Abstract
Runt-related transcription factor 1 (RUNX1) is generally considered to function as a tumor suppressor in the development of leukemia, but a growing body of evidence suggests that it has pro-oncogenic properties in acute myeloid leukemia (AML). Here we have demonstrated that the antileukemic effect mediated by RUNX1 depletion is highly dependent on a functional p53-mediated cell death pathway. Increased expression of other RUNX family members, including RUNX2 and RUNX3, compensated for the antitumor effect elicited by RUNX1 silencing, and simultaneous attenuation of all RUNX family members as a cluster led to a much stronger antitumor effect relative to suppression of individual RUNX members. Switching off the RUNX cluster using alkylating agent–conjugated pyrrole-imidazole (PI) polyamides, which were designed to specifically bind to consensus RUNX-binding sequences, was highly effective against AML cells and against several poor-prognosis solid tumors in a xenograft mouse model of AML without notable adverse events. Taken together, these results identify a crucial role for the RUNX cluster in the maintenance and progression of cancer cells and suggest that modulation of the RUNX cluster using the PI polyamide gene-switch technology is a potential strategy to control malignancies.
Keywords: Hematology, Oncology
Introduction
Runt-related transcription factor 1 (RUNX1), also known as acute myeloid leukemia 1 protein (AML1), is an essential master transcription factor implicated in the differentiation of hematopoietic stem cells (1). RUNX1 is a member of RUNX family proteins (RUNX1, RUNX2, and RUNX3), and each member forms a heterodimeric complex with core-binding factor β subunit (CBFB) to exert its sequence-specific transactivation function (2). RUNX modulates the transcription of its target genes through recognizing the core consensus binding sequence 5′-TGTGGT-3′, or very rarely, 5′-TGCGGT-3′, within their regulatory regions via its runt domain, while CBFB is a non–DNA-binding regulatory subunit that allosterically enhances the sequence-specific DNA-binding capacity of RUNX (3, 4). According to previous studies, RUNX1 plays a pivotal role in definitive hematopoiesis, as examined by the phenotypes of Runx1-deficient mice (5). On the other hand, the conditional knockout of Runx1 in adult mice showed marginal changes in platelet counts without hematopoietic stem cell exhaustion (6), implying that RUNX1 is dispensable for adult hematopoiesis. With respect to tumorigenicity, RUNX1 has been considered to act as an oncosuppressor during the development of leukemia owing to its relatively higher frequency of mutations and translocations (7, 8). This classical viewpoint has been challenged by our recent observations showing that wild-type RUNX1 is strongly required for the development of AML with inv(16) or with mixed lineage leukemia (MLL) fusions (9–11). These findings first shed light on the oncogenic property of RUNX1 in the initiation of leukemia. Unfortunately, despite these discoveries, the precise molecular basis for how RUNX1 as well as the other RUNX family members could contribute to the maintenance and/or progression of leukemia has been largely unknown.
p53 (TP53), structurally a distant relative of RUNX, is one of the most famous and intensely studied tumor suppressors in human cancers (12). p53 deficiency generally enhances the initiation or progression of cancers, and tumors lacking p53 elicit more malignant phenotypes characterized by poorer cellular differentiation and increased genetic instability and metastatic potential (13). Frequency of p53 mutations varies according to the tumor background, from less than 10% among de novo AML patients to nearly 50% in lung adenocarcinoma patients (14, 15). Although a few reports have discussed the interactions between RUNX and p53 transcription factors in the context of radiation sensitivity with overexpression experiments (16), their functional cross-talk in the development and maintenance of cancer cells has so far been poorly defined at the physiological level.
Pyrrole-imidazole (PI) polyamides are noncovalent, synthetic oligomers that recognize specific DNA sequences located within the minor groove by virtue of their pyrrole and imidazole pairs interlocked by a hairpin linkage (17). Designing the order of PI pairs enables in vivo delivery of PI polyamides to the targeted site of the genome in a controllable manner. Despite their relatively large molecular weight, PI polyamides are membrane permeant, localize to the cell nucleus, and then affect endogenous gene transcription at nanomolar levels (18). We have recently succeeded in generating potent histone deacetylase (HDAC) inhibitors, suberoylanilide hydroxamic acid–conjugated (SAHA-conjugated) PI polyamides, and demonstrated that they have the ability to specifically stimulate the expression of target genes through enhanced acetylation of their regulatory regions (19, 20). We have also successfully conjugated the nitrogen mustard alkylating agent chlorambucil (Chb) to PI polyamides, and showed that they have a much stronger sequence-specific DNA-binding capacity and reduce target gene expression (21, 22). Taking advantage of these attractive technologies, we have newly synthesized PI polyamides that target the consensus RUNX-binding sequences and efficiently inhibit the recruitment of RUNX family members to their binding sites, and evaluated their therapeutic efficacy against AML cells as well as several types of solid tumors originating from different organs in vivo.
Results
RUNX1 depletion–mediated antileukemic effect requires functional p53.
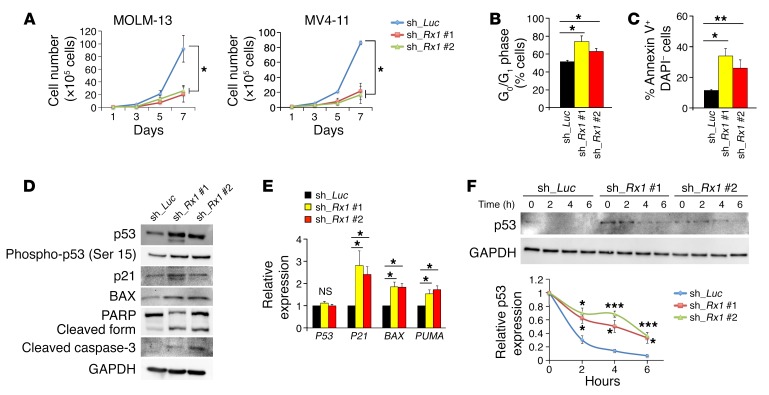
We first investigated whether depletion of RUNX1 could have an antileukemic effect on AML cells by taking advantage of the tetracycline-inducible shRNA-mediated RUNX1-knockdown system. As shown in Figure 1, A–C, and Supplemental Figure 1, A–D (supplemental material available online with this article; https://doi.org/10.1172/JCI91788DS1), silencing of RUNX1 stimulated cell cycle arrest at the G0/G1 phase and simultaneously induced apoptosis in MV4-11, MOLM-13, and OCI-AML3 cells bearing wild-type P53. These results prompted us to examine whether depletion of RUNX1 could affect the proapoptotic p53-mediated cell death pathway. As expected, a remarkable induction of p53 as well as its target gene products such as p21, BAX, and PUMA, and proteolytic cleavage of caspase-3 and its substrate PARP were detectable in RUNX1-depleted MV4-11 cells (Figure 1D). We also observed increased formation of γ-H2AX foci in RUNX1-depleted AML cells, possibly reflecting a cellular response to the activated proapoptotic p53 signaling (Supplemental Figure 2, A and B). Consistent with these results, real-time quantitative reverse transcription PCR (RT-qPCR) analysis revealed that silencing of RUNX1 causes upregulation of p53-target genes (Figure 1E). Intriguingly, the expression of P53 at the mRNA level remained unchanged regardless of RUNX1 depletion (Figure 1E), raising the possibility that RUNX1 could affect the protein stability of p53. To address this issue, we performed cycloheximide chase experiments. As shown in Figure 1F, the half-life of p53 was prolonged in RUNX1-knockdown cells relative to control ones, indicating that RUNX1 negatively regulates p53 protein in AML cells.
Figure 1. p53-dependent antitumor effect of RUNX1 depletion.
(A) Growth curves of MOLM-13 and MV4-11 cells transduced with control (sh_Luc) or with RUNX1 shRNAs (sh_Rx1 #1 and sh_Rx1 #2) in the presence of 3 μM doxycycline (n = 3). (B) RUNX1 depletion–mediated increase in number of cells with G0/G1 DNA content. Nondepleted and RUNX1-depleted MV4-11 cells were cultured in the presence of 3 μM doxycycline. Forty-eight hours after treatment, cells were harvested and analyzed by flow cytometry (n = 3). (C) Early apoptotic cell death induced by RUNX1 silencing. Nondepleted and RUNX1-depleted MV4-11 cells were treated as in B, and the early apoptotic cells (annexin V+ DAPI–) were scored by flow cytometric analysis (n = 3). (D) RUNX1 depletion–mediated stimulation of p53-dependent cell death pathway. Nondepleted and RUNX1-depleted MV4-11 cells were treated as in B. Cell lysates were analyzed by immunoblotting with the indicated antibodies. (E) Induction of p53-target genes in RUNX1-depleted MV4-11 cells. Nondepleted and RUNX1-depleted MV4-11 cells were treated as in B. Total RNA was prepared and then analyzed by real-time RT-PCR for the indicated p53-target genes. Values are normalized to that of control vector–transduced cells (n = 3). (F) Increase in half-life of p53 in RUNX1-silenced MV4-11 cells. MV4-11 cells were treated with 3 μM doxycycline as in B. Forty-eight hours after treatment, cells were exposed to cycloheximide (CHX). Cell lysates were then prepared at the indicated time points, and analyzed for p53 by immunoblotting. Results are normalized to that of control vector–transduced cells (n = 3). Data are the mean ± SEM values. *P < 0.05, **P < 0.01, ***P < 0.001, NS, not significant, by 2-tailed Student’s t test.
