Abstract
Chronic liver disease with cirrhosis is the 12th leading cause of death in the United States, and alcoholic liver disease accounts for approximately half of all cirrhosis deaths. Chronic alcohol consumption is associated with intestinal bacterial dysbiosis, yet we understand little about the contribution of intestinal fungi, or mycobiota, to alcoholic liver disease. Here we have demonstrated that chronic alcohol administration increases mycobiota populations and translocation of fungal β-glucan into systemic circulation in mice. Treating mice with antifungal agents reduced intestinal fungal overgrowth, decreased β-glucan translocation, and ameliorated ethanol-induced liver disease. Using bone marrow chimeric mice, we found that β-glucan induces liver inflammation via the C-type lectin–like receptor CLEC7A on Kupffer cells and possibly other bone marrow–derived cells. Subsequent increases in IL-1β expression and secretion contributed to hepatocyte damage and promoted development of ethanol-induced liver disease. We observed that alcohol-dependent patients displayed reduced intestinal fungal diversity and Candida overgrowth. Compared with healthy individuals and patients with non–alcohol-related cirrhosis, alcoholic cirrhosis patients had increased systemic exposure and immune response to mycobiota. Moreover, the levels of extraintestinal exposure and immune response correlated with mortality. Thus, chronic alcohol consumption is associated with an altered mycobiota and translocation of fungal products. Manipulating the intestinal mycobiome might be an effective strategy for attenuating alcohol-related liver disease.
Keywords: Hepatology
Introduction
Liver cirrhosis is the 12th leading cause of mortality worldwide (1). Approximately 50% of cirrhosis-related deaths are due to alcohol abuse (2). Chronic alcoholism can lead to intestinal bacterial overgrowth and dysbiosis, a leaky gut barrier, and increased systemic levels of bacterial products (3–7). Although the intestinal microbiome contains bacteria, fungi, and viruses, research in the field of alcohol-associated disease has almost exclusively focused on the interaction between the host and bacteria. Changes in the composition of the commensal intestinal fungi, also called the mycobiome, and their interaction with human tissues have been associated with other diseases (8).
The predominant commensal fungal species in the human intestine are Candida species, Saccharomyces cerevisiae, and Malassezia species (9). Like commensal bacteria in the intestine, fungi interact with their host. Although the host immune system develops tolerance to colonization with commensal fungi, it must contain the spread, and, in particular, invasion, of fungi (10). The human intestine can serve as a source of systemic fungal products or fungal infection when the gut barrier is disrupted (11). Patients with cirrhosis frequently either are exposed to fungal products (12) or develop fungal infections, with high mortality (13–15). Spontaneous fungal peritonitis is mainly caused by Candida species — in particular by Candida albicans in patients with cirrhosis (16).
C-type lectin domain family 7 member A (CLEC7A; also known as DECTIN1) is a pattern recognition receptor. CLEC7A recognizes a variety of 1,3-β-glucans (10) — cell wall polysaccharides found in and released from most fungi, including species of Candida, Aspergillus, and Pneumocystis (17). Upon ligand binding, CLEC7A activates caspase recruitment domain family member 9 (CARD9) and NF-κB, leading to transcription of several cytokines, including pro–interleukin-1β (pro–IL-1β) (18). The CLEC7A signaling pathway results in cleavage of pro–IL-1β and production of mature IL-1β by activating the NLR family pyrin domain–containing 3 (NLRP3) and caspase-1 inflammasome pathways (19).
We investigated the role of the mycobiota and CLEC7A signaling in development of alcoholic liver disease.
Results
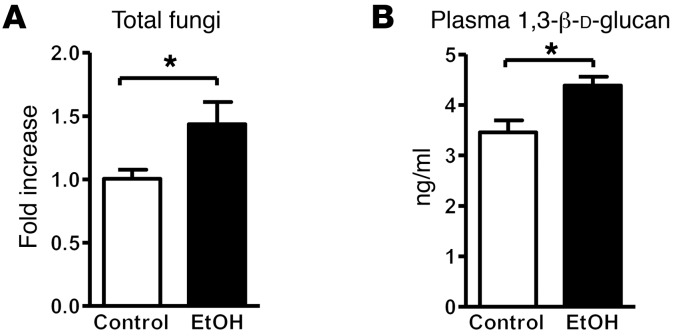
Chronic ethanol administration results in intestinal fungal overgrowth and increased plasma levels of β-glucan in mice.
Intestinal bacterial overgrowth is common in alcohol-dependent patients (3). To determine whether chronic ethanol administration increases commensal fungi in mice, fecal numbers of fungi were measured by quantitative PCR (qPCR). Significantly increased fungal populations were observed after 8 weeks of ethanol administration in C57BL/6 WT mice (Figure 1A); a nonsignificant trend toward increased fungal populations was already seen after 5 weeks of ethanol feeding (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI90562DS1). Fungal dysbiosis was determined in mice fed an ethanol-containing diet for 8 weeks using internal transcribed spacer (ITS) sequencing. Ethanol consumption resulted in an increase in both fungal species richness (average of 189.2 ± 94.7 operational taxonomic units [OTUs] for ethanol vs. 74.7 ± 17.5 OTUs for control diet) and diversity (average Simpson index of 0.256 ± 0.097 for ethanol vs. 0.664 ± 0.224 for control diet). Compositional changes in the intestinal mycobiome were characterized by significantly increased proportions of Humicola species (P = 0.0017), Fusarium (P = 0.0190), and Aspergillus (P = 0.0258), while proportions of Candida species decreased (P = 0.0007) with ethanol consumption (Supplemental Figure 1B). Candida species constitute between 40% and nearly 90% of all fungal reads, with the majority being Candida parapsilosis and Candida albicans (Supplemental Figure 1B). There was a significant decrease in relative abundance of C. parapsilosis (P = 0.0007) with ethanol consumption. The apparent increase in C. albicans relative abundance was not significantly different.
Figure 1. Chronic alcohol feeding increases the number of fungi in the intestine and translocation of fungal products.
C57BL/6 mice were fed an oral control diet (n = 6–8) or ethanol diet (n = 8–14). (A) Total fungi in feces were assessed by qPCR. (B) Mean plasma levels of 1,3-β-d-glucan. Unpaired Student’s t test. *P < 0.05.
Development of alcoholic liver disease involves increased translocation of microbial products from the intestinal lumen to the systemic circulation, facilitated by dysfunctional intestinal epithelial tight junctions, and increased paracellular permeability (20). The mean plasma level of 1,3-β-d-glucan was slightly higher in WT mice fed an ethanol diet than in WT mice fed an isocaloric control diet for 8 weeks (Figure 1B). A shorter ethanol feeding period of 5 weeks did not increase plasma 1,3-β-d-glucan (Supplemental Figure 1C). These results indicate that chronic ethanol feeding promotes expansion of intestinal fungi and translocation of fungal cell products to the systemic circulation. Future studies are required to assess not only the quantity of intestinal fungi, but also compositional changes at different time points after ethanol feeding.
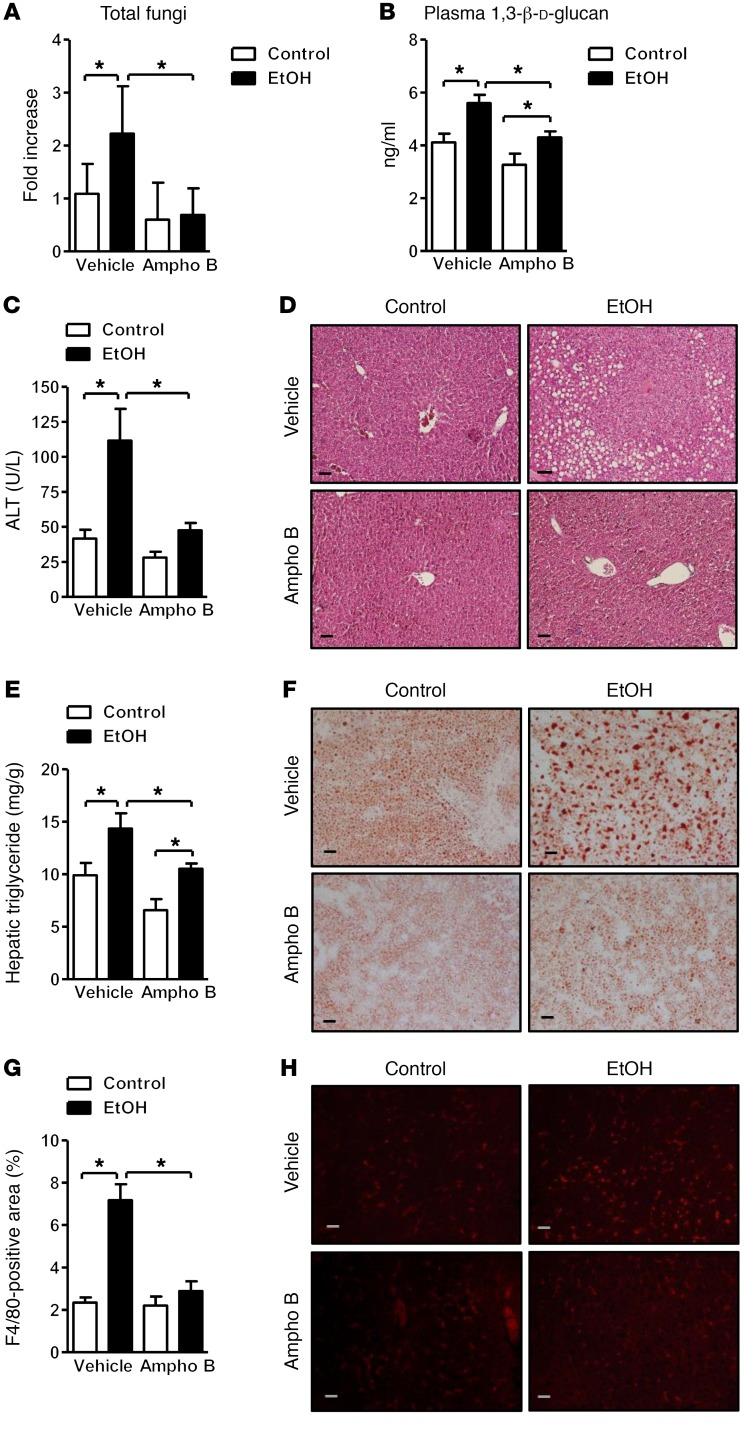
Reducing an alcohol-associated increase in intestinal fungi attenuates features of ethanol-induced liver disease in mice.
