Abstract
Biological membranes contain a broad variety of lipid species whose individual physicochemical properties and collective interactions ultimately determine membrane organization. A key aspect of the organization of cellular membranes is their lateral subdivision into domains of distinct structure and composition. The most widely studied membrane domains are lipid rafts, which are the biological manifestations of liquid-ordered phases that form in sterol-containing membranes. Detailed studies of biomimetic membrane mixtures have yielded wide-ranging insights into the physical principles behind lipid rafts; however, these simplified models do not fully capture the diversity and complexity of the mammalian lipidome, most notably in their exclusion of polyunsaturated lipids. Here, we assess the role of lipid acyl chain unsaturation as a driving force for phase separation using coarse-grained molecular dynamics (CGMD) simulations validated by model membrane experiments. The clear trends in our observations and good qualitative agreements between simulations and experiments support the conclusions that highly unsaturated lipids promote liquid-liquid domain stability by enhancing the differences in cholesterol content and resulting lipid chain order between the coexisting domains. These observations reveal the important role of non-canonical biological lipids in the physical properties of membranes, showing that lipid polyunsaturation is a driving force for liquid-liquid phase separation.
Graphical Abstract
INTRODUCTION
The functionality of the mammalian plasma membrane is believed to be enhanced by lateral segregation of its lipid and protein constituents into domains of distinct structure and composition.1–2 The most widely studied membrane domains are called lipid rafts because they arise from preferred interactions between specific membrane lipids. Such ‘rafts’ could potentially modulate cellular processes and signaling pathways by establishing local micro-environments in the plasma membrane that facilitate or inhibit particular protein-protein and protein-lipid interactions3–4. For example, the stability of lipid rafts is tightly correlated to the clustering dynamics of the oncogenes H-Ras5–6 and K-Ras7, which are in turn critical for the activity of Ras-regulated pathways7. Membrane rafts have also been shown to be involved in subcellular sorting and membrane traffic8–9 and trafficking to the neuronal synapse10. Although there are currently few insights into specific molecular mechanisms by which rafts regulate cellular processes, it is certain that the functions of membrane domains are dependent on their physical properties, including size, lifetime, packing, and dynamics. The mechanisms by which such properties are regulated remain largely unexplored, although recent reports have shown that domains in biomimetic and biological membranes are dependent on properties such as bilayer thickness11, lipid order12 and cholesterol content13–16.
The study of membrane domains is facilitated by lipid model membranes that provide access to the full experimental and computational toolbox of biophysics in well controlled systems whose composition and properties are designed to address specific questions related to the physicochemical basis of domain formation17–18. In such model membranes, the inclusion of cholesterol often yields large lateral membrane domains due to the formation of a liquid-ordered (Lo) phase distinguished from the bulk liquid-disordered (Ld) phase by more ordered lipids, lower diffusivity, and tighter lipid packing, among other differences19–20. The separation of biomimetic model membranes into coexisting Lo and Ld domains is believed to underlie lipid raft formation in vivo. A useful tool for interrogating molecular-scale behavior in such domains is molecular dynamics simulation, which has been used to elucidate the detailed compositions of membrane domains, the mechanisms of domain formation21–23, and the behavior of membrane proteins therein24. It should be noted that the macro-scale domains observed in synthetic25, computational26, and isolated biological bilayers27–28 are not observable in the plasma membrane of living cells, and were not detected in simulations of a complex bilayer system that was designed to mimic the composition of the plasma membrane29. Domains in vivo are believed to be nanoscopic and highly dynamic, with the mechanisms governing the relationships between macro- and nano-domains being an area of active debate30–33.
Although both experiments and computations have yielded far-reaching insights into model membrane behavior, both approaches incompletely recapitulate the diversity and complexity of the mammalian lipidomes. One notable difference between model and biological membranes is that while model membranes are usually composed of lipids with saturated and monounsaturated acyl chains, natural membranes contain a sizeable fraction of polyunsaturated lipids34–38 (Fig. 1). Such polyunsaturated lipids have been shown to have distinct properties from their more saturated counterparts39, and these properties could play an important role in modulating the structure and dynamics of membrane domains40–48. For example, increasing levels of polyunsaturated lipids have been shown to progressively promote domain formation in ternary model membranes by CGMD simulations.49–50 However, the specific structural mechanisms that determine the extent and nature of domain formationin complex membranes remain incompletely understood.
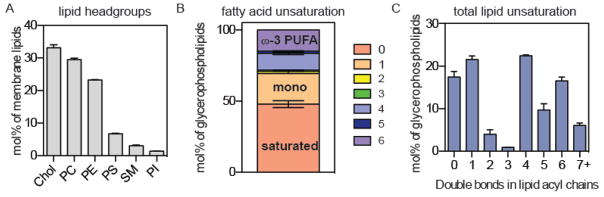
Figure 1. Synaptic membrane lipidome is rich in polyunsaturated lipids.
Synaptic lipids were isolated from synaptosomes51 prepared from adult rat hippocampus and subjected to lipidomic analysis. (a) Cholesterol and the neutral glycerophospholipids PC and PE were the major lipid classes. (b) Acyl chain unsaturation in glycerophospholipids (GPLs). ~50% of all GPL acyl chains were fully saturated, while >28% were polyunsaturated. (c) Total lipid unsaturation for both acyl chains of GPLs. More than half of GPLs have at least 4 unsaturations, with ~30% having 6 or more. Detailed lipidome was shown in Table S1. Mean ± s.d. was obtained from four independent preparations.
To address the role of polyunsaturated lipids in determining the properties of domains in biomimetic membranes, we applied both CGMD and experimental approaches to model membranes containing lipids with varying numbers of cis double bonds in their acyl chains. We measured the effects of lipid unsaturation on lipid chain order, membrane thickness, cholesterol interactions, and ultimately separation into ordered and disordered domains. We observed that increasing both abundance and extent of lipid unsaturation yielded larger, more stable membrane domains, likely due to enhancement of the differences between coexisting domains with respect to lipid order and cholesterol partitioning. These observations show that polyunsaturated lipids are important determinants of membrane physical properties and organization and begin to define the effects on non-canonical lipids on domains formation in biomimetic membranes.
METHODS
Molecular Dynamics Simulations
All simulations in this work were performed with the Martini CG force field (version 2.1)52 and the GROMACS program v4.5.453. Parameterization of the Martini model is based on united-atom MD simulations (GROMOS force field) and experiments. With respect to modeling lipid unsaturation, an important aspect is that lipid acyl chain disorder is an entropic effect that arises from reduced energy of conformational transition around the double bond, which leads on average to kinked/bent acyl chains. This effect is modeled in MARTINI by an enthalpic component via an enforced bond angle and force constant for bending between beads. Although this model recapitulates many molecular and mesoscopic membrane behaviors (including lipid domain formation24, 26, 29, 54–56), it is nevertheless important to emphasize that the basis for the disordering effect of unsaturation is different. Therefore, this coarse-grained approximation may be susceptible to artifacts. For example, the Martini force field does not reproduce experimentally observed57–58 large scale domain formation of ternary DPPC/DOPC/Chol (DPPC: 1,2-dipalmitoyl-sn-glycero-3-phosphocholine, DOPC: 1,2-dioleoyl-sn-glycero-3-phosphocholine, Chol: Cholesterol) mixtures, instead typically relying on doubly-unsaturated DLiPC to recapitulate experimentally observed lipid collective behavior24, 26, 59. We used standard setup protocols along with topologies of lipids, water, and ions.52 Our simulation systems consisted of 600 saturated lipids (DPPC), 360 unsaturated lipids (DOPC, DLiPC: 1,2-dilinoleoyl-sn-glycero-3-phosphocholine, DAPC: 1,2-diarachidonoyl-sn-glycero-3-phosphocholine, or DDPC: 1,2-didocosahexaenoyl-sn-glycero-3-phosphocholine), 240 Cholesterol, 20516 CG waters, plus Na+ and Cl− ions to achieve an ionic strength of 0.15 M. The simulation box was ~ 18 × 18 × 12 nm. The chemical structures and Martini-based configurations of all lipids used in this work are shown in Fig. S1. Initial symmetric bilayers were built using the tool insane developed by Wassenaar et al.60. To homogenize the distribution of molecules, the system was initially simulated at temperature T = 400 K and the isobaric isothermal (NPT) ensemble for 100ns simulation time (4×100ns = 400ns effective time)52. A speed-up factor of 4 was used in the current work to approximately estimate the effective time from the simulation time, as suggested by Martini force field developers52. Subsequent production simulations were performed for 5 μs (4×5 = 20 μs effective time) with a time step of 20 fs and periodic boundary conditions. For all simulations, a cutoff of 1.2 nm was used for van der Waals (vdW) interactions, and the Lennard-Jones potential was smoothly shifted to zero between 0.9 – 1.2 nm to reduce cutoff noise. For electrostatic interactions, the Columbic potential was smoothly shifted from 0 – 1.2 nm, with a cutoff at 1.2 nm. The relative dielectric constant was 15, which is the default value of the force field52. Lipids, and water plus ions, were coupled separately to V-rescale heat baths61 at T = 298K (coupling constant τ = 1ps), which is below the expected Lo/Ld miscibility transition temperature of all assayed mixtures25. The systems were simulated at 1 bar pressure using a semi-isotropic Parrinello-Rahman pressure coupling scheme62 with a coupling constant τ = 5ps and compressibility of 3 × 10−4 bar−1. The neighbor list for non-bonded interactions was updated every 10 steps with a cut-off of 1.4 nm. Snapshots of the simulation system were rendered by VMD63.
