Abstract
Epidemiologic and animal studies implicate overconsumption of fructose in the development of nonalcoholic fatty liver disease, but the molecular mechanisms underlying fructose-induced chronic liver diseases remain largely unknown. Here, we have presented evidence supporting the essential function of the lipogenic transcription factor carbohydrate response element–binding protein (ChREBP) in mediating adaptive responses to fructose and protecting against fructose-induced hepatotoxicity. In WT mice, a high-fructose diet (HFrD) activated hepatic lipogenesis in a ChREBP-dependent manner; however, in Chrebp-KO mice, a HFrD induced steatohepatitis. In Chrebp-KO mouse livers, a HFrD reduced levels of molecular chaperones and activated the C/EBP homologous protein–dependent (CHOP-dependent) unfolded protein response, whereas administration of a chemical chaperone or Chop shRNA rescued liver injury. Elevated expression levels of cholesterol biosynthesis genes in HFrD-fed Chrebp-KO livers were paralleled by an increased nuclear abundance of sterol regulatory element–binding protein 2 (SREBP2). Atorvastatin-mediated inhibition of hepatic cholesterol biosynthesis or depletion of hepatic Srebp2 reversed fructose-induced liver injury in Chrebp-KO mice. Mechanistically, we determined that ChREBP binds to nuclear SREBP2 to promote its ubiquitination and destabilization in cultured cells. Therefore, our findings demonstrate that ChREBP provides hepatoprotection against a HFrD by preventing overactivation of cholesterol biosynthesis and the subsequent CHOP-mediated, proapoptotic unfolded protein response. Our findings also identified a role for ChREBP in regulating SREBP2-dependent cholesterol metabolism.
Keywords: Hepatology, Metabolism
Introduction
Nonalcoholic fatty liver disease (NAFLD), which affects almost one-quarter of the population in the United States (1), is a slowly progressive metabolic liver disorder, ranging from simple steatosis and steatohepatitis to fibrosis and eventual cirrhosis. Overconsumption of saturated fat has been considered a major trigger for NAFLD (2). More recently, with the wide use of high-fructose corn syrup in diets, chronic overconsumption of fructose has been attributed to the incidence of NAFLD (2, 3). Primarily metabolized in the liver, fructose intake has been linked to increased liver steatosis, inflammation, and mitochondrial dysfunction (4, 5). So far, a clear understanding of how fructose consumption is involved in the pathogenesis of NAFLD has been lacking. In particular, specific cellular pathways that protect hepatocytes from the adverse metabolic effects of fructose are unknown.
Carbohydrate response element–binding protein (ChREBP) is a glucose-sensing transcription factor that potently activates a panel of de novo lipogenesis (DNL) genes upon glucose excess (6, 7). Chrebp mRNA expression increases in the liver of patients with NAFLD (8). ChREBP deficiency in diabetic mice alleviates liver steatosis and improves glucose metabolism (9, 10). In addition, ChREBP was found to regulate ethanol metabolism and modulate the degree of ethanol-induced liver injury (11). These studies suggest that ChREBP may regulate liver pathophysiology in a context-dependent manner. Whether ChREBP can regulate genes beyond lipogenesis in response to fructose intake has not been explored.
The endoplasmic reticulum (ER) responds to the burden of unfolded proteins in its lumen by activating the unfolded protein response (UPR). Under normal conditions, the duration and degree of UPRs are tightly controlled to ensure cell viability while reducing the burden of misfolded proteins. However, prolonged or overactivated UPRs leads to apoptosis and organ damage (12) and has been implicated in the pathogenesis of nonalcoholic steatohepatitis (NASH) in rodents (13). Another important function of the ER is to control cellular cholesterol levels by fine-tuning cholesterol synthesis and clearance (14). Accumulation of free cholesterol in the ER membrane can induce ER stress and hepatocyte apoptosis (15). In patients with NASH, elevated levels of free cholesterol instead of saturated fatty acids have been reported in the liver (16–18). Hepatocytes overloaded with free cholesterol are highly susceptible to TNF-α or FAS-induced apoptosis (19). SREBP2 (sterol regulatory element–binding protein) is the major transcription activator of liver cholesterol biosynthesis (14). SREBP2-dependent function is abnormally elevated in NAFLD and may contribute to excessive cholesterol overload and trigger hepatocyte death (17, 20). Therefore, SREBP2-mediated free cholesterol accumulation and ER stress create a forward feedback loop to promote the progression toward NASH. How this feedback loop is involved in fructose-associated NAFLD has not been examined.
Here, we sought to determine how Chrebp deficiency sensitizes fructose-rich diet–induced liver steatosis and injury. We uncovered an intricate pathway through which ChREBP activates de novo lipogenesis but suppresses the UPR and cholesterol biosynthesis in fructose-overloaded hepatocytes. Collectively, our study highlights a metabolic adaptation pathway mainly driven by ChREBP to protect the liver in response to a fructose-rich diet.
Results
A short-term high-fructose diet activates ChREBP-dependent hepatic lipogenesis.
Given the prevalence of fructose-rich diets in many countries, we investigated the metabolic response to a short-term high-fructose diet (HFrD) (OpenSource Diets; 20% calories provided by protein, 10% by fat, 70% by free fructose) in WT mice. Compared with regular chow (26.8% calories provided by protein, 16.7% by fat, 56.4% by starch), HFrD feeding did not induce obesity, hyperglycemia, or systemic insulin resistance (Supplemental Figure 1, A–D; supplemental material available online with this article; https://doi.org/10.1172/JCI89934DS1). However, HFrD-fed mice displayed elevated serum triglycerides, hepatomegaly (Supplemental Figure 1, E and F), and increased liver triglyceride content (Supplemental Figure 2A). Oil Red O staining revealed neutral lipid accumulation and the formation of lipid droplets within hepatocytes (Supplemental Figure 2B). Liver histology analysis showed normal liver structure without evident injury (Supplemental Figure 2B), indicating that WT mice develop simple liver steatosis in response to a HFrD.
To investigate the primary cause of liver steatosis upon short-term fructose intake, we analyzed the expression profile of the major lipid metabolic pathways that could contribute to liver steatosis (Supplemental Figure 2, C and D) (21). Both mRNA and protein expression levels of the major lipogenic enzymes (fatty acid synthase [FASN], stearoyl-coenzyme A desaturase 1 [SCD1], and glucokinase [GCK]) were markedly induced by a HFrD, along with the induction of the lipogenic transcription factor ChREBP (2.7-fold increase in mRNA levels and 2.2-fold increase in protein levels) (Supplemental Figure 2, D and E), pointing to ChREBP-dependent lipogenesis as an early metabolic adaptation to acute fructose intake. Of note, we did not observe a similar induction of SREBP1c protein, although the mRNA level of Srebp1c increased (Supplemental Figure 2, D and E), indicating that hepatocytes selectively activate the ChREBP-dependent lipogenic pathway upon fructose intake.
Since hepatocytes are the major cell type capable of metabolizing fructose, we investigated whether hepatocytes from HFrD-fed mice have increased levels of lipogenesis when cultured ex vivo. We isolated primary hepatocytes from mice fed regular chow or a HFrD for 1 week and 2 weeks and then measured the mRNA and protein levels of major DNL factors and the rate of DNL. The induction of FASN, SCD1, and ChREBP in hepatocytes was detected as early as 1 week after a HFrD and further increased after 2 weeks of a HFrD (Supplemental Figure 3, A and B). Acetylation of ChREBP is known to increase ChREBP transcriptional activity (22). Indeed, we detected increased ChREBP acetylation in primary hepatocytes isolated from HFrD-fed mice in comparison with regular chow-fed mice (Supplemental Figure 3C), suggesting that fructose feeding also enhances the ChREBP function. Furthermore, using 3H-acetate as a substrate for DNL, we detected a 2-fold increase in the rate of DNL in primary hepatocytes from mice after 1 week of HFrD feeding (Supplemental Figure 3D). Thus, these results support a cell-autonomous activation of ChREBP-promoted DNL in mouse hepatocytes following HFrD feeding.