To determine whether reducing intestinal fungi protects against ethanol-induced liver disease, the commensal mycobiota was decreased during ethanol feeding with the nonabsorbable antifungal agent amphotericin B. Intestinal fungal overgrowth was observed in mice fed ethanol, but not in mice given amphotericin B with the ethanol diet (Figure 2A). Reducing the alcohol-associated increase in intestinal fungi significantly lowered the mean plasma level of 1,3-β-d-glucan produced with chronic ethanol feeding (Figure 2B).
Figure 2. Decreased ethanol-induced liver disease in mice treated with antifungals.
C57BL/6 mice were fed an oral control diet (n = 4–14) or ethanol diet (n = 10–23), and also given vehicle or amphotericin B (Ampho B). (A) Total fungi in feces were assessed by qPCR. (B) Plasma levels of 1,3-β-d-glucan. (C) Plasma levels of alanine aminotransferase (ALT). (D) Representative liver sections after H&E staining. (E) Hepatic triglyceride content. (F) Representative Oil Red O–stained liver sections. (G and H) Representative liver sections of F4/80 immunofluorescence staining; positively stained area was quantified by image analysis software (n = 2–5). Scale bars: 50 μm. Unpaired Student’s t test. *P < 0.05.
Ratios of liver to body weight were similar in mice given vehicle versus amphotericin B and fed ethanol for 8 weeks (Supplemental Figure 2A). Mice receiving amphotericin B developed less severe ethanol-associated liver disease; these mice had lower levels of liver injury (based on plasma level of alanine aminotransferase) and hepatic steatosis than mice receiving vehicle (Figure 2, C and D). This was confirmed by measurement of hepatic triglycerides and staining for hepatic steatosis (Figure 2, E and F). Amphotericin B also prevented a hepatic increase of the macrophage marker F4/80, indicating a lower number of inflammatory Kupffer cells (Figure 2, G and H).
To determine whether treatment with amphotericin B affects intestinal absorption and hepatic metabolism of ethanol, we measured plasma levels of ethanol and analyzed hepatic ethanol metabolism. Plasma levels of ethanol were comparable between mice receiving vehicle versus amphotericin B following chronic ethanol administration (Supplemental Figure 2B). Alcohol dehydrogenase (ADH) and cytochrome P450 enzyme 2E1 (CYP2E1) are main hepatic enzymes that metabolize ethanol and convert it to acetaldehyde (5). Hepatic ADH activity did not differ significantly between groups given vehicle versus amphotericin B while fed ethanol (Supplemental Figure 2C). Hepatic levels of CYP2E1 protein increased to a similar extent following ethanol administration to mice given vehicle versus amphotericin B (Supplemental Figure 2D). These findings suggest that amphotericin B prevents liver disease without affecting intestinal absorption or hepatic metabolism of ethanol.
To determine whether a reduction of fungi in the intestine alters the bacterial composition, the fecal microbiota was analyzed by 16S rRNA gene sequencing. Chronic ethanol administration induced bacterial dysbiosis in mice given vehicle (4, 21, 22) (Supplemental Figure 2E). Antifungal treatment did not produce major changes in the bacterial composition in isocaloric or ethanol diet–fed mice by comparison of bacterial 16S rRNA data sets (Supplemental Figure 2E). In addition, total numbers of bacteria did not differ significantly between mice given vehicle versus amphotericin B while fed ethanol (Supplemental Figure 2F).
Chronic ethanol administration is associated with a breach in the gut barrier and lower expression of intestinal tight junction proteins (23). We confirmed that occludin expression decreased in the jejunum after chronic ethanol feeding of mice given vehicle, similar to the reduced expression in mice receiving amphotericin B (Supplemental Figure 2G). Consistent with occludin expression data, paracellular permeability, based on systemic levels of translocated lipopolysaccharide (LPS), did not differ between vehicle- and amphotericin B–treated mice after ethanol feeding (Supplemental Figure 2H). These findings indicate that a reduction of ethanol-associated fungal overgrowth does not alter bacterial dysbiosis, increased intestinal permeability, and translocation of LPS. Interestingly, hepatic IL-1R–associated kinase 3 (Irak3, also called IRAK-M) expression was significantly higher in control and ethanol-fed mice after amphotericin B treatment (Supplemental Figure 2I). IRAK3 is a negative regulator of Toll-like receptor (TLR) signaling through inhibition of MYD88 signaling (24), which might decrease LPS/TLR4 signaling during chronic ethanol feeding.
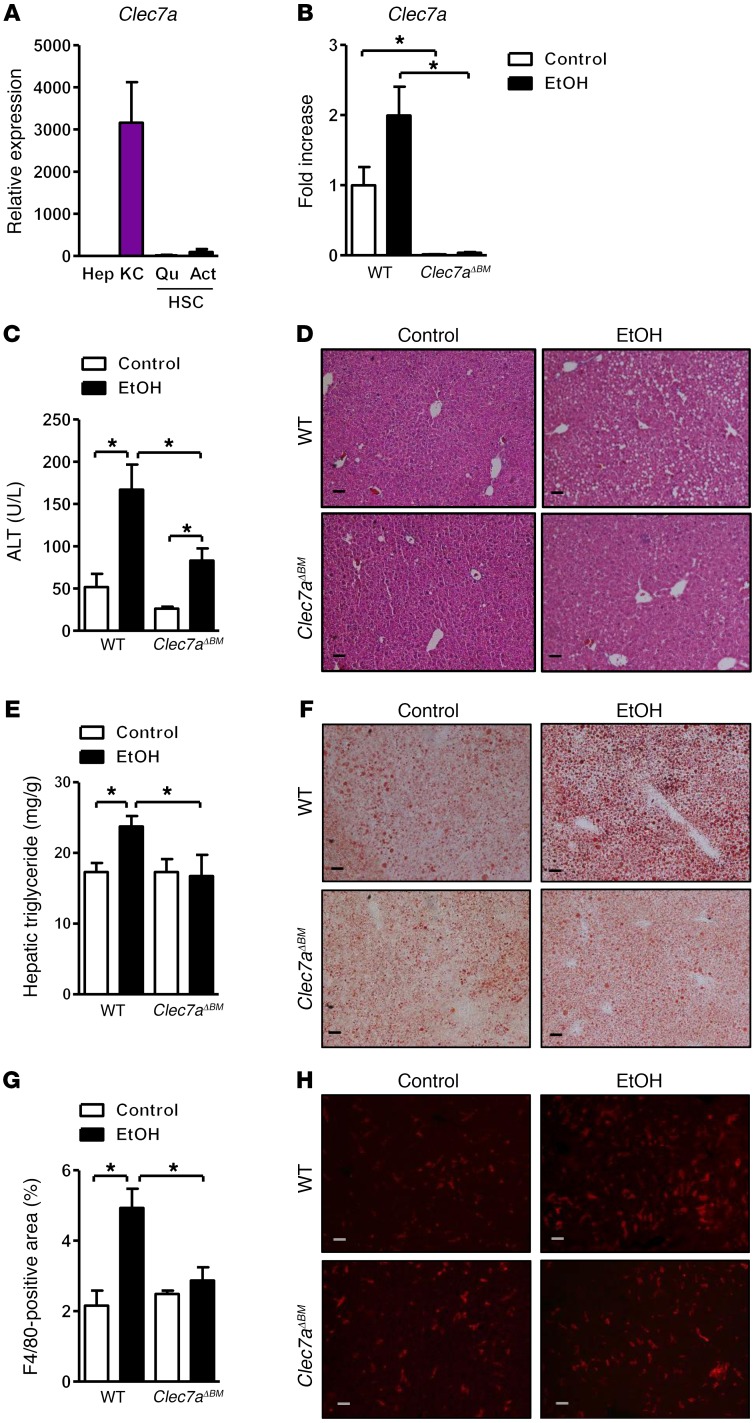
Loss of CLEC7A from bone marrow–derived cells decreases ethanol-induced steatohepatitis.
