Abstract
Sequencing and comparative analyses of genomes from multiple vertebrates are providing insights about the genetic basis for biological diversity. To date, these efforts largely have focused on eutherian mammals, chicken, and fish. In this article, we describe the generation and study of genomic sequences from noneutherian mammals, a group of species occupying unusual phylogenetic positions. A large sequence data set (totaling >5 Mb) was generated for the same orthologous region in three marsupial (North American opossum, South American opossum, and Australian tammar wallaby) and one monotreme (platypus) genomes. These ancient mammalian genomes are characterized by unusual architectural features with respect to G + C and repeat content, as well as compression relative to human. Approximately 14% and 34% of the human sequence forms alignments with the orthologous sequence from platypus and the marsupials, respectively; these numbers are distinctly lower than that observed with nonprimate eutherian mammals (45–70%). The alignable sequences between human and each marsupial species are not completely overlapping (only 80% common to all three species) nor are the platypus-alignable sequences completely contained within the marsupial-alignable sequences. Phylogenetic analysis of synonymous coding positions reveals that platypus has a notably long branch length, with the human–platypus substitution rate being on average 55% greater than that seen with human–marsupial pairs. Finally, analyses of the major mammalian lineages reveal distinct patterns with respect to the common presence of evolutionarily conserved vertebrate sequences. Our results confirm that genomic sequence from noneutherian mammals can contribute uniquely to unraveling the functional and evolutionary histories of the mammalian genome.
Keywords: comparative genomics, genome sequencing, genome analysis, phylogenetics, mammalian evolution
Comparisons of genome sequences from evolutionarily diverse species are central to decoding the functions of vertebrate genomes (1). Of particular interest is the use of highly diverged species for detecting and characterizing sequences under purifying selection (2). Large-scale sequence comparisons have been reported for eutherian (commonly referred to as “placental”) mammals (3) or fish (4), with the most detailed studies to date emphasizing human–rodent comparisons (5, 6).
We previously described our efforts to sequence the same orthologous regions from large collections of vertebrates (7, 8) and to perform multispecies sequence comparisons (9). These analyses have helped to refine phylogenetic relationships (7), to gain insight about the mutational process (10, 11), and to reveal differences between eutherian mammals and other vertebrates (e.g., birds and fish) with respect to their utility for detecting highly conserved regions in the human genome (9). However, these studies also demonstrate that for comparative sequence analyses, the optimal phylogenetic distances among species vary, depending on the question(s) being addressed [with the distance between humans and eutherian mammals sometimes being too close, and that between humans and birds (or fish) sometimes being too far].
Within this large phylogenetic gap between eutherian mammals and birds reside the marsupials and monotremes (12, 13). These metatherian and prototherian mammals diverged before the eutherian radiation, estimated at 185 and 200 million years ago (mya), respectively (14). Indeed, these divergence dates, as well as the origins of prototherian mammals relative to metatherian mammals, remain a source of scientific debate, in part because of insufficient molecular data (13, 15–17). Until recently, very little marsupial or monotreme DNA sequence was available in public databases. Although comparative studies involving small amounts of genomic sequence from a marsupial species [the stripe-faced dunnart (Sminthopsis macroura)] have been described (18), no comparisons involving large, contiguous blocks of marsupial or monotreme sequence have been reported to date.
In this article, we present the results of comparative sequence analyses involving >5 Mb of sequence from four noneutherian mammals. Specifically, we describe the features of their genomes, provide insights about their phylogenetic relationships, and reveal similarities and differences among mammalian lineages with respect to the presence of evolutionarily conserved vertebrate sequences.
Materials and Methods
Genomic Sequence Data Set. Genomic segments orthologous to a 1.9-Mb region on human chromosome 7q31.3, encompassing the cystic fibrosis transmembrane conductance regulator (CFTR) gene (referred to as the “greater CFTR region”), were isolated from the North American (N.A.) opossum, South American (S.A.) opossum, Australian tammar wallaby, and duckbilled platypus, and the segments were subjected to shotgun sequencing, as detailed in the supporting information, which is published on the PNAS web site. Sequences from an additional 23 vertebrates were generated and used for comparative analyses; the sequence data [including a listing of individual GenBank records for each bacterial artificial chromosome (BAC), assimilated and annotated sequences for each species, and multispecies sequence alignments (see below)] are available in the supporting information and at www.nisc.nih.gov/data.
Repeat Identification. Repetitive elements in noneutherian mammalian sequences were identified by using a recon-based approach (19), as described in the supporting information. Importantly, this approach was tuned to correctly detect repetitive elements in the human sequence at high specificity (99.8%) but at the cost of a lower sensitivity (63%). In turn, the identified repeats were used with repeatmasker (July 13, 2002; www.repeatmasker.org) and the standard repeatmasker mammalian repeat libraries to detect and mask all repetitive sequences. This process involved adding the identified repeats in the noneutherian mammalian sequence to the standard artiodactyl repeat library and then running repeatmasker with the -cow option.
