Abstract
TGF-β is considered a master switch in the pathogenesis of organ fibrosis. The primary mediators of this activity are the SMAD proteins, particularly SMAD3. In the current study, we have developed a cell-penetrating peptide (CPP) conjugate of the HIV TAT protein that is fused to an aminoterminal sequence of sorting nexin 9 (SNX9), which was previously shown to bind phosphorylated SMAD3 (pSMAD3). We determined that specifically preventing the nuclear import of pSMAD3 using the TAT-SNX9 peptide inhibited profibrotic TGF-β activity in murine cells and human lung fibroblasts as well as in vivo with no demonstrable toxicity. TGF-β signaling mediated by pSMAD2, bone morphogenetic protein 4 (BMP4), EGF, or PDGF was unaffected by the TAT-SNX9 peptide. Furthermore, while the TAT-SNX9 peptide prevented TGF-β’s profibrotic activity in vitro as well as in 2 murine treatment models of pulmonary fibrosis, a 3–amino acid point mutant that was unable to bind pSMAD3 proved ineffective. These findings indicate that specifically targeting pSMAD3 can ameliorate both the direct and indirect fibroproliferative actions of TGF-β.
Keywords: Cell Biology
Introduction
Normal cellular proliferation is a complex process that requires the coordinated integration of both stimulatory and inhibitory growth factors. TGF-β, a 25-kDa homodimer prototypic of a family of proteins that regulate cell growth and differentiation, is unique in this regard since it is capable of both stimulating and inhibiting cell growth (1–3). The response of cultured cells to TGF-β is divergent; mesenchymal cells are routinely growth stimulated by TGF-β, while the majority of other cell types are inhibited. Although there is some evidence for the activation of distinct signaling pathways in mesenchymal versus epithelial cultures (4–6), the mechanism or mechanisms regulating this response are relatively unknown (7–9).
The majority of cell types express 3 TGF-β–binding species referred to as type 1 (TβRI), type 2 (TβRII), and type 3 (TβRIII) receptors (10, 11). Ligand binding to TβRII results in complex formation with TβRI, activation of TβRI Ser/Thr kinase activity, and subsequent downstream signaling (12, 13). The primary mediators of TGF-β action are the SMAD proteins (14–16) SMAD2 and SMAD3, although non-SMAD responses are often observed (17, 18). Once phosphorylated by TβRI, they most often form a heterotrimer consisting of 2 molecules of either SMAD2 or SMAD3 complexed with SMAD4 prior to undergoing nuclear translocation, where they function as comodulators of gene expression. Recently, we have extended this model and provide evidence that sorting nexin 9 (SNX9), a member of the PX/BAR subfamily of intracellular trafficking proteins (19–23), has an obligate role in mediating phosphorylated SMAD3 (pSMAD3) nuclear import following ligand treatment (24).
The sorting nexins consist of a large family of proteins that regulate a variety of intracellular trafficking activities (19, 21, 25). We have recently determined that SNX9 (domain structure provided in Figure 1A) binds to pSMAD3 and, together with importin 8 and importin β, promotes pSMAD3 (but not pSMAD2) nuclear import, soft agar colony formation, migration, and the induction of pSMAD3 target genes (24). Although these actions are dependent upon SNX9’s ability to bind phosphoinositides, they are independent of SNX9 homodimerization, differentiating canonical SNX9 function from this recently defined activity in regulating TGF-β signaling (24, 26, 27).
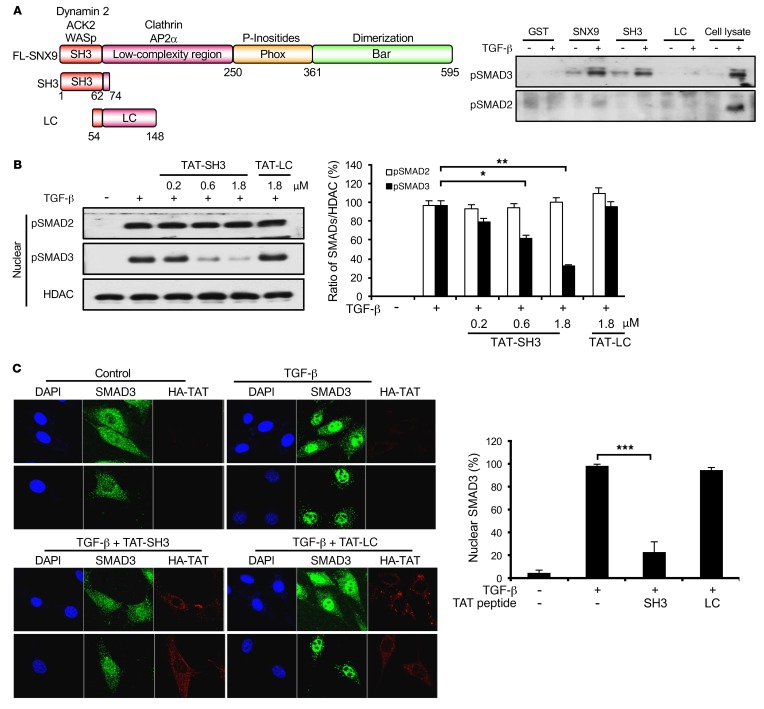
Figure 1. SH3 domain of SNX9 specifically binds pSMAD3 and prevents nuclear import.
(A) Schematic depicting domains in FL SNX9 and constructs used for GST pull-down assays (left). Lysates from AKR-2B cells untreated (–) or stimulated (+) for 45 minutes with 5 ng/ml TGF-β were incubated with GST beads or the indicated fusion proteins immobilized on GST beads. Bound proteins were eluted and assessed by Western blot analysis for pSMAD3 or pSMAD2. Cell lysate reflects signal obtained from 10 μg total protein; the slower migrating band in the pSMAD3 lane is a nonspecific protein (right, representative of 3 separate experiments). (B) AKR-2B cells were transduced for 90 minutes with the indicated concentration of TAT peptide. Following washing and 1 hour TGF-β (5 ng/ml) treatment, nuclear fractions were isolated and assessed by Western blot analysis for pSMAD2, pSMAD3, or histone deacetylase 1 (HDAC) (left, representative of 3 separate experiments). Quantitation of nuclear pSMADs was performed with ImageJ software (NIH) and represents the mean ± SEM of 3 experiments (right). (C) Left panels: AKR-2B cells were incubated with vehicle (top panels) or transduced (bottom panels) as in B with TAT-SH3 or TAT-LC (1.8 μM). Following treatment with or without TGF-β (5 ng/ml) for 1 hour, immunofluorescence for SMAD3 or the HA-tagged TAT peptide was performed as described in Methods and nuclei were stained with DAPI. Upper and lower panels show 2 distinct microscopic fields for each condition. Original magnification in C: ×100. Right: quantitation of nuclear SMAD3 from 30 cells in each of 3 experiments. *P < 0.05; **P < 0.005; ***P < 0.0005, 1-way ANOVA followed by Dunnett’s multiple comparisons test.
Given that TGF-β has a fundamental role in normal tissue homeostasis/development as well as organ fibrosis and tumor development/progression (28–30), it provides an opportune target to affect these actions and/or pathologies. Most of these approaches, however, have targeted either TGF-β receptor activity or the TGF-β ligand directly (31–33). While both will clearly affect the cellular and/or organismal response to TGF-β, neither provides any discriminatory ability, as all TGF-β actions are affected. This concern is best exemplified in kidney fibrosis and skin cancer, where it has been documented that, while SMAD3 is profibrotic/procarcinogenic, SMAD2 is antifibrotic/anticarcinogenic (34, 35). Thus, for some conditions, the aforementioned global approaches may be unwarranted and a strategy that specifically targets the causal pathway or pathways more efficacious. In the current study, we do just that by using a cell-penetrating peptide (CPP) (36–38) consisting of the HIV TAT protein fused to an N-terminal 31–amino acid SNX9 fragment found to bind and specifically inhibit pSMAD3 nuclear import and biologic action. The results show that, while this TAT-SNX9 peptide had no effect on TGF-β signaling mediated via pSMAD2 or luciferase activity stimulated by bone morphogenetic protein 4 (BMP4), EGF, or PDGF, it inhibited (a) TGF-β induction of pSMAD3 target genes, soft agar colony formation, and cell migration; and (b) fibrotic changes associated with the bleomycin (BLM) and adenoviral models of lung fibrosis. Moreover, a 3–amino acid point mutant of the TAT-SNX9 peptide unable to bind pSMAD3 was ineffective in analogous in vitro and in vivo assays.
Results
A TAT-SNX9 peptide specifically blocks pSMAD3 nuclear import and profibrotic TGF-β signaling.
We previously determined that SNX9 has an obligate role in mediating profibrotic TGF-β signaling dependent upon SMAD3 (24). In order to investigate whether pSMAD3 bound to a defined region or regions in SNX9, glutathione S-transferases (GST) fusion constructs encoding either the amino (e.g., SH3 and low-complexity [LC] domains) or carboxyl (e.g., Phox and BAR domains) half of SNX9 were generated and pull-down assays for pSMAD3 performed. While elements within the Phox and BAR domains were unable to bind pSMAD3, equivalent pSMAD3 binding was observed with GST–full-length (GST-FL) SNX9 and the amino terminal fragment (data not shown). To define this interaction further, we generated 2 overlapping amino terminal fragments. As shown in Figure 1A, while equivalent pSMAD3 binding was observed with constructs expressing FL SNX9 or the first 74 amino acids encoding the SH3 domain, the LC domain showed only minimal binding. Of note, no pSMAD2 association was observed for any of the constructs, nor did FL SNX9 bind BMP4-stimulated pSMAD1/5/8 (Figure 1A and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI88696DS1).
