In addition to high miR159-target gene complementarity, conserved RNA secondary structures abutting the miR159 binding sites are required for strong miR159-mediated silencing of MYB target genes in Arabidopsis.
Abstract
In plants, microRNA (miRNA)-target complementarity has long been considered the predominant factor determining the silencing outcome of the miRNA-target interaction, although the efficacy of such interactions have rarely been appraised in plants. Here, we perform in planta silencing efficacy assays on seven Arabidopsis MYB genes, all of which contain conserved miR159-binding sites of analogous complementarity. These genes were found to be differentially silenced by miR159; MYB81, MYB97, MYB101, MYB104, and DUO1 were all poorly silenced, whereas MYB33 and MYB65 were strongly silenced. Curiously, this is consistent with previous genetic analysis defining MYB33 and MYB65 as the major functional targets of miR159. Neither the free energy of miR159-target complementarity, nor miRNA binding site accessibility, as determined by flanking region AU content, could fully explain the discrepancy of miR159 silencing efficacy. Instead, we found that MYB33 and MYB65 were both predicted to contain a distinctive RNA secondary structure abutting the miR159 binding site. The structure is composed of two stem-loops (SLs) that are predicted to form in MYB33/65 homologs of species as evolutionary distant as gymnosperms. Functional analysis found that the RNA structure in MYB33 correlated with strong silencing efficacy; introducing mutations to disrupt either SL attenuated miR159 efficacy, while introducing complementary mutations to restore the SLs, but not the sequence, restored strong miR159-mediated silencing. Therefore, it appears that this RNA secondary structure demarcates MYB33/65 as sensitive targets of miR159, which underpins the narrow functional specificity of Arabidopsis miR159.
MicroRNAs (miRNAs) are small 20- to 24-nucleotide (nt) RNAs that guide the RNA-Induced Silencing Complex to target mRNAs and mediate their silencing through a combination of transcript degradation and translational repression (Axtell, 2013). In plants, miRNAs have been shown to be involved in a multitude of critical developmental events and stress responses, and are often referred to as master regulators of gene expression. Central to understanding miRNA function has been identifying their target mRNAs (Sun et al., 2014). In plants, it is clear that high sequence complementarity between a miRNA and its target mRNA is compulsory for a miRNA-target interaction (Mallory et al., 2004; Schwab et al., 2005; Addo-Quaye et al., 2008; German et al., 2008), with most experimentally validated miRNA-target pairs having very few mismatches (Schwab et al., 2005; Liu et al., 2014). Consequently, miRNA-target complementarity has been the cornerstone of plant miRNA biology, determining miRNA target prediction (Dai and Zhao, 2011), the design of artificial miRNAs (amiRNAs; Schwab et al., 2005), the design of artificial miRNA decoys such as target MIMICs (Todesco et al., 2010), or the identification of endogenous target MIMICs (Karakülah et al., 2016). However, bioinformatic prediction of target genes often fails to accurately predict functionally relevant targets, where from numerous predicted targets, only a select few appear functionally significant (for review, see Li et al., 2014a). Likewise, it has been reported that amiRNAs with high complementarity to their intended targets perform with considerable variability in plants (Li et al., 2013; Deveson et al., 2013). Finally, different miRNA decoys that contain identical miRNA binding sites work with widely varying efficacies (Reichel et al., 2015). These and other observations argue that miRNA-target interaction is not simply a product of complementarity, but additional factors are required for functional miRNA-target interactions (Wang et al., 2015).
In animals, it has long been known that the contextual sequence features in which a miRNA-binding site resides can strongly impact silencing. For example, miRNA-binding site accessibility was shown to be important, where introduction of mutations to decrease predicted accessibility disrupted efficient regulation, with impacts being as strong as mutations within the binding site itself (Kertesz et al., 2007). Furthermore, it has been shown that for certain animal miRNA-target interactions, strong regulation only occurs when the binding sites are within specific sequence arrangements or contexts (Didiano and Hobert, 2006; Vella et al., 2004). Other factors impacting the efficiency of regulation include RNA-binding proteins (RBPs), which can either attenuate or facilitate the access of the miRNA to its binding site (Kedde et al., 2010). Factors such as these have been studied less in plants, but evidence is accumulating that sequence context of miRNA binding sites may also be important in plants. First, Gu et al. (2012) found a synonymous codon bias favoring AU-richness, and hence reduced RNA secondary structure, around predicted miRNA target sites in several plant species. Second, Li et al. (2012) have found that miRNA binding sites in Arabidopsis (Arabidopsis thaliana) are generally less structured than their flanking regions, indicating a preference for high accessibility. Indeed, Fei et al. (2015) found that target site accessibility may explain select regulation of only a few targets from a large number of predicted target genes. Therefore, it would be of interest to functionally test these potential factors on miRNA-target interactions.
In plants, the Arabidopsis miR159 family has been extensively studied as a model for plant miRNA-mediated gene regulation (Palatnik et al., 2003, 2007; Allen et al., 2007, 2010). The family has two major isoforms, miR159a and miR159b, which are strongly expressed throughout Arabidopsis (Palatnik et al., 2007; Li et al., 2016). Such expression is consistent with a loss-of-function mir159ab double mutant that displays strong pleiotropic developmental defects. In Arabidopsis, miR159 is bioinformatically predicted to regulate more than 20 targets, including eight genes encoding conserved R2R3 domain MYB transcription factors (Palatnik et al., 2007). Despite this, genetic analysis revealed that miR159-mediated regulation of only two of the predicted target genes, MYB33 and MYB65, account for the developmental defects of mir159ab, as all defects are suppressed in a myb33.myb65.mir159ab quadruple mutant (Allen et al., 2007). This defined the functional specificity of Arabidopsis miR159 being restricted to MYB33 and MYB65, but also raised the question of the functional significance of miR159-mediated regulation of the additional bioinformatically predicted targets, including those that have a strongly conserved miR159 binding site (Allen et al., 2007, 2010). Curiously, this narrower functional specificity as defined by genetics has also been found in other plant and animal miRNA systems, suggesting the functional scope of miRNA-mediated silencing is narrower than generally assumed (Seitz, 2009; Li et al., 2014a).
For miR159, it is likely that multiple factors contribute to this apparent narrow functional specificity, including nonoverlapping transcriptional domains of the miR159 and MYB target genes (Allen et al., 2007), whether regulation of other targets is important under certain untested growth conditions or certain miR159 isoforms have become obsolete (Allen at al., 2010). However, one untested hypothesis is that MYB33 and MYB65 are more sensitive to miR159 regulation than the other MYB target genes. Recently, we have shown that factors beyond complementary govern the efficacy of the miR159-MYB33 silencing outcome (Li et al., 2014b). This not only included the miR159:MYB33 transcript stoichiometry, but also the sequence context of the miR159 binding site in MYB33. We showed that mutation of nts that immediately flank the miR159-binding site attenuated silencing to a similar extent to mutating nts within the binding site itself (Li et al., 2014b). This is further evidence that sequence complementarity alone does not guarantee strong miRNA regulation and that additional factor(s) are at play impacting miRNA-mediated regulation in plants.
Here, by carrying out in planta miR159 efficacy assays, we show that MYB33 and MYB65 are indeed much more sensitive to miR159 regulation than the other MYB genes with conserved miR159 binding sites. Neither flanking AU content nor predicted accessibility of the miR159 binding site underlie this sensitivity. Rather, MYB33 and MYB65 are shown to share a predicted RNA secondary structure consisting of two stem-loops that partially overlap with the miR159 binding site. Structure-function analysis demonstrates that both these stem-loops are required for efficient miR159 mediated silencing of MYB33. We hypothesize that having strong RNA stem-loops adjacent to a miRNA binding site may facilitate accessibility of the binding site to the miRNA, which in turn promotes efficient silencing of the target gene.
RESULTS
Conserved MYB Gene Family Members Have Different Sensitivities to miR159 Silencing
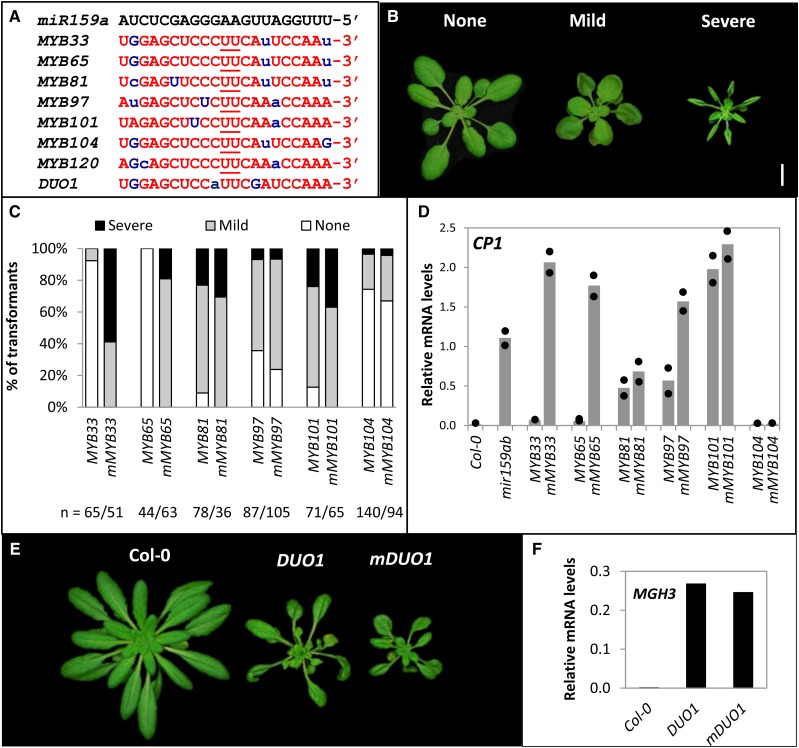
Despite bioinformatics predicting miR159 to target approximately 20 genes in Arabidopsis, including eight MYB genes that contain conserved, highly complementary miR159 binding sites, miR159 appears functionally specific for MYB33 and MYB65 (Allen et al., 2007; Fig. 1A). Although many possible factors likely underlie this apparent functional specificity (Allen et al., 2010), here we investigate whether MYB33 and MYB65 are more sensitive to miR159-mediated regulation compared to these other conserved MYB targets. To investigate this, we carried out in planta miR159-silencing efficacy assays. Here, each target was fused to the 35S promoter to constitutively transcribe each target individually in Arabidopsis. Multiple primary transformants were then obtained for each 35S-MYB transgene and their phenotype scored. If the MYB transcripts are efficiently silenced by miR159, no developmental abnormalities will be observed, as endogenous miR159 appears to be widely expressed throughout Arabidopsis (Palatnik et al., 2007; Allen et al., 2007; Li et al., 2016). Conversely, if the MYB transcripts are poorly silenced by miR159, constitutive MYB expression will result in developmental abnormalities (Fig. 1B), with their frequency and severity in the primary transformant populations acting as a measure of silencing. As these MYB genes differ in their amino acid sequence, scoring for each 35S-MYB (wild-type miRNA binding site) transgenic population was compared to a corresponding miR159-resistant 35S-mMYB (mutated miRNA binding site) transgenic population, with the difference in frequency and severity of phenotypic abnormalities acting as a measure of miR159 silencing efficacy.
