Nine morphologically altered new BRI1 mutants were identified from 83 independent TILLING mutations in Arabidopsis.
Abstract
The identification and characterization of a mutational spectrum for a specific protein can help to elucidate its detailed cellular functions. BRASSINOSTEROID INSENSITIVE1 (BRI1), a multidomain transmembrane receptor-like kinase, is a major receptor of brassinosteroids in Arabidopsis (Arabidopsis thaliana). Within the last two decades, over 20 different bri1 mutant alleles have been identified, which helped to determine the significance of each domain within BRI1. To further understand the molecular mechanisms of BRI1, we tried to identify additional alleles via targeted induced local lesions in genomes. Here, we report our identification of 83 new point mutations in BRI1, including nine mutations that exhibit an allelic series of typical bri1 phenotypes, from subtle to severe morphological alterations. We carried out biochemical analyses to investigate possible mechanisms of these mutations in affecting brassinosteroid signaling. A number of interesting mutations have been isolated via this study. For example, bri1-702, the only weak allele identified so far with a mutation in the activation loop, showed reduced autophosphorylation activity. bri1-705, a subtle allele with a mutation in the extracellular portion, disrupts the interaction of BRI1 with its ligand brassinolide and coreceptor BRI1-ASSOCIATED RECEPTOR KINASE1. bri1-706, with a mutation in the extracellular portion, is a subtle defective mutant. Surprisingly, root inhibition analysis indicated that it is largely insensitive to exogenous brassinolide treatment. In this study, we found that bri1-301 possesses kinase activity in vivo, clarifying a previous report arguing that kinase activity may not be necessary for the function of BRI1. These data provide additional insights into our understanding of the early events in the brassinosteroid signaling pathway.
Brassinosteroids (BRs) are known to play critical roles in regulating many aspects of plant growth and development, including germination, cell elongation, flowering time control, reproduction, photomorphogenesis, and skotomorphogenesis (Clouse and Sasse, 1998; Gudesblat and Russinova, 2011; Wang et al., 2012; Zhu et al., 2013). Recent studies revealed that BRs also are involved in many additional physiological processes, such as root meristem maintenance (González-García et al., 2011; Hacham et al., 2011; Vilarrasa-Blasi et al., 2014; Chaiwanon and Wang, 2015; Wei and Li, 2016), stomatal development (Gudesblat et al., 2012; Kim et al., 2012), root hair differentiation (Cheng et al., 2014), leaf angle regulation in monocots (Zhang et al., 2012), shoot branching control (Wang et al., 2013), and organ boundary determination (Bell et al., 2012; Gendron et al., 2012).
In Arabidopsis (Arabidopsis thaliana), BRs are perceived by their major receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) and coreceptor BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1), both of which are single-pass transmembrane Leu-rich repeat receptor-like protein kinases (LRR-RLKs; Li and Chory, 1997; Li et al., 2002; Nam and Li, 2002; Gou et al., 2012). There are 25 total extracellular LRRs in BRI1. Between the 21st and 22nd LRRs, there is an amino acid sequence containing 68 residues that is termed an island. Genetic and structural analyses indicated that the island and the 22nd LRR are involved directly in BR binding. In the absence of BRs, the kinase activity of BRI1 is inhibited by its C terminus (Wang et al., 2005) and by its associated inhibitory protein, named BRI1 KINASE INHIBITOR1 (BKI1; Wang and Chory, 2006). When BRs are present, on the other hand, they can interact directly with the extracellular pocket of BRI1, causing BKI1 to dissociate from the complex (Wang and Chory, 2006; Hothorn et al., 2011; She et al., 2011; Wang et al., 2014). The newly formed BRI1-brassinolide (BL) surface can interact directly with the extracellular residues of BAK1 via several hydrogen bonds (Santiago et al., 2013; Sun et al., 2013). The BRI1-BL-BAK1 complex can then initiate early BR signaling events and activate its downstream signaling cascade (Wang et al., 2008). Genetic and structural analyses demonstrated the essential roles of both BRI1 and BAK1 in controlling BR signal transduction (Clouse et al., 1996; Li and Chory, 1997; Hothorn et al., 2011; She et al., 2011; Gou et al., 2012; Santiago et al., 2013; Sun et al., 2013).
During the past two decades, over 20 unique bri1 alleles have been isolated (Clouse et al., 1996; Li and Chory, 1997; Noguchi et al., 1999; Friedrichsen et al., 2000; Xu et al., 2008; Belkhadir et al., 2010; Shang et al., 2011; Gou et al., 2012). Analyses of these mutants have contributed significantly to our better understanding of BRI1 in regulating plant growth and development (Vert et al., 2005). These alleles share similar phenotypes in various degrees, including dwarfism, dark-green, compact rosette leaves with short petioles, delayed flowering time and senescence, reduced male fertility, and altered photomorphogenesis and skotomorphogenesis (Clouse, 1996; Kwon and Choe, 2005). Most alleles exhibiting the strongest morphological defects contain point mutations in the island or kinase domain (Friedrichsen et al., 2000). BRI1 was first cloned via a map-based strategy (Li and Chory, 1997). Many of the early isolated bri1 alleles are strong alleles with largely reduced male sterility, preventing their use in large-scale genetic transformation assays. Fertile weak bri1 alleles, such as bri1-5 and bri1-9, on the other hand, are excellent genetic tools used for characterizing the entire BR signaling pathway via extragenic modifier screens (Noguchi et al., 1999). For example, several key components regulating BR signal transduction or BR homeostasis were identified via activation-tagging genetic screens using bri1-5 as background, including BRS1 (Li et al., 2001a), BAK1 (Li et al., 2002), BSU1 (Mora-García et al., 2004), BRL1 (Zhou et al., 2004), BEN1 (Yuan et al., 2007), and TCP1 (Guo et al., 2010). EBS1 to EBS7, a series of functionally related proteins mediating endoplasmic reticulum (ER) quality control, were isolated via a suppressor screen from ethyl methanesulfonate (EMS)-mutagenized bri1-9 (Jin et al., 2007, 2009; Su et al., 2011, 2012; Hong et al., 2012; Liu et al., 2015). BES1, a key downstream transcription factor in the BR signaling pathway, was identified via a suppressor screen utilizing an EMS-mutagenized bri1-119 library (Yin et al., 2002). Identification of these key regulatory components using weak bri1 mutants has contributed significantly to our current knowledge of the BR signaling pathway, from BR perception to downstream-responsive gene regulation.