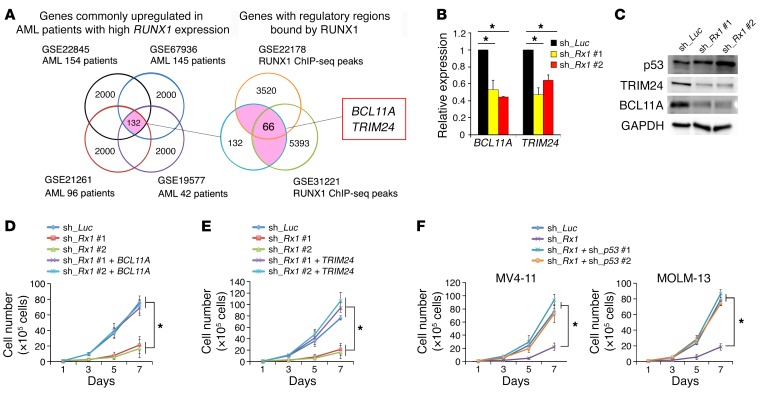
Since RUNX1 is a vital transcription factor, we hypothesized that RUNX1 downregulates p53 through the transcriptional activation of genes implicated in its degradation. To identify genes required for RUNX1-dependent degradation of p53, we first employed 4 independent clinical gene expression array datasets of AML patients and divided their samples into 2 groups based on the expression levels of RUNX1 in their bone marrow. Next, we extracted the top 2,000 commonly upregulated genes in AML patients highly expressing RUNX1 from these array datasets (Supplemental Table 1). We then compared these datasets with the results obtained from extensive chromatin immunoprecipitation using an anti-RUNX1 antibody followed by DNA sequencing (ChIP-seq) and finally identified 66 candidate genes (Figure 2A and Supplemental Table 2). Among them, we focused on BCL11A and TRIM24, both of which have been reported to promote p53 degradation either directly or indirectly (23–26). Moreover, we found that expression levels of BCL11A and TRIM24 were positively correlated to those of RUNX1 among AML patient cohorts, underpinning the possible transcriptional regulation of these genes by RUNX1 (Supplemental Figure 3, A and B). In accordance with these observations, knockdown of RUNX1 resulted in a significant downregulation of BCL11A and TRIM24 at both the mRNA and protein levels (Figure 2, B and C). During close inspection of the transcriptional regulatory regions of the BCL11A and TRIM24 genes, we found several RUNX1-binding consensus sites. ChIP-qPCR experiments using anti-RUNX1 antibody revealed that RUNX1 binds to 4 major consensus sites (5′-TGTGGT-3′) located within 3 kb upstream of the transcription start site (TSS) of BCL11A, and 1 major consensus site found within 1.5 kb downstream of the TSS of TRIM24 (Supplemental Figure 4, A and B). Notably, forced expression of BCL11A or TRIM24 in RUNX1-knockdown MV4-11 cells increased their proliferation rates together with the reduction of p53 amount, indicating that BCL11A and TRIM24 play important roles in the RUNX1-mediated pro-oncogenic pathway (Figure 2, D and E, and Supplemental Figure 5, A and B).
Figure 2. RUNX1 inhibition activates the p53 pathway by transcriptionally controlling BCL11A and TRIM24 expression.
(A) Identification of genes under the control of RUNX1. One hundred and thirty-two genes were commonly upregulated in 4 independent AML patients with a higher RUNX1 expression (GSE22845, GSE67936, GSE21261, and GSE19577). Among them, RUNX1 bound to 66 gene regulatory regions (GSE22178 and GSE31221). See Supplemental Tables 1 and 2 for the list of the indicated genes. (B) Expression levels of BCL11A and TRIM24 in MV4-11 cells transduced with control or RUNX1 shRNAs. Cells were treated with 3 μM doxycycline for 48 hours. Values are normalized to that of control vector–transduced cells (n = 3). (C) Downregulation of BCL11A and TRIM24 in RUNX1-silenced MV4-11 cells. Nondepleted and RUNX1-depleted MV4-11 cells were treated as in B. Cell lysates were analyzed by immunoblotting with the indicated antibodies. (D and E) Rescue of cell proliferation rate by forced expression of BCL11A (D) or TRIM24 (E) in RUNX1-depleted MV4-11 cells. The indicated MV4-11 cells were cultured in the presence of 3 μM doxycycline (n = 3). (F) p53-dependent growth suppression of RUNX1-depleted cells. Nondepleted and RUNX1-depleted MV4-11 (left panel) and MOLM-13 cells (right panel) were transduced with lentivirus vector encoding shRNA targeting p53 (sh_p53 #1 or sh_p53 #2) (n = 3). Data are the mean ± SEM values. *P < 0.05, by 2-tailed Student’s t test.
As mentioned above, knockdown of RUNX1 potentiated the p53-dependent proapoptotic pathway. To further confirm this notion, we knocked down RUNX1 in additional AML cells including P53-deficient KG1a and HL60 cells and P53-mutated MV4-11NR cells. MV4-11NR cells bearing an R248W point mutation in p53 were derived from their parental MV4-11 cells (27). Since nutlin-3 treatment did not induce expression of the p53 target p21 in these AML cells (Supplemental Figure 6A), it suggests that the cell cycle regulatory function of wild-type p53 is diminished. As expected, silencing of RUNX1 did not affect their proliferation rates up to 7 days after induction (Supplemental Figure 6B). Accordingly, knockdown of wild-type P53 in RUNX1-depleted MV4-11 and MOLM13 cells augmented their proliferation potential relative to the control level, implying that RUNX1 depletion–mediated growth inhibition in AML cells is dependent on a functional p53 pathway (Figure 2F and Supplemental Figure 6C).
Regulation of the RUNX family as a cluster exerts a much stronger antileukemic effect relative to the individual silencing of each family member.
Although shRNA-based RUNX1 depletion was highly effective against the proliferation of AML cells, a small subpopulation of leukemia cells retained their proliferation potential even after the silencing of RUNX1 (Figure 1A and Supplemental Figure 1B). Considering that drug-mediated downregulation of target gene expression appears to be incomplete in vivo (28), it is likely that our knockdown experiments with shRNAs might be more likely to exemplify the practical output of RUNX1 inhibitory drugs in vivo compared with the experiments analyzing biallelic Runx1 knockout, which delivers completely depleted Runx1 expression (9, 11).
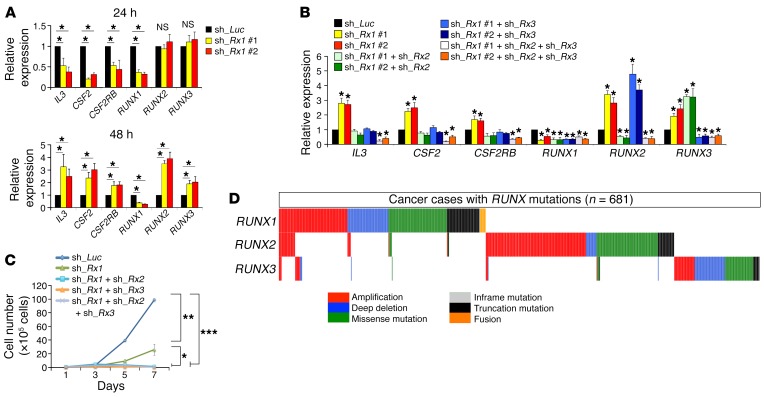
Since it has been shown that RUNX family members have redundant functions (9, 29, 30), we sought to examine the expression levels of the other RUNX family members, RUNX2 and RUNX3, in RUNX1-knockdown AML cells. Under our tetracycline-inducible shRNA expression system, the expression levels of RUNX1 started to decline at 24 hours after doxycycline treatment, when the expression levels of RUNX2 and RUNX3 were unchanged. Notably, the doxycycline-dependent increase in the expression levels of RUNX2 and RUNX3 was detectable at 48 hours of incubation (Figure 3A and Supplemental Figure 7A), suggesting that RUNX2 and RUNX3 might compensate for the loss of RUNX1 expression. Although the expression levels of well-established RUNX1-target genes (IL3, CSF2, and CSF2RB) (31–33) were decreased at 24 hours after RUNX1 knockdown, their expression levels were reciprocally increased at 48 hours after RUNX1 knockdown and accompanied by RUNX2 and RUNX3 stimulation (Figure 3A). Forced expression of RUNX1, RUNX2, or RUNX3 suppressed the expression of RUNX2 and RUNX3, RUNX1 and RUNX3, or RUNX1 and RUNX2, respectively (Supplemental Figure 7B). Careful observations in the proximal regulatory regions of RUNX1, RUNX2, and RUNX3 revealed the existence of recurrent consensus RUNX-binding sequences. As expected, a ChIP-qPCR assay confirmed that all of the RUNX family members could bind to their respective binding sites (Supplemental Figure 8, A–C). Moreover, individual RUNX family members consistently suppressed the promoter activity of the other RUNX members (Supplemental Figure 8D), indicating that each RUNX family member physiologically suppresses the function of the other members, probably owing to their functionally redundant roles in leukemia cells. In accordance with these findings, additional knockdown of RUNX2, RUNX3, or both of them in RUNX1-depleted MV4-11 cells effectively repressed RUNX1-target gene expression and completely suppressed their proliferation (Figure 3, B and C, and Supplemental Figure 8E). These results collectively indicate that the expression of all 3 RUNX family members is required for the maintenance of the expanding AML cells, and thus the simultaneous targeting of all RUNX family members as a cluster achieves much more stringent control of leukemia cells. In addition, we found that all RUNX family members bind to the regulatory regions of BCL11A and TRIM24 (Supplemental Figure 4, A and B) and that additive inhibition of RUNX2 and RUNX3 to RUNX1 depletion proportionally suppressed the expression of BCL11A and TRIM24 and potentiated p53 activity in AML cells (Supplemental Figure 9A), underscoring the importance of functional redundancy among RUNX family members in the maintenance of tumor cells. Furthermore, deceleration of tumor proliferation rate through p53 pathway activation mediated by RUNX family depletion was observed not only in AML cells, but also in solid tumors of different origins as well (Supplemental Figure 9, B and C). Moreover, considering that the functional alterations of RUNX family members exist in a mutually exclusive manner in AML cells as well as in various cancers (Figure 3D and Supplemental Figure 10), their functional redundancy in the maintenance of AML cells might be generally accepted. If this were the case, regulation of RUNX family as a cluster could be an ideal strategy to control not only leukemia, but also other malignant tumors of different origins. With this in mind, we next addressed this possibility of using the synthetic PI polyamides that could target all RUNX family members.