Generation and Characterization of Sequence Alignments. A multisequence alignment of the assembled sequences from 27 vertebrates was generated by using the threaded blockset aligner (TBA) (20). The resulting alignment then was “projected” onto the human reference sequence for subsequent analyses (see the supporting information for details). A portion of the sequenced interval (541 kb distributed across nine distinct regions; see the supporting information) was selected where there was complete sequence coverage in a subset of species (chimpanzee, cat, cow, mouse, wallaby, N.A. opossum, S.A. opossum, platypus, and chicken). For each human–species pair-wise combination, the number of human-referenced positions of TBA-aligned bases was determined; these data then were used to calculate the number of bases in alignments for each human–species combination.
Estimating Phylogenetic Branch Lengths. A “virtual” multisequence alignment consisting solely of synonymous [4-fold degenerate (4D)] coding positions was generated by using the human-referenced annotations. Sites that fell within sequence gaps or that were no longer synonymous (because of changes in the first two bases) were treated as missing data. Substitution rates were estimated from this multisequence alignment by maximum likelihood with the phast package (21). A generally accepted tree topology for the analyzed species was used (7, 22). The most general reversible substitution model (REV) was used, and no molecular clock was assumed. Errors associated with the resulting branch length calculations were estimated by bootstrapping (both nonparametric and parametric methods; see the supporting information), with the tree topology fixed.
Examining Lineage Specificity of Multispecies Conserved Sequences (MCSs). MCSs were identified by using the multisequence alignment generated with sequences from 27 vertebrate species (8). A portion of the sequenced interval (571 kb distributed across seven separate regions; see the supporting information) was selected where there was complete sequence coverage in a subset of species (cat, dog, cow, pig, rat, mouse, N.A. opossum, wallaby, and platypus). Note that this limited data set is distinct from the one above used for characterizing the multisequence alignments. Each of the nine species' sequences was analyzed for the presence of the above-identified MCSs; specifically, each MCS in the relevant interval was scored as being present or absent based on blastz analysis (see the supporting information).
Results
Comparative Sequence Data Set. We generated large blocks of high-quality sequence from three marsupial species (N.A. opossum, S.A. opossum, and wallaby) and one monotreme species (platypus). All sequences correspond to genomic segments orthologous to the greater CFTR region on human chromosome 7q31.3 (7), with 1.17–1.63 Mb of nonredundant sequence generated from each species (Table 1). Based on comparisons with available genome-wide human (23), mouse (5), and rat (6) sequence, the greater CFTR region is close to average with respect to general genomic properties (e.g., repeat content, G + C content, fraction of coding sequence, and synonymous substitution rate). The resulting sequences from the four noneutherian mammals were analyzed individually and also compared with corresponding sequences from 23 additional vertebrates (7, 8).
Table 1. General characteristics of comparative sequence data set.
| Species | No. sequenced BACs | No. sequencing gaps* | No. mapping gaps† | Total nonredundant sequence, Mb | Amount relative to human,‡ Mb |
|---|---|---|---|---|---|
| N.A. opossum | 12 | 3 | 3 | 1.63 | 1.36 |
| S.A. opossum | 8 | 7 | 7 | 1.17 | 1.19 |
| Wallaby | 10 | 5 | 5 | 1.35 | 1.18 |
| Platypus | 13 | 0 | 0 | 1.26 | 1.65 |
Gaps reflecting missing sequence in the assembly of shotgun sequence data from an individual BAC; these are typically 100 bp or less. See the supplement in ref. 7 for details
Gaps reflecting the lack of BAC coverage across an interval. See the supplement in ref. 7 for details
The amount of human sequence in or between pair-wise alignments for the covered portions of each species' sequence; this value includes an estimate of sequence that might be proximal to the first and distal to the last alignment (utilizing the estimated degree of compression relative to human for that species)
Genomic Architecture. Analysis of the orthologous genes in this region reveals no gross differences in the content, order, orientation, or intron-exon structure between human and the noneutherian mammals (note that there are two instances of a missing exon within noneutherian sequence, but these appear to be due to gaps in sequence coverage; data not shown). However, examination of several architectural features associated with each species' sequence uncovered a number of differences. For example, the size of this genomic region (relative to human) varies by as much as 24% among the noneutherian mammals (Table 2). Specifically, evidence of both genome compression (e.g., 24% in platypus) and expansion (e.g., 17% and 15% in N.A. opossum and wallaby, respectively) is seen; these findings are generally consistent with previous estimates of genome sizes (refs. 24 and 25; also see www.genomesize.com).
