a number of target genes, including antiapoptotic and cell cycle control genes, as well as genes encoding cytokines and chemokines (38). We found that the p65 subunit of NF-κB is also a non-histone substrate of SMYD2 in cystic renal epithelial cells (Figure 5). Methylation of p65 at lysine 310 and partially at lysine 221 via SMYD2 resulted in its phosphorylation and activation, which might regulate cystic renal epithelial cell apoptosis, as suggested recently (24).
STAT3, as a transcription factor, is involved in the IL-6 family of cytokines and growth factor–induced expression of target genes responsible for the acute-phase response, cell cycle progression, and cell transformation (39). Cytokine-induced JAK/STAT3 signaling has emerged as a positive regulator of injured axon regrowth and cancer (40). NF-κB, as another transcription factor, can be activated to induce expression of chemokines and cytokines within premalignant cells, leading to the recruitment and activation of immune cells. Activated immune cells in turn produce more proinflammatory cytokines/chemokines and growth factors, such as IL-1, IL-6, and TNF-α, in an autocrine and/or paracrine manner to further stimulate NF-κB activation within the premalignant cells, in a positive feedback loop (41). Renal inflammation has been associated with the expression and accumulation of multiple cytokines, including TNF-α, IL-1, IL-6, and MCP-1 in cyst fluid (42–44). These cytokines, whether endogenously induced or further upregulated by exogenous treatment, promote cyst growth in PKD (42). We found that IL-6 and TNF-α treatment induced the phosphorylation and activation of STAT3 and p65, and increased the expression of SMYD2 (Figure 6). In addition, STAT3 and p65 bound to the Smyd2 promoter (Figure 6), and treatments with the inhibitors of STAT3 and NF-κB decreased the expression of SMYD2 in cystic renal epithelial cells (Figure 6), suggesting that there are two positive feedback loops: SMYD2/IL-6/STAT3/SMYD2 and SMYD2/TNF-α/NF-κB/SMYD2 (Figure 8). These results integrate renal inflammation and epigenetic regulation of renal cyst development and provide one of the mechanisms explaining how the elevation of SMYD2 might be induced by cyst fluid IL-6 and TNF-α in the clinical setting.
Activation of ERK, mTOR, and Akt has been associated with PKD (45, 46). However, the mediators between PKD mutations and the phosphorylation and activation of these signaling pathways are uncertain. Cyclic AMP has been proposed as one (47). We identified that PTPN13 as a transcriptional target of SMYD2. PTPN13 is one of the members of the PTP family, which regulates a variety of cellular processes and oncogenic transformation by removing phosphate groups from phosphorylated residues on proteins, including ERK and STAT (31). SMYD2 is able to downregulate the expression of PTPN13 in cystic renal epithelial cells (Supplemental Figure 9). Knockdown of Ptpn13 increased the phosphorylation of ERK, mTOR, and Akt in mIMCD3 cells (Figure 7D), which suggested that SMYD2-mediated downregulation of PTPN13 might be one of the other mechanisms responsible for the phosphorylation and activation of these PKD-associated signaling pathways in cystic renal epithelial cells (Figure 7D).
Several protein lysine methyltransferases are being pursued as drug targets for cancer and/or other diseases (48). Our study provides evidence that SMYD2 regulates multiple physiological and pathological processes in a cellular context–dependent manner in cystic renal epithelial cells. The small molecule inhibitor of SMYD2, AZ505, identified from a high-throughput screen (HTS) (20), has been shown to bind to the lysine pocket of SMYD2 via a co-crystal structure and is quite selective toward SMYD2 (>700-fold) over 6 other methyltransferases. Our study tested for the first time to our knowledge the effect of AZ505 in an in vivo disease model, and found that it delayed renal cyst growth in a preclinical ADPKD mouse model.
In conclusion, we identified SMYD2 as an epigenetic regulator of ADPKD. The SMYD2-mediated methylation regulates STAT3 and NF-κB function by inducing their phosphorylation, which enhances cystic renal epithelial cell growth and survival. SMYD2-mediated histone methylation regulates the expression of genes associated with a number of PKD signaling pathways. Given that SMYD2 is overexpressed in cystic renal epithelial cells as well as in cancer, it is likely to be a promising target for development of a novel ADPKD therapy (49).