Analysis of trajectories
Normalized Lateral Contacts of Unsaturated Lipids for Estimating Relative Domain Sizes
In the current simulations, Lo domains consisted almost entirely of saturated lipids and cholesterol while Ld domains are almost completely made up of unsaturated lipids. Therefore, relative domain sizes can be evaluated by proxy, using the number of lateral contacts between lipids of the same type (e.g. unsaturated contacts with unsaturated). In light of inter-leaflet cholesterol flip-flop during CG simulations54, 64, we chose unsaturated lipids for contact calculation. To this end, we first obtained the total number of lateral contacts, NLd, among unsaturated lipids in the phase separated bilayer, averaged over the two leaflets; contact was defined based on a distance cutoff of 0.6 nm between any two CG beads of the specified lipid type.65–66 Then we normalized NLd by Npure, the total number of lateral contacts in a pure bilayer of the same unsaturated lipid type obtained from the last 400 ns trajectory of a 1μs run. Npure thus represents the maximum number of contacts of unsaturated lipids in a fluid bilayer at the same temperature. The normalized number of Ld lipid contacts (N = NLd/Npure) was used as a proxy for quantifying relative domain size.
Lipid Chain Order Parameter
Lipid chain order parameter (Sz) was calculated using the formula
where θn is the angle between the vector connecting the n − 1 and n + 1 sites of the tail and the bilayer normal z. To compare lipid chain order parameters among different systems we averaged Sz over the two lipid chains and the entire bilayer.
Lipid Height
Lipid height, h, was defined as the distance along the membrane normal between the head-group PO4 bead (colored in tan in Fig. S1) and the center-of-mass of the terminal lipid tail beads, averaged over same type lipids.
Cholesterol Preference
Cholesterol preference was determined based on the number of contacts (cutoff 0.6 nm) of cholesterol wit h saturated (Ns) and unsaturated lipids(Nus) 65 as
where χs and χus are the fraction of cholesterol in contact with saturated and unsaturated lipids, and ns and nus are the total number of CG beads of saturated and unsaturated lipids, respectively.
Giant Unilamellar Vesicle (GUV) experiments
GUVs were prepared by electroformation67. Briefly, a lipid mixture of dipalmitoyl phosphatidylcholine (DPPC), dioleoyl PC (DOPC) or didocosahexaenoyl PC (DDPC), and cholesterol was prepared from the lipid solution in chloroform/methanol (2:1) with a molar ratio of 5:3:2 (lipids purchased from Avanti). The fluorescent lipid marker DiO-C18 (Molecular Probes, Invitrogen) dissolved in chloroform was added to the lipid mixture at 0.1 mol %. The lipid mixture was applied to Pt-wires of a custom electroformation chamber and solvents were evaporated in a vacuum chamber. The electroformation chamber was filled with sucrose solution (0.1 M) and heated to 53°C. An alternating sinusoidal current was applied across the cell unit with 2.5 V, 10 Hz and 53 °C for 2 hours and 15 minutes. Fluorescence microscopy of GUV samples was done on a commercial fluorescence microscope. The percentage of phase separation at a given temperature was defined as the number of vesicles presenting microscopic phase separation divided by the total number of vesicles counted. A total of 200 GUVs were counted at each temperature. For each system, at least three independent GUV preparations were performed. Miscibility transition temperature Tmisc is defined as the temperature at which 50% of vesicles are phase separated, as determined from sigmoidal fits to phase separation versus temperature data68.
Synaptosomes and lipidomics
Synaptosomes were prepared from 4 separate preparations of adult (60 days post-natal) hippocampus as previously described34, 69. Briefly, both hippocampi from three female Sprague-Dawley rats were harvested within 30 sec of decapitation and placed in ice-cold isotonic sucrose solution (5 mM Hepes, pH 7.4, 0.32 M sucrose, 1 mM MgCl2, 0.5 mMCaCl2, and 1 μg/ml leupeptin) followed by disruption in a motor-driven glass/Teflon homogenizer. Nuclei and debris were removed by centrifugation at 1,400g for 10 min and then a crude synaptosomal fraction prepared by centrifugation at 13,800g for 10 min. The pellet was suspended in isotonic solution (5 mM Hepes, pH 7.4, 0.32 M sucrose and 1 ug/ml leupeptin) and synaptosomes were purified by centrifugation through a discontinuous sucrose density gradient (0.8/1.0/1.4 M) at 110,000g for 2 hr. The synaptosomal fraction was recovered from the 1.0 M/1.4 M interface and stored as aliquots at −80°C.
Lipidomics analysis was performed by Lipotype, GmbH, according to established protocols70. Briefly, membrane samples are spiked with internal standards for all relevant lipid classes, followed by extraction of lipids from synaptosomes using a two-step extraction procedure35, 44. Lipids are detected by a mass spectrometer instrument (Q Exactive, Thermo Scientific) then identified and quantified by an automated processing protocol using commercial software.28 Only lipid identifications with a signal -to-noise ratio >5, an absolute abundance of at least 1 pmol, and a signal intensity 5-fold higher than in corresponding blank samples were considered for further data analysis.
RESULTS AND DISCUSSION
Polyunsaturated lipids are abundant in rat synaptic membranes
Although investigations of biomimetic membrane typically, though not exclusively29, 41–44, focus on mixtures of saturated and monounsaturated phospholipids, polyunsaturated lipids are abundant in all biological membranes. In particular, such lipids are highly enriched in the excitable membranes of cells of the mammalian nervous system35–36. To justify our focus on polyunsaturated lipids, we performed detailed lipidomic analysis of the lipid composition of adult rat neuronal synapse membranes, isolated as “synaptosomes”71. These synaptosomes are an effective preparation of synaptic membranes, composed mainly of the post- and pre-synaptic plasma membranes and the neurotransmitter-loaded synaptic vesicles. Mass spectrometry coupled to electron spray ionization (ESI-MS) provides comprehensive, quantitative data on the abundance of hundreds of membrane lipid species, identified by the polar headgroup as well as the length and unsaturation of both hydrophobic tails. Consistent with a plasma membrane enriched preparation9, 27, the major lipid class detected was cholesterol, comprising 33 mol% of all lipids. The neutral phospholipids PC (30 mol%) and phosphatidylethanolamine (PE; 23 mol%) were the other major lipid classes (Fig. 1a). Analysis of the fatty acids in glycerophospholipids revealed that although 48% were fully saturated (mostly palmitate and stearate), another 28% were polyunsaturated (PUFA). Of these, 12% had four double bonds (e.g. arachidonate) and 15% were ω-3 DHA with six double bonds (Fig. 1b). In lipids, these PUFA chains were often combined with saturated acyl chains to make mixed/hybrid lipids (Fig. 1c; see detailed lipidome in Table S1). This analysis confirmed the high abundance of polyunsaturated lipids in synaptic membranes, consistent with previous reports on the lipid composition of synaptic vesicles34 and post-synaptic plasma membranes69, and prompted our study of the effects of these on the properties of biomimetic membranes. For the simulations and experiments below, we have focused on lipids with two polyunsaturated acyl chains because they have not been as widely studied as their hydrid counterparts and because they provide a direct comparison to the most widely-studied domain forming membrane models25–26. Such di-polyunsaturated lipids are widely found in biological membranes, most notably in retinal membranes of mammals72–73, but also in fish74–75, amphibians76–77, and other aquatic organisms78.