In order to deeply understand the molecular mechanisms of BRI1 in regulating the BR signaling pathway, we analyzed all BRI1 point mutations generated from a targeted induced local lesions in genomes (TILLING) project (McCallum et al., 2000). Among 83 total BRI1 point mutations isolated, we identified nine mutants, including four subtle, one weak, and four strong alleles. For example, bri1-702 is a new weak allele with a mutation in the activation loop of the BRI1 kinase domain. Several previously identified bri1 activation loop point mutants are all strong alleles. Therefore, bri1-702 represents the first weak allele bearing a point mutation in the activation loop. bri1-705 is a new subtle allele containing a point mutation in the extracellular portion. Structural analysis suggests that this mutation likely disrupts the formation of hydrogen bonds among BRI1, BL, and BAK1. A third interesting mutant is bri1-706, containing a point mutation in the eighth LRR of the extracellular portion of BRI1. bri1-706 shows a subtle aerial part defective phenotype compared with wild-type plants, but its root growth is largely insensitive to exogenously applied BL. The new mutants identified from these studies can be used as genetic tools to further dissect the functions of BRI1 as well as utilized as references to study the biological functions of other RLKs in plants.
RESULTS
A Series of New bri1 Alleles Were Identified via TILLING Analyses
BRI1, the major receptor of BRs in Arabidopsis, contains multiple functional domains (Li and Chory, 1997; Friedrichsen et al., 2000). Mutational analyses have helped to characterize the biological significance of each of these domains. To further dissect the roles of BRI1 in the BR signaling pathway, we tried to identify all possible BRI1 mutations and identify the mutants with altered phenotypes.
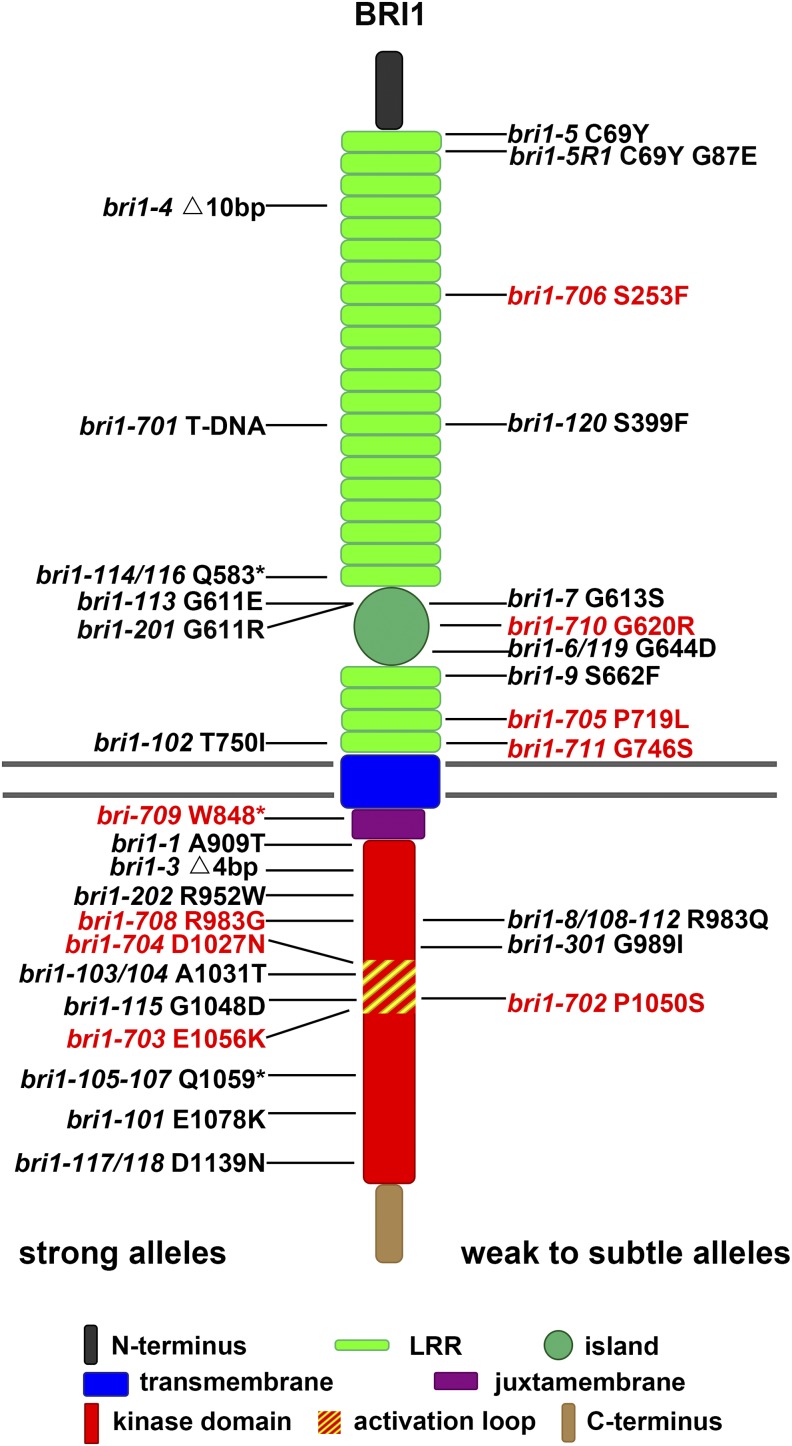
TILLING is a reverse genetic method using a high-throughput screening technology to detect point mutations from an EMS-mutagenized library (McCallum et al., 2000). Using this approach, a large number of mutations can be isolated for a specific target gene. TILLING has been used to generate mutations for a number of important genes, including BRI1 in Arabidopsis (Till et al., 2003). All 87 BRI1 TILLING mutations were obtained from the Arabidopsis Biological Resource Center, and 83 mutations were confirmed by a derived cleaved-amplified polymorphic sequence (dCAPS) method (Neff et al., 1998). These mutations are distributed throughout the entire BRI1 sequence, especially in LRR1 to LRR10 of its extracellular portion, island, and kinase domain (Supplemental Fig. S1). These mutations were generated in the Columbia-0 (Col-0) background containing an er mutation. A high dose of EMS could introduce mutations in other genes that could result in BRI1-unrelated phenotypes. In order to remove those additional mutations, we backcrossed the mutants with Col-0 for at least three generations. Offspring plants containing BRI1 mutations were selected using the dCAPS approach (Neff et al., 1998). After backcrossing three times, most mutations were found to exhibit no morphological differences from wild-type plants. Only nine mutations consistently exhibit typical bri1-like phenotypes. These mutants were named in the order of their identification, from bri1-702 to bri1-711 (Fig. 1; Table I).