Table 2. Architectural features of different species' sequences.
| G + C content*
|
|||||
|---|---|---|---|---|---|
| Species | Total | Nonrepetitive sites | Synonymous 4D sites | Relative size† | Percentage repetitive‡ |
| Human | 0.384 | 0.369 | 0.432 | NA | 40.3 |
| Cat | 0.383 | 0.372 | 0.434 | 0.95 | 36.4 |
| Pig | 0.377 | 0.366 | 0.455 | 0.92 | 31.9 |
| Mouse | 0.401 | 0.391 | 0.479 | 0.90 | 32.6 |
| N.A. opossum | 0.358 | 0.358 | 0.415 | 1.17 | 43.2 |
| S.A. opossum | 0.358 | 0.358 | 0.380 | 0.99 | 34.2 |
| Wallaby | 0.373 | 0.374 | 0.412 | 1.15 | 37.0 |
| Platypus | 0.459 | 0.457 | 0.642 | 0.76 | 44.9 |
| Chicken | 0.412 | 0.407 | 0.423 | 0.44 | 6.0 |
| Fugu | 0.486 | 0.485 | 0.721 | 0.16 | 2.3 |
Boldface indicates the data for noneutherian mammals.
Fraction of G + C bases in the entire sequence (total), the nonrepetitive portion of sequence (i.e., sequence not masked by repeatmasker), and synonymous 4D sites (the third position of codons that can be any base and still code for the same amino acid)
Ratio of sequence length in each species to the amount of corresponding human sequence (as defined in Table 1)
Percentage of sequence masked by repeatmasker
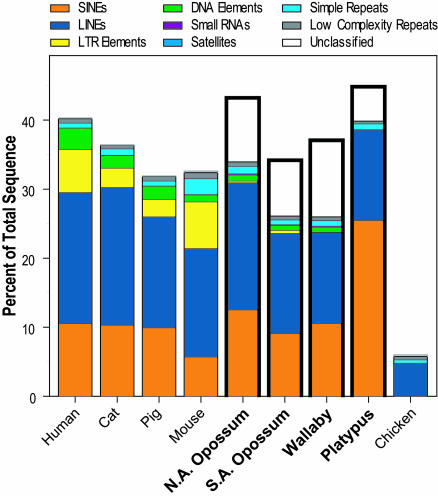
The asserted correlation between genome size and repeat content (4, 26) prompted us to investigate the amount and composition of repetitive elements within each species' sequence. Because repetitive sequences in noneutherian mammals have not been fully characterized, this analysis first required assembling repeat libraries for each marsupial and monotreme species (see Materials and Methods). Fig. 1 shows a summary of the content and types of repeats in each species' sequence, with data from several other vertebrates provided for comparison. Note the considerable variation in total repeat content among these species and the lack of correlation with genome size (relative to human; see Table 2). Specifically, the orthologous platypus genomic region is smaller than the human region yet contains a larger proportion of repetitive sequences; similarly, the wallaby genomic region is larger than the human region yet contains a smaller proportion of repetitive sequences. Another finding is the relatively large proportion of short interspersed nucleotide elements (SINEs) in the platypus sequence (27, 28), markedly different from other vertebrate sequences. The latter is consistent with the PCR-based identification of an abundant SINE repeat within monotreme genomes (J. A. M. Graves and P. J. Kirby, personal communication).
Fig. 1.
Comparison of the content and types of repetitive elements among different species' sequences. Sequences from the orthologous regions of the indicated species' genomes were analyzed by repeatmasker, allowing detection and quantification of the indicated types of repetitive elements. The data for the noneutherian mammals are highlighted for emphasis. SINEs, short interspersed nucleotide elements; LINEs, long interspersed nucleotide elements.
The overall G + C content is similar among the three marsupial sequences (35.8–37.3%; see Table 2), which is slightly lower than that of the orthologous human genomic region (38.4%). In contrast, the overall G + C content of the platypus sequence is notably high (45.9%), more like that seen with the orthologous Fugu genomic region (48.6%). A similarly high G + C content for platypus is seen in the nonrepetitive sites and at synonymous 4D sites (see Table 2). Examining the distribution of G + C content in 1-kb windows across the noneutherian sequences reveals the same general trends (see the supporting information).