Increasing the proportion of polyunsaturated lipids in model membranes enhances domain stability
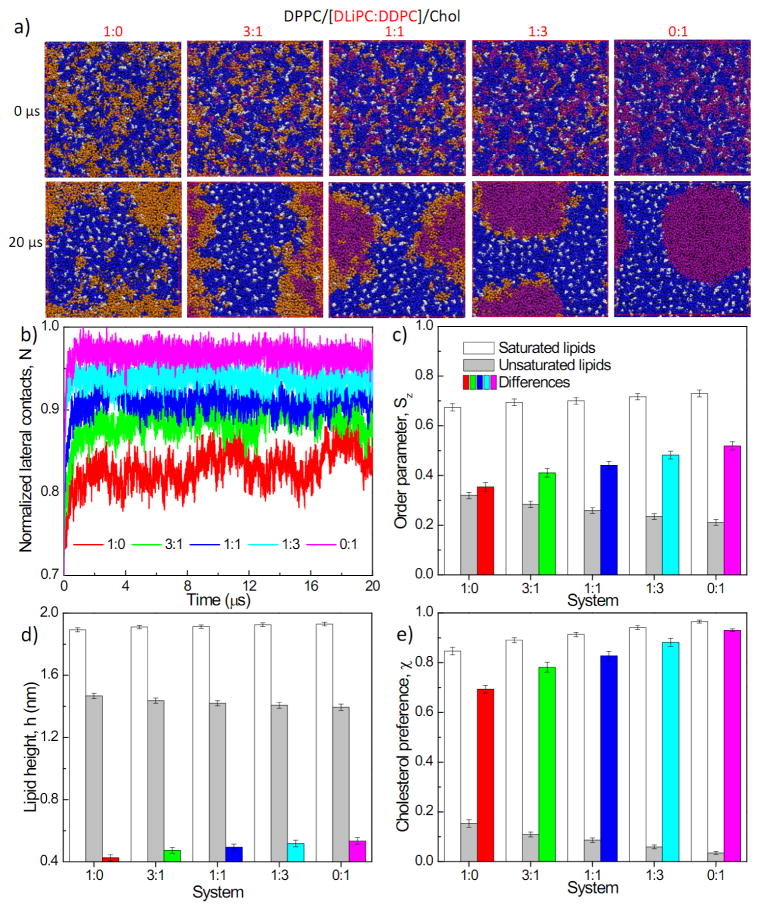
We performed 20 μs CG molecular dynamics simulations for four-component model membranes composed of DPPC/[DLiPC+DDPC]/Chol with varying molar ratios of di-unsaturated (DLiPC) to highly polyunsaturated (DDPC) lipids (full lipid names, chemical structures, and Martini-based configurations are shown in Fig. S1). Snapshots at the end of the simulation (Fig. 2a) reveal that all systems evolved toward stable liquid domains, with Lo domains enriched in DPPC and Chol (blue and white regions in Fig. 2a) coexisting with Ld domains rich in unsaturated lipids (orange and purple). However, the detailed features of the domains were dependent on the specific composition of unsaturated lipids. Namely, large domains were clearly stabilized by increasing the molar ratio of polyunsaturated DDPC relative to DLiPC, as assayed by the number of lateral contacts between unsaturated lipids, a proxy for domain size (see Materials and Methods) (Fig. 2b). Surprisingly, the lipids in the disordered phase were not well-mixed, instead showing subdomains enriched in the two different unsaturated lipids (Fig. 2a). Sub-structure has been recently described in the Lo phase by atomistic simulations79, and it remains to be determined whether the Ld substructure observed here is a real effect or an artifact of the simulation. The promotion of liquid-liquid domain segregation by polyunsaturated lipids is consistent with previous MD simulations of di-unsaturated lipids reported by Ackerman and Feigenson32 and experimental work by Shaikh and Wassall42. Lipids containing polyunsaturated fatty acids like docosahexaenoic acid (DHA, 22 carbons and 6 double bonds) do not interact readily with cholesterol80–81 due to the intrinsic disorder of their acyl chains39. This behavior is likely responsible for the enhanced lateral segregation we observe. It is important to point out that our model lipids contain polyunsaturated acyl chains in both the sn-1 and sn-2 positions, distinct from previous experimental investigations of “hybrid” lipids in which only the sn-2 position is polyunsaturated while the sn-1 bears a saturated acyl chain. This discrepancy explains why we did not observe penetration of the ordered phase by polyunsaturated PC 41, 43.
Figure 2.
Four-component lipid systems with increasing polyunsaturated lipid ratios: (a) Snapshots at the end of 20 μs simulations (DPPC: blue, DLi PC: orange, DDPC: purple, Chol: white); (b) time evolution of normalized lateral contact N of unsaturated lipids; (c) lipid chain order parameter, Sz; (d) lipid height, h; and (e) percentage contact with cholesterol, χ, of saturated lipids, unsaturated lipids and their differences. Results in (c–e) are obtained by averaging the last 4 μs of a 20 μs trajectory; error bars represent standard deviations. (d) and (e) share the same figure legend as (c). The datasets corresponding to (c–e) can be found in Table S2.
Interestingly, analysis of the order parameter (Fig. 2c) and two-dimensional radial pair distribution of DPPC (Fig. S2a) shows that the presence of polyunsaturated lipids also affects the packing of the saturated lipids, with order and packing of DPPC progressively enhanced by increasing concentrations of DDPC. This observation suggested that PUFA-containing lipids potentially affect the saturated lipid-rich ordered domains concomitant with their effects on disordered domains. To characterize this behavior, we analyzed several membrane properties (lipid chain order parameter, lipid height, bilayer thickness, and cholesterol preference) for both saturated and unsaturated lipids across all simulations (Table S2). For all these properties, we observed a progressive enhancement of the differences between Lo-preferring saturated lipids and Ld-preferring unsaturated lipids with increasing proportions of polyunsaturated DDPC (Fig. 2c–e and Fig. S2b–c). We note that all reported observables for disordered lipids are a combination of DLiPC and DDPC, and appear to scale linearly with the relative DLiPC/DDPC fraction, suggesting that these effects are not an emergent property of mixed Ld phases, but rather simply the result of substituting lipids with one type of behavior (i.e. polyunsaturated) for those of another (less unsaturated).