Figure 1.
MYB target genes are differentially silenced by miR159. A, Sequence of miR159-binding sites in GAMYB-like and DUO1 transcripts. Nucleotides underlined represent the cleavage site, lower-case blue letters represent mismatches to miR159a, and G:U pairing is shown in uppercase blue letters. B, Different phenotypic categories based on 35S-GAMYB-like expression; none (indistinguishable from wild-type), mild (display of some leaf curl), and severe (two or more leaves showing the abaxial side from an aerial view). Scale bar represents 1 cm. C, Percentage of 35S:MYB and 35S:mMYB primary transformants showing None, Mild, and Severe phenotypes. n = the number of primary transformants. D, Transcript levels of the MYB-downstream gene CP1 in 35S:MYB and 35S:mMYB primary transformants measured by qRT-PCR. RNA was extracted from two independent biological pools of 30 to 50 randomly selected, 15-d-old primary transformants. Col-0 and mir159ab were used as controls, and mRNA levels were normalized to CYCLOPHILIN. The two measurements are shown as dots, with the bar representing the mean. E, Phenotypic characteristics of 40-d-old 35S:DUO1 and 35S:mDUO1 primary transformants. Scale bar represents 1 cm. F, Transcript levels of the DUO1-downstream gene MGH3 in 35S:DUO1 and 35S:mDUO1 primary transformants. Measurements are the average of three technical replicates.
35S-MYB and 35S-mMYB transgenes were generated for MYB33, MYB65, MYB81, MYB97, MYB101, MYB104, and DUO1, and then individually transformed into Arabidopsis. Multiple Arabidopsis primary transformants were selected for each construct, followed by phenotyping to assess developmental abnormalities. Phenotypic severities of 35S:MYB rosettes were placed into three broad categories; No Phenotypic (N) abnormalities, where the plant appears indistinguishable from wild-type; Mild (M), where the plant contains one or more moderately curled leaves; and Severe (S), where the plant has strongly curled leaves with the abaxial (bottom) side of two or more leaves visible from an aerial view (Fig. 1B). Multiple primary transformants (>35) were scored as to negate the impact of transgene position effects.
The majority of 35S-MYB33 and all of 35S-MYB65 primary transformants were phenotypically indistinguishable from wild type. By contrast, all 35S-mMYB33 and 35S-mMYB65 transformants resulted in developmental defects (Fig. 1C). Additionally, CP1, a marker gene of MYB protein activity (Alonso-Peral et al., 2010; Li et al., 2016), was measured in pooled primary transformant lines, as miR159-mediated silencing of MYB targets has a strong translational repression component, such that MYB mRNA levels do not accurately reflect silencing (Li et al., 2014a, 2014b). A strong difference in CP1 mRNA levels were found for the MYB33/mMYB33 and the MYB65/mMYB65 pairs (Fig. 1D). The stark difference in both developmental phenotypes and CP1 levels demonstrated that both MYB33 and MYB65 are strongly silenced by miR159.
For the other four GAMYB-like genes, MYB81, MYB97, MYB101, and MYB104, no such stark difference was seen for the MYB/mMYB pairs. First, expression of all four of these 35S-MYB genes could result in plants with severe phenotypic defects, resembling mMYB33 and mMYB65 transgenic lines or the mir159ab mutant (Supplemental Fig. S1). For 35S-MYB81, 35S-MYB97, and 35S-MYB101 primary transformants, the majority displayed phenotypic defects. For 35S-MYB104 primary transformants lines, the majority displayed no phenotypic defects; however, some 35S-MYB104 transformants displayed severe phenotypes (Fig. 1C). More importantly, for each MYB gene, the frequency and the severity of phenotypes in the 35S-MYB primary transformants populations were only slightly lower than the corresponding 35S-mMYB populations. Although this difference implies that MYB81, MYB97, MYB101, and MYB104 are still miR159-regulated, such small differences would argue that these genes are poorly silenced by miR159. Supporting this notion for MYB81, MYB97, and MYB101 are the CP1 levels, which are strongly up-regulated in the 35S-MYB populations compared to wild type (Col-0), and which have smaller differences between the MYB/mMYB pairs, relative to the MYB33/mMYB33 and MYB65/mMYB65 pairs (Fig. 1D). For MYB104, the low frequency of severe phenotypes in the 35S-mMYB populations and low CP1 levels indicate that this gene does not trigger this pathway as strongly as the other genes. However, again the lack of difference between the 35S-MYB104 and 35S-mMYB104 populations strongly argues that this gene is poorly silenced by miR159 (Fig. 1, C and D). In 35S-MYB81/97/101 transformants, MYB33 and MYB65 levels remained unchanged, confirming that the observed phenotypes were due to the poor miR159-mediated silencing of MYB81/97/101 (Supplemental Fig. S2).
Finally, DUO1 encodes an atypical R2R3 MYB transcription factor that does not belong in the GAMYB-like family clade, but nevertheless its miR159 binding site is highly conserved (Palatnik et al., 2007). All 35S-mDUO1 primary transformants (n = 55) exhibited phenotypic defects of reduced rosette size but with downward curled leaves (Fig. 1E), suggesting DUO1 activates different developmental pathways compared to the GAMYB-like family members. Interestingly, all 35S-DUO1 plants (n = 54) exhibited similar phenotypic defects to the 35S-mDUO1 plants and similar activation of a downstream gene, MGH3 (Fig. 1F). The lack of a large difference between the 35S-DUO1 and 35S-mDUO1 primary transformant populations again implies that DUO1 is also poorly silenced by miR159 relative to MYB33/MYB65.
In summary, the seven MYB genes that contain conserved miR159-binding sites appear to fall into two distinct categories regarding the efficacy of miR159 silencing; MYB33 and MYB65 appear to be very efficiently silenced, whereas MYB81, MYB97, MYB101, MYB104, and DUO1 appear poorly silenced. Given that the latter five genes have highly conserved miR159 binding sites that appear to be of equivalent complementarity to miR159 binding sites of MYB33/65, the difference in silencing efficacies seems surprising.
The ΔG Free Energy of a miRNA-target Interaction Is Not an Absolute Determinant of Silencing Efficacy
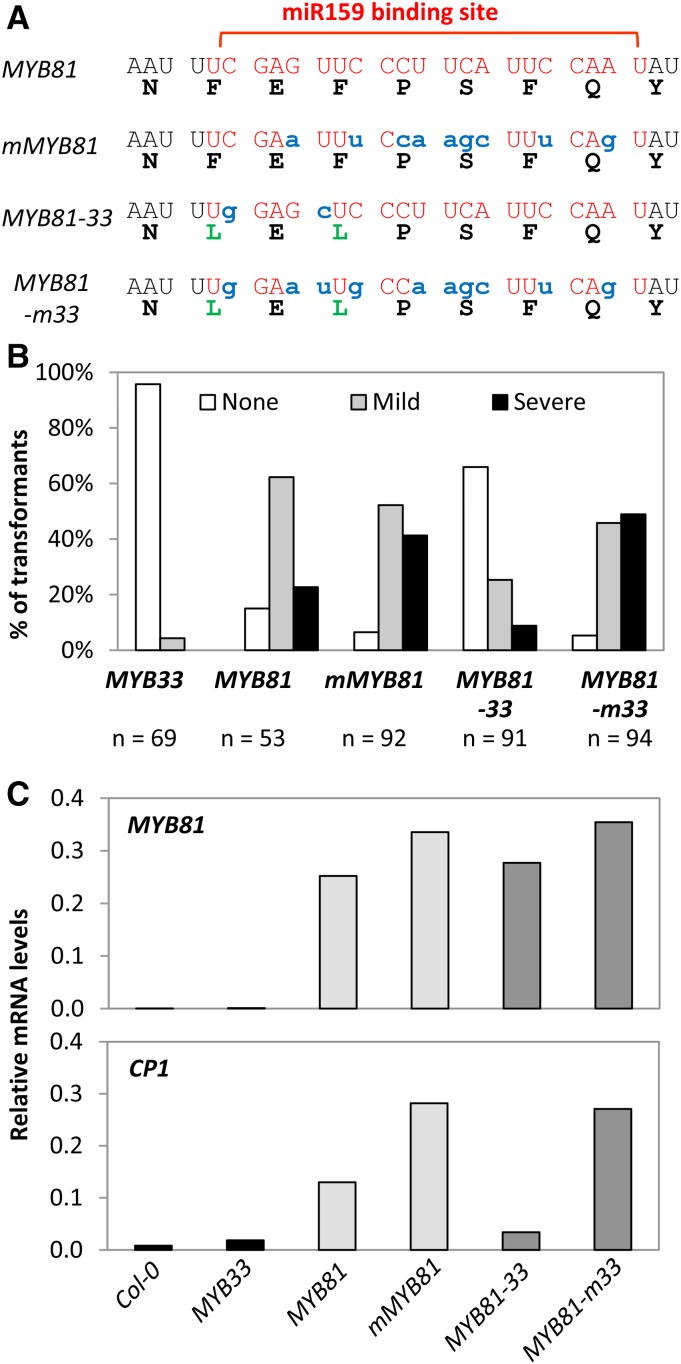
Possibly explaining the stronger silencing of MYB33/MYB65 is their stronger ΔG free energy interaction with miR159 compared to the other MYB genes (Supplemental Fig. S3), with the exception of MYB104, which has the strongest free energy, but is poorly silenced. To investigate this, we mutated the miR159 binding site of MYB81 to make it identical to the miR159 binding site of MYB33, and investigated whether this modified 35S-MYB81 transgene (MYB81-33) was now as strongly silenced as MYB33. As the protein sequence of MYB81-33 differs from MYB81, a miR159-resistant version (MYB81-m33) that was identical in amino acid sequence to MYB81-33 was used as a control to ensure these amino acid changes did not impact protein activity (Fig. 2A).