CLEC7A is primarily expressed on myeloid cells (18, 25). To determine the expression pattern of CLEC7A in liver, parenchymal and nonparenchymal hepatic cells were isolated from WT mice. Clec7a mRNA was highly expressed on Kupffer cells but not on hepatocytes or hepatic stellate cells in mice (Figure 3A). Using immunofluorescence analyses, we found that in livers of mice, CLEC7A colocalized with F4/80 (Supplemental Figure 3A). These results were confirmed in human liver cells and tissue sections (Supplemental Figure 3, B and C). CLEC7A gene expression was not significantly different in liver and duodenal biopsies of alcohol-dependent patients as compared with controls (Supplemental Figure 3, D and E). To investigate whether innate immune signaling through CLEC7A on bone marrow–derived cells and Kupffer cells mediates the effect of translocated fungal products following chronic ethanol administration, we generated CLEC7A bone marrow chimeric mice using a combination of clodronate-mediated Kupffer cell depletion, irradiation, and bone marrow transplantation; WT or CLEC7A-deficient bone marrow was transplanted into WT recipient mice. This protocol achieves full reconstitution of Kupffer cells (26). To confirm successful bone marrow transplantation, we measured whole-liver expression of Clec7a mRNA; we detected low levels of Clec7a mRNA in livers of WT mice that received Clec7a–/– bone marrow (Figure 3B). Chimeric mice with bone marrow–derived cells that lacked Clec7a expression had slightly lower ratios of liver to body weight (Supplemental Figure 4A) and developed less severe ethanol-induced liver injury (Figure 3, C and D) and steatosis (Figure 3, E and F) than chimeric mice with bone marrow–derived cells that expressed CLEC7A.
Figure 3. Mice lacking CLEC7A in bone marrow–derived cells are protected from ethanol-induced liver disease.
(A) Expression of Clec7a in primary mouse hepatocytes (Hep), Kupffer cells (KC), and quiescent and activated hepatic stellate cells (QuHSC and ActHSC) was measured by qPCR (n = 2–3 independent experiments). (B–H) C57BL/6 mice underwent transplantation of WT or Clec7a–/– bone marrow (Clec7aΔBM) and were fed an oral control diet (n = 5) or ethanol diet (n = 8–10; 3 technical replicates). (B) Hepatic Clec7a mRNA expression. (C) Plasma levels of ALT. (D) Representative liver sections after H&E staining. (E) Hepatic triglyceride content. (F) Representative Oil Red O–stained liver sections. (G and H) Representative liver sections of F4/80 immunofluorescence staining; positively stained area was quantified by image analysis software (n = 5–7). Scale bars: 50 μm. Unpaired Student’s t test. *P < 0.05.
Chimeric mice lacking Clec7a on bone marrow–derived cells also had fewer F4/80-positive cells in the liver following ethanol feeding than mice with WT bone marrow (Figure 3, G and H). Plasma levels of ethanol and hepatic ethanol metabolism did not differ significantly with ethanol feeding in chimeric mice (Supplemental Figure 4, B–D). Following ethanol administration, paracellular intestinal permeability (as quantified by plasma level of LPS and 1,3-β-d-glucan) did not differ between mice with WT versus Clec7a–/– bone marrow (Supplemental Figure 4, E and F). These findings indicate that CLEC7A expression on Kupffer cells and possibly other bone marrow–derived cells contributes to liver disease following chronic ethanol administration without altering ethanol metabolism or affecting ethanol-induced gut barrier dysfunction.
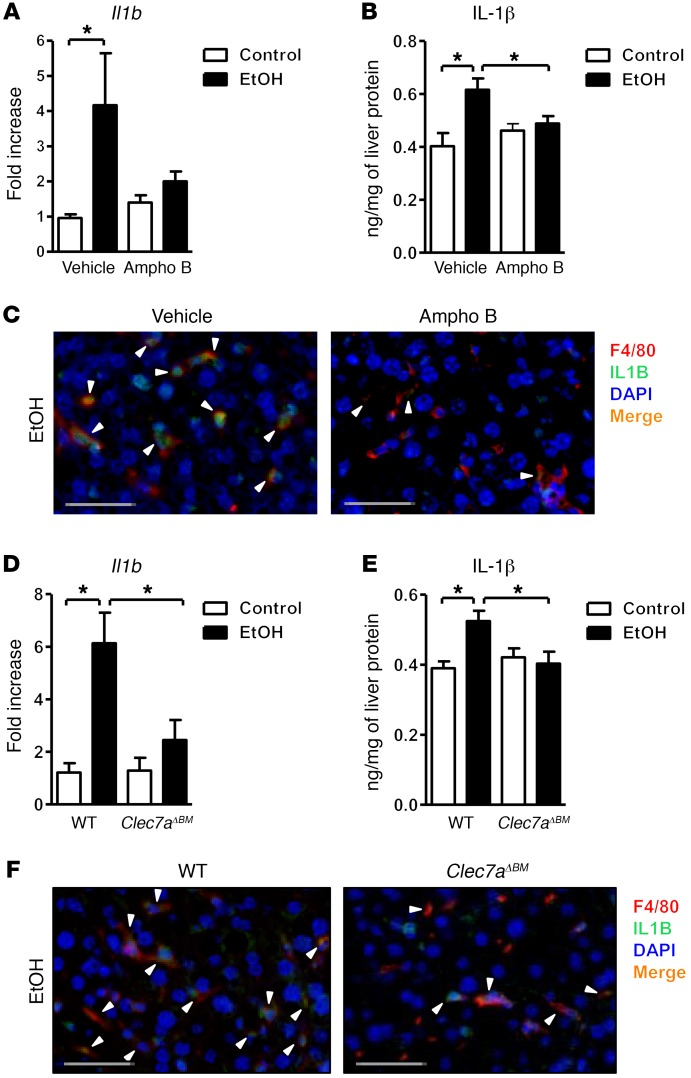
In mice fed ethanol, fungal products induce secretion of IL-1β from Kupffer cells via CLEC7A signaling.
CLEC7A signaling leads to upregulation of the inflammatory cytokine IL-1β (19). Development of alcoholic liver disease requires IL-1β (27). Blocking IL-1β prevents alcoholic steatohepatitis in mice (27). We confirmed that levels of Il1b mRNA and IL-1β protein significantly increased after chronic administration of ethanol to WT mice (Figure 4, A and B). Amphotericin B prevented increases in hepatic Il1b mRNA and IL-1β protein following ethanol feeding (Figure 4, A and B). Inflammation, mediated by Kupffer cells, is required for ethanol-induced liver disease (28). It is therefore possible that CLEC7A-mediated signals in Kupffer cells are required for induction of IL-1β and liver inflammation.
Figure 4. Fungal products induce IL-1β in Kupffer cells.
(A–C) C57BL/6 mice were fed an oral control diet (n = 5–11) or ethanol diet (n = 13–15) and given vehicle or amphotericin B (Ampho B). (A) Hepatic expression of Il1b mRNA. (B) Hepatic levels of IL-1β protein. (C) Immunofluorescence analysis of F4/80 (red) and IL-1β (green) (representative liver sections); nuclei are blue. (D–F) C57BL/6 mice underwent transplantation of WT or Clec7a–/– bone marrow (Clec7aΔBM) and were fed an oral control diet (n = 5) or ethanol diet (n = 8–10). (D) Hepatic expression of Il1b mRNA. (E) Hepatic levels of IL-1β protein. (F) Immunofluorescence analysis of F4/80 (red) and IL-1β (green) (representative liver sections); nuclei are blue. Arrowheads indicate double-positive cells. Scale bars: 50 μm. Unpaired Student’s t test. *P < 0.05.
Using immunofluorescence analysis we found that in livers of ethanol-fed mice, F4/80 colocalized with IL-1β (Figure 4C), indicating that Kupffer cells are the main liver cell type to express and secrete IL-1β during ethanol-induced liver disease. This colocalization disappeared from livers of mice given amphotericin B. Consistent with our data from mice given amphotericin B, chimeric mice lacking Clec7a on Kupffer cells expressed lower levels of Il1b mRNA and IL-1β protein (Figure 4, D and E), due to reduced expression by Kupffer cells (Figure 4F). Plasma IL-1β did not significantly differ between mice lacking CLEC7A on bone marrow–derived cells following ethanol feeding and mice with WT bone marrow (data not shown). To demonstrate that soluble β-glucan can activate CLEC7A in vivo, carboxymethyl-β-1,3-d-glucan (CM-curdlan) was injected into WT and Clec7a-deficient mice. CM-curdlan significantly induced hepatic Il1b in WT but not in Clec7a–/– mice (Supplemental Figure 4G). Plasma IL-1β was undetectable in vehicle-treated WT mice and in WT mice 2, 8, and 24 hours after CM-curdlan injection (data not shown). These findings suggest that alcohol-induced translocation of fungal products, such as β-glucans, induces liver rather than systemic inflammation because these molecules bind to and activate CLEC7A on Kupffer cells. The subsequent increase in expression and secretion of IL-1β promotes ethanol-induced liver disease.
1,3-β-Glucan–induced secretion of IL-1β from Kupffer cells contributes to hepatocyte damage.
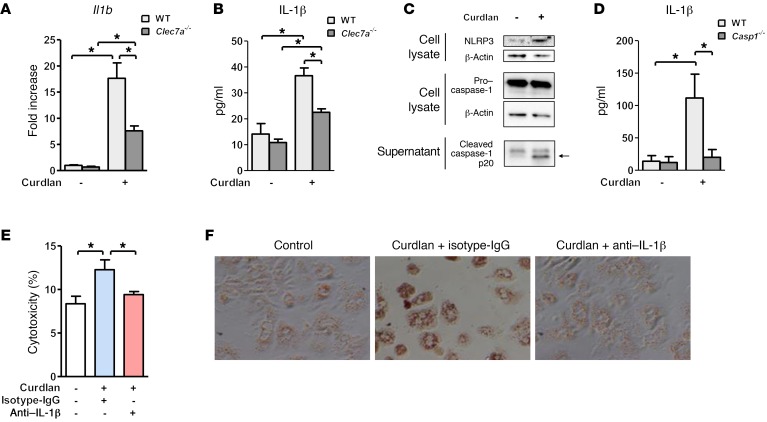
To further define a mechanism by which β-glucan induces an inflammatory response that is mediated by Kupffer cells, we isolated Kupffer cells from WT and Clec7a–/– mice and stimulated them with curdlan, a linear form of 1,3-β-d-glucan. Curdlan induced Il1b mRNA expression and IL-1β secretion in WT Kupffer cells, and at much lower levels in Clec7a–/– Kupffer cells (Figure 5, A and B). Curdlan-stimulated Kupffer cells that lacked CLEC7A still expressed higher levels of Il1b mRNA and IL-1β protein than unstimulated Clec7a–/– Kupffer cells. This could be because β-glucan induces caspase-1 activation and IL-1β production through other receptors, such as complement receptor 3 (CR3) (29). Curdlan-induced expression of chemokine (C-X-C motif) ligand 1 (Cxcl1), Cxcl2, and tumor necrosis factor (Tnf) was independent of CLEC7A (Supplemental Figure 5A).