The correlation between differences of lipid height/bilayer thickness and relative domain size in our simulations is consistent with neutron scattering measurements of another quatenary system (DSPC/[POPC+DOPC]/Chol, DSPC: 1,2-distearoyl-sn-glycero-3-phosphocholine, POPC: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) where increasing molar ratio of DOPC was used to tune thickness differences11. Similarly, the correlation between increased interdomain order differences and enhanced phase separation are supported by experimental results12–13. We interpret these observations based on cholesterol-mediated lipid ordering82 and the effective “repulsion” of cholesterol from PUFA-containing lipids80, 83 – i.e. disordered domains rich in DDPC essentially exclude cholesterol leading to thinner, less ordered, and more fluid membranes84. Exclusion of cholesterol from the disordered domain leads to its concentration in the DPPC-rich domains, enhancing its ordering effect on DPPC. This interpretation is similar to the “push-pull” mechanism for ordered domain formation, based on measurements of pairwise interaction parameters between lipids85–86 and NMR experiments 48, 87. Together, these effects enhance the difference between ordered and disordered lipids, which ultimately promotes their separation into stable domains.
Increasing unsaturation of disordered lipids enhances interdomain differences to promote domain stability
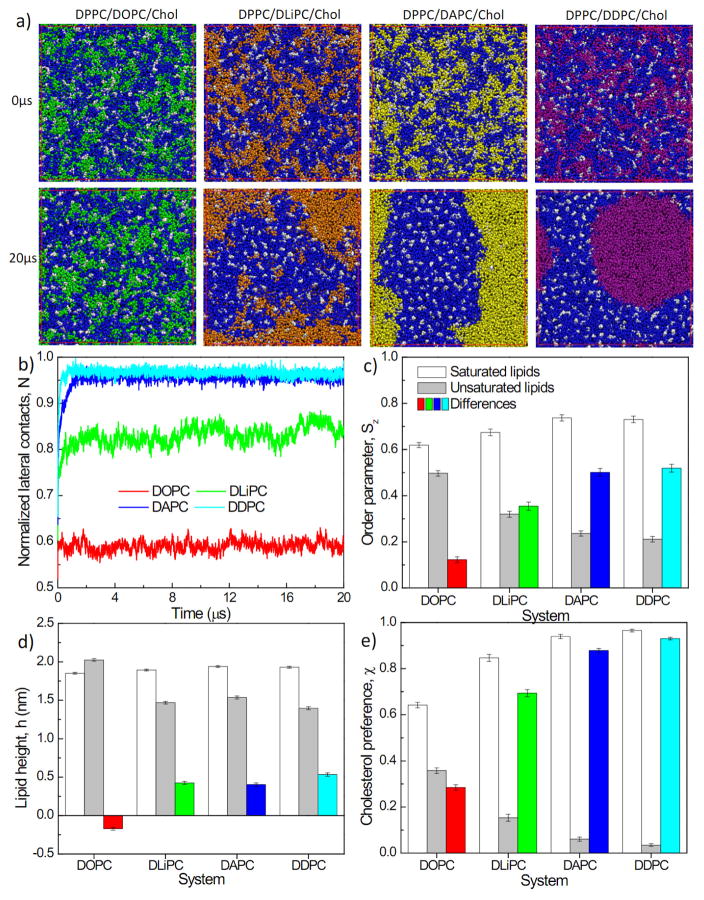
The observations described above suggest that increased levels of polyunsaturated lipids promote membrane domain stability. To analyze the effects of the degree of polyunsaturation on membrane domains, we investigated membrane systems wherein the concentration of unsaturated lipids was kept constant but the extent of unsaturation was varied. Specifically, we compared three-component model membranes composed of DPPC/DXPC/Chol (5:3:2), with DXPC being one of several different lipids containing varying numbers of double bonds in both lipid chains: DOPC (1); DLiPC (2); DAPC (4); and DDPC (6). The snapshots and analyses in Fig. 3a–b and Fig. S3a confirm the above observations that the presence of polyunsaturated lipids promotes the formation of larger, more stable domains. This effect was progressive, with domain size in DLiPC-containing membranes being approximately intermediate between the monounsaturated DOPC and the highly polyunsaturated DAPC and DDPC (Fig. 3a–b). It should be noted that DOPC-containing bilayers did not evolve to stable phase separation, rather small, transient, unsaturated lipid-rich clusters persisted throughout the simulation time course. This observation is in conflict with well validated experimental results25 and our own observations (see Fig. 5), revealing that the Martini model does not perfectly capture the collective behavior of DOPC, as previously noted57.
Figure 3.
Three-component lipid systems with varying degree of lipid unsaturation: (a) Snapshots at the end of 20 μs simulations (DPPC: blue, DOPC: green, DLiPC: orange, DAPC: yellow, DDPC: purple, Chol: white); (b) time evolution of normalized lateral contact N of unsaturated lipids; (c) lipid chain order parameters, Sz; (d) lipid height, h; and (e) percentage contact with cholesterol χ of saturated lipids, unsaturated lipids and their differences. Results in (c–g) are obtained by averaging the last 4 μs of a 20 μs trajectory. Error bars represent standard deviations. In (c–e) white bars represent observables measured for DPPC, gray bars are unsaturated lipids, and the colored bars are the differences between DPPC and unsaturated lipid. The datasets corresponding to (c–e) are presented in Table S3.
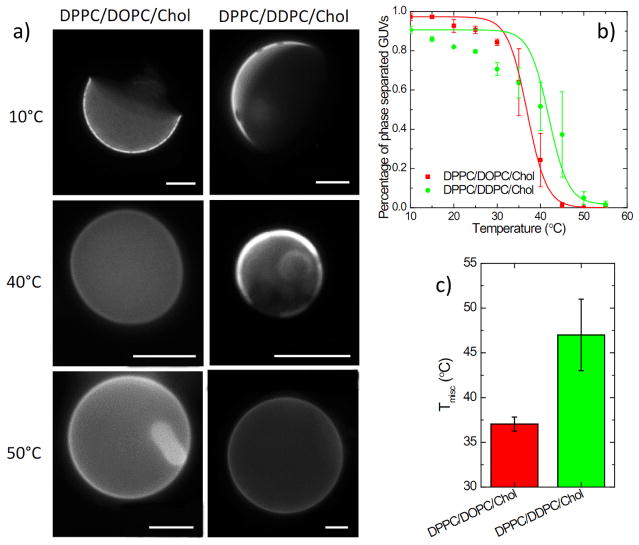
Figure 5. Experimental validation of domain stabilization by polyunsaturated lipids.
(a) Exemplary fluorescence micrographs of ternary GUVs show that Lo / Ld phase separation is observable at 40°C in DDPC- but not DOPC-containing membranes; (b) Representative curves show that DDPC produces a rightward shift of the phase separation curve in GUVs, indicative of more stable phase separation; (c) Effect confirmed by quantification of miscibility transition temperature Tmisc. Average ± s.d. of ~200 vesicles, representative of two different trials. Scale bar: 10 μm.
As above, domain size was tightly correlated with the interdomain differences in lipid chain order (Fig. 3c) and cholesterol preference (Fig. 3f), and to a lesser extent, lipid height (Fig. 3d) and bilayer thickness (Fig. S3c). We also observed increased DPPC ordering resulting from increasing the extent of Ld lipid poyunsaturation (Fig. 3c). Interestingly, we find only slight differences in all measured membrane parameters induced by increasing lipid unsaturation from 4 (DAPC) to 6 (DDPC) double bonds per acyl chain, suggesting an upper limit on the effects observed in this computational model. Both highly polyunsaturated lipids (DAPC and DDPC) produced Ld domains that were nearly devoid of saturated DPPC and cholesterol. This finding contrasts with conclusions from detailed experimental evaluations of phase diagrams of DOPC/DPPC/Cholesterol and similar systems88–89, which suggest that Ld phases have substantial levels of cholesterol, and to a lesser extent, saturated lipid. Unfortunately, the phase diagrams of mixtures with highly polyunsaturated lipids have not been detailed and therefore our observations of near-complete exclusion of DPPC and cholesterol from their Ld phases remains to be experimentally verified. Finally, although the shapes of the domains in DAPC- versus DDPC-containing membranes were different across multiple independent simulations runs (i.e. circular in DDPC, stripes in DAPC), these may be affected by periodic boundary conditions and limited system size.