Methods
Cell culture and reagents.
mIMCD3 cells were maintained at 37°C in 5% CO2 in DMEM (Invitrogen) supplemented with 10% FBS. Pkd1 WT and Pkd1-null MEK cells, derived from collecting ducts and sorted by the collecting duct marker dolichos biflorus agglutinin (DBA) from kidneys of WT and Pkd1-null mice, were maintained as previously described (50, 51). PH2 and PN24 cells (provided by S. Somlo through the George M. O’Brien Kidney Center, Yale University, New Haven, Connecticut, USA) were cultured as described previously (52). Primary human ADPKD and NHK cells were provided by the PKD Research Biomaterials and Cellular Models Core at the University of Kansas Medical Center (KUMC) and were cultured as described previously (42, 53). AZ505 was purchased from MedChem Express and dissolved in DMSO (Sigma-Aldrich) at a stock solution of 20 mM. S3I-201 was purchased from Sigma-Aldrich and dissolved in DMSO (Sigma-Aldrich) at a stock solution of 100 mM. BAY-11-7085 was purchased from Cayman Chemical and dissolved in DMSO (Sigma-Aldrich) at a stock solution of 40 mM. All the stock solutions were stored at –20°C.
The antibodies used for Western blot analysis included (a) anti-SMYD2 (sc-79084), anti-STAT3 (sc-482), anti-p65 (sc372 and sc-8008), anti-p53 (sc-6423), and anti-PTPN13 (sc-15356), which were purchased from Santa Cruz Biotechnology Inc.; (b) anti-SMYD2 (no. 9734), anti-ERK (no. 4696), anti-S6 (no. 2217), anti-Rb (no. 9309), anti-GFP (no. 2956), anti-AKT (no. 9272), and anti–lamin A/C (no. 2032), purchased from Cell Signaling Technology; and (c) the phosphorylated antibodies for STAT3-Y705 (no. 9131), p65-S536 (no. 3031), ERK-T202/Y204 (no. 9101), S6-S235/236 (no. 2211), AKT-S473 (no. 9271), and Rb-S780 (no. 9307), also purchased from Cell Signaling Technology. All of the anti–methylated histone antibodies, including H3K4-mono (ab8895), H3K4-di (ab7766), H3K4-tri (ab8580), H3K36-mono (ab9048), H3K36-di (ab9049), and H3K36-tri (ab9050), anti-Ki67 antibody (ab15580), anti-T7 antibody (ab9138), and anti–pan–methyl lysine antibody (ab23366) were purchased from Abcam. Anti-actin antibody (A5316) and anti-FLAG antibody (F3165) were purchased from Sigma-Aldrich. The secondary antibodies, including donkey anti-rabbit IgG–horseradish peroxidase (sc-2313), donkey anti-goat IgG–horseradish peroxidase (sc-2020), and goat anti-mouse IgG–horseradish peroxidase (sc-2005) were purchased from Santa Cruz Biotechnology Inc.
Plasmids.
The Flag-tagged WT STAT3 plasmid and mutant STAT3 plasmids, including the K49R, K140R, and K685R mutants, and T7-tagged p65 (RelA) and its mutants were purchased from Addgene. The GFP-tagged SMYD2 plasmid was constructed by cloning full-length Smyd2 into the pAcGFP-C1 vector (Clontech). Each Flag-tagged STAT3 construct was transfected into HEK293T cells along with packaging plasmids, using Lipofectamine (Invitrogen). The virus-containing supernatant was harvested 72 hours later. In the meantime, GFP-tagged SMYD2 expression plasmid DNA was transfected into the HEK293T cells together with the virus-containing supernatants. After 48 hours, the cells were harvested for further analysis. Each T7-tagged p65 (RelA) construct, was transfected into HEK293T cells with or without GFP-tagged SMYD2 expression plasmid. After 24–36 hours, the cells were harvested for further analysis.
Subcellular fractionation.
The cytosol and nuclear fractions from Pkd1-null MEK cells or Pkd1-homozygous PN24 cells were prepared according to the instructions of the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (ThermoFisher Scientific).
Western blot analysis and immunoprecipitation.
Cell pellets were collected and resuspended in lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM Na3VO4, 25 mM β-glycerolphosphate, 0.1 mM PMSF, Roche complete protease inhibitor set, and Sigma-Aldrich phosphatase inhibitor set). The resuspended cell pellet was vortexed for 20 seconds and then incubated on ice for 30 minutes and centrifuged at 20,000 g for 30 minutes. The supernatants were collected for Western blot analysis or immunoprecipitation.