SNX9 was previously shown to be required for the nuclear import of pSMAD3, but not pSMAD2 (24). Since the SNX9 SH3 domain was capable of binding pSMAD3 similarly to the FL protein (Figure 1A), we next determined whether expression of this fragment could act in trans as a dominant inhibitor of pSMAD3 nuclear uptake. To address this issue, constructs were prepared expressing either the SNX9 SH3 or LC domains fused to the cell-penetrating TAT peptide from HIV (39). Subsequent to TAT peptide transduction, cultures were treated with TGF-β and nuclear accumulation of pSMAD2 or pSMAD3 determined. As shown in Figure 1B and Supplemental Figure 2, while the TAT-SH3 peptide inhibited nuclear import of pSMAD3 in a dose-dependent manner and increased cytoplasmic pSMAD3, it had no effect on pSMAD2. Furthermore, consistent with the inability of the LC domain to bind receptor-regulated SMADs (R-SMADs) (Figure 1A), it was similarly ineffective in modulating nuclear translocation (Figure 1B). These biochemical findings were independently confirmed using immunofluorescence (Figure 1C and Supplemental Figure 3).
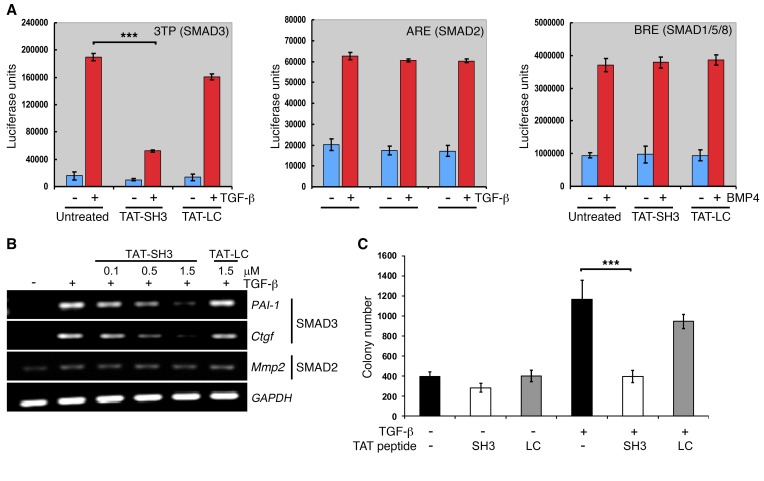
While the results of Figure 1 and Supplemental Figure 2 show that TAT-SH3 peptides are capable of inhibiting the nuclear accumulation of pSMAD3 following TGF-β addition, they do not address the functional impact of this response. To investigate whether this loss was sufficient to inhibit SMAD3-mediated responses, the studies shown in Figure 2 were performed. AKR-2B cells were transfected with luciferase constructs responsive to either SMAD3, SMAD2, or BMPs and the impact of inhibiting pSMAD3 nuclear import by TAT-SH3 determined. While SMAD3-dependent luciferase activity was inhibited by approximately 70%, no discernible effect on either SMAD2 or SMAD1/5/8 (e.g., BMP) signaling was observed (Figure 2A). These luciferase results were extended both transcriptionally and biologically, as shown in Figure 2, B and C, respectively. While SMAD3 targets and TGF-β–stimulated anchorage-independent growth in soft agar (anchorage-independent cell growth [AIG]) were inhibited by transduction with TAT-SH3, induction of the SMAD2-regulated matrix metalloproteinase 2 (Mmp2) gene was unaffected and, as expected, the negative control TAT-LC peptide was inert for all responses.
Figure 2. TAT-SH3 inhibits SMAD3-dependent responses.
(A) AKR-2B cells were transiently transfected with a SMAD3 (3TP), SMAD2 (ARE), or SMAD1/5/8 (BRE) reporter construct and luciferase activity determined following 12-hour incubation in the absence (–) or presence (+) of the indicated ligand (5 ng/ml TGF-β; 10 ng/ml BMP4) or TAT peptide (1.5 μM). Data represent the mean ± SEM of duplicate samples from 3 separate experiments. (B) RT-PCR analysis of SMAD3-responsive (PAI-1 and Ctgf) and SMAD2-responsive (Mmp2) genes following 24-hour treatment of AKR-2B cells with the indicated concentration of TAT peptide with or without 5 ng/ml TGF-β. Data are representative of 3 separate experiments. (C) Soft agar colony formation was performed as described in Methods following 7 days growth in the absence (–) or presence (+) of TGF-β (20 ng/ml) or the indicated TAT peptide (1.5 μM). Data reflect the mean ± SEM of triplicate wells from 3 experiments. ***P < 0.0005, 1-way ANOVA followed by Dunnett’s multiple comparisons test.
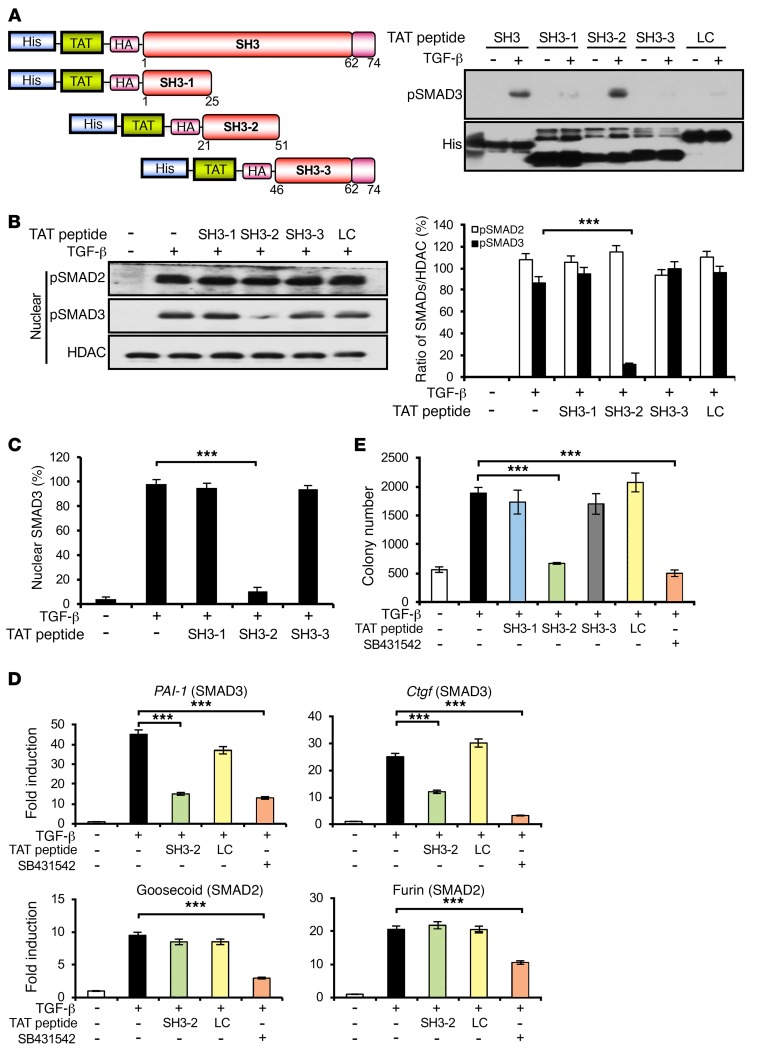
The preceding figures show that expression of the SH3 domain from SNX9 can function in trans as a specific inhibitor of SMAD3-regulated responses. We extended these findings by, first, further defining the functional motif or motifs in the SH3 peptide regulating SMAD3 signaling and, second, generating and testing, both in vitro and in vivo, a mutant SH3 peptide unable to bind pSMAD3. To address the first of these issues, 3 overlapping 25–31 mer TAT-SH3 peptides were constructed and tested for their ability to bind pSMAD3 in cell lysates prepared from TGF-β–treated cultures. As shown in Figure 3, A–C, and Supplemental Figure 4, TAT–SH3-2 (encoding SNX9 amino acids 21–51) specifically bound pSMAD3 to a degree similar to that of the FL SH3 peptide, prevented pSMAD3 nuclear import, and increased cytoplasmic pSMAD3. Moreover, TAT–SH3-2 not only decreased SMAD3-dependent transcriptional and biological responses, but the inhibition was shown to be dose dependent (Supplemental Figure 5) and analogous to that observed with the TβRI kinase inhibitor SB431542 (Figure 3, D and E).
Figure 3. Inhibition of SMAD3 signaling by a defined region of the SH3 domain in SNX9.
