Abstract
Cleidocranial dysplasia (CCD) is an autosomal dominant human disorder characterized by abnormal bone development that is mainly due to defective intramembranous bone formation by osteoblasts. Here, we describe a mouse strain lacking the E3 ubiquitin ligase RNF146 that shows phenotypic similarities to CCD. Loss of RNF146 stabilized its substrate AXIN1, leading to impairment of WNT3a-induced β-catenin activation and reduced Fgf18 expression in osteoblasts. We show that FGF18 induces transcriptional coactivator with PDZ-binding motif (TAZ) expression, which is required for osteoblast proliferation and differentiation through transcriptional enhancer associate domain (TEAD) and runt-related transcription factor 2 (RUNX2) transcription factors, respectively. Finally, we demonstrate that adipogenesis is enhanced in Rnf146–/– mouse embryonic fibroblasts. Moreover, mice with loss of RNF146 within the osteoblast lineage had increased fat stores and were glucose intolerant with severe osteopenia because of defective osteoblastogenesis and subsequent impaired osteocalcin production. These findings indicate that RNF146 is required to coordinate β-catenin signaling within the osteoblast lineage during embryonic and postnatal bone development.
Keywords: Bone Biology, Cell Biology
Introduction
Cleidocranial dysplasia (CCD) is an autosomal dominant skeletal disorder characterized by short stature, hypoplasia of facial bones and clavicles, and open fontanelles due to defective bone formation (1, 2). Previous studies have demonstrated that loss-of-function mutations in a single allele of the CBFA1 gene encoding runt-related transcription factor 2 (RUNX2), which is an osteoblastogenic master transcription factor, are detected in 60% of the CCD patients as a consequence of impaired osteoblast-mediated ossification (3–8). However, the genetic origin of 40% of the CCD patients is still unknown (8), suggesting that mutations in other genes can lead to this syndrome.
Bone formation, which is orchestrated by osteoblasts, can occur through 2 different processes, intramembranous and endochondral ossification (5). Mesenchymal progenitor cells directly differentiate into osteoblasts required for intramembranous ossification in the calvarium and clavicles, whereas during longitudinal endochondral bone formation, cartilaginous template is eventually replaced by bone deposited by osteoblasts (5). The cell-autonomous osteoblast defect caused by dysfunction of RUNX2 results in impairment of both endochondral and intramembranous bone formation, although the latter is usually the dominant phenotypic feature in CCD patients (3–7), suggesting that dysregulation of the RUNX2 activation pathway may cause CCD.
The osteoblast developmental program is promoted by the essential transcription factor complex formed by RUNX2 and transcriptional coactivator with PDZ-binding motif (TAZ) (9). The Hippo pathway effector TAZ is a mesenchymal fate-determining factor that controls lineage commitment depending on its target substrate. TAZ forms a complex with and increases the activity of RUNX2, and this drives osteoblast differentiation while it represses PPARγ activity that is important for adipocyte lineage commitment (9). However, the regulatory mechanism of TAZ expression during osteoblastogenesis is still unknown.
The E3 ubiquitin ligase RNF146 controls the Wnt/β-catenin pathway through ubiquitylation of its substrate AXIN, a member of the β-catenin destruction complex (10, 11). RNF146 recognizes AXIN as its substrate following the addition of ADP-ribose chains by tankyrase (10). Following binding and activation of RNF146, AXIN is ubiquitylated and destroyed through proteasome-mediated degradation (10). RNF146 thus promotes Wnt/β-catenin signaling through suppression of its inhibitor AXIN (10). The Wnt/β-catenin pathway plays essential roles in embryonic development and, in particular, osteoblast differentiation during embryonic skeletal formation (12, 13), suggesting a potential link between RNF146 and human developmental disorders.
Here, we provide evidence that RNF146 is critical for normal embryonic and postnatal bone formation. Mice with conditional knockout of Rnf146 driven by the osterix promoter (Rnf146fl/fl Osx-Cre) have short stature, fail to close their fontanelles, demonstrate general hypomineralization of the calvarium, have small clavicles, and are generally osteopenic, with low serum levels of osteocalcin. This phenotype is reminiscent of some features observed in CCD patients. While RUNX2 protein expression appears to be normal in these mice, they have low levels of TAZ. We determined that loss of RNF146 results in a defective signaling pathway, defined by reduced β-catenin activity due to stabilization of AXIN1 and reduced expression of Fgf18 required for TAZ protein expression. We show that FGF18 enhances TAZ expression through activation of the MAPK pathway and that defective proliferation and mineralization observed in RNF146-null osteoblasts can be rescued by exogenous FGF18. Finally, we found that Rnf146fl/fl Osx-Cre mice had increased fat stores and were glucose intolerant due to defective insulin secretion as a consequence of reduced osteocalcin production. These data demonstrate that RNF146 is a mesenchymal fate–determining factor required for the development of the osteoblast lineage while antagonizing adipocyte differentiation and provide a molecular mechanism by which hypoplastic bone formation causes abnormal energy metabolism.
Results
RNF146 deficiency in osteoblasts causes a CCD-like syndrome in mice.
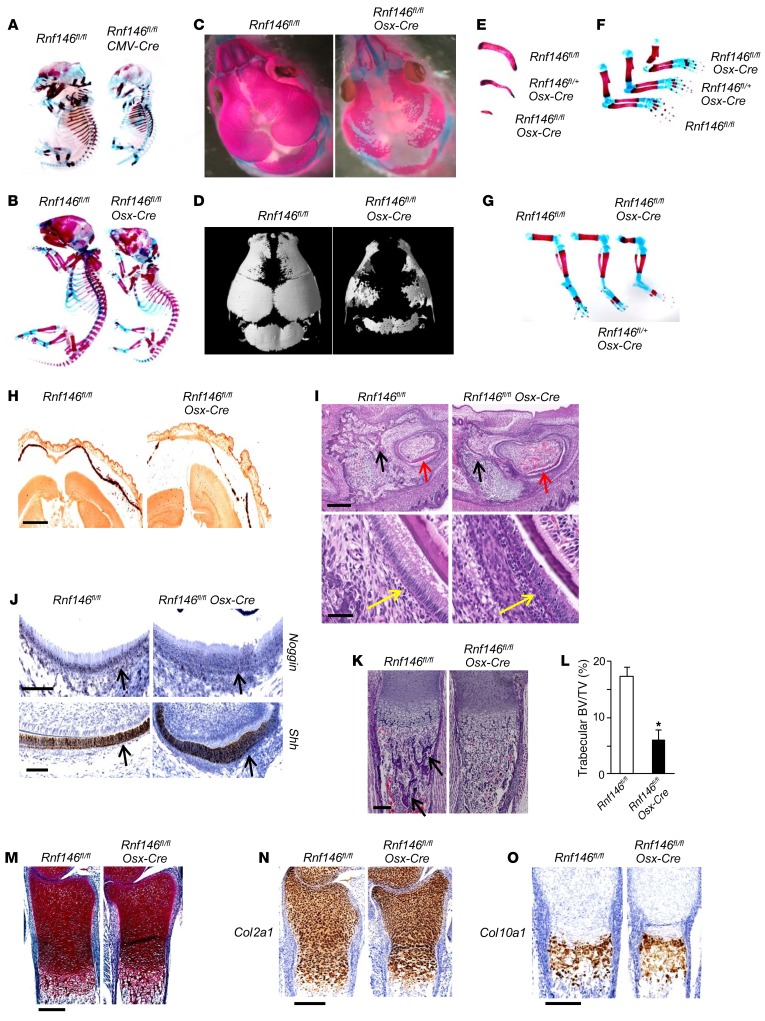
To elucidate the role of RNF146 during embryonic development, we first generated Rnf146fl/fl CMV-Cre mice whose floxed Rnf146 alleles were excised in all tissues, including germ cells (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI92233DS1) (14), and observed that deletion of Rnf146 resulted in embryonic lethality. We analyzed embryonic skeletal development in E15.5 Rnf146fl/fl CMV-Cre embryos and observed delayed bone formation in the calvarium (Figure 1A). In view of these results, we generated Rnf146 conditional knockout mice (Rnf146fl/fl Osx-Cre) in which endogenous RNF146 was deleted in cells of the osteoblast lineage. Rnf146fl/fl Osx-Cre newborn pups were shorter than Rnf146fl/fl control littermates and died shortly after birth due to respiratory failure (Figure 1B and Supplemental Figure 1B). Alizarin red/Alcian blue staining and 3D reconstruction using μCT of skeletal preparations showed that Rnf146fl/fl Osx-Cre pups displayed enlarged anterior and posterior fontanelles with calvarial hypomineralization as a consequence of impaired intramembranous ossification (Figure 1, C and D). This phenotype was more severe than that of Osx-Cre pups (Supplemental Figure 1C), which have previously been reported to have mild hypomineralization of the calvarium (15). Other skeletal structures, including clavicles, ribs, and limb bones, were severely hypoplastic in Rnf146fl/fl Osx-Cre pups (Figure 1, E–G). Alizarin red staining of the calvarium from Rnf146fl/fl Osx-Cre pups revealed defective mineralization compared with Rnf146fl/fl control pups (Figure 1H). H&E staining of mandibular incisor from Rnf146fl/fl Osx-Cre pups showed hypoplasia of the periodontal alveolar bone and enamel and a loss of polarized ameloblasts with occasional pyknotic nuclei (Figure 1I). We further investigated expression levels of noggin and sonic hedgehog (SHH), which are essential for normal calvarial and teeth development (16), by RNA ISH and observed that expression of both noggin and SHH was similar in Rnf146fl/fl and Rnf146fl/fl Osx-Cre pups (Figure 1J), suggesting that perturbation of noggin and SHH expression does not account for the abnormal phenotype observed in the Rnf146fl/fl Osx-Cre pups. Finally, H&E staining of tibiae from Rnf146fl/fl Osx-Cre pups showed a 65% reduction of trabecular bone volume compared with Rnf146fl/fl control pups (Figure 1, K and L). In contrast, safranin O staining and ISH of type II and X collagen of tibiae from Rnf146fl/fl and Rnf146fl/fl Osx-Cre pups showed no difference between these groups (Figure 1, M–O), demonstrating that chondrocyte differentiation during endochondral ossification was normal in Rnf146fl/fl Osx-Cre pups. These data demonstrate that impairment of endochondral ossification in Rnf146fl/fl Osx-Cre pups is principally due to defective osteoblastogenesis. Thus, RNF146 is required for embryonic bone development through both intramembranous and endochondral ossification.