So far, one striking similarity in both liver and primary mouse hepatocytes following fructose intake is the upregulation of ChREBP protein, ChREBP acetylation, and its lipogenic targets, including Fasn, pyruvate kinase liver and red blood cell (Pklr), and Scd1, along with a slight increase in Srebp1c mRNA levels. We therefore hypothesized that ChREBP is required for acute activation of hepatic lipogenesis upon HFrD feeding. To test this hypothesis, Chrebp–/– mice from the Uyeda group (6) and with their WT littermates were fed a HFrD for 2 weeks. Notably, Chrebp–/– mice lost approximately 20% of their BW, whereas the WT mice maintained their BW (Supplemental Figure 4A). Interestingly, while WT and Chrebp–/– mice consumed similar amounts of regular chow, Chrebp–/– mice consumed only about 62% of the HFrD compared with WT mice 72 hours after switching to a HFrD (Supplemental Figure 4, B and C), suggesting that Chrebp–/– mice become intolerant to a fructose-rich diet and tend to avoid fructose. The livers of Chrebp–/– mice weighed 39% less (Supplemental Figure 4D) and displayed a higher glycogen content, an observation that was similarly found in chow-fed and starch-fed Chrebp–/– mice (6) (Supplemental Figure 4E). However, Chrebp–/– mice showed lower triglyceride accumulation in the liver (Figure 1, A and B). The protein and mRNA levels of major lipogenic genes including Fasn, Scd1, and acetyl-CoA carboxylase 1 (Acc1) were also significantly reduced in the livers of Chrebp–/– mice following HFrD feeding (Figure 1C and Supplemental Figure 4, F and G). These data confirmed the essential role of ChREBP in the activation of the lipogenic pathway and lipid accumulation in the liver upon fructose influx.
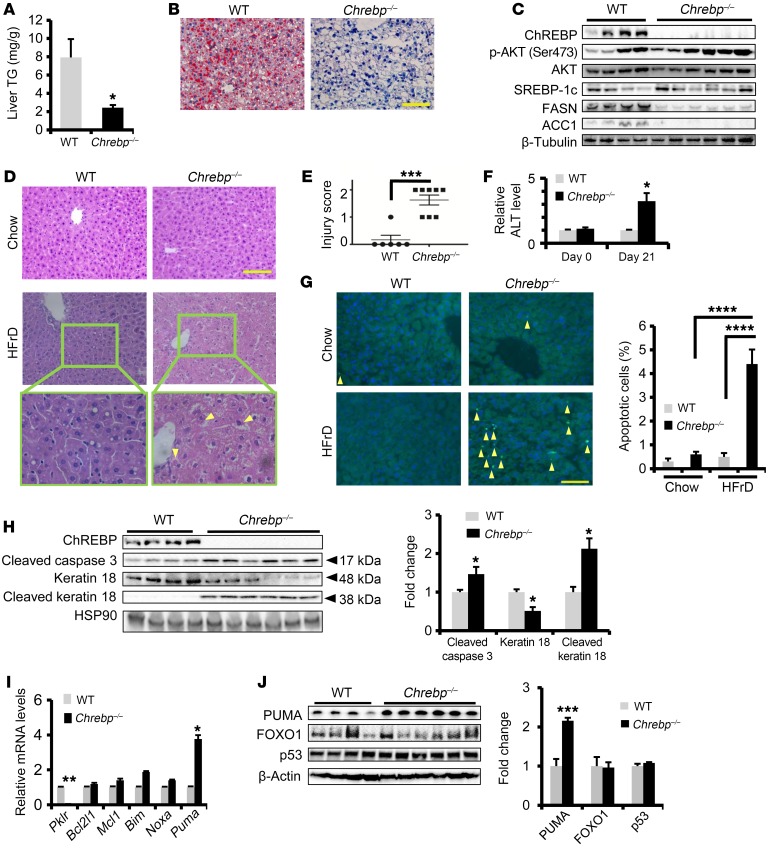
Figure 1. Loss of Chrebp sensitizes mice to HFrD-induced liver injury.
Eight-week-old WT and Chrebp–/– mice were fed a HFrD (70% calories from free fructose) for two weeks (n = 4 for WT, n = 6 for Chrebp–/– mice). (A–C) Loss of Chrebp blocked HFrD-induced hepatic lipogenesis. After 2 weeks of HFrD feeding, (A) a liver triglyceride (TG) assay and (B) Oil Red O staining were performed to assess lipid accumulation in the livers of Chrebp–/– mice and their WT littermates. (C) Protein levels of lipogenic enzymes were assessed by Western blotting (protein quantification is shown in Supplemental Figure 4G). (D–F) HFrD feeding induced liver injury in Chrebp–/– mice. (D) After H&E staining of livers from Chrebp–/– mice and their WT littermates fed either regular chow or a HFrD, (E) liver injury was scored blindly on a scale of 0 to 2. Mallory-Denk bodies are indicated by yellow arrows. (F) Serum ALT levels were measured at the start and end of HFrD feeding. (G and H) HFrD feeding induced apoptosis in Chrebp–/– mouse livers. Apoptosis was determined by (G) TUNEL staining and (H) Western blotting for apoptotic markers. Apoptotic cells are indicated by arrowheads. (I and J) HFrD feeding increased the expression of PUMA at both (I) mRNA and (J) protein levels. *P < 0.05, **P < 0.01, and ***P < 0.001, by 2-tailed Student’s t test; ****P < 0.0001, by 1-way ANOVA with Tukey’s post-hoc test. Data represent the mean ± SEM. Scale bars: 100 μm; original magnification, ×400 (enlarged images in bottom panels of D).
Chrebp–/– mice show severe liver injury following HFrD feeding.
As shown in Figure 1D (top panels), the liver histology of regular chow–fed Chrebp–/– mice was indistinguishable from that of WT liver. However, upon fructose diet challenge, the liver histology of Chrebp–/– mice showed characteristics of liver injury including loss of nuclei, hepatocyte ballooning, and the formation of Mallory-Denk bodies (Figure 1D, bottom panels). When blindly scored in terms of NASH grade, the livers of Chrebp–/– mice showed a significantly higher degree of liver injury (Figure 1E). Furthermore, fructose-challenged Chrebp–/– mice showed a 3-fold increase in serum alanine aminotransferase (ALT) (Figure 1F). Fructose feeding also augmented apoptosis in the livers of Chrebp–/– mice, as determined by TUNEL staining (Figure 1G), and increased the levels of cleaved caspase 3 and keratin 18 (Figure 1H), without significantly increasing the levels of serum fibroblast growth factor 21, a biomarker of NAFLD and NASH (23, 24) (Supplemental Figure 5).
To test whether Chrebp–/– mice mount a maladaptive response to intermediate levels of free fructose, we fed Chrebp–/– mice and their WT littermates a 34% fructose diet (20% calories from protein, 10% from fat, and 34% from free fructose) for 4 weeks. Compared with WT mice on a 34% fructose diet, Chrebp–/– mice consistently developed elevated levels of serum ALT and hepatocyte apoptosis (Supplemental Figure 6), a phenotype similar to that of Chrebp–/– mice fed a 70% fructose diet. These results suggest that Chrebp deficiency renders mice intolerant to fructose feeding.
Hepatocyte apoptosis results from an imbalance in the expression of proapoptotic versus antiapoptotic factors. We examined the mRNA expression of proapoptotic factors (BCL-2–interacting mediator of cell death [Bim], NADPH oxidase activator 1 [Noxa], and p53-upregulated modulator of apoptosis [Puma]) and antiapoptotic factors (BCL2-like protein 1 [Bcl2l1] and MCL1, BCL2 family apoptosis regulator [Mcl1]) in both WT and Chrebp–/– livers following HFrD feeding. As a BH3-only protein critical for hepatocyte apoptosis in response to various insults (25, 26), PUMA mRNA and protein levels were elevated by approximately 4-fold and 2-fold, respectively (Figure 1, I and J). We also detected the induction of PUMA mRNA and protein in the livers of Chrebp–/– mice fed a 34% fructose diet (Supplemental Figure 6, E and F). Meanwhile, FOXO1 and p53 protein levels were comparable between WT and Chrebp–/– livers (Figure 1J), indicating that the induction of PUMA was probably not due to FOXO1 and p53, two known transcription activators of Puma (26, 27).
Given the prominent role of the liver in fructose metabolism, we tested whether restoring Chrebp expression in the livers of Chrebp–/– mice could reduce liver injury following HFrD feeding. To this end, we administered Ad-Flag-Chrebp virus to mice via the tail vein prior to switching them to a 2-week-long HFrD. In Ad-Flag-Chrebp–injected mice, we observed that the severity of liver injury was markedly reduced, as shown by TUNEL staining and an increase in cleaved caspase 3 and keratin 18 (Figure 2). In summary, these findings indicate that Chrebp deficiency sensitizes hepatocytes to apoptosis upon consumption of a fructose-rich diet, possibly via upregulation of the proapoptotic pathway.
Figure 2. Restoring Chrebp expression rescues liver injury in HFrD-fed Chrebp–/– mice.