(A) Schematic depicting constructs used for His pull-down assays (left). AKR-2B lysates were prepared as in Figure 1, incubated with the indicated TAT peptide immobilized on TALON Metal Affinity Resin, and immunoblotted for bound pSMAD3 or total TAT peptide (His) (right, representative of 3 separate experiments). (B) Nuclear fractions were prepared and assessed as in Figure 1B following transduction with the indicated TAT peptides (left, representative of 3 separate experiments). Quantitation (mean ± SEM) of nuclear pSMAD2 or pSMAD3 from 3 experiments (right). (C) Immunofluorescence of nuclear SMAD3 was determined as in Figure 1C from 30 cells in each of 3 experiments in the absence (–) or presence (+) of 5 ng/ml TGF-β and the indicated TAT peptide (1.5 μM). (D) qPCR of SMAD3-responsive (PAI-1 and Ctgf) and SMAD2-responsive (goosecoid and furin) genes following 24 hours in the absence (–) or presence (+) of TGF-β (5 ng/ml), SB431542 (10 μM; TβRI inhibitor), or the indicated TAT peptide. Data reflect mean ± SEM from 3 experiments. (E) TGF-β–stimulated (20 ng/ml) soft agar colony formation as in Figure 2C in the absence or presence of the indicated TAT peptide (1.5 μM) or SB431542 (10 μM). Data reflect the mean ± SEM of triplicate wells from 3 experiments. ***P < 0.0005, 1-way ANOVA followed by Dunnett’s multiple comparisons test.
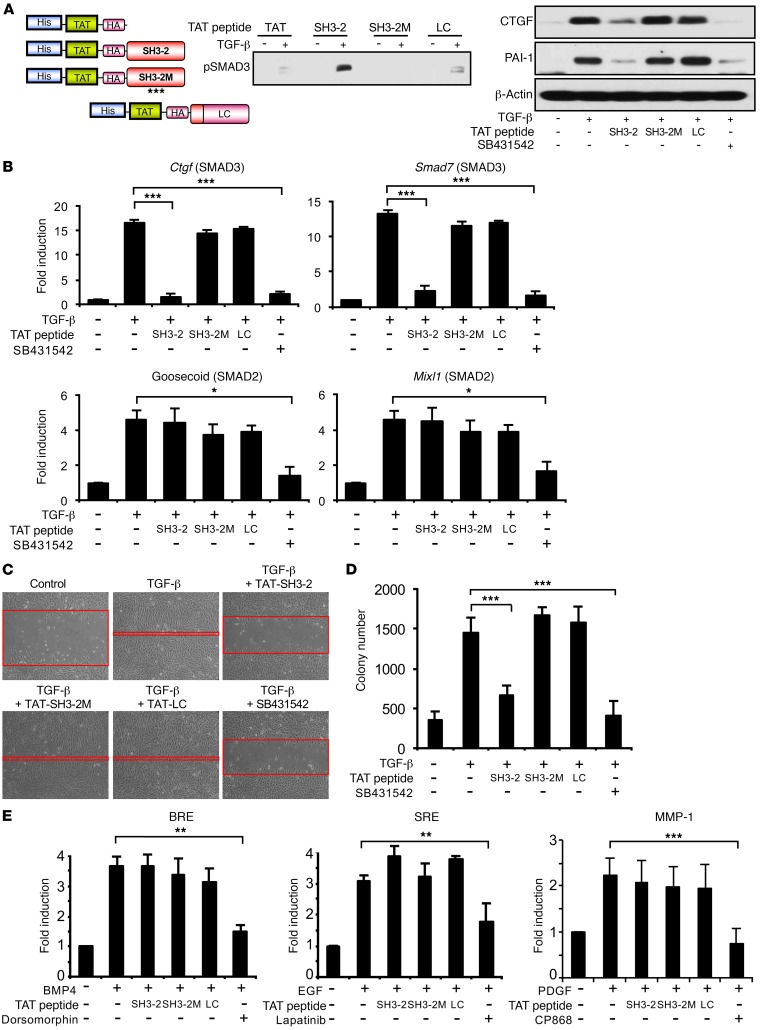
To define the element further, 3 additional overlapping 15- or 16-mer TAT–SH3-2 peptides were generated and were assessed for whether they could inhibit pSMAD3 nuclear translocation. While activity similar to that obtained with SB431542 was observed with the TAT peptide encoding amino acids 27–42 (data not shown), additional studies revealed that the 16 mer did not provide as consistent responses as the 31 mer encoded by TAT–SH3-2. These studies did, however, suggest that point mutations in a highly conserved glycine-rich region previously shown to mediate protein/protein interactions (40–42) might similarly be effective in blocking the inhibitory actions of TAT–SH3-2. This was directly tested, as shown in Figure 4. While TAT–SH3-2, as expected (Figure 3), bound pSMAD3 and prevented TGF-β induction of SMAD3 targets, such as connective tissue growth factor (Ctgf), plasminogen activator inhibitor 1 (PAI-1), and Smad7, to a degree similar to that of SB431542 inhibition of TβRI, point mutations in amino acids 36–38 (TAT–SH3-2M) abolished both pSMAD3 binding and inhibition of target genes (Figure 4, A and B). Identical results were observed when SMAD3-regulated biological responses were examined in the presence of TAT–SH3-2 (inhibits) or TAT–SH3-2M (no effect, not inhibitory) (Figure 4, C and D).
Figure 4. A point mutant of TAT–SH3-2 abolishes the inhibitory action on SMAD3 responses.
(A) Schematic depicting TAT peptide constructs (left). Asterisks in SH3-2M (mutant) reflect G to V mutation at amino acids 36–38. His pull-down of pSMAD3 bound to TAT peptides was performed as in Figure 3A (middle, representative of 3 separate experiments). AKR-2B cells were transduced for 90 minutes with 1.5 μM TAT–SH3-2, TAT–SH3-2M, or TAT-LC. Western blot analysis was performed for the indicated proteins following 24-hour treatment in the absence (–) or presence (+) of TGF-β (5 ng/ml) or SB431542 (10 μM) (right, representative of 3 separate experiments). (B) qPCR of SMAD3-responsive (Ctgf and Smad7) and SMAD2-responsive (goosecoid and Mixl1) genes as in Figure 3D. Data reflect mean ± SEM from 3 experiments. (C) Scratch assays were performed on AKR-2B cells as described in Methods following transduction with the indicated TAT peptides (1.5 μM) or addition of SB431542 (10 μM). Red bands indicate the leading edge following 24 hours in the absence (control) or presence of 5 ng/ml TGF-β and are representative of 3 separate experiments. Original magnification: ×100. (D) Soft agar colony formation as in Figure 3E. Data reflect the mean ± SEM of triplicate wells from 3 experiments. (E) AKR-2B cells were transiently transfected with a BMP-responsive (BRE), EGF-responsive (SRE), or PDGF-responsive (MMP-1) reporter construct and luciferase activity determined following 12-hour incubation in the absence (–) or presence (+) of the indicated ligand (10 ng/ml BMP4, EGF, or PDGF), inhibitor (10 μM dorsomorphin; 3 μM lapatinib; 2 μM CP868), or TAT peptide (1.5 μM). Data represent the mean ± SEM of duplicate samples from 3 separate experiments.*P < 0.05; **P < 0.005; ***P < 0.0005, 1-way ANOVA followed by Dunnett’s multiple comparisons test.
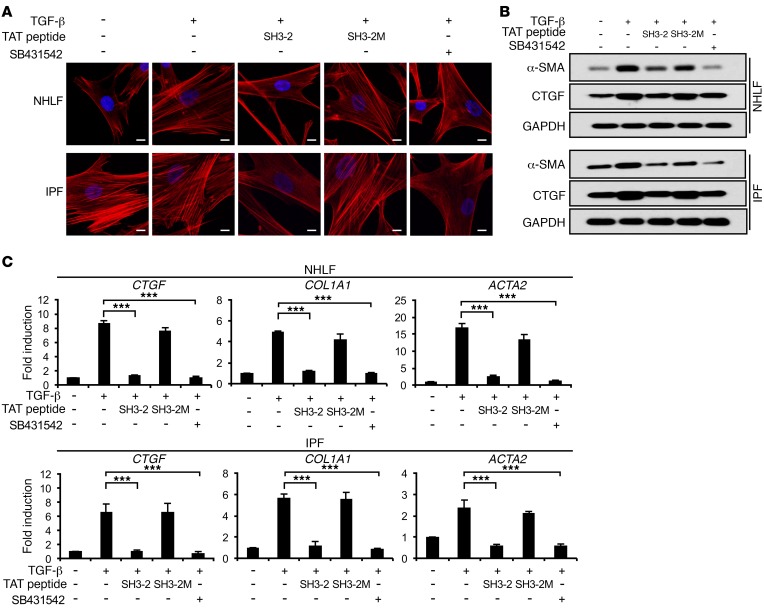
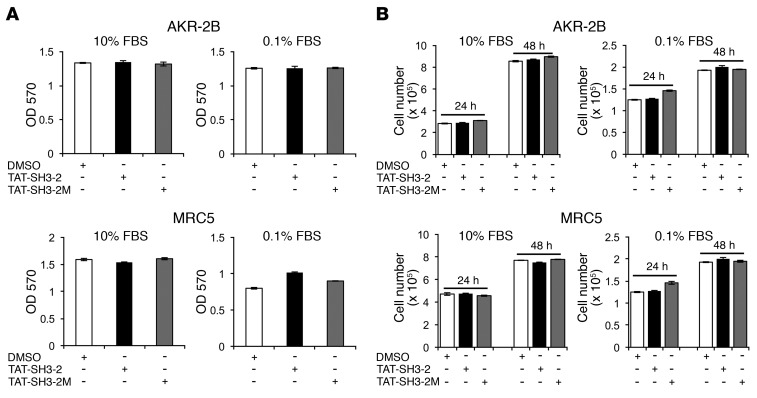
To further document the specific action of TAT–SH3-2 on SMAD3 action, we next addressed the impact of the TAT-SNX9 peptides on (a) luciferase activity stimulated by BMP4-, EGF-, or PDGF-responsive reporters (Figure 4E); (b) the kinetics and extent of R-SMAD phosphorylation (Supplemental Figure 6); (c) the response of normal human lung fibroblasts (NHLF) and idiopathic pulmonary fibrosis (IPF) fibroblasts to TGF-β stimulation (Figure 5); and (d) serum-dependent proliferation of murine and human lung fibroblast lines (Figure 6). Consistent with the data shown in Figure 3, Figure 4, A–D, and ref. 24, there was no detectable impact of either TAT peptide on TGF-β–independent signaling (Figure 4E) or SMAD2/SMAD3 phosphorylation (Supplemental Figure 6) and profibrotic TGF-β signaling in primary human lung fibroblasts (normal and IPF) was similarly inhibited by TAT–SH3-2, but not TAT–SH3-2M (Figure 5). Finally, to further document that the aforementioned in vitro actions of the TAT peptides were specific and did not reflect a toxic response, MTT and cell proliferation assays were performed on AKR-2B cells and MRC5 human lung fibroblasts under optimal (e.g., 10% FBS) as well as stressful (e.g., 0.1% FBS) growth conditions. As shown in Figure 6, there was no effect of either the SH3-2 or SH3-2M peptide on any of these responses.