For immunoprecipitation, anti-SMYD2, anti-STAT3, or anti-p65 antibodies and their isotype control antibodies were coupled to protein A agarose beads (Pierce) in PBS containing 5 mg/ml bovine serum albumin (Sigma-Aldrich) for 6 hours at 4°C on a rotating platform. The cell lysates were then incubated with the beads coupled with antibodies overnight at 4°C. The next day, beads were washed with lysis buffer containing an additional 300 mM NaCl, and the immune complexes were eluted off the beads using loading buffer with boiling for 5 minutes and then subjected to Western blot analysis.
Histology and immunohistochemistry.
Paraffin-embedded sections (6 μm) were subjected to H&E staining and immunohistochemistry. For SMYD2 staining, a polyclonal goat anti-SMYD2 antibody (Santa Cruz Biotechnology Inc.; 1:100 dilution), biotinylated secondary antibody (Santa Cruz Biotechnology Inc.; 1:100 dilution), and DAB substrate system were used. Kidney sections were counterstained by hematoxylin. Images were analyzed with a Nikon Eclipse 80i microscope.
Immunofluorescence staining.
For Ki67 staining, a rabbit anti-Ki67 antibody (ab15580; Abcam) and Alexa Fluor 488 anti-rabbit IgG secondary antibody were used. Images were analyzed using a Nikon Eclipse 80i microscope.
qRT-PCR.
Total RNA was extracted using the RNeasy Plus Mini Kit (QIAGEN). Total RNA (1 μg) was used for RT reactions in a 20-μl reaction to synthesize cDNA using an iScript cDNA Synthesis Kit (Bio-Rad). RNA expression profiles were analyzed by real-time PCR using iTaq SYBR Green Supermix with ROX (Bio-Rad) in an iCycler iQ Real-Time PCR Detection System. The complete reactions were subjected to the following program of thermal cycling: 40 cycles of 10 seconds at 95°C and 20 seconds at 60°C. A melting curve was run after the PCR cycles, followed by a cooling step. Each sample was run in triplicate in each experiment, and each experiment was repeated 3 times. Expression levels of target genes were normalized to the expression level of actin. All primers used are listed in Supplemental Table 1.
ChIP assay.
ChIP assay was performed according to the protocol described in ref. 54. Chromatin DNA was subjected to immunoprecipitation with anti-SMYD2, anti-STAT3, and anti-p65 antibodies and anti-H3K4me2 antibody, or normal rabbit IgG, and then washed, after which the DNA-protein crosslinks were reversed. The recovered DNA was analyzed by PCR for the binding of STAT3, p65, and H3K4me at the mouse Smyd2 promoter.
ChIP-seq analysis.
ChIP-seq was performed using an anti-SMYD2 antibody. Each sample with 10 ng of DNA generated from the ChIP assay was used to construct the sequencing libraries with the ChIP-Seq Sample Prep Kit (Illumina). These libraries were sequenced by an Illumina HiSeq2500 Sequencing System at the Genome Sequencing Facility of KUMC. The sequencing data sets from ChIP-seq analyses have been deposited in the in the NCBI’s Gene Expression Omnibus (GSE97708).
TUNEL assay.
TUNEL assay for AZ505-treated Pkd1 WT MEK, Pkd1-null MEK, or Smyd2-knockdown renal epithelial cells and for AZ505-treated kidneys was performed according to the manufacturer’s protocols (In Situ Death Detection Kit; Roche). ProLong Gold Antifade reagent with DAPI (Invitrogen) was used. Immunofluorescence images were obtained with a Nikon Eclipse 80i microscope.
RNA interference.
The RNA oligonucleotides that specifically targeted mouse Smyd2 and mouse P53 were purchased from Santa Cruz Biotechnology Inc. The RNA oligonucleotides were transfected with DharmaFECT siRNA transfection reagent (Dharmacon). 24 hours and 48 hours after transfection, cells were harvested and analyzed by Western blotting.
Cell cycle analysis.