Figure 3. Functional redundancy among RUNX family members.
(A) Stimulation of RUNX2 and RUNX3 in RUNX1-depleted MV4-11 cells. MV4-11 cells were transduced with lentivirus encoding shRNA targeting luciferase (sh_Luc.) or shRNAs against RUNX1 (sh_Rx1 #1 or sh_Rx1 #2) and incubated with 3 μM doxycycline. At the indicated time periods after treatment, total RNA was prepared and analyzed by real-time RT-PCR. Values are normalized to that of control vector–transduced cells (n = 3). (B) Mutual regulation of RUNX family members. MV4-11 cells were transduced with the indicated combinations of lentivirus vectors and incubated with 3 μM doxycycline. Forty-eight hours after treatment, total RNA was prepared and processed for real-time RT-PCR analysis (n = 3). (C) A simultaneous knockdown of RUNX family members leads to a massive decline of proliferation rate of MV4-11 cells. MV4-11 cells were transduced and treated as in (B). Forty-eight hours after treatment, the number of viable cells was scored. Values are normalized to that of control vector–transduced cells (n = 3). (D) Mutually exclusive gene alterations of RUNX family members in 681 human cancer samples out of 18,754 patients with various cancer types. Data were analyzed and visualized by the cBioPortal for Cancer Genomics software (50). Data are the mean ± SEM values. *P < 0.05, **P < 0.01, ***P < 0.001, NS, not significant, by 2-tailed Student’s t test.
Cluster regulation of the RUNX family with PI polyamides.
PI polyamides have been considered to selectively bind to the designated target sites in the minor groove of the genome sequence at nanomolar levels (17). Taking advantage of the nature of this small molecule, we have designed PI polyamides that can specifically recognize and bind to RUNX-binding consensus sites (5′-TGTGGT-3′ and 5′-TGCGGT-3′) by introducing 4 PI pairs. As shown in Supplemental Figure 11A, we first screened the HDAC inhibitors, SAHA-conjugated PI polyamides targeting RUNX consensus binding sequences (SAHA-M′ targets 5′-TGTGGT-3′ and SAHA-50 targets 5′-TGCGGT-3′), which can enhance the transcriptional machinery around the binding sites, as we have previously described (19). We discovered that SAHA-M′ and SAHA-50 potently upregulated the target genes of RUNX1 (Supplemental Figure 11B). We next modified the indicated PI polyamides by switching the conjugate from SAHA to the potent alkylating agent Chb (Chb-M′ and Chb-50; Chb-M′ targets 5′-TGTGGT-3′ and Chb-50 targets 5′-TGCGGT-3′), which could confer stronger and irreversible DNA-binding ability to PI polyamides (Figure 4A). As expected, Chb-M′ and Chb-50 efficiently repressed RUNX1-target gene expression (Figure 4B), and potentiated the p53-dependent apoptotic pathway through downregulation of BCL11A and TRIM24 (Figure 4, C and D, and Supplemental Figure 12). We also confirmed that Chb-M′, which theoretically recognizes the major RUNX consensus binding sequence 5′-TGTGGT-3′, specifically binds and alkylates the 5′-TGTGGT-3′ sequence of DNA (Figure 4E). Of note, RUNX family–mediated regulation of gene expression by these PI polyamides was observed at a genome-wide level (Figure 4F and Supplemental Figures 13–16), further indicating that Chb-M′ and Chb-50 modulate gene expression patterns through regulation of the RUNX family.
Figure 4. Cluster regulation of RUNX family by PI polyamides.
(A) Chemical structures of the synthetic chlorambucil/PI polyamide conjugates targeting 5′-TGTGGT-3′ (Chb-M′) and 5′-TGCGGT-3′ (Chb-50). (B) Chb-50 and Chb-M′ reduce RUNX1-target gene expression. MV4-11 cells were treated with 1 μM Chb-50 or Chb-M′ for 6 hours, and then total RNA was prepared and analyzed by real-time RT-PCR. Values are normalized to that of DMSO–treated cells (n = 3). (C) Chb-50 and Chb-M′ stimulate p53-target gene expression. MV4-11 cells were treated with 1 μM Chb-50 or Chb-M′ for 6 hours, and then total RNA was prepared and analyzed by real-time RT-PCR. Values are normalized to that of DMSO-treated cells (n = 3). (D) Chb-50 and Chb-M′ augment the p53-dependent proapoptotic pathway. MV4-11 cells were treated with 1 μM Chb-50 or Chb-M′ for 24 hours, and then cell lysates were prepared and immunoblotted. (E) Schematic drawing of thermally induced cleavage at the 5′-TGTGGT-3′ sites. Filled circles represent imidazole and open circles represent pyrrole moieties. Arrow indicates the position of Chb-M′–dependent alkylation and cleavage. (F) Heatmap showing top 500 upregulated and downregulated genes in response to 1 μM Chb-M′ in MV4-11 cells compared with those in cells transduced with the indicated combinations of lentivirus vectors. Lentivirus vector–transduced cells were incubated with 3 μM doxycycline for 48 hours before total RNA extractions. Data are the mean ± SEM values. *P < 0.05, NS, not significant, by 2-tailed Student’s t test.
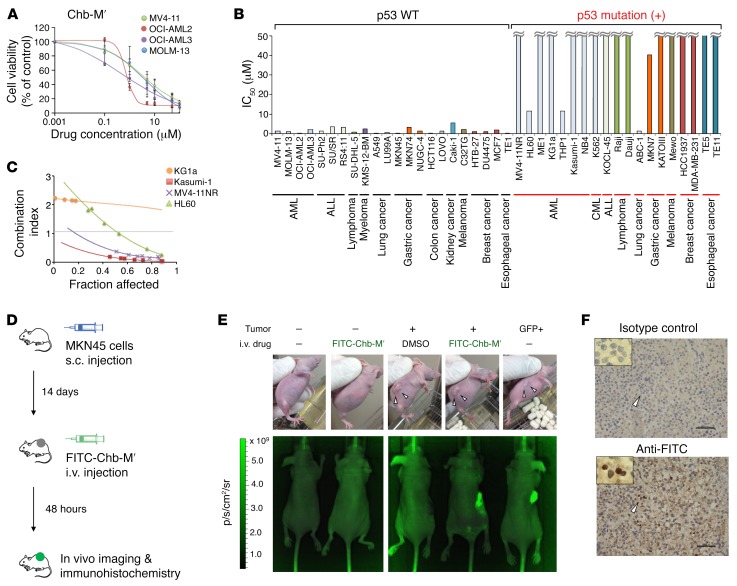
We next calculated the half maximal inhibitory concentration (IC50) of these PI polyamides in several AML-derived cell lines with wild-type P53. As clearly shown in Figure 5A, Chb-M′ was highly effective against these AML cells at nanomolar levels. It is worth noting that Chb-M′ had a remarkable effect on cancer cells of several different origins including acute lymphoblastic leukemia (ALL), lymphoma, myeloma, lung cancer, gastric cancer, esophageal cancer, breast cancer, melanoma, kidney cancer, and colon cancer (Figure 5B). Of note, Chb itself or Chb-conjugated PI polyamides that target the 5′-WGGCCW-3′ sequence (Chb-S) were totally ineffective for AML cell lines in vitro, making a sharp contrast to Chb-M′ (Supplemental Figure 17, A–C). Considering that Chb-M′ targets the major RUNX-binding sequences, while Chb-50 targets the minor ones (Supplemental Figure 17D), it is likely that Chb-M′ has a higher efficacy than Chb-50. Additionally, we found that our PI polyamides lack their growth-suppressive capability against P53-mutated cancer cells, which is consistent with the results obtained from RUNX-knockdown experiments. We have further confirmed that knockdown of p53 in p53-proficient AML cells decreases their sensitivity to Chb-M′ and Chb-50 (Supplemental Figure 18, A and B). In addition, these polyamides lost their potency when p53 expression was indirectly downregulated with additive MDM2 expression or p14ARF suppression in AML cells (Supplemental Figure 18, C–F). In spite of this limitation, the simultaneous treatment of Chb-M′ with the p53 inducer PRIMA-1 synergistically reduced the proliferation rates of P53-deficient AML cells (Figure 5C).