Figure 1.
Diagram showing all bri1 mutants identified thus far, including nine new alleles identified by this study. New mutants are labeled in red, and known mutants are labeled in black. Known bri1 mutants include bri1-1 (Clouse et al., 1996; Friedrichsen et al., 2000), bri1-3, bri1-4, bri1-5, bri1-6, bri1-7, bri1-8, bri1-9 (Noguchi et al., 1999), bri1-5R1 (Belkhadir et al., 2010), bri1-101, bri1-102, bri1-103/104, bri1-105-107, bri1-108-112, bri1-113, bri1-114/116, bri1-115, bri1-117/118, bri1-119 (Li and Chory, 1997; Friedrichsen et al., 2000), bri1-120 (Shang et al., 2011), bri1-201, bri1-202 (Domagalska et al., 2007), bri1-301 (Xu et al., 2008), and bri1-701 (Gou et al., 2012).
Table I. bri1 alleles (for references, see Fig. 1).
| Allele | Base Pair Change | Accession | Allelic Strength | Possible Mechanism |
|---|---|---|---|---|
| bri1-1 | G2725A | Col-0 | Strong | Unknown |
| bri1-3 | 4-bp deletion after 2,745 | Wassilewskija-2 (WS2) | Strong | Premature stop |
| bri1-4 | 10-bp deletion after 459 | WS2 | Strong | Premature stop |
| bri1-5 | G206A | WS2 | Weak | ER retention |
| bri1-5R1 | G260A | bri1-5 | Weak | Partially restore ER retention of bri1-5 |
| bri1-6/119 | G1931A | Enkheim-2 (En-2) | Weak | Unknown |
| bri1-7 | G1838A | WS2 | Weak | Unknown |
| bri1-8/108-112 | G2948A | WS2/Col-0 | Intermediate | Autophosphorylation cannot be detected in vitro |
| bri1-9 | C1985T | WS2 | Weak | ER retention |
| bri1-101 | G3232A | Col-0 | Strong | Autophosphorylation cannot be detected in vitro |
| bri1-102 | C2249T | Col-0 | Strong | Unknown |
| bri1-103/104 | G3091A | Col-0 | Strong | Unknown |
| bri1-105-107 | C3175T | Col-0 | Strong | Unknown |
| bri1-113 | G1832A | Col-0 | Strong | Unknown |
| bri1-114/116 | C1747T | Col-0 | Strong | Unknown |
| bri1-115 | G3143A | Col-0 | Strong | Unknown |
| bri1-117/118 | G3415A | Col-0 | Strong | Unknown |
| bri1-120 | T1196C | Landsberg erecta | Weak | Unknown |
| bri1-201 | G1831A | WS2 | Strong | Unknown |
| bri1-202 | C2854T | WS2 | Strong | Unknown |
| bri1-301 | GG2965/6AT | Col-0 | Weak | Autophosphorylation of bri1-301 cannot be detected in vitro, but transphosphorylation of BAK1 can be detected in vivo |
| bri1-701 | T-DNA insertion | Col-0 | Strong | Knockout of BRI1 |
| bri1-702 | C3148T | Col-0 | Weak | Reduced autophosphorylation in vitro |
| bri1-703 | G3166A | Col-0 | Strong | Autophosphorylation cannot be detected in vitro |
| bri1-704 | G3079A | Col-0 | Strong | Autophosphorylation cannot be detected in vitro |
| bri1-705 | C2156T | Col-0 | Subtle | Disrupt the formation of hydrogen bonds among BRI1, BL, and BAK1 |
| bri1-706 | C758T | Col-0 | Subtle | Unknown |
| bri1-708 | C2947G | Col-0 | Strong | Autophosphorylation cannot be detected in vitro |
| bri1-709 | G2543A | Col-0 | Strong | Premature stop |
| bri1-710 | G1858A | Col-0 | Subtle | Unknown |
| bri1-711 | G2236A | Col-0 | Subtle | Unknown |
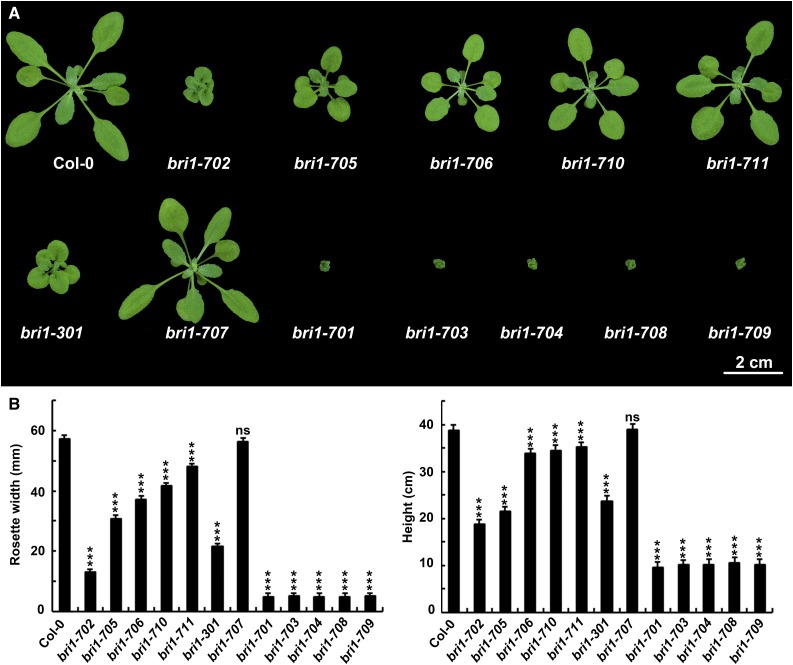