A total of 150,000 cells were seeded in 6-well plates and allowed to recover for 16–24 hours; cells were then starved with serum-free medium for 24 hours. Then cells were left untreated or treated with AZ505 for 24 hours. The cells were pelleted and washed by adding 1 ml PBS; the cells were centrifuged at 500 g for 5 minutes, and the supernatant was aspirated; 1 ml of cold 70% ethanol was added drop by drop into the cell pellet to suspend the cells. The cells were then incubated at –20°C overnight. After that, the cells were washed twice in 1× PBS to remove the ethanol, then centrifuged for 5 minutes at 1,000 g, and the supernatant was aspirated. For propidium iodide (PI) staining, the cells were suspended in 0.5 ml PI buffer (containing RNase and Triton X-100) for 10–15 minutes at room temperature. The cells were kept away from light at 4°C before further analysis on the flow cytometer, which took place within 1 hour. FACS data were analyzed with FlowJo software 5.7.2 (Tree Star).
MTT assays.
Cell proliferation was measured using an MTT-based kit (Sigma-Aldrich), according to the manufacturer’s instructions.
Transfection assays.
mIMCD3 cells were cultured to confluence in DMEM medium containing 10% FCS (Fetalclone III, Clontech). The control pACGFPC1 vector or SMYD2 constructs were transfected into cells using the Lipofectamine 2000 Kit (Invitrogen) according to the manufacturer’s recommendations. Transfected cells were maintained at 37°C for 24 or 48 hours. For knockdown of Pkd1 and Smyd2, IMCD3 cells were transduced with pVIRHD/P/siLuc and pVIRHD/P/siPKD13297 lentivectors (55). Twenty-four hours following transduction, cells were selected in puromycin (10 μg/ml) for 48 hours, and on day 4, the siRNA-Smyd2 oligonucleotides were transfected with DharmaFECT siRNA transfection reagent (Dharmacon) for 24 hours. Cells were harvested and analyzed by Western blotting and qRT-PCR.
Mouse strains and treatments.
Hypomorphic Pkd1nl/nl mice, generated by cross-breeding Pkd1nl/+ females with Pkd1nl/+ males, were used to test the effect of AZ505 on cyst progression at P28 (21). Pkd1nl/nl pups were injected i.p. with AZ505 (5 mg/kg, dissolved in DMSO, with a final DMSO concentration of 10% [v/v] in PBS) or DMSO (control) daily from P7 to P27, and kidneys from 6 female mice and 6 male mice per group were harvested and analyzed at P28.
Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre mice were used to observe the effect of SMYD2 on cyst formation and mouse survival.
Pkd1fl/fl mice and Ksp-Cre transgenic mice were generated as described previously (19). Pkd1fl/fl mice (B6; 129S4-Pkd1tm2Ggg/J; stock 010671; Jackson Laboratory) possess loxP sites on either side of exons 2–4 of Pkd1 (52). Ksp-Cre mice express Cre recombinase under the control of the Ksp-cadherin promoter (19). Pkd1fl/fl:Ksp-Cre mice were generated by cross-breeding Pkd1fl/fl female mice with Pkd1fl/+:Ksp-Cre male mice. Smyd2fl/fl mice were provided by Julien Sage (Department of Pediatrics and Genetics, Stanford University Medical Center, Stanford, California, USA). The Smyd2fl/fl mice developed normally in size and behavior, and were fertile. First, we generated Pkd1fl/+:Smyd2fl/+:Ksp-Cre mice by crossing Pkd1fl/+:Ksp-Cre mice with Smyd2fl/fl mice. Second, we generated Pkd1fl/fl:Smyd2fl/fl:Ksp-Cre mice by crossing Pkd1fl/+:Smyd2fl/+:Ksp-Cre female mice with Pkd1fl/+:Smyd2 fl/+:Ksp-Cre male mice.
Pkd1fl/fl:tamoxifen-Cre mice were injected i.p. with tamoxifen (125 mg/kg body weight, formulated in corn oil) on 2 sequential postnatal days (P31 and P32) to induce Pkd1 deletion in the test group. Pkd1fl/fl tamoxifen-Cre mice were treated with AZ505 for about 5 months, starting 10 days (P42) after Pkd1 deletion with a dosage of 10 mg/kg body weight. Treatment began with a 3-times-per-week schedule up to 3.5 months of age, following by a once-per-week schedule from 3.5–6 months of age. We collected blood samples 24 hours after the last dose and harvested kidneys from 6-month-old AZ505- and vehicle-treated (DMSO-treated) mice for histopathological and biochemical analysis. 7 male and 7 female animals were used for all experiments, and comparable experimental results were obtained with each sex.