Figure 5. Antitumor activity of Chb-M′ in multiple cancer cell lines.
(A) Dose-response curves of Chb-M′ in AML cells (MV4-11, OCI-AML2, OCI-AML3, and MOLM-13 cells). Cells were treated with the indicated concentrations of Chb-M′. Forty-eight hours after treatment, cell viability was examined by WST assay (n = 3). (B) IC50 values of Chb-M′ against human cancer cell lines established from various origins. Forty-eight hours after Chb-M′ treatment, cell viability was examined by WST assay (n = 3). (C) Combination index plots of Chb-M′ and PRIMA-1 in P53-deficient (KG1a and HL60) and P53-mutated (MV4-11NR and Kasumi-1) AML cells (n = 3). (D) Schematic representation of treatment and monitoring schedule in MKN45-transplanted BALB/c-nu mice. (E) Live animal biofluorescence images at 48 hours after treatment with a single dose of FITC-Chb-M′ (320 μg/kg body weight). Scale represents relative fluorescence units, in photons per second per cm2 per steradian (p/s/cm2/sr) detected through a GFP filter. (F) Immunohistochemistry detecting FITC in the MKN45-derived tumor treated with FITC-Chb-M′ in E. Samples were stained either by isotype-matched control antibody (upper) or anti-FITC antibody (lower). Scale bars: 50 μm. Original magnification, ×40 and ×100 (insets). Data are the mean ± SEM values.
To examine the antitumor efficacy as well as the pharmacological safety of Chb-M′, we next sought to examine the in vivo distribution of Chb-M′. As shown in Supplemental Figure 19, A and B, we prepared FITC-labeled Chb-M′ (FITC-Chb-M′) and visualized its cellular uptake in tumor cells both in vitro and in vivo. FITC-Chb-M′ was readily incorporated in the tumor cells in vitro (Supplemental Figure 19C), and to our surprise, this molecule seemed to accumulate at much higher concentrations in tumor cells than in normal tissue cells when administered intravenously in vivo (Figure 5, D–F), potentially creating a therapeutic window and minimizing the toxic side effects of Chb-M′ in antitumor treatment.
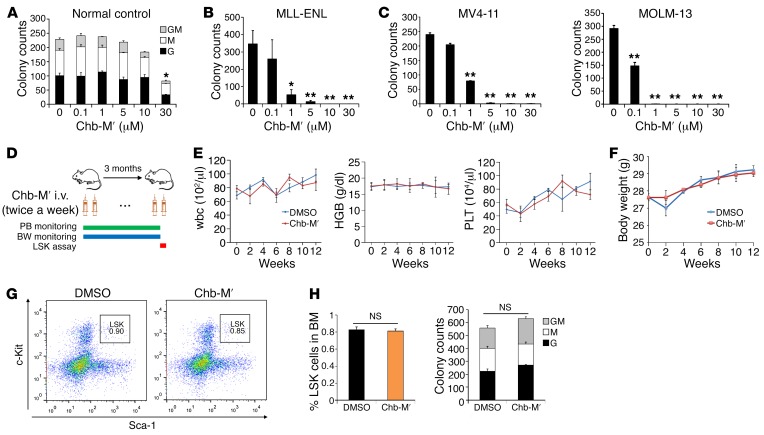
To further justify the use of Chb-M′ in vivo, we examined its acute toxicological effects in mice. As seen in Supplemental Figure 20, A–C, a marginal decrease in the number of platelets was detectable in mice that received up to 10 mg/kg body weight of Chb-M′. This concentration of Chb-M′ was more than 30 times higher than that of the generally accepted dose of PI polyamides (34, 35), indicating that Chb-M′ is highly tolerable even in vivo. We also examined the pharmacokinetic profile of Chb-M′ in vivo in mice and determined the elimination half-life of this molecule as about 6 hours (Supplemental Figure 21). While Chb-M′ treatment significantly reduced the colony-forming capacity in mouse AML cells with MLL-ENL or in human AML cell lines MV4-11 and MOLM-13 at 1 μM, the c-Kit+ fraction of bone marrow cells extracted from wild-type mice was insensitive to Chb-M′ at this concentration and could tolerate up to 10 μM Chb-M′ without a significant reduction in colony-forming capacity (Figure 6, A–C). Treatment with Chb-M′ (320 μg/kg body weight injections twice per week) in mice for 3 months did not influence the complete blood counts of peripheral blood, and no significant change in body weight was observed between Chb-M′– or vehicle-treated cohorts (Figure 6, D–F). In addition, no change in the frequency of the LSK (Lin–Sca-1+c-Kit+) fraction was detected in mouse bone marrow cells after a 3-month treatment with Chb-M′ compared with vehicle-treated mice (Figure 6, G and H). Moreover, the colony-forming ability of these immature c-Kit+ cells was unchanged after 3-month treatment with Chb-M′ (Figure 6I), suggesting that the quality and quantity of normal hematopoietic stem cells were not influenced by 3-month treatment with Chb-M′ at this dose in mice.
Figure 6. In vivo tolerability of Chb-M′.
(A) The number of granulocyte (G), macrophage (M), and G/M colonies were scored based on their morphology in a colony-forming cell assay. c-Kit+ bone marrow cells derived from wild-type C57BL/6 mice were cultured in the presence of indicated concentrations of Chb-M′ (n = 3). (B) The number of colonies was scored in a colony-forming cell assay with MLL-ENL–transduced mouse immortalized bone marrow cells in the presence of indicated concentrations of Chb-M′ (n = 3). (C) The number of colonies was scored in a colony-forming cell assay with MV4-11 or MOLM-13 cells in the presence of indicated concentrations of Chb-M′ (n = 3). (D) Schematic representation of treatment and monitoring schedule in wild-type C57BL/6 mice. PB, peripheral blood; BW, body weight. (E and F) Peripheral blood cell counts (E) and body weight (F) in mice receiving Chb-M′ or DMSO treatment for 3 months (n = 3). HGB, hemoglobin; PLT, platelets. (G and H) Frequency of LSK fraction was determined in bone marrow cells extracted from mice after a 3-month treatment with Chb-M′ or DMSO (n = 6). Representative FACS plots (G) and cumulative data (H) are shown. (I) The number of GM, G, and M colonies were scored based on their morphology in a colony-forming cell assay with c-Kit+ bone marrow cells extracted from mice after a 3-month treatment with Chb-M′ or DMSO (n = 6). Data are the mean ± SEM values. *P < 0.05, **P < 0.01, NS, not significant, by 2-tailed Student’s t test.
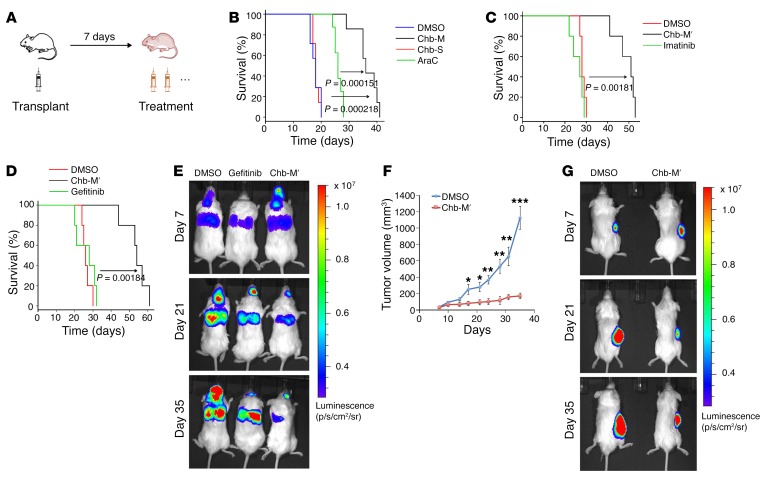
To further examine the antitumor potency of Chb-M′ in vivo, we generated a xenograft AML mouse model by transplanting MV4-11 cells onto immunodeficient mice and treated these mice with Chb-M′ (320 μg/kg body weight injections twice per week) (Figure 7A). Under our experimental conditions, DMSO, Chb, Chb-S, and cytarabine (Ara-C) were injected as controls. As shown in Figure 7B and Supplemental Figure 22, A–C, Chb-M′ treatment significantly prolonged the overall survival periods in this fms-related tyrosine kinase 3 (FLT3) mutation–positive poor-prognosis AML model (36). Chb-M′ was also highly effective against a Philadelphia chromosome–positive (Ph+) ALL mouse model with SU/SR cells. Although SU/SR cells harbor the T315I mutation, which confers substantial resistance to tyrosine kinase inhibitors currently in use (37, 38), Chb-M′ exhibited a higher efficacy in vivo (Figure 7C and Supplemental Figure 23, A–C). We next sought to extend our study to a lung cancer mouse model. As described previously (39), non–small cell lung cancer A549 cells are highly resistant to gefitinib or erlotinib, both of which are clinically available standard tyrosine kinase inhibitors for EGFR mutation–positive lung cancer patients. Indeed, gefitinib was ineffective in our lung cancer model. In sharp contrast to gefitinib, Chb-M′ obviously suppressed A549 cell proliferation in vivo and finally extended the overall survival periods compared with those of vehicle- and gefitinib-treated experimental groups (Figure 7, D and E, and Supplemental Figure 24, A and B). Lastly, we examined its possible effect on gastric cancer cells. For this purpose, human gastric cancer MKN45 cells were subcutaneously injected into immunodeficient mice. We monitored the size of tumors up to 35 days after engraftment and observed that Chb-M′ efficiently controls the growth of gastric tumors (Figure 7, F and G, and Supplemental Figure 24C). Collectively, our results strongly indicate that Chb-M′ is a highly tolerable and potentially novel anticancer drug that targets multiple cancer types through cluster regulation of the RUNX family in vivo.