To further confirm their bri1-related phenotypes, we carried out two additional backcrosses for the nine new mutants isolated via TILLING. In the F2 generation of the third backcrossing, the phenotypes of bri1-702, bri1-705, bri1-706, bri1-710, and bri1-711 were segregated out as single recessive alleles with a ratio close to 3:1. However, for strong alleles, the ratio is less than 4:1, which might be caused by lower germination rates, as observed previously (Steber and McCourt, 2001). Thus, the phenotype appears to be caused by a single locus for all nine different mutants. bri1-702 shows a weak bri1 phenotype similar to bri1-5, bri1-9, and bri1-301 (Fig. 2A; Noguchi et al., 1999; Xu et al., 2008). In comparison, bri1-705, bri1-706, bri1-710, and bri1-711 show even weaker phenotypes than bri1-702, which, therefore, were named as subtle alleles in this study. On the other hand, bri1-703, bri1-704, bri1-708, and bri1-709 show severe phenotypes similar to a previously reported bri1 null allele, bri1-701 (Fig. 2A; Gou et al., 2012). Interestingly, bri1-707 and bri1-301 both have mutations in Gly-989, but with different amino acid substitutions. As a consequence, bri1-301 shows a weak bri1 phenotype (Xu et al., 2008), whereas bri1-707 does not show any defective phenotypes. Similarly, bri1-708 and bri1-8/108-112 bear different mutations at the same residue. While bri1-8/108-112 are intermediate bri1 mutants (Noguchi et al., 1999), bri1-708 shows a null bri1 phenotype (Fig. 2A). Statistically, all nine of these new bri1 mutants show significantly reduced growth (Fig. 2B).
Figure 2.
New bri1 mutants identified in this study and their phenotypes. A, Phenotypes of mutants and wild-type seedlings. The photograph was taken 3 weeks after germination. B, Rosette width and height of the mutants and wild-type seedlings. Rosette width was measured 3 weeks after germination. Height was measured when growth was completely finished. Student’s t tests were carried out to show the significance between each allele and Col-0. Each data point shown is the average and sd (n ≥ 20). ***, P < 0.001; and ns, not significant.
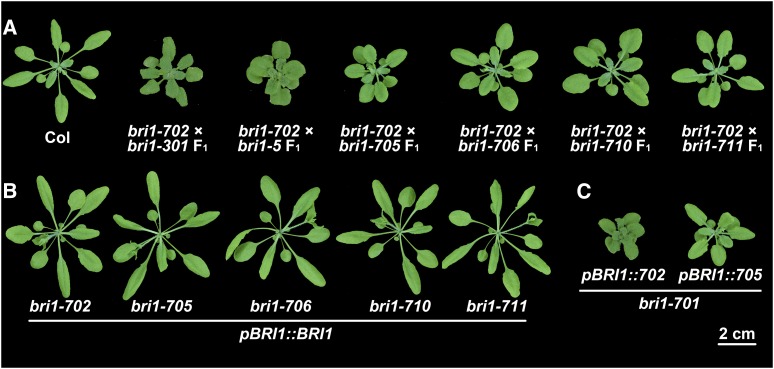
To exclude the possibility that the phenotypes are caused by mutations of other genes closely linked to BRI1, which are not easily segregated out by backcrossing, we crossed these mutants with a number of known bri1 weak alleles. If the phenotype is caused by a mutation in a different gene, the F1 seedlings should show a wild-type phenotype due to functional complementation. All the obtained F1 seedlings show bri1-like phenotypes (Fig. 3A). In addition, we transformed the wild-type BRI1 sequence driven by its native promoter into these nine different bri1 mutant alleles. The transgenic plants showed BRI1-overexpressing phenotypes in the wild-type background (Fig. 3B; Wang et al., 2001; Nam and Li, 2002). We next transformed the genomic DNA sequences (containing the BRI1 promoter and its coding sequences) of bri1-702 and bri1-705 to the null mutant, bri1-701. The resulting transgenic plants resembled the phenotypes of bri1-702 and bri1-705, respectively (Fig. 3C). These genetic and transgenic results demonstrated that all nine new mutants are truly allelic to bri1.
Figure 3.
Complementation analyses confirm that all mutants are alleles of bri1. A, The phenotypes of new bri1 weak or subtle alleles cannot be complemented by other bri1 weak alleles. B, BRI1 driven by its native promoter can completely rescue the phenotypes of new bri1 weak or subtle alleles. C, The phenotypes of bri1-702 and bri1-705 can be recapitulated by expressing their corresponding genomic DNA in a null BRI1 mutant allele, bri1-701. Photographs were taken 4 weeks after germination.