The route of AZ505 administration to animals was compared with oral administration; i.p. injection was more efficient, and the amount of drug administered to animals was more controllable. Thus, we used i.p. injection of AZ505 for treating hypomorphic Pkd1nl/nl mice and Pkd1fl/fl:tamoxifen-Cre mice.
Measurement of cyst area.
The cyst volume was quantified in whole kidney after H&E staining using Image-Pro Plus v5 software (Media Cybernetics). The cyst area was calculated as (cyst area/total area) × 100%. Three sections from both kidneys were analyzed for each mouse.
Quantitative BUN determination.
Serum samples were first diluted 5-fold in distilled water prior to assay. Next, 5 μl water (blank), 5 μl standard (50 mg/dl), and 5 μl samples were transferred in triplicate into wells of a clear-bottom 96-well plate. 200 μl of working reagent was added and tapped lightly to mix, and the samples were incubated 20 minutes at room temperature. Optical density was read at 520 nm.
Statistics.
All data are presented as mean ± SEM. All statistical analyses were performed using SPSS Statistics 22 software. P values were calculated by 2-tailed unpaired Student’s t-test and 1-way ANOVA; and a P value less than 0.05 was considered significant.
Study approval.
All animal protocols were approved by and conducted in accordance with Laboratory Animal Resources of KUMC and Institutional Animal Care and Use Committee regulations (protocol 2015-2290).
Author contributions
LXL performed most experiments. LXF performed some of the animal experiments. JXZ performed some of the ChIP-seq experiments and data analysis. JJG and JPC assisted in data analysis and manuscript preparation. JS assisted in animal experiments. XL supervised the entire project, analyzed the data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We are grateful to S. Somlo for providing cell lines PH2 and PN24 through the George M O’Brien Kidney Center at Yale University (NIH P30 DK079310). We are also grateful to Peter Igarashi for providing Ksp-Cre mice through the George M. O’Brien Kidney Research Core Center at UT Southwestern (NIH P30 DK079328). We are grateful to Dorien J.M. Peters for providing the hypomorphic Pkd1nl/nl mouse through the Leiden University Medical Center. XL acknowledges support from NIH R01 DK084097 and NIH P30 DK106912. JS acknowledges support from NIH grant R01 CA114102, and JPC acknowledges support from the PKD Foundation and from the Kansas Research and Translation Core Center (NIH P30 DK106912). LXL acknowledges support from a postdoctoral fellowship funded by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences (P20 GM103418). JXZ acknowledges support from the PKD Foundation and NIH K01 DK107729.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(7):2751–2764.https://doi.org/10.1172/JCI90921.
Contributor Information
Linda Xiaoyan Li, Email: xli@kumc.edu.
Jared J. Grantham, Email: jgrantha@kumc.edu.
Julien Sage, Email: julsage@stanford.edu.
Xiaogang Li, Email: xli3@kumc.edu.
References
- 1.Torres VE, Harris PC. Polycystic kidney disease in 2011: connecting the dots toward a polycystic kidney disease therapy. Nat Rev Nephrol. 2011;8(2):66–68. doi: 10.1038/nrneph.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters DJ, Sandkuijl LA. Genetic heterogeneity of polycystic kidney disease in Europe. Contrib Nephrol. 1992;97:128–139. doi: 10.1159/000421651. [DOI] [PubMed] [Google Scholar]
- 3.Torres VE, Harris PC. Strategies targeting cAMP signaling in the treatment of polycystic kidney disease. J Am Soc Nephrol. 2014;25(1):18–32. doi: 10.1681/ASN.2013040398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Y, Zhou H, Yang BX. Drug discovery for polycystic kidney disease. Acta Pharmacol Sin. 2011;32(6):805–816. doi: 10.1038/aps.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X. Epigenetics and autosomal dominant polycystic kidney disease. Biochim Biophys Acta. 2011;1812(10):1213–1218. doi: 10.1016/j.bbadis.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou X, Fan LX, Sweeney WE, Jr, Denu JM, Avner ED, Li X. Sirtuin 1 inhibition delays cyst formation in autosomal-dominant polycystic kidney disease. J Clin Invest. 2013;123(7):3084–3098. doi: 10.1172/JCI64401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamamoto R, Toyokawa G, Nakakido M, Ueda K, Nakamura Y. SMYD2-dependent HSP90 methylation promotes cancer cell proliferation by regulating the chaperone complex formation. Cancer Lett. 2014;351(1):126–133. doi: 10.1016/j.canlet.