Figure 5. Curdlan induction of IL-1β by Kupffer cells contributes to hepatocyte injury and steatosis.
(A and B) Primary mouse WT and Clec7a–/– Kupffer cells were stimulated with curdlan; graphs show expression of Il1b mRNA (A) and secretion of IL-1β (B) (n = 3–6 independent experiments). (C) Levels of cellular NLRP3 and caspase-1 and cleaved caspase-1 in supernatants of Kupffer cells with and without curdlan stimulation. (D) IL-1β secretion by WT or caspase-1–deficient Kupffer cells following curdlan stimulation (n = 4–8 independent experiments). (E and F) Conditioned medium from Kupffer cells (stimulated or not stimulated with curdlan) was transferred to primary mouse hepatocytes in the presence of control (IgG) or IL-1β neutralizing antibody. Hepatocyte cytotoxicity (E) and lipid accumulation, determined by Oil Red O staining (F) (n = 3 independent experiments performed in triplicate). Unpaired Student’s t test (A and B); Mann-Whitney U-statistic test (D); 1-way ANOVA with Newman-Keuls post-test (E). *P < 0.05.
Together, our results suggest that curdlan activates the transcription of Il1b and processing of pro–IL-1β into mature IL-1β in cultured Kupffer cells. The CLEC7A signaling pathway results in cleavage of pro–IL-1β and production of mature IL-1β by activating canonical NLRP3 and caspase-1 inflammasome pathways (19). We confirmed that curdlan leads to increased levels of cellular NLRP3 and cleaved caspase-1 in supernatant of Kupffer cells (see immunoblot, Figure 5C). Caspase-1–deficient Kupffer cells exposed to curdlan secrete lower levels of IL-1β than WT Kupffer cells (Figure 5D). Consistent with these findings, cleaved caspase-1 was lower in the liver of chimeric mice that received Clec7a–/– bone marrow versus mice with WT bone marrow following ethanol administration (Supplemental Figure 5B).
We transferred conditioned medium from Kupffer cells stimulated with curdlan to cultured primary mouse hepatocytes. The supernatant from Kupffer cells stimulated with curdlan significantly increased death of the primary hepatocytes. This cytotoxic effect was reduced with a neutralizing antibody against IL-1β (Figure 5E).
Oil Red O staining showed marked lipid accumulation in cultured hepatocytes following incubation with conditioned medium from Kupffer cells stimulated with curdlan. Steatosis was also blocked when a neutralizing antibody against IL-1β was added to the cell culture experiment (Figure 5F). Direct stimulation of hepatocytes with curdlan did not induce cell death or steatosis (Supplemental Figure 5, C and D). IL-1β alone is not sufficient to induce cell death in cultured hepatocytes (Supplemental Figure 5E). These results suggest that β-glucan–induced IL-1β and other inflammatory mediators by secreted Kupffer cells synergize to promote hepatocyte injury and steatosis.
Immune response to fungi correlates with survival of patients with chronic alcohol abuse.
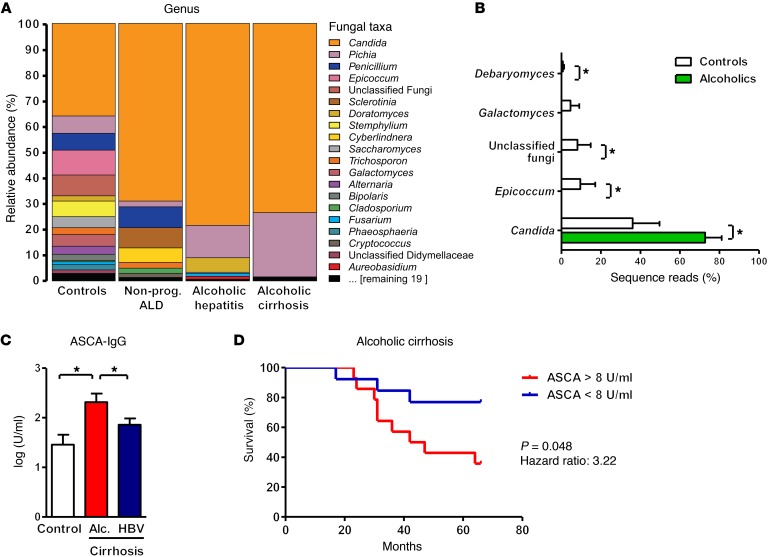
We investigated whether patients who abuse alcohol have disturbances in their intestinal mycobiome and extraintestinal exposure to fungal products. Using ITS sequencing, we observed changes in the abundance and composition of the fecal mycobiome between alcohol-dependent patients and healthy individuals (controls). Fungal species richness (average number of OTUs, 5 ± 3 vs. 9 ± 6) and diversity (average Simpson index, 0.74 ± 0.25 vs. 0.42 ± 0.24) were lower in alcoholics compared with controls. There was also a dramatic overgrowth of Candida, with concomitant decreases in Epicoccum, unclassified fungi, Galactomyces, and Debaryomyces (Figure 6, A and B, and Supplemental Figure 6, A and B). Interestingly, overgrowth of Candida was independent of the stage of liver disease; patients with nonprogressive alcoholic liver disease, alcoholic hepatitis, or alcoholic cirrhosis had similar levels of fungal dysbiosis in the intestine. The opportunistic pathogen Candida albicans was among the most abundant Candida species in feces of patients abusing alcohol; however, its average relative abundance diminished with increasing severity of liver disease. Instead, there was an increase in the average relative abundance of Candida dubliniensis (Supplemental Figure 6C).
Figure 6. Fungal dysbiosis and immune response in alcohol-dependent patients.
(A and B) ITS sequencing of fecal samples from controls (n = 8), alcohol-dependent patients with nonprogressive alcoholic liver disease (Nonprog. ALD; n = 10), alcoholic hepatitis (n = 6), or alcoholic cirrhosis (n = 4). (A) The graph demonstrates the average relative abundance of sequence reads in each genus for controls and for each of the 3 liver disease stages. (B) Green bars indicate the average relative abundance of sequence reads in a combined alcohol-dependent group (independent of liver disease stage, n = 20) that were significantly different from the controls (white bars, n = 8). Kruskal-Wallis rank-sum statistical test with YAP. *P < 0.05. (C) Serum level of ASCA in healthy individuals (controls, n = 14) or patients with alcohol-related cirrhosis (Alc. cirrhosis; n = 28) or chronic HBV-related cirrhosis (n = 43). One-way ANOVA with Newman-Keuls post-test. *P < 0.05. (D) Kaplan-Meier curve of liver-related mortality for patients with alcoholic cirrhosis. Patients were grouped according to their serum levels of ASCA. Fourteen patients had serum concentrations of ASCA greater than 8 U/ml (the median value), and 13 had concentrations below this value. One patient was lost during the follow-up period.
We next determined systemic exposure and immune response to intestinal fungi by measuring serum anti–Saccharomyces cerevisiae IgG antibodies (ASCA). Although S. cerevisiae mannan is used to detect ASCA, C. albicans is an important immunogen for ASCA and is likely the origin of an aberrant immune response in humans (30). Serum samples from patients with cirrhosis due to alcohol abuse had significantly higher levels of ASCA compared with controls or patients with cirrhosis due to chronic hepatitis B virus (HBV) infection (Figure 6C). These results indicate fungal dysbiosis and increased systemic exposure and immune response to intestinal fungi in patients with alcohol abuse. Moreover, levels of ASCA correlated with mortality in patients with alcoholic cirrhosis. A significantly smaller proportion of patients with high levels of ASCA survived for 5.5 years than patients with low levels (Figure 6D). The 2 groups were similar with respect to model for end-stage liver disease (MELD) score at inclusion (12.9 ± 2 for patients with low ASCA levels vs. 11.9 ± 1.2 for patients with high ASCA levels).