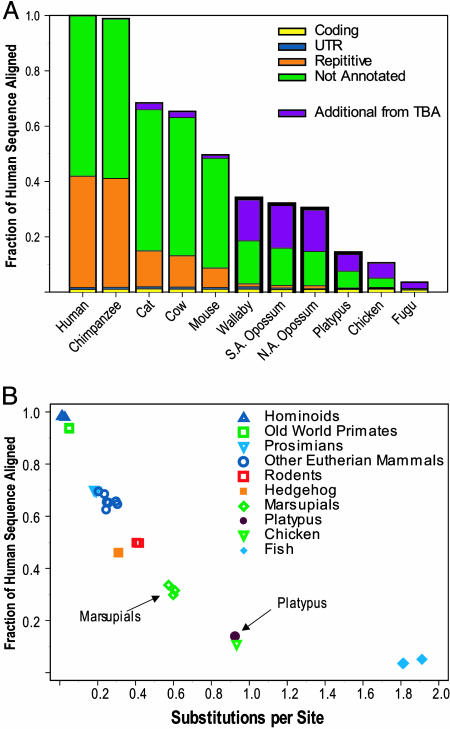
Multispecies Sequence Comparisons. Analyses of a multisequence alignment generated by using data from 27 vertebrates revealed notable patterns of sequence conservation. For example, the fraction of the human sequence forming alignments with nonprimate eutherian mammals is typically 45–70% (Fig. 2A) (7); these alignments include both neutrally evolving and functionally constrained portions of the sequence. This fraction of alignable sequence is significantly lower for the noneutherian mammals (14–34%), with the decrease mostly reflecting fewer alignments within nonannotated regions (i.e., those reflecting sequences not thought to be genes or repeats). A substantially larger amount of noneutherian sequence could be aligned to the human sequence by generating a true multisequence alignment with the program TBA (20) as opposed to simple pair-wise alignments (Fig. 2 A, purple bars). In the case of eutherian mammals (where no such difference is seen), it is thought that both pair-wise and multisequence alignments contain virtually all neutrally evolving sequence (5). However, with the noneutherian mammals, the dramatic difference likely reflects a larger amount of neutrally evolving sequence within the multisequence alignment; it remains to be determined whether this accounts for all neutrally evolving sequence.
Fig. 2.
Patterns of sequence conservation among different vertebrates. (A) The fraction of human sequence forming alignments with sequences from each of the indicated species is shown, broken down for four annotated categories. The additional alignable sequence (indicated in purple, see text for details) found exclusive to the TBA-generated multisequence alignment (20) falls largely within nonexonic regions. For data that include a larger set of vertebrates, see the supporting information. (B) The relationships between the fraction of human sequence aligned and estimated branch length from human (calculated as substitutions per site) are shown for the indicated vertebrate species.
We examined more closely the relationships among the human-alignable portions of each species' sequence, focusing our analyses on a 571-kb portion of the region with complete sequence coverage in a representative subset of species (see Materials and Methods). Although each marsupial sequence individually aligns with ≈34% of the human sequence, only 27% of the human sequence aligns with all three marsupial sequences, indicating that the human-alignable portions of each marsupial sequence are not completely overlapping. Similarly, whereas the platypus sequence aligns with ≈14% of the human sequence, only 11% of the human sequence aligns with all four noneutherian sequences, indicating that 21% of the human sequence that aligns with the platypus sequence is distinct from that aligning to all three marsupial sequences. These results demonstrate that the human-alignable sequence from more distantly related species is not fully contained within that from more closely related species. This finding also was observed with additional combinations of species (i.e., cat and mouse, but not cow; see the supporting information). These nonoverlapping alignable sequences may represent neutrally evolving, lineage-specific insertions and deletions.
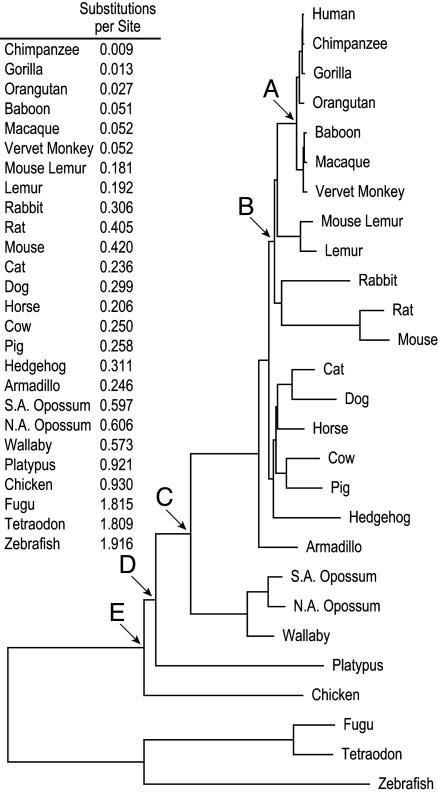
To better understand the phylogenetic relationships among the noneutherian mammals, as well as their relationship to other vertebrate species, we calculated the substitution rates at synonymous coding positions within the multisequence alignment. These rates then were used to scale the branch lengths of the phylogenetic tree depicted in Fig. 3; note that the total branch lengths between human and each species also are indicated (with all possible pair-wise branch lengths provided as supporting information). The synonymous substitution rate (per site) between the two opossum species is 0.09, whereas that between wallaby and either opossum species is 0.18. These rates are similar to those observed with primate–primate comparisons. Interestingly, platypus has a notably long branch length, with the platypus–marsupial substitution rate averaging 0.85. Also note that the human–platypus substitution rate is 55% higher (on average) than that for all human–marsupial pairs, providing further evidence for the considerable divergence of monotremes relative to both the marsupial and eutherian mammals (16). The synonymous substitution rates we calculated for the mouse and rat sequences are similar to the genome-wide estimates (5, 6), whereas that for the chicken sequence is substantially lower than the genome-wide estimate (29). The latter is likely attributable to differences in the methods and assumptions used and/or characteristics of the respective data sets (i.e., pair-wise whole-genome analyses vs. multisequence targeted analyses).