Eight-week-old male Chrebp–/– mice were injected with either Chrebp-expressing adenovirus (Ad-Flag-Chrebp, n = 9) or GFP-expressing control adenovirus (Ad-GFP, n = 7). Three mice from each group were dissected three days after adenovirus injection, and Chrebp overexpression was confirmed by (A) Western blotting with anti-Flag and (B) RT-qPCR for Fasn and Pklr. The remaining mice were fed a 70% HFrD for 2 weeks before dissection. (C) Effects of restoring ChREBP on liver triglyceride levels. Liver injury was assessed by (D) H&E staining, (E) TUNEL staining (apoptotic cells are indicated by arrowheads), (F) ALT assay, and (G) Western blotting for apoptotic markers. *P < 0.05, by 2-tailed Student’s t test. Data represent the mean ± SEM. Scale bars: 100 μm.
A HFrD activates the C/EBP homologous protein–mediated apoptotic UPR and leads to liver injury in Chrebp–/– mice.
To determine the mechanisms of hepatocyte death and liver injury in fructose-fed Chrebp–/– mice, we focused on the UPR-associated cell death pathways, since these pathways have been shown to play a critical role in the pathogenesis of alcohol- and drug-induced liver injury (28). We first evaluated the UPR pathway in WT mice with or without a HFrD. Compared with mice fed regular chow, fructose-fed WT mice showed increased levels of the UPR sensors endoribonuclease/protein kinase IRE1-like protein (IRE1), activating transcription factor 6 (ATF6), and glucose-regulated protein 78 (GRP78) as well as increased mRNA levels of spliced X-box binding protein 1 (Xbp1) in the liver (Supplemental Figure 7, A and B). In contrast, protein kinase R–like endoplasmic reticulum kinase (PERK) and phosphorylated PERK (p-PERK) levels were comparable in both groups. As expected, the protein expression of C/EBP homologous protein (CHOP) in the liver was similar between regular chow–fed mice and HFrD-fed mice (Supplemental Figure 7A). These changes in the UPR pathway were also observed in primary mouse hepatocytes following 1 week of a HFrD (Supplemental Figure 7, C and D), indicating that WT hepatocytes are fully capable of activating the adaptive UPR pathway in response to a fructose-rich diet.
We hypothesized that an uncontrolled UPR in the Chrebp–/– mouse liver may account for hepatocyte death and subsequent liver injury after HFrD feeding. Notably, we detected comparable expression levels of major UPR factors in regular chow–fed WT and Chrebp–/– mice (Figure 3A). However, the UPR pathway was largely reprogrammed in HFrD-fed Chrebp–/– mice. Specifically, we observed a reduction in nuclear ATF6, even though the total levels of IRE1α were similar between WT and Chrebp–/– groups after HFrD feeding (Figure 3B). Furthermore, the UPR chaperone GRP78 was reduced by 50% in Chrebp–/– livers (Figure 3B and Supplemental Figure 8). The most significant change in the UPR pathway was the activation of the PERK/ATF4/CHOP pathway, including the elevation of p-PERK (Figure 3B and Supplemental Figure 8). Increased levels of CHOP might be responsible for the induction of PUMA in the fructose-fed Chrebp–/– mice (Figure 1, I and J).
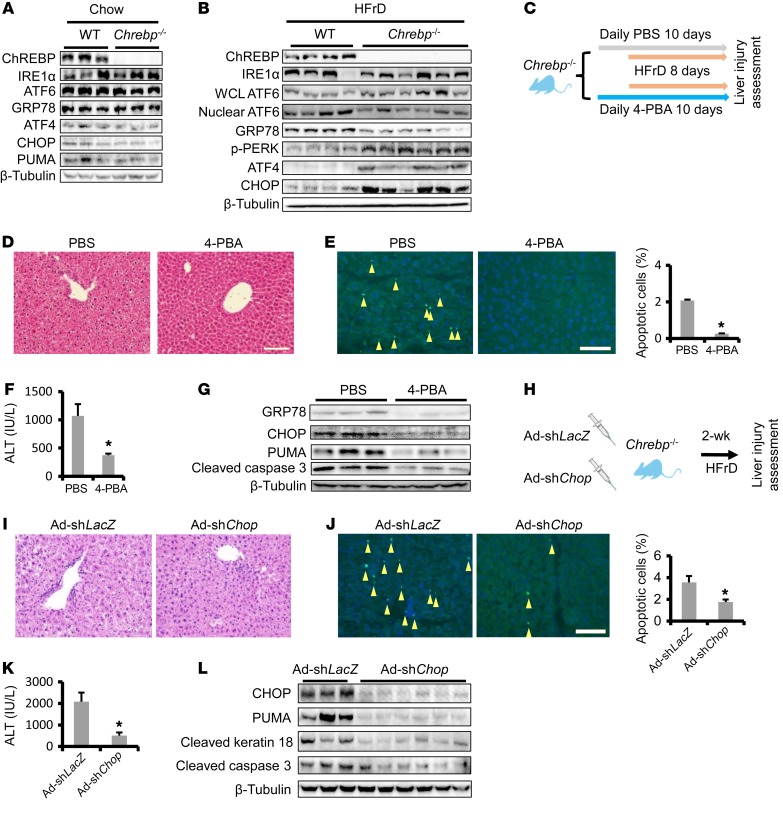
Figure 3. HFrD activates the proapoptotic branch of the UPR in Chrebp–/– mouse liver.
(A) Comparable levels of ER stress in the livers of regular chow–fed Chrebp–/– mice and their WT littermates. Livers of 8-week-old male Chrebp–/– mice and their WT littermates (n = 3) on a regular chow diet were subjected to Western blotting for ER stress markers. (B) HFrD feeding activated the proapoptotic branch of the UPR in Chrebp–/– mouse livers. Male and female Chrebp–/– mice and their WT littermates were fed a 70% HFrD for 2 weeks before dissection (n = 4 for WT, n = 6 for Chrebp–/– mice). Protein levels of components of adaptive and proapoptotic branches of ER stress in the liver were assessed with Western blotting (protein level quantification is shown in Supplemental Figure 8). WCL, whole-cell lysate. (C–G) Administration of the chemical chaperone 4-BPA protected Chrebp–/– mice from HFrD-induced liver injury. (C) Male 8-week-old Chrebp–/– mice were pretreated with 4-PBA (1 g/kg BW/day) or PBS by oral gavage for 2 days (n = 3/group), followed by 8 days of HFrD feeding plus 4-PBA or PBS gavage. Liver injury was determined by (D) H&E staining, (E) TUNEL staining (apoptotic cells are indicated by arrowheads), (F) ALT assay, and (G) Western blotting for apoptotic markers. (H–L) Blocking the proapoptotic branch of the UPR protected Chrebp–/– mice from HFrD-induced liver injury. (H) Male and female 8-week-old Chrebp–/– mice were injected with either Chop-knockdown adenovirus (Ad-shChop, n = 6) or control adenovirus (Ad-shLacZ, n = 3) and then fed a HFrD for 2 weeks. Liver injury was assessed by (I) H&E staining, (J) TUNEL staining (apoptotic cells are indicated by arrowheads; original magnification, ×200), (K) ALT assay, and (L) Western blotting for apoptotic markers. *P < 0.05, by 2-tailed Student’s t test. Data represent the mean ± SEM. Scale bars: 100 μm.
It is noted that this major shift in the UPR was absent in Chrebp–/– mice on regular chow (Figure 3A and Supplemental Figure 8). We found that neither Chrebp deficiency nor Chrebp overexpression affected the Chop mRNA levels in the chow-fed condition (Supplemental Figure 9), suggesting that ChREBP does not participate in the regulation of the UPR under normal conditions.
Activity of calcium/calmodulin-dependent protein kinase II (CaMKII) has been shown to link calcium release from severe ER stress to participate in CHOP-mediated apoptosis (29, 30). Indeed, we observed increased Camk2d mRNA, total CaMKII abundance, as well as CaMKII phosphorylation at Thr286 in the livers of Chrebp–/– mice (Supplemental Figure 10). Taken together, our observations suggest that chronic activation of the proapoptotic UPR branch might be a major cause of the hepatocyte death and liver injury observed in Chrebp–/– mice upon fructose feeding.
