Mutational analysis of SACPD-C reveals an impact of stearic acid accumulation in leaf and nodule structure and morphology.
Abstract
Stearoyl-acyl carrier protein desaturase (SACPD-C) has been reported to control the accumulation of seed stearic acid; however, no study has previously reported its involvement in leaf stearic acid content and impact on leaf structure and morphology. A subset of an ethyl methanesulfonate mutagenized population of soybean (Glycine max) ‘Forrest’ was screened to identify mutants within the GmSACPD-C gene. Using a forward genetics approach, one nonsense and four missense Gmsacpd-c mutants were identified to have high levels of seed, nodule, and leaf stearic acid content. Homology modeling and in silico analysis of the GmSACPD-C enzyme revealed that most of these mutations were localized near or at conserved residues essential for diiron ion coordination. Soybeans carrying Gmsacpd-c mutations at conserved residues showed the highest stearic acid content, and these mutations were found to have deleterious effects on nodule development and function. Interestingly, mutations at nonconserved residues show an increase in stearic acid content yet retain healthy nodules. Thus, random mutagenesis and mutational analysis allows for the achievement of high seed stearic acid content with no associated negative agronomic characteristics. Additionally, expression analysis demonstrates that nodule leghemoglobin transcripts were significantly more abundant in soybeans with deleterious mutations at conserved residues of GmSACPD-C. Finally, we report that Gmsacpd-c mutations cause an increase in leaf stearic acid content and an alteration of leaf structure and morphology in addition to differences in nitrogen-fixing nodule structure.
Soybean (Glycine max) is the most widely consumed legume crop in the world, providing 56% of the world’s oilseed production. Soybean oil can be found in food products such as margarine, salad dressings, and cooking oils as well as in industrial products such as plastics and biodiesel fuel. There are five major fatty acids in soybean oil: two saturated fatty acids, palmitic acid (16:0) and stearic acid (18:0), and three unsaturated fatty acids, oleic acid (18:1), linoleic acid (18:2), and linolenic acid (18:3).
Oxidatively stable oil with a high melting temperature is necessary for solid fat application. Highly saturated soybean seed oil would be suitable for this end use (Clemente and Cahoon, 2009). Palmitic acid not only improves the oxidative stability of soybean oil but also can be used to produce trans-fat-free shortenings, margarines, and cosmetic products. However, this saturated short-chain fatty acid is undesirable for nutrition because its consumption results in an unfavorable lipoprotein profile in blood serum (Mensink and Katan, 1990). Stearic acid does not exhibit these cholesterolemic effects on human health (Kris-Etherton et al., 1997; Kris-Etherton and Yu, 1997). Additionally, stearic acid is less likely to be incorporated into cholesterol esters and has a neutral effect on the concentration of blood serum low-density lipoprotein cholesterol (Byfield et al., 2006). Soybean oils are typically subjected to hydrogenation to increase stearic acid content. When partially hydrogenated, parts of the soybean unsaturated fatty acids are converted into unhealthy trans-fats (Clemente and Cahoon, 2009). Consumers are increasingly avoiding products that contain trans-fats, and governments have begun to ban trans-fats in food products. Because of its high melting temperature and oxidative stability, soybean oil with a high stearic acid content may be useful in reducing the need for chemical hydrogenation in the production of trans-fat-free margarines and shortenings (List et al., 1997; Kok et al., 1999; Knowlton, 2001). Genetic manipulation of stearic acid is more efficient in reducing trans-fats introduced by the hydrogenation process (Clemente and Cahoon, 2009). Therefore, the development of soybean germplasm with higher seed stearic acid (greater than 20%) and oleic acid (greater than 50%) and lower linolenic acid (less than 3%) content is a main objective of the edible oil industry (Ruddle et al., 2013, 2014).
Stearic acid content in soybean seeds is typically between 2% and 5% of the total fatty acids. The enzyme stearoyl-acyl carrier protein desaturase (SACPD-C) is responsible for the conversion of stearic acid to oleic acid in the developing soybean seed. However, no study has previously reported SACPD-C involvement in leaf stearic acid content and impact on leaf structure and morphology. Spontaneously occurring mutants of GmSACPD-C, like FAM94-41 (9% stearic acid), have been found to have a high seed stearic acid phenotype (Zhang et al., 2008). Mutagenesis has been used in some soybean cultivars to reduce the catalytic activity of GmSACPD-C and accumulate stearic acid within the seed (Ruddle et al., 2013; Carrero-Colón et al., 2014). In this study, soybean lines with high seed stearic acid content were isolated using a forward genetic screen, and the correlation of GmSACPD-C mutations with an alteration of seed stearic acid content was analyzed. Consequently, five Gmsacpd-c mutants were identified by target sequencing with high levels of seed stearic acid, all of which will be used as new high stearic acid soybean germplasm. It has been reported that high stearic acid mutants carrying deletions on GmSACPD-C resulted in nodule cell senescence and the formation of necrotic cavities (Gillman et al., 2014). Interestingly, in this study, homology modeling and mutational analysis revealed that mutations within nonconserved residues around the iron-binding pocket resulted in relatively high stearic acid content but no cell senescence and necrotic cavities in nodules. Finally, the impact of stearic acid accumulation due to GmSACPD-C mutations has been identified to affect leaf structure and morphology.
RESULTS
Forward Genetic Screening Identifies High Seed Stearic Acid Content Mutants
The distribution of the seed stearic acid of the M3 ethyl methanesulfonate (EMS) cv Forrest mutagenized population ranges from 0.41% to 11% (Supplemental Fig. S1). Traditionally, the average seed stearic acid content of the wild-type cv Forrest, PI 88788, Essex, and Williams 82 range between 2.4% and 4.1% (Table I). Interestingly, in the developed cv Forrest mutagenized M3 population, 19 lines presented seed stearic acid content that was twice that of the wild type. However, among those 19 M3 lines, only five lines (F605, F620, F714, F813, and F869) retained the high stearic acid phenotype in two successive generations (Table I). The average seed stearic acid content in the wild-type cv Forrest was 3.25%, whereas the five M3 mutants F605, F620, F714, F813, and F869 presented 5.88%, 6.06%, 6.49%, 6.9%, and 6.52% content, respectively (Supplemental Table S1). Interestingly, the seed stearic acid content of the mutants in the M4 generation increased to 9.48%, 10.04%, 7.24%, and 6.15%, respectively. Unfortunately, because the F869 mutant was not advanced to the M4 and M5 generations, no stearic acid content was available for these generations.
Table I. Stearic acid content in the seed oil of the five screened high stearic acid mutants averaged for n homozygous M3 (2012), M4 (2013), or M5 (2014) lines.
The content of five fatty acids in seed oil of the four screened cv Forrest, Essex, Williams 82, and PI 88788 wild types also was averaged for n lines. Averages and sd are shown.