Figure 5. TAT–SH3-2 inhibits profibrotic responses in human lung fibroblasts.
(A) NHLF or lung fibroblasts from IPF patients were transduced with the indicated TAT peptide (1.5 μM) as in Figure 1. TGF-β (5 ng/ml) or SB431542 (10 μM) was then added for an additional 24 hours. Images were obtained on an LSM510 confocal microscope following F-actin labeling with phalloidin-TRITC and DAPI nuclei staining. Photographs are representative of 3 separate experiments. Scale bars: 10 μm. (B) Western blot analysis (representative of 3 separate experiments) for the indicated proteins subsequent to TAT peptide transduction (1.5 μM) and 24-hour treatment in the absence (–) or presence (+) of TGF-β (5 ng/ml) or SB431542 (10 μM). (C) NHLF or IPF fibroblasts were treated as in A and qPCR performed as described in Methods and Figure 3D. Results represent mean ± SEM from 3 independent experiments. ***P < 0.0005, 1-way ANOVA followed by Dunnett’s multiple comparisons test.
Figure 6. TAT peptides do not inhibit in vitro cell proliferation.
(A) AKR-2B (10% DMEM/FBS) and MRC5 (10% EMEM/FBS) cells were seeded at 2.5 × 103 or 1 × 104 cells/96-well plate, respectively. Twenty-four hours after seeding, the medium was removed and replaced with DMEM or EMEM containing vehicle (0.1% DMSO), TAT–SH3-2 (1.5 μM), or TAT–SH3-2M (1.5 μM) either in 10% or 0.1% FBS for 24 hours prior to MTT assay. Absorbance was measured at 570 nm. (B) AKR-2B (1.25 × 104/well) or MRC5 (5 × 104/well) cells were seeded in 24-well plates for 24 hours. Cultures were treated as in A and cell counts determined following an additional 24-hour and 48-hour incubation. Results represent mean ± SEM from 3 independent experiments.
CPPs that block pSMAD3 action are effective in treatment models of pulmonary fibrosis.
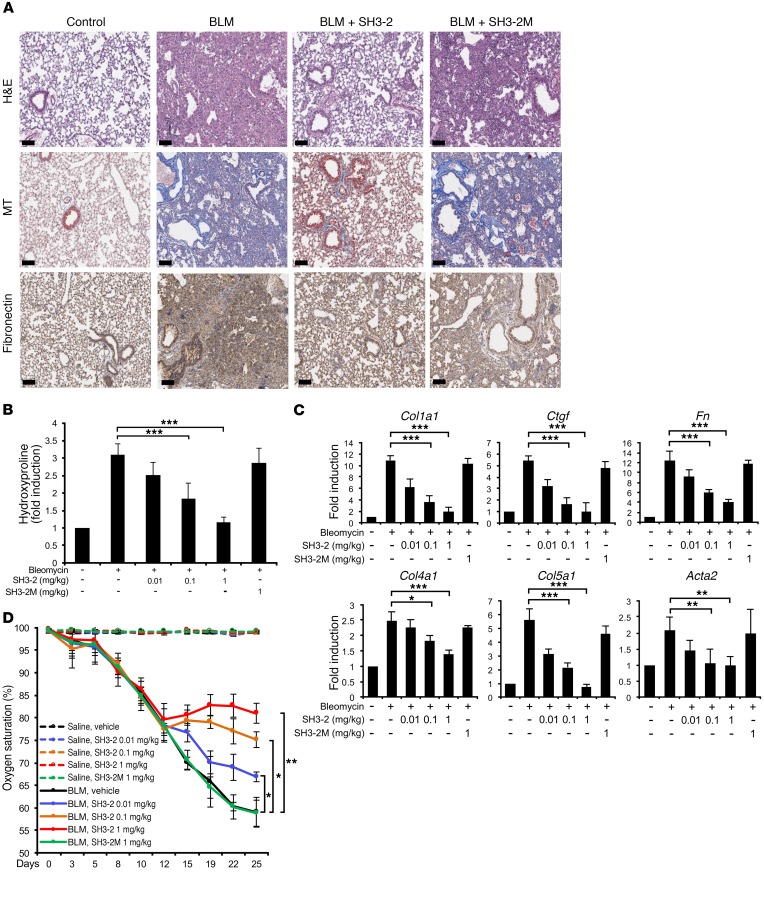
The preceding data (Figures 1–6) document that a CPP encoding a defined region of SNX9 inhibits TGF-β–stimulated biochemical, transcriptional, and biological actions dependent upon pSMAD3. Furthermore, (a) the degree of inhibition is analogous to that obtained with a small molecule inhibitor of the TβRI kinase; (b) SMAD2-, SMAD1/5/8- (e.g., BMP), EGF-, and/or PDGF-stimulated responses are unaffected; and (c) this occurs independently of any general inhibition of cell growth. In that the profibrotic actions of TGF-β are primarily mediated via SMAD3 (29, 35, 43), we extended these findings in vivo using treatment models of lung fibrosis initiated by intratracheal administration of BLM or adenovirus expressing active TGF-β1 where intervention was started following the resolution of inflammation and initiation of fibrotic changes (44–46).
Initial studies were done using BLM, as it is the most common and robust method to induce fibrotic changes. As shown in Figure 7, A and B, and Supplemental Figure 7, while intraperitoneal administration of TAT–SH3-2 improved lung histology, reduced interstitial fibronectin to essentially basal levels, and showed a dose-dependent diminution in total collagen production induced by BLM, the mutant TAT peptide (TAT–SH3-2M) unable to bind and inactivate pSMAD3 actions had no significant effect and neither peptide showed any demonstrable toxicity when administered to control (e.g., intratracheal saline) lungs. Analogous findings on lung fibrosis were observed by quantitative PCR (qPCR) when the expression of 6 profibrotic gene targets (e.g., collagen Iα1 [Col1a1], Ctgf, fibronectin [Fn], collagen IVα1 [Col4a1], collagen Vα1 [Col5a1], and α-smooth muscle actin [Acta2]) was examined (Figure 7C). While the preceding data provide evidence that by inhibiting pSMAD3 action, various targets mediating the fibroproliferative actions of TGF-β can be inhibited, a critical question is whether these biochemical and molecular changes reflect similar improvements in lung function. To address this issue, peripheral blood oxygen saturation (SpO2, on room air) was determined (Figure 7D). While vehicle- or SH3-2M–treated (e.g., the inactive TAT-SNX9 peptide) animals showed an approximate 25% decrease in SpO2 during the treatment regime, a dose-dependent improvement in gas exchange was observed with TAT–SH3-2 such that animals receiving the highest concentration displayed no additional loss of lung function. Thus, by inhibiting a critical signaling component of TGF-β action, a substantial aspect of normal lung physiology is maintained.
Figure 7. BLM-induced lung remodeling is attenuated by TAT–SH3-2.
(A) C57BL/6 mice were intratracheally treated with an equal volume of saline (control) or BLM (0.075 U). Beginning on day 14, mice were daily administered vehicle (METHOCEL/saline; control and BLM) or 1.0 mg/kg of the indicated TAT peptide (SH3-2 or SH3-2M) intraperitoneally. Lung tissue was harvested at day 28 and subjected to H&E staining for histology, Masson’s trichrome (MT) for collagen, or with anti-fibronectin and hematoxylin. Representative images from 5 animals are shown. Scale bars: 100 μm. (B) Mice were treated with saline or BLM as in A and beginning on day 14 administered daily vehicle (–) or the indicated concentration of TAT peptide. Animals were sacrificed on day 28 and hydroxyproline content determined as described in Methods. Data reflect mean ± SEM of n = 5. (C) Mice were treated as in B, and qPCR of the indicated genes assessed in lung tissue harvested on day 28. Data reflect mean ± SEM of n = 5. (D) Time-dependent fluctuation of oxygen saturation (SpO2) levels (determined on room air) in mice challenged with BLM (or saline) for 28 days and treated 1×/d with vehicle (METHOCEL/saline) or the indicated concentration of TAT–SH3-2 or TAT–SH3-2M beginning 14 days after initial BLM insult. Error bars reflect mean ± SEM from n = 5. *P < 0.05; **P < 0.005; ***P < 0.0005, 1-way ANOVA followed by Dunnett’s multiple comparisons test.