Fig. 3.
Phylogenetic tree of vertebrate species. By using the generated 27-species multisequence alignment, branch lengths were calculated based on analysis of synonymous coding positions. The branch lengths (as substitutions per synonymous site) between human and each species are listed (with additional pair-wise branch lengths provided in the supporting information). The last common ancestor among the catarrhine primates (A) is estimated at 25 mya (36, 37), between the rodents and primates (B) at 75 mya (5, 6), between eutherians and metatherians (C) at 185 mya (14), between monotremes and other therians (D) at 200 mya (14), and between mammals and birds (E) at 310 mya (13).
These findings reinforce the distinct phylogenetic positions of marsupials and monotremes within the vertebrate and mammalian radiations (12, 13). In addition, the simultaneous examination of alignment and branch length properties of each species' sequence compared to human (Fig. 2B) reveals a clear grouping of the marsupials at an intermediate position between the eutherian mammals and birds, consistent with the purported phylogenetic relationships. In contrast, the grouping of platypus and chicken in this analysis is surprising based on the significant evolutionary distance thought to separate these species (30, 31).
Presence of Evolutionarily Conserved Sequences in Different Lineages. The unique genomic properties of marsupials and monotremes make their sequences of particular interest for identifying and characterizing the small portion of the mammalian genome under purifying selection (5, 32, 33). We previously described an approach for using sequences from multiple vertebrates to detect evolutionarily conserved sequences in the human genome (called MCSs) and demonstrated that different species' sequences vary greatly in their relative contribution to the identification of MCSs (7–9).
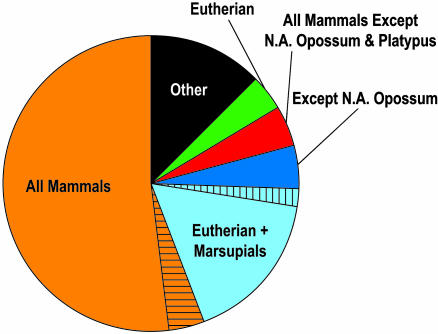
Given the diverse representation of mammalian species in our sequence data set, especially with the inclusion of metatherian and prototherian sequences, we next investigated the presence of MCSs among the different mammalian lineages. For this analysis, we studied a set of 418 MCSs falling within a 571-kb portion of the targeted genomic region where there was complete sequence coverage from cat and dog (carnivores), cow and pig (artiodactyls), rat and mouse (rodents), N.A. opossum and wallaby (marsupials), and platypus (monotreme). Note that S.A. opossum sequence was not included in this analysis, so that each lineage would be represented by two species (except monotremes, where only one species was available). The presence or absence of each of the 418 MCSs in each species' sequence was determined based on whether there was a human–species sequence alignment that overlapped that MCS in the human sequence (note that virtually all such alignments reflect high levels of sequence identity). Although virtually all 58 MCSs overlapping coding regions and 46 MCSs overlapping UTRs are present in all species, the remaining noncoding MCSs show interesting patterns of conservation (Fig. 4; also see the supporting information for additional details).
Fig. 4.
Lineage specificity of MCSs. The proportion of nonexonic MCSs found in the sequences of species in each category is indicated. Note that virtually all MCSs overlapping known exonic sequences are present in all mammals (data not shown). All Mammals: cat, dog, cow, pig, rat, mouse, N.A. opossum, wallaby, and platypus; Eutherian: cat, dog, cow, pig, rat, and mouse; Marsupials: N.A. opossum and wallaby; and Other: species combinations containing <2% of the analyzed MCSs (see the supporting information for the complete data set). Hashed areas of “All Mammals” reflect portions lacking one or both rodents, and hashed portions of “Eutherian + Marsupials” reflect portions lacking both rodents.