Figure 1. RNF146 deficiency in osteoblasts causes a CCD-like syndrome in mice.
(A) Whole-mount skeletons of Rnf146fl/fl and Rnf146fl/fl CMV-Cre E15.5 embryos stained by alizarin red and Alcian blue. (B and C) Whole-mount skeletons (B) or the calvarium (C) of Rnf146fl/fl and Rnf146fl/fl Osx-Cre newborn pups stained by alizarin red and Alcian blue. (D) μCT reconstruction of the calvarium of Rnf146fl/fl and Rnf146fl/fl Osx-Cre newborn pups. (E–G) Clavicles (E) or limb bones (F and G) of Rnf146fl/fl, Rnf146fl/+ Osx-Cre and Rnf146fl/fl Osx-Cre newborn pups stained by alizarin red and Alcian blue. (H) Alizarin red staining of the calvarium from Rnf146fl/fl and Rnf146fl/fl Osx-Cre newborn pups. Scale bar: 1 mm. The calcified tissue appears red. (I) H&E staining of mandibular incisor from Rnf146fl/fl and Rnf146fl/fl Osx-Cre newborn pups. Scale bars: 300 μm (top panels); 50 μm (bottom panels). Black, red, and yellow arrows indicate periodontal alveolar bone, enamel, and ameloblasts, respectively. (J) ISH of noggin (top panel) and Shh (bottom panel) in mandibular incisor from Rnf146fl/fl and Rnf146fl/fl Osx-Cre newborn pups. Scale bars: 100 μm. Black arrows indicate ameloblasts. (K) H&E staining of tibiae from Rnf146fl/fl and Rnf146fl/fl Osx-Cre newborn pups. Scale bar: 150 μm. Black arrows indicate trabecular bone. (L) Histomorphometric analysis of tibial trabecular bone volume per total volume (BV/TV) of Rnf146fl/fl and Rnf146fl/fl Osx-Cre newborn pups shown in K. n = 3. P values were determined by unpaired t test. Data are presented as mean ± SEM. *P < 0.05. (M) Safranin O staining of tibiae from Rnf146fl/fl and Rnf146fl/fl Osx-Cre newborn pups. Scale bar: 250 μm. (N and O) ISH of Col2a1 (N) and Col10a1 (O) in tibiae from Rnf146fl/fl and Rnf146fl/fl Osx-Cre newborn pups. Scale bars: 250 μm.
Rnf146fl/fl Osx-Cre mice cause osteopenia due to the osteoblast defect.
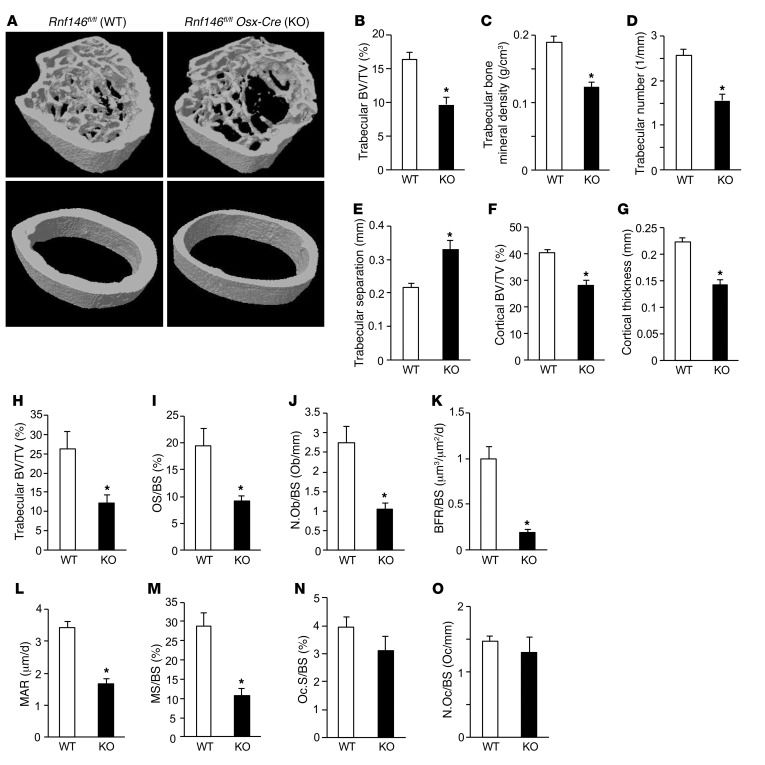
We next queried whether RNF146 was required for postnatal bone development and generated Rnf146fl/fl Osx-Cre adult mice using a doxycycline Tet-off Cre system (17). In contrast with pups in which endogenous RNF146 was deleted during embryonic development, excision of RNF146 within the osteoblast lineage after birth produced viable pups. 3D reconstruction of the femora using μCT showed a 40% reduction of trabecular bone volume and trabecular bone mineral density in Rnf146fl/fl Osx-Cre (KO) mice compared with Rnf146fl/fl (WT) mice (Figure 2, A–C). Rnf146fl/fl Osx-Cre mice demonstrated decreased trabecular numbers (Figure 2D) and increased trabecular separation (Figure 2E), though trabecular thickness was near normal (Supplemental Figure 2A). Moreover, loss of cortical bone volume and reduced cortical thickness were observed in Rnf146fl/fl Osx-Cre mice (Figure 2, A, F, and G). Major and minor bone diameter and periosteal and endosteal perimeter of Rnf146fl/fl Osx-Cre mice were comparable to those of Rnf146fl/fl controls (Supplemental Figure 2, B–E).
Figure 2. Osteoblast defect causes osteopenia in Rnf146fl/fl Osx-Cre mice.
(A) μCT reconstruction of the trabecular region below the distal femur growth plate (top panels) and the cortical region of midshaft femurs (bottom panels) of 12-week-old Rnf146fl/fl (WT) and Rnf146fl/fl Osx-Cre (KO) mice. (B–G) μCT-derived measurements of trabecular bone volume (BV/TV) (B), trabecular bone mineral density (C), trabecular number (D), trabecular separation (E), cortical bone volume (BV/TV) (F), and cortical thickness (G) of 12-week-old Rnf146fl/fl and Rnf146fl/fl Osx-Cre mice. n = 5–6. (H–J) Histomorphometric analysis of tibial trabecular bone volume per total volume (H), osteoid surface per bone surface (OS/BS) (I), and osteoblast number per bone surface (N.Ob/BS) (J) of 12-week-old Rnf146fl/fl and Rnf146fl/fl Osx-Cre mice. n = 6–7. (K–M) Dynamic histomorphometric analysis of tibial trabecular bone formation rate (BFR/BS) (K), mineral apposition rate (MAR) (L), and mineralizing surface (MS/BS) (M) of 12-week-old Rnf146fl/fl and Rnf146fl/fl Osx-Cre mice. n = 5–7. (N and O) Histomorphometric analysis of osteoclast surface per bone surface (Oc.S/BS) (N) and osteoclast number per bone surface (N.Oc/BS) (O) of 12-week-old Rnf146fl/fl and Rnf146fl/fl Osx-Cre mice. n = 6–7. P values were determined by unpaired t test. Data are presented as mean ± SEM. *P < 0.05.