Discussion
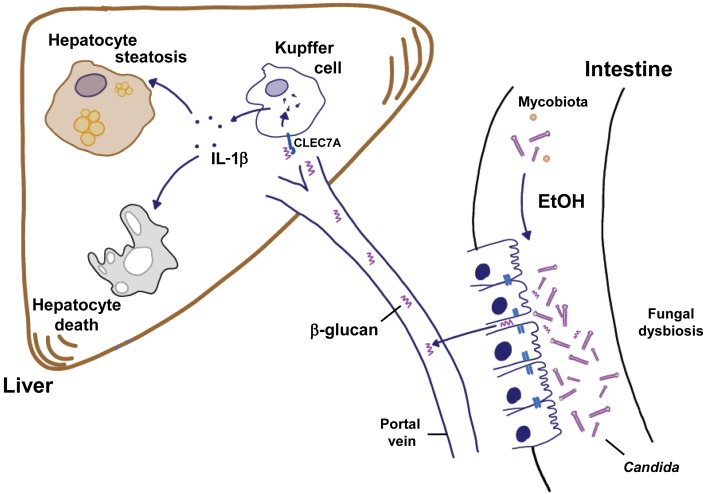
Commensal fungi have important functions in health, but alterations in the intestinal mycobiota have been associated with disease (8, 31). Our study links compositional changes of the intestinal mycobiota to progression of alcoholic liver disease. Chronic alcohol abuse is associated with fungal dysbiosis in mice and humans. Fungal cell wall components, mainly β-glucan, translocate from the intestinal lumen to extraintestinal spaces that include the liver. Binding of β-glucan to CLEC7A on Kupffer cells and possibly other bone marrow–derived cells promotes alcoholic liver disease by inducing hepatic inflammation, ultimately leading to steatosis and cell death of hepatocytes. Preventing fungal overgrowth protects mice from ethanol-induced liver disease (Figure 7). In our human cohort, the degree of exposure to fungal products correlates with mortality of patients with cirrhosis from alcohol abuse but not viral hepatitis. Although a larger, prospective study is required to confirm data from our retrospective analysis of a human cohort with a relatively small number of patients, our findings support the importance of the mycobiota for patients with alcoholic liver disease.
Figure 7. Contribution of the intestinal mycobiota to alcoholic liver disease.
Chronic consumption of ethanol increases the fungal population and causes dysbiosis of the mycobiota in the intestine (right). The fungal dysbiosis results in higher amounts of β-glucan translocating across a damaged gut barrier to the liver (left). Increased β-glucan binds to CLEC7A on hepatic Kupffer cells and induces the expression and secretion of IL-1β. This cytokine contributes to ethanol-induced liver inflammation, hepatocyte injury, and steatosis.
We confirmed that ethanol-induced liver disease is associated with an increase in hepatic F4/80-positive cells (22). CLEC7A is primarily expressed on myeloid cells (18). Due to technical experimental limitations, there is a possibility that CLEC7A-positive bone marrow–derived cells other than Kupffer cells mediate the effects of β-glucan in the liver. However, CLEC7A on bone marrow–derived cells does not alter gut barrier function and systemic inflammation. Elimination of resident Kupffer cells and infiltrating macrophages prevents ethanol-induced liver disease in rodents (28), suggesting that these myeloid cells are required for progression of alcoholic liver disease. Activation of pathogen recognition receptors of the innate immune system induces the appropriate host defense response to pathogens. Receptors such as CLEC7A detect fungal infections and activate a strong innate immune response. Once the infection is cleared, the inflammatory response is turned off. However, chronic exposure of CLEC7A to fungal products continuously activates the cellular inflammasome pathway, causing a continuous inflammatory response and tissue damage. Similar processes might contribute to fungus allergies following chronic exposure in mice (32).
Alcoholic liver disease is associated with a dysfunctional gut barrier that allows translocation of microbial products from the intestine to the liver (20, 33, 34). We have previously demonstrated that dysbiosis-induced intestinal inflammation increases intestinal permeability (23, 35). Reducing bacteria with nonabsorbable antibiotics and restoration of bacterial eubiosis abolishes the onset of an intestinal barrier dysfunction (23, 36). Interestingly, decreasing intestinal fungal overgrowth with nonabsorbable antifungals did not alter gut barrier function, and intestinal permeability remained increased. These findings suggest that a main pathogenic mechanism for mycobiota-associated progression of liver disease is an increase in intestinal fungal populations. Overgrowth of fungi produces more fungal products such as β-glucan, which can escape from the intestinal lumen through already dysfunctional enteric tight junctions to the liver. Thus, fungal dysbiosis and overgrowth do not seem to modulate the intestinal barrier function.
The mycobiome of mice is dominated by Candida parapsilosis; following alcohol feeding its abundance decreased, while other fungal populations increased. In humans, Candida albicans is the most abundant species in nonalcoholics, and its abundance is further increasing in alcoholics with nonprogressive alcoholic liver disease. Given these differences between alcohol-associated changes in the human and mouse mycobiome, our data indicate that compositional changes might not be causatively linked to progression of alcoholic liver disease. Rather overgrowth of intestinal fungi in combination with a dysfunctional gut barrier results in increased systemic levels of β-glucan, eliciting a chronic inflammatory response in the liver. Future studies are required to determine whether single fungal species contribute to liver disease progression more than others by possibly generating larger amounts of β-glucan. Candida dubliniensis is increasing in patients with alcoholic hepatitis and is the most abundant Candida species in patients with end-stage liver disease due to alcohol abuse. C. dubliniensis is an opportunistic fungal pathogen recently shown to cause meningitis in a patient with hepatitis C virus–related cirrhosis (37). Chronic liver disease is an independent risk factor for C. dubliniensis bloodstream infections (38).
We have identified a cross-talk between bacterial and fungal pathogen-associated molecular patterns (PAMPs) during alcoholic liver disease. Hepatic IRAK3, a negative regulator of TLR signaling (24), was increased after reduction of an alcohol-associated increase in intestinal fungi. IRAK3-deficient mice are more susceptible to alcohol-induced liver injury (39). This indicates that in the absence of fungal PAMPs, bacterial signaling through TLRs and MYD88 is inhibited. A synergism between bacterial and fungal PAMPs is necessary for development of ethanol-induced liver disease.
Our study demonstrates an important role of fungal dysbiosis in the development of alcoholic liver disease. We found that antifungal drugs prevented ethanol-induced steatohepatitis in mice. This therapeutic strategy can easily be translated into clinical practice, because oral amphotericin B has no systemic side effects and is well tolerated by patients. Alcohol-associated fungal dysbiosis in humans is characterized by a large increase in Candida, which is susceptible to amphotericin B. The effects of this drug should be tested in patients with alcohol-related liver disease, who are in urgent need of new therapeutics. Manipulation of the intestinal mycobiome might be an effective strategy for attenuation of alcohol-related liver disease.
Methods
Mice.
Clec7a–/– mice have been described (40). Heterozygous (Clec7a+/–) mice on a C57BL/6 genetic background were bred to generate mice without disruption of Clec7a (WT) and mice with homozygous disruption (Clec7a–/– mice). Female mice (age, 8 weeks) were fed ethanol for 8 weeks (the Lieber-DeCarli diet model of chronic alcohol consumption), as previously described (23). The Lieber-DeCarli diet comprises Micro-Stabilized Alcohol Rodent Liquid Diet (LD101A, irradiated; TestDiet), maltodextrin IRR (9598; TestDiet), and 200-proof ethanol (Gold Shield). The caloric intake from ethanol was 0 on day 1, 10% of total calories on days 2–4, 20% on days 5–7, 30% from day 8 until the end of 6 weeks, and 36% for the last 2 weeks. Control mice received an isocaloric amount of isomaltose instead of ethanol.
To produce bone marrow chimeras, mice were given lethal doses of radiation (600 rad) twice, using a 137Cs source. Two weeks after bone marrow transplantation, mice were injected i.p. with 200 μl of clodronate liposomes (5 mg/ml; Vrije Universiteit, Amsterdam, Netherlands) to deplete radioresistant Kupffer cells. The Lieber-DeCarli diet began 4 weeks after bone marrow transplantation.
To reduce intestinal commensal fungi, nonabsorbable amphotericin B (60 mg/kg/d) (41) (Medisca) was added to the liquid Lieber-DeCarli alcohol-containing and control diets. Female C57BL/6 mice were purchased from Charles River Laboratories for this experiment.
To prove whether soluble β-glucan can activate CLEC7A in vivo, carboxymethyl-β-1,3-d-glucan (2 mg; CM-curdlan, Wako) was injected i.p. into WT and Clec7a–/– mice. Control animals received a comparable vehicle (0.01 M NaOH, saline; pH 7) treatment. Mice were sacrificed 2, 8, and 24 hours after CM-curdlan injection and investigated for plasma IL-1β levels and hepatic Il1b expression.
Real-time qPCR.
RNA of mouse tissues was extracted using Trizol (Invitrogen). DNase treatment of RNA was performed using the DNA-free Kit (Ambion), and RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription kit (ABI). Real-time qPCR was performed with iTaq Universal SYBR Green Supermix (Bio-Rad) using primer sequences obtained from NIH qPrimerDepot (mouse and human) and a StepOnePlus thermocycler (ABI). The qPCR value was normalized to mouse 18S. Gene expression results are expressed relative to the levels of control (WT) mice or cells. DNA was isolated from feces as previously described (4, 21, 42). Published microbial primer sequences for 16S rRNA gene (43) and fungal-specific 18S rDNA gene were used (44). To quantify the total bacterial or fungal load present in feces, the qPCR value of fungal 18S rRNA gene or the bacterial 16S rRNA gene, respectively, was multiplied by the total amount of DNA (micrograms) per milligram of feces for each sample.
Sequencing and analysis.
We performed deep DNA pyrosequencing of fecal DNA, targeting the hypervariable V4 region of prokaryotic 16S rRNA loci, using a dual indexing strategy on the Illumina MiSeq platform, as previously described (45). Microbial community profiles were generated using species-level (97% similarity) operational taxonomic unit–based (OTU-based) classification and analysis, as previously described by us (22). Sequence data were registered at NCBI under BioProject PRJNA317649. Sequence reads are available at NCBI under the following consecutive BioSample IDs: SAMN04874715–SAMN04874755.