| Lines | Palmitic Acid (16:0) | Stearic Acid (18:0) | Oleic Acid (18:1) | Linoleic Acid (18:2) | Linolenic Acid (18:3) | Stearic Acid Change | n |
|---|---|---|---|---|---|---|---|
| % | |||||||
| Forrest | 10.93 ± 0.59 | 3.25 ± 0.15 | 18.89 ± 2.34 | 53.71 ± 3.28 | 6.89 ± 0.79 | – | 33 |
| F605 | 6.93 ± 1.08 | 20.11 ± 1.68 | 20.77 ± 7.70 | 31.39 ± 4.99 | 4.32 ± 0.50 | 636.63 | 4 |
| F620 | 9.60 ± 0.27 | 10.04 ± 2.08 | 12.91 ± 1.50 | 53.03 ± 0.94 | 7.52 ± 0.36 | 267.76 | 5 |
| F714 | 10.09 ± 0.53 | 7.24 ± 1.89 | 14.88 ± 1.57 | 53.73 ± 2.41 | 8.23 ± 1.09 | 165.20 | 7 |
| F813 | 9.78 ± 1.33 | 6.90 ± 1.27 | 14.7 ± 3.49 | 42.91 ± 3.63 | 6.7 ± 1.81 | 152.74 | 5 |
| F869 | 8.50 ± 1.62 | 6.52 ± 2.07 | 11.79 ± 1.76 | 41.44 ± 4.33 | 8.29 ± 1.55 | 138.82 | 3 |
| Essex | 10.91 ± 0.42 | 4.11 ± 0.41 | 18.27 ± 1.61 | 52.8 ± 0.9 | 8.16 ± 0.67 | 50.54 | 4 |
| Williams 82 | 9.96 ± 0.7 | 3.74 ± 0.36 | 16.29 ± 5.18 | 50.78 ± 1.88 | 7.08 ± 0.96 | 36.99 | 4 |
| PI 88788 | 9.05 ± 0.71 | 2.42 ± 0.33 | 15.59 ± 6.44 | 41.05 ± 2.96 | 8.36 ± 1.75 | −11.35 | 4 |
Fatty acid analyses show that the mutant lines F714 and F813 presented consistent stearic acid content among the three tested generations: M3 (2012), M4 (2013), and M5 (2015). However, the stearic acid content of the mutant lines F605 and F620 was significantly different among the M3 and M4 generations (Supplemental Tables S1 and S2). In the M3 generation, the stearic acid content of F605 and F620 was 5.88% and 6.06%, whereas in the M4 generation, the stearic acid content of F605 and F620 increased to 9.48% and 10.04%, respectively. Surprisingly, in the M5 generation, the mutant line F605 presented an extremely high seed stearic acid content of 20.11% (Table I). This variation within the F605 and F620 mutant families may have been due to allelic segregation during these three generations. In addition to measuring stearic acid, the content of palmitic acid, oleic acid, linoleic acid, and linolenic acid of each mutant was measured. We observed that, between these mutants and the wild type, the contents of the other four types of fatty acids only showed a significant difference in oleic acid (a decrease of up to 5% compared with the wild-type cv Forrest; Table I). Knowing the role of SACPD-C in converting stearic acid to oleic acid in the fatty acid biosynthetic pathway, the decrease in oleic acid may be due to a reduced activity of SACPD-C.
Identification of Five Novel SACPD-C Alleles Conferring Higher Stearic Acid Content in Soybean Seeds
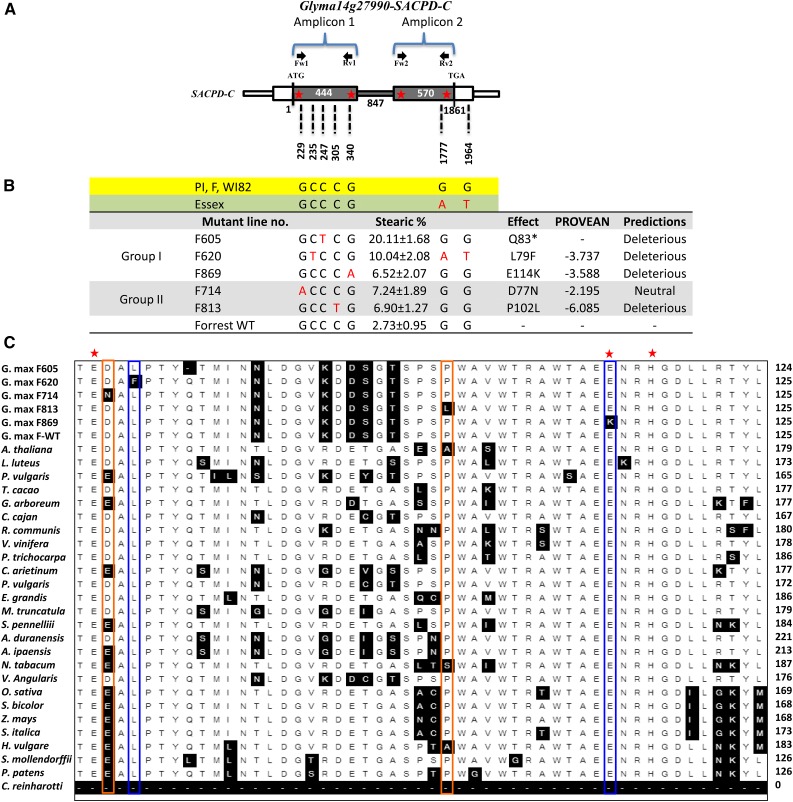
We analyzed the genomic sequence of GmSACPD-C of the five mutants (F605, F620, F714, F813, and F869) and the wild types (cv Forrest, Essex, and Williams 82). Interestingly, F605, F620, F714, F813, and F869 were identified to each carry a mutation within GmSACPD-C (C247T, C235T, G229A, C305T, and G340A, respectively; Fig. 1B). In addition, F620 contains G1777A and G1964T single-nucleotide polymorphisms in the second exon and 3′ untranslated region, respectively. These two single-nucleotide polymorphisms are present in the Essex cultivar. Therefore, F620 may have crossed with cv Essex (Fig. 1B). These mutations produced five new GmSACPD-C alleles, including one nonsense mutation (Q83* in F605) and four missense mutations (D77N in F714, L79F in F620, P102L in F813, and E114K in F869; Fig. 1B). PROVEAN predictions were performed on all mutations. Interestingly, four of the five missense mutations identified had PROVEAN scores of less than −2.5 (Fig. 1B). The mutations SACPD-CQ83*, SACPD-CD77N (PROVEAN score = −2.19), SACPD-CP102L (PROVEAN score = −6.08), and SACPD-CE114K (PROVEAN score = −3.58) were predicted to be deleterious to the protein (Fig. 1B).
Figure 1.
SACPD-C mutations of the five screened high stearic acid mutants. A, SACPD-C gene model and polymorphisms (G1777A and G1964T) between cv PI 88788, Forrest, Essex, and Williams 82. Primers used for genotyping are indicated by arrows. B, Predicted SACPD-C protein sequences showing amino acid differences between the five mutants identified by forward genetics: four missense mutations (D77N, L79F, P102L, and E114K) and one nonsense mutation (Q83*) within SACPD-C. PROVEAN predictions of the four missense mutations identified are included. WT, Wild type. C, Protein sequence alignment of the five SACPD-C screened mutants showing the identified mutations, SACPD-CD77N, SACPD-CL79F, SACPD-CQ83*, SACPD-CP102L, and SACPD-CE114K, in addition to SACPD-C orthologs from 26 other plant species. SACPD-C proteins identified from the following model plants, S. mollendorffii (lycophyte), P. patens (moss), C. reinhardtii (alga), rice (monocot), and Arabidopsis (eudicot), in addition to other eudicots and monocots, were included in the alignment and showed striking sequence conservation. SACPD-C mutations within residues conserved in all species analyzed are in blue boxes (group I). Mutations within nonconserved residues are in orange boxes (group II). Red stars represent the conserved Glu-105/76, Glu-143/114, and His-146/117 residues important for iron ion binding.
Identification of Conserved GmSACPD-C Residues across Several Dicot Species
In order to study if the new Gmsacpd-c mutations are located in conserved positions along the GmSACPD-C predicted protein sequence, an alignment was performed between the five Gmsacpd-c screened mutants, the four wild-type cultivars (Essex, Forrest, Williams 82, and PI 88788), and orthologous SACPD-C proteins of several plant species obtained from the National Center for Biotechnology Information database. The alignment of the soybean GmSACPD-C protein sequence with the orthologous SACPD-C proteins from 26 other species, including SACPD-C from a dicot model (Arabidopsis [Arabidopsis thaliana]), a monocot model (rice [Oryza sativa]), other monocot and dicot species, and primitive land plant models: a lycophyte (Selaginella mollendorffii), a moss (Physcomitrella patens), and a chlorophyte (Chlamydomonas reinhardtii), showed striking sequence conservation (Fig. 1C). From the alignment, the amino acid changes in the four identified missense mutants affected residues that were classified within two groups. Group I presented mutations in conserved residues: SACPD-CL79F and SACPD-CE114K. However, group II contained mutations within nonconserved residues: SACPD-CD77N and SACPD-CP102L. Interestingly, the seed stearic acid content correlated with both groups. Mutations in conserved residues increased to up to 3 times the levels of seed stearic acid content (group I). However, mutations in nonconserved residues resulted in a limited increase in seed stearic acid, presenting only 2 times the level contained in the wild-type cv Forrest seeds (group II).