To determine the cause of the severe osteopenia observed in Rnf146fl/fl Osx-Cre mice, we analyzed tibiae stained with Goldner’s trichrome. Histomorphometric analysis of tibiae from Rnf146fl/fl Osx-Cre mice revealed a severe reduction in trabecular bone volume compared with Rnf146fl/fl mice (Figure 2H), consistent with the results of μCT analysis shown in Figure 2, A and B. Additionally, osteoid surface per bone surface and osteoblast number per bone surface were decreased in Rnf146fl/fl Osx-Cre mice (Figure 2, I and J). Moreover, dynamic bone histomorphometric analysis after calcein green labeling confirmed a substantial decrease in the bone formation rate (Figure 2K), mineral apposition rate (Figure 2L), and mineralizing surface (Figure 2M) in Rnf146fl/fl Osx-Cre mice compared with Rnf146fl/fl mice, indicating an intrinsic defective osteoblast-mediated bone formation. In contrast, tartrate-resistant acid phosphatase (TRAP) staining of tibiae showed that osteoclast surface per bone surface and osteoclast number per bone surface were similar in Rnf146fl/fl and Rnf146fl/fl Osx-Cre mice (Figure 2, N and O).
These results demonstrate that Rnf146fl/fl Osx-Cre mice are osteopenic due to defective osteoblast-mediated bone formation, but not enhanced osteoclast-mediated bone resorption.
RNF146 is required for osteoblast differentiation.
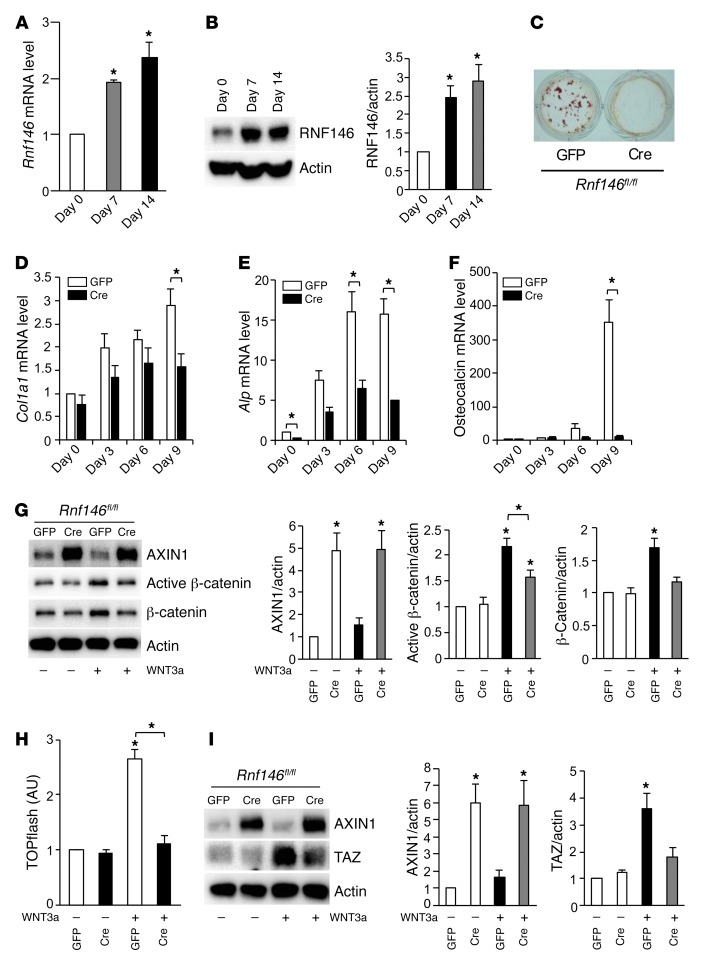
RNF146 is required for both osteoblast-mediated embryonic and postnatal bone development. We queried whether RNF146 expression was normally modulated during osteoblast differentiation and observed that RNF146 transcripts and protein levels increased in primary murine osteoblasts cultured in osteogenic medium (Figure 3, A and B). To investigate the molecular mechanism by which RNF146 deficiency in osteoblasts causes a CCD-like syndrome and osteopenia, we isolated murine calvarial osteoblasts from Rnf146fl/fl mice and infected cells with GFP- or Cre-expressing adenovirus to excise the Rnf146 gene (18) (Supplemental Figure 3, A and B). We observed defective mineralization in RNF146-null osteoblasts and reduced mRNA expression of collagen type I α 1 chain (Col1a1), alkaline phosphatase (Alp), and osteocalcin (Figure 3, C–F). We next examined the protein levels of AXIN, a known substrate of RNF146, in RNF146-null osteoblasts and observed that AXIN1 protein was markedly increased and that this led to impaired activation of β-catenin (Figure 3, G and H). Our observation that Rnf146fl/fl Osx-Cre mice phenocopy features of CCD patients suggested a potential genetic link between RNF146 and RUNX2 (3–7). However, we observed no effect on Runx2 expression in murine osteoblasts following treatment with WNT3a (Supplemental Figure 3C). Moreover, the expression of Runx2 transcripts was similar in WT and RNF146-null calvarial osteoblasts cultured in osteogenic medium (Supplemental Figure 3D). We therefore examined the expression level of TAZ, a RUNX2-binding partner that is an essential cotranscription factor required for mediating the osteoblastogenic transcriptional program. The induction of TAZ protein levels, induced by WNT3a, were noted to be severely reduced in RNF146-null osteoblasts compared with WT controls (Figure 3I), which may explain in part the CCD-like phenotype associated with RUNX2 loss of function.
Figure 3. RNF146 is required for osteoblast differentiation.
(A) qPCR analysis of Rnf146 mRNA expression in primary murine osteoblasts cultured in osteogenic medium for 7–14 days. (B) Whole cell lysates from cells in A were probed with the indicated antibodies for Western blot analysis. (C) Rnf146fl/fl mouse–derived calvarial osteoblasts infected with GFP- or Cre-expressing adenovirus to excise endogenous RNF146 protein were cultured in osteogenic medium and stained with alizarin red S solution. (D–F) qPCR analysis of Col1a1 (D), Alp (E), and osteocalcin (F) mRNA expression in cells in C cultured in osteogenic medium for 3–9 days. (G) Rnf146fl/fl mouse–derived calvarial osteoblasts infected with GFP- or Cre-expressing adenovirus were cultured in serum-free medium in the presence or absence of WNT3a (40 ng/ml). Whole cell lysates were probed with the indicated antibodies for Western blot analysis. Right panels: quantification of Western blot analysis. (H) Luciferase activity from a TOPflash reporter assay in cells in G cultured in serum-free medium in the presence or absence of WNT3a (40 ng/ml). (I) Western blot analysis of cells in G cultured in serum-free medium in the presence or absence of WNT3a (40 ng/ml). n = 3. P values were determined by ANOVA with Tukey-Kramer’s post-hoc test (A, B and G–I) or unpaired t test (D–F). Data are presented as mean ± SEM. *P < 0.05.
RNF146 regulates osteoblast differentiation through the FGF18-TAZ axis.