When administered in ischemia-reperfusion liver injury, the chemical chaperone 4-phenylbutyric acid (4-BPA) was shown to reduce the severity of liver injury in rodents (31, 32). We hypothesized that alleviation of ER stress using 4-BPA might reverse liver injury in fructose-fed Chrebp–/– mice. To test this hypothesis, we pretreated Chrebp–/– mice with vehicle control or 4-PBA for 2 days and followed this with 8 days of HFrD feeding in the presence of vehicle or 4-PBA (Figure 3C). Compared with controls, we found that 4-PBA treatment indeed normalized the histological alterations and reduced serum ALT levels, apoptotic hepatocyte numbers, as detected by TUNEL staining, and caspase 3 cleavage (Figure 3, D–G). The improvement in liver injury correlated with reduced levels of CHOP and its target PUMA (Figure 3G).
Upon HFrD feeding, GRP78 expression was upregulated to accommodate the adaptive UPR in WT mouse livers (Supplemental Figure 7, A and C). However, such a response was not observed in fructose-fed Chrebp–/– mouse livers (Figure 3B). In a complementary experiment, we tested whether restoring GRP78 expression reduces the degree of liver injury in fructose-fed Chrebp–/– mice. We generated recombinant adenovirus to restore GRP78 expression in Chrebp–/– mice (Supplemental Figure 11). Consistent with the results from 4-PBA treatment, Chrebp–/– mice injected with Ad-Grp78 showed a marked improvement in liver histology and a reduction in TUNEL-positive hepatocytes and serum ALT levels (P = 0.08) (Supplemental Figure 11, B–D). Taken together, our data suggest that restoring the adaptive UPR via either treatment with 4-PBA or GRP78 overexpression reduces liver injury in HFrD-challenged Chrebp–/– mice.
Chrebp deficiency reprogrammed the UPR in the liver following fructose feeding, suppressing the adaptive UPR, while activating the proapoptotic UPR (Figure 3B). As a critical mediator of ER stress–induced apoptosis, CHOP was significantly elevated in HFrD-fed Chrebp–/– mouse livers (Figure 3B). Activation of CHOP triggers hepatocyte apoptosis by altering apoptotic and survival proteins (28). Loss of Chop was shown to protect mice from liver injury induced by acetaminophen, endotoxin, and alcohol (33, 34). We therefore hypothesized that knockdown of hepatic Chop in fructose-fed Chrebp–/– mice might prevent hepatocyte death and liver injury. To test this hypothesis, we depleted Chop in Chrebp–/– mice with Ad-shChop injection prior to 2 weeks of HFrD feeding (Figure 3H). While Chrebp–/– mice with Ad-shLacZ still exhibited liver injury after a fructose diet challenge, administration of Ad-shChop led to significantly lower levels of serum ALT, fewer apoptotic hepatocytes, improved liver histology, decreased caspase 3 cleavage, and reduced PUMA protein (Figure 3, I–L).
In summary, our findings highlight the critical role of ChREBP in handling an influx of fructose by activating the adaptive UPR pathway in hepatocytes. In the case of Chrebp deficiency, hepatocytes develop persistent ER stress and activate the CHOP-dependent UPR when challenged with HFrD. Suppressing ER stress via 4-PBA gavage, Grp78 overexpression, or Chop knockdown protects Chrebp–/– mice from HFrD-induced liver injury.
Elevated HMGCR and cholesterol biosynthesis drive liver injury in HFrD-fed Chrebp–/– mice.
To gain insights into the origin of persistent ER stress in fructose-fed Chrebp–/– mice, we performed a genome-wide expression analysis in the livers of WT (regular chow vs. 70% HFrD) and Chrebp–/– (regular chow vs. 70% HFrD) mice. As expected, lipogenic target genes of ChREBP such as Fasn, Acc1, and Me1 were all downregulated in HFrD-fed Chrebp–/– mouse livers (Figure 4A). However, the cholesterol biosynthesis pathway was significantly altered in HFrD-fed Chrebp–/– mouse livers (Figure 4, A and B). Notably, the levels of 6 genes (3-hydroxy-3-methylglutaryl-CoA reductase [Hmgcr], squalene epoxidase [Sqle], lanosterol synthase [Lss], mevalonate [diphospho] decarboxylase [Mvd], 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1 [Hmgcs1], and isopentenyl-diphosphate delta isomerase [Idi1]) directly involved in cholesterol biosynthesis were decreased in HFrD-fed WT mouse livers but increased by more than 2-fold in HFrD-fed Chrebp–/– livers (Figure 4A). The induction of cholesterol metabolic genes was further confirmed by reverse transcription quantitative PCR (RT-qPCR) (Figure 4C), with a 3-fold increase in Srebp2 mRNA levels and an 8-fold increase in Sqle mRNA levels. HMGCR, the rate-limiting enzyme in cholesterol biosynthesis, was elevated by 7-fold at the mRNA level and by 2-fold at the protein level (Figure 4, C and D). Consistent with the induction of HMGCR, total cholesterol levels in the livers of Chrebp–/– mice on a HFrD were elevated, even though the change was not statistically significant (P = 0.18) (Figure 4G). mRNA expression of Srebp2, Hmgcr, and Sqle was also induced by a 34% fructose diet in the livers of Chrebp–/– mice (Supplemental Figure 12A). In contrast, restoring ChREBP expression in the livers of Chrebp–/– mice suppressed the induction of all 3 cholesterol biosynthesis genes and HMGCR protein following HFrD feeding (Figure 4, E and F). We noted that Chrebp deficiency did not affect cholesterol biosynthesis in regular chow feeding conditions (Supplemental Figure 13). Taken together, our Chrebp–/– mouse study supports the idea that ChREBP regulates hepatic cholesterol biosynthesis in response to a HFrD.
Figure 4. ChREBP protects mice from HFrD-induced liver injury via suppression of cholesterol biosynthesis.
Microarray analysis was performed with RNA samples pooled from the livers of regular chow–fed WT mice (n = 7) versus 70% HFrD–fed WT mice (n = 7) or from the livers of HFrD-fed WT mice (n = 4) versus HFrD-fed Chrebp–/– mice (n = 6). Both male and female mice were used in all 4 groups. (A) List of ChREBP-regulated genes in mouse livers in response to high-fructose feeding (genes involved in de novo cholesterol biosynthesis are highlighted in red). (B) Enrichment of genes in cholesterol biosynthesis in HFrD-fed Chrebp–/– mouse liver by PANTHER pathway analysis. (C and D) Elevation of genes in cholesterol biosynthesis in the livers of HFrD-fed Chrebp–/– mice. Increased levels of cholesterol biosynthesis genes in HFrD-fed Chrebp–/– mouse livers were confirmed by (C) RT-qPCR and (D) Western blotting. (E and F) Restoring hepatic ChREBP expression suppressed cholesterol biosynthesis genes and HMGCR protein in the livers of HFrD-fed Chrebp–/– mice. (G and H) Increased free cholesterol content in Chrebp–/– mouse livers after HFrD feeding. (G) Total cholesterol and (H) free cholesterol levels in the liver were assessed with a cholesterol quantification kit and filipin staining, respectively. (I) Restoration of hepatic ChREBP expression suppressed free cholesterol loading in the livers of HFrD-fed Chrebp–/– mice. *P < 0.05 and **P < 0.01; an unpaired, 2-tailed Student’s t test was used to determine the P values in C–E and G–I. Data represent the mean ± SEM. Scale bars: 100 μm.
Free cholesterol accumulation has been linked to liver injury and NASH, in part by disrupting the membrane integrity of mitochondria and ER, triggering mitochondrial oxidative damage and inducing unresolved ER stress (15). We measured hepatic free cholesterol accumulation in HFrD-fed Chrebp–/– and WT mouse livers by filipin staining. Compared with HFrD-fed WT livers, the free cholesterol level of HFrD-fed Chrebp–/– livers was elevated 3-fold (Figure 4H). In contrast, restoring ChREBP expression in the liver of Chrebp–/– mice suppressed hepatic free cholesterol loading induced by a HFrD (Figure 4I). To our knowledge, this is the first report linking Chrebp deficiency and the derangement in cholesterol metabolism.