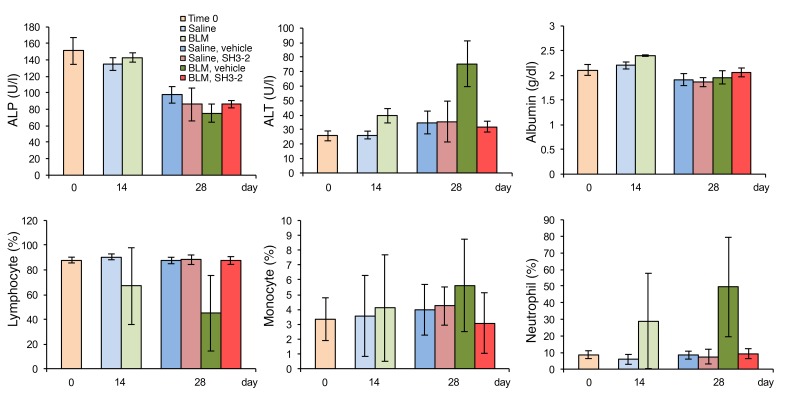
The previous data show that inhibition of SMAD3 action using CPP technology markedly affects profibrotic TGF-β signaling both in vitro and in vivo independently of any effect on cell proliferation. To further characterize the generality of these findings, we examined the systemic in vivo toxicity of the effective TAT-SNX9 peptide and extended our BLM results in another model of murine lung fibrosis. As shown in Figure 8, TAT–SH3-2 treatment had no toxic effect on either inflammatory cell recruitment or liver function in the BLM model. In fact, the TAT peptide restored inflammatory cell and alanine aminotransferase levels to control values when given to BLM-treated mice. Finally, to further confirm our in vivo findings, we employed intratracheal adenoviral delivery of active TGF-β1 as developed by Sime et. al. (46). In that this model is not as robust as using BLM, the only difference (e.g., besides the initiating agent) is that we waited 3 weeks (instead of 2 weeks) prior to beginning treatment with the TAT peptides. This simply reflected the aforementioned robustness, as additional time was needed to see a significant impairment in lung function (e.g., as assessed by the decrease in peripheral blood oxygenation). Data are provided (Figure 9) documenting similar physiologic and molecular effectiveness of TAT–SH3-2 (but not the control TAT–SH3-2M) in treating pulmonary fibrosis in the adeno model as seen with BLM.
Figure 8. TAT–SH3-2 has no demonstrable effect on murine liver enzymes or inflammatory cell recruitment.
C57BL/6 mice were treated as described in Methods and Figure 7. On day 14, all animals began daily treatment with either vehicle (METHOCEL/saline) or 1.0 mg/kg of TAT–SH3-2. Blood samples were obtained at days 0, 14, and 28 from the facial vein of unanesthetized animals and assessed for effect on the indicated liver enzymes and inflammatory cells. Serum levels of alkaline phosphatase (ALP), alanine aminotransferase (ALT), and albumin were determined using a Piccolo Xpress Chemistry Analyzer. Quantification of lymphocytes, monocytes, and neutrophils was measured using a VetScan HM5 Analyzer. The Toxicology and Pharmacology Laboratory in the Department of Molecular Medicine at the Mayo Clinic performed all analyses. Data are presented as mean ± SEM of n = 5.
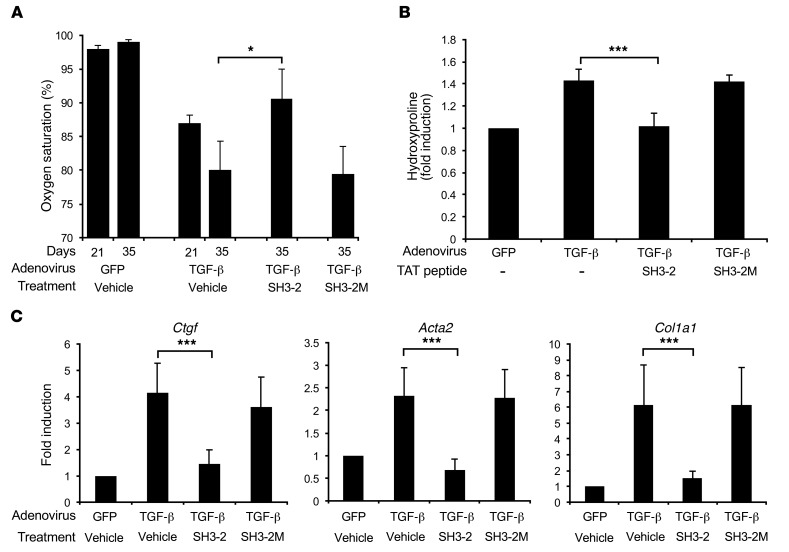
Figure 9. TAT–SH3-2 stabilizes lung function in adenovirus–TGF-β model of pulmonary fibrosis.
Mice were infected with 1 × 108 PFU adenovirus particles expressing control (GFP) or active TGF-β1 by tracheal instillation as described in Methods. On day 21, all animals began daily treatment with either vehicle (METHOCEL/saline) or 1.0 mg/kg of TAT–SH3-2 or the control TAT-SH3-2M peptide. (A) Peripheral blood oxygen determined on days 21 and 35. (B and C) Mice treated as indicated were sacrificed on day 39 and processed for lung hydroxyproline content (B) or qPCR expression (C). Data reflect mean ± SEM of n = 8 and n = 16 for adenovirus-GFP and adenovirus–TGF-β, respectively. *P < 0.05, ***P < 0.0005, 1-way ANOVA followed by Dunnett’s multiple comparisons test.
Discussion
Fibroproliferative disorders are a leading cause of high morbidity and mortality and surround a wide spectrum of pathological conditions featuring localized or systemic tissue/organ fibrosis. Management of these diseases remains an ongoing challenge due to their inherently complex and often undefined etiology (47–50). Although significant advances in delineating the molecular events leading to fibrosis have recently occurred (50–52), attempts to devise efficient therapeutic treatments in clinical contexts have generally met with limited success (48, 50, 53). In that upwards of nearly 45% of all deaths in the developed world are attributed to some chronic fibroproliferative disease (49), new approaches are clearly needed.
TGF-β plays a significant role in the generation and/or maintenance of organ fibrosis primarily via the actions of pSMAD3 (4, 35, 44, 54–56). While we are unaware of any patient-based data for abnormal SMAD3 or TGF-β signaling in IPF, the ability to specifically affect pSMAD3 function could provide a means for modulating the pathophysiologic aspects of TGF-β signaling while maintaining other homeostatic activities. To that end, our recent report showing an obligate role for SNX9 in promoting pSMAD3 nuclear import provides such an opportunity (24). In the current study, we extend this finding by first, identifying the domain within SNX9 required for binding pSMAD3 and second, determining whether a cell-permeable construct expressing this peptide could prevent pSMAD3 nuclear translocation and subsequent profibrotic TGF-β action in vitro as well as in vivo.
Peptides as drugs have advantages as well as disadvantages compared with other small molecules (37, 57). Most importantly, peptides can be designed to bind with high specificity to their in vivo targets and have relatively few off-target side effects. While the market for peptide-based drugs is currently estimated at more than 40 billion per year (57), a major limiting factor is the permeability barrier imposed by the plasma membrane (36–38). To overcome that limitation, over 100 peptide sequences have been identified capable of transporting bioactive molecules (38, 58). One of the most promising delivery vehicles is the TAT protein transduction domain of HIV-1. It has been used to deliver a wide variety of cargo into cells and has shown efficacy in preclinical models in vivo (38, 59, 60). In addition, several clinical trials are currently underway using TAT-mediated delivery (57, 60–64), although no therapy as of yet has received FDA approval, as issues associated with efficacy, bioavailablilty, and targeting remain primary challenges.
In this study, we generated TAT fusion peptides from the SH3 domain of SNX9 shown to bind pSMAD3 (Figure 1A, Figure 3A, and Figure 4A) and examined their effect on profibrotic TGF-β signaling. While pSMAD3 nuclear import, targets, and dependent biologies were inhibited (Figures 1–5 and Supplemental Figure 5), pSMAD2 signaling was unaffected (Figure 1B; Figure 2, A and B; Figure 3, B and D; and Figure 4B). Furthermore, a 3–amino acid mutant (TAT–SH3-2M) unable to bind pSMAD3 was similarly unable to affect pSMAD3 actions (Figure 4, Figure 5, and Supplemental Figure 5). As the SNX9 SH3 domain is known to have a number of binding partners including dynamin 2, CDC42-associated kinase, WASp, and ITCH (19, 20, 22, 23), we further addressed this issue by demonstrating a lack of SNX9 association with pSMAD1/5/8 (Supplemental Figure 1) and no inhibitory effect or effects of TAT-SH3 peptides on BMP4-, PDGF-, or EGF-responsive reporters (Figure 2A and Figure 4E) as well as SMAD2 or SMAD3 phosphorylation (Supplemental Figure 6) or serum-dependent growth (Figure 6).