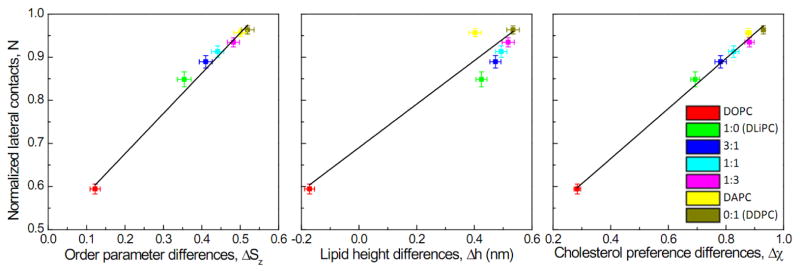
Our extended simulations allow access to molecular-level information that can inform the physical mechanisms underlying the enhancement of domain size/stability by polyunsaturated lipids. Specifically, across all our simulations, we observed striking correlations between the differentials in properties - including lipid chain order (ΔSz), lipid height (Δh), and cholesterol preference (Δχ) - between coexisting domains and the stability of the domains estimated by normalized lateral contacts between unsaturated lipids (Fig. 4). In other words, domain stability in biomimetic membranes is driven by the differences between domains, consistent with several experimental observations11–13. In our experiments, the extent and abundance of unsaturation is a major determinant of these differences, and therefore domain size.
Figure 4.
Normalized lateral contact N of unsaturated lipids (relative domain size) is tightly correlated to differences between saturated and unsaturated lipids in lipid chain order ΔSz (a) and cholesterol preference Δχ (c), and to a lesser extent, lipid height Δh (b). Results are averages of the last 4 μs of 20 μs runs and error bars represent standard deviation. Different colors represent different systems as described in Fig. 2 and Fig. 3.
To verify our simulation results, we performed model membrane experiments in Giant Unilamellar Vesicles (GUVs) composed of DPPC, Chol, and an unsaturated lipid component being either monounsaturated DOPC or polyunsaturated DDPC (5:3:2 with 0.1 mol% DiO-C18 for fluorescent visualization). As expected25, GUVs with these compositions showed Lo / Ld phase separation across a wide temperature range (Fig. 5a–b). The miscibility transition temperature (Tmisc) is an estimate of domain stability and was quantified by counting the number of phase-separated vesicles as a function of temperature and sigmoid-fitting the resulting data to obtain the 50% phase separation temperature (Fig. 5b). The Tmisc of DOPC-containing GUVs was approximately 37°C, within range of published observations25. Strikingly, the presence of DDPC as the unsaturated component increased Tmisc by almost ~10 °C (Fig. 5c), confirming more stable domains in membranes containing polyunsaturated lipids and supporting our simulations.
Domain stability is tightly correlated with interdomain mismatch in lipid chain order
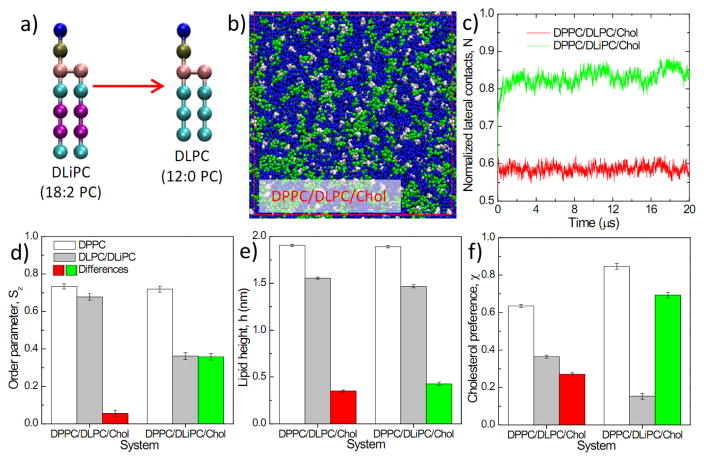
The simulations above show that the differences in physical properties between coexisting domains correlated with domain size / stability (Fig. 4). Moreover, these differences in domain properties (ΔSz, Δh, and Δχ) were also correlated with each other – the best correlations were between interdomain cholesterol partitioning Δχ and lipid chain order differences ΔSz (Fig. S4), consistent with recent model membrane experiments11, 38. To disentangle these effects, we computationally compared model membranes comprised of lipids either with different heights but similar predicted order parameters(Fig. 6), or with different order parameters but similar predicted heights( Fig. 7). For the former, we compared ternary membranes containing DPPC and Chol with either di-unsaturated lipid (DLiPC) or a length-matched saturated lipid (DLPC: 1,2-dilauroyl-sn-glycero-3-phosphocholine; Fig. 6a). As predicted, DLiPC in these mixtures was much more disordered than DLPC (whose order parameter was similar to DPPC; Fig. 6d), whereas the lipid height for DLiPC and DLPC were very similar (Fig. 6e). DPPC and DLPC also had more similar cholesterol affinity compared to DPPC/DLiPC (Fig. 6f). Strikingly, the DPPC/DLPC/Chol system did not show any macroscopic lipid demixing (Fig. 6b–c), in contrast to the clearly separated DPPC/DLiPC/Chol (Fig. 3). This result is consistent with experiments on the same system, which found no domains formation in DPPC/DLPC/Chol GUVs90. These simulations and experiments suggest that differences in lipid melting temperature (DPPC ~ 42°C; DLPC ~1°C) and lipid height (Fig. 6e) are insufficient for liquid-liquid domain separation.
Figure 6.
(a) Swapping DLiPC for DLPC in a lipid mixture to remove unsaturation, thus maintaining lipid height but changing lipid order; (b) A snapshot at the end of a 20 μs simulation of DPPC/DLPC/Chol; (c) time evolution of the normalized lateral contact N of unsaturated lipids; (d) lipid chain order parameter Sz, (e) lipid height h, (f) percentage contacts with cholesterol χ of saturated lipids, unsaturated lipids and their differences. Results in (d–f) are obtained from averaging the last 4 μs trajectory of a 20 μs run. Error bars represent standard deviation. (e) and (f) share the same figure legend as (d).
Figure 7.
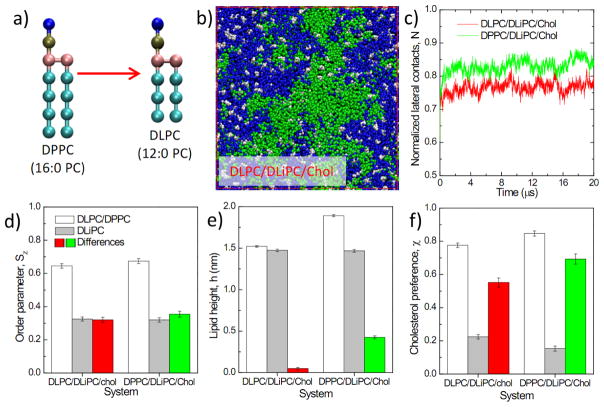
(a) Illustration of DPPC and DLPC with different lipid height but same tail saturation; (b) A snapshot at the end of a 20 μs simulation of a DLPC/DLiPC/Chol system; (c) time evolution of the normalized lateral contact N of unsaturated lipids; (d) lipid chain order parameter Sz; (e) lipid height h; (f) percentage contacts with cholesterol χ of saturated lipids, unsaturated lipids and their differences. Results in (d–f) are obtained from averaging the last 4 μs of a 20 μs run. Error bars represent standard deviation. (e) and (f) share the same figure legend as (d).