Characterization of Newly Identified bri1 Mutants
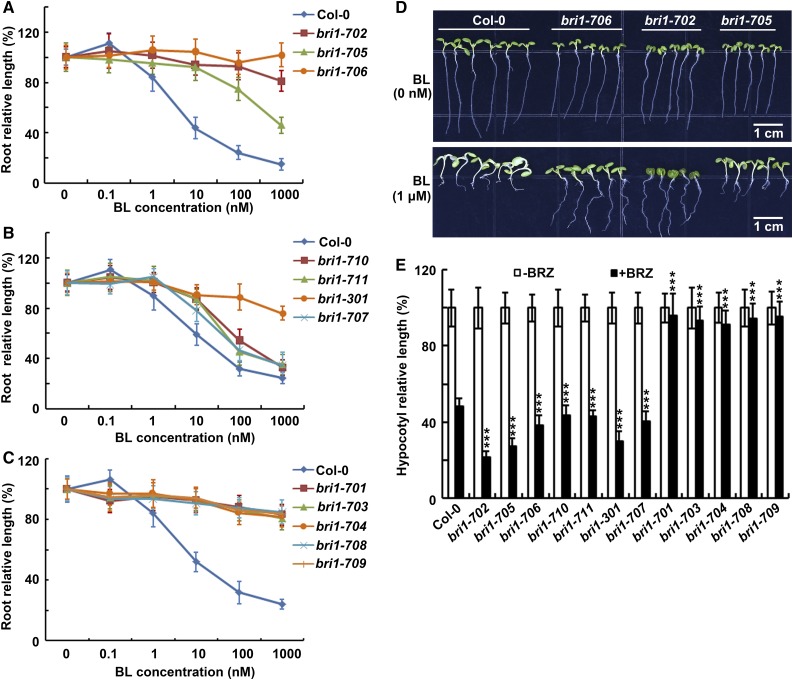
Root inhibition analysis indicated that bri1-702 and bri1-705 are less sensitive to exogenously applied BL (Fig. 4A). It is interesting that the rosette of bri1-706 is significantly bigger than those of bri1-702 and bri1-705 (Fig. 2A), but its root growth is almost completely insensitive to BL treatment, similar to that of null bri1 mutants (Fig. 4, A and D). bri1-707, bri1-710, and bri1-711 show slightly reduced sensitivity to BL compared with Col-0 (Fig. 4B). All new bri1 strong mutants, including bri1-703, bri1-704, bri1-708, and bri1-709, are insensitive to BL, similar to bri1-701 (Fig. 4C; Gou et al., 2012). Previous studies indicated that bri1 weak mutants show reduced sensitivity to BL but increased sensitivity to brassinazole (BRZ), a specific BR biosynthetic inhibitor (Asami et al., 2000; Yin et al., 2002). Therefore, we tested the sensitivity of these new bri1 alleles to BRZ under darkness. As expected, the hypocotyl growth of weak and subtle alleles exhibited hypersensitivity to BRZ, whereas all strong alleles are insensitive to the BRZ treatment (Fig. 4E). These data indicated the new bri1 alleles exhibit typical bri1 phenotypes and reduced sensitivity to exogenously applied BL.
Figure 4.
All mutants show either insensitivity or reduced sensitivity to exogenously applied BL. A to C, Responses of root growth to treatment with BL. Seven-day-old seedlings were grown on one-half-strength Murashige and Skoog (MS) medium supplemented with different concentrations of BL. D, bri1-706 is almost completely insensitive to BL. Seven-day-old seedlings were grown on one-half-strength MS medium supplemented with or without 1 μm BL. The seedlings were grown under long-day light conditions in a 22°C growth chamber for A to D. E, Responses of hypocotyl growth to treatment with BRZ. Four-day-old seedlings were grown on one-half-strength MS medium with or without BRZ application under darkness in a 22°C growth chamber. Student’s t tests were performed to show significance between each allele and Col-0. Each data point represents the average and sd (n ≥ 20). ***, P < 0.001.
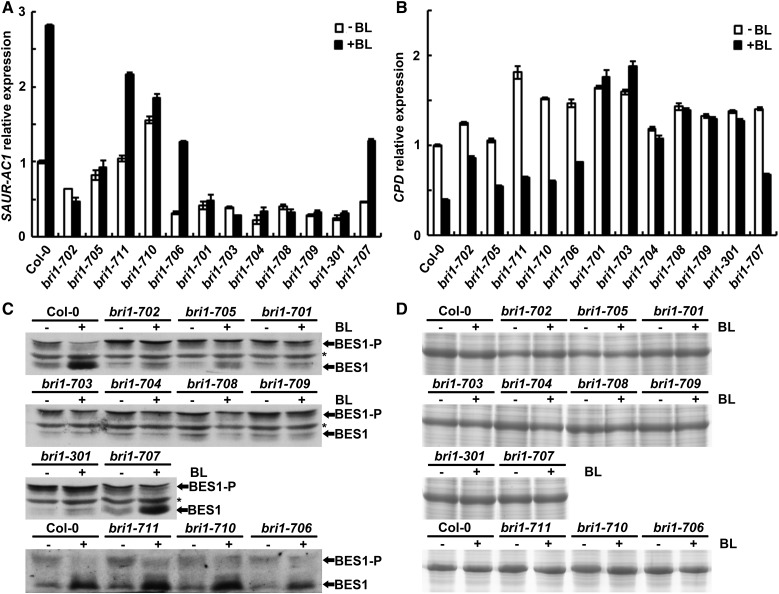
Previous studies also indicated that, upon application of exogenous BL, the expression of the BR biosynthesis gene CPD is down-regulated and the expression of a BR response gene, SAUR-AC1, is up-regulated (Li et al., 2001b; Yin et al., 2005). In addition, the phosphorylation status of BES1 also can be used as an indicator of a downstream signaling response to the BL treatment (Yin et al., 2002; Sun et al., 2010; Yu et al., 2011). Quantitative real-time PCR was performed to examine the expression levels of CPD and SAUR-AC1 in the wild type and newly identified bri1 mutants treated with or without 1 μm BL (Fig. 5, A and B). Similar to previous reports, the expression of SAUR-AC1 was increased dramatically in Col-0 but had no significant change in bri1-701 after the BL treatment. The expression of SAUR-AC1 in the newly identified weak and subtle bri1 alleles exhibited reduced sensitivity to the BL treatment, whereas in newly identified strong bri1 alleles, the expression of SAUR-AC1 is insensitive to treatment with BL and remained at a lower level of expression with or without the treatment (Fig. 5A). Similar to the SAUR-AC1 response, the CPD responses of bri1-702 and bri1-705 showed reduced sensitivity to the BL treatment (Fig. 5B). Although the expression levels of CPD in BR-treated bri1-706, bri1-707, bri1-710, and bri1-711 decreased relatively to the same degree compared with Col-0, basal expression levels of CPD without BL treatment were higher than in the wild type, suggesting that BR signaling is indeed partially altered in these mutants (Fig. 5B).