To evaluate the human intestinal mycobiome, fungal-specific internal transcribed spacer (ITS) amplicon sequencing and analysis were conducted as previously described (4, 46–48). In brief, ITS sequence data were generated with Illumina MiSeq V2 kit using primers BITS and B58S3 specific for ITS1 (47), by a sequencing dual indexing strategy similar to that used for 16S rRNA. Specifically, the 3 primers used are as follows (pad in regular type, linker in brackets, and targeting primer sequence in boldface): read 1 (ITSFWD, containing BITS), CATTATAGCT[CA]ACCTGCGGARGGATCA; I7 index, AACTTTYARCAAYGGATCTC[TG]CTGTATGACT; and read 2 (ITSREV, containing B58S3), AGTCATACAG[CA]GAGATCCRTTGYTRAAAGTT. Index 2 was the standard Illumina Index 2 sequencing primer. PCR parameters included a 95°C denaturing step followed by 35 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, followed by a final extension step of 72°C for 5 minutes. Amplicons were purified using the GeneJet PCR purification system (Thermo Fisher Scientific). Purified amplicons were then quantified using SYBR Gold (Thermo Fisher Scientific), normalized, and pooled to generate a library of equimolar DNA molecules per sample.
To evaluate the mouse intestinal mycobiome, which has a lower abundance of fungi than that of humans, we optimized DNA extraction, quantitation, and amplification methods. Samples were vortexed in Qbiogene lysing matrix B tubes using a Mini-Beadbeater instrument (Biospec Products). We then followed a published DNA isolation protocol (8). DNA samples were quantified using PicoGreen dsDNA assay (Life Technologies); 100 ng DNA was used for PCR with Q5 High Fidelity Polymerase (NEB) with BITS and B58S3 primers specific for ITS1 containing partial dual-indexed Illumina adapters as for 16S rRNA (47) under the following conditions: 98°C denaturing step followed by 39 cycles of 98°C for 10 seconds, 55°C for 20 seconds, and 72°C for 30 seconds, followed by a final extension step of 72°C for 2 minutes. Amplicons were purified twice with Agencourt AMPure beads (Beckman Coulter) at 0.9 beads to DNA ratio. To assess whether amplicons were free of adapter dimers and to verify insert size, amplicons were analyzed and quantified on the Bioanalyzer, using the high-sensitivity dsDNA chip (Agilent), and pooled. The final pool was quantified with Library Quant kit (Kapa Biosystems). Sequences were generated using the MiSeq 2*150 V2 kit (Illumina) with 35% PhiX library added and custom primers spiked in. The ITS and index primer sequences were the same as those used for the human mycobiome.
Sequence processing and analysis steps were performed by our open-access, mothur-based (49) workflow software, YAP (46, 50). Mothur version 1.34 was used in this study. To achieve acceptable memory and run-time scalability with large MiSeq data sets, the preclustering and clustering stages of the pipeline were implemented with CD-HIT (51, 52) sequence clustering software instead of the corresponding built-in mothur commands. Taxonomic classification of assembled forward and reverse reads was performed by mothur’s implementation of the RDP Naïve Bayesian Classifier (53) using version 1.2 of the manually curated targeted host-associated fungi (THF) database (54), trimmed to include only the ITS1 region amplified by the BITS/B58S3 primers, removal of non-ITS sequences, and trimmed sequences less than 45 nucleotides in length, and then formatted for use with mothur. Fungal species-level OTUs were defined at 97% sequence identity (47). Candida species-level classification of ITS sequences was performed using STIRRUPS (55) with a custom sequence library of 31 nonredundant representative sequences pulled from the trimmed THF database. Sequence data were registered at NCBI under BioProject PRJNA317653. Sequence reads are available from the NCBI under the following consecutive BioSample IDs: SAMN04874830–SAMN04874861.
Protein analysis.
IL-1β (14-7012, clone B122; eBioscience) and albumin (E90-134; Bethyl Labs) were measured by ELISA. Human serum samples were assessed for the presence of ASCA-IgG using ELISA (KA1270; Abnova). Immunoblot analysis was performed using antibodies against cytochrome P450 family 2 subfamily E polypeptide 1 (AB1252; CYP2E1; Millipore), β-actin (A5441; Sigma), occludin (71-1500; Invitrogen), NLRP3 (MAB7578; R&D Systems), and caspase-1 p20 (AG-20B-0042-C100; AdipoGen). For detection of cleaved caspase-1 in culture supernatant, proteins were precipitated using methanol/chloroform, and whole extracts were subjected to immunoblotting. Immunoblots were analyzed by densitometry, using NIH ImageJ.
Biochemical analysis.
Plasma levels of alanine aminotransferase (ALT) and hepatic levels of triglyceride were measured using the Infinity ALT kit (Thermo Fisher Scientific) and Triglyceride Liquid Reagents Kit (Pointe Scientific), respectively. Plasma levels of ethanol were measured using the Ethanol Assay Kit (BioVision). Hepatic alcohol dehydrogenase (ADH) activity was determined using the ADH Assay Kit (BioVision). Plasma LPS (CEB526Ge; Cloud-Clone Corp.) and 1,3-β-d-glucan (MBS730464; MyBioSource) levels were measured by ELISA. Plasma was diluted 1:4 for 1,3-β-d-glucan measurements.
Staining procedures.
For immunofluorescence staining, antigens of formalin-fixed slides were retrieved with a retrieval solution (Dako) containing 1% Triton and blocked using tissue blocking solution (Dako). F4/80 antibody (14-4801; eBioscience) and antibody against IL-1β (ab9722; Abcam) were used as primary antibodies. Nuclei were stained with DAPI (blue). For histologic analysis of liver tissues, formalin-fixed liver sections were stained with H&E. Frozen liver sections were analyzed by Oil Red O staining. Samples were analyzed by densitometry, using NIH ImageJ. The results are presented as percentage area positively stained. Immunofluorescent staining of mouse and human liver was performed using frozen sections with primary antibodies against CLEC7A (MCA2289FT; conjugated with FITC; Bio-Rad), mouse F4/80 (14-4801; eBioscience), and human CD68 (IS613; Dako).
Mouse cell culture studies.
Primary mouse Kupffer cells, hepatic stellate cells, and hepatocytes were isolated as previously described (56) with minor modifications. RNA was extracted from hepatic stellate cells directly after the isolation. Quiescent hepatic stellate cells were isolated from C57BL/6 mice, and activated hepatic stellate cells were purified from mice exposed to carbon tetrachloride. C57BL/6 mice were gavaged with 200 μl carbon tetrachloride (25% vol/vol in corn oil) twice weekly for 6 weeks (57). Kupffer cells were isolated from C57BL/6, Clec7a–/–, or Casp1–/– mice (which are also Casp11-deficient) (generated by R. Flavell, Yale University, New Haven, CT, USA) (58) and selected by magnetic cell sorting using anti-CD11b Micro Beads (Miltenyi Biotec). Kupffer cells were cultured with RPMI 1640 (Thermo Fisher Scientific) containing 10% FBS for 4 hours. After an overnight starvation in medium without FBS, Kupffer cells were stimulated with curdlan (100 μg/ml; InvivoGen) for 4 hours (gene expression) or 8–9 hours (protein secretion experiments, immunoblotting experiments).
Primary mouse hepatocytes were cultured with Medium 199 (Thermo Fisher Scientific) containing 10% FBS for 4 hours. After overnight starvation in medium without FBS, cells were stimulated with curdlan (100 μg/ml) for 8 hours, or IL-1β (10 ng/ml; R&D Systems) for 24 hours. Hepatocyte death was quantified using the Pierce LDH Cytotoxicity Detection Kit (Thermo Fisher Scientific). Oil Red O staining was used to identify lipid accumulation in cultured hepatocytes.
For conditioned medium and IL-1β neutralization experiments, Kupffer cells were cultured with RPMI 1640 medium containing 10% FBS for 4 hours and serum starved in RPMI 1640 and Medium 199 (50% vol/vol) overnight. Kupffer cells were then stimulated with curdlan for 8 hours. Cell supernatants were transferred to hepatocytes, and incubated for 24 hours. A neutralizing antibody against IL-1β (10 ng/ml; ab9722; Abcam) or isotype IgG (10 ng/ml, control) was added to the conditioned medium after the transfer. Hepatocyte cytotoxicity and intracellular lipid staining experiments were performed.
Isolation of human liver cells.
Primary human hepatocytes, Kupffer cells, and hepatic stellate cells were isolated as previously described (59, 60). RNA extraction and quantitative reverse transcriptase PCR were performed as previously described (60) using specific primers for human CLEC7A (NIH human qPrimerDepot) and 18S RNA (forward: 5′-AAACGGCTACCACATCCAAG-3′; reverse: 5′-CCTCCAATGGATCCTCGTTA-3′). 18S was used for normalization. Liver tissues used in this study were provided anonymized by the Biobank under the Administration of the Human Tissue and Cell Research (HTCR) Foundation at the Hospital of the LMU Munich (http://www.klinikum.uni-muenchen.de/Chirurgische-Klinik-und-Poliklinik-Grosshadern/de/0800-gewebebank/index.html). The framework of HTCR Foundation (http://www.htcr.org) (61) includes obtaining written informed consent from all donors after the nature and possible consequences of the donation were explained. This framework has been approved by the ethics commission of the Faculty of Medicine in the University of Munich (no. 025-12) as well as the Bavarian State Medical Association (no. 11142).
Human serum samples.