While the aforementioned response of specifically inhibiting pSMAD3 action is important, a critical question is whether such in vitro findings are similarly manifested in vivo. To address that issue, we utilized 2 distinct treatment models of lung fibrosis where test agents are administered following the initiation of fibrotic changes dependent upon pSMAD3 (44–46). Consistent with our in vitro findings, administration of the TAT–SH3-2 peptide not only diminished the expression of profibrotic genes (Figure 7, A–C, Supplemental Figure 7B, and Figure 9, B and C) in the absence of any demonstrable systemic toxicity (Figure 8 and Supplemental Figure 7A), but stabilized lung function as assessed by peripheral blood oxygenation in a pSMAD3-binding and dose-dependent manner (Figure 7D and Figure 9A). These results not only provided a physiologic readout consistent with our biochemical and molecular data, but in addition, pulse-oximetry has been shown to accurately predict lung pathology in numerous animal models (65–68). It is currently unclear whether these findings will reflect lung function in humans. Whether such levels of oxygenation indicate improvement in humans in IPF will need to be tested in clinical trials.
In conclusion, the present study demonstrates that biologically active peptides can be effectively delivered to inhibit a defined component of profibrotic TGF-β signaling in murine AKR-2B cells, human MRC5 fibroblasts, primary NHLF and IPF cultures, and 2 murine models of lung fibrosis. Such findings provide proof of concept that by specifically targeting pSMAD3, the profibrotic actions of TGF-β can be decreased. In order for such an approach to be extended further, additional cell line assessment as well as pharmacokinetic data on the CPPs are required. If, however, such information is forthcoming and analogous findings are observed in other disease models, this would provide the preclinical support/justification to target specific pathophysiologic mechanisms.
Methods
Cell culture.
AKR-2B cells (Sigma-Aldrich) were grown in DMEM supplemented with 10% FBS. Prior to processing for Western blotting or PCR, 2.5 × 105 cells were seeded on 6-well plates and incubated overnight at 37°C. The next day, the medium was replaced with 0.5% FBS/DMEM and cells were transduced for 90 minutes with the indicated TAT peptide. After transduction, cells were incubated with or without TGF-β (5 ng/ml) for the indicated times in 0.5% FBS/DMEM. Primary NHLF and IPF fibroblasts were obtained from Lonza and grown in FGM-2 medium containing 2% FBS. Cultures were treated as above following overnight incubation in 0.5% FBS/FGM-2 medium and used within the first 2 passages. MRC5 human lung fibroblasts (ATCC) were grown in EMEM (ATCC) supplemented with 10% FBS.
Generation of TAT fusion proteins.
TAT fusion proteins were prepared from BL21(DE3)pLysS E. coli (OD600 of 0.4) as previously described (24). Following addition of isopropyl-d-thiogalactopyranoside to a final concentration of 0.5 mM and 4 hours induction at 37°C, lysates were prepared and the precleared supernatant poured into a TALON Metal Affinity Resin Column (Clontech). Proteins were eluted (50 mM sodium phosphate, 300 NaCl, and 150 mM imidazole, pH 7.4) and dialyzed against PBS using a Slide-A Lyzer MINI Dialysis Unit (Thermo Scientific). Results are shown in Figures 1–5 and Supplemental Figures 1–6. For toxicity and animal studies (Figures 6–9 and Supplemental Figure 7), peptides were synthesized and purified by the Mayo Proteomics Core.
Soft agar and scratch assay.
Soft agar assays were performed as previously described (69). Briefly, 1 × 104 cells were transduced for 90 minutes with the indicated TAT peptide and seeded in a 6-well plate in the presence or absence of 20 ng/ml TGF-β (R&D Systems). Following 7 days growth at 37°C, the number of colonies more than 50 μm in diameter were counted using an Optronix Gelcount (Oxford Optronix). For scratch assays, cells were seeded into 6-well plates (4 × 105 cells/well) in 10% FBS/DMEM and incubated overnight at 37°C. Following monolayer disruption using a sterile 200-μl pipette tip, the medium was changed to 0.5% FBS/DMEM and transduced for 90 minutes with the indicated TAT peptide. After transduction, cells were incubated in the presence or absence of TGF-β (5 ng/ml) for 24 hours. Images (×100) were taken at 24 hours.
Luciferase reporter assays.
For luciferase assays, 2 × 105 cells were plated in 6-well plates. The next day, cells were transfected with 2 μg (3TP, BRE, SRE, and MMP1) or 1.5 μg (ARE; plus 1.5 μg FAST1) of the indicated luciferase constructs together with 0.5 μg CMV–β-galactosidase with TransIT-2020 reagent (Mirus Bio). Following 18 to 24 hours of incubation, the medium was changed to 0.5% FBS/DMEM containing the indicated TAT fusion proteins for 90 minutes. After transduction, cultures were incubated in the presence or absence of 5 ng/ml TGF-β for 12 hours. Cells were harvested in 200 μl of reporter lysis buffer (Promega) and luciferase activity determined in a Berthold Lumat 9507 luminometer after normalization for transfection efficiency by detecting β-galactosidase activity with chlorophenol red–β-d-galactopyranoside (CPRG) and measuring substrate conversion at 562 nm.
Immunofluorescent microscopy.
AKR-2B cells (2 × 104) were plated onto coverslips in 6-well plates with 10% FBS/DMEM and incubated overnight at 37°C. Cultures were placed in 0.5% FBS/DMEM and transduced with TAT fusion peptides for 90 minutes. After transduction and addition of TGF-β (5 ng/ml) for 1 hour, cells were processed as previously described (24). SMAD3 was detected using anti-AF488 (green; A11001), while the TAT peptide HA tag was visualized with anti–AF-594 (red; A11012) secondary antibody, both from Invitrogen. Fluorescence images were collected on an LSM510 confocal microscope (Carl Zeiss Microimage Inc.). For primary human lung fibroblasts, 1 × 105 cells were seeded onto coverslips in 0.5% FBS/FGM-2 medium. Following overnight incubation at 37°C, cultures were treated with TAT peptides and TGF-β as indicated prior to phalloidin staining according to the manufacturer’s recommendation (Sigma-Aldrich).
Quantitative RT-PCR analysis.
Following TGF-β stimulation, total RNA was isolated using Rneasy Plus Mini Kit (QIAGEN) and 1 μg reverse transcribed with the SuperScript III Reverse Transcriptase System (Invitrogen). Complementary DNAs were subjected to qPCR with Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) or TaqMan gene expression analysis (Thermo Scientific). Primers are presented in Supplemental Table 1. Sample induction for qPCR was normalized to histone H3 (in vitro samples) or β-actin (in vivo samples) while reverse-transcription PCR (RT-PCR) (Figure 2B) was normalized to GAPDH.
Western blotting.
Cells were lysed for 30 minutes on ice in RIPA buffer (50 mM Tris, pH 7.4, 1% Triton-X 100, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, pH 8, and 10 mM NaF) containing cOmplete Protease Inhibitor (Roche). Insoluble material was removed by centrifugation at 18,000 g for 10 minutes and 10–25 μg protein separated by 10% SDS-PAGE. Commercial antibodies are provided in Supplemental Table 2. Rabbit anti–phospho-SMAD3 and anti–phospho-SMAD2 antibodies were generated in our laboratory to the peptides COOH-GSPSIRCSpSVpS and COOH-CGSPSVRCSpSMpS, respectively, and used at 1/2,000 dilution. See complete unedited blots in the supplemental material.
GST and His pull-down.
GST- and TAT-His fusion proteins were purified using Glutathione Superflow or TALON Metal Affinity Resin following the manufacturer’s instructions (Clontech). To assess SNX9 binding to GST or His constructs, cells were incubated in the presence or absence of 5 ng/ml TGF-β for 45 minutes. Following RIPA buffer lysis, 500 μg of protein was precleared with GST Resin or TALON Metal Affinity Resin for 2 hours at 4°C. Precleared cell lysates were then treated with 5 μg purified GST or His fusion proteins and incubated overnight at 4°C with gentle shaking. Following addition of GST or TALON Metal Affinity Resin and 2-hour incubation, the pelleted (3,000 g; 10 minutes) resin was washed several times with PBS and the remaining proteins eluted using 1× Laemmli sample buffer. Western blotting was as described above.
Isolation of cellular fractions.
Nuclear extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Kits for Cultured Cells (Thermo Fisher Scientific) with the addition of cOmplete Protease Inhibitor (Roche) to the lysis buffers. Following removal of the cytoplasmic extract, the nuclear pellet was washed 3 times in PBS containing cOmplete before nuclear lysis and Western blot analysis.
Murine BLM model of pulmonary fibrosis.
Animals were maintained under standard laboratory conditions on a 12-hour light/12-hour dark cycle supplied with chow and water ad libitum, and interventions were performed during the dark cycle. Female 18–20 g C57BL/6 mice (Charles River Laboratories) were administered BLM (0.075 U diluted in 50 μl 0.9% normal saline) or 50 μl 0.9% normal saline alone by tracheal instillation using an intratracheal aerosolizer (Penn-Century Inc.) as previously described (44). At this time, animals were shaved around the collar region to allow monitoring of dissolved oxygen levels (every third day) on room air using a MouseOX Collar Clip Monitoring System (Starr Life Science Corp.). TAT peptides were solubilized with DMSO and prepared by thoroughly blending with METHOCEL (Sigma-Aldrich) at a ratio of 1:7. Mice were treated daily by intraperitoneal injection (100 μl) with the indicated concentration (mg/kg) of TAT peptide or an equal volume of METHOCEL/saline (vehicle) beginning on day 14. On day 28, mice were euthanized, lungs dissected, and samples prepared for immunohistochemistry and other analyses as described (44, 70, 71).