Figure 7. Antitumor activity of Chb-M′ in vivo.
(A) Schematic representation of treatment schedule in xenotransplanted mice. (B) Overall survival of NOG mice transplanted with MV4-11 cells followed by treatment with DMSO, Ara-C, Chb-S, or with Chb-M′ (n = 7). P < 0.001, by log-rank (Mantel-Cox) test. (C) Overall survival of NOG mice transplanted with SU/SR cells followed by treatment with DMSO, imatinib, or with Chb-M′ (n = 5). P < 0.01, by log-rank (Mantel-Cox) test. (D and E) Human lung cancer xenotransplant model in NOG mice transplanted with A549 cells followed by treatment with DMSO, gefitinib, or with Chb-M′. (D) Overall survival of NOG mice (n = 5). P < 0.01, by log-rank (Mantel-Cox) test. (E) Live animal bioluminescence images at 7, 14, and 21 days after transplantation of A549 cells. Rainbow scale represents relative light units. (F and G) Human stomach cancer xenotransplant model in NOG mice transplanted with MKN45 cells followed by treatment with DMSO or with Chb-M′. (F) Tumor volume of NOG mice (n = 8). (G) Live animal bioluminescence images at 7, 14, and 21 days after transplantation of MKN45 cells. Rainbow scale represents relative light units. Data are the mean ± SEM values. *P < 0.05, **P < 0.01, ***P < 0.001, by 2-tailed Student’s t test.
RUNX-related CBFB is a potentially novel molecular marker for pan-cancer.
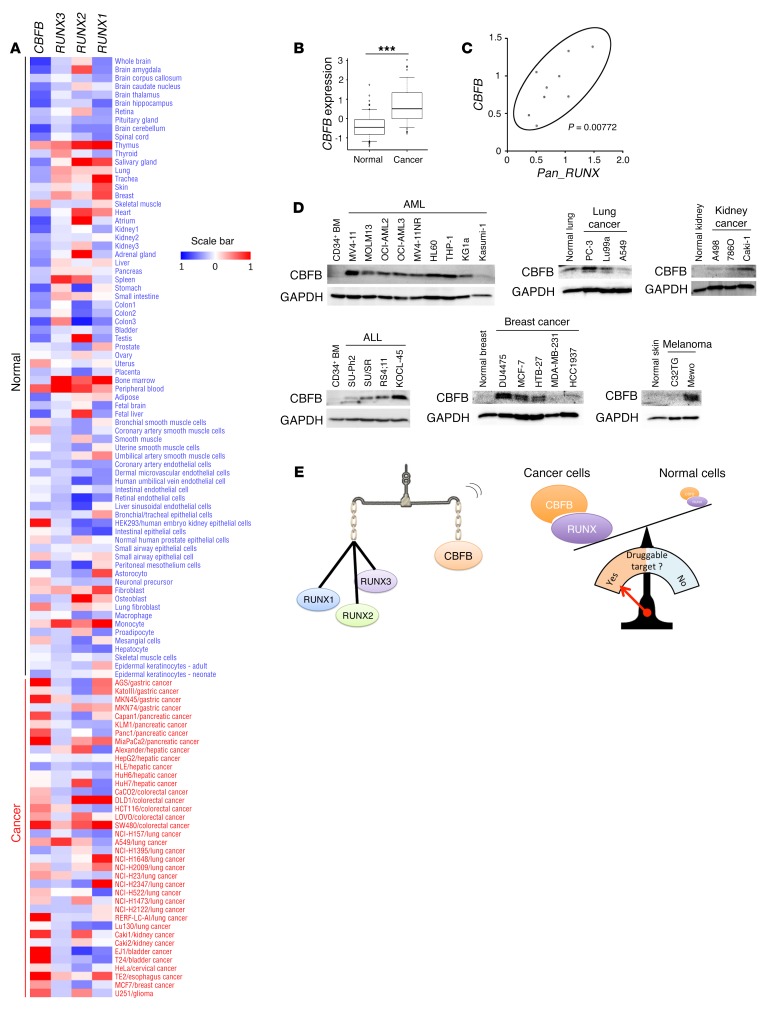
To establish the antitumor strategy targeting all of the RUNX family members, we first checked the expression levels of RUNX1, RUNX2, RUNX3, and CBFB in cancer tissues and in their normal counterparts. Based on previously reported array datasets (reference database for gene expression analysis, RefExA; see ref. 40), we found that the expression levels of CBFB are consistently higher in cancer tissues compared with their normal counterparts, while the expression level of each RUNX family member varied between cancer tissues and their normal counterparts (Figure 8, A and B). Intriguingly, however, the amounts of total RUNX mRNAs (RUNX1 + RUNX2 + RUNX3), as examined by primers selectively amplifying the common region of RUNX family members (Pan_RUNX), were positively correlated to those of CBFB in AML cells, which might be attributable to the heterodimeric complex formation between RUNX family members and CBFB (Figure 8C). Similar results were also obtained by immunoblotting (Supplemental Figure 25, A and B), suggesting that the expression level of CBFB is a molecular marker for predicting the expression of all 3 RUNX family members. Since CBFB is one of the most highly upregulated genes in various cancer tissues, it is likely that CBFB is a reliable molecular marker for a wide variety of cancers (Supplemental Figure 25C).
Figure 8. Total RUNX expression, equivalent to that of CBFB, is an ideal target for anticancer strategy.
(A) Heatmap showing relative expression levels of RUNX1, RUNX2, RUNX3, and CBFB in normal tissues and cancer cell lines of various origins. (B) Expression levels of CBFB in normal tissues (n = 74) and cancer cell lines (n = 38). (C) Correlation between the expression levels of CBFB and RUNX1 + RUNX2 + RUNX3 (Pan_RUNX) (n = 9). P < 0.01, by Spearman’s correlation. (D) Immunoblotting of CBFB and GAPDH in various cancer cells and their corresponding normal tissues. (E) Graphical abstract showing the balanced expression of total RUNX and CBFB (left). Consistent difference in the expression levels of total RUNX or CBFB in between malignant and benign cells provides a pharmacological window to be targeted (right). Data are the mean ± SEM values. ***P < 0.001, by 2-tailed Student’s t test.
We also assessed the expression level of CBFB protein in cancer cell lines of various origins and in their normal counterparts. Immunoblotting experiments demonstrated that CBFB is expressed at higher levels in cancer cells than their normal counterparts (Figure 8D). This tendency was further confirmed in the patient-derived samples by immunohistochemistry (Supplemental Figure 26). Finally, we compared the expression of all 3 RUNX family members and CBFB in AML cells and in their normal counterpart CD34+ hematopoietic cells from healthy donors (Supplemental Figure 27, A and B). Through this experiment, we have confirmed that the total expression of RUNX family members as well as that of CBFB is consistently higher in malignant cells than in normal ones. Taken together, these results suggest that the entire RUNX family, the total expression of which is equivalent to that of CBFB, is a promising druggable target of antitumor therapy in multiple cancer types (Figure 8E).
Discussion
Mammalian RUNX transcription factors mediate the homeostasis of both benign and malignant cells through their ability to promote gene activation (2). By further exploring our prior finding that RUNX is a prerequisite for the initiation of leukemia development (10, 11), we now reveal that RUNX plays a pivotal role in the maintenance of leukemia cells as well as tumor cells derived from different origins. According to a previous report, the frequency of RUNX1 point mutations in the Runt domain among de novo adult AML cases is around 10% (41). Some of these mutant RUNX1 (such as D171N and S291fsX300 mutations) and RUNX1 fusion genes (such as RUNX1-ETO and RUNX1-ETO9a) are proven oncogenes in mice with a p53-proficient environment (42). Although these observations remind us of the oncogenic property of RUNX1 alterations, mutations and translocations involving RUNX1 are usually monoallelic and AML cases with homozygous dysfunction of RUNX1 are rarely encountered in humans (43, 44). In addition, we and other groups have previously reported that wild-type RUNX1 is stringently required in leukemogenesis, both in RUNX1-mutated cases (9) and in translocated cases (11, 45). These results suggest that wild-type RUNX1 still exists and could be targeted in RUNX1-mutated leukemia. In addition, we have found and report in this study that functional alterations of RUNX1, RUNX2, and RUNX3 do exist in a mutually exclusive manner in various cancers including AML, indicating that RUNX1-mutated tumors do express wild-type RUNX2 and RUNX3. We believe that these findings collectively rationalize targeting these remaining wild-type RUNX1 or other RUNX family members, even in malignancies harboring RUNX1 mutations.