We analyzed data collected from 28 patients with alcoholic liver cirrhosis from July 1, 2010, through October 31, 2010, as previously described (62). In brief, our analysis excluded patients who tested positive for hepatitis C antibody or hepatitis B surface antigen or had any other liver diseases. Liver cirrhosis was detected by ultrasonography (based on presence of cirrhotic liver parenchyma), presence of esophageal or gastric varices (based on upper endoscopy examination), or splenomegaly (based on imaging studies). Continuous alcohol drinking was defined as ingestion of more than 60 g/d of alcohol for more than 10 years in men and more than 20 g/d for more than 10 years in women. We also analyzed data from 2 control cohorts of similar age and sex distribution: 14 healthy individuals and 43 patients with HBV-related liver cirrhosis who did not consume alcohol. Patient characteristics are presented in Supplemental Table 1.
Patients did not take antibiotics or immunosuppressive agents during the 2 months preceding enrollment. Other exclusion criteria were diabetes, inflammatory bowel disease, known liver disease of any other etiology, and clinically significant cardiovascular, pulmonary, or renal comorbidities. The study protocol was approved by the Ethics Committee of En Chu Kong Hospital, and a written informed consent was signed by each participant after the nature and possible consequences of the studies were explained. The percentages of patients with alcoholic liver cirrhosis surviving for a 66-month period were determined from chart review. The cutoff value (8 U/ml) was the median serum level of ASCA-IgG, and the endpoint was liver-related mortality. During the follow-up period (5.5 years), 13 of 28 patients died. The cause of death was liver-related disease in 12 patients, and 1 patient died from metastatic nasopharyngeal carcinoma. One patient was lost to follow-up and excluded from the survival analysis.
Human fecal samples.
Healthy individuals without chronic disease (controls, n = 8), alcohol-dependent patients without evidence of progressive liver disease (nonprogressive liver disease, n = 10), patients with alcoholic hepatitis (alcoholic hepatitis, n = 6), and patients with alcoholic liver cirrhosis (alcoholic cirrhosis, n = 4) were included in the study. Diagnosis of alcoholic hepatitis was made by biopsy in 3 of 6 patients. The remaining patients with alcoholic hepatitis and all patients with alcoholic cirrhosis were diagnosed based on a combination of clinical, biochemical, and imaging parameters. All patients included were actively drinking. Baseline features of this cohort are provided in Supplemental Table 2. Stool samples were collected, and DNA was extracted from fecal samples (21). Written informed consent was signed by each participant after the nature and possible consequences of the studies were explained. The study protocol was approved by the IRB of each institution involved.
Human liver and duodenal biopsies.
Patients with alcohol dependence and with active alcohol consumption were compared with individuals without alcohol dependency (controls). Duodenal biopsies were taken and immediately snap-frozen from otherwise normal subjects who underwent outpatient upper gastrointestinal endoscopy for gastroesophageal reflux symptoms. Only biopsies from subjects with endoscopically and histologically normal duodenum were finally retained as true controls. Alcohol-dependent patients who followed an inpatient alcohol withdrawal program underwent routine upper gastrointestinal endoscopy with duodenal biopsy on the second day of admission. Patients with disturbed liver function tests and suspicion of significant fibrosis determined noninvasively by Fibroscan were proposed for a liver biopsy. Liver biopsy was performed on the third day of admission in patients. Liver specimens from control patients were obtained from surgical resections or from size-reduced donor livers before transplantation. Written informed consent was signed by each participant after the nature and possible consequences of the studies were explained. The study protocol was approved by the Ethics Committee of the Université Catholique de Louvain.
Statistics.
Results are expressed as mean ± SEM. Significance was evaluated using the unpaired Student’s t test except when stated otherwise. For comparison of 3 groups, 1-way ANOVA with Newman-Keuls post-test was used. Not normally distributed (Shapiro-Wilk test) serum levels of ASCA were transformed by taking the logarithm to the base of 10. Significance of mouse microbiome data was determined using the Kruskal-Wallis rank-sum statistical test with YAP. Kaplan-Meier curves were used to compare survival between groups; significance was assessed using the log-rank test. A P value less than 0.05 was considered to be statistically significant.
Study approval.
All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee of UCSD. Studies using human materials were reviewed and approved by an appropriate IRB: (a) ethics commission of the Faculty of Medicine in the University of Munich (no. 025-12) as well as the Bavarian State Medical Association (no. 11142) (human liver cell isolation); (b) Ethics Committee of En Chu Kong Hospital (human serum samples, human cohort study); (c) Human Research Protections Program of UCSD and of the VA San Diego Healthcare System; IRB at Rush University Medical Center; Yale University IRB; University of Wisconsin–Madison Health Sciences IRBs (Madison, Wisconsin, USA); University of North Carolina at Chapel Hill IRB; Ethics Committee of the Université Catholique de Louvain (human fecal samples); (d) Ethics Committee of the Université Catholique de Louvain (human duodenal and liver biopsies). All subjects provided informed consent prior to their participation in the study.
Author contributions
AMY and TI were responsible for acquisition of data, analysis and interpretation of data, and drafting of the manuscript. PC, LW, CL, SB, KH, PH, JX, and YK assisted with experiments and data analysis. MGT, KM, KB, and DEF were responsible for deep sequencing and analysis. TK, CSC, HMH, and GDB provided experimental resources. KF, CH, and SMLL analyzed human liver cells. WZM, GGT, EAM, AK, SBH, RB, and PS were responsible for collection of human samples. BS was responsible for the study concept and design, writing of the manuscript, and study supervision.
Supplementary Material
Acknowledgments
This study was supported in part by NIH grants R01 AA020703, U01 AA24726 and U01 AA021856 and by Award I01BX002213 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development (to BS). KH was supported by a DFG fellowship (HO/5690/1-1). SB was supported by a grant from the Swiss National Science Foundation (P2SKP3_158649). GGT received funding from the Yale Liver Center NIH P30 DK34989 and RB from National Institute on Alcohol Abuse and Alcoholism grant U01 AA021908. AK received support from NIH grants RC2 AA019405, R01 AA020216, and R01 AA023417. GDB is supported by funds from the Wellcome Trust. We acknowledge the Human Tissue and Cell Research (HTCR) Foundation for making human tissue available for research, and Hepacult GmbH (Munich, Germany) for providing primary human hepatocytes for in vitro analyses. We thank Chien-Yu Lin, Department of Medicine, Fu-Jen Catholic University, Taiwan, for statistical analysis.
Footnotes
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(7):2829–2841.https://doi.org/10.1172/JCI90562.
Contributor Information
An-Ming Yang, Email: elwins5840@gmail.com.
Tatsuo Inamine, Email: inaminet@nagasaki-u.ac.jp.
Katrin Hochrath, Email: khochrath@ucsd.edu.
Peng Chen, Email: perchen@smu.edu.cn.
Lirui Wang, Email: liw049@ucsd.edu.
Sena Bluemel, Email: sbluemel@ucsd.edu.
Phillipp Hartmann, Email: phhartmann@ucsd.edu.
Jun Xu, Email: jux004@ucsd.edu.
Yukinori Koyama, Email: ykoyama@ucsd.edu.
Tatiana Kisseleva, Email: tkisseleva@ucsd.edu.
Kelvin Moncera, Email: KMONCERA@jcvi.org.
Karen Beeri, Email: kbeeri@jcvi.org.
Chien-Sheng Chen, Email: cchen103@gmail.com.
Kim Freese, Email: Kim.Freese@fau.de.
Claus Hellerbrand, Email: claus.hellerbrand@fau.de.
Hal M. Hoffman, Email: hahoffman@ucsd.edu.
Guadalupe Garcia-Tsao, Email: guadalupe.garcia-tsao@yale.edu.
Ali Keshavarzian, Email: ali_keshavarzian@rush.edu.
Samuel B. Ho, Email: Samuel.ho2@va.gov.
Ramon Bataller, Email: bataller@med.unc.edu.
Peter Stärkel, Email: peter.starkel@uclouvain.be.
Bernd Schnabl, Email: beschnabl@ucsd.edu.
References
- 1.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. WHO. Global Status Report On Alcohol And Health 2014. World Health Organization website. http://www.who.int/substance_abuse/publications/global_alcohol_report/en/ Accessed April 10, 2017.