Mutational Analysis of GmSACPD-C
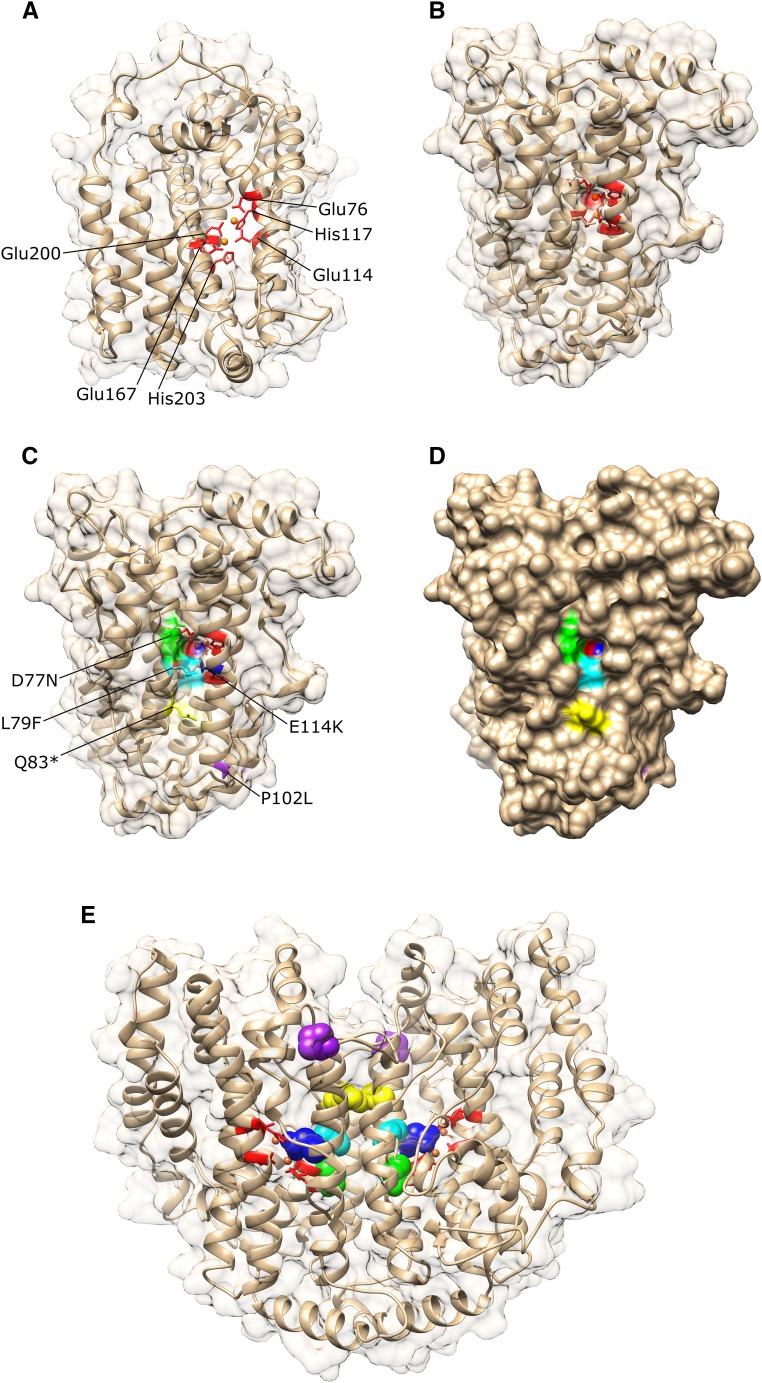
In order to study the impact of the missense mutations on the catalytic activity of the enzyme, homology modeling of GmSACPD-C was carried out. Three missense mutants (D77N, L79F, and E114K) were localized directly at the surface of the iron ion-binding pocket (Fig. 2), with one mutation directly altering the negatively charged bridging ligand Glu-114 into a positively charged Lys (SACPD-CE114K). Because of the iron ion pocket localization, the alteration of charge in the missense mutants SACPD-CD77N and SACPD-CL79F, and the presence of steric hindrance by L79F, these mutations are predicted to affect iron ion-binding kinetics and stability. The missense mutant SACPD-CP102L was not localized at the iron ion-binding pocket but was positioned at the first residue of the α4 chain, which holds the ligands Glu-114 and His-117 in place. Considering Pro’s cyclic conformation, in which the secondary amine binds to the α-carbon of the protein backbone, a disruption of this conformational rigidity may impact the ability of the α4 chain to be in its proper location, disrupting the enzymatic activity of GmSACPD-C.
Figure 2.
Homology modeling of GmSACPD-C from cv Forrest with important catalytic residues and the five identified sacpd-c missense mutations mapped. A and B, One subunit containing a diiron coordination complex. The first iron has highly conserved ligands Glu-76 and His-117, the second iron has ligands Glu-167 and His-203, and the two residues Glu-114 and Glu-200 act as bridging ligands in the coordination space. Iron ions are highlighted in orange. C and D, One subunit with the five missense mutants highlighted in different colors showing an iron ion pocket localization. E, The predicted homodimeric GmSACPD-C enzyme.
Analysis of Nodule Structure Reveals Morphological Differences between Group I and Group II Gmsacpd-c Mutants
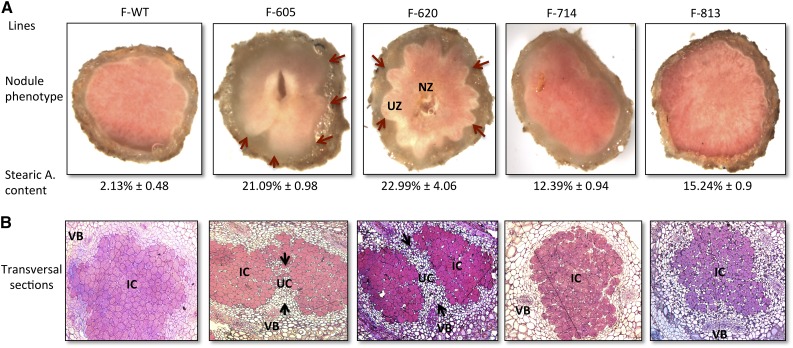
It has been demonstrated that a homolog of the soybean GmSACPD-C gene also was identified as a nodulin gene in yellow lupine (Lupinus luteus; Swiderski et al., 2000). Therefore, the effect of the GmSACPD-C mutation on stearic acid content in the nitrogen-fixing nodules was analyzed. Unfortunately, because the F869 mutant was not advanced in the M4 and M5 generations, from here on, only four mutants (F605, F620, F714, and F813) will be characterized further. The average nodule stearic acid content in the wild-type cv Forrest was 2.13%. Surprisingly, the four M4 mutants F605, F620, F714, and F813 presented a large increase in the nodule stearic acid content, showing 21.09%, 22.99%, 12.39%, and 15.24%, respectively. These data are in accordance with the two groups cited previously and correlated with the stearic acid levels obtained. In fact, members belonging to group I presented the highest nodule stearic acid content (greater than 21%); however, group II members presented a limited stearic acid content (less than 16%). No significant root stearic acid content increase was observed in the mutants (Supplemental Fig. S2).