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Huang J, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444(7119):629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 9.Saddic LA, et al. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem. 2010;285(48):37733–37740. doi: 10.1074/jbc.M110.137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seeger-Nukpezah T, et al. Inhibiting the HSP90 chaperone slows cyst growth in a mouse model of autosomal dominant polycystic kidney disease. Proc Natl Acad Sci U S A. 2013;110(31):12786–12791. doi: 10.1073/pnas.1301904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Bodegom D, et al. The polycystic kidney disease-1 gene is a target for p53-mediated transcriptional repression. J Biol Chem. 2006;281(42):31234–31244. doi: 10.1074/jbc.M606510200. [DOI] [PubMed] [Google Scholar]
- 12.Gottlieb PD, et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31(1):25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- 13.Wu J, et al. Biochemical characterization of human SET and MYND domain-containing protein 2 methyltransferase. Biochemistry. 2011;50(29):6488–6497. doi: 10.1021/bi200725p. [DOI] [PubMed] [Google Scholar]
- 14.Brown MA, Sims RJ, Gottlieb PD, Tucker PW. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer. 2006;5:26. doi: 10.1186/1476-4598-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamamoto R, Saloura V, Nakamura Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat Rev Cancer. 2015;15(2):110–124. doi: 10.1038/nrc3884. [DOI] [PubMed] [Google Scholar]
- 16.Spellmon N, Holcomb J, Trescott L, Sirinupong N, Yang Z. Structure and function of SET and MYND domain-containing proteins. Int J Mol Sci. 2015;16(1):1406–1428. doi: 10.3390/ijms16011406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abu-Farha M, Lambert JP, Al-Madhoun AS, Elisma F, Skerjanc IS, Figeys D. The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol Cell Proteomics. 2008;7(3):560–572. doi: 10.1074/mcp.M700271-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Komatsu S, et al. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis. 2009;30(7):1139–1146. doi: 10.1093/carcin/bgp116. [DOI] [PubMed] [Google Scholar]
- 19.Shao X, Somlo S, Igarashi P. Epithelial-specific Cre/lox recombination in the developing kidney and genitourinary tract. J Am Soc Nephrol. 2002;13(7):1837–1846. doi: 10.1097/01.ASN.0000016444.90348.50. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson AD, et al. Structural basis of substrate methylation and inhibition of SMYD2. Structure. 2011;19(9):1262–1273. doi: 10.1016/j.str.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Lantinga-van Leeuwen IS, et al. Lowering of Pkd1 expression is sufficient to cause polycystic kidney disease. Hum Mol Genet. 2004;13(24):3069–3077. doi: 10.1093/hmg/ddh336. [DOI] [PubMed] [Google Scholar]
- 22.Piontek K, Menezes LF, Garcia-Gonzalez MA, Huso DL, Germino GG. A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat Med. 2007;13(12):1490–1495. doi: 10.1038/nm1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takakura A, et al. Pyrimethamine inhibits adult polycystic kidney disease by modulating STAT signaling pathways. Hum Mol Genet. 2011;20(21):4143–4154. doi: 10.1093/hmg/ddr338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan LX, Zhou X, Sweeney WE, Jr, Wallace DP, Avner ED, Grantham JJ, Li X. Smac-mimetic-induced epithelial cell death reduces the growth of renal cysts. J Am Soc Nephrol. 2013;24(12):2010–2022. doi: 10.1681/ASN.2013020176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwensen KG, et al. Early cyst growth is associated with the increased nuclear expression of cyclin D1/Rb protein in an autosomal-recessive polycystic kidney disease rat model. Nephron Exp Nephrol. 2011;117(4):e93–103. doi: 10.1159/000320149. [DOI] [PubMed] [Google Scholar]
- 26.Torres VE. Treatment strategies and clinical trial design in ADPKD. Adv Chronic Kidney Dis. 2010;17(2):190–204. doi: 10.1053/j.ackd.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X, Fan LX, Peters DJ, Trudel M, Bradner JE, Li X. Therapeutic targeting of BET bromodomain protein, Brd4, delays cyst growth in ADPKD. Hum Mol Genet. 2015;24(14):3982–3993. doi: 10.1093/hmg/ddv136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H, et al. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15(4):283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, van Boxel-Dezaire AH, Cheon H, Yang J, Stark GR. STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor. Proc Natl Acad Sci U S A. 2013;110(42):16975–16980. doi: 10.1073/pnas.1315862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oeckinghaus A, Ghosh S. The NF-κB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1(4):a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sardina JL, et al. PTPN13 regulates cellular signalling and β-catenin function during megakaryocytic differentiation. Biochim Biophys Acta. 2014;1843(12):2886–2899. doi: 10.1016/j.bbamcr.