Figure 5.
Responses of downstream BR signaling components of the new mutants to BL. A and B, Transcriptional responses of SAUR-AC1 and CPD to treatment with BL. ACTIN2 was used as a reference gene. Error bars represent sd (n = 3). C, Responses of BES1 phosphorylation status to treatment with BL. Total protein was extracted and immunoblotted with an anti-BES1 antibody. D, Coomassie Blue staining showing equal loading between each pair of samples in C. Seven-day-old long-day-grown seedlings treated with or without 1 μm BL for 4 h were used. −, Without BL treatment; +, with BL treatment; *, nonspecific band. BES1-P, Phosphorylated BES1; BES1, unphosphorylated BES1.
We next investigated the phosphorylation status of BES1 in the wild type and in the newly identified bri1 mutants with or without 1 μm BL application (Fig. 5, C and D). In Col-0, two almost equal intensity bands, representing phosphorylated and unphosphorylated forms of BES1, were detected without the BL treatment. After the BL treatment, phosphorylated BES1 was reduced greatly and unphosphorylated BES1 was accumulated significantly in Col-0, suggesting that the BR signaling pathway is functional. In bri1-702 and bri1-705, only a small amount of unphosphorylated BES1 was detected after the BL treatment. In the strong bri1 alleles bri1-701, bri1-703, bri1-704, bri1-708, and bri1-709, the accumulation of unphosphorylated BES1 could not be easily observed after the treatment with BL. In subtle alleles, bri1-711, bri1-710, and bri1-706, the accumulation of unphosphorylated BES1 is similar to that in Col-0 upon treatment with 1 μm BL. It is intriguing that bri1-301 and bri1-707 contain different missense mutations at the same residue, but the BES1 response to BL is totally different. bri1-301 remains partially sensitive, while bri1-707 is completely sensitive to the treatment with BL (Fig. 5, C and D). These molecular and biochemical results confirmed that bri1-706, bri1-710, and bri1-711 are subtle alleles of bri1.
bri1-702, with a Point Mutation in the Activation Loop of BRI1, Shows Reduced Autophosphorylation Activity
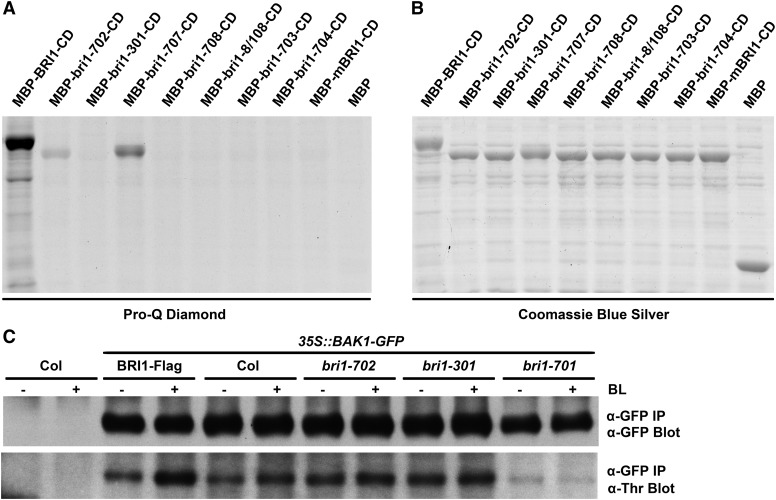
Autophosphorylation, especially the pocket opening of the activation loop, is a common activation mechanism for protein kinases (Oh et al., 2000). In order to understand whether the mutations in the newly identified bri1 mutants affect the kinase activity of BRI1, we used the phospho-amino acid stain Pro-Q Diamond to detect the autophosphorylation activity of the Escherichia coli-expressed mutant BRI1 cytoplasmic domain (CD) fused with a maltose-binding protein (MBP-BRI1-CD; Fig. 6, A and B). After sequential staining with the Pro-Q Diamond and Coomassie Blue Silver, the E. coli cell lysate, which contains the expressed bri1-301-CD, exhibited an undetectable kinase activity in vitro, similar to the results from a previous report (Xu et al., 2008). However, bri1-707-CD, which bears a different substitution at the same residue with bri1-301-CD, showed reduced but detectable autophosphorylation activity (Fig. 6, A and B). Interestingly, compared with the undetectable kinase activities of other intracellular bri1 mutants, bri1-702, a weak allele with a mutation in the activation loop, still shows autophosphorylation activity, although it is partially reduced compared with the wild type (Fig. 6, A and B). This result indicated that mutation of bri1-702 leads to a reduced kinase activity of BRI1 and, therefore, attenuated BR signal transduction.
Figure 6.
In vitro and in vivo phosphorylation analyses of BRI1, bri1, and BAK1. A, Pro-Q Diamond analyses of BRI1 and various bri1 cytoplasmic domains. The recombinant proteins were expressed in E. coli, and total protein extracts were separated by 10% SDS-PAGE. The gel was stained by Pro-Q Diamond. B, Coomassie Blue Silver staining to show equal loading of the samples. mBRI1 contains a mutation within the ATP-binding site (Lys-911Glu). C, In vivo phosphorylation analyses of BAK1-GFP from various bri1 mutants in response to treatment with BL. Membrane proteins were extracted from 10-d-old long-day-grown seedlings treated with or without 100 nm BL for 1.5 h. BAK1-GFP proteins from different bri1 alleles were immunoprecipitated (IP) with an anti-GFP antibody. The phosphorylation levels of BAK1 were detected with a phospho-Thr antibody. The immunoprecipitated BAK1-GFP was immunoblotted with an anti-GFP antibody to show equal loading. −, Without BL treatment; +, with BL treatment.