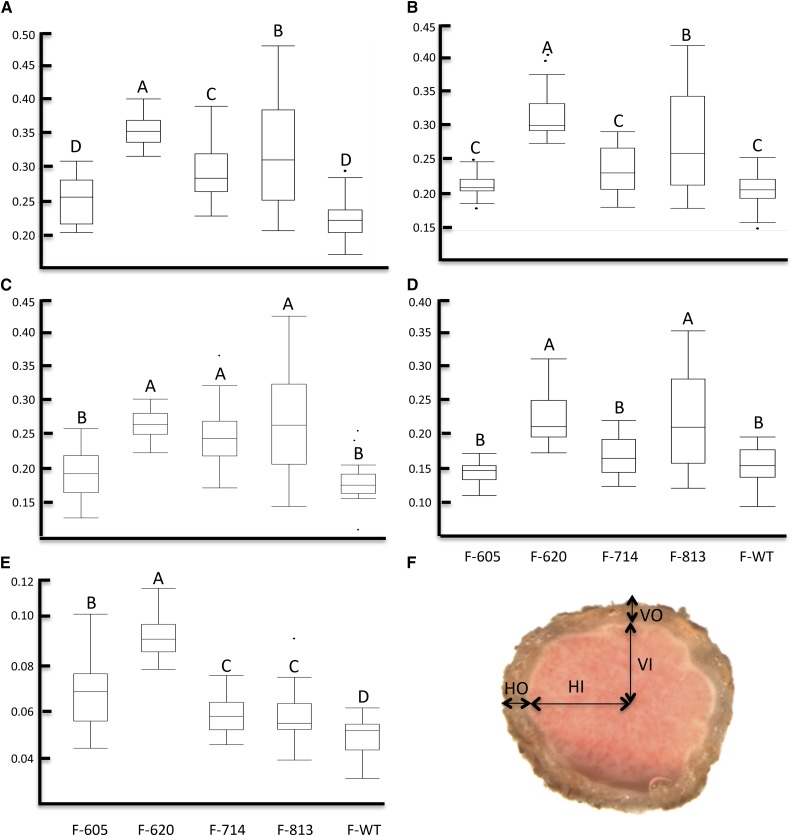
To test the effect of the nodule stearic acid content increase between the two groups, light microscopy of nodule cross sections was carried out. Group I exhibited an undulate coat phenotype and the presence of a central necrotic cavity. Surprisingly, group II did not show any undulate coat phenotype or central necrotic cavity. To elucidate the effect of each mutation on nodule structure, biometric measures were collected from the nodule coat (Fig. 3F). Interestingly, the nodule average diameter showed an increase in coat size across the four Gmsacpd-c mutants analyzed (Fig. 3).
Figure 3.
Comparison of the cortex and cavity measurements between the Gmsacpd-c mutants and the cv Forrest wild type (F-WT). A, Nodule horizontal outer diameter. B, Nodule vertical outer diameter. C, Nodule horizontal inner diameter. D, Nodule vertical inner diameter. E, Biometric measures of the nodule coat size using the formula (HO − HI) + (VO − VI)/2. F, Dissecting light microscope image of a hand-sectioned nodule from the cv Forrest wild type showing details of horizontal outer (HO), horizontal inner (HI), vertical outer (VO), and vertical inner (VI). Numbers are as follows: F605 (n = 21), F620 (n = 22), F714 (n = 20), F813 (n = 25), and wild type (n = 20).
Furthermore, in order to gain insight into changes that caused the central cavity formation at a cellular level, nodule cross sections in the wild type and the four identified mutants were compared. Microscopy analysis revealed a reduced differentiation of vascular bundles observed in group I (Fig. 4). The nodules showed a significant increase in size of all cell layers, indicating that all nodule cell types were affected by the deleterious GmSACPD-C mutations. In addition, a very strong reduction of bacteroids was observed in group I, with an increased cortex size and an expanded cavity (Fig. 4B). However, no differences in radial organization or defects between the wild-type Forrest and the group II mutants were observed. These results indicate that GmSACPD-C mutations not only control seed and nodule stearic acid content but also may ultimately impact nodule structure and morphology.
Figure 4.
Comparison of nodule structure and morphology of the cv Forrest wild type (F-WT) and the four sacpd-c mutants. A, Dissecting light microscope images of hand-sectioned nodules showing the necrotic central cavity in the deleterious group I Gmsacpd-c mutants (F605 and F620) and its absence in the case of the group II mutants (F714 and F813). B, Nodule cross sections stained with Methylene Blue and picrofushin showing typical nodule cortex and vascular tissue in the cv Forrest wild type compared with the central cavity, reduced infected cells, less vascular bundle differentiation, and decreased bacteroids of the group I mutants (magnification, 100×). IC, Infected cells; NZ, necrotic zone; UC, uninfected cells; UZ, undulated zone; VB, vascular bundles. Red arrows present the undulated phenotype presented by group I mutants. Black arrows represent a separation between infected cell zones and uninfected cells in group I mutants. This separation was absent in the wild-type cv Forrest and group II mutants.
Analysis of Leaf Morphology in Gmsacpd-c Mutant Lines
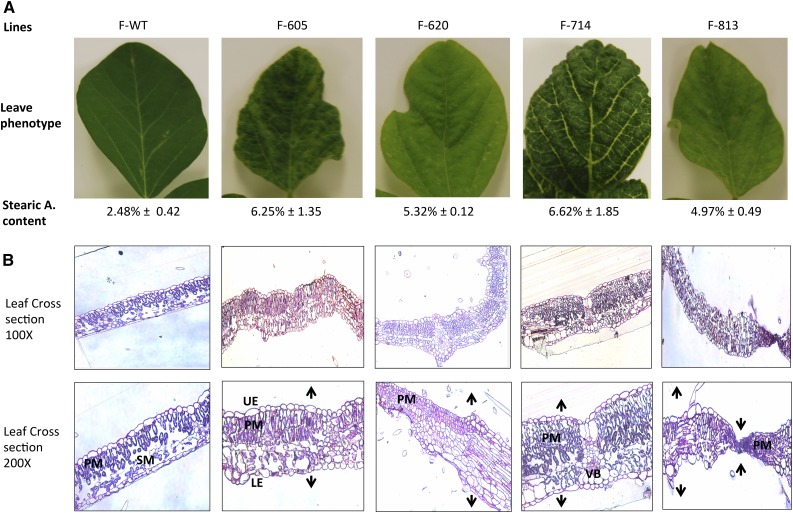
Phenotypic analysis of plant development of the Gmsacpd-c mutants revealed a common undulate morphology in the leaves (Fig. 5A). To investigate the possible effect of the GmSACPD-C mutation on the observed leaf phenotype, leaf fatty acid content was analyzed. Leaf stearic acid content was increased significantly from 2.48% in the cv Forrest wild type to 6.25%, 5.32%, 6.62%, and 4.97% in the four M4 mutants F605, F620, F714, and F813, respectively (Supplemental Fig. S2). To gain insight into the changes that caused the undulate leaf formation at the cellular level, leaf cross sections in the wild type and the four identified mutants were analyzed (Fig. 5).
Figure 5.
Comparison of leaf structure and morphology of the cv Forrest wild type (F-WT) and the four Gmsacpd-c mutants. A, Leaf images showing typical soybean leaf structure in the cv Forrest wild type compared with the rigid undulated phenotype presented by the Gmsacpd-c mutants. B, Leaf cross sections stained with Methylene Blue showing typical upper and lower epidermis (UE and LE), palisade and spongy mesophyll (PM and SM), and vascular bundle (VB; xylem and phloem) organization. This structure was drastically altered in the four Gmsacpd-c mutants.
In the wild-type cross sections, the leaf anatomy observed beneath the adaxial epidermis includes a palisade mesophyll layer that is one to two cells thick. The cells in this region are more uniform in shape, size, and spacing. The spongy mesophyll layer is made up of more irregularly shaped cells and contains several air spaces throughout. These air spaces often correspond with the stomata found on the abaxial epidermal region of the leaf. In the Gmsacpd-c mutants, the palisade layer appears to be three to five cells thick. In addition, the cells are more densely packed and are more elongated than in the wild type. Interestingly, the air spaces found in the spongy mesophyll layer were reduced in the mutants, with the spaces occupied by an increase in either the spongy or palisade mesophyll cells. The organization of the cell layers, including the upper and lower epidermis and vascular bundles, also was found to be distorted (Fig. 5B). Thus, the high leaf stearic acid content of the Gmsacpd-c mutants (F605, F620, F714, and F813) may have contributed to the drastic leaf cell disorganization, reflected by a stronger undulate phenotype.