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 32.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13(5):343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joshi CP, Chiang VL. Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferases. Plant Mol Biol. 1998;37(4):663–674. doi: 10.1023/A:1006035210889. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen H, et al. LLY-507, a Cell-active, potent, and selective inhibitor of protein-lysine methyltransferase SMYD2. J Biol Chem. 2015;290(22):13641–13653. doi: 10.1074/jbc.M114.626861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talbot JJ, et al. Polycystin-1 regulates STAT activity by a dual mechanism. Proc Natl Acad Sci U S A. 2011;108(19):7985–7990. doi: 10.1073/pnas.1103816108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim E, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23(6):839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci U S A. 2010;107(50):21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grivennikov SI, Karin M. Dangerous liaisons: STAT3 and NF-κB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010;21(1):11–19. doi: 10.1016/j.cytogfr.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3(11):900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 40.Qin S, Zou Y, Zhang CL. Cross-talk between KLF4 and STAT3 regulates axon regeneration. Nat Commun. 2013;4:2633. doi: 10.1038/ncomms3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol. 2009;1(5):a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, et al. A tumor necrosis factor-α-mediated pathway promoting autosomal dominant polycystic kidney disease. Nat Med. 2008;14(8):863–868. doi: 10.1038/nm1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merta M, Tesar V, Zima T, Jirsa M, Rysava R, Zabka J. Cytokine profile in autosomal dominant polycystic kidney disease. Biochem Mol Biol Int. 1997;41(3):619–624. doi: 10.1080/15216549700201651. [DOI] [PubMed] [Google Scholar]
- 44.Karihaloo A, et al. Macrophages promote cyst growth in polycystic kidney disease. J Am Soc Nephrol. 2011;22(10):1809–1814. doi: 10.1681/ASN.2011010084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shillingford JM, et al. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc Natl Acad Sci U S A. 2006;103(14):5466–5471. doi: 10.1073/pnas.0509694103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boca M, Distefano G, Qian F, Bhunia AK, Germino GG, Boletta A. Polycystin-1 induces resistance to apoptosis through the phosphatidylinositol 3-kinase/Akt signaling pathway. J Am Soc Nephrol. 2006;17(3):637–647. doi: 10.1681/ASN.2005050534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calvet JP, Grantham JJ. The genetics and physiology of polycystic kidney disease. Semin Nephrol. 2001;21(2):107–123. doi: 10.1053/snep.2001.20929. [DOI] [PubMed] [Google Scholar]
- 48.Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8(9):724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 49.Cho HS, et al. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia. 2012;14(6):476–486. doi: 10.1593/neo.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nauli SM, et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet. 2003;33(2):129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- 51.Li X, Luo Y, Starremans PG, McNamara CA, Pei Y, Zhou J. Polycystin-1 and polycystin-2 regulate the cell cycle through the helix-loop-helix inhibitor Id2. Nat Cell Biol. 2005;7(12):1202–1212. doi: 10.1038/ncb1326. [DOI] [PubMed] [Google Scholar]
- 52.Shibazaki S, et al. Cyst formation and activation of the extracellular regulated kinase pathway after kidney specific inactivation of Pkd1. Hum Mol Genet. 2008;17(11):1505–1516. doi: 10.1093/hmg/ddn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pinto CS, et al. Phosphodiesterase isoform regulation of cell proliferation and fluid secretion in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2016;27(4):1124–1134. doi: 10.1681/ASN.2015010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48(3):240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Battini L, et al. Loss of polycystin-1 causes centrosome amplification and genomic instability. Hum Mol Genet. 2008;17(18):2819–2833. doi: 10.1093/hmg/ddn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.