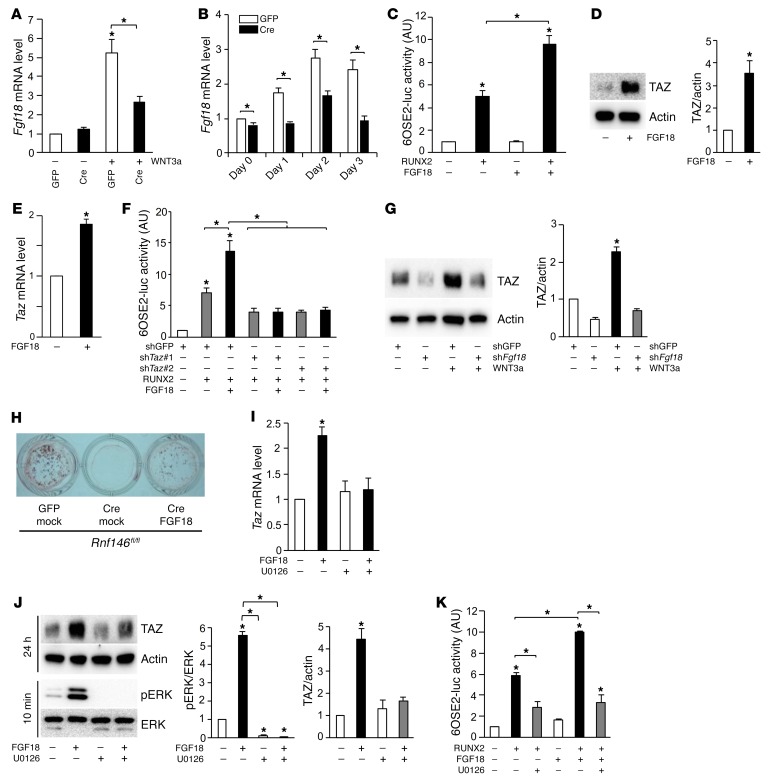
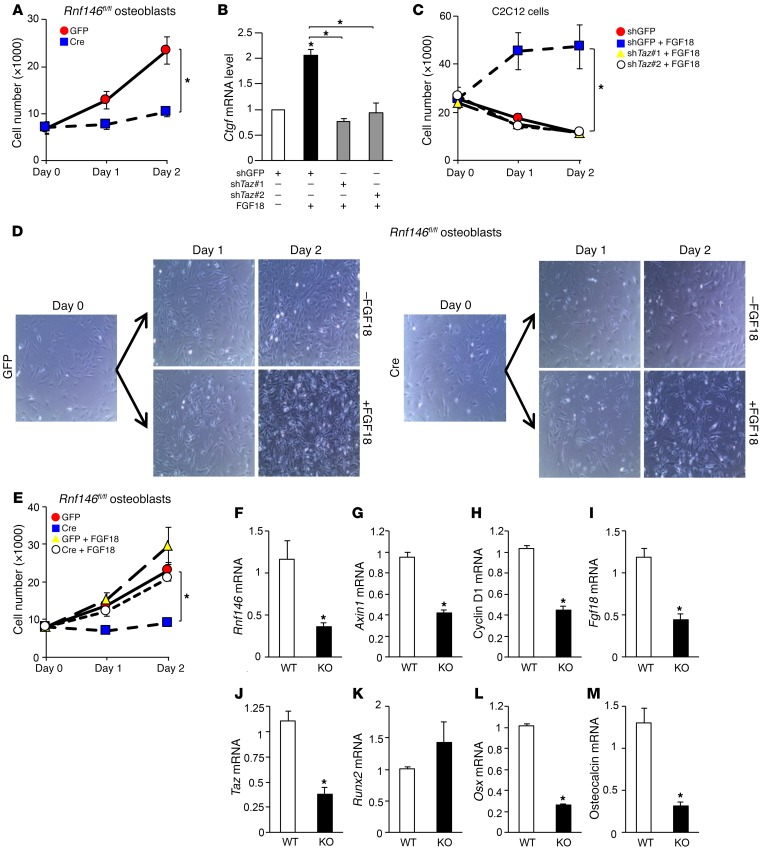
We further investigated the molecular mechanism by which RNF146 controls osteoblast differentiation through regulation of TAZ expression. Previous studies showed that Fgf18 is a β-catenin target (19) and that Fgf18–/– mice phenocopy CCD patients (20), suggesting that Fgf18 may be dysregulated in RNF146-null osteoblasts. We observed that both WNT3a-mediated induction of Fgf18 transcripts (Figure 4A) and expression of Fgf18 mRNAs during osteoblastogenesis (Figure 4B) were impaired in RNF146-null osteoblasts, suggesting that defective mineralization observed in RNF146-null osteoblasts may be due to impaired WNT3a-mediated Fgf18 expression. We next examined whether FGF18 regulates the expression of osteocalcin, a target gene of the RUNX2-TAZ osteoblastogenic master transcription complex (9). We observed that FGF18 enhances the RUNX2-mediated osteocalcin promoter activity (Figure 4C). Since we found that WNT3a was not able to induce Runx2 transcripts (Supplemental Figure 3C), we hypothesized that FGF18 may regulate endogenous TAZ expression. Consistent with this hypothesis, we observed that FGF18 enhanced expression of TAZ protein and transcripts in murine osteoblasts and myoblast C2C12 cells (Figure 4, D and E, and Supplemental Figure 4, A and B). In contrast, RUNX2 expression was unchanged following FGF18 stimulation of murine osteoblasts (Supplemental Figure 4C). To confirm that FGF18 requires the presence of TAZ to enhance osteocalcin expression and osteoblast differentiation, we analyzed osteocalcin promoter activity in TAZ-depleted C2C12 cells and observed that the enhancement of RUNX2 activity by FGF18 was suppressed by Taz deletion (Figure 4F). Consistent with these results, depletion of FGF18 blocked the induction of TAZ by WNT3a (Figure 4G). Conversely, ectopic expression of FGF18 rescued the mineralization defect observed in RNF146-null osteoblasts (Figure 4H and Supplemental Figure 4D). Finally, FGF18-induced TAZ expression and RUNX2-TAZ transcriptional activity were abolished by the MEK inhibitor U0126 in murine osteoblasts and C2C12 cells (Figure 4, I–K, and Supplemental Figure 4, E and F). These data demonstrate that FGF18 controls osteoblast differentiation through transcriptional regulation of TAZ and that the mineralization defect observed in RNF146-null osteoblasts is due in part to impaired signaling through the FGF18/MAPK/TAZ pathway.
Figure 4. RNF146 regulates osteoblast differentiation through the FGF18-TAZ axis.
(A and B) qPCR analysis of Fgf18 mRNA expression in cells in Figure 3I (A) or C (B), cultured in osteogenic medium for 1–3 days. (C) Luciferase activity from an osteocalcin reporter assay in C2C12 cells transfected with or without RUNX2, and cultured in serum-free medium in the presence or absence of FGF18 (50 ng/ml). (D) Western blot analysis in primary murine osteoblasts cultured in serum-free medium in the presence or absence of FGF18 (50 ng/ml). (E) qPCR analysis of Taz mRNA expression in cells in D. (F) Luciferase activity from an osteocalcin reporter assay in C2C12 cells infected with shGFP or shTaz, transfected with or without RUNX2 and cultured in serum-free medium in the presence or absence of FGF18 (50 ng/ml). (G) Western blot analysis in primary murine osteoblasts infected with shGFP or shFgf18 and cultured in serum-free medium in the presence or absence of WNT3a (40 ng/ml). (H) Cells in Figure 3C, infected with an empty vector control (mock) or an FGF18-expressing retroviral vector were cultured in osteogenic medium and stained with alizarin red S solution. (I) qPCR analysis of Taz mRNA expression in primary murine osteoblasts cultured in serum-free medium in the presence or absence of FGF18 (50 ng/ml) and U0126 (10 μM). (J) Western blot analysis in cells in I cultured for 10 minutes or 24 hours. (K) Luciferase activity from an osteocalcin reporter assay in C2C12 cells transfected with or without RUNX2 and cultured in serum-free medium in the presence or absence of FGF18 (50 ng/ml) and U0126 (10 μM). n = 3. P values were determined by ANOVA with Tukey–Kramer’s post-hoc test (A, C, F, G, and I–K) or unpaired t test (B, D, and E). Data are presented as mean ± SEM. *P < 0.05.
RNF146 is required for osteoblast proliferation.
In addition to defective mineralization, we noted that osteoblast proliferation was reduced in RNF146-null osteoblasts (Figure 5A and Supplemental Figure 5A). TAZ in complex with transcriptional enhancer associate domain (TEAD) is a regulator of cell proliferation in part through the induction of its target gene connective tissue growth factor (Ctgf) (21–24). Our observation that RNF146 controls TAZ expression through regulation of FGF18 prompted us to investigate whether RNF146 plays a role in osteoblast proliferation through the FGF18/TAZ pathway. We found that FGF18 induced Ctgf expression and that this effect was abolished in TAZ-depleted C2C12 cells (Figure 5B and Supplemental Figure 5B). Moreover, recombinant FGF18 enhanced proliferation of WT, but not TAZ-depleted, C2C12 cells cultured in serum-free medium (Figure 5C and Supplemental Figure 5C), demonstrating that FGF18-mediated Ctgf induction and subsequent cell proliferation are contingent on TAZ expression. Accordingly, the defective proliferation observed in RNF146-null osteoblasts was rescued by treatment with FGF18 (Figure 5, D and E). These results demonstrate that RNF146 is required for both osteoblast differentiation and proliferation through the modulation of the WNT3a-FGF18-TAZ axis.
Figure 5. RNF146 is required for osteoblast proliferation.
(A) Growth curves of Rnf146fl/fl mouse–derived calvarial osteoblasts infected with GFP- or Cre-expressing adenovirus and cultured in growth medium for 2 days. (B) qPCR analysis of Ctgf mRNA expression in C2C12 cells infected with shGFP or shTaz and cultured in serum-free medium in the presence or absence of FGF18 (50 ng/ml). (C) Growth curves of C2C12 cells infected with shGFP or shTaz and cultured in serum-free medium in the presence or absence of FGF18 (50 ng/ml) for 2 days. (D) Bright-field images of Rnf146fl/fl mouse–derived calvarial osteoblasts infected with GFP- or Cre-expressing adenovirus and cultured in growth medium in the presence or absence of FGF18 (50 ng/ml) for 2 days. Original magnification: ×200. (E) Growth curves of cells in D cultured for 2 days. (F–M) qPCR analysis of Rnf146 (F), Axin1 (G), cyclin D1 (H), Fgf18 (I), Taz (J), Runx2 (K), Osx (L), and osteocalcin (M) mRNA expression in the calvarium derived from Rnf146fl/fl and Rnf146fl/fl Osx-Cre newborn pups. n = 3. P values were determined by ANOVA with Tukey-Kramer’s post-hoc test (B, C, and E) or unpaired t test (A and F–M). Data are presented as mean ± SEM. *P < 0.05.