To further test the effect of order difference on domain size, we compared DLPC/DLiPC/Chol with DPPC/DLiPC/Chol (Fig. 7a). Both systems formed domains with similar time scales and stabilities (Fig. 7b). DLPC was chosen to match the height of DLiPC, and consistently, no lipid height difference was observed in DLPC/DLiPC/Chol (Fig. 7e). In contrast, there were major differences in lipid chain order and cholesterol preference between DLPC and DLiPC, which were similar to those observed for DPPC/DLiPC/Chol( Fig. 7d, f). Thus, disparities in lipid order and cholesterol preference, rather than lipid height, correlated with domain stability. This observation is consistent with recent experiments that did not find a strong relationship between domain stability and thickness mismatch 91, and others that observed a correlation between order disparity and domain stability 38. These observations do not rule out lipid height as a factor in phase separation, as Heberle et al. clearly show that thickness mismatch correlates strongly with domain size11. However, for the PUFA lipid-containing model membranes that we have tested, domain size and stability are more strongly correlated with differences in lipid chain order, rather than lipid height (Fig. S5).
CONCLUSION
Using coarse-grained molecular dynamics simulations and experiments, we explored the effect of biologically important polyunsaturated lipids on liquid domain formation in biomimetic model membranes. The clear trends in our observations and good qualitative agreements between simulations and experiments support the conclusions that the abundance and degree of lipid polyunsaturation promote domain stability by enhancing the differences in lipid chain order and cholesterol preference between the coexisting domains. These observations elucidate the important role of non-canonical biological lipids on the physical properties of membranes, showing that lipid polyunsaturation is a driving force for liquid-liquid phase separation. For all computationally tested systems, differences in cholesterol preference between saturated and unsaturated lipids were tightly correlated to difference in chain order, which were in turn correlated to domain stability (Fig. 4). Using model systems in which lipid chain orders or lipid heights were independently varied, we observed that membrane domain size and stability are tightly correlated with differences in lipid chain order (Fig. S5). Further systematic studies are needed to definitively uncouple the relationships between these properties and resolve the key question of the causative physical drivers of membrane domain properties.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Grant (RO1 GM100078, GM114282) and the Cancer Prevention and Research Institute of Texas (CPRIT) New Investigator Recruitment Grant (R1215). We thank the Texas Advanced Computing Center (TACC) and the Extreme Science and Engineering Discovery Environment (XSEDE)allocation ( project: TG-MCB150054) for generous computational resources.
Footnotes
Notes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Additional gures are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 2.Simons K, Sampaio JL. Membrane organization and lipid rafts. Cold Spring Harbor Perspect Biol. 2011;3:a004697. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 4.Kusumi A, Fujiwara TK, Chadda R, Xie M, Tsunoyama TA, Kalay Z, Kasai RS, Suzuki KG. Dynamic organizing principles of the plasma membrane that regulate signal transduction: Commemorating the fortieth anniversary of singer and nicolson’s fluid-mosaic model. Annu Rev Cell Dev Biol. 2012;28:215–250. doi: 10.1146/annurev-cellbio-100809-151736. [DOI] [PubMed] [Google Scholar]
- 5.Li Z, Janosi L, Gorfe AA. Formation and domain partitioning of h-ras peptide nanoclusters: Effects of peptide concentration and lipid composition. J Am Chem Soc. 2012;134:17278–17285. doi: 10.1021/ja307716z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin X, Li Z, Gorfe AA. Reversible effects of peptide concentration and lipid composition on h-ras lipid anchor clustering. Biophys J. 2015;109:2467–2470. doi: 10.1016/j.bpj.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y, Maxwell KN, Sezgin E, Lu M, Liang H, Hancock JF, Dial EJ, Lichtenberger LM, Levental I. Bile acids modulate signaling by functional perturbation of plasma membrane domains. J Biol Chem. 2013;288:35660–35670. doi: 10.1074/jbc.M113.519116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz-Rohrer B, Levental KR, Levental I. Rafting through traffic: Membrane domains in cellular logistics. Biochim Biophys Acta-Biomembr. 2014;1838:3003–3013. doi: 10.1016/j.bbamem.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 9.Diaz-Rohrer BB, Levental KR, Simons K, Levental I. Membrane raft association is a determinant of plasma membrane localization. Proc Natl Acad Sci U S A. 2014;111:8500–8505. doi: 10.1073/pnas.1404582111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hering H, Lin CC, Sheng M. Lipid rafts in the maintenance of synapses, dendritic spines, and surface ampa receptor stability. J Neurosci. 2003;23:3262–3271. doi: 10.1523/JNEUROSCI.23-08-03262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heberle FA, Petruzielo RS, Pan J, Drazba P, Ku erka N, Standaert RF, Feigenson GW, Katsaras J. Bilayer thickness mismatch controls domain size in model membranes. J Am Chem Soc. 2013;135:6853–6859. doi: 10.1021/ja3113615. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser HJ, Lingwood D, Levental I, Sampaio JL, Kalvodova L, Rajendran L, Simons K. Order of lipid phases in model and plasma membranes. Proc Natl Acad Sci U S A. 2009;106:16645–16650. doi: 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levental I, Byfield FJ, Chowdhury P, Gai F, Baumgart T, Janmey PA. Cholesterol-dependent phase separation in cell-derived giant plasma membrane vesicles. Biochem J. 2009;424:163–167. doi: 10.1042/BJ20091283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheidt HA, Meyer T, Nikolaus J, Baek DJ, Haralampiev I, Thomas L, Bittman R, Müller P, Herrmann A, Huster D. Cholesterol’s aliphatic side chain modulates membrane properties. Angew Chem Int Edit. 2013;52:12848–12851. doi: 10.1002/anie.201306753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin X, Zhang S, Ding H, Levental I, Gorfe AA. The aliphatic chain of cholesterol modulates bilayer interleaflet coupling and domain registration. FEBS Lett. 2016;590:3368–3374. doi: 10.1002/1873-3468.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X, London E. The effect of sterol structure on membrane lipid domains reveals how cholesterol can induce lipid domain formation. Biochemistry. 2000;39:843–849. doi: 10.1021/bi992543v. [DOI] [PubMed] [Google Scholar]
- 17.Sezgin E, Levental I, Grzybek M, Schwarzmann G, Mueller V, Honigmann A, Belov VN, Eggeling C, Coskun Ü, Simons K. Partitioning, diffusion, and ligand binding of raft lipid analogs in model and cellular plasma membranes. Biochim Biophys Acta-Biomembr. 2012;1818:1777–1784. doi: 10.1016/j.bbamem.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 18.van Zanten TS, Mayor S. Current approaches to studying membrane organization. F1000 Research. 2015;4:1380. doi: 10.12688/f1000research.6868.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.London E. How principles of domain formation in model membranes may explain ambiguities concerning lipid raft formation in cells. Biochim Biophys Acta-Mol Cell Res. 2005;1746:203–220. doi: 10.1016/j.bbamcr.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Simons K, Vaz WL. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 21.Bennett WD, Tieleman DP. Computer simulations of lipid membrane domains. Biochim Biophys Acta-Biomembr. 2013;1828:1765–1776. doi: 10.1016/j.bbamem.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 22.Sodt AJ, Pastor RW, Lyman E. Hexagonal substructure and hydrogen bonding in liquid-ordered phases containing palmitoyl sphingomyelin. Biophys J. 2015;109:948–955. doi: 10.1016/j.bpj.