Just over one-half (52%) of the human-referenced noncoding MCSs are present in all nine nonhuman mammals analyzed. These regions thus represent the most anciently constrained sequences in the mammalian lineage. An additional 3.8% of the MCSs are present in all mammals except one or both rodents; this could be due to the known high deletion rate in the rodent lineage (5) or imprecision of current MCS-detection methods. An additional 17% of MCSs are present in all mammals except monotremes, with an additional 2% present in all mammals except monotremes and both rodents. The other major combinations are MCSs in all mammals except N.A. opossum (4.5%), in all mammals except N.A. opossum and platypus (4.5%), and in all eutherian mammals (4.0%). Together, these data provide evidence for lineage specificity with respect to the presence of evolutionarily conserved sequences in the human genome.
Discussion
Phylogenetic diversity is an important component of comparative genomic studies (8, 34). To date, the comparative sequencing of mammalian genomes largely has involved species within the eutherian radiation, each contributing relatively short branch lengths. Although short branch lengths allow for accurate sequence alignments, many species' sequences then are needed to identify those bases under purifying selection. The more diverged metatherian and prototherian mammals contribute longer branch lengths, making their sequences particularly valuable for identifying genomic regions under purifying selection, while still allowing for reliable alignments to the human sequence. The latter has been challenging with nonmammalian vertebrates, such as chicken and fish (W. Miller, personal communication).
Here, we report the large-scale generation and comparative studies of genome sequences from noneutherian mammals. This initial in-depth glimpse revealed several intriguing properties of these species' genomes. The platypus genome, which, at least for the region studied, shows: (i) ≈25% compression relative to the human genome; (ii) an unusually high G + C content for a mammal; (iii) a disproportionately high fraction of SINEs among its repetitive sequences; (iv) a notably low fraction of human-alignable sequence (14% compared with 34% for marsupials); and (v) a markedly long branch length revealed by phylogenetic analyses. Interestingly, these last two properties of platypus are quite similar to those of chicken (see Fig. 2B), despite the large difference in their evolutionary distances from human [estimated at 200 versus 310 mya, respectively (12–14)]. Although the long branch length for platypus is intriguing, it was calculated by using the reversible substitution model (REV), which assumes similar nucleotide composition among analyzed sequences. Because this is not the case for platypus (Table 2), and because the synonymous 4D sites analyzed in this study might not be entirely neutrally evolving, caution should be used in making strong claims about the phylogenetic position of monotremes based on our data. Finally, it is interesting to note that the observed compression of the platypus genome (relative to human) cannot be explained fully by differences in gene or repeat content. The evolutionary events that led to this relative compression are not obvious from the analyses performed here; however, more detailed examination of larger data sets of platypus sequence, with particular emphasis on cataloging repetitive versus nonrepetitive sequences and searching for evidence of insertions and deletions, should shed light on this issue.
It is interesting to note that we were able to align a greater amount of sequence by using a multisequence alignment tool [TBA (20)] compared to simpler pair-wise alignment methods. Importantly, this enhancement was most evident with the sequences from the noneutherian mammals, which showed roughly a 2-fold increase in the fraction of human-alignable sequence (purple portion of bars in Fig. 2 A). Similar improvements likely would enhance comparative sequence analyses involving more distantly related, nonmammalian vertebrates (e.g., birds, reptiles, and fish). At the same time, the observed increase in alignability in part reflected the large number of species' sequences being studied (a total of 27); the minimal number and phylogenetic characteristics of mammalian species required for such enhanced alignments remain to be established.
Analyses of the multisequence alignment revealed that the 14% of the human sequence that aligns with the platypus sequence is not completely contained within the larger fraction of the human sequence that aligns with all three marsupial sequences. Similar situations were encountered among the similarly diverged marsupials as well as other combinations of eutherians and nonmammals (see the supporting information). Although there is a general trend that alignments of more diverged sequences are contained within the alignments of more closely related sequences, significant exceptions emerge that may point to lineage-specific aspects of genome evolution.
Our studies confirm that sequences from noneutherian mammals will play an important role in identifying evolutionarily conserved regions of the human genome, which is important for establishing a comprehensive catalog of all functional genomic elements. Our previous work (8, 9) demonstrated that such highly conserved regions (MCSs) could be identified by comparative sequence analyses, but that sequences from large numbers of species (e.g., >12) are needed to maximize their detection. This requirement is particularly true for attaining high specificities in the detection of conserved noncoding sequences. Indeed, a problem with the currently available set of genome-wide mammalian sequences [e.g., mouse (5) and rat (6)] is the low specificity they provide in detecting functionally constrained sequences. As we show here, the alignment properties of marsupial and monotreme sequences make them particularly well suited for detecting and characterizing the most ancient conserved regions in mammalian genomes, reinforcing the notion that noneutherian mammals can be exploited in comparative genomic studies aiming to identify functional genomic elements. Of course, noneutherian sequences actually will be ineffective at identifying eutherian- or primate-specific genomic elements.