Murine adenoviral TGF-β model of pulmonary fibrosis.
Studies analogous to those described above for the BLM model were used with the sole exceptions being the following: (a) 1 × 108 PFUs of replication-deficient adenovirus encoding active porcine TGF-β1 (Ad5TGF-β) or GFP as a control protein (Ad5GFP) in 50 μl 0.9% normal saline was administered into the distal trachea on day 0; (b) treatment began on day 21; and (c) mice were sacrificed on day 35. Christopher H. Evans (Mayo Clinic) provided adenovirus expressing GFP and active porcine TGF-β1.
Hydroxyproline assay.
The Hydroxyproline Assay Kit from Sigma-Aldrich was used to assess total lung collagen levels. Briefly, following sacrifice, lung tissue was washed in PBS and 30 mg homogenized in 300 μl water. Samples of 100 μl were hydrolyzed in 12 M HCl and duplicate samples of 50 μl analyzed according to the manufacturer’s recommendations.
Statistics.
All data were evaluated for statistical significance between experimental and control groups using 1-way ANOVA (with Dunnett’s multiple comparisons) and/or Student’s t test (2-tailed). P values of less than 0.05 were considered statistically significant, and all values are reported in Supplemental Table 3. Analyses were all performed with GraphPad Prism software (version 7.02).
Study approval.
All animal studies were approved by the Mayo Clinic Institutional Animal Care and Use Committee (Rochester, MN) (A15714-14 for the BLM studies and A00002036-16 for the adenoviral lung fibrosis model).
Author contributions
JHK and EBL designed the research and wrote the paper. JHK, MYJ, XY, MA, and DMH performed the experiments, with the majority done by JHK. All authors analyzed the data and revised the paper.
Supplementary Material
Acknowledgments
The binding of pSMAD3 to the amino half of SNX9 was initially determined by Mark C. Wilkes. The authors would like to thank Andrew Limper for clinical insights. This work was supported by Public Health Service grants GM-55816 and GM-54200 from the National Institute of General Medical Sciences, a pilot grant from the Mayo Brain SPORE (CA-108961), the Caerus Foundation, and the Mayo Foundation (to EBL).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(7):2541–2554.https://doi.org/10.1172/JCI88696.
Contributor Information
Jeong-Han Kang, Email: Kang.JeongHan@mayo.edu.
Mi-Yeon Jung, Email: Jung.MiYeon@mayo.edu.
Xueqian Yin, Email: yin.xueqian@mayo.edu.
Mahefatiana Andrianifahanana, Email: andrianifahanana.mahefatiana@mayo.edu.
Edward B. Leof, Email: leof.edward@mayo.edu.
References
- 1.Akhurst RJ. TGF beta signaling in health and disease. Nat Genet. 2004;36(8):790–792. doi: 10.1038/ng0804-790. [DOI] [PubMed] [Google Scholar]
- 2.Elliott RL, Blobe GC. Role of transforming growth factor Beta in human cancer. J Clin Oncol. 2005;23(9):2078–2093. doi: 10.1200/JCO.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 3.Roberts AB, Wakefield LM. The two faces of transforming growth factor beta in carcinogenesis. Proc Natl Acad Sci U S A. 2003;100(15):8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrianifahanana M, et al. ERBB receptor activation is required for profibrotic responses to transforming growth factor beta. Cancer Res. 2010;70(19):7421–7430. doi: 10.1158/0008-5472.CAN-10-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahimi RA, Leof EB. TGF-beta signaling: a tale of two responses. J Cell Biochem. 2007;102(3):593–608. doi: 10.1002/jcb.21501. [DOI] [PubMed] [Google Scholar]
- 6.Wilkes MC, Murphy SJ, Garamszegi N, Leof EB. Cell-type-specific activation of PAK2 by transforming growth factor beta independent of Smad2 and Smad3. Mol Cell Biol. 2003;23(23):8878–8889. doi: 10.1128/MCB.23.23.8878-8889.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkes MC, et al. Erbin and the NF2 tumor suppressor Merlin cooperatively regulate cell-type-specific activation of PAK2 by TGF-beta. Dev Cell. 2009;16(3):433–444. doi: 10.1016/j.devcel.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heldin CH, Landström M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol. 2009;21(2):166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 10.Gatza CE, Oh SY, Blobe GC. Roles for the type III TGF-beta receptor in human cancer. Cell Signal. 2010;22(8):1163–1174. doi: 10.1016/j.cellsig.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massagué J, et al. TGF-beta receptors. Mol Reprod Dev. 1992;32(2):99–104. doi: 10.1002/mrd.1080320204. [DOI] [PubMed] [Google Scholar]
- 12.Anders RA, Leof EB. Chimeric granulocyte/macrophage colony-stimulating factor/transforming growth factor-beta (TGF-beta) receptors define a model system for investigating the role of homomeric and heteromeric receptors in TGF-beta signaling. J Biol Chem. 1996;271(36):21758–21766. doi: 10.1074/jbc.271.36.21758. [DOI] [PubMed] [Google Scholar]
- 13.Wrana JL, et al. TGF beta signals through a heteromeric protein kinase receptor complex. Cell. 1992;71(6):1003–1014. doi: 10.1016/0092-8674(92)90395-S. [DOI] [PubMed] [Google Scholar]
- 14.Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 15.Hill CS. Nucleocytoplasmic shuttling of Smad proteins. Cell Res. 2009;19(1):36–46. doi: 10.1038/cr.2008.325. [DOI] [PubMed] [Google Scholar]
- 16.Macias MJ, Martin-Malpartida P, Massagué J. Structural determinants of Smad function in TGF-β signaling. Trends Biochem Sci. 2015;40(6):296–308. doi: 10.1016/j.tibs.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 18.Moustakas A, Heldin CH. Non-Smad TGF-beta signals. J Cell Sci. 2005;118(Pt 16):3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 19.Badour K, et al. Interaction of the Wiskott-Aldrich syndrome protein with sorting nexin 9 is required for CD28 endocytosis and cosignaling in T cells. Proc Natl Acad Sci U S A. 2007;104(5):1593–1598. doi: 10.1073/pnas.0610543104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumann C, Lindholm CK, Rimoldi D, Lévy F. The E3 ubiquitin ligase Itch regulates sorting nexin 9 through an unconventional substrate recognition domain. FEBS J. 2010;277(13):2803–2814. doi: 10.1111/j.1742-4658.2010.07698.x. [DOI] [PubMed] [Google Scholar]
- 21.Carlton J, Bujny M, Rutherford A, Cullen P. Sorting nexins-unifying trends and new perspectives. Traffic. 2005;6(2):75–82. doi: 10.1111/j.1600-0854.2005.00260.x. [DOI] [PubMed] [Google Scholar]
- 22.Lundmark R, Carlsson SR. SNX9 - a prelude to vesicle release. J Cell Sci. 2009;122(Pt 1):5–11. doi: 10.1242/jcs.037135. [DOI] [PubMed] [Google Scholar]
- 23.Worby CA, Dixon JE. Sorting out the cellular functions of sorting nexins. Nat Rev Mol Cell Biol. 2002;3(12):919–931. doi: 10.1038/nrm974. [DOI] [PubMed] [Google Scholar]
- 24.Wilkes MC, Repellin CE, Kang JH, Andrianifahanana M, Yin X, Leof EB. Sorting nexin 9 differentiates ligand-activated Smad3 from Smad2 for nuclear import and transforming growth factor β signaling. Mol Biol Cell. 2015;26(21):3879–3891. doi: 10.1091/mbc.E15-07-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verges M. Retromer and sorting nexins in development. Front Biosci. 2007;12:3825–3851. doi: 10.2741/2355. [DOI] [PubMed] [Google Scholar]
- 26.Yarar D, Surka MC, Leonard MC, Schmid SL. SNX9 activities are regulated by multiple phosphoinositides through both PX and BAR domains. Traffic. 2008;9(1):133–146. doi: 10.1111/j.1600-0854.2007.00675.x. [DOI] [PubMed] [Google Scholar]
- 27.Yarar D, Waterman-Storer CM, Schmid SL. SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev Cell. 2007;13(1):43–56. doi: 10.1016/j.devcel.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 28.Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10(6):415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 29.Leask A. Potential therapeutic targets for cardiac fibrosis: TGFbeta, angiotensin, endothelin, CCN2, and PDGF, partners in fibroblast activation. Circ Res. 2010;106(11):1675–1680. doi: 10.1161/CIRCRESAHA.110.217737. [DOI] [PubMed] [Google Scholar]
- 30.Wu MY, Hill CS. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16(3):329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 31.Giannelli G, Villa E, Lahn M. Transforming growth factor-β as a therapeutic target in hepatocellular carcinoma. Cancer Res. 2014;74(7):1890–1894. doi: 10.1158/0008-5472.CAN-14-0243. [DOI] [PubMed] [Google Scholar]