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eggeling C, Honigmann A. Closing the gap: The approach of optical and computational microscopy to uncover biomembrane organization. Biochim Biophys Acta-Biomembr. 2016 doi: 10.1016/j.bbamem.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 24.Schäfer LV, de Jong DH, Holt A, Rzepiela AJ, de Vries AH, Poolman B, Killian JA, Marrink SJ. Lipid packing drives the segregation of transmembrane helices into disordered lipid domains in model membranes. Proc Natl Acad Sci U S A. 2011;108:1343–1348. doi: 10.1073/pnas.1009362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veatch SL, Keller SL. Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys J. 2003;85:3074–3083. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Risselada HJ, Marrink SJ. The molecular face of lipid rafts in model membranes. Proc Natl Acad Sci U S A. 2008;105:17367–17372. doi: 10.1073/pnas.0807527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sezgin E, Kaiser HJ, Baumgart T, Schwille P, Simons K, Levental I. Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat Protoc. 2012;7:1042–1051. doi: 10.1038/nprot.2012.059. [DOI] [PubMed] [Google Scholar]
- 28.Levental KR, Levental I. Giant plasma membrane vesicles: Models for understanding membrane organization. Curr Top Membr. 2015;75:25–57. doi: 10.1016/bs.ctm.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 29.Ingólfsson HI, Melo MN, van Eerden FJ, Arnarez C, Lopez CA, Wassenaar TA, Periole X, De Vries AH, Tieleman DP, Marrink SJ. Lipid organization of the plasma membrane. J Am Chem Soc. 2014;136:14554–14559. doi: 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- 30.Veatch SL, Cicuta P, Sengupta P, Honerkamp-Smith A, Holowka D, Baird B. Critical fluctuations in plasma membrane vesicles. ACS Chem Biol. 2008;3:287–293. doi: 10.1021/cb800012x. [DOI] [PubMed] [Google Scholar]
- 31.Goh SL, Amazon JJ, Feigenson GW. Toward a better raft model: Modulated phases in the four-component bilayer, dspc/dopc/popc/chol. Biophys J. 2013;104:853–862. doi: 10.1016/j.bpj.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ackerman DG, Feigenson GW. Multiscale modeling of four-component lipid mixtures: Domain composition, size, alignment, and properties of the phase interface. J Phys Chem B. 2015;119:4240–4250. doi: 10.1021/jp511083z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shlomovitz R, Schick M. Model of a raft in both leaves of an asymmetric lipid bilayer. Biophys J. 2013;105:1406–1413. doi: 10.1016/j.bpj.2013.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takamori S, Holt M, Stenius K, Lemke EA, Grønborg M, Riedel D, Urlaub H, Schenck S, Brügger B, Ringler P. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 35.Sampaio JL, Gerl MJ, Klose C, Ejsing CS, Beug H, Simons K, Shevchenko A. Membrane lipidome of an epithelial cell line. Proc Natl Acad Sci U S A. 2011;108:1903–1907. doi: 10.1073/pnas.1019267108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerl MJ, Sampaio JL, Urban S, Kalvodova L, Verbavatz JM, Binnington B, Lindemann D, Lingwood CA, Shevchenko A, Schroeder C. Quantitative analysis of the lipidomes of the influenza virus envelope and mdck cell apical membrane. J Cell Biol. 2012;196:213–221. doi: 10.1083/jcb.201108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zech T, Ejsing CS, Gaus K, de Wet B, Shevchenko A, Simons K, Harder T. Accumulation of raft lipids in t-cell plasma membrane domains engaged in tcr signalling. EMBO J. 2009;28:466–476. doi: 10.1038/emboj.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levental KR, Lorent JH, Lin X, Skinkle AD, Surma MA, Stockenbojer EA, Gorfe AA, Levental I. Polyunsaturated lipids regulate membrane domain stability by tuning membrane order. Biophys J. 2016;110:1800–1810. doi: 10.1016/j.bpj.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feller SE, Gawrisch K, MacKerell AD. Polyunsaturated fatty acids in lipid bilayers: Intrinsic and environmental contributions to their unique physical properties. J Am Chem Soc. 2002;124:318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 40.Feller SE, Gawrisch K. Properties of docosahexaenoic-acid-containing lipids and their influence on the function of rhodopsin. Curr Opin Struct Biol. 2005;15:416–422. doi: 10.1016/j.sbi.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Shaikh SR, Kinnun JJ, Leng X, Williams JA, Wassall SR. How polyunsaturated fatty acids modify molecular organization in membranes: Insight from nmr studies of model systems. Biochim Biophys Acta-Biomembr. 2015;1848:211–219. doi: 10.1016/j.bbamem.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 42.Shaikh SR, Dumaual AC, Castillo A, LoCascio D, Siddiqui RA, Stillwell W, Wassall SR. Oleic and docosahexaenoic acid differentially phase separate from lipid raft molecules: A comparative nmr, dsc, afm, and detergent extraction study. Biophys J. 2004;87:1752–1766. doi: 10.1529/biophysj.104.044552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams JA, Batten SE, Harris M, Rockett BD, Shaikh SR, Stillwell W, Wassall SR. Docosahexaenoic and eicosapentaenoic acids segregate differently between raft and nonraft domains. Biophys J. 2012;103:228–237. doi: 10.1016/j.bpj.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wassall SR, Stillwell W. Docosahexaenoic acid domains: The ultimate non-raft membrane domain. Chem Phys Lipids. 2008;153:57–63. doi: 10.1016/j.chemphyslip.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 45.Konyakhina TM, Feigenson GW. Phase diagram of a polyunsaturated lipid mixture: Brain sphingomyelin/1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine/cholesterol. Biochim Biophys Acta-Biomembr. 2016;1858:153–161. doi: 10.1016/j.bbamem.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filippov A, Orädd G, Lindblom G. Domain formation in model membranes studied by pulsed-field gradient-nmr: The role of lipid polyunsaturation. Biophys J. 2007;93:3182–3190. doi: 10.1529/biophysj.107.111534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hyvönen MT, Kovanen PT. Molecular dynamics simulations of unsaturated lipid bilayers: Effects of varying the numbers of double bonds. Eur Biophys J. 2005;34:294–305. doi: 10.1007/s00249-004-0455-7. [DOI] [PubMed] [Google Scholar]
- 48.Brzustowicz MR, Cherezov V, Caffrey M, Stillwell W, Wassall SR. Molecular organization of cholesterol in polyunsaturated membranes: Microdomain formation. Biophys J. 2002;82:285–298. doi: 10.1016/S0006-3495(02)75394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosetti C, Pastorino C. Comparison of ternary bilayer mixtures with asymmetric or symmetric unsaturated phosphatidylcholine lipids by coarse grained molecular dynamics simulations. J Phys Chem B. 2012;116:3525–3537. doi: 10.1021/jp212406u. [DOI] [PubMed] [Google Scholar]
- 50.Ferraro M, Masetti M, Recanatini M, Cavalli A, Bottegoni G. Modeling lipid raft domains containing a mono-unsaturated phosphatidylethanolamine species. RSC Advances. 2015;5:37102–37111. [Google Scholar]
- 51.Swulius MT, Kubota Y, Forest A, Waxham MN. Structure and composition of the postsynaptic density during development. J Comp Neurol. 2010;518:4243–4260. doi: 10.1002/cne.22451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, De Vries AH. The martini force field: Coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 53.Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D. Gromacs 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bennett WD, MacCallum JL, Hinner MJ, Marrink SJ, Tieleman DP. Molecular view of cholesterol flip-flop and chemical potential in different membrane environments. J Am Chem Soc. 2009;131:12714–12720. doi: 10.1021/ja903529f. [DOI] [PubMed] [Google Scholar]
- 55.Lin X, Bai T, Zuo YY, Gu N. Promote potential applications of nanoparticles as respiratory drug carrier: Insights from molecular dynamics simulations. Nanoscale. 2014;6:2759–2767. doi: 10.1039/c3nr04163h. [DOI] [PubMed] [Google Scholar]