We finally confirmed the molecular mechanism by which loss of RNF146 in osteoblasts results in impairment of the Wnt/β-catenin pathway and expression of its target gene Fgf18, leading to suppression of Taz and osteocalcin expression in vivo. We extracted mRNA from the calvarium of Rnf146fl/fl Osx-Cre pups and observed that expression levels of Rnf146 and the β-catenin target genes Axin1, cyclin D1, and Fgf18 mRNAs were reduced in Rnf146fl/fl Osx-Cre pups compared with Rnf146fl/fl pups (Figure 5, F–I). Moreover, expression of Taz, Osx, and osteocalcin, but not Runx2, mRNAs was decreased in Rnf146fl/fl Osx-Cre pups (Figure 5, J–M), consistent with the in vitro data of primary murine osteoblasts shown in Figures 3 and 4. These in vivo and in vitro data conclusively demonstrate that RNF146 is required for osteoblast differentiation and proliferation through regulation of the FGF18/MAPK/TAZ pathway, leading to both embryonic and postnatal bone formation.
RNF146 represses adipocyte development and fat stores.
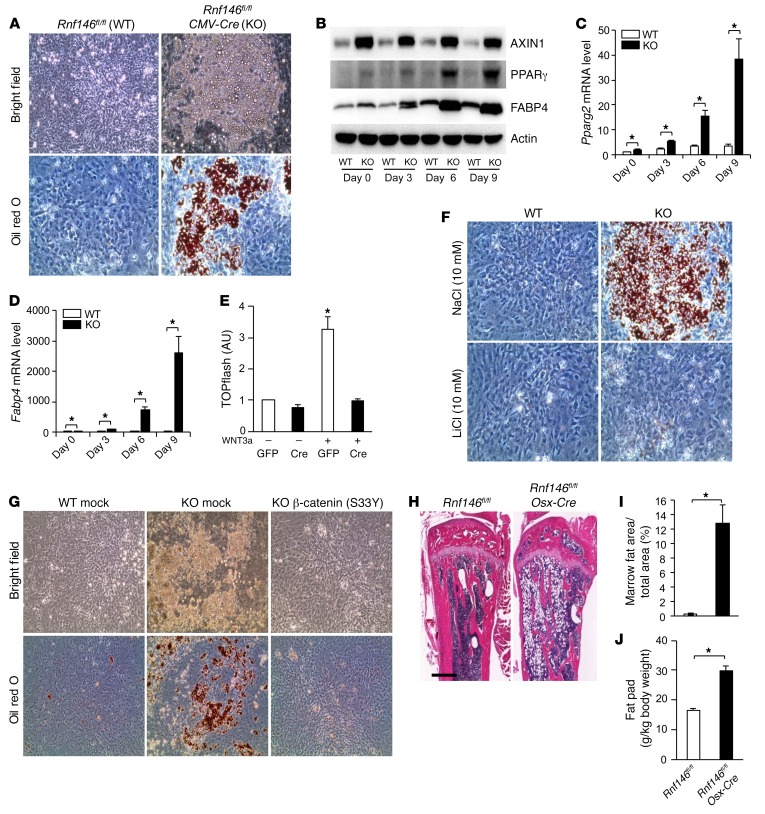
During the course of these studies, we noted that RNF146-null osteoblast progenitor cells cultured in osteogenic medium differentiated into adipocytes instead of mature osteoblasts (Supplemental Figure 6A). The expression levels of both Pparg2 and its target gene fatty acid binding protein 4 (Fabp4), which are required for adipogenesis, were elevated in RNF146-null osteoblast progenitor cells cultured in osteogenic medium (Supplemental Figure 6, B and C). In support of a role of RNF146 as an inhibitor of adipogenesis, we observed that adipocyte differentiation was strongly enhanced in RNF146-deficient mouse embryonic fibroblasts (MEFs) cultured in troglitazone-containing growth medium (Figure 6A and Supplemental Figure 6, D and E) and that this was associated with elevated expression of PPARγ and FABP4 proteins and transcripts (Figure 6, B–D, and Supplemental Figure 6F). As the Wnt/β-catenin pathway inhibits adipogenesis through repression of PPARγ expression (25), we determined whether the adipocyte differentiation observed in RNF146-deficient MEFs was dependent on reduced β-catenin activity. We observed that WNT3a-mediated β-catenin activity was suppressed in RNF146-deficient MEFs as a consequence of AXIN1 stabilization (Figure 6, B and E). Moreover, inhibition of glycogen synthase kinase 3β (GSK-3β) by lithium chloride (LiCl) reversed the enhanced adipogenic phenotype observed in RNF146-deficient MEFs (Figure 6F and Supplemental Figure 6, G–I). Similarly, ectopic expression of a constitutively active β-catenin (S33Y) mutant blocked the enhanced adipogenesis observed in RNF146-deficient MEFs (Figure 6G and Supplemental Figure 6J). These results support a model whereby RNF146 suppresses adipogenesis while enhancing osteoblast differentiation through regulation of the Wnt/β-catenin pathway. Consistent with these observations, tibial bone marrow fat stores and fat pad weight were elevated 40-fold and 2-fold in Rnf146fl/fl Osx-Cre mice compared with controls, respectively (Figure 6, H–J). These results indicate that RNF146 is a mesenchymal fate–determining factor that accelerates osteoblastogenesis while suppressing adipogenesis through regulation of the Wnt/β-catenin pathway.
Figure 6. RNF146 represses adipocyte development and fat stores.
(A) MEFs isolated from Rnf146fl/fl (WT) and Rnf146fl/fl CMV-Cre (KO) embryos were cultured in adipogenic medium and stained with oil red O. Top panels, bright field images; bottom panels, oil red O staining. Original magnification: ×200. (B) Whole cell lysates from cells in A cultured in adipogenic medium for 3–9 days were probed with the indicated antibodies for Western blot analysis. (C and D) qPCR analysis of Pparg2 (C) or Fabp4 (D) mRNA expression in cells in A cultured in adipogenic medium for 3–9 days. n = 3. (E) Luciferase activity from a TOPflash reporter assay in cells in A cultured in serum-free medium in the presence or absence of WNT3a (40 ng/ml). n = 3. (F) Oil red O staining of MEFs in A cultured in adipogenic medium in the presence or absence of NaCl (10 mM, top panels) and LiCl (10 mM, bottom panels). Original magnification: ×200. (G) MEFs in A infected with an empty vector control (mock) or a β-catenin–expressing (S33Y) retroviral vector were cultured in adipogenic medium and stained with oil red O. Top panels, bright field images; bottom panel, oil red O staining. Original magnification: ×200. (H) H&E staining of tibiae from 12-week-old Rnf146fl/fl and Rnf146fl/fl Osx-Cre mice. Scale bar: 500 μm. (I) Quantification of trabecular bone marrow fat area in tibiae in H. n = 5–6. (J) Fat pad mass (fat pad weight over body weight) from 6-month-old Rnf146fl/fl and Rnf146fl/fl Osx-Cre mice. n = 3. P values were determined by ANOVA with Tukey-Kramer’s post-hoc test (E) or unpaired t test (C, D, I, and J). Data are presented as mean ± SEM. *P < 0.05.
Rnf146fl/fl Osx-Cre mice have defects in glucose metabolism.
Previous studies showed that osteocalcin-deficient mice are glucose intolerant due to impaired insulin sensitivity and secretion (26, 27). Osteocalcin, which is secreted by osteoblasts through regulation of the RUNX2-TAZ transcription complex, is posttranslationally carboxylated in a vitamin K–dependent manner, while uncarboxylated osteocalcin enhances insulin production and sensitivity in vivo (27, 28). Our observation that loss of RNF146 suppressed osteoblastogenesis and was associated with the reduced promoter activity of osteocalcin suggested a potential link between RNF146 and glucose homeostasis. Consistent with the reduction of osteocalcin mRNA in RNF146-null osteoblasts in vitro shown in Figure 3F, we observed that levels of undercarboxylated osteocalcin in serum were reduced in Rnf146fl/fl Osx-Cre mice compared with those in Rnf146fl/fl mice (Figure 7A), leading to transcriptional reduction of adiponectin, an osteocalcin target gene (Figure 7B) (26). Moreover, we observed increased triglyceride and glucose levels in Rnf146fl/fl Osx-Cre mice compared with Rnf146fl/fl mice (Figure 7, C and D). To further investigate glucose regulation in Rnf146fl/fl Osx-Cre mice, we performed glucose tolerance tests and insulin secretion tests following intraperitoneal injection of glucose and observed impaired glucose tolerance (Figure 7E) as a consequence of defective glucose-stimulated insulin secretion (Figure 7F), indicating that loss of RNF146 in osteoblasts leads to abnormal glucose tolerance in vivo. Histological and immunochemical analyses showed a decrease in number of islets, islet size, and pancreatic insulin production in Rnf146fl/fl Osx-Cre mice (Figure 7G and Supplemental Figure 7, A and B). Finally, to determine whether the defective islet development and insulin production were due to a failure to produce physiologic levels of osteocalcin, we treated mice with recombinant uncarboxylated osteocalcin and improved the abnormal glucose tolerance observed in Rnf146fl/fl Osx-Cre mice (Figure 7H). These data demonstrate that abnormal glucose tolerance observed in the Rnf146fl/fl Osx-Cre mice is due in part to impaired production of osteocalcin.