During our workup, we have also noticed that RUNX inhibition is solely effective against tumors with functional p53. In this regard, RUNX inhibition therapy should be given according to the pretreatment status of p53, and potentially permits the emergence of resistant cells harboring acquired p53 mutations in future patients. Despite this potential disadvantage, Chb-M′ is highly efficient against p53-proficient tumor cells, and furthermore, we believe this concern could be overcome through adopting the concurrent use of p53 modifiers such as PRIMA-1 with Chb-M′ since this combined therapy was indeed effective against p53-mutated AML cells as we have shown in this report. We are currently preparing for the first clinical trial of Chb-M′ in AML patients, and future tumor cell samples obtained after courses of Chb-M′ treatment might exhibit p53 dysfunctions and further elucidate the close relationships between RUNX family members and p53.
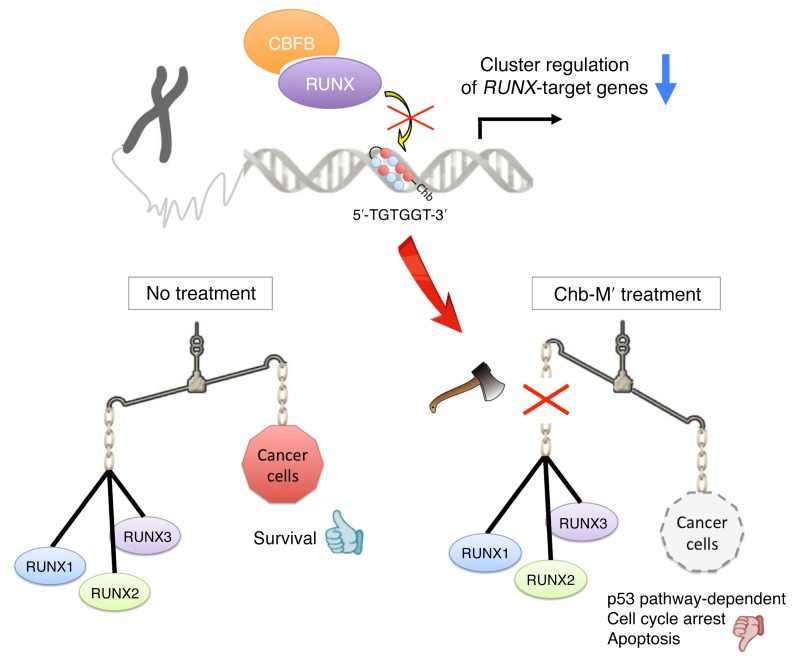
The fact that the total amount of RUNX expression, which is estimated by the expression of CBFB, is considerably higher in cancer tissues of different origins than in their normal counterparts rationalizes our strategy to target the RUNX family by cluster in a variety of cancer patients. In addition, the finding that Chb-M′ is well tolerated in mice even at high doses makes Chb-M′ a safe lead compound for the potentially novel anticancer drug category we refer to as “cluster regulation of RUNX (CROX).” Further studies involving clinical trials with this drug are awaited to validate the CROX strategy against human cancers (Figure 9).
Figure 9. Chb-M′ as a safe lead compound of a potentially novel anticancer drug category.
Chb-M′ targets the consensus RUNX binding sequences on the genome DNA and achieves cluster regulation of RUNX target genes, which offers efficient antitumor activity through potentiating a proapoptotic p53-mediated cell death pathway.
Methods
Cell proliferation.
To assess cell proliferation, 1 × 105 cells of the indicated AML-derived cells were seeded in 6-well plates. For the tetracycline-inducible gene or shRNA expression, doxycycline was added to the culture at a final concentration of 3 μM. Trypan blue dye exclusion assays were performed every other day.
RT-qPCR.
Total RNA was isolated with an RNeasy Mini Kit (Qiagen) and reverse transcribed with a ReverTra Ace kit (TOYOBO) to generate cDNA. RT-qPCR was carried out with a 7500 Real-Time PCR System (Applied Biosystems) according to the manufacturer’s instructions. The results were normalized to GAPDH levels. Relative expression levels were calculated using the 2–ΔΔCt method. Primers used for RT-qPCR are listed in Supplemental Table 3.
ChIP-qPCR.
ChIP was performed using a SimpleChIP Plus Enzymatic Chromatin IP Kit (Cell Signaling Technology) according to the manufacturer’s instructions. In brief, cells were cross-linked in 1% formaldehyde in PBS for 10 minutes at room temperature. After glycine quenching, cell pellets were collected, lysed, and then sonicated with a Q55 sonicator system (QSONICA). The supernatant was diluted with the same sonication buffer, and processed for immunoprecipitation with the following antibodies at 4°C overnight: anti-RUNX1 (Abcam, ab23980), anti-RUNX2 (Cell Signaling Technology, D1L7F), and anti-RUNX3 (Abcam, ab11905). The beads were then washed and DNA was reverse cross-linked and purified. Following ChIP, DNA was quantified by qPCR using the standard procedures for the 7500 Real-Time PCR System. Primers used for ChIP-qPCR are listed in Supplemental Table 4.
shRNA interference.
Specific shRNAs targeting human RUNX1, RUNX2, RUNX3, and TP53 were designed and subcloned into pENTR4-H1tetOx1, CS-RfA-ETBsd, CS-RfA-ETV, and CS-RfA-ETR vectors (RIKEN BioResource Center). Nontargeting control shRNA was designed against luciferase (sh_Luc.). The target sequences are provided in Supplemental Table 5.
Expression plasmids.
We amplified cDNAs for human RUNX1, RUNX2, RUNX3, MDM2, BCL11A, and TRIM24, and then inserted them into CSII-EF-MCS-IRES2-Venus, CSII-EF-MCS-IRES2-hKO1, and CSIV-TRE-Ubc-KT expression vectors, which were a gift from Hiroyuki Miyoshi (RIKEN BioResource Center) . We express our gratitude to Takuro Nakamura and Takeshi Kuwata (Japanese Foundation For Cancer Research) for providing the BCL11A cDNA construct. All of the PCR products were verified by DNA sequencing.
Production and transduction of lentivirus.
For the production of lentivirus, HEK293T cells were transiently cotransfected with lentivirus vectors such as psPAX2 and pMD2.G by polyethylenimine (PEI, Sigma-Aldrich). Forty-eight hours after transfection, viral supernatants were collected and immediately used for infection, and then successfully transduced cells were sorted using a FACSAria III (BD Biosciences).
Immunoblotting.
Cells were washed twice in ice-cold PBS and lysed in lysis buffer (50 mM Tris [pH 7.4], 100 mM NaCl, 0.1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM β-glycerophosphate, 2.5 mM sodium pyrophosphate, 1 mM Na3VO4, 1× protease inhibitor [Roche], and PhosSTOP [Roche]). Whole-cell extracts were separated by SDS-PAGE and electrotransferred onto polyvinylidene difluoride membranes. Membranes were probed with the following primary antibodies: anti-BCL11A (Abcam, catalog ab19487), anti-TRIM24 (Bethyl Laboratories, catalog A300-815A), anti-RUNX1 (Santa Cruz Biotechnology, clone A-2), anti-GAPDH (Santa Cruz Biotechnology, clone FL-335), anti-p21 (Santa Cruz Biotechnology, clone C-19), anti-BAX (Santa Cruz Biotechnology, clone N-20), anti-RUNX2 (Cell Signaling Technology, clone D1L7F), anti-RUNX3 (Cell Signaling Technology, clone D6E2), anti-CBFB (Santa Cruz Biotechnology, clone FL-182), anti-p14ARF (Cell Signaling Technology, catalog 2407), anti-MDM2 (Santa Cruz Biotechnology, clone SMP14), anti-PUMA (Cell Signaling Technology, catalog 4976), anti–cleaved caspase-3 (Cell Signaling Technology, clone 5A1E), anti-PARP (Cell Signaling Technology, clone 46D11), anti-p53 (Cell Signaling Technology, clone 1C12), and anti–phosphorylated p53 (Ser-15) (Cell Signaling Technology, clone 16G8). For secondary antibodies, HRP-conjugated anti-rabbit IgG and anti-mouse IgG (Cell Signaling Technology, catalog 7074 and 7076) were used. Blots were visualized using Chemi-Lumi One Super (Nacalai Tesque) and the ChemiDoc XRS+ Imager (Bio-Rad Laboratories) according to the manufacturers’ recommendations. Protein levels were quantified with Image Lab Software (Bio-Rad Laboratories).