To confirm these results in vivo, we generated transgenic plants overexpressing BAK1-GFP in Col-0 and various genotypic backgrounds. A phospho-Thr antibody was used to detect the phosphorylation on Thr residues of immunoprecipitated BAK1-GFP from different backgrounds applied with or without BL (Fig. 6C). Similar to a previous report (Wang et al., 2008), the phosphorylation level of BAK1 was strongly enhanced by the BL treatment or by overexpressing BRI1 in Col-0. The phosphorylation of BAK1 was reduced to an almost undetectable level and exhibited no response to BL treatment in bri1 null alleles. In bri1-702 and bri1-301, the phosphorylation level of BAK1 was induced slightly by exogenously applied BL. Interestingly, without BL, the phosphorylation level of BAK1 was much higher in these two weak alleles than even in the wild type (Fig. 6C). These results suggested that bri1-702, the only weak allele in the activation loop identified so far, exhibited not only a reduced autophosphorylation activity of BRI1 in vitro but also a reduced sensitivity of the BAK1 phosphorylation response to exogenously applied BL in vivo. Even though in vitro bri1-301 autophosphorylation could not be detected, as reported previously (Xu et al., 2008), BAK1 phosphorylation in the bri1-301 background is much higher than that in bri1-701 and is induced by the addition of BL. This result suggests that bri1-301 is indeed partially functional toward its substrate in vivo, clarifying a previous argument regarding whether the kinase activity of BRI1 is even critical to the BR signaling transduction (Xu et al., 2008).
Overexpression of mBAK1 Failed to Generate a Dominant Negative Phenotype in bri1-705
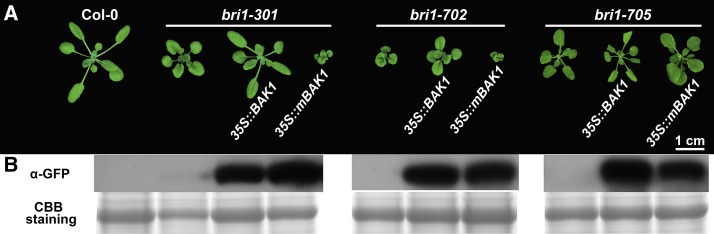
Structural analysis suggested that the extracellular portions of BRI1 and BAK1 can form a BL-induced complex (Santiago et al., 2013; Sun et al., 2013). Several crucial surfaces for the interaction of BRI1 and BAK1 also were identified, among which were two residues of BRI1 (Thr-726 and Met-727) associated directly with BAK1 via hydrophobic interactions (Santiago et al., 2013; Sun et al., 2013). Coincidentally, bri1-705, a new subtle allele with a typical BR-deficient phenotype, contained a nearby mutation at Pro-719 to Leu, which may affect the interaction between BRI1 and BAK1. In order to test this hypothesis, a series of transgenic analyses were carried out. Based on previous studies, overexpression of BAK1 can suppress the phenotype of bri1-5, while overexpression of a kinase-inactive version of BAK1 (mBAK1), in which a point mutation is introduced on a conserved Lys residue within the ATP-binding site, results in a dominant negative phenotype (Li et al., 2002; Gou et al., 2012). Both genetic suppression and dominant negative phenotypes rely on interactions between the extracellular portion of BRI1 and BAK1. If a mutation in BRI1 disrupts the interaction with BAK1 or mBAK1, overexpression of mBAK1 may not dramatically alter the phenotype of the bri1 mutant. BAK1-GFP and mBAK1-GFP were transformed to a series of weak bri1 alleles. Consistent with previous reports (Li et al., 2002), the defective phenotype of bri1-5 was partially rescued by overexpressing BAK1 but greatly enhanced by overexpressing mBAK1 (Supplemental Fig. S2). Overexpression of BAK1 or mBAK1 in bri1-301 and bri1-702 resulted in phenotypes similar to those in bri1-5 (Fig. 7; Li et al., 2002). However, overexpression of BAK1 or mBAK1 failed to dramatically alter the phenotype of bri1-705 (Fig. 7), suggesting that the mutation in bri1-705 may interfere with the interaction between bri1-705 and BAK1 or mBAK1. These genetic data demonstrated that the BL-dependent extracellular heterodimerization of BRI1 and BAK1 is an important step in the initiation of BR signaling.
Figure 7.
Overexpression of mBAK1 in bri1-301 and bri1-702 but not in bri1-705 resulted in dominant negative phenotypes. A, Phenotypes of the transgenic plants and their backgrounds. The photograph was taken 3 weeks after germination. B, Western-blot analyses showing the expressed BAK1-GFP and mBAK1-GFP fusion proteins in transgenic plants. Total proteins from rosette leaves were extracted and immunoblotted with an anti-GFP antibody. mBAK1 contains a mutation within the ATP-binding site (Lys-317Glu). CBB, Coomassie Blue Silver.