Figure 7. Rnf146fl/fl Osx-Cre mice have defects in glucose metabolism.
(A) Undercarboxylated osteocalcin (ucOC) levels in serum from 6-month-old Rnf146fl/fl and Rnf146fl/fl Osx-Cre mice were measured by ELISA. n = 6. (B) qPCR analysis of adiponectin mRNA expression in fat in Figure 6J. n = 3. (C) Serum triglyceride levels after an overnight fast. n = 7–8. (D) Blood glucose levels after random feeding. n = 7–8. (E and F) Glucose tolerance test (E) and glucose-stimulated insulin secretion test (F) in 6-month-old Rnf146fl/fl and Rnf146fl/fl Osx-Cre mice. n = 7–8. (G) H&E staining (top panels) and insulin immunostaining (bottom panels) of pancreas from 6-month-old Rnf146fl/fl and Rnf146fl/fl Osx-Cre mice. Scale bars: 500 μm. (H) Glucose tolerance test in 6-month-old Rnf146fl/fl and Rnf146fl/fl Osx-Cre mice injected with glucose and recombinant uncarboxylated osteocalcin (1 ng/g) or vehicle. n = 7. P values were determined by unpaired t test. Data are presented as mean ± SEM. *P < 0.05.
Discussion
Mice lacking the E3 ubiquitin ligase RNF146 develop a syndrome that phenocopies features of CCD.
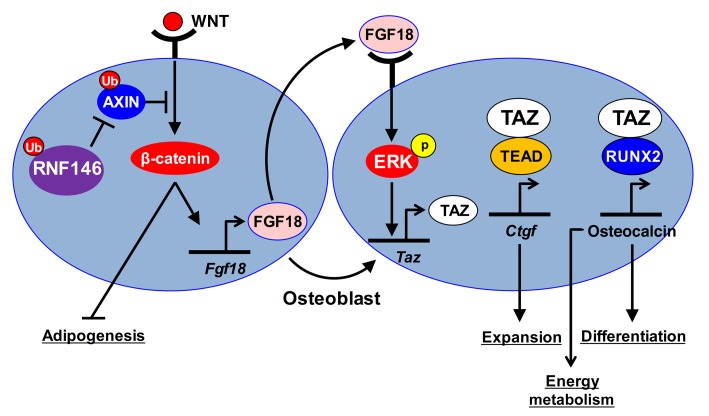
Our study provides evidence suggesting the existence of a link between the E3 ubiquitin ligase RNF146 and the development of a CCD-like syndrome in mice. We show that Rnf146fl/fl Osx-Cre mice have defective intramembranous and endochondral ossification during embryonic bone development. Loss of RNF146 in osteoblasts results in stabilization of AXIN1 and attenuation of β-catenin, leading to impaired FGF18 expression (Figure 8). FGF18 in turn induces TAZ expression through activation of the MAPK pathway (Figure 8).
Figure 8. Schematic model showing that RNF146 controls bone, glucose, and lipid metabolism through regulation of the Wnt/β-catenin and Hippo pathways.
FGF18, a member of the FGF family, plays key roles in morphogenesis, angiogenesis, and development of cellular identity (29–32). Like Runx2+/– mice, Fgf18–/– mice suffer from defective calvarium due to the reduction of osteoblast proliferation (20, 33), a phenotype similar to what we have observed in Rnf146fl/fl Osx-Cre mice. These data support the idea that the skeletal abnormality observed in Rnf146fl/fl Osx-Cre mice is due in part to the attenuation of FGF18 expression resulting from the impaired activation of β-catenin (19).
We have recently shown that the Hippo pathway transcriptional coactivator TAZ is stabilized and activated by the nonreceptor tyrosine kinase ABL following disruption of the interaction between TAZ and the E3 ubiquitin ligase F-box/WD repeat-containing protein 1A (β-TrCP) during osteoblast differentiation (34). TAZ stabilization and activation by ABL lead to activation of the transcription factors TEAD and RUNX2, required for osteoblast proliferation, differentiation, and mineralization (34). Our present study provides insights into an additional mode of TAZ regulation controlled by the RNF146–Wnt/β-catenin–FGF18 axis during osteoblastogenesis. Loss of RNF146 reveals the importance of the FGF18/TAZ pathway in a syndrome that phenocopies features of the human hereditary disease CCD.
RNF146 is the critical E3 ubiquitin ligase modulating bone dynamics.
Bone undergoes continuous synthesis and remodeling through osteoblast-mediated bone formation and osteoclast-mediated bone resorption. We propose that RNF146 is a switch required for homeostatic bone remodeling. We have recently shown that RNF146 similarly integrates both SRC and β-catenin signaling pathways downstream of RANK ligand within the osteoclast lineage (35). RANKL-mediated suppression of RNF146 synchronizes the stabilization of 3BP2, an adapter protein required for the activation of SRC, and the stabilization of AXIN1 to shut off β-catenin activity. Excision of Rnf146 within the macrophage lineage in mice (Rnf146fl/fl LysM-Cre) sensitizes osteoclast progenitors to RANKL; as a result, Rnf146fl/fl LysM-Cre mice develop osteopenia due to highly activated osteoclasts. Our observation that RNF146 is required for osteoblast development while antagonizing osteoclast differentiation highlights the importance of this E3 ubiquitin ligase in regulating bone dynamics. Further studies will be required to elucidate a signaling network that controls RNF146 induction during osteoblastogenesis.
RNF146 is a mesenchymal fate–determining factor.
Our results indicate that loss of RNF146 in osteoblasts and MEFs enhances adipogenesis. During embryonic bone development, the active RUNX2-TAZ complex is required for osteoblastogenesis, whereas TAZ suppresses PPARγ transcriptional activity and adipogenesis (9). Similarly, activation of the Wnt/β-catenin pathway is required for osteoblastogenesis, but is antagonistic to adipogenesis (13, 25). In the present study, we have unveiled the molecular mechanism by which RNF146 controls osteoblast and adipocyte lineage commitment through regulation of the Wnt/β-catenin and Hippo pathways required for development of cellular identity.
RNF146 is a regulator for energy metabolism.
Bone has emerged as a regulator of energy metabolism (36). Osteocalcin, which is secreted by osteoblasts, controls glucose and lipid metabolism (26). Osteocalcin-deficient mice exhibit abnormal glucose tolerance as a consequence of impaired insulin production and sensitivity due to decreased β cell proliferation and adiponectin secretion from fat tissue, respectively (26).
We show in the present study that the FGF18-TAZ-osteocalcin axis is controlled by RNF146, which is required for normal glucose homeostasis. Treatment of the uncarboxylated and active form of osteocalcin improved the glucose intolerance observed in Rnf146fl/fl Osx-Cre mice. Importantly, osteocalcin-deficient mice display increased bone mass (37) while Rnf146fl/fl Osx-Cre mice show severe osteopenia and phenocopy CCD patients, demonstrating that our study using Rnf146fl/fl Osx-Cre mice provides a genetic link between hypoplastic bone formation and abnormal glucose and lipid metabolism in vivo.
RNF146 coordinates bone, cellular identity, and glucose and lipid metabolism. Our study suggests that CCD patients may be at risk for glucose intolerance and highlights a genetic link among RNF146, osteopenia, and metabolic syndrome.
Methods
Mice.
The generation of Rnf146 conditional knockout mice has been described previously (35). Rnf146fl/fl mice were crossed with CMV-Cre (14) or Osx-Cre mice (17). Since Rnf146fl/fl Osx-Cre pups died after birth, we generated Rnf146fl/fl Osx-Cre adult mice using a doxycycline Tet-off Cre system (17). We treated pregnant breeder mice with doxycycline during gestation to prevent Cre expression. After birth, these offspring were fostered to another mother without doxycycline.
Genotyping of Rnf146fl/fl Cre mice.
Genotyping of Rnf146fl/fl Cre mice has been described previously (35). Floxed alleles and Cre were genotyped by PCR using the following primers: Rnf146 (forward primer, 5′-CCAGTCAGACCGATCAGGAACTGAC-3′; reverse primer, 5′-TACTGAACAGCTCTCAACTATCAACACC-3′), and Cre (forward primer, 5′-GCATTACCGGTCGATGCAACGAGTGATGAG-3′; reverse primer, 5′-GAGTGAACGAACCTGGTCGAAATCAGTGCG-3′).
Cell cultures.
HEK293T cells (ATCC), C2C12 cells (ATCC), and MEFs were cultured in DMEM (Wisent) supplemented with 10% FBS (Wisent). All cultures were maintained in a 5% CO2 environment at 37°C.
Osteoblast cultures.
Neonatal calvaria-derived osteoblasts were harvested and cultured in growth medium as described previously (38). Osteoblast differentiation was induced by culturing cells in osteogenic medium (α-MEM containing 10% FBS, 100 μg/ml ascorbic acid, and 10 mM β-glycerophosphate) for 21 days. Alizarin red staining of mineralization was performed by cell fixation in 4% formaldehyde for 30 minutes followed by staining with 0.1% alizarin red S solution (pH 4.8) for 20 minutes.