Figure 2.
Lowering ΔG free energy of a miRNA-target interaction can improve silencing efficacy. A, Nucleotide and amino acid changes introduced to generate mMYB81, MYB81-33, and MYB81-m33 compared to MYB81. The miR159 binding sequences are marked in red. Amino acid changes are marked in green, and nucleotide changes in blue. B, The number of primary transformants of MYB33, MYB81, mMYB81, MYB81-33, and MYB81-m33 falling into each phenotypic category (none, mild, or severe), as a percentage of the total number of transformants generated for each construct (n). C, Transcript levels of MYB81 and CP1 in primary transformants of MYB33, MYB81, mMYB81, MYB81-33, and MYB81-m33 measured by qRT-PCR. RNA was extracted from pools of rosette tissue of 10 to 15 randomly selected, 3- to 4-week-old primary transformants. Col-0 was used as wild-type control and mRNA levels were normalized to CYCLOPHILIN. Measurements are the average of three technical replicates.
The incorporation of the miR159 binding site from MYB33 into MYB81 improved silencing, as greater than 60% of MYB81-33 primary transformants had no phenotype as compared to less than 20% of MYB81 primary transformants (Fig. 2B). As the proportion of phenotypic categories was highly similar between MYB81-m33 and mMYB81 (Fig. 2B), this difference between MYB81-33 and MYB81 cannot be attributed to an altered protein activity. However, silencing of MYB81-33 was still not as strong as for MYB33, as a much greater proportion of MYB81-33 plants exhibited mild and severe defects when compared to MYB33 transformants (Fig. 2B).
To support this data, we performed transcript profiling on the MYB81 transgenes and the downstream marker gene of MYB protein activity, CP1 (Alonso-Peral et al., 2010). Consistent with miR159 having a strong translational repression component of silencing its MYB targets (Li et al., 2014a, 2014b), MYB81 transcript levels do not reflect the differences in phenotypic severities of the different transgenic lines (Figs. 2C and S3B). By contrast, CP1 levels tightly correlated with phenotypic severity, being highest in the lines carrying miR159-resistant versions (mMYB81 and MYB81-m33), followed by MYB81, and lowest in MYB81-33 (Figs. 2C and S3B). Therefore, consistent with the phenotypic severities, the high MYB81 transcript levels in MYB81-33 plant are being strongly repressed, as indicated by CP1 levels.
Finally, MYB33 transcript levels in MYB81, mMYB81, MYB81-33, and MYB81-m33 transformants remained unchanged, confirming that the observed phenotypes were due to miR159-mediated silencing of the respective MYB81 transgenes (Supplemental Fig. S3). Therefore, although incorporation of the MYB33 miR159 binding site into MYB81 has improved silencing, both morphological and molecular data indicated that MYB81-33 is still not being as strongly silenced as MYB33, implying there are factors other than complementarity impacting silencing.
Flanking AU Content Does Not Correlate with Efficacy of miR159-mediated Silencing of MYB33
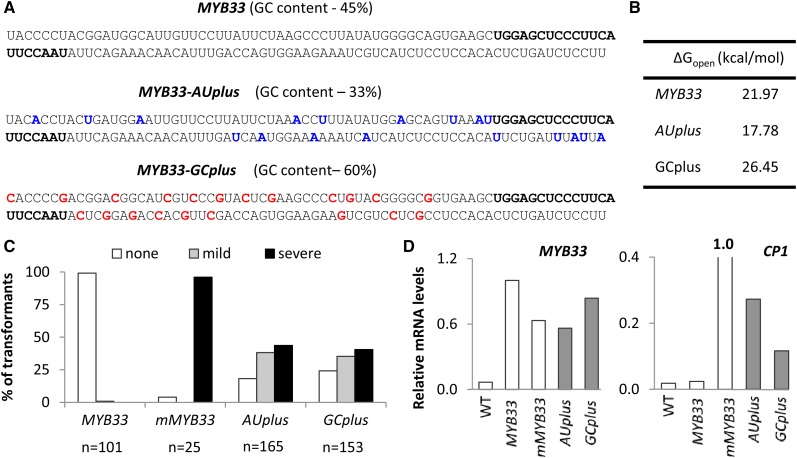
One possible factor impacting silencing is target site accessibility. Gu et al. (2012) previously established a link between the AU-richness of a target site’s local region and its predicted accessibility. Based on this, we generated variants of a MYB33 genomic clone with either elevated AU content (MYB33-AUplus) or elevated GC content (MYB33-GCplus) within the vicinity (approximately 50 nts both 5′ and 3′) of the miR159 target site, changing only bases in wobble positions so that the protein sequence remains unaltered (Fig. 3A). To predict the accessibility of the miR159 binding site within these MYB33 variants, the theoretical energy cost of unpairing all nucleotides in a 51-nucleotide window comprising each miRNA-binding site plus 17-nucleotides upstream and 13-nucleotides downstream (∆Gopen) was used, because the predicted accessibility of this window was shown to correlate most faithfully with animal miRNA performances (Kertesz et al., 2007) and is used by the psRNATarget server (Dai and Zhao, 2011). As anticipated, ∆Gopen values for the miR159 target site in these constructs were decreased for MYB33-AUplus (17.78 kcal/mol) and increased for MYB33-GCplus (26.45 kcal/mol) relative to wild-type MYB33 (21.97 kcal/mol; Fig. 3B). Each construct was transformed into wild-type Arabidopsis, as was the MYB33 transgene and the miR159-resistant version, mMYB33 as controls. As done previously, the efficacy of miR159-mediated silencing was measured by scoring the frequency and severity of developmental abnormalities in primary transformants (Fig. 1B).
Figure 3.
Changes to AU content perturb miR159 silencing of MYB33. A, MYB33 transgenes with elevated AU content (MYB33-AUplus) and elevated GC content (MYB33-GCplus) in the vicinity of the miR159 target site (bold) were generated, changing only nucleotides occupying wobble positions. B, As a measure of their accessibility, ∆Gopen values were calculated for the miR159 target site in each construct using the Vienna RNAup web server. These values are a prediction of the energy required to unpair all nucleotides in a 51-nucleotide window comprising the miR159 binding site plus 17 nts 5′ and 13 nts 3′. C, The number of 24-d-old primary transformants of AUplus and GCplus falling into each phenotypic category (none, mild, or severe), as a percentage of the total number of transformants generated for each construct (n). Primary transformants of an endogenous MYB33 transgene (MYB33) or a miR159-resitant MYB33 transgene (mMYB33) were grown in parallel as controls. D, MYB33 and CP1 transcript levels, normalized to CYCLOPHILIN mRNA abundance, were measured in total RNA samples derived from 30 to 50 randomly selected 8-d-old transformants for each construct. Data are the averaged of three technical replicates.
Consistent with previous experiments (Fig. 1C; Allen et al., 2007), the vast majority of MYB33 primary transformants were indistinguishable from wild type, whereas the majority of mMYB33 primary transformants had severe developmental abnormalities (Fig. 3C). For both the MYB33-AUplus and MYB33-GCplus constructs, the majority of primary transformants exhibited mild or severe developmental abnormalities, indicating poor silencing of MYB33. Surprisingly, there was little difference between the two constructs in terms of the frequency and severity of phenotypes retrieved, with a slightly higher proportion of affected individuals obtained for MYB33-AUplus (Fig. 3C). Although MYB33 transcript levels were elevated in all transgenic lines, CP1 levels were significantly higher in both MYB33-AUplus and MYB33-GCplus compared to MYB33 plants (Fig. 3D). However, compared to mMYB33 plants, both the phenotypic scoring and CP1 levels indicate that both MYB33-AUplus and MYB33-GCplus are still being miR159 regulated, albeit poorly compared to MYB33. In all cases, MYB65 levels were unchanged, confirming that the observed deregulation of miR159-mediated silencing was specific to MYB33 (Supplemental Fig. S4).