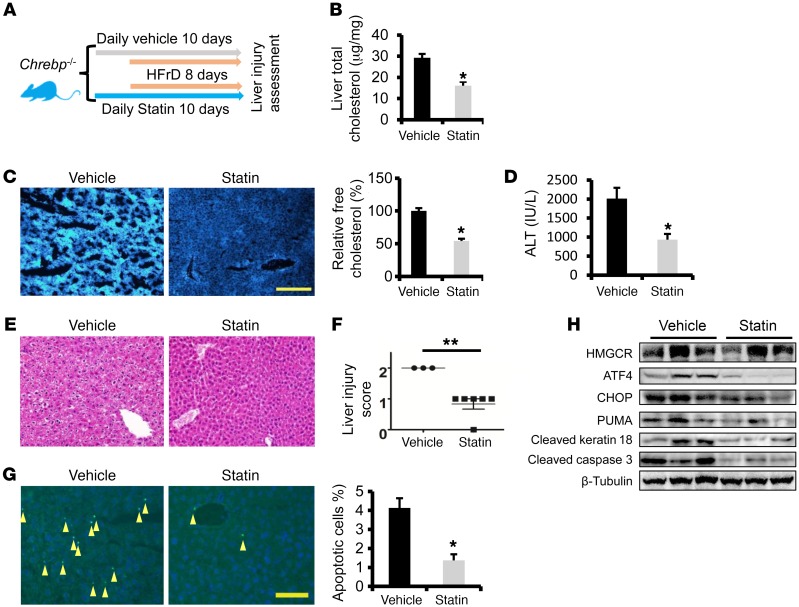
Although we observed a substantial increase in cholesterol biosynthesis and accumulation of free cholesterol in the livers of HFrD-fed Chrebp–/– mice, it is unclear whether cholesterol accumulation contributes to liver injury in these mice. HMGCR, which catalyzes the first reaction of cholesterol biosynthesis, has been successfully targeted using a pharmacological approach (15). We postulated that inhibition of HMGCR and its mediated cholesterol biosynthesis in Chrebp–/– mice during the course of fructose feeding might reduce hepatocyte death and alleviate liver injury. To this end, Chrebp–/– mice were given the HMGCR inhibitor atorvastatin once daily for 2 days prior to HFrD feeding as well as for the entire duration of the HFrD feeding (Figure 5A). At the end of 8 days of HFrD feeding, atorvastatin treatment elevated mRNA levels of Srebp2, Hmgcr, and Sqle, while reducing not only serum cholesterol and ALT levels but also total and free hepatic cholesterol levels and hepatocyte apoptosis in Chrebp–/– mice (Figure 5, B–H and Supplemental Figure 14). Thus, we showed evidence that pharmacological suppression of cholesterol biosynthesis mitigates HFrD-induced liver injury in Chrebp–/– mice.
Figure 5. Blocking cholesterol biosynthesis protects Chrebp–/– mice from fructose-induced liver injury.
(A) Male 8-week-old Chrebp–/– mice were pretreated by oral gavage with atorvastatin (20 mg/kg BW/day) or vehicle for 2 days, followed by 8 days of 70% HFrD feeding plus atorvastatin or vehicle gavage (n = 6/group). (B and C) Liver total cholesterol and free cholesterol levels were determined. Liver injury was assessed by (D) ALT assay, (E and F) H&E staining, and (G) TUNEL staining (arrowheads indicate apoptotic cells). (H) HMGCR and apoptotic markers were measured by Western blotting. *P < 0.05 and **P < 0.01, by 2-tailed Student’s t test.
ChREBP suppresses cholesterol biosynthesis by degrading nuclear SREBP2 upon HFrD feeding.
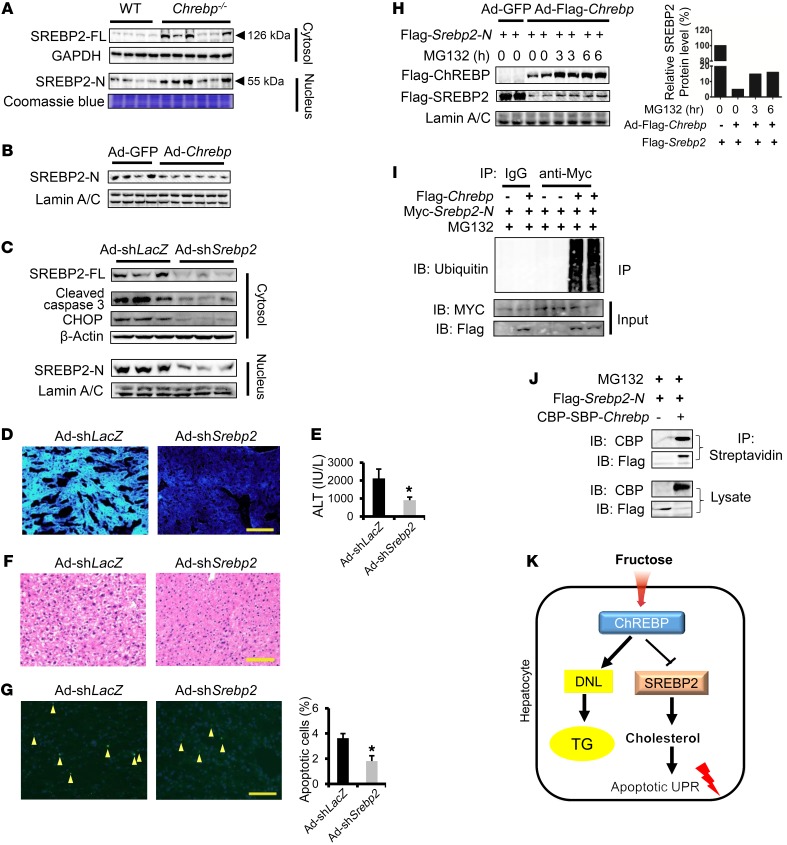
It has been well established that SREBP2 acts as the major transcription factor that regulates hepatic cholesterol biosynthesis, uptake, secretion, and transport. SREBP2 drives the expression of major enzyme genes for cholesterol metabolism such as Hmgcr and Sqle (14, 35, 36). Given that 7 cholesterol metabolism genes were elevated in the Chrebp–/–mice following HFrD feeding, we postulate that SREBP2 could play a critical role in this process. The full-length SREBP2 (SREBP-FL) protein is synthesized and retained in the ER and is then processed into the mature form, which translocates into the nucleus as nuclear SREBP (SREBP-N) to activate transcription (35). We observed that in the liver of Chrebp–/– mice, both SREBP-FL and SREBP2-N levels were robustly elevated by HFrD feeding (Figure 6A, Supplemental Figure 12B, and Supplemental Figure 15). On the contrary, restoring ChREBP expression suppressed SREBP2-N accumulation in nuclear fractions of HFrD-fed Chrebp–/– mouse liver (Figure 6B). To test whether hepatic Srebp2 depletion in Chrebp–/– mice might suppress cholesterol biosynthesis and prevent liver injury upon fructose feeding, we generated Ad-shSrebp2 to deplete mouse Srebp2 expression in vivo. We confirmed the efficiency of Srebp2 knockdown by detecting reduced levels of both SREBP2-FL and SREBP2-N in the liver (Figure 6C). As expected, Srebp2 depletion resulted in a reduction of its targets, Hmgcr, Hmgcs1, and Sqle, and subsequently lowered serum cholesterol levels (Supplemental Figure 16, A and B). More important, Srebp2 depletion significantly reduced free cholesterol accumulation in the liver and mitigated HFrD-induced liver injury (Figure 6, C–G and Supplemental Figure 16C). Collectively, our data suggest that SREBP2 overactivation acts as a major driver to promote liver injury via cholesterol biosynthesis in Chrebp–/– mice following HFrD feeding. Thus, our data uncovered what we believe to be a novel link between ChREBP and SERBP2-driven cholesterol biosynthesis in the livers of fructose diet–fed mice.
Figure 6. ChREBP blocks HFrD-induced liver injury in part by degrading SREBP2-N to suppress cholesterol biosynthesis.
(A) HFrD feeding induced both SREBP2-FL and SREBP2-N protein expression in the livers of 70% HFrD–fed Chrebp–/– mice. (B) Restoring ChREBP expression lowered SREBP2-N in the nuclear extract from the livers of HFrD-fed Chrebp–/– mice. (C–G) Depletion of Srebp2 by shRNA ameliorated HFrD-induced liver injury in Chrebp–/– mice. Chrebp–/– mice (8 weeks of age) were injected with either Srebp2-knockdown adenovirus (Ad-shSrebp2, n = 3) or control adenovirus (Ad-shLacZ, n = 3) and then fed a HFrD for 2 weeks. (C) Srebp2-knockdown efficiency was confirmed by Western blotting, and (D) its targets expression and filipin staining detected free cholesterol. Liver injury was assessed by (C) Western blotting with antibodies against apoptotic markers, (E) a serum ALT assay, (F) liver H&E staining, and (G) TUNEL staining (arrowheads indicate apoptotic cells). (H) ChREBP promoted SREBP2 protein degradation. U2OS cells were transfected with Flag-Srebp2-N and cotransduced with Ad-GFP or Ad-Flag-Chrebp and then treated with 10 μM MG132 for 3 or 6 hours. ChREBP and SREBP2-N expression levels were assessed by Western blotting with anti-Flag antibody. (I) ChREBP promoted SREBP2 protein ubiquitination. 293A cells were transfected with Myc-Srebp2-N and cotransduced with Ad-GFP or Ad-Flag-Chrebp. Polyubiquitinated SREBP2-N was pulled down by denaturing immunoprecipitation with anti-Myc antibody and detected by Western blotting with antiubiquitin. (J) ChREBP interacted with SREBP2. 293A cells were transfected with Flag-Srebp2-N and cotransfected with pNTAP-CBP-SBP-Chrebp or pNTAP empty vector. The lysate was subjected to immunoprecipitation with streptavidin beads and to Western blotting with CBP or Flag antibodies. (K) Working model: high-fructose–induced ChREBP suppresses free cholesterol loading and protects mice from liver injury via the promotion of SREBP2 degradation. Data shown in H, I, and J are representative results of 3 independent experiments. *P < 0.05, by 2-tailed Student’s t test. Data represent the mean ± SEM. Scale bars: 100 μm. IB, immunoblot; IP, immunoprecipitation.