Adipocyte differentiation.
Adipocyte differentiation was induced by culturing cells in adipogenic medium (DMEM containing 10% FBS and 10 μM troglitazone). Adipocytes were evaluated by oil red O staining accomplished by cell fixation in 4% formaldehyde for 15 minutes followed by staining with filtered 0.6% oil red O solution in 60% isopropanol. Oil red O–positive cells were determined by cytoplasmic fat globule cells.
μCT.
Pups’ calvarium and femurs from 12-week-old male mice were scanned using μCT (SkyScan 1076 or 1174, Bruker) at a resolution of 18.2 μm or 9.1 μm, respectively. Morphometric analysis was performed on a volume of interest (VOI) of trabecular bone 1 mm in length beginning 1 mm beneath the first instance of the formation of the mineralized cartilage bridge in the developing metaphysis and on a VOI of cortical bone 0.9 mm in length in the midshaft. 3D images were reconstructed from 0.5-mm regions within the trabecular and cortical VOIs identified above. Trabecular and cortical morphometric parameters were determined according to standard protocols (39).
Skeletal analysis.
For skeletal alizarin red and Alcian blue staining, newborn pups were fixed in 95% ethanol and transferred to acetone followed by staining, as described previously (7).
Histomorphometry.
For dynamic histomorphometric analysis, calcein green was injected on days 0 and 7, and mice were sacrificed 3 days after the second calcein injection. In vivo osteoblast parameters were generated from undecalcified tibial sections embedded in Spurr’s resin and stained with Goldner’s trichrome. In vivo osteoclast parameters were generated from decalcified tibial sections embedded in paraffin and stained for TRAP activity. Images were analyzed using BIOQUANT software (40).
Histology.
Pancreatic tissues and pups’ tibiae were fixed in 10% neutral formalin, embedded in paraffin, sectioned, and stained with H&E or safranin O. IHC was performed using guinea pig anti-insulin antibody (Invitrogen) and the VECTASTAIN Elite ABC HRP Kit (Vector).
ISH.
ISH of calvarium and tibiae was performed using the RNAscope 2.5 High Definition (HD) — BROWN (Advanced Cell Diagnostics) according to the manufacturer’s protocol.
Metabolic studies.
For glucose tolerance test, glucose (2 g/kg body weight) was injected intraperitoneally after an overnight fast, and blood glucose was monitored using the Accu-Chek glucometer (Roche). For glucose-stimulated insulin secretion test, glucose (2 g/kg body weight) was injected intraperitoneally after an overnight fast, and serum insulin levels were measured with the Ultra Sensitive Mouse Insulin ELISA Kit (Crystal Chem).
Construction and expression of β-catenin (S33Y) and FGF18 retroviral vectors.
pMXs–β-catenin (S33Y) vector was a gift from Shinya Yamanaka (Department of Life Science Frontiers, Center for iPS Cell Research and Application, Kyoto University, Kyoto, Japan) (41). FGF18 cDNA was purchased from Origene Technologies, and pMXs-FGF18 vector was constructed as described previously (40). HEK293T cells were cotransfected with an empty vector control (mock), pMXs–β-catenin (S33Y), or pMXs-FGF18 with pMCV-Ecopac (EcoPak) using a CalPhos Mammalian Transfection Kit (Clontech). Osteoblasts were infected and then cultured in osteogenic medium as described previously (40).
Lentiviral transduction.
pLKO.1 lentiviral vectors expressing shRNAs targeting Taz (shTaz), Fgf18 (shFgf18), or nonspecific GFP sequence (shGFP) were cotransfected into HEK293T cells with p-PAX2 and p-VSVG (Addgene) using X-tremeGENE 9 transfection reagent (Roche). The virus was collected 48 hours after transfection, and cells were infected as described previously (40).
Reagents and antibodies.
Unless stated otherwise, all chemicals were purchased from Sigma-Aldrich. Antibodies were obtained as follows: anti-AXIN1 (catalog 2087), anti–non–p–β-catenin (active; catalog 8814), anti–β-catenin (catalog 9562), anti–p-ERK (catalog 9101), anti-ERK (catalog 9102), anti-PPARγ (catalog 2443), anti-FABP4 (catalog 3544) (Cell Signaling Technology), anti-RNF146 (catalog SAB1408054), anti-FGF18 (catalog SAB1406731), and anti-actin (catalog sc-47778) (Santa Cruz Biotechnology Inc.). Recombinant osteocalcin was from the laboratory of Gerard Karsenty (26). Halt Protease and Phosphatase Inhibitor Cocktail was from Thermo Fisher Scientific. Undercarboxylated osteocalcin levels in serum were measured with the Mouse Undercarboxylated Osteocalcin ELISA Kit (MyBioSource.com).
Promoter assays.
To assess the modulation of Wnt/β-catenin–driven gene expression by WNT3a, we used TOPflash (TCF Reporter Plasmid, TOPflash-luciferase reporter system). To assess the modulation of RUNX2-TAZ complex-driven gene expression by FGF18, we used the osteocalcin promoter–luciferase reporter construct (p6OSE2-luc) that contains 6 tandem repeats of the OSE2 element of the osteocalcin promoter (42). C2C12 cells, primary murine osteoblasts, and MEFs were transiently cotransfected with p6OSE2-luc or TOPflash reporter and pRL Renilla plasmids (Promega) using Lipofectamine 2000 (Invitrogen) or FuGENE 6 (Promega), respectively. Cells were lysed and assayed for firefly and renilla luciferase activity using the Dual-Luciferase Reporter System (Promega). The data were expressed as the ratio of firefly to renilla activity.
RNA extraction and quantitative real-time PCR analysis.
Total cellular RNA was extracted using an RNeasy Plus Mini Kit (QIAGEN). An ImProm-II Reverse Transcription System (Promega) was used for reverse transcription, and quantitative real-time PCR (qPCR) was performed on a StepOne Plus Real-Time PCR System (Applied Biosystems) using TaqMan Gene Expression assays (Applied Biosystems) for Gapdh (Mn99999915_g1), Rnf146 (Mn00509629_m1), and Runx2 (Mn00501584_m1). The sequences of primers were as follows: mouse Rpl19 (forward primer, 5′-CTGAAGGTCAAAGGGAATGTG-3′; reverse primer, 5′-GGACAGAGTCTTGATGATCTC-3′), mouse Actb (forward primer, 5′-AAGCAGGAGTATGACGAGTCCG-3′; reverse primer, 5′-CGGAACTAAGTCATAGTCCGCG-3′), mouse Alp (forward primer, 5′-GCTGATCATTCCCACGTTTTC-3′; reverse primer, 5′-CTGGGCCTGGTAGTTGTTGT-3′), mouse osteocalcin (forward primer, 5′-CTGACAAAGCCTTCATGTCCAA-3′; reverse primer, 5′-GCGCCGGAGTCTGTTCACTA-3′), mouse Runx2 (forward primer, 5′-GCTATTAAAGTGACAGTGGACG-3′; reverse primer, 5′-CACGTCAGTGATGGCAGGTAGC-3′), mouse Col1a1 (forward primer, 5′-GGAAGAGCGGAGAGTACTGG-3′; reverse primer, 5′-TTGCAGTAGACCTTGATGGC-3′), mouse Fgf18 (forward primer, 5′-ACTGCTGTGCTTCCAGGTTC-3′; reverse primer, 5′-CCCAGGACTTGAATGTGCTT-3′), mouse Ctgf (forward primer, 5′-ACTGGAAGACACATTTGGCC-3′; reverse primer, 5′-GTCTTCACACTGGTGCAGCC-3′), mouse Pparg2 (forward primer, 5′-GCCAGTTTCGATCCGTAGAAGCC-3′; reverse primer, 5′-CCTGGGCGGTCTCCACTGAG-3′), mouse Fabp4 (forward primer, 5′-GTGGGAACCTGGAAGCTTGTC-3′; reverse primer, 5′-CTTCACCTTCCTGTCGTCTGC-3′), mouse Taz (forward primer, 5′-CAGCCTCTGAATCATGTGAACCTC-3′; reverse primer, 5′-GGCTAGTGGCCACGACTTGCTGGT-3′), mouse Axin1 (forward primer, 5′-CCTCTACCTCACATTCCTCGCACTT-3′; reverse primer, 5′-TCAACCGTTCCTCCAACTTTTCT-3′), mouse cyclin D1 (forward primer, 5′-AGGCTACAGAAGAGTATTTATGGGAAA-3′; reverse primer, 5′-TGCGTTTGAATCAAGGGAGAT-3′), and mouse osterix (forward primer, 5′-CGCTTTGTGCCTTTGAAAT-3′; reverse primer, 5′-CCGTCAACGACGTTATGC-3′). The relative expression of each mRNA was calculated by the ΔCt method.
Western blot analysis.