Analysis of gene expression microarray.
MV4-11 cells were treated with 1 μM Chb-M′, Chb-50, or with DMSO for 6 hours, or transduced with control shRNA (sh_Luc.) or with shRNAs targeting RUNX1 (sh_Rx1), RUNX2 (sh_Rx2), and RUNX3 (sh_Rx3), and subsequently incubated with 3 μM doxycycline for 24 hours. After the indicated time of incubation, total RNA was prepared and its quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies). Cyanine 3–labeled cRNA was generated in the presence of T7 polymerase, purified, and its concentration was measured using a Nanodrop ND1000 v3.5.2 (Thermo Scientific). The resultant cRNA (825 ng) was fragmented and subsequently hybridized with a Human Gene 2.1 ST Array Strip (Affymetrix). The raw data together with the associated sample information were analyzed by GeneSpring GX v12.1.0 (Agilent Technologies), and our microarray data have been deposited in NCBI’s Gene Expression Omnibus (GEO GSE94835). Gene set enrichment analysis (GSEA) was utilized to analyze the microarray data obtained in the present study (46). Gene ontology enrichment analysis was conducted by Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources 6.7 software according to the provider’s instructions (47, 48).
Statistics.
Statistical significance of differences between control and experimental groups was assessed by a 2-tailed unpaired Student’s t test, and was declared if the P value was less than 0.05. Equality of variances in 2 populations was calculated with an F test. The results are represented as the average ± SEM values obtained from 3 independent experiments. In transplantation experiments, animals were randomly allocated to each experimental group, and the treatments were given with blinding. The overall survival of mice is shown in a Kaplan-Meier curve. Survival between the indicated groups was compared using the log-rank test. For the measurement of correlation between mRNA or protein expression, Spearman’s rank correlation coefficient was used. To quantify the synergism of Chb-M′ and PRIMA-1, the combination index theorem of Chou-Talalay was calculated using COMPUSYN software. A combination index value less than 1 was considered statistically significant (49).
Mice.
C57BL/6J mice were purchased from CLEA Japan, Inc. and NOD/Shi-scid,IL-2RγKO (NOG) mice were from the Central Institute for Experimental Animals, Japan. Crl:CD1(ICR) mice and BALB/c-nu mice were purchased from Charles River Laboratories International, Inc. Littermates were used as controls in all experiments.
Xenograft mouse model.
Xenograft mouse models of human cancer cell lines were developed using NOG mice. For leukemia mouse models, 2.5 × 106 cells/body of MV4-11 or SU/SR cells were intravenously injected. Peripheral blood (PB) was then collected every week and chimerism was checked by flow cytometer using an anti–human CD45 antibody (BD Biosciences). Seven days after injection, mice were treated with PI polyamides (320 μg/kg body weight, twice per week intravenous injections) or with the equivalent amount of DMSO. For Chb injections, solution was delivered intravenously at 320 μg/kg body weight, twice per week. Administration of cytarabine (Ara-C, Wako Pure Chemical Industries, Ltd.) was performed intraperitoneally (100 mg/kg body weight) for the first 5 consecutive days (from day 7 through day 11) as a standard induction therapy. Oral administration of imatinib mesylate (Focus Biomolecules) was carried out (100 mg/kg body weight) twice per day (beginning at day 7 until the recipient mice succumb to their disease). For lung cancer mouse models, 1 × 106 cells/body of A549 cells were injected intravenously. Cells were then transduced with lentivirus encoding luciferase (Addgene, pLenti-luciferase vector). Mice were subsequently injected with D-luciferin (150 mg/kg body weight, Wako Pure Chemical Industries, Ltd.) and their tumor volumes were assessed using an IVIS Spectrum In Vivo Imaging System (PerkinElmer) every week. For gastric cancer xenograft mouse models, 1 × 106 cells/body of MKN45 cells were injected subcutaneously at the right dorsal flank. Cells were marked with luciferase, and tumor growth was monitored by IVIS. For biofluorescence assay, BALB/c-nu mice were subcutaneously transplanted with 2 × 106 cells/body of MKN45 cells, followed by intravenous injection of FITC-labeled Chb-M′ 320 μg/kg body weight. Biofluorescence was detected by IVIS through the GFP filter.
Synthesis of PI polyamides.
Reagents and solvents were purchased from standard suppliers and used without further purification. Flash column purifications were performed by a CombiFlash Rf (Teledyne Isco, Inc.) with a C18 RediSep Rf Flash Column. Electrospray ionization time-of-flight mass spectrometry (ESI-TOF MS) was performed on a Bio-TOF II (Bruker Daltonics) mass spectrometer using positive ionization mode. Machine-assisted polyamide syntheses were performed on a PSSM-8 (Shimadzu) system with computer-assisted operation. Proton nuclear magnetic resonance (1H NMR) spectra were recorded with a JEOL JNM ECA-600 spectrometer operating at 600 MHz and in parts per million (ppm) downfield relative to tetramethylsilane used as an internal standard. The following abbreviations apply to spin multiplicity: s (singlet), d (doublet), t (triplet), q (quartet), quint (quintet), m (multiplet). The results of NMR and ESI-TOF-MS are available in Supplemental Figures 28 and 29.
Study approval.
All animal studies were properly conducted in accordance with the Regulation on Animal Experimentation at Kyoto University, based on International Guiding Principles for Biomedical Research Involving Animals. All procedures employed in this study were approved by Kyoto University Animal Experimentation Committee (permit number: Med Kyo 14332).
See supplemental data for more information.
Author contributions
KM designed research, performed experiments, analyzed data, and wrote the manuscript. KS, SM, AM, GK, JT, YM, AY, YY, HK, CT, HM, RM, MN, MH, and ST helped to collect data. MT, YO, MM, KN, KI, T. Kataoka, T. Kitamura, Y. Kaneda, PL, and SA participated in discussions and interpretation of the data and results, and commented on research direction. TB and HS synthesized and designed the PI polyamides. Y. Kamikubo initiated and designed the study and led the entire project.
Supplementary Material
Acknowledgments
This work was supported by a Grant-in-Aid from the Japan Agency for Medical Research and Development (Basic Science and Platform Technology Program for Innovative Biological Medicine); a Grant-in-Aid for Scientific Research (KAKENHI); a grant from the Joint Research Project of the Institute of Medical Science, the University of Tokyo; an Extramural Collaborative Research grant from the Cancer Research Institute, Kanazawa University; grants from the Kobayashi Foundation for Cancer Research, the Foundation for Promotion of Cancer Research, and National Cancer Center Research; and a Grant in-Aid from the Agency for Medical Research and Development (Project for Development of Innovative Research on Cancer Therapeutics and Practical Research for Innovative Cancer Control).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(7):2815–2828.https://doi.org/10.1172/JCI91788.
Contributor Information
Ken Morita, Email: kenkenken710@yahoo.co.jp.
Kensho Suzuki, Email: monokenp102020@yahoo.co.jp.
Shintaro Maeda, Email: grclown4050@gmail.com.
Akihiko Matsuo, Email: akihiko123454321@yahoo.co.jp.
Yoshihide Mitsuda, Email: mitsuda.yoshihide.36z@st.kyoto-u.ac.jp.
Chieko Tokushige, Email: mr.r0ck.children@gmail.com.
Gengo Kashiwazaki, Email: kashigen@kuchem.kyoto-u.ac.jp.
Junichi Taniguchi, Email: j.tani@kuchem.kyoto-u.ac.jp.
Rina Maeda, Email: r.maeda32@chemb.kuchem.kyoto-u.ac.jp.
Mina Noura, Email: mina4059@gmail.com.
Masahiro Hirata, Email: hiratama@kuhp.kyoto-u.ac.jp.
Tatsuki Kataoka, Email: trkataoka@yahoo.co.jp.
Ayaka Yano, Email: ayaka.yano.225@gmail.com.
Yoshimi Yamada, Email: yamada.yoshimi.52a@st.kyoto-u.ac.jp.
Hiroki Kiyose, Email: kiyose.hiroki.82z@st.kyoto-u.ac.jp.
Mayu Tokumasu, Email: tokusan@kuhp.kyoto-u.ac.jp.
Hidemasa Matsuo, Email: matsuo@kuhp.kyoto-u.ac.jp.
Sunao Tanaka, Email: sunao@kuhp.kyoto-u.ac.jp.
Yasushi Okuno, Email: okuno.yasushi.4c@kyoto-u.ac.jp.
Manabu Muto, Email: mmuto@kuhp.kyoto-u.ac.jp.
Kazuhito Naka, Email: kanaka55@hiroshima-u.ac.jp.
Kosei Ito, Email: itok@nagasaki-u.ac.jp.
Toshio Kitamura, Email: kitamura@ims.u-tokyo.ac.jp.
Yasufumi Kaneda, Email: kaneday@gts.med.osaka-u.ac.jp.
Toshikazu Bando, Email: bando@kuchem.kyoto-u.ac.jp.
Souichi Adachi, Email: adachiso@kuhp.kyoto-u.ac.jp.
Hiroshi Sugiyama, Email: hs@kuchem.kyoto-u.ac.jp.