So far it remains unclear how ChREBP deficiency leads to the induction of the SREBP2 pathway and especially to elevated SREBP2-N protein levels. We hypothesize that ChREBP could negatively regulate SREBP2 at both transcriptional and posttranscriptional levels. After searching through our microarray data and screening reported cholesterol biosynthesis regulators using RT-qPCR (Supplemental Figure 17), we were unable to identify any obvious targets shown to inhibit the transcription of Srebp2. However, we obtained evidence that ChREBP could directly regulate SERBP2-N stability. In U20S cells, overexpression of ChREBP reduced Flag-SREBP2-N abundance by approximately 95%, an effect that could be partially reversed by the treatment of the proteasome inhibitor MG132 (Figure 6H). Moreover, we showed that ChREBP overexpression led to polyubiquitination of SREBP2-N, indicating that ChREBP may control SREBP2-N protein turnover through ubiquitin-dependent degradation (Figure 6I and Supplemental Figure 18). Last, we detected a strong protein-protein interaction between tagged ChREBP and SREBP2-N in the presence MG132 in cultured cells (Figure 6J). In summary, our in vitro overexpression data suggest, for the first time to our knowledge, that ChREBP could destabilize SREBP2-N by promoting its ubiquitination. This inhibitory action of ChREBP could play a crucial role in hepatoprotection in response to fructose influx.
Discussion
Given that a fructose-rich diet has been widely adopted in many countries and that it is closely associated with metabolic disorders, it is crucial to understand the molecular basis of the metabolic responses to fructose at the cellular and tissue levels. Here, for the first time to our knowledge, we uncovered a ChREBP transcription factor–mediated adaptive pathway that protects the liver from HFrD-induced hepatocyte apoptosis and injury. In the absence of ChREBP, mice on a HFrD develop severe liver injury due to overactivation of ER stress and CHOP-mediated hepatocyte apoptosis. Administration of the chemical chaperone 4-PBA or acute depletion of Chop mitigates liver injury in HFrD-fed Chrebp–/– mice. Increased cholesterol biosynthesis probably contributes to hepatocyte apoptosis in fructose-challenged Chrebp–/– mice, since inhibition of cholesterol biosynthesis by the HMGCR inhibitor or Srebp2 knockdown rescues Chrebp–/– mice from HFrD-induced liver injury (Figure 6K).
Fructose has become one of the most abundant sources of dietary carbohydrates in the United States (37). The liver is the primary site of fructose uptake and metabolism (37). Although it has been observed that a fructose-rich diet is closely linked to liver steatosis (1), how chronic fructose feeding reprograms hepatic metabolism and leads to liver steatosis remains elusive. In hepatocytes, ChREBP and SREBP1c are the major players driving the transcription of lipogenic genes (38, 39). Our data show that HFrD feeding selectively induces ChREBP protein instead of SREBP1c protein in the mouse liver, although the mRNA levels of both of these proteins are elevated. In the Chrebp–/– mouse liver, we detected lower levels of Fasn and other lipogenic genes, indicating that there is a lack of compensation in the activity of SREBP1c. It will be of great interest to investigate how hepatocytes activate specific lipogenic programs in response to different sources of nutrient inputs.
The unexpected finding from our study is that ChREBP is required to protect hepatocytes from apoptosis and preserve liver function in response to fructose influx. What are the causes of hepatocyte apoptosis in the Chrebp–/– mice upon HFrD feeding? It has been reported that adaptive and transient hepatic ER stress alleviates misfolded protein stress without triggering hepatocyte death, whereas prolonged ER stress activates the expression of proapoptotic proteins, inhibits prosurvival factors, and eventually triggers apoptosis via the PERK/CHOP pathway (12, 40). Our data provide evidence that dysregulation of hepatic ER stress is the major culprit of hepatocyte death in fructose-fed Chrebp–/– mice. Fructose feeding increases the expression of GRP78, IRE1, and ATF6, without hepatocyte death in WT mice, indicative of the induction of adaptive ER stress. However, in the case of Chrebp–/– mice, fructose feeding activates the proapoptotic PERK-CHOP branch and the CHOP target PUMA, while suppressing the genes in the adaptive branch in the mouse liver. We suspect, in certain cases of human NAFLD, that the impaired ChREBP pathway may be one of the underlying causes of the progression from simple steatosis to NASH.
We further showed that the chemical chaperone 4-BPA alleviates liver injury and reduces apoptosis. Acute depletion of Chop also reverses the liver injury phenotype in Chrebp–/– mice after HFrD feeding. These data highlight the importance of the proper control of UPR and ER stress activity in hepatocytes in the presence of fructose influx and highlight the essential role of ChREBP in maintaining normal liver functions by suppressing the PERK-CHOP branch of the UPR. Recent work from the Postic group demonstrated that ChREBP is important for protecting the liver by regulating ethanol metabolism via sirtuin 1 (SIRT1) (11). It will be interesting to examine the role of the ChREBP-controlled UPR pathway in alcohol-induced liver injury models.
Several reports have documented free cholesterol accumulation in the livers of patients with NAFLD (16, 18). Lipidomic analysis revealed a positive correlation between free cholesterol accumulation in the livers of NAFLD patients and the histological severity of NASH (18). Induction of hepatic free cholesterol accumulation has been shown to promote steatohepatitis and liver fibrosis, whereas the reduction of hepatic free cholesterol alleviates liver injury in NASH (41). Our current study supports the notion that free cholesterol accumulation is a major cause of the proapoptotic ER stress response and hepatocyte apoptosis in Chrebp–/– mouse livers after HFrD feeding. The rate-limiting enzyme HMGCR and its regulator SREBP2 are drastically elevated in Chrebp–/– liver at both the mRNA and protein levels. Treatment with the HMGCR inhibitor atorvastatin or depletion of Srebp2 in the liver reduces cholesterol synthesis and rescues liver injury in HFrD-fed Chrebp–/– mice. Thus, for the first time to our knowledge, we provide data supporting the idea that ChREBP protects against HFrD-induced liver injury, a protection that likely occurs through the repression of cholesterol biosynthesis and free cholesterol accumulation.
We postulate that there are 2 potential mechanisms by which ChREBP suppresses liver cholesterol biosynthesis during fructose diet feeding. One possibility is that ChREBP directly blocks the transcription of enzymes of cholesterol metabolism. We speculate that this mechanism might be specific for HFrD feeding, since we did not observe altered HMGCR expression in Chrebp–/– mice fed a regular chow diet. Alternatively, ChREBP might activate the expression of miR species that specifically target cholesterol metabolism. Several liver-rich miRs, including miR-21 (42, 43), miR-33 (44), and miR-34a (16), have been shown to suppress HMGCR expression. Our microarray data showed that several miR species were selectively downregulated in HFrD-fed Chrebp–/– mouse livers. Future studies will use ChIP-sequencing (ChIP-seq) with an anti-ChREBP antibody to uncover how HFrD feeding affects the genomic footprints of ChREBP on target genes involved in cholesterol metabolism.