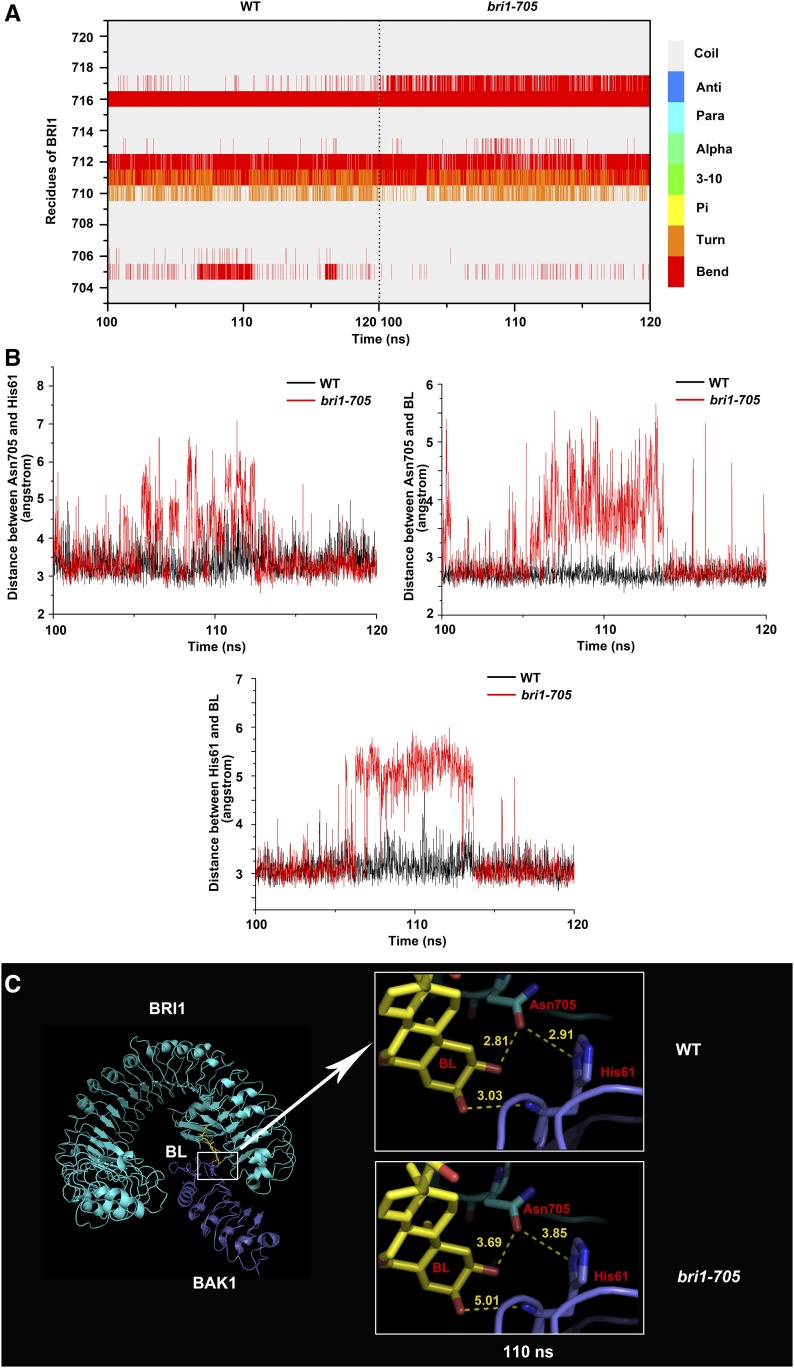
To further confirm this notion, we performed a molecular dynamic (MD) simulation analysis of the BRI1-BL-BAK1 structure. The fluctuation of 120-ns MD trajectories obtained from two simulation systems (the wild-type complex and the BRI1-Pro-719Leu complex) was monitored using the root mean square deviation of the backbone atoms of BRI1, BAK1, and BL (Supplemental Fig. S3). It can be seen clearly that the root mean square deviation values of both BRI1-Pro-719Leu and wild-type BRI1 oscillate at ∼4 Å after 20 ns, whereas the structural fluctuations of BAK1 and BL are significantly higher in the BRI1-Pro-719Leu complex compared with the wild-type complex, implying that the Pro-719Leu mutation of BRI1 may destabilize the interactions of BRI1-BL and BRI1-BAK1. The simulation analysis also showed that the mutation leads to significant conformation alterations of Arg-717, which precedes Pro-719 and has the propensity to change from a random coil to a β-turn in the mutant complex. Meanwhile, the β-turn structure formed by Asn-705 and Ile-706 in the wild-type complex seems to be destabilized and prone to show a random coil conformation in the mutant complex (Fig. 8A). Analysis of the interaction interfaces revealed that the hydrogen bonding of the residue Asn-705, which bridges the side chain of His-61 from BAK1 and the hydroxyl group of BL, was interrupted continuously in the BRI1-Pro-719Leu complex during the last 20-ns simulation (Fig. 8B). Additionally, the hydrogen bond between the main-chain oxygen atom of His-61 from BAK1 and the hydroxyl group of BL also breaks up more frequently in the BRI1-Pro-719Leu complex compared with the wild-type complex (Fig. 8B). Taken together, these results suggest that the mutation in bri1-705 results in significant local conformational changes involving the residue Asn-705 that interacts with both BAK1 and BL, thus destabilizing the pairwise interactions among BRI1, BAK1, and BL (Fig. 8C).
Figure 8.
Conformational changes monitored during the last 20-ns MD simulation. A, Time evolution of the secondary structures of residues 703 to 721 of BRI1 in the wild-type (WT) complex (left) and the mutant complex (right). Compared with the wild-type complex, the residue Arg-717 preceding Pro-719 has the propensity to change from a random coil to a β-turn, while Asn-705 and Ile-706 seem to be destabilized and prone to adopt a random coil conformation in the mutant complex. B, Monitoring time evolution of the distances between the atom OD1 of BRI1-Asn 705 and ND1 of BAK1-His-61 (top left), atom OD1 of BRI1-Asn-705 and O02 from C2 of BL (top right), and atom ND1 of BAK1-His-61 and O03 from C3 of BL (bottom). The distances in the wild-type complex and the mutant complex are colored in black and red, respectively. C, Interaction modes of BRI1-Asn-705 (cyan), BAK1-His-61 (purple), and BL (yellow). At 110 ns, as shown in the top right inset, Asn-705, His-61, and BL form pairwise hydrogen bonds, indicated by the close distances of 2.91, 2.81, and 3.03 Å between Asn-705, His-61, and BL, respectively. However, in the bri1-705 mutant complex, as shown in the bottom right inset, these hydrogen bonds are disrupted, as evidenced by the much larger pairwise distances between Asn-705, His-61, and BL.
bri1 Point Mutants Exhibiting Defective Phenotypes All Bear Point Mutations within Conserved Residues among BRI1s and BRLs