- 56.Lin X, Gu N. Surface properties of encapsulating hydrophobic nanoparticles regulate the main phase transition temperature of lipid bilayers: A simulation study. Nano Res. 2014;7:1195–1204. [Google Scholar]
- 57.Davis RS, Sunil Kumar P, Sperotto MM, Laradji M. Predictions of phase separation in three-component lipid membranes by the martini force field. J Phys Chem B. 2013;117:4072–4080. doi: 10.1021/jp4000686. [DOI] [PubMed] [Google Scholar]
- 58.Veatch SL, Soubias O, Keller SL, Gawrisch K. Critical fluctuations in domain-forming lipid mixtures. Proc Natl Acad Sci U S A. 2007;104:17650–17655. doi: 10.1073/pnas.0703513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perlmutter JD, Sachs JN. Interleaflet interaction and asymmetry in phase separated lipid bilayers: Molecular dynamics simulations. J Am Chem Soc. 2011;133:6563–6577. doi: 10.1021/ja106626r. [DOI] [PubMed] [Google Scholar]
- 60.Wassenaar TA, Ingólfsson HI, Böckmann RA, Tieleman DP, Marrink SJ. Computational lipidomics with insane: A versatile tool for generating custom membranes for molecular simulations. J Chem Theory Comput. 2015;11:2144–2155. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- 61.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 62.Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 63.Humphrey W, Dalke A, Schulten K. Vmd: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 64.Janosi L, Li Z, Hancock JF, Gorfe AA. Organization, dynamics, and segregation of ras nanoclusters in membrane domains. Proc Natl Acad Sci U S A. 2012;109:8097–8102. doi: 10.1073/pnas.1200773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Noel JK, Whitford PC, Onuchic JN. The shadow map: A general contact definition for capturing the dynamics of biomolecular folding and function. J Phys Chem B. 2012;116:8692–8702. doi: 10.1021/jp300852d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Gorfe AA. Aggregation of lipid-anchored full-length h-ras in lipid bilayers: Simulations with the martini force field. PloS one. 2013;8:e71018. doi: 10.1371/journal.pone.0071018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dimitrov D, Angelova M. Lipid swelling and liposome formation mediated by electric fields. Bioelectrochem Bioenerg. 1988;19:323–336. [Google Scholar]
- 68.Levental I, Grzybek M, Simons K. Raft domains of variable properties and compositions in plasma membrane vesicles. Proc Natl Acad Sci U S A. 2011;108:11411–11416. doi: 10.1073/pnas.1105996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tulodziecka K, Diaz-Rohrer BB, Farley MM, Chan RB, Di Paolo G, Levental KR, Waxham MN, Levental I. Remodeling of the postsynaptic plasma membrane during neural development. Molecular Biology of the Cell. 2016 doi: 10.1091/mbc.E16-06-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Surma MA, Herzog R, Vasilj A, Klose C, Christinat N, Morin-Rivron D, Simons K, Masoodi M, Sampaio JL. An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur J Lipid Sci Technol. 2015;117:1540–1549. doi: 10.1002/ejlt.201500145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miljanich GP, Sklar LA, White DL, Dratz EA. Disaturated and dipolyunsaturated phospholipids in the bovine retinal rod outer segment disk membrane. Biochim Biophys Acta-Biomembr. 1979;552:294–306. doi: 10.1016/0005-2736(79)90284-0. [DOI] [PubMed] [Google Scholar]
- 73.Stinson AM, Wiegand RD, Anderson RE. Fatty acid and molecular species compositions of phospholipids and diacylglycerols from rat retinal membranes. Exp Eye Res. 1991;52:213–218. doi: 10.1016/0014-4835(91)90261-c. [DOI] [PubMed] [Google Scholar]
- 74.Bell M, Dick J, Buda C. Molecular speciation of fish sperm phospholipids: Large amounts of dipolyunsaturated phosphatidylserine. Lipids. 1997;32:1085–1091. doi: 10.1007/s11745-997-0140-y. [DOI] [PubMed] [Google Scholar]
- 75.Bell MV, Batty RS, Dick JR, Fretwell K, Navarro JC, Sargent JR. Dietary deficiency of docosahexaenoic acid impairs vision at low light intensities in juvenile herring (clupea harengus l.) Lipids. 1995;30:443–449. doi: 10.1007/BF02536303. [DOI] [PubMed] [Google Scholar]
- 76.Wiegand RD, Anderson RE. Phospholipid molecular species of frog rod outer segment membranes. Exp Eye Res. 1983;37:159–173. doi: 10.1016/0014-4835(83)90075-1. [DOI] [PubMed] [Google Scholar]
- 77.Li F, Chen H, Anderson RE. Biosynthesis of docosahexaenoate-containing glycerolipid molecular species in the retina. J Mol Neurosci. 2001;16:205–214. doi: 10.1385/JMN:16:2-3:205. [DOI] [PubMed] [Google Scholar]
- 78.Bell MV, Henderson RJ. Molecular species composition of phosphatidylcholine fromcrypthecodinium cohnii in relation to growth temperature. Lipids. 1990;25:115–118. [Google Scholar]
- 79.Sodt AJ, Sandar ML, Gawrisch K, Pastor RW, Lyman E. The molecular structure of the liquid ordered phase of lipid bilayers. J Am Chem Soc. 2014;136:725–732. doi: 10.1021/ja4105667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pitman MC, Suits F, MacKerell AD, Feller SE. Molecular-level organization of saturated and polyunsaturated fatty acids in a phosphatidylcholine bilayer containing cholesterol. Biochemistry. 2004;43:15318–15328. doi: 10.1021/bi048231w. [DOI] [PubMed] [Google Scholar]
- 81.Eldho NV, Feller SE, Tristram-Nagle S, Polozov IV, Gawrisch K. Polyunsaturated docosahexaenoic vs docosapentaenoic acid differences in lipid matrix properties from the loss of one double bond. J Am Chem Soc. 2003;125:6409–6421. doi: 10.1021/ja029029o. [DOI] [PubMed] [Google Scholar]
- 82.Ipsen JH, Karlström G, Mourtisen O, Wennerström H, Zuckermann M. Phase equilibria in the phosphatidylcholine-cholesterol system. Biochim Biophys Acta-Biomembr. 1987;905:162–172. doi: 10.1016/0005-2736(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 83.Harroun TA, Katsaras J, Wassall SR. Cholesterol is found to reside in the center of a polyunsaturated lipid membrane. Biochemistry. 2008;47:7090–7096. doi: 10.1021/bi800123b. [DOI] [PubMed] [Google Scholar]
- 84.Brzustowicz MR, Cherezov V, Zerouga M, Caffrey M, Stillwell W, Wassall SR. Controlling membrane cholesterol content. A role for polyunsaturated (docosahexaenoate) phospholipids. Biochemistry. 2002;41:12509–12519. doi: 10.1021/bi0262808. [DOI] [PubMed] [Google Scholar]
- 85.Wang C, Krause MR, Regen SL. Push and pull forces in lipid raft formation: The push can be as important as the pull. J Am Chem Soc. 2015;137:664–666. doi: 10.1021/ja5115437. [DOI] [PubMed] [Google Scholar]
- 86.Krause MR, Daly TA, Almeida PF, Regen SL. Push–pull mechanism for lipid raft formation. Langmuir. 2014;30:3285–3289. doi: 10.1021/la500510s. [DOI] [PubMed] [Google Scholar]
- 87.Kucerka N, Marquardt D, Harroun TA, Nieh MP, Wassall SR, de Jong DH, Schäfer LV, Marrink SJ, Katsaras J. Cholesterol in bilayers with pufa chains: Doping with dmpc or popc results in sterol reorientation and membrane-domain formation. Biochemistry. 2010;49:7485–7493. doi: 10.1021/bi100891z. [DOI] [PubMed] [Google Scholar]
- 88.Veatch SL, Gawrisch K, Keller SL. Closed-loop miscibility gap and quantitative tie-lines in ternary membranes containing diphytanoyl pc. Biophys J. 2006;90:4428–4436. doi: 10.1529/biophysj.105.080283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Feigenson GW, Buboltz JT. Ternary phase diagram of dipalmitoyl-pc/dilauroyl-pc/cholesterol: Nanoscopic domain formation driven by cholesterol. Biophys J. 2001;80:2775–2788. doi: 10.1016/S0006-3495(01)76245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Korlach J, Schwille P, Webb WW, Feigenson GW. Characterization of lipid bilayer phases by confocal microscopy and fluorescence correlation spectroscopy. Proc Natl Acad Sci U S A. 1999;96:8461–8466. doi: 10.1073/pnas.96.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bleecker JV, Cox PA, Keller SL. Mixing temperatures of bilayers not simply related to thickness differences between l o and l d phases. Biophys J. 2016;110:2305–2308. doi: 10.1016/j.bpj.2016.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.