Our data also point to another possible mechanism by which ChREBP controls cholesterol biosynthesis via the destabilization of SREBP2 protein during HFrD feeding. In the liver, ChREBP deficiency resulted in highly elevated SREBP2-N, which can be reversed by restoring ChREBP expression. Despite substantial efforts made to understand how SREBP2-FL is processed and matured during cholesterol metabolism, little is known regarding the fate of matured SREBP2-N. The Ericson group suggested that SREBP2 is targeted for ubiquitination and proteasome degradation by GSK3β and the E3 ligase FBW7(36, 45). Other studies suggest that the NAD-dependent deacetylases SIRT1 and SIRT6 could potentially promote SREBP2 protein turnover (46, 47). In our studies, we unexpectedly found that ChREBP could be a potent factor promoting ubiquitination and destabilization of the mature SREBP2 protein. The interaction between ChREBP and SREBP2-N was detected in cultured cells overexpressing both proteins, suggesting that such an interaction might be required for ChREBP to promote ubiquitination of SREBP2-N. Our future work will systematically map the region of ChREBP that is critical for its interaction with SREBP2-N and identify ChREBP mutants that fail to interact with SREBP2-N. We will test the functional consequences of such mutants on SREBP-N degradation and SREBP2-driven cholesterol biosynthesis in the mouse liver following HFrD feeding. Because of the nature of ChREBP as a transcription factor, it remains to be addressed whether such an interaction is sufficient to drive ubiquitination-dependent degradation of SREBP2-N. We speculate that, upon fructose diet feeding, there could be the possible formation of a novel ChREBP-interacting protein network that contains specific ubiquitin E3 ligases. Identification of such a ChREBP-specific protein network in our future work would shed light on how hepatocytes cope with fructose influx by degrading SREBP2-N protein.
Our current study provides a classical example of gene and nutrition interaction as a survival mechanism throughout human evolution. This ChREBP-mediated adaptive mechanism allows animals to efficiently store energy from fructose, which was rare in ancient diets. It is conceivable that, after consuming large amounts of fructose, humans might develop liver steatosis without severe liver injury caused by a highly activated ChREBP pathway. However, NAFLD patients with genetic defects in Chrebp might undergo an accelerated transition from simple liver steatosis to NASH. Our findings suggest that the treatment of these patients with statins might curb the progression of NAFLD and restore liver function. Our findings also underscore the need for an in-depth understanding of the regulation and action of the ChREBP pathway upon fructose diet feeding.
Methods
Reagents.
Filipin (catalog F-9765) and 4-BPA (catalog P21005) were purchased from Sigma-Aldrich. Atorvastatin was obtained from Cayman Chemical (catalog 10493). The high-fructose diet was purchased from Research Diets, and the ingredients are included in the Supplemental Experimental Procedures.
Animal experiments.
All animal experiments were approved by the Institutional Animal Care and Research Advisory Committee of the University of Michigan. C57BL/6 mice were maintained on a 12-hour light/12-hour dark cycle with ad libitum access to food and water. Chrebp–/– mice were provided by Jiandie Lin of the University of Michigan. For food intake, mice were kept in separate cages, and their food intake was measured for 3 successive days.
For liver-specific knockdown or overexpression, Ad-shLacZ versus Ad-shChop, Ad-GFP versus Ad-Flag-Chrebp, or Ad-Grp78 adenoviruses were delivered via tail-vein injection at a dose of 1 × 1012 PFU. 4-PBA (Sigma-Aldrich) was dissolved in Milli-Q water, titrated with 5 M NaOH, and given by oral gavage to 8-week-old mice (1 g/kg BW/day). Atorvastatin (Cayman Chemical) was dissolved in 0.1% sodium carboxymethyl cellulose solution and administered to mice by oral gavage (20 mg/kg BW/day), with CMC-Na solution as a control. Mice were gavaged with 4-PBA or atorvastatin for 10 days and fed a HFrD starting on the third day, until dissection. Liver tissues were harvested for mRNA and protein analysis. Frozen sections were used for TUNEL staining and Oil Red O staining. Paraffin-embedded tissue was used for section and H&E staining. After H&E staining, the slides were transferred to GFB, with only the mouse number indicated but no genotype or treatment information, for liver injury scoring.
Primary mouse hepatocyte isolation and culture.
Primary mouse hepatocytes (PMHs) were isolated by digesting mice liver with type I collagenase. Detailed protocols are available in the Supplemental Experimental Procedures.
Immunoblotting.
Whole-cell lysates of hepatocytes or frozen liver were prepared in a modified RIPA buffer. The cytosolic and nuclear fraction was separated with the hypotonic buffer, and nuclei were suspended with radio immunoprecipitation assay (RIPA) buffer to prepare nuclear protein. Detailed procedures and antibody information are available in the Supplemental Experimental Procedures. The complete, unedited blots are provided in the supplemental material.
cDNA synthesis and qPCR.
Total cellular RNA extraction was performed using TRIzol Reagent (Invitrogen, Thermo Fisher Scientific) and chloroform. cDNA was synthesized and subjected to qPCR using the SYBR Green dye-based assay. qPCR primer sequences are listed in the Supplemental Experimental Procedures.
Microarray.
Total RNA was extracted from the livers of WT mice fed a chow diet (n = 7) or a HFrD (n = 7), or from the livers of Chrebp–/– mice (n = 6) or their WT littermates fed a HFrD (n = 4). Total RNA from the livers of mice of the same genotype and treatment was pooled. A microarray assay was performed in the DNA Sequencing Core at the University of Michigan using Mouse Gene ST 2.1 Strip of Affymetrix. The original data have been deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE 96820; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE96820).
De novo lipogenesis assay.
De novo lipogenesis rates in primary hepatocytes were assessed by measuring the radioactivity of newly synthesized lipids from H3-labeled substrate. Detailed procedures are available in the Supplemental Experimental Procedures.
Serum assays.
Serum ALT and cholesterol assays were performed with kits from Pointe Scientific (A7525 and A7510, respectively), according to the manufacturer’s instruction.
Liver triglyceride and cholesterol measurements.
Detailed liver triglyceride and cholesterol assays are available in the Supplemental Experimental Procedures.
TUNEL staining.
TUNEL staining was performed with an in situ cell death detection kit from Roche. Briefly, frozen sections of liver were fixed with 4% paraformaldehyde, permeabilized with the buffer containing 0.5% Triton X-100 and 0.05% SDS, and then incubated with reaction buffer. The slides were then counterstained with DAPI and visualized under a microscope. TUNEL-positive cells were quantified in ×200-magnified fields of 3 mice livers from each group and normalized to DAPI staining, with numbers counted using ImageJ software (NIH).
Statistics.
Statistical analysis was performed using GraphPad Prism version 6.0 (GraphPad Software). Statistical significance was determined either by unpaired 2-tailed Student’s t test for comparisons between 2 groups or by 1-way ANOVA with Tukey’s or Dunnett’s post-hoc test for multiple group comparisons. All results are given as the mean ± SEM. Results were considered statistically significant with a P value of less than 0.05.
Study approval.
All animal experiments were approved by the Institutional Animal Care and Research Advisory Committee of the University of Michigan. All animal care and use was in accordance with IACUC guidelines of the University of Michigan.
Author contributions
LY and DZ conceived the project and designed the research. DZ, XT, and KV performed the studies. NG and KS helped with Srebp2 overexpression and knockdown constructs and adenovirus concentrations. GFB performed H&E staining scoring and provided input regarding the assessment of liver injury. LY, DZ, MBO, LR, and JL analyzed and interpreted the data and reviewed and made edits to the manuscript. LY and DZ wrote the first draft of the manuscript and finalized it. ZM helped with generating the Chrebp-KO mice.
Supplementary Material
Acknowledgments
We thank Howard C. Towle (University of Minnesota, Minneapolis, Minnesota, USA) for providing the pCMV-Flag-Chrebp expression vector and Richard Austin (McMaster University, Hamilton, Ontario, Canada) for providing the Ad-Grp78 virus. This work was funded by NIH grants DK099593 (to LY) and DK52951 (to MBO).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(7):2855–2867.https://doi.org/10.1172/JCI89934.
See the related Commentary at ChREBP refines the hepatic response to fructose to protect the liver from injury.
Contributor Information
Xin Tong, Email: xintong@umich.edu.
Kyle VanDommelen, Email: kvando@umich.edu.
Neil Gupta, Email: ngupes@umich.edu.
Kenneth Stamper, Email: ktstamp@umich.edu.
Lei Yin, Email: leiyin@umich.edu.