Data resulting from sequencing the S.A. opossum, wallaby, and platypus genomes (see www.intlgenome.org) should reveal the molecular basis for the unique genetic and physiologic features of noneutherian mammals, including their unusual anatomy and reproductive systems (12, 13, 35). At the same time, the additional data will augment the ever-growing list of vertebrate sequences that can be used for comparative analyses, paving the way toward reconstructing the evolutionary history of the mammalian genome.
Supplementary Material
Acknowledgments
We thank P. Green, W. Miller, N. Hansen, and D. Haussler for helpful discussions and critical review of the manuscript, A. Siepel for assistance with phylogenetic analyses, members of the NISC Comparative Sequencing Program (particularly B. Blakesley, G. Bouffard, J. McDowell, B. Maskeri, and N. Hanson), and R. Wing and J.-F. Cheng for BAC library construction. National Heart, Lung, and Blood Institute Grant HL66728 supported the construction of the N.A. opossum BAC library.
Author contributions: E.H.M. and E.D.G. designed research; E.H.M., N.C.S.P., V.V.B.M., and P.J.T. performed research; E.H.M., J.P.T., C.T.A., M.L., and E.D.G. contributed new reagents/analytic tools; E.H.M. and P.J.T. analyzed data; and E.H.M. and E.D.G. wrote the paper.
Abbreviations: NISC, National Institutes of Health Intramural Sequencing Center; mya, million years ago; N.A., North American; S.A., South American; BAC, bacterial artificial chromosome; TBA, threaded blockset aligner; MCS, multispecies conserved sequence; 4D, 4-fold degenerate; SINEs, short interspersed nucleotide elements; LINEs, long interspersed nucleotide elements.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AC127465, AC129065, AC129066, AC129885, AC142561, AC144364, AC144365, AC144600, AC144690, AC144691, AC144755, and AC144756 (N.A. opossum); AC147869, AC147870, AC147871, AC147872, AC147873, AC147874, AC148151, and AC148214 (S.A. opossum); AC127464, AC129882, AC129883, AC129884, AC130185, AC138553, AC144363, AC144689, AC144753, AC144754, AC144788, AC146535, and AC146754 (platypus); and AC145041, AC145042, AC145183, AC145184, AC145249, AC145250, AC145407, AC145408, AC145409, and AC145841 (wallaby)]. See Table 3, which is published as supporting information on the PNAS web site for specific versions of all GenBank accession nos. used in this study.
References
- 1.Nobrega, M. A. & Pennacchio, L. A. (2004) J. Physiol. 554, 31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper, G. M. & Sidow, A. (2003) Curr. Opin. Genet. Dev. 13, 604-610. [DOI] [PubMed] [Google Scholar]
- 3.Ureta-Vidal, A., Ettwiller, L. & Birney, E. (2003) Nat. Rev. Genet. 4, 251-262. [DOI] [PubMed] [Google Scholar]
- 4.Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.-M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. (2002) Science 297, 1301-1310. [DOI] [PubMed] [Google Scholar]
- 5.International Mouse Genome Sequencing Consortium (2002) Nature 420, 520-562. [DOI] [PubMed] [Google Scholar]
- 6.Rat Genome Sequencing Project Consortium (2004) Nature 428, 493-521.15057822 [Google Scholar]
- 7.Thomas, J. W., Touchman, J. W., Blakesley, R. W., Bouffard, G. G., Beckstrom-Sternberg, S. M., Margulies, E. H., Blanchette, M., Siepel, A. C., Thomas, P. J., McDowell, J. C., et al. (2003) Nature 424, 788-793. [DOI] [PubMed] [Google Scholar]
- 8.Margulies, E. H., NISC Comparative Sequencing Program & Green, E. D. (2004) Cold Spring Harbor Symp. Quant. Biol. 68, 255-263. [DOI] [PubMed] [Google Scholar]
- 9.Margulies, E. H., Blanchette, M., NISC Comparative Sequencing Program, Haussler, D. & Green, E. D. (2003) Genome Res. 13, 2507-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green, P., Ewing, B., Miller, W., Thomas, P. J., NISC Comparative Sequencing Program & Green, E. D. (2003) Nat. Genet. 33, 514-517. [DOI] [PubMed] [Google Scholar]