Quantitative Reverse Transcription-PCR Analysis Reveals That Group I Gmsacpd-c Mutants Have a Greater Expression of Nodule Leghemoglobins Than Group II
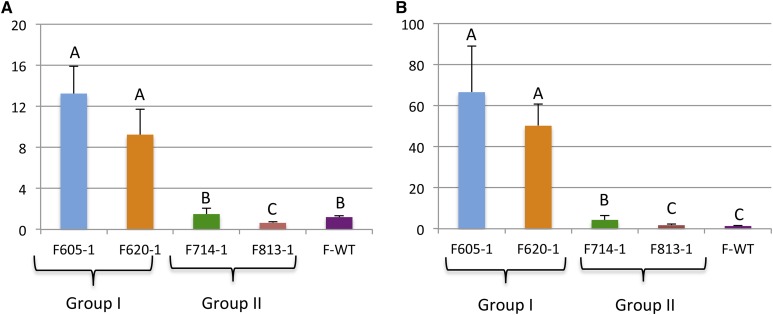
Investigation of the cv Williams 82 genome indicates that the leghemoglobin (GmLegh) gene family is composed of three members located on chromosomes 10 (Glyma.10G199000), 20 (Glyma.20G191200), and 11 (Glyma.11G121700), named GmLegh-C1, GmLegh-C2, and GmLegh-C3, respectively. First, the expression data of the three GmLegh genes was compiled using the public RNA sequencing data available at Soybase (www.soybase.org). Two distinct gene expression patterns were identified for the GmLegh gene members (Supplemental Fig. S3). Whereas GmLegh-C1 and GmLegh-C2 were both highly expressed only in nodules, GmLegh-C3 had almost no expression in most tissues analyzed, except for a minor expression in the roots. Next, the expression of the three GmLegh gene family members was analyzed by quantitative reverse transcription (qRT)-PCR. As expected, no expression of GmLegh-C3 was detected in any of the soybean lines analyzed. Furthermore, both GmLegh-C1 and GmLegh-C2 transcripts were found to be significantly more abundant in group I than in group II mutants (Fig. 6).
Figure 6.
qRT-PCR analysis of the Leghemoglobin gene family in the four Gmsacpd-c mutant soybean lines. Differences in expression are shown for the Legh-C1 (A) and Legh-C2 (B) genes between group I and group II mutants. The Legh-C3 gene was not expressed. Expression values were normalized using Ubiquitin as a reference. The gene-specific primers designed for qRT-PCR analysis are detailed in Supplemental Table S3. Results were analyzed with ANOVA (P < 0.0001). F-WT, The cv Forrest wild type.
DISCUSSION
Random Mutagenesis as a Rheostat for Agronomically Important Traits
Many of the developed high seed stearic acid soybean germplasm suffer agronomic problems (Hammond and Fehr, 1983; Lundeen et al., 1987; Rahman et al., 1995, 1997; Gillman et al., 2014). A sodium azide-induced mutant called A6 presented extremely high stearic acid content (28%) but was described as nondesirable, since it was associated with poor germination and low seed yield (Hammond and Fehr, 1983; Lundeen et al., 1987; Rahman et al., 1997). An extraordinarily large deletion representing one-eighth of chromosome 14 downstream from the GmSACPD-C gene, including at least 56 genes, was identified recently and may explain the mutant A6’s extremely poor agronomic characteristics, which has delayed the commercialization of high seed stearic acid lines (Gillman et al., 2014). Therefore, developing mutants with a large genomic deletion is agronomically impractical. Consequently, producing new EMS mutants with a single base pair mutation seems to be a better strategy for developing lines with agronomically important traits without unintended poor agronomic characteristics. Additionally, an increase of certain traits over a threshold of plant tolerance may negatively impact its agronomic performance, such as the appearance of a central necrotic cavity in the root nodules of high stearic acid soybeans (Gillman et al., 2014). Thus, fine-tuning the level of a certain trait is pertinent to the development of improved crop plants. This can be reached by studying the impact of mutations on important catalytic sites within the enzymatic structure.
SACPD is a homodimeric enzyme with each homomer containing a diiron coordination complex (Lindqvist et al., 1996). The crystal structure of SACPD in the genus Ricinus reveals that the two iron ions of the diiron complex are liganded by residues on four of the α-helices (Lindqvist et al., 1996). The first iron has highly conserved ligands Glu-105/76 and His-146/117 and the second iron has ligands Glu-196/167 and His-232/203, with the amino acid positions corresponding to the genus Ricinus and soybean protein sequences, respectively. The two residues, Glu-143/114 and Glu-229/200, act as bridging ligands in the coordination space. From here on, only soybean amino acid positions will be noted. By mapping the high stearic acid Gmsacpd-c mutations onto a homology model based on the wild-type cv Forrest, we can begin to understand how the mutations affect the enzymatic activity of GmSACPD-C. Due to the steric hindrance of the missense L79F on the diiron core and the nonsense truncated Q83* mutation, a strong reduction of enzymatic activity was reflected by the highest seed stearic acid contents of 10% and 20%, respectively (group I; Figs. 1 and 2). However, the D77N and P102L mutations did not have as much of a deleterious impact on the protein activity, which is reflected by the limited stearic acid contents of 7% and 6%, respectively (group II), observed through the M5 generation. In the case of P102L, although the mutation was not localized at the iron ion-binding pocket, due to a disruption of the conformational rigidity on the α4 chain, a decrease in enzymatic activity is observed. Such mutations allow for a fine-tuning of the enzymatic kinetics, leading to an increase in the desired trait without the drawback of poor agronomic characteristics. This is supported by the observation that only mutations localized within conserved residues (group I) presented unhealthy nodules with cell senescence, a necrotic cavity, reduced differentiation of vascular bundles, and a drastic decrease in infected cells. However, no differences in radial organization between the cv Forrest wild-type nodules and the lines carrying mutations localized outside of conserved residues (group II) were observed.
Effects of Gmsacpd-c Mutations on Leaf Structure and Morphology
Studies on metabolomic profiling showed a differential accumulation of fatty acids (precursors to membrane phospholipids) between roots and nodules and between infected and uninfected root hairs (Colebatch et al., 2004). Mutants with high levels of saturated fatty acids within the leaves of Arabidopsis have been described. The fad5 mutant, with an increased palmitic acid content, due to a deficiency in the activity of the chloroplast 16:O-MGD desaturase, contains 24% palmitic acid and 1% stearic acid in leaves (25% total saturates; Kunst et al., 1989). A fabl mutant had a reduced activity of 3-ketoacyl-ACP synthase11 and contained 22% palmitic acid and 1% stearic acid in leaves (23% total saturates; Wu et al., 1994). Both fabl and fad5 mutants had no visible phenotype at normal growth temperatures (Wu et al., 1994). However, a fab2 mutant, due to a mutation partially affecting the action of the stromal SACPD, contains 12% palmitic acid and 20% stearic acid in leaves (31% total saturates; Lightner et al., 1994). This mutant has a profoundly altered growth and morphology characterized by a reduction in the size of almost all organs, including leaves. In this study, the F605 nonsense Gmsacpd-c mutant presented similar saturated fatty acid phenotypes (27% total saturates), with 6.9% and 20.1% contributed by palmitic and stearic acids, respectively (Table I). Interestingly, mutations of GmSACPD-C exhibited different morphological phenotypes of the leaves than the stromal Arabidopsis fab2 mutant. In fact, the Gmsacpd-c mutations in soybeans affected the organization of the leaf cell layers, conferring a strong undulate leaf phenotype. This is exhibited by the increased abundance and elongation of the palisade mesophyll layer, the reduction of air spaces in the spongy mesophyll layer, and the abnormal differentiation of the vascular bundles within the mutant leaves.