That the mutation of nucleotides flanking the miR159 target site leads to attenuated silencing in both the MYB33-AUplus and MYB33-GCplus constructs further reiterates the importance of target site context for miRNA efficacy. However, this attenuation occurred regardless of whether G/C or A/U composition was increased or decreased. As such, it suggests that a particular sequence of nucleotides is required for strong silencing of MYB33, rather than a general increase of A/U nts around the miR159 binding site.
The miR159 Binding Site of MYB33 and MYB65 Abuts a Strongly Predicted RNA Secondary Structure
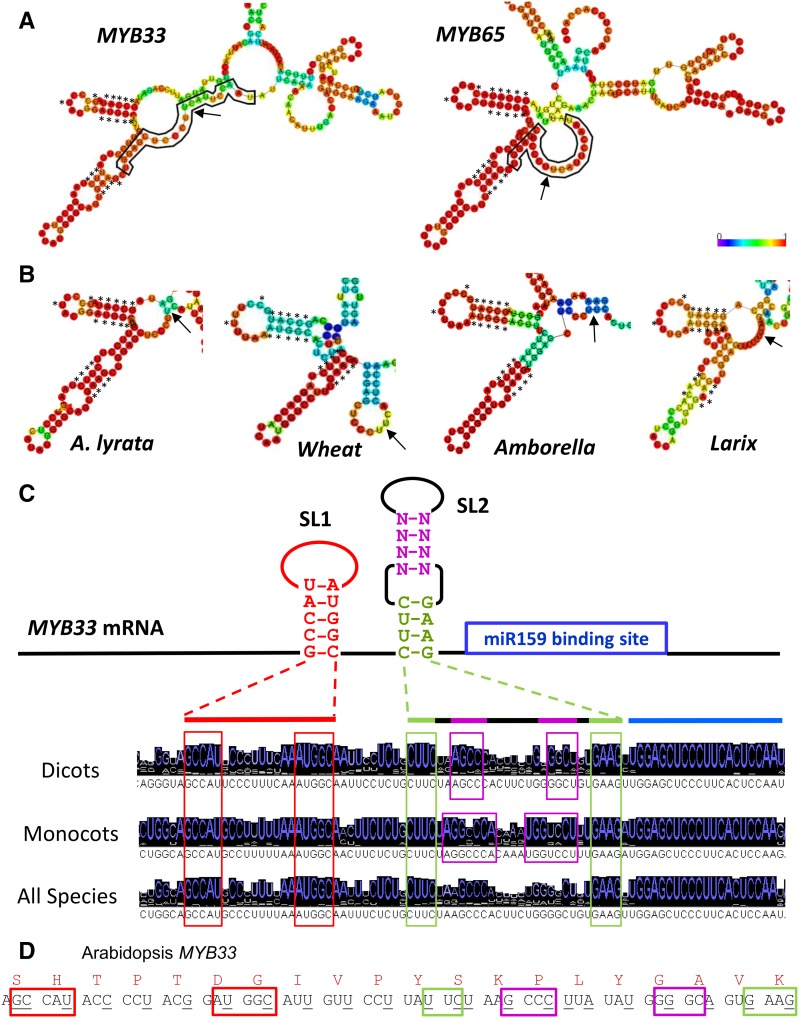
Another factor that is known to influence miRNA regulation in animals, but has not been given much attention to in plants, is RNA secondary structure. Comparison of the predicted RNA secondary structure of MYB33 and MYB65 using the Vienna RNA fold server revealed that these genes have highly similar RNA structures (Fig. 4A). Of particular interest are two stem-loop structures immediately upstream of the miR159 binding site (termed “SL1” and “SL2”). These structures, which have high base-pairing probabilities, are predicted to be present in MYB homologs of closely related species, such as Arabidopsis lyrata, and in that of more distantly related species such as wheat (Triticum aestivum), the basal angiosperm Amborella and the gymnosperm Larix (Fig. 4B). To investigate this further, nucleotide sequence alignments were performed on MYB33 homologs from dicotyledonous and monocotyledonous species separately, and then all species combined (Supplemental Fig. S5). From these alignments, consensus sequences were determined for the region of the MYB33 homologs that are predicted to form the stem-loop regions (Figs. 4C and S5). The nucleotides forming the stems of these structures were found to be the most strongly conserved nts in this region, and among the most highly conserved nucleotides in the entire gene (Supplemental Fig. S5D), with the sequences forming the stem of SL1 and the sequences forming the base of SL2 appearing very strongly conserved (Figs. 4C and S5). However, the consensus sequence of the upper part of SL2 varies between MYB33 homologs from dicotyledonous and monocotyledonous species of plants, but in both instances, the sequences are still able to form stems, arguing that RNA secondary structure is being selected for in this region (Fig. 4C). Supporting this, of the 25 conserved nts predicted to form stems in this predicted Arabidopsis MYB33 RNA structure, 12 nts correspond to synonymous codon position regarding the amino acid sequence of the MYB33 protein (Fig. 4D). Together, this argues that selection of these conserved nts is occurring mainly at the RNA secondary structure level, not at the protein level. Given their proximity to the miR159 binding site, it possibly suggests that these predicted structures have functional significance regarding miR159-mediated silencing. By contrast, SL1 and SL2 are not predicted to form in the other Arabidopsis GAMYB-like genes and DUO1 (Supplemental Fig. S6), suggesting their presence may be associated with strong silencing.
Figure 4.
MYB33/65 is predicted to contain highly similar RNA stem-loop structures abutting the miR159 binding site that appear strongly conserved. A, MYB33 and MYB65 transcripts contain two highly similar predicted stem-loop structures (SL1 and SL2) as determined by the Vienna RNAfold web server using a 221-bp window of the coding regions (the miR159 binding site plus 100 bp immediately 5′ and 3′). The miR159 binding sites are outlined in black and the cleavage sites are indicated with arrows. Conserved nucleotides are labeled with asterisks. B, RNA secondary structures similar to SL1 and SL2 are also predicted to form in MYB33 homologs of Arabidopsis lyrata, wheat (Triticum aestivum), the basal angiosperm, Amborella trichopoda, and the gymnosperm, Larix kaempferi (Lamb.). The miR159 cleavage sites are indicated with arrows. Conserved nucleotides are marked with asterisks. The heat maps indicate the probability of second structure formation, from low (purple) to high (red). C, MYB33/MYB65 homologs from a diverse range of species contain conserved nucleotides corresponding to the predicted stem regions. Consensus sequences of MYB33 homologs from dicots, monocots, and all species combined, as indicated by alignments and histograms generated using Jalview (Waterhouse et al., 2009). D, Many of the conserved nucleotide positions correspond to both synonymous and nonsynonymous nucleotides, implying conservation is not selected for at the protein level. Boxed nucleotides correspond to conserved stem positions as shown in C, and underlined nucleotides correspond to synonymous positions.
The Predicted RNA Secondary Structure of the MYB33 mRNA Correlates with Strong miR159-mediated Silencing
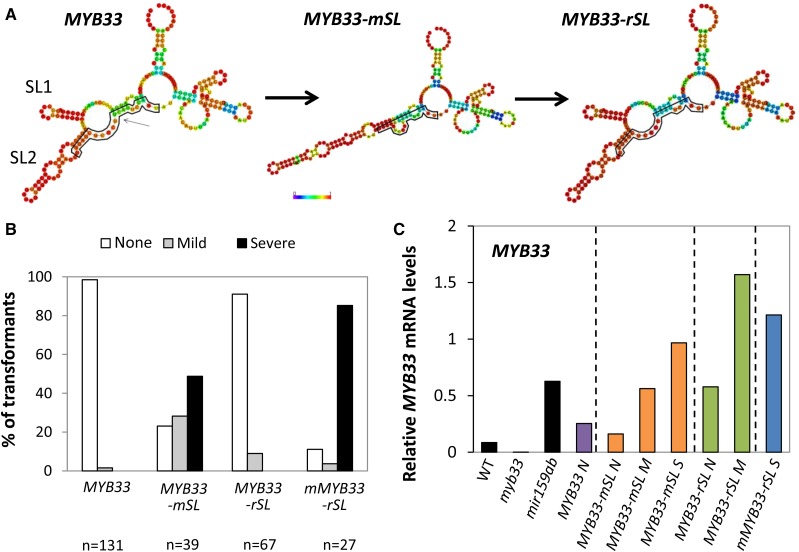
To test whether these predicted SL1 and SL2 structures have an impact on miR159-mediated silencing, mutations disrupting these structures, but not changing the amino acid sequence or the miR159 binding site, were introduced into a MYB33 genomic clone (MYB33-mSL). Moreover, another construct containing mutations to restore SL1 and SL2 (MYB33-rSL) was generated (Fig. 5A). This construct, even though containing predicted SL1 and SL2 structures identical to MYB33, has a different nucleotide and amino acid sequence (Supplemental Fig. S7). To ascertain that these changes are not affecting MYB33 protein activity, a control construct with a miR159-resistant binding site (mMYB33-rSL) was generated (Supplemental Fig. S7). These constructs, as well as a wild-type MYB33 positive control, were individually transformed into myb33 plants and multiple primary transformants were selected and classified according to their phenotypic defects shown as defined above [None (N), Mild (M), and Severe (S)]. Consistent with previous results, the vast majority (98%) of MYB33 plants did not show any phenotypic abnormalities, demonstrating that MYB33 is strongly silenced by miR159 (Fig. 5, B and C). However, the majority of MYB33-mSL plants showed either mild (28%) or severe (49%) phenotypes (Fig. 5, B and C), indicating that miR159 regulation of this construct has been strongly disrupted. In contrast, 91% (61/67) of MYB33-rSL plants displayed a wild-type phenotype, suggesting that restoring SL1 and SL2 leads to the restoration of strong silencing. Analysis of mMYB33-rSL plants revealed that 85% (23/27) of plants had severe phenotypes, confirming that the low level of phenotypic defects in MYB33-rSL plants is not due to an inactive protein. Therefore, the prediction of SL1 and SL2 formation in MYB33 correlates with its strong silencing by miR159.