Yasuhiko Kamikubo, Email: amikubo.yasuhiko.7u@kyoto-u.ac.jp.
References
- 1.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84(2):321–330. doi: 10.1016/S0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 2.Ito Y, Bae SC, Chuang LS. The RUNX family: developmental regulators in cancer. Nat Rev Cancer. 2015;15(2):81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- 3.Warren AJ, Bravo J, Williams RL, Rabbitts TH. Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBFbeta. EMBO J. 2000;19(12):3004–3015. doi: 10.1093/emboj/19.12.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan J, Liu Y, Lukasik SM, Speck NA, Bushweller JH. CBFbeta allosterically regulates the Runx1 Runt domain via a dynamic conformational equilibrium. Nat Struct Mol Biol. 2004;11(9):901–906. doi: 10.1038/nsmb819. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996;93(8):3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ichikawa M, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10(3):299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 7.Liu P, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261(5124):1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 8.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci U S A. 1991;88(23):10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goyama S, et al. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J Clin Invest. 2013;123(9):3876–3888. doi: 10.1172/JCI68557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamikubo Y, et al. Accelerated leukemogenesis by truncated CBF beta-SMMHC defective in high-affinity binding with RUNX1. Cancer Cell. 2010;17(5):455–468. doi: 10.1016/j.ccr.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyde RK, Zhao L, Alemu L, Liu PP. Runx1 is required for hematopoietic defects and leukemogenesis in Cbfb-MYH11 knock-in mice. Leukemia. 2015;29(8):1771–1778. doi: 10.1038/leu.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berardi MJ, et al. The Ig fold of the core binding factor alpha Runt domain is a member of a family of structurally and functionally related Ig-fold DNA-binding domains. Structure. 1999;7(10):1247–1256. doi: 10.1016/S0969-2126(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 13.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14(5):359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renneville A, et al. Cooperating gene mutations in acute myeloid leukemia: a review of the literature. Leukemia. 2008;22(5):915–931. doi: 10.1038/leu.2008.19. [DOI] [PubMed] [Google Scholar]
- 16.Ozaki T, Nakagawara A, Nagase H. RUNX family participates in the regulation of p53-dependent DNA damage response. Int J Genomics. 2013;2013:271347. doi: 10.1155/2013/271347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trauger JW, Baird EE, Dervan PB. Recognition of DNA by designed ligands at subnanomolar concentrations. Nature. 1996;382(6591):559–561. doi: 10.1038/382559a0. [DOI] [PubMed] [Google Scholar]
- 18.Best TP, Edelson BS, Nickols NG, Dervan PB. Nuclear localization of pyrrole-imidazole polyamide-fluorescein conjugates in cell culture. Proc Natl Acad Sci U S A. 2003;100(21):12063–12068. doi: 10.1073/pnas.2035074100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandian GN, et al. Distinct DNA-based epigenetic switches trigger transcriptional activation of silent genes in human dermal fibroblasts. Sci Rep. 2014;4:3843. doi: 10.1038/srep03843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saha A, et al. Synthesis and biological evaluation of a targeted DNA-binding transcriptional activator with HDAC8 inhibitory activity. Bioorg Med Chem. 2013;21(14):4201–4209. doi: 10.1016/j.bmc.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Bando T, Sugiyama H. Synthesis and biological properties of sequence-specific DNA-alkylating pyrrole-imidazole polyamides. Acc Chem Res. 2006;39(12):935–944. doi: 10.1021/ar030287f. [DOI] [PubMed] [Google Scholar]
- 22.Minoshima M, Bando T, Shinohara K, Sugiyama H. Molecular design of sequence specific DNA alkylating agents. Nucleic Acids Symp Ser (Oxf) 2009;(53):69–70. doi: 10.1093/nass/nrp035. [DOI] [PubMed] [Google Scholar]
- 23.Allton K, et al. Trim24 targets endogenous p53 for degradation. Proc Natl Acad Sci U S A. 2009;106(28):11612–11616. doi: 10.1073/pnas.0813177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain AK, Allton K, Duncan AD, Barton MC. TRIM24 is a p53-induced E3-ubiquitin ligase that undergoes ATM-mediated phosphorylation and autodegradation during DNA damage. Mol Cell Biol. 2014;34(14):2695–2709. doi: 10.1128/MCB.01705-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain AK, Barton MC. Regulation of p53: TRIM24 enters the RING. Cell Cycle. 2009;8(22):3668–3674. doi: 10.4161/cc.8.22.9979. [DOI] [PubMed] [Google Scholar]
- 26.Yu Y, et al. Bcl11a is essential for lymphoid development and negatively regulates p53. J Exp Med. 2012;209(13):2467–2483. doi: 10.1084/jem.20121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kojima K, et al. Selective FLT3 inhibitor FI-700 neutralizes Mcl-1 and enhances p53-mediated apoptosis in AML cells with activating mutations of FLT3 through Mcl-1/Noxa axis. Leukemia. 2010;24(1):33–43. doi: 10.1038/leu.2009.212. [DOI] [PubMed] [Google Scholar]
- 28.Rautio J, et al. Prodrugs: design and clinical applications. Nat Rev Drug Discov. 2008;7(3):255–270. doi: 10.1038/nrd2468. [DOI] [PubMed] [Google Scholar]
- 29.Kuo YH, Zaidi SK, Gornostaeva S, Komori T, Stein GS, Castilla LH. Runx2 induces acute myeloid leukemia in cooperation with Cbfbeta-SMMHC in mice. Blood. 2009;113(14):3323–3332. doi: 10.1182/blood-2008-06-162248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klunker S, et al. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J Exp Med. 2009;206(12):2701–2715. doi: 10.1084/jem.20090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uchida H, Zhang J, Nimer SD. AML1A and AML1B can transactivate the human IL-3 promoter. J Immunol. 1997;158(5):2251–2258. [PubMed] [Google Scholar]
- 32.Bowers SR, Calero-Nieto FJ, Valeaux S, Fernandez-Fuentes N, Cockerill PN. Runx1 binds as a dimeric complex to overlapping Runx1 sites within a palindromic element in the human GM-CSF enhancer. Nucleic Acids Res. 2010;38(18):6124–6134. doi: 10.1093/nar/gkq356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyde RK, et al. Cbfb/Runx1 repression-independent blockage of differentiation and accumulation of Csf2rb-expressing cells by Cbfb-MYH11. Blood. 2010;115(7):1433–1443. doi: 10.1182/blood-2009-06-227413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiraoka K, et al. Inhibition of KRAS codon 12 mutants using a novel DNA-alkylating pyrrole-imidazole polyamide conjugate. Nat Commun. 2015;6:6706. doi: 10.1038/ncomms7706. [DOI] [PubMed] [Google Scholar]
- 35.Kamei T, et al. Quantitation of pyrrole-imidazole polyamide in rat plasma by high-performance liquid chromatography coupled with UV detection. J Biomed Biotechnol. 2012;2012:715928. doi: 10.1155/2012/715928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakao M, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10(12):1911–1918. [PubMed] [Google Scholar]
- 37.Hirase C, Maeda Y, Takai S, Kanamaru A. Hypersensitivity of Ph-positive lymphoid cell lines to rapamycin: Possible clinical application of mTOR inhibitor. Leuk Res. 2009;33(3):450–459. doi: 10.1016/j.leukres.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 38.Redaelli S, et al. Activity of bosutinib, dasatinib, and nilotinib against 18 imatinib-resistant BCR/ABL mutants. J Clin Oncol. 2009;27(3):469–471. doi: 10.1200/JCO.2008.19.8853. [DOI] [PubMed] [Google Scholar]
- 39.Kurtze I, Sonnemann J, Beck JF. KRAS-mutated non-small cell lung cancer cells are responsive to either co-treatment with erlotinib or gefitinib and histone deacetylase inhibitors or single treatment with lapatinib. Oncol Rep. 2011;25(4):1021–1029. doi: 10.3892/or.2011.1160. [DOI] [PubMed] [Google Scholar]
- 40.Ge X, et al. Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics. 2005;86(2):127–141. doi: 10.1016/j.ygeno.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 41.Gaidzik VI, et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia. 2016;30(11):2282. doi: 10.1038/leu.2016.207. [DOI] [PubMed] [Google Scholar]
- 42.Chin DW, Watanabe-Okochi N, Wang CQ, Tergaonkar V, Osato M. Mouse models for core binding factor leukemia. Leukemia. 2015;29(10):1970–1980. doi: 10.1038/leu.2015.181. [DOI] [PubMed] [Google Scholar]
- 43.Tang JL, et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: prognostic implication and interaction with other gene alterations. Blood. 2009;114(26):5352–5361. doi: 10.1182/blood-2009-05-223784. [DOI] [PubMed] [Google Scholar]
- 44.Schnittger S, et al. RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis. Blood. 2011;117(8):2348–2357. doi: 10.1182/blood-2009-11-255976. [DOI] [PubMed] [Google Scholar]
- 45.Ben-Ami O, et al. Addiction of t(8;21) and inv(16) acute myeloid leukemia to native RUNX1. Cell Rep. 2013;4(6):1131–1143. doi: 10.1016/j.celrep.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 46.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 48.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 50.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.