It has been reported previously that fatty acid composition and saturation level affect membrane integrity, allowing plants to tolerate heat stress (Murakami et al., 2000). In fact, increasing the degree of saturation enhances lipid heat stability, positively impacting plant tolerance to high temperature in various species (Raison et al., 1982; Grover et al., 2000). However, double bonds present in unsaturated fatty acids are easily attacked by hydroxyl free radicals under stress environments (Murakami et al., 2000). The presence of these free radicals can cause membrane lipid peroxidation and, thus, membrane disruption (Smirnoff, 1993; Foyer et al., 1994). Moreover, it has been shown that heat stress associated with cellular membrane damage causes physiological injuries and leaf senescence, leading to cell solute leakage and the disruption of cell function (Blum and Ebercon, 1981; Marcum, 1998). Therefore, increasing saturated fat content in leaves, like stearic acid, may positively impact plant tolerance to heat stress and avoid cellular membrane damage.
Effects of Gmsacpd-c Mutations on Leghemoglobin Expression
It has been shown that products of SACPD, such as oleic acid precursors, were found to increase significantly in root hairs in response to rhizobial infection by Bradyrhizobium japonicum (Brechenmacher et al., 2010). Although the sacpd-c mutants have shown large increases in stearic acid, only oleic acid was found to have a reduction from the wild-type levels. The association between legumes and rhizobia results in the formation of root nodules, where symbiotic nitrogen fixation occurs (Herridge et al., 2008).
Leghemoglobin is an abundant hemeprotein of legume nodules and plays an essential role as an oxygen carrier. Leghemoglobin is generally produced by legumes in response to the colonization of roots by nitrogen-fixing bacteria (O’Brian et al., 1987). However, roots not colonized by rhizobia do not accumulate leghemoglobin. Leghemoglobin buffers the concentration of free oxygen in the infected plant cells to ensure the proper function of root nodules (O’Brian et al., 1987). Expression analysis revealed that group I, exhibiting deleterious Gmsacpd-c mutations, had the highest abundance of leghemoglobin transcripts. Deleterious Gmsacpd-c mutations around conserved catalytic residues in group I may negatively affect iron ion binding, which may result in an increase in iron ion availability within the nodules. Since leghemoglobin also uses an iron cofactor, we speculate that the decrease of nodule GmSACPD-C iron binding may allow for an increase in leghemoglobin iron binding and expression (Brear et al., 2013). This is supported by the greater abundance of both Legh-C1 and Legh-C2 transcripts in group I mutants as compared with the group II mutants or the cv Forrest wild type.
In this study, we have shown that independent GmSACPD-C mutations confer high stearic acid content in seeds, nodules, and leaves. These mutations also affected leaf and nodule morphology, cell organization, and vasculature. Using random mutagenesis coupled with homology modeling and mutational analysis, high seed stearic acid content can be achieved while the associated negative agronomic characteristics are circumvented. Although the impact of stearic acid accumulation on leaf and nodule structure and morphology has been revealed, its mechanism of action needs to be studied further and the cross talk between SACPD and leghemoglobin still needs to be elucidated.
MATERIALS AND METHODS
Development of an EMS Mutagenesis cv Forrest Population
The soybean (Glycine max) ‘Forrest’ seed was obtained from the Southern Illinois University Carbondale Agricultural Research Center and used to develop an EMS-mutagenized population. The wild-type cv Forrest seed was mutagenized with 0.6% (v/v) EMS as described previously (Meksem et al., 2008) and planted to harvest 1,588 M2 families of seed in 2011. This population was advanced successively to the M3 generation in 2012, the M4 generation in 2013, and the M5 generation in 2014 at Southern Illinois University Carbondale. In total, more than 1,000 mutant families (lines) of M3/M4/M5 were developed.
Soybean Cyst Nematode Infection Phenotyping
All M3 lines were screened with the soybean cyst nematode infection phenotyping as described previously (Liu et al., 2011). Seedlings were inoculated with infective eggs from the PA3 population (HG type 0). Briefly, cysts were extracted from cv Essex infested roots and soil by flotation in water and collected on a 250-μm sieve. Harvested cysts were gently crushed using a drill press, and the eggs were collected on a 25-μm sieve (Faghihi and Ferris, 2000). The eggs were further diluted to 1,000 eggs per mL of water. Individual seedlings were inoculated with 1 mL of the egg suspension. Plants were maintained in the growth chamber at 27°C. Cyst counts were performed at 30 d postinoculation.
Forward Genetic Analysis of Seed Fatty Acid
All M3, M4, and M5 lines were analyzed for seed fatty acid composition using the two-step procedure as outlined previously (Kramer et al., 1997). Five seeds per mutant from the M3, M4, and M5 generations: 30 wild-type cv Forrest seeds and four cv Essex, Williams 82, and PI 88788 wild-type seeds, were randomly selected for fatty acid analysis. Briefly, each seed was scarified and then loaded into one 16- × 200-mm tube with a Teflon-lined screw cap. Next, 2 mL of sodium methoxide was added into each tube, and then the tube was incubated in a water bath at 50°C for 10 min. Afterward, 3 mL of 5% methanolic HCl was added into each tube after the tube was cooled for 5 min. Subsequently, the tube was incubated in a water bath at 80°C for 10 min and cooled for 7 min, and then 7.5 mL of 6% (w/v) potassium carbonate and 1 mL of hexane were added. Finally, the tubes were centrifuged at 1,200g for 5 min to separate the layers. The aqueous layer was then transferred into vials and used for the fatty acid analysis using a Shimadzu GC-2010 gas chromatograph equipped with a flame ionization detector and a Supelco 60-m SP-2560 fused silica capillary famewax column (0.25 mm i.d. × 0.25 µm film thickness). The helium carrier gas was maintained at a linear velocity of 23 cm s−1. The oven temperature was programmed for 175°C for 22 min, then increased at 15°C min−1 to 225°C, and maintained for 3 min. The injector and detector temperatures were set at 255°C. Peaks were identified by comparing the retention times with those of the corresponding standards (Nu-Chek-Prep and Supelco). The contents of palmitic acid, stearic acid, oleic acid, linoleic acid, and linolenic acid were calculated using the statistical computing package JMP Pro 12.
Detection of Genotypes and Mutations of the Mutagenized Population
Young leaf tissue of the Gmsacpd-c mutants and four soybean lines (wild-type cv Forrest, Essex, Williams 82, and PI 88788) were used to extract DNA using the cetyltrimethylammonium bromide method (Rogers and Bendich, 1994) with small modifications. The specific primers (Supplemental Table S3) were designed to amplify the fragments covering all GmSACPD-C exons from the extracted DNAs with 38 cycles of PCR amplification at 94°C for 30 s, 51°C for 30 s, and 72°C for 1 min. The PCR products were purified by enzymatic cleanup, or the specific bands were recovered from the agarose gels after electrophoresis using the Qiagen gel extraction kit (QIAquick). Then, the purified PCR fragments were sequenced by GENEWIZ (www.genewiz.com). The genotypes of the mutants were determined by comparison of the genomic, known cDNA, and predicted protein sequences of the genes with those of cv Forrest, Essex, Williams 82, and PI 88788 using DNASTAR Lasergene software. PROVEAN predictions were performed on all mutations. PROVEAN predicts whether an amino acid substitution or insertion/deletion has an impact and affects the biological protein function based on sequence homology and the physical properties of amino acids (Choi, 2012; Choi et al., 2012; Choi and Chan, 2015). PROVEAN variant predictions with a score equal to or below −2.5 are predicted to be deleterious.