Figure 5.
Predicted stem-loop structures correlate with strong miR159-mediated silencing of MYB33. A, Synonymous mutations were introduced into MYB33 to disrupt the stem-loop structures (SL1 and SL2) resulting in MYB33-mSL. Complimentary mutations were then introduced to restore SL1 and SL2, generating MYB33-rSL. RNA secondary structures were predicted using the Vienna RNAfold web server. The heat maps indicate the probability of second structure formation, from low (purple) to high (red). The miR159 binding site is outlined in black. B, Number of primary transformants of MYB33, MYB33-mSL, MYB33-rSL, and mMYB33-rSL falling into each phenotypic category, as a percentage of the total number of transformants analyzed for each construct (n). C, qRT-PCR analysis of MYB33 mRNA levels in plants expressing MYB33-mSL, MYB33-rSL, and mMYB33-rSL. Wild-type (Col-0), myb33, mir159ab, and MYB33 plants were used as controls. All measurements are relative to CYCLOPHILIN. RNA was extracted from four-week-old primary transformants using tissue from whole plants. Measurements are the average of three technical replicates.
Next, MYB33 mRNA levels were measured in the different phenotypic categories of MYB33, MYB33-mSL, and MYB33-rSL transgenic plants. Levels of MYB33 transcript will dependent on at least two factors: first, the strength of transcription of the transgene; second, the strength of miR159-mediated silencing, which includes a transcript cleavage/degradation mechanism, as well as a translational repression mechanism (Li et al., 2014b). Consistent with the transcript cleavage mechanism, mMYB33-rSL plants with a severe phenotype have high levels of miR159-resistant mMYB33-rSL transcript. However, MYB33-rSL plants with a mild phenotype have an even higher level of MYB33 transcript, and much higher than the corresponding levels in MYB33-mSL plants (Fig. 5D). This supports the notion that the MYB33-rSL transcript is being much more strongly miR159-regulated than the MYB33-mSL transcript, as much higher transcript levels are required to result in the same phenotypic outcome. Together, these data demonstrate that the potential SL1 and SL2 structures promote strong silencing of MYB33.
Mutations of Either Putative SL1 and SL2 Structure Attenuates Strong MYB33 Silencing
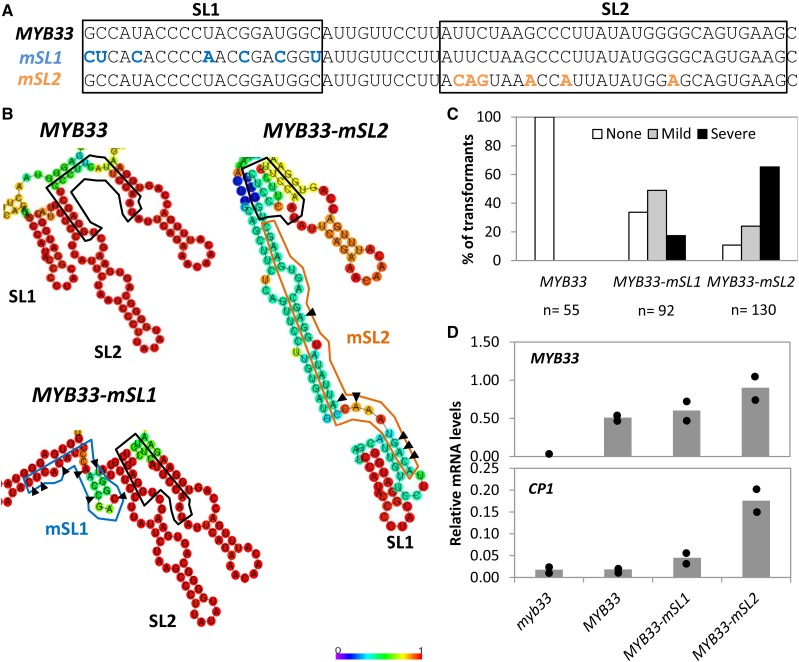
To investigate which of the conserved nts are required for strong silencing, the nts corresponding to SL1 or SL2 were synonymously mutated individually. For SL1, this corresponded to seven nts in a region that was 40 nts to 60 nts upstream of the miR159 binding site, and the construct was termed MYB33-mSL1 (Fig. 6A). The mutations for SL2 corresponded to six nts that were more proximal, being 11 nts to 29 nts upstream of the miR159 binding site, and the construct was termed MYB33-mSL2 (Fig. 6A). Both MYB33-mSL1 and MYB33-mSL2 encoded a MYB33 protein identical to wild-type MYB33. Additionally, the mutations were predicted to result in a RNA secondary structure that does not disrupt the nonmutated SL (Fig. 6B). Constructs were transformed into Arabidopsis and transformants planted out alongside MYB33 controls.
Figure 6.
Mutation of nucleotides corresponding to SL1 or SL2 attenuate miR159-mediated silencing of MYB33. A, Synonymous mutations were introduced into the MYB33 gene to generate MYB33-mSL1 (blue) and MYB33-mSL2 (orange). The SL1 and SL2 sequences are indicated by black boxes. B, Predicted RNA secondary structure of MYB33, MYB33-mSL1, and MYB33-mSL2. Structures were predicted via the Vienna RNAfold web server. The heat map indicates the probability of second structure formation, from low (purple) to high (red). The miR159 binding site is outlined in black. The altered mSL1 and mSL2 structure are outlined in blue and orange, respectively. The mutated nucleotides in mSL1 and mSL2 as shown in Figure 6A, are indicated with arrows. C, Number of primary transformants falling into each phenotypic category, as a percentage of the total number of transformants analyzed for each construct (n). D, qRT-PCR measurement of MYB33 and CP1 mRNA levels in primary transformants of MYB33, MYB33-mSL1, and MYB33-mSL2. myb33 plants were used as a control. All measurements are relative to CYCLOPHILIN. RNA was extracted from two independent biological replicates, each being composed of rosette tissue sampled from 30 to 50 primary transformants. The two measurements are shown as dots, with the bar representing the mean.
For MYB33-mSL1, despite the distal nature of the mutations to the miR159 binding site, these mutations were able to perturb miR159 regulation. Compared to MYB33 primary transformants that displayed no phenotypic defects, greater than 60% of MYB33-mSL1 primary transformants displayed mild or severe phenotypic defects (Fig. 6C). For the more proximal mutations in SL2, a more severe perturbation of miR159 regulation was observed, where approximately 90% of transformants displayed mild or severe phenotypes (Fig. 6C). Consistent with this, both MYB33 and CP1 transcript levels were higher in MYB33-mSL2 than in MYB33-mSL1 plants (Fig. 6D). Together, these morphological and molecular data argue that both SL1 and SL2 are required for strong miR159-regulation of MYB33.