- 11.Hwang, D. G. & Green, P. (2004) Proc. Natl. Acad. Sci. USA 101, 13994-14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graves, J. A. M. & Westerman, M. (2002) Trends Genet. 18, 517-521. [DOI] [PubMed] [Google Scholar]
- 13.Grützner, F., Deakin, J., Rens, W., El-Mogharbel, N. & Graves, J. A. M. (2003) Comp. Biochem. Physiol. A Mol. Integr. Physiol. 136, 867-881. [DOI] [PubMed] [Google Scholar]
- 14.Woodburne, M. O., Rich, T. H. & Springer, M. S. (2003) Mol. Phylogenet. Evol. 28, 360-385. [DOI] [PubMed] [Google Scholar]
- 15.Janke, A., Magnell, O., Wieczorek, G., Westerman, M. & Arnason, U. (2002) J. Mol. Evol. 54, 71-80. [DOI] [PubMed] [Google Scholar]
- 16.Phillips, M. J. & Penny, D. (2003) Mol. Phylogenet. Evol. 28, 171-185. [DOI] [PubMed] [Google Scholar]
- 17.Killian, J. K., Buckley, T. R., Stewart, N., Munday, B. L. & Jirtle, R. L. (2001) Mamm. Genome 12, 513-517. [DOI] [PubMed] [Google Scholar]
- 18.Chapman, M. A., Charchar, F. J., Kinston, S., Bird, C. P., Grafham, D., Rogers, J., Grützner, F., Graves, J. A. M., Green, A. R. & Göttgens, B. (2003) Genomics 81, 249-259. [DOI] [PubMed] [Google Scholar]
- 19.Bao, Z. & Eddy, S. R. (2002) Genome Res. 12, 1269-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanchette, M., Kent, W. J., Riemer, C., Elnitski, L., Smit, A. F. A., Roskin, K. M., Baertsch, R., Rosenbloom, K., Clawson, H., Green, E. D., et al. (2004) Genome Res. 14, 708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siepel, A. & Haussler, D. (2004) Mol. Biol. Evol. 21, 468-488. [DOI] [PubMed] [Google Scholar]
- 22.Murphy, W. J., Eizirik, E., O'Brien, S. J., Madsen, O., Scally, M., Douady, C. J., Teeling, E., Ryder, O. A., Stanhope, M. J., de Jong, W. W., et al. (2001) Science 294, 2348-2351. [DOI] [PubMed] [Google Scholar]
- 23.International Human Genome Sequencing Consortium (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 24.Bick, Y. A. E. & Jackson, W. D. (1967) Nature 215, 192-193. [DOI] [PubMed] [Google Scholar]
- 25.Garagna, S. & Formenti, D. (1981) Bollettino di Zoologia 48, 255-261. [Google Scholar]
- 26.Crollius, H. R., Jaillon, O., Dasilva, C., Ozouf-Costaz, C., Fizames, C., Fischer, C., Bouneau, L., Billault, A., Quetier, F., Saurin, W., et al. (2000) Genome Res. 10, 939-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert, N. & Labuda, D. (1999) Proc. Natl. Acad. Sci. USA 96, 2869-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbert, N. & Labuda, D. (2000) J. Mol. Biol. 298, 365-377. [DOI] [PubMed] [Google Scholar]
- 29.International Chicken Genome Sequencing Consortium (2004) Nature 432, 695-716. [DOI] [PubMed] [Google Scholar]
- 30.Kumar, S. & Subramanian, S. (2002) Proc. Natl. Acad. Sci. USA 99, 803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar, S. & Hedges, S. B. (1998) Nature 392, 917-920. [DOI] [PubMed] [Google Scholar]
- 32.Elnitski, L., Hardison, R. C., Li, J., Yang, S., Kolbe, D., Eswara, P., O'Connor, M. J., Schwartz, S., Miller, W. & Chiaromonte, F. (2003) Genome Res. 13, 64-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolbe, D., Taylor, J., Elnitski, L., Eswara, P., Li, J., Miller, W., Hardison, R. & Chiaromonte, F. (2004) Genome Res. 14, 700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper, G. M., Brudno, M., Stone, E. A., Dubchak, I., Batzoglou, S. & Sidow, A. (2004) Genome Res. 14, 539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wakefield, M. J. & Graves, J. A. M. (2003) EMBO Rep. 4, 143-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodman, M., Porter, C. A., Czelusniak, J., Page, S. L., Schneider, H., Shoshani, J., Gunnell, G. & Groves, C. (1998) Mol. Phylogenet. Evol. 9, 585-598. [DOI] [PubMed] [Google Scholar]
- 37.Thomas, J. W. & Touchman, J. W. (2002) Trends Genet. 18, 104-108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.