- 3.Bode JC, Bode C, Heidelbach R, Dürr HK, Martini GA. Jejunal microflora in patients with chronic alcohol abuse. Hepatogastroenterology. 1984;31(1):30–34. [PubMed] [Google Scholar]
- 4.Yan AW, et al. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology. 2011;53(1):96–105. doi: 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartmann P, et al. Deficiency of intestinal mucin-2 ameliorates experimental alcoholic liver disease in mice. Hepatology. 2013;58(1):108–119. doi: 10.1002/hep.26321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keshavarzian A, et al. Evidence that chronic alcohol exposure promotes intestinal oxidative stress, intestinal hyperpermeability and endotoxemia prior to development of alcoholic steatohepatitis in rats. J Hepatol. 2009;50(3):538–547. doi: 10.1016/j.jhep.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mutlu EA, et al. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol. 2012;302(9):G966–G978. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iliev ID, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336(6086):1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang ZK, Yang YS, Stefka AT, Sun G, Peng LH. Review article: fungal microbiota and digestive diseases. Aliment Pharmacol Ther. 2014;39(8):751–766. doi: 10.1111/apt.12665. [DOI] [PubMed] [Google Scholar]
- 10.Iliev ID, Underhill DM. Striking a balance: fungal commensalism versus pathogenesis. Curr Opin Microbiol. 2013;16(3):366–373. doi: 10.1016/j.mib.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miranda LN, et al. Candida colonisation as a source for candidaemia. J Hosp Infect. 2009;72(1):9–16. doi: 10.1016/j.jhin.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Papp M, et al. Presence of anti-microbial antibodies in liver cirrhosis — a tell-tale sign of compromised immunity? PLoS One. 2010;5(9):e12957. doi: 10.1371/journal.pone.0012957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park WB, et al. Spontaneous cryptococcal peritonitis in patients with liver cirrhosis. Am J Med. 2006;119(2):169–171. doi: 10.1016/j.amjmed.2005.06.070. [DOI] [PubMed] [Google Scholar]
- 14.Bartoletti M, et al. Epidemiology and outcomes of bloodstream infection in patients with cirrhosis. J Hepatol. 2014;61(1):51–58. doi: 10.1016/j.jhep.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 15.Gustot T, et al. Invasive aspergillosis in patients with severe alcoholic hepatitis. J Hepatol. 2014;60(2):267–274. doi: 10.1016/j.jhep.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Hwang SY, et al. Spontaneous fungal peritonitis: a severe complication in patients with advanced liver cirrhosis. Eur J Clin Microbiol Infect Dis. 2014;33(2):259–264. doi: 10.1007/s10096-013-1953-2. [DOI] [PubMed] [Google Scholar]
- 17.Theel ES, Doern CD. β-D-glucan testing is important for diagnosis of invasive fungal infections. J Clin Microbiol. 2013;51(11):3478–3483. doi: 10.1128/JCM.01737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dambuza IM, Brown GD. C-type lectins in immunity: recent developments. Curr Opin Immunol. 2015;32:21–27. doi: 10.1016/j.coi.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kankkunen P, Teirilä L, Rintahaka J, Alenius H, Wolff H, Matikainen S. (1,3)-β-glucans activate both dectin-1 and NLRP3 inflammasome in human macrophages. J Immunol. 2010;184(11):6335–6342. doi: 10.4049/jimmunol.0903019. [DOI] [PubMed] [Google Scholar]
- 20.Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146(6):1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen P, et al. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology. 2015;148(1):203–214.e16. doi: 10.1053/j.gastro.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, et al. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe. 2016;19(2):227–239. doi: 10.1016/j.chom.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen P, Stärkel P, Turner JR, Ho SB, Schnabl B. Dysbiosis-induced intestinal inflammation activates tumor necrosis factor receptor I and mediates alcoholic liver disease in mice. Hepatology. 2015;61(3):883–894. doi: 10.1002/hep.27489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi K, Hernandez LD, Galán JE, Janeway CA, Medzhitov R, Flavell RA. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110(2):191–202. doi: 10.1016/S0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 25.Taylor PR, et al. The β-glucan receptor, dectin-1, is predominantly expressed on the surface of cells of the monocyte/macrophage and neutrophil lineages. J Immunol. 2002;169(7):3876–3882. doi: 10.4049/jimmunol.169.7.3876. [DOI] [PubMed] [Google Scholar]
- 26.Seki E, et al. TLR4 enhances TGF-β signaling and hepatic fibrosis. Nat Med. 2007;13(11):1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- 27.Petrasek J, et al. IL-1 receptor antagonist ameliorates inflammasome-dependent alcoholic steatohepatitis in mice. J Clin Invest. 2012;122(10):3476–3489. doi: 10.1172/JCI60777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niemelä O, Parkkila S, Bradford B, Iimuro Y, Pasanen M, Thurman RG. Effect of Kupffer cell inactivation on ethanol-induced protein adducts in the liver. Free Radic Biol Med. 2002;33(3):350–355. doi: 10.1016/S0891-5849(02)00894-8. [DOI] [PubMed] [Google Scholar]
- 29.Ganesan S, et al. Caspase-8 modulates dectin-1 and complement receptor 3-driven IL-1β production in response to β-glucans and the fungal pathogen, Candida albicans. J Immunol. 2014;193(5):2519–2530. doi: 10.4049/jimmunol.1400276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Standaert-Vitse A, et al. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn’s disease. Gastroenterology. 2006;130(6):1764–1775. doi: 10.1053/j.gastro.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Sokol H, et al. Fungal microbiota dysbiosis in IBD. Gut. [published online ahead of print February 3, 2016]. https://doi.org/10.1136/gutjnl-2015-310746. [Google Scholar]
- 32.Lilly LM, et al. The β-glucan receptor dectin-1 promotes lung immunopathology during fungal allergy via IL-22. J Immunol. 2012;189(7):3653–3660. doi: 10.4049/jimmunol.1201797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szabo G. Gut-liver axis in alcoholic liver disease. Gastroenterology. 2015;148(1):30–36. doi: 10.1053/j.gastro.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirpich IA, et al. Saturated and unsaturated dietary fats differentially modulate ethanol-induced changes in gut microbiome and metabolome in a mouse model of alcoholic liver disease. Am J Pathol. 2016;186(4):765–776. doi: 10.1016/j.ajpath.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du Plessis J, et al. Activated intestinal macrophages in patients with cirrhosis release NO and IL-6 that may disrupt intestinal barrier function. J Hepatol. 2013;58(6):1125–1132. doi: 10.1016/j.jhep.2013.01.038. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, et al. Lactobacillus rhamnosus GG treatment potentiates intestinal hypoxia-inducible factor, promotes intestinal integrity and ameliorates alcohol-induced liver injury. Am J Pathol. 2011;179(6):2866–2875. doi: 10.1016/j.ajpath.2011.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamahiro A, Lau KH, Peaper DR, Villanueva M. Meningitis caused by Candida Dubliniensis in a patient with cirrhosis: a case report and review of the literature. Mycopathologia. 2016;181(7–8):589–593. doi: 10.1007/s11046-016-0006-7. [DOI] [PubMed] [Google Scholar]
- 38.Chen SC, et al. Candidaemia with uncommon Candida species: predisposing factors, outcome, antifungal susceptibility, and implications for management. Clin Microbiol Infect. 2009;15(7):662–669. doi: 10.1111/j.1469-0691.2009.02821.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, et al. Role of IRAK-M in alcohol induced liver injury. PLoS One. 2013;8(2):e57085. doi: 10.1371/journal.pone.0057085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor PR, et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol. 2007;8(1):31–38. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner M, Srivastava KK. Decontamination of gnotobiotic mice experimentally monoassociated with Candida albicans. Infect Immun. 1975;12(6):1401–1404. doi: 10.1128/iai.12.6.1401-1404.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fouts DE, Torralba M, Nelson KE, Brenner DA, Schnabl B. Bacterial translocation and changes in the intestinal microbiome in mouse models of liver disease. J Hepatol. 2012;56(6):1283–1292. doi: 10.1016/j.jhep.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maeda H, et al. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol Med Microbiol. 2003;39(1):81–86. doi: 10.1016/S0928-8244(03)00224-4. [DOI] [PubMed] [Google Scholar]
- 44.Liu CM, et al. FungiQuant: a broad-coverage fungal quantitative real-time PCR assay. BMC Microbiol. 2012;12:255. doi: 10.1186/1471-2180-12-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fouts DE, et al. Next generation sequencing to define prokaryotic and fungal diversity in the bovine rumen. PLoS One. 2012;7(11):e48289. doi: 10.1371/journal.pone.0048289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bokulich NA, Mills DA. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl Environ Microbiol. 2013;79(8):2519–2526. doi: 10.1128/AEM.03870-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bokulich NA, Thorngate JH, Richardson PM, Mills DA. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci U S A. 2014;111(1):E139–E148. doi: 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fouts DE, et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med. 2012;10:174. doi: 10.1186/1479-5876-10-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 53.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang J, Iliev ID, Brown J, Underhill DM, Funari VA. Mycobiome: Approaches to analysis of intestinal fungi. J Immunol Methods. 2015;421:112–121. doi: 10.1016/j.jim.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fettweis JM, et al. Species-level classification of the vaginal microbiome. BMC Genomics. 2012;13(suppl 8):S17. doi: 10.1186/1471-2164-13-S8-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwaisako K, et al. Protection from liver fibrosis by a peroxisome proliferator-activated receptor δ agonist. Proc Natl Acad Sci U S A. 2012;109(21):E1369–E1376. doi: 10.1073/pnas.1202464109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kisseleva T, et al. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc Natl Acad Sci U S A. 2012;109(24):9448–9453. doi: 10.1073/pnas.1201840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brydges SD, Broderick L, McGeough MD, Pena CA, Mueller JL, Hoffman HM. Divergence of IL-1, IL-18, and cell death in NLRP3 inflammasomopathies. J Clin Invest. 2013;123(11):4695–4705. doi: 10.1172/JCI71543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee SM, Schelcher C, Demmel M, Hauner M, Thasler WE. Isolation of human hepatocytes by a two-step collagenase perfusion procedure. J Vis Exp. 2013;(79):50615. doi: 10.3791/50615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steiling H, Mühlbauer M, Bataille F, Schölmerich J, Werner S, Hellerbrand C. Activated hepatic stellate cells express keratinocyte growth factor in chronic liver disease. Am J Pathol. 2004;165(4):1233–1241. doi: 10.1016/S0002-9440(10)63383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thasler WE, Weiss TS, Schillhorn K, Stoll PT, Irrgang B, Jauch KW. Charitable state-controlled foundation human tissue and cell research: ethic and legal aspects in the supply of surgically removed human tissue for research in the academic and commercial sector in Germany. Cell Tissue Bank. 2003;4(1):49–56. doi: 10.1023/A:1026392429112. [DOI] [PubMed] [Google Scholar]
- 62.Yang AM, Wen LL, Yang CS, Wang SC, Chen CS, Bair MJ. Interleukin 10 promoter haplotype is associated with alcoholic liver cirrhosis in Taiwanese patients. Kaohsiung J Med Sci. 2014;30(6):291–298. doi: 10.1016/j.kjms.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.