References
- 1.Nomura K, Yamanouchi T. The role of fructose-enriched diets in mechanisms of nonalcoholic fatty liver disease. J Nutr Biochem. 2012;23(3):203–208. doi: 10.1016/j.jnutbio.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Tiniakos DG, Vos MB, Brunt EM. Nonalcoholic fatty liver disease: pathology and pathogenesis. Annu Rev Pathol. 2010;5:145–171. doi: 10.1146/annurev-pathol-121808-102132. [DOI] [PubMed] [Google Scholar]
- 3.Neuschwander-Tetri BA. Carbohydrate intake and nonalcoholic fatty liver disease. Curr Opin Clin Nutr Metab Care. 2013;16(4):446–452. doi: 10.1097/MCO.0b013e328361c4d1. [DOI] [PubMed] [Google Scholar]
- 4.Dekker MJ, Su Q, Baker C, Rutledge AC, Adeli K. Fructose: a highly lipogenic nutrient implicated in insulin resistance, hepatic steatosis, and the metabolic syndrome. Am J Physiol Endocrinol Metab. 2010;299(5):E685–E694. doi: 10.1152/ajpendo.00283.2010. [DOI] [PubMed] [Google Scholar]
- 5.Ren LP, et al. Differing endoplasmic reticulum stress response to excess lipogenesis versus lipid oversupply in relation to hepatic steatosis and insulin resistance. PLoS ONE. 2012;7(2):e30816. doi: 10.1371/journal.pone.0030816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci USA. 2004;101(19):7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamashita H, et al. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc Natl Acad Sci USA. 2001;98(16):9116–9121. doi: 10.1073/pnas.161284298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benhamed F, et al. The lipogenic transcription factor ChREBP dissociates hepatic steatosis from insulin resistance in mice and humans. J Clin Invest. 2012;122(6):2176–2194. doi: 10.1172/JCI41636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dentin R, et al. Liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in ob/ob mice. Diabetes. 2006;55(8):2159–2170. doi: 10.2337/db06-0200. [DOI] [PubMed] [Google Scholar]
- 10.Iizuka K, Miller B, Uyeda K. Deficiency of carbohydrate-activated transcription factor ChREBP prevents obesity and improves plasma glucose control in leptin-deficient (ob/ob) mice. Am J Physiol Endocrinol Metab. 2006;291(2):E358–E364. doi: 10.1152/ajpendo.00027.2006. [DOI] [PubMed] [Google Scholar]
- 11.Marmier S, et al. Novel role for carbohydrate responsive element binding protein in the control of ethanol metabolism and susceptibility to binge drinking. Hepatology. 2015;62(4):1086–1100. doi: 10.1002/hep.27778. [DOI] [PubMed] [Google Scholar]
- 12.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 13.Lee JS, Zheng Z, Mendez R, Ha SW, Xie Y, Zhang K. Pharmacologic ER stress induces non-alcoholic steatohepatitis in an animal model. Toxicol Lett. 2012;211(1):29–38. doi: 10.1016/j.toxlet.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstein JL, Brown MS. A century of cholesterol and coronaries: from plaques to genes to statins. Cell. 2015;161(1):161–172. doi: 10.1016/j.cell.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musso G, Gambino R, Cassader M. Cholesterol metabolism and the pathogenesis of non-alcoholic steatohepatitis. Prog Lipid Res. 2013;52(1):175–191. doi: 10.1016/j.plipres.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Min HK, et al. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 2012;15(5):665–674. doi: 10.1016/j.cmet.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caballero F, Fernández A, De Lacy AM, Fernández-Checa JC, Caballería J, García-Ruiz C. Enhanced free cholesterol, SREBP-2 and StAR expression in human NASH. J Hepatol. 2009;50(4):789–796. doi: 10.1016/j.jhep.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 18.Puri P, et al. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 2007;46(4):1081–1090. doi: 10.1002/hep.21763. [DOI] [PubMed] [Google Scholar]
- 19.Marí M, et al. Mitochondrial free cholesterol loading sensitizes to TNF- and Fas-mediated steatohepatitis. Cell Metab. 2006;4(3):185–198. doi: 10.1016/j.cmet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Van Rooyen DM, et al. Hepatic free cholesterol accumulates in obese, diabetic mice and causes nonalcoholic steatohepatitis. Gastroenterology. 2011;141(4):1393–1403. doi: 10.1053/j.gastro.2011.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawano Y, Cohen DE. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J Gastroenterol. 2013;48(4):434–441. doi: 10.1007/s00535-013-0758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bricambert J, Miranda J, Benhamed F, Girard J, Postic C, Dentin R. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J Clin Invest. 2010;120(12):4316–4331. doi: 10.1172/JCI41624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dushay J, et al. Increased fibroblast growth factor 21 in obesity and nonalcoholic fatty liver disease. Gastroenterology. 2010;139(2):456–463. doi: 10.1053/j.gastro.2010.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, et al. Fibroblast growth factor 21 levels are increased in nonalcoholic fatty liver disease patients and are correlated with hepatic triglyceride. J Hepatol. 2010;53(5):934–940. doi: 10.1016/j.jhep.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 25.Kim H, et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol. 2006;8(12):1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 26.Jeffers JR, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4(4):321–328. doi: 10.1016/S1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 27.Liu ZQ, et al. Expression of PUMA in follicular granulosa cells regulated by FoxO1 activation during oxidative stress. Reprod Sci. 2015;22(6):696–705. doi: 10.1177/1933719114556483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaplowitz N, Than TA, Shinohara M, Ji C. Endoplasmic reticulum stress and liver injury. Semin Liver Dis. 2007;27(4):367–377. doi: 10.1055/s-2007-991513. [DOI] [PubMed] [Google Scholar]
- 29.Ozcan L, Tabas I. Pivotal role of calcium/calmodulin-dependent protein kinase II in ER stress-induced apoptosis. Cell Cycle. 2010;9(2):223–224. doi: 10.4161/cc.9.2.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Timmins JM, et al. Calcium/calmodulin-dependent protein kinase II links ER stress with Fas and mitochondrial apoptosis pathways. J Clin Invest. 2009;119(10):2925–2941. doi: 10.1172/JCI38857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vilatoba M, et al. Sodium 4-phenylbutyrate protects against liver ischemia reperfusion injury by inhibition of endoplasmic reticulum-stress mediated apoptosis. Surgery. 2005;138(2):342–351. doi: 10.1016/j.surg.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 32.Qi X, Hosoi T, Okuma Y, Kaneko M, Nomura Y. Sodium 4-phenylbutyrate protects against cerebral ischemic injury. Mol Pharmacol. 2004;66(4):899–908. doi: 10.1124/mol.104.001339. [DOI] [PubMed] [Google Scholar]
- 33.Uzi D, et al. CHOP is a critical regulator of acetaminophen-induced hepatotoxicity. J Hepatol. 2013;59(3):495–503. doi: 10.1016/j.jhep.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 34.Endo M, Oyadomari S, Suga M, Mori M, Gotoh T. The ER stress pathway involving CHOP is activated in the lungs of LPS-treated mice. J Biochem. 2005;138(4):501–507. doi: 10.1093/jb/mvi143. [DOI] [PubMed] [Google Scholar]
- 35.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109(9):1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bengoechea-Alonso MT, Ericsson J. SREBP in signal transduction: cholesterol metabolism and beyond. Curr Opin Cell Biol. 2007;19(2):215–222. doi: 10.1016/j.ceb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Vos MB, Lavine JE. Dietary fructose in nonalcoholic fatty liver disease. Hepatology. 2013;57(6):2525–2531. doi: 10.1002/hep.26299. [DOI] [PubMed] [Google Scholar]
- 38.Rui L. Energy metabolism in the liver. Compr Physiol. 2014;4(1):177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang D, Yin L. Transcriptional Regulation of De Novo Lipogenesis in Liver. In: Ntambi, JM, ed. Hepatic De Novo Lipogenesis and Regulation of Metabolism. New York, NY; Springer; 2016:1–31. [Google Scholar]
- 40.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13(3):184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Rooyen DM, et al. Pharmacological cholesterol lowering reverses fibrotic NASH in obese, diabetic mice with metabolic syndrome. J Hepatol. 2013;59(1):144–152. doi: 10.1016/j.jhep.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 42.Sun C, et al. miR-21 regulates triglyceride and cholesterol metabolism in non-alcoholic fatty liver disease by targeting HMGCR. Int J Mol Med. 2015;35(3):847–853. doi: 10.3892/ijmm.2015.2076. [DOI] [PubMed] [Google Scholar]
- 43.Larsen L, et al. Expression and localization of microRNAs in perinatal rat pancreas: role of miR-21 in regulation of cholesterol metabolism. PLoS ONE. 2011;6(10):e25997. doi: 10.1371/journal.pone.0025997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rayner KJ, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328(5985):1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sundqvist A, et al. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell Metab. 2005;1(6):379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 46.Tao R, Xiong X, DePinho RA, Deng CX, Dong XC. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J Lipid Res. 2013;54(10):2745–2753. doi: 10.1194/jlr.M039339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker AK, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24(13):1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.