Multiple Sequence Alignment of SACPD-C
To evaluate the positions of substitutions, multiple sequence alignments were performed with the ClustalW program, and mutations were overlaid on the protein structure of the wild-type lines. In silico analyses of the five identified cv Forrest GmSACPD-C mutants, SACPD-CD77N, SACPD-CL79F, SACPD-CQ83*, SACPD-CP102L, and SACPD-CE114K, in addition to SACPD-C orthologs from 26 other plant species, were performed using the MegAlign (DNASTAR Lasergene 8) software package and the ClustalW algorithm. All parameter values correspond to default definitions.
Fixation and Infiltration of Nodules and Leaves
Soybean root nodules and leaves from the Gmsacpd-c mutants and the cv Forrest wild type were washed with triple distilled nano-pure water to remove soil and then placed in a sodium phosphate buffer (0.05 m, pH 7.2) for 24 h. The samples were sectioned longitudinally, transferred to an Eppendorf tube, and fixed in a 2% gluteraldehyde solution buffered with a sodium phosphate buffer (0.05 m, pH 7.2) for 1 h at 22°C, then overnight at 4°C. The samples were washed four times (30 min each) in the same phosphate buffer described above. After the fourth buffer rinse, the samples were placed in fresh buffer solution and held overnight at 4°C. The samples were postfixed in a 2% osmium tetraoxide water solution (triple distilled nano-pure water) for 10 min. Three consecutive water rinses (30 min each) were immediately performed postfixation. The osmicated specimens were serially dehydrated with water and ethanol (25%–100%), with each dehydration lasting 20 min. The ethanol was serially replaced with proplyn oxide (50%–100%). The samples were then serially infiltrated with a proplyn oxide Spurr’s resin (25%–100%) solution for 5 d. The infiltrated nodules and leaves were placed in BEEM capsules with fresh 100% Spurr’s resin and baked for 2 d at 60°C. Samples were sectioned on an ultramicrotome at a 1,000-µm thickness. Staining was performed using picrofushin and 1% Methylene Blue in a 50% ethanol and 50% water solution. For the hand-sectioned nodules, sections were placed on a glass slide and stained with 1% Toluidine Blue. Prepared slides were viewed with a Leica DM5000B compound light microscope and imaged with a Q-Imaging Retiga 2000R digital camera.
Homology Modeling of GmSACPD-C Proteins
Homology modeling of a putative GmSACPD-C protein structure was conducted with Deepview and Swiss Model Workspace software using the protein sequence from cv Forrest and an available SACPD crystal structure from Ricinus communis as a template (Protein Data Bank accession no. 1OQ4; Moche et al., 2003). All residues were modeled against this template with a sequence identity of 69%. Mutation mapping and visualizations were performed using the UCSF Chimera package (Pettersen et al., 2004).
Rhizobia Inoculation Assay of the Identified High Stearic Acid Gmsacpd-c Mutants
Soybean plants were grown on an autoclaved sandy and organic soil mixture (50:50) and watered as needed. U.S. Department of Agriculture Bradyrhizobium japonicum strain 110 was grown in a modified arabinose gluconate medium for 6 d at 28°C until it reached the exponential growth phase. Bacterial inoculation was done at the time of sowing (10% volume by seed weight). Inoculated plants were grown in the greenhouse at 27°C and 70% humidity, with 16 h of artificially supplemented light. Root, nodule, and leaf samples were frozen dry for fatty acid analysis as described previously or macerated in liquid nitrogen for qRT-PCR analysis.
qRT-PCR Analysis of the Leghemoglobin Gene Family in Soybean
Soybean seedlings from the four EMS mutants and the cv Forrest wild type were grown in a growth chamber for 1 week and then inoculated with B. japonicum strain 110 from the U.S. Department of Agriculture. After 5 weeks, total RNA was isolated from root samples using a Qiagen RNeasy Plant Mini Kit (catalog no. 74904). Total RNA was DNase treated and purified using a Turbo DNA-free Kit (QAmbion/Life Technologies; AM1907). RNA was quantified using a Nanodrop 1000 (version 3.7), then a total of 400 ng of treated RNA was used to generate cDNA using a cDNA synthesis kit (Thermoscript, Life Technologies; 11146-025) with random hexamers. One-tenth of a 20-μL reverse transcription reaction was used in gene-specific quantitative PCR with the Power SYBR Green PCR Master Mix kit (Applied Biosystems; 4368706). A list of primers used in this work is found in Supplemental Table S3. For each genotype, RNA from three biological replicates was used for quantification and then normalized using the ΔCq method with Ubiquitin used as a reference gene [ΔCq = Cq(TAR) – Cq(REF)]. Each gene’s expression was exponentially transformed to the expression level using the following formula: ΔCq expression = 2−ΔCq. A negative control (without reverse transcription) reaction also was performed in all the samples.
Statistical Analysis
All the presented results were performed with ANOVA comparison using the statistical computing package JMP Pro 12.
Accession Numbers
The SACPD (EC 1.14.19.2) nucleotide and amino acid sequences of the newly identified high stearic acid alleles in addition to the cv Forrest wild type are deposited at the National Center for Biotechnology Information under the following GenBank accession numbers: SACPD-CQ83*(F605), KX856376; SACPD-CL79F(F620), KX856377; SACPD-CD77N(F714), KX856378; SACPD-CP102L(F813), KX856379; SACPD-CE114K(F869), KX856380; and SACPD-C_Forrest_WT, KX856375.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Distribution of stearic acid contents in the seed oil of the cv Forrest M3 population.
Supplemental Figure S2. Levels of stearic acid of the screened high stearic acid sacpd-c mutants and the wild type cv Forrest in the M4 (2013) generation.
Supplemental Figure S3. Expression levels of soybean leghemoglobin genes in planta based on Soyseq resource available from RNA sequencing data.
Supplemental Table S1. Levels of stearic acid in seed oil of the five screened high stearic mutants and the cv Forrest wild type were averaged for n M3 (2012) lines for each mutation.
Supplemental Table S2. Levels of stearic acid in seed oil of the four screened high stearic mutants and the cv Forrest wild type were averaged for n M4 (2013) lines for each mutation.
Supplemental Table S3. Primers used for genotyping, sequencing, and qRT-PCR analysis.
Acknowledgments
We thank Darcie Hastings for excellent technical assistance with gas chromatography analysis, Dr. Judy Davie for providing the qRT-PCR machine, Abhinav Adhikari for assistance, and all the student workers who contributed to the EMS-mutagenized cv Forrest population development.
Glossary
- EMS
ethyl methanesulfonate
- qRT
quantitative reverse transcription
Footnotes
This work was supported by the United Soybean Board (grant nos. 1520-532-5607 and 0253 to K.M.).
Articles can be viewed without a subscription.