Cells were lysed with RIPA buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP40, 0.1% SDS, 0.25% sodium deoxycholate, 1 mM EDTA) supplemented with protease and phosphatase inhibitors. Lysates were cleared by centrifugation at 18,000 g for 10 minutes at 4°C. The protein was resolved by SDS-PAGE and transferred to a PVDF membrane (Immobilon; Millipore). Membranes were blocked in 5% BSA or 5% nonfat dried milk in PBST (PBS + 0.1% Tween-20). The images are representative of 3 independent experiments. The relative integrated density of each protein band was digitized by ImageJ (NIH) and shown in figures.
Statistics.
All results are shown as mean ± SEM of data from at least 3 separate experiments. The data were subjected to ANOVA with Tukey-Kramer’s post-hoc test or the unpaired t test with JMP 7 (SAS Institute Inc.) to determine differences. P values of less than 0.05 were accepted as statistically significant.
Study approval.
All animal studies were approved by the Animal Research Council at University Health Network, Toronto, Ontario.
Author contributions
YM and RR designed the experiments. YM performed experiments and analyzed the results. JLR, ML, HAA, NL, AC, LZ, MS, MDG, DG, and CB performed specific experiments and analyzed the results. XM, FC, PM, and GK contributed to materials. YM and RR wrote the manuscript with helpful comments from HAA, GK, DG, and LZ .
Supplementary Material
Acknowledgments
This work was supported by grants from the Canadian Institute for Health Research (CIHR). RR is supported by the Princess Margaret Cancer Center and the Ontario Institute for Cancer Research. YM is supported by the Postdoctoral Fellowship for Research Abroad, contributed by the Japan Society for the Promotion of Science, the Japan Rheumatism Foundation, the Sumitomo Life Social Welfare Services Foundation, and the Nakayama Science Foundation. The authors would like to acknowledge Lily Morikawa (The Pathology Core at the Toronto Centre for Phenogenomics, Toronto, Ontario, Canada) for histology services.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(7):2612–2625.https://doi.org/10.1172/JCI92233.
Contributor Information
Yoshinori Matsumoto, Email: yossy.m0629@nifty.com.
Jose La Rose, Email: larosa@uhnres.utoronto.ca.
Melissa Lim, Email: melissal@uhnresearch.ca.
Hibret A. Adissu, Email: Hibret.Adissu@covance.com.
Napoleon Law, Email: napoleon.law@rmp.uhn.ca.
Xiaohong Mao, Email: Xiaohong.Mao@novartis.com.
Feng Cong, Email: feng.cong@novartis.com.
Paula Mera, Email: pm2698@columbia.edu.
Gerard Karsenty, Email: gk2172@cumc.columbia.edu.
David Goltzman, Email: david.goltzman@mcgill.ca.
Adele Changoor, Email: adele.changoor@gmail.com.
Lucia Zhang, Email: lucia.zhang@mail.utoronto.ca.
Megan Stajkowski, Email: mstajkow@mail.uoguelph.ca.
Carsten Bergmann, Email: Carsten.Bergmann@bioscientia.de.
Robert Rottapel, Email: rottapel@uhnresearch.ca.
References
- 1.Mundlos S. Cleidocranial dysplasia: clinical and molecular genetics. J Med Genet. 1999;36(3):177–182. [PMC free article] [PubMed] [Google Scholar]
- 2.Marie P, Sainton P. Sur la dysostose cléido-cranienne héréditaire. Rev Neurol. 1898;6:835–838. [PubMed] [Google Scholar]
- 3.Mundlos S, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89(5):773–779. doi: 10.1016/S0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 4.Lee B, et al. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat Genet. 1997;16(3):307–310. doi: 10.1038/ng0797-307. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi T, Kronenberg HM. Overview of skeletal development. Methods Mol Biol. 2014;1130:3–12. doi: 10.1007/978-1-62703-989-5_1. [DOI] [PubMed] [Google Scholar]
- 6.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–754. doi: 10.1016/S0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 7.Komori T, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755–764. doi: 10.1016/S0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 8.Ott CE, et al. Deletions of the RUNX2 gene are present in about 10% of individuals with cleidocranial dysplasia. Hum Mutat. 2010;31(8):E1587–E1593. doi: 10.1002/humu.21298. [DOI] [PubMed] [Google Scholar]
- 9.Hong JH, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309(5737):1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol. 2011;13(5):623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- 11.Zeng L, et al. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell. 1997;90(1):181–192. doi: 10.1016/S0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 12.Krishnan V, Bryant HU, Macdougald OA. Regulation of bone mass by Wnt signaling. J Clin Invest. 2006;116(5):1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med. 2013;19(2):179–192. doi: 10.1038/nm.3074. [DOI] [PubMed] [Google Scholar]
- 14.Baskar JF, et al. Developmental analysis of the cytomegalovirus enhancer in transgenic animals. J Virol. 1996;70(5):3215–3226. doi: 10.1128/jvi.70.5.3215-3226.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Mishina Y, Liu F. Osterix-Cre transgene causes craniofacial bone development defect. Calcif Tissue Int. 2015;96(2):129–137. doi: 10.1007/s00223-014-9945-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pummila M, et al. Ectodysplasin has a dual role in ectodermal organogenesis: inhibition of Bmp activity and induction of Shh expression. Development. 2007;134(1):117–125. doi: 10.1242/dev.02708. [DOI] [PubMed] [Google Scholar]
- 17.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133(16):3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 18.Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26(2):99–109. [PubMed] [Google Scholar]
- 19.Reinhold MI, Naski MC. Direct interactions of Runx2 and canonical Wnt signaling induce FGF18. J Biol Chem. 2007;282(6):3653–3663. doi: 10.1074/jbc.M608995200. [DOI] [PubMed] [Google Scholar]
- 20.Ohbayashi N, et al. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16(7):870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21(21):2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao B, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25(1):51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284(20):13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai D, Ho KC, Hao Y, Yang X. Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 2011;71(7):2728–2738. doi: 10.1158/0008-5472.CAN-10-2711. [DOI] [PubMed] [Google Scholar]
- 25.Ross SE, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289(5481):950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 26.Lee NK, et al. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130(3):456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferron M, Hinoi E, Karsenty G, Ducy P. Osteocalcin differentially regulates beta cell and adipocyte gene expression and affects the development of metabolic diseases in wild-type mice. Proc Natl Acad Sci U S A. 2008;105(13):5266–5270. doi: 10.1073/pnas.0711119105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ducy P. The role of osteocalcin in the endocrine cross-talk between bone remodelling and energy metabolism. Diabetologia. 2011;54(6):1291–1297. doi: 10.1007/s00125-011-2155-z. [DOI] [PubMed] [Google Scholar]
- 29.Haque T, Nakada S, Hamdy RC. A review of FGF18: Its expression, signaling pathways and possible functions during embryogenesis and post-natal development. Histol Histopathol. 2007;22(1):97–105. doi: 10.14670/HH-22.97. [DOI] [PubMed] [Google Scholar]
- 30.Hu MC, et al. FGF-18, a novel member of the fibroblast growth factor family, stimulates hepatic and intestinal proliferation. Mol Cell Biol. 1998;18(10):6063–6074. doi: 10.1128/MCB.18.10.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimoaka T, et al. Regulation of osteoblast, chondrocyte, and osteoclast functions by fibroblast growth factor (FGF)-18 in comparison with FGF-2 and FGF-10. J Biol Chem. 2002;277(9):7493–7500. doi: 10.1074/jbc.M108653200. [DOI] [PubMed] [Google Scholar]
- 32.Ishibe T, et al. Disruption of fibroblast growth factor signal pathway inhibits the growth of synovial sarcomas: potential application of signal inhibitors to molecular target therapy. Clin Cancer Res. 2005;11(7):2702–2712. doi: 10.1158/1078-0432.CCR-04-2057. [DOI] [PubMed] [Google Scholar]
- 33.Liu Z, Xu J, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 2002;16(7):859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumoto Y, et al. Reciprocal stabilization of ABL and TAZ regulates osteoblastogenesis through transcription factor RUNX2. J Clin Invest. 2016;126(12):4482–4496. doi: 10.1172/JCI87802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumoto Y, et al. RANKL coordinates multiple osteoclastogenic pathways by regulating expression of ubiquitin ligase RNF146. J Clin Invest. 2017;127(4):1303–1315. doi: 10.1172/JCI90527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karsenty G, Olson EN. Bone and muscle endocrine functions: unexpected paradigms of inter-organ communication. Cell. 2016;164(6):1248–1256. doi: 10.1016/j.cell.2016.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ducy P, et al. Increased bone formation in osteocalcin-deficient mice. Nature. 1996;382(6590):448–452. doi: 10.1038/382448a0. [DOI] [PubMed] [Google Scholar]
- 38. Ralston S, Helfrich MH. Bone Research Protocols. New York, NY: Humana Press; 2003. [Google Scholar]
- 39.Parfitt AM, et al. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 1987;2(6):595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- 40.Levaot N, et al. 3BP2-deficient mice are osteoporotic with impaired osteoblast and osteoclast functions. J Clin Invest. 2011;121(8):3244–3257. doi: 10.1172/JCI45843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 42.Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15(4):1858–1869. doi: 10.1128/MCB.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.