An Artificial Predicted RNA Stem-Loop Abutting the miR159 Binding Site Promotes Silencing
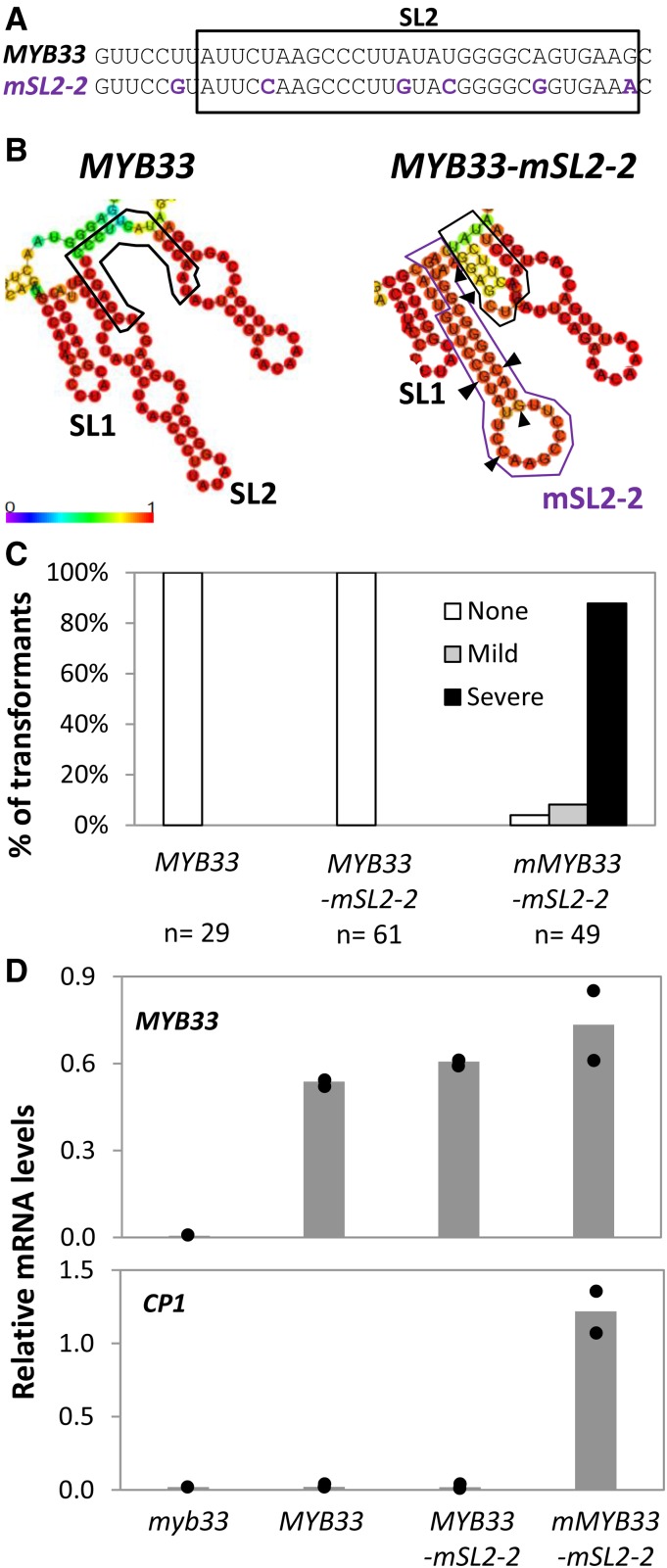
The above experiments raise the question as to what is the function of these sequences in promoting silencing. We speculate that if nts that are adjacent to a miRNA-binding site form a stem structure, these nts are then unavailable to base-pair with nts of the miRNA-binding site, thus making the miRNA-binding site highly accessible for the miRNA. In an attempt to gain some insight into this possibility, we made an additional SL2 mutant in which six nts were changed, but an alternative long stem-loop was predicted to be formed, which we called MYB33-mSL2-2 (Fig. 7, A and B). Interestingly, despite the nt changes that alter the predicted RNA secondary structure, this transgene was found to be as strongly silenced as wild-type, as no phenotypic defects were seen in greater than 100 primary transformants (Fig. 7C). Even though all six nt changes resulted in synonymous amino acid substitutions, a miR159-resistant version was made (mMYB33-SL2-2; Fig. 7C) to ensure a functional protein is being made. As most mMYB33-SL2-2 transformants had a severe phenotype, the lack of phenotypes seen in MYB33-SL2-2 transformants is due to strong miR159-silencing. Supporting this strong silencing of MYB33-SL2-2, even though high levels of its transcript accumulated, it appears to be totally silenced as CP1 mRNA levels remained low in MYB33-SL2-2 plants (Fig. 7D).
Figure 7.
An artificial stem-loop structure replacing mSL2 in MYB33 promotes silencing. A, Synonymous mutations were introduced into the MYB33 gene to generate MYB33-mSL2-2 (purple). B, Predicted RNA secondary structure of MYB33 and MYB33-mSL2-2. Structures were predicted via the Vienna RNAfold web server. The heat map indicates the probability of second structure formation, from low (purple) to high (red). The miR159 binding site is outlined in black. The altered mSL2-2 structure is outlined in purple. The mutated nucleotides in mSL2-2 as shown in Figure 7A, are indicated with arrows. C, Number of primary transformants falling into each phenotypic category, as a percentage of the total number of transformants analyzed for each construct (n). D, qRT-PCR measurement of MYB33 and CP1 mRNA levels in primary transformants of MYB33, MYB33-mSL2-2, and mMYB33-mSL2-2. myb33 plants were used as controls. All measurements are relative to CYCLOPHILIN. RNA was extracted from two independent biological replicates, each being composed of rosette tissue sampled from 10 to 15 primary transformants. The two measurements are shown as dots, with the bar representing the mean.
DISCUSSION
Here, we have assessed the efficacy of miR159-mediated silencing in planta against multiple MYB target genes in Arabidopsis that contain conserved miR159 binding sites. Contrary to the possibility that miR159 regulates conserved MYB targets with equivalent efficacies, we show that there are dramatic differences in silencing. Although slight differences in miR159-binding sites can in part explain these differential efficacies, a predicted conserved RNA secondary structure, consisting of two stem-loops that is associated with the miR159-binding site, was shown to confer efficient silencing of MYB33. This provides the rationale of the why the nts flanking the miR159-binding site of MYB33 are important in miRNA-mediated regulation (Li et al., 2014b), as the GAAG motif just upstream of the miR159 binding site was mutated, and this sequence composes the base of SL2 (Fig. 4). Moreover, this study represents, to our knowledge, the first evidence that RNA secondary structure can strongly impact miRNA-mediated silencing in plants. Such a factor beyond complementary likely contributes to the narrow functional specificity of miR159 (Allen et al., 2007).
Differential miR159-mediated Silencing of MYB Family Members
Many plant miRNAs are predicted to target multiple paralogous members of gene families (Jones-Rhoades, 2012), including that of multiple GAMYB-like family members in many different species (Tsuji et al., 2006; Yang et al., 2014). However, how silencing varies among family members has not been extensively studied. Here, we have performed in planta efficacy assays on the GAMYB-like family members, as well as another MYB gene with a conserved miR159-binding site, DUO1. Despite all these target genes containing miR159-binding sites that satisfy the widely accepted empirical parameters required for strong regulation (Schwab et al., 2005; Liu et al., 2014), we found that the strength of silencing varied dramatically, with MYB33 and MYB65 being much more sensitive than the other MYB genes. As miR159-guided cleavage products have been detected for MYB81, MYB97, MYB101, MYB104, and DUO1 (Palatnik et al., 2007; Alves et al.; 2009; Allen et al., 2010), these are experimentally validated targets of miR159. However, given that only subtle differences were observed between the wild-type and miR159-resistant transgenes for all of these MYB targets, our in planta assays argue that these genes are weakly regulated by miR159, supporting genetic experiments that they are not major functional targets. Given that all these MYB targets have retained strong complementarity to miR159, the variability in silencing must be considered surprising.
Several studies have previously assessed the variation in silencing of related target genes. First, quantitative transient assays in Nicotiana benthamiana to assess silencing efficacy of multiple family members of targets genes found that efficacy can vary dramatically among different family members, where subtle differences in complementarity can dramatically change efficacy (Liu et al., 2014). However, in this system the miRNA binding sites were taken out of their endogenous sequence context and placed in the ORF or UTR of luciferase; therefore, what impact sequence context is having on these targets will be lost. In another study, different Medicago miRNAs were overexpressed in Arabidopsis, with each miRNA potentially able to target a large number of NB-LRR family members (Fei et al., 2015). However, all were found to only target a surprisingly few NB-LRR genes (< 4%) that differences in complementarity (or target score) could not explain (Fei et al., 2015). Such an observation adds further weight that factors in addition to complementarity have a large impact on a miRNA-target interaction.
AU Content and Predicted Accessibility Does Not Correlate with Silencing Efficacy
Previously, a synonymous codon bias favoring AU-richness (and hence reduced secondary structure) around predicted miRNA target sites has been identified in several plant species (Gu et al., 2012). Indeed, an enrichment of single-stranded RNA at predicted miRNA target sites was found in Arabidopsis, indicating a preference for high accessibility (Li et al., 2012). However, increasing the AU content and predicted target accessibility around the miR159-binding site of MYB33 attenuated silencing to a similar extent as increasing the GC content. This argues that a specific nucleotide sequence flanking the miR159 binding site is important for regulation, not just the AU content per se. Previously, no correlation between amiRNA efficacy and predicted target site accessibility was observed (Li et al., 2013; Deveson et al., 2013), arguing that existing evidence is insufficient to support the notion that this parameter is a reliable predictor of miRNA efficacy in plants. Supporting this, the mainstream plant miRNA target prediction tool (psRNAtarget; http://plantgrn.noble.org/psRNATarget/) incorporates target accessibility into its algorithm, which then ranks miR159a targets (MYB101, DUO1, MYB104, MYB33, and MYB97 lowest) and miR159b targets (MYB101, DUO1, MYB104, MYB97, MYB120, and MYB33 lowest) in an order not consistent with our in planta miRNA silencing efficacy assays. So, although programs such as this can predict the functional targets of miRNAs where high complementarity appears a prerequisite to strong silencing, the ranking of the targets, with regard to silencing efficacy, may have little meaning.
A Conserved RNA Secondary Structure Element Promotes Silencing Of MYB33
We present two lines of evidence for the existence of a predicted RNA secondary structure that is important for miR159 efficacy. First, experimental, where we have carried out a structure/function analysis in which strong miR159-regulation correlates with the predicted formation of the RNA secondary structure. Secondly, bioinformatic, where these structures, or variations thereof, are strongly predicted to reside in MYB33 homologs of numerous angiosperm and gymnosperm plant species (Fig. 4, B and C), arguing that these structures have been integral in the miR159-MYB regulatory relationship over a long period of time. Interestingly, even though SL1 is more distal to the miR159 binding site, it appears more conserved than SL2, which varies in its consensus sequence between monocot and dicot species at the top of the predicted stem (Fig. 4C). However, the both consensus sequences are still predicted to form stems (5′-AGCC and 5′-GGCU for dicots and 5′-AGGCCCA and 5′-UGGUCCU for monocots; Fig. 4C). This variation again argues that a RNA secondary structure is being strongly selected-for immediately adjacent to the miR159 binding site. Currently, no other miRNA binding site has been associated with a potential RNA structure, and very little is known about the role of RNA secondary structure in controlling plant gene expression.
Consistent with the conservation of nts corresponding to the stems of the RNA structures, we found that disrupting either predicted SL1 or SL2 disrupted efficient silencing of MYB33. Curiously, although more distal, the nts corresponding to SL1 appear more tightly conserved than the nts of the stem of SL2 (Fig. 4). The impact of the SL1 mutations must be considered surprising: seven synonymous mutations 40 nts to 60 nts upstream of the binding site would not be predicted to impact miRNA silencing with any accessibility program; for instance, the widely used target site accessibility program of Kertesz et al. (2007) assesses 17 nts and 13 nts upstream and downstream of the binding site, respectively. Our result alone would suggest that a miRNA target site (binding site plus flanking nts) can be much larger than previously considered, and again highlights that current bioinformatic accessibility prediction programs likely need refinement to generate more accurate predictions.
Possible Function of the Predicted RNA Structural Element of MYB33
We have not determined how these predicted structures are promoting silencing. One obvious possibility is that the stem-loop structures in the vicinity of the miR159 binding site maintain binding site accessibility for the miRNA, where if adjacent sequences are forming strong stem structures, they are less likely to base-pair with binding site nts, thereby keeping the binding site accessible. Supporting such a notion is the MYB33-mSL2-2 construct, which is predicted to form a different SL2, but appears as strongly silenced as the wild-type MYB33 gene. This would argue that the presence of a stem per se is important, rather than the precise structure of the stem. However, this does not dismiss the possibility that RBPs interact with the stem-loop structures that potentially promote silencing. In humans, the RBP Pumilio1 binds to the p27 mRNA, which introduces a local change in p27 RNA structure and thereby enables efficient binding of miR-221 and miR-222 leading to p27 silencing (Kedde et al., 2010). Whether such regulatory processes occur for miR159-mediated regulation of MYB33 will require further investigation, but it is interesting to note that the efficacy of miR159-mediated regulation of MYB33 appears tissue dependent (Alonso-Peral et al., 2012), raising the possibility that an RBP-RNA interaction may mediate such differential regulation. Also uncertain is whether engineering these RNA secondary structures into the poorly regulated MYB genes would improve their silencing or if these structures only promote silencing in the MYB33/65 context. If the former, the use of such structures may possibly have biotechnological applications, where engineering their presence into a miRNA-binding site may increase the sensitivity of the target to miRNA-mediated gene silencing.
MiRNA Target Recognition Is Still Poorly Understood
Presently, studies that have investigated the efficacy of miRNA-target interaction have focused solely on complementarity (Schwab et al., 2005; Iwakawa and Tomari, 2013; Liu et al., 2014). However, this article reinforces that miRNA target recognition is a complex process involving multiple factors, which are far from fully appreciated. Consistent with strong sequence complementarity being a prerequisite for miRNA recognition, we have shown that replacing the miR159 binding site in MYB81 with that of MYB33, which increases the free energy when bound to miR159, results in more efficient regulation. However, this change alone does not render MYB81 as strongly silenced as MYB33, where it appears that both strong free energy of miR159 binding and the stem-loop structures is required for strong silencing. Therefore, our study adds to the evidence that factors beyond complementarity, including target site structure, play a vital role in miRNA regulation. Just as miRNA-target complementarity or miRNA abundance can impact silencing efficacy and directly affect major plant traits (Todesco et al., 2012; Houston et al., 2013), our data would predict that sequence context likely has such impacts.
How common such potential RNA secondary structures are in controlling plant miRNA-mediated gene regulation is yet to be determined. One possible constraint on such structures arising is that most canonical miRNA-binding sites reside in coding regions, where the amino acid sequence may limit the possibility of such structures occurring. Thereby, RNA secondary structures may be more likely to arise for miRNA-binding sites that reside in noncoding regions, or regions where the amino acid sequence conservation is not critical. Nevertheless, it is becoming clearer that the flanking sequences of miRNA binding sites can strongly impact miRNA-mediated silencing. Although algorithms used to predict targets and design amiRNAs take accessibility into account, it is clear that such analysis is rudimentary and will need much refinement to enable the development of more accurate miRNA target prediction programs.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0) was used in all experiments and is referred to as wild type. The myb33 mutant is in ecotype Col-6 with a glaborous1 background mutation leading to a no-trichome phenotype (Millar and Gubler, 2005). The mir159ab mutant is in a Col-0 background and represents a T-DNA insertion loss-of-function mutant, which was described in Allen et al. (2007).
After harvesting, seeds were dried and sterilized using a vapor-phase method, where seeds were placed into a desiccator jar and exposed to chlorine gas, which was prepared by mixing 100 mL commercial bleach with 3 mL concentrated HCl, for 3 to 4 h. Sterilized seeds were either sown on soil (Debco Plugger soil mixed with Osmocote Extra Mini fertilizer at 3.5 g/L), or on agar plates containing Murashige & Skoog Basal Medium (2.2 g/L, pH 5.8), and stratified for 48 h at 4°C in the dark. Plants were grown in 22°C growth cabinets under long day conditions (16-h light/8-h dark, 150 μmol/m2/s).
Generation of 35S:MYB and 35S:mMYB Constructs
MYB gene sequences were amplified with gene-specific primers that contained attB1 and attB2 recombination sites to allow integration into a Gateway donor vector. PCR amplification was performed using high-fidelity KOD Hot Start DNA Polymerase (Novagen) with the following cycling conditions: 1 cycle of 95°C/2 min, 35 cycles of 95°C/20 s, 55°C/10 s, 70°C for 20 s/kb extension time according to amplicon size, and 1 cycle of 70°C for 10 min. PCR products were analyzed by agarose gel electrophoresis and products of the desired sizes were excised from the gel and purified using the Wizard SV Gel and PCR Clean-Up System (Promega). The BP reactions were performed with Gateway BP Clonase II enzyme mix (Invitrogen), to integrate the purified PCR products into the pDONOR/Zeo vector (Invitrogen) to generate pENTRY-MYB vectors. Entry vectors were screened by restriction enzyme digestion and verified by sequencing.
The pENTRY-mMYB, MYB81-33 and MYB81-m33 constructs were obtained by performing site-directed mutagenesis of the miR159 binding site on the confirmed pENTRY-MYB vectors. A mutagenesis approach based on Liu and Naismith (2008) was used, where each pair of forward and reverse primers contained nonoverlapping sequences at the 3′ end and complementary sequences at the 5′ end to minimize primer dimerization and enable primers to use the PCR products as the template. The nonoverlapping sequences were larger than the complementary sequences and had a 5°C to 10°C higher Tm. PCR reactions were then performed using KOD Hot Start DNA Polymerase (Novagen) with 50 ng of plasmid template, 100 ng of forward primer, and 100 ng of reverse primer, at the cycling conditions recommended by the manufacturer. The PCR products were analyzed by agarose gel electrophoresis and digested with 2 μL DpnI enzyme at 37°C for 3 to 4 h. After digestion, the PCR products were purified using the Wizard SV Gel and PCR Clean-Up System (Promega) and transformed into Escherichia coli Alpha-Select Gold Efficiency competent cells (Bioline), and clones were confirmed by restriction-enzyme digestion and sequencing.
All entry clones were recombined into the Gateway destination vector pGWB602Ω (Nakamura et al., 2010) via LR reactions using Gateway LR Clonase II enzyme mix (Invitrogen), to generate the corresponding binary vectors expressing 35S:MYB. Binary vectors were verified by restriction-enzyme digestion.
Generation of MYB33-AU/GCplus and (m)MYB33-m/rSL Constructs
To generate MYB33-AU/GCplus and MYB33-m/rSL, a 787-bp genomic fragment of MYB33 containing the respective mutations was synthesized by Integrated DNA Technologies. Synthesized fragments were sequenced to verify their integrity and then cloned into a 4356-bp MYB33 genomic fragment in the pDONR/Zeo vector (Li et al., 2014b) using the restriction enzymes BspEI and StuI. For the construction of MYB33-mSL1, MYB33-mSL2, and MYB33-mSL2-2, site-directed mutagenesis with primers containing the respective mutations was performed on confirmed MYB33 entry vectors (MYB33 genomic fragment in pDONR/Zeo). For mMYB33-rSL and mMYB33 mSL2-2, site-directed mutagenesis using primers that contain mutations in the miR159 binding site was performed on confirmed MYB33-rSL and MYB33-mSL2-2 entry vectors, respectively.
Entry vectors were confirmed by restriction-enzyme digestion and sequencing and recombined into the destination vector pMDC123 (Curtis and Grossniklaus, 2003) through Gateway LR reaction. Binary vectors were verified through restriction-enzyme digest.
Transformation of Arabidopsis
All binary vectors were transformed into the Agrobacterium tumefaciens strain GV3101 by electroporation (Hellens et al., 2000), and the integrity of the vectors was confirmed by restriction-enzyme digestion on the extracted plasmids. Using the floral dip method (Clough and Bent, 1998), 35S:MYB and 35S:mMYB constructs were transformed into Col-0, whereas MYB33-AU/GCplus, MYB33 m/rSL, mMYB33 rSL, MYB33-mSL1, MYB33-mSL2-1, MYB33-mSL2-2, and mMYB33-mSL2-2 constructs were transformed into myb33.
Seeds were harvested after about 6 weeks and sterilized as described above. Transformants were selected by growing seeds on agar plates containing Murashige & Skoog Basal Medium and antibiotics for selection. After 7 to 10 d of growth, transformants were identified and transplanted onto soil.
Transcript Analysis
TRIzol (Invitrogen) was used for RNA extraction of tissues from plants at different growth stages. The extraction procedure was carried out as per manufacturer’s instructions except for the following modifications: (1) approximately 500 mg of plant material was used with 1 mL of TRIzol reagent for each extraction, (2) homogenization of tissues was carried out using a mortar and pestle, (3) the chloroform extraction step was repeated once, and (4) precipitation of RNA was carried out overnight at −20°C to maximize the recovery of RNAs. The quality and integrity of purified RNA was then examined by agarose gel electrophoresis. RQ1 RNase-Free DNase (Promega) was used to treat RNA samples for qRT-PCR. A quantity of 20 μg to 100 μg of total RNA was treated for each sample following the protocol provided, with the addition of RNaseOut Recombinant RNase Inhibitor (Invitrogen) at a concentration of 1 μL/10 μg RNA. Treated RNA was then purified using the RNeasy Plant Mini Kit (QIAGEN) following the manufacturer’s instructions and the integrity of RNA was examined on a 1% agarose gel. cDNA synthesis was carried out using SuperScript III Reverse Transcriptase (Invitrogen) and an oligo(dT) primer according to the manufacturer’s protocol. For each sample, 500 ng to 5 μg of total RNA was used. The 20 μL reaction was then diluted 50 times in nuclease-free distilled water and used for subsequent qRT-PCR. For qRT-PCR, 10 μL of SensiFAST SYBR No-Rox mix (Bioline) with 0.8 μL of forward and reverse primers at 10 μm each was added to 9.2 μL of cDNA. All qRT-PCR reactions (for both reference and genes of interests) were carried out on a Rotor-Gene Q real-time PCR machine (QIAGEN) in triplicate, under the following cycling conditions: 1 cycle of 95°C/5 min, 45 cycles of 95°C/15 s, 60°C/15 s, and 72°C/20 s of fluorescence was acquired at the 72°C step. A 55°C to 99°C melting cycle was then carried out. CYCLOPHILIN5 (At2g29960) was used to normalize mRNA levels using the comparative quantitation program in the Rotor-Gene Q software package provided by QIAGEN. The value for each gene represents the average of triplicate assays.
Accession Numbers
Sequence information of the genes studies in this article can be found at The Arabidopsis Information Resource database using the following accession numbers; MYB33 (At5g06100), MYB65 (At3g11440), MYB101 (At2g32460), MYB104 (At2g26950), MYB97 (At4g26930), MYB81 (At2g26960), DUO1 (At3g60460).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression of all GAMYB-like genes in rosettes result in similar phenotypic defects.
Supplemental Figure S2. MYB33 and MYB65 transcript levels are unaltered in MYB81, MYB97, and MYB101 plants.
Supplemental Figure S3. ΔG free energy of miR159-MYB target interactions and MYB81, CP1, and MYB33 transcript levels in various MYB81 transgenic lines.
Supplemental Figure S4. MYB65 transcript levels remain unaltered in MYB33-AUplus and MYB33-GCplus transgenic lines.
Supplemental Figure S5. Sequences corresponding to the stems of SL1 and SL2 are strongly conserved among MYB33 homologs of diverse plant species.
Supplemental Figure S6. Predicted RNA secondary structures of MYB genes.
Supplemental Figure S7. The generation of the MYB33-mSL, MYB33-rSL, and mMYB33-rSL constructs and their analysis in transgenic plants.
Footnotes
This work was supported by a Research School of Biology International student Ph.D. scholarship to Z.Z., an International ANU Ph.D. scholarship to M.R., and an Australian Research Council Discovery Grant (No. DP110103493) to A.A.M.
Articles can be viewed without a subscription.
References
- Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18: 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves L Jr., Niemeier S, Hauenschild A, Rehmsmeier M, Merkle T (2009) Comprehensive prediction of novel microRNA targets in Arabidopsis thaliana. Nucleic Acids Res 37: 4010–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RS, Li J, Alonso-Peral MM, White RG, Gubler F, Millar AA (2010) MicroR159 regulation of most conserved targets in Arabidopsis has negligible phenotypic effects. Silence 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RS, Li J, Stahle MI, Dubroué A, Gubler F, Millar AA (2007) Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc Natl Acad Sci USA 104: 16371–16376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Peral MM, Li J, Li Y, Allen RS, Schnippenkoetter W, Ohms S, White RG, Millar AA (2010) The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol 154: 757–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Peral MM, Sun C, Millar AA (2012) MicroRNA159 can act as a switch or tuning microRNA independently of its abundance in Arabidopsis. PLoS One 7: e34751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64: 137–159 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39: W155−W159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveson I, Li J, Millar AA (2013) MicroRNAs with analogous target complementarities perform with highly variable efficacies in Arabidopsis. FEBS Lett 587: 3703–3708 [DOI] [PubMed] [Google Scholar]
- Didiano D, Hobert O (2006) Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol 13: 849–851 [DOI] [PubMed] [Google Scholar]
- Fei Q, Li P, Teng C, Meyers BC (2015) Secondary siRNAs from Medicago NB-LRRs modulated via miRNA-target interactions and their abundances. Plant J 83: 451–465 [DOI] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, De Paoli E, Lu C, et al. (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26: 941–946 [DOI] [PubMed] [Google Scholar]
- Gu W, Wang X, Zhai C, Xie X, Zhou T (2012) Selection on synonymous sites for increased accessibility around miRNA binding sites in plants. Mol Biol Evol 29: 3037–3044 [DOI] [PubMed] [Google Scholar]
- Hellens R, Mullineaux P, Klee H (2000) Technical Focus: a guide to Agrobacterium binary Ti vectors. Trends Plant Sci 5: 446–451 [DOI] [PubMed] [Google Scholar]
- Houston K, McKim SM, Comadran J, Bonar N, Druka I, Uzrek N, Cirillo E, Guzy-Wrobelska J, Collins NC, Halpin C, Hansson M, Dockter C, et al. (2013) Variation in the interaction between alleles of HvAPETALA2 and microRNA172 determines the density of grains on the barley inflorescence. Proc Natl Acad Sci USA 110: 16675–16680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa HO, Tomari Y (2013) Molecular insights into microRNA-mediated translational repression in plants. Mol Cell 52: 591–601 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW. (2012) Conservation and divergence in plant microRNAs. Plant Mol Biol 80: 3–16 [DOI] [PubMed] [Google Scholar]
- Karakülah G, Yücebilgili Kurtoğlu K, Unver T (2016) PeTMbase: a database of plant endogenous target mimics (eTMs). PLoS One 11: e0167698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R (2010) A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol 12: 1014–1020 [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284 [DOI] [PubMed] [Google Scholar]
- Li F, Zheng Q, Vandivier LE, Willmann MR, Chen Y, Gregory BD (2012) Regulatory impact of RNA secondary structure across the Arabidopsis transcriptome. Plant Cell 24: 4346–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Reichel M, Li Y, Millar AA (2014a) The functional scope of plant microRNA-mediated silencing. Trends Plant Sci 19: 750–756 [DOI] [PubMed] [Google Scholar]
- Li J, Reichel M, Millar AA (2014b) Determinants beyond both complementarity and cleavage govern microR159 efficacy in Arabidopsis. PLoS Genet 10: e1004232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-F, Chung HS, Niu Y, Bush J, McCormack M, Sheen J (2013) Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25: 1507–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Alonso-Peral M, Wong G, Wang M-B, Millar AA (2016) Ubiquitous miR159 repression of MYB33/65 in Arabidopsis rosettes is robust and is not perturbed by a wide range of stresses. BMC Plant Biol 16: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Naismith JH (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 8: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang F, Axtell MJ (2014) Analysis of complementarity requirements for plant microRNA targeting using a Nicotiana benthamiana quantitative transient assay. Plant Cell 26: 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J 23: 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Gubler F (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17: 705–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Mano S, Tanaka Y, Ohnishi M, Nakamori C, Araki M, Niwa T, Nishimura M, Kaminaka H, Nakagawa T, Sato Y, Ishiguro S (2010) Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci Biotechnol Biochem 74: 1315–1319 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R, Warthmann N, Allen E, Dezulian T, Huson D, Carrington JC, Weigel D (2007) Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell 13: 115–125 [DOI] [PubMed] [Google Scholar]
- Reichel M, Li Y, Li J, Millar AA (2015) Inhibiting plant microRNA activity: molecular SPONGEs, target MIMICs and STTMs all display variable efficacies against target microRNAs. Plant Biotechnol J 13: 915–926 [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Seitz H. (2009) Redefining microRNA targets. Curr Biol 19: 870–873 [DOI] [PubMed] [Google Scholar]
- Sun X, Zhang Y, Zhu X, Korir NK, Tao R, Wang C, Fang J (2014) Advances in identification and validation of plant microRNAs and their target genes. Physiol Plant 152: 203–218 [DOI] [PubMed] [Google Scholar]
- Todesco M, Balasubramanian S, Cao J, Ott F, Sureshkumar S, Schneeberger K, Meyer RC, Altmann T, Weigel D (2012) Natural variation in biogenesis efficiency of individual Arabidopsis thaliana microRNAs. Curr Biol 22: 166–170 [DOI] [PubMed] [Google Scholar]
- Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D (2010) A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Aya K, Ueguchi-Tanaka M, Shimada Y, Nakazono M, Watanabe R, Nishizawa NK, Gomi K, Shimada A, Kitano H, Ashikari M, Matsuoka M (2006) GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J 47: 427–444 [DOI] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ (2004) The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3'UTR. Genes Dev 18: 132–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Polydore S, Axtell MJ (2015) More than meets the eye? Factors that affect target selection by plant miRNAs and heterochromatic siRNAs. Curr Opin Plant Biol 27: 118–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang N, Mi X, Wu L, Ma R, Zhu X, Yao L, Jin X, Si H, Wang D (2014) Identification of miR159s and their target genes and expression analysis under drought stress in potato. Comput Biol Chem 53PB: 204–213 [DOI] [PubMed] [Google Scholar]