References
- Blum A, Ebercon A (1981) Cell membrane stability as a measure of drought and heat tolerance in wheat. Crop Sci 21: 43–47 [Google Scholar]
- Brear EM, Day DA, Smith PMC (2013) Iron: an essential micronutrient for the legume-rhizobium symbiosis. Front Plant Sci 4: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechenmacher L, Lei Z, Libault M, Findley S, Sugawara M, Sadowsky MJ, Sumner LW, Stacey G (2010) Soybean metabolites regulated in root hairs in response to the symbiotic bacterium Bradyrhizobium japonicum. Plant Physiol 153: 1808–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byfield GE, Xue H, Upchurch RG (2006) Two genes from soybean encoding soluble Δ9 stearoyl-ACP desaturases. Crop Sci 46: 840–846 [Google Scholar]
- Carrero-Colón M, Abshire N, Sweeney D, Gaskin E, Hudson K (2014) Mutations in SACPD-C result in a range of elevated stearic acid concentration in soybean seed. PLoS ONE 9: e97891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. (2012) A fast computation of pairwise sequence alignment scores between a protein and a set of single-locus variants of another protein. In Proceedings of the ACM Conference on Bioinformatics, Computational Biology and Biomedicine. ACM, New York: pp. 414–417 [Google Scholar]
- Choi Y, Chan AP (2015) PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31: 2745–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 7: e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente TE, Cahoon EB (2009) Soybean oil: genetic approaches for modification of functionality and total content. Plant Physiol 151: 1030–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebatch G, Desbrosses G, Ott T, Krusell L, Montanari O, Kloska S, Kopka J, Udvardi MK (2004) Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J 39: 487–512 [DOI] [PubMed] [Google Scholar]
- Faghihi J, Ferris JM (2000) An efficient new device to release eggs from Heterodera glycines. J Nematol 32: 411–413 [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Lelandais M, Kunert KJ (1994) Photooxidative stress in plants. Physiol Plant 92: 696–717 [Google Scholar]
- Gillman JD, Stacey MG, Cui Y, Berg HR, Stacey G (2014) Deletions of the SACPD-C locus elevate seed stearic acid levels but also result in fatty acid and morphological alterations in nitrogen fixing nodules. BMC Plant Biol 14: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover A, Agarwal M, Katiyar-Agarwal S, Sahi C, Agarwal S (2000) Production of high temperature tolerant transgenic plants through manipulation of membrane lipids. Curr Sci 79: 557–559 [Google Scholar]
- Hammond EG, Fehr WR (1983) Registration of A6 germplasm line of soybean. Crop Sci 23: 192–193 [Google Scholar]
- Herridge DF, Peoples MB, Boddey RM (2008) Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311: 1–18 [Google Scholar]
- Knowlton S, inventor. May 8, 2001. Fat products from high stearic soybean oil and a method for the production thereof. US Patent Application No. US6229033 [Google Scholar]
- Kok LL, Fehr WR, Hammond EG, White PJ (1999) Trans-free margarine from highly saturated soybean oil. J Am Oil Chem Soc 76: 1175–1181 [Google Scholar]
- Kramer JK, Fellner V, Dugan ME, Sauer FD, Mossoba MM, Yurawecz MP (1997) Evaluating acid and base catalysts in the methylation of milk and rumen fatty acids with special emphasis on conjugated dienes and total trans fatty acids. Lipids 32: 1219–1228 [DOI] [PubMed] [Google Scholar]
- Kris-Etherton PM, Yu S (1997) Individual fatty acid effects on plasma lipids and lipoproteins: human studies. Am J Clin Nutr (Suppl) 65: 1628S–1644S [DOI] [PubMed] [Google Scholar]
- Kris-Etherton PM, Yu S, Etherton TD, Morgan R, Moriarty K, Shaffer D (1997) Fatty acids and progression of coronary artery disease. Am J Clin Nutr 65: 1088–1090 [DOI] [PubMed] [Google Scholar]
- Kunst L, Browse J, Somerville C (1989) A mutant of Arabidopsis deficient in desaturation of palmitic acid in leaf lipids. Plant Physiol 90: 943–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightner J, Wu J, Browse J (1994) A mutant of Arabidopsis with increased levels of stearic acid. Plant Physiol 106: 1443–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist Y, Huang W, Schneider G, Shanklin J (1996) Crystal structure of delta9 stearoyl-acyl carrier protein desaturase from castor seed and its relationship to other di-iron proteins. EMBO J 15: 4081–4092 [PMC free article] [PubMed] [Google Scholar]
- List GR, Mounts TL, Orthoefer F, Neff WE (1997) Effect of interesterification on the structure and physical properties of high-stearic acid soybean oils. J Am Oil Chem Soc 74: 327–329 [Google Scholar]
- Liu X, Liu S, Jamai A, Bendahmane A, Lightfoot DA, Mitchum MG, Meksem K (2011) Soybean cyst nematode resistance in soybean is independent of the Rhg4 locus LRR-RLK gene. Funct Integr Genomics 11: 539–549 [DOI] [PubMed] [Google Scholar]
- Lundeen PO, Fehr WR, Hammond EG, Cianzio SR (1987) Association of alleles for high stearic acid with agronomic characters of soybean. Crop Sci 27: 1102–1105 [Google Scholar]
- Marcum KB. (1998) Cell membrane thermostability and whole-plant heat tolerance of Kentucky bluegrass. Crop Sci 38: 1214–1218 [Google Scholar]
- Meksem K, Liu S, Liu XH, Jamai A, Mitchum MG, Bendahmane A, El-Mellouki T(2008) TILLING: a reverse genetics and a functional genomics tool in soybean. In The Handbook of Plant Functional Genomics: Concepts and Protocols, G Kahl and K Meksem, eds, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany. http://doi.org/10.1002/9783527622542.ch12 [Google Scholar]
- Mensink RP, Katan MB (1990) Effect of dietary trans fatty acids on high-density and low-density lipoprotein cholesterol levels in healthy subjects. N Engl J Med 323: 439–445 [DOI] [PubMed] [Google Scholar]
- Moche M, Shanklin J, Ghoshal A, Lindqvist Y (2003) Azide and acetate complexes plus two iron-depleted crystal structures of the di-iron enzyme delta9 stearoyl-acyl carrier protein desaturase: implications for oxygen activation and catalytic intermediates. J Biol Chem 278: 25072–25080 [DOI] [PubMed] [Google Scholar]
- Murakami Y, Tsuyama M, Kobayashi Y, Kodama H, Iba K (2000) Trienoic fatty acids and plant tolerance of high temperature. Science 287: 476–479 [DOI] [PubMed] [Google Scholar]
- O’Brian MR, Kirshbom PM, Maier RJ (1987) Bacterial heme synthesis is required for expression of the leghemoglobin holoprotein but not the apoprotein in soybean root nodules. Proc Natl Acad Sci USA 84: 8390–8393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Rahman SM, Takagi Y, Kinoshita T (1997) Genetic control of high stearic acid content in seed oil of two soybean mutants. Theor Appl Genet 95: 772–776 [Google Scholar]
- Rahman SM, Takagi Y, Miyamoto K, Kawakita T (1995) High stearic acid soybean mutant induced by X-ray irradiation. Biosci Biotechnol Biochem 59: 922–923 [Google Scholar]
- Raison JK, Roberts JKM, Berry JA (1982) Correlations between the thermal stability of chloroplast (thylakoid) membranes and the composition and fluidity of their polar lipids upon acclimation of the higher plant, Nerium oleander, to growth temperature. Biochim Biophys Acta 688: 218–228 [Google Scholar]
- Rogers SO, Bendich AJ (1994) Extraction of total cellular DNA from plants, algae and fungi. In Gelvin SB, Schilperoort RA, eds, Plant Molecular Biology Manual. Springer, Dordrecht, The Netherlands, pp. 183–190 [Google Scholar]
- Ruddle P, Whetten R, Cardinal A, Upchurch R, Miranda L (2013) Effect of a novel mutation in a Δ9-stearoyl-ACP-desaturase on soybean seed oil composition. Theor Appl Genet 126: 241–249 [DOI] [PubMed] [Google Scholar]
- Ruddle P II, Whetten R, Cardinal A, Upchurch RG, Miranda L (2014) Effect of Δ9-stearoyl-ACP-desaturase-C mutants in a high oleic background on soybean seed oil composition. Theor Appl Genet 127: 349–358 [DOI] [PubMed] [Google Scholar]
- Smirnoff N. (1993) The role of active oxygen in the response of plants to water deficit and desiccation. New Phytol 125: 27–58 [DOI] [PubMed] [Google Scholar]
- Swiderski MR, Zaborowska Z, Legocki AB (2000) Identification of new nodulin cDNAs from yellow lupine by differential display. Plant Sci 151: 75–83 [Google Scholar]
- Wu J, James DW Jr, Dooner HK, Browse J (1994) A mutant of Arabidopsis deficient in the elongation of palmitic acid. Plant Physiol 106: 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Burton JW, Upchurch RG, Whittle E, Shanklin J, Dewey RE (2008) Mutations in a Δ-stearoyl-ACP-desaturase gene are associated with enhanced stearic acid levels in soybean seeds. Crop Sci 48: 2305–2313 [Google Scholar]