- 32.Rice LM, et al. Fresolimumab treatment decreases biomarkers and improves clinical symptoms in systemic sclerosis patients. J Clin Invest. 2015;125(7):2795–2807. doi: 10.1172/JCI77958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodón J, et al. Pharmacokinetic, pharmacodynamic and biomarker evaluation of transforming growth factor-β receptor I kinase inhibitor, galunisertib, in phase 1 study in patients with advanced cancer. Invest New Drugs. 2015;33(2):357–370. doi: 10.1007/s10637-014-0192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoot KE, et al. Keratinocyte-specific Smad2 ablation results in increased epithelial-mesenchymal transition during skin cancer formation and progression. J Clin Invest. 2008;118(8):2722–2732. doi: 10.1172/JCI33713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng XM, et al. Smad2 protects against TGF-beta/Smad3-mediated renal fibrosis. J Am Soc Nephrol. 2010;21(9):1477–1487. doi: 10.1681/ASN.2009121244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kauffman WB, Fuselier T, He J, Wimley WC. Mechanism matters: a taxonomy of cell penetrating peptides. Trends Biochem Sci. 2015;40(12):749–764. doi: 10.1016/j.tibs.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramsey JD, Flynn NH. Cell-penetrating peptides transport therapeutics into cells. Pharmacol Ther. 2015;154:78–86. doi: 10.1016/j.pharmthera.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Rizzuti M, Nizzardo M, Zanetta C, Ramirez A, Corti S. Therapeutic applications of the cell-penetrating HIV-1 Tat peptide. Drug Discov Today. 2015;20(1):76–85. doi: 10.1016/j.drudis.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 39.Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55(6):1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 40.Harrison CF, et al. Conservation of a glycine-rich region in the prion protein is required for uptake of prion infectivity. J Biol Chem. 2010;285(26):20213–20223. doi: 10.1074/jbc.M109.093310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jang HS, Greenwood JA. Glycine-rich region regulates cysteine-rich protein 1 binding to actin cytoskeleton. Biochem Biophys Res Commun. 2009;380(3):484–488. doi: 10.1016/j.bbrc.2009.01.125. [DOI] [PubMed] [Google Scholar]
- 42.Shaw DJ, Morse R, Todd AG, Eggleton P, Lorson CL, Young PJ. Identification of a self-association domain in the Ewing’s sarcoma protein: a novel function for arginine-glycine-glycine rich motifs? J Biochem. 2010;147(6):885–893. doi: 10.1093/jb/mvq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verrecchia F, Mauviel A. Transforming growth factor-beta and fibrosis. World J Gastroenterol. 2007;13(22):3056–3062. doi: 10.3748/wjg.v13.i22.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrianifahanana M, et al. Profibrotic TGFβ responses require the cooperative action of PDGF and ErbB receptor tyrosine kinases. FASEB J. 2013;27(11):4444–4454. doi: 10.1096/fj.12-224907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaudhary NI, Schnapp A, Park JE. Pharmacologic differentiation of inflammation and fibrosis in the rat bleomycin model. Am J Respir Crit Care Med. 2006;173(7):769–776. doi: 10.1164/rccm.200505-717OC. [DOI] [PubMed] [Google Scholar]
- 46.Sime PJ, Xing Z, Graham FL, Csaky KG, Gauldie J. Adenovector-mediated gene transfer of active transforming growth factor-beta1 induces prolonged severe fibrosis in rat lung. J Clin Invest. 1997;100(4):768–776. doi: 10.1172/JCI119590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steele MP, Schwartz DA. Molecular mechanisms in progressive idiopathic pulmonary fibrosis. Annu Rev Med. 2013;64:265–276. doi: 10.1146/annurev-med-042711-142004. [DOI] [PubMed] [Google Scholar]
- 48.Todd NW, Luzina IG, Atamas SP. Molecular and cellular mechanisms of pulmonary fibrosis. Fibrogenesis Tissue Repair. 2012;5(1):11. doi: 10.1186/1755-1536-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wynn TA. Common and unique mechanisms regulate fibrosis in various fibroproliferative diseases. J Clin Invest. 2007;117(3):524–529. doi: 10.1172/JCI31487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med. 2012;18(7):1028–1040. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beyer C, Distler O, Distler JH. Innovative antifibrotic therapies in systemic sclerosis. Curr Opin Rheumatol. 2012;24(3):274–280. doi: 10.1097/BOR.0b013e3283524b9a. [DOI] [PubMed] [Google Scholar]
- 52.Rosenbloom J, Castro SV, Jimenez SA. Narrative review: fibrotic diseases: cellular and molecular mechanisms and novel therapies. Ann Intern Med. 2010;152(3):159–166. doi: 10.7326/0003-4819-152-3-201002020-00007. [DOI] [PubMed] [Google Scholar]
- 53.Sivakumar P, Ntolios P, Jenkins G, Laurent G. Into the matrix: targeting fibroblasts in pulmonary fibrosis. Curr Opin Pulm Med. 2012;18(5):462–469. doi: 10.1097/MCP.0b013e328356800f. [DOI] [PubMed] [Google Scholar]
- 54.Sato M, Muragaki Y, Saika S, Roberts AB, Ooshima A. Targeted disruption of TGF-beta1/Smad3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction. J Clin Invest. 2003;112(10):1486–1494. doi: 10.1172/JCI19270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor LM, Khachigian LM. Induction of platelet-derived growth factor B-chain expression by transforming growth factor-beta involves transactivation by Smads. J Biol Chem. 2000;275(22):16709–16716. doi: 10.1074/jbc.275.22.16709. [DOI] [PubMed] [Google Scholar]
- 56.Zhao J, et al. Smad3 deficiency attenuates bleomycin-induced pulmonary fibrosis in mice. Am J Physiol Lung Cell Mol Physiol. 2002;282(3):L585–L593. doi: 10.1152/ajplung.00151.2001. [DOI] [PubMed] [Google Scholar]
- 57.Craik DJ, Fairlie DP, Liras S, Price D. The future of peptide-based drugs. Chem Biol Drug Des. 2013;81(1):136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 58.Koren E, Torchilin VP. Cell-penetrating peptides: breaking through to the other side. Trends Mol Med. 2012;18(7):385–393. doi: 10.1016/j.molmed.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 59.Nagahara H, et al. Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat Med. 1998;4(12):1449–1452. doi: 10.1038/4042. [DOI] [PubMed] [Google Scholar]
- 60.Bolhassani A, Jafarzade BS, Mardani G. In vitro and in vivo delivery of therapeutic proteins using cell penetrating peptides. Peptides. 2017;87:50–63. doi: 10.1016/j.peptides.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 61.Domashenko AD, Danet-Desnoyers G, Aron A, Carroll MP, Emerson SG. TAT-mediated transduction of NF-Ya peptide induces the ex vivo proliferation and engraftment potential of human hematopoietic progenitor cells. Blood. 2010;116(15):2676–2683. doi: 10.1182/blood-2010-03-273441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo J, Chen Y, Yuan B, Liu S, Rao P. Effects of intracellular superoxide removal at acupoints with TAT-SOD on obesity. Free Radic Biol Med. 2011;51(12):2185–2189. doi: 10.1016/j.freeradbiomed.2011.09.036. [DOI] [PubMed] [Google Scholar]
- 63.Lönn P, Dowdy SF. Cationic PTD/CPP-mediated macromolecular delivery: charging into the cell. Expert Opin Drug Deliv. 2015;12(10):1627–1636. doi: 10.1517/17425247.2015.1046431. [DOI] [PubMed] [Google Scholar]
- 64.Rapoport M, Saada A, Elpeleg O, Lorberboum-Galski H. TAT-mediated delivery of LAD restores pyruvate dehydrogenase complex activity in the mitochondria of patients with LAD deficiency. Mol Ther. 2008;16(4):691–697. doi: 10.1038/mt.2008.4. [DOI] [PubMed] [Google Scholar]
- 65.Foskett AM, Bazhanov N, Ti X, Tiblow A, Bartosh TJ, Prockop DJ. Phase-directed therapy: TSG-6 targeted to early inflammation improves bleomycin-injured lungs. Am J Physiol Lung Cell Mol Physiol. 2014;306(2):L120–L131. doi: 10.1152/ajplung.00240.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghosh S, et al. Extracellular hemin crisis triggers acute chest syndrome in sickle mice. J Clin Invest. 2013;123(11):4809–4820. doi: 10.1172/JCI64578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurotsu S, et al. Ameliorative effect of mepenzolate bromide against pulmonary fibrosis. J Pharmacol Exp Ther. 2014;350(1):79–88. doi: 10.1124/jpet.114.213009. [DOI] [PubMed] [Google Scholar]
- 68.Verhoeven D, Teijaro JR, Farber DL. Pulse-oximetry accurately predicts lung pathology and the immune response during influenza infection. Virology. 2009;390(2):151–156. doi: 10.1016/j.virol.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rahimi RA, et al. Distinct roles for mammalian target of rapamycin complexes in the fibroblast response to transforming growth factor-beta. Cancer Res. 2009;69(1):84–93. doi: 10.1158/0008-5472.CAN-08-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Daniels CE, et al. Imatinib mesylate inhibits the profibrogenic activity of TGF-beta and prevents bleomycin-mediated lung fibrosis. J Clin Invest. 2004;114(9):1308–1316. doi: 10.1172/JCI19603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang S, Wilkes MC, Leof EB, Hirschberg R. Imatinib mesylate blocks a non-Smad TGF-beta pathway and reduces renal fibrogenesis in vivo. FASEB J. 2005;19(1):1–11. doi: 10.1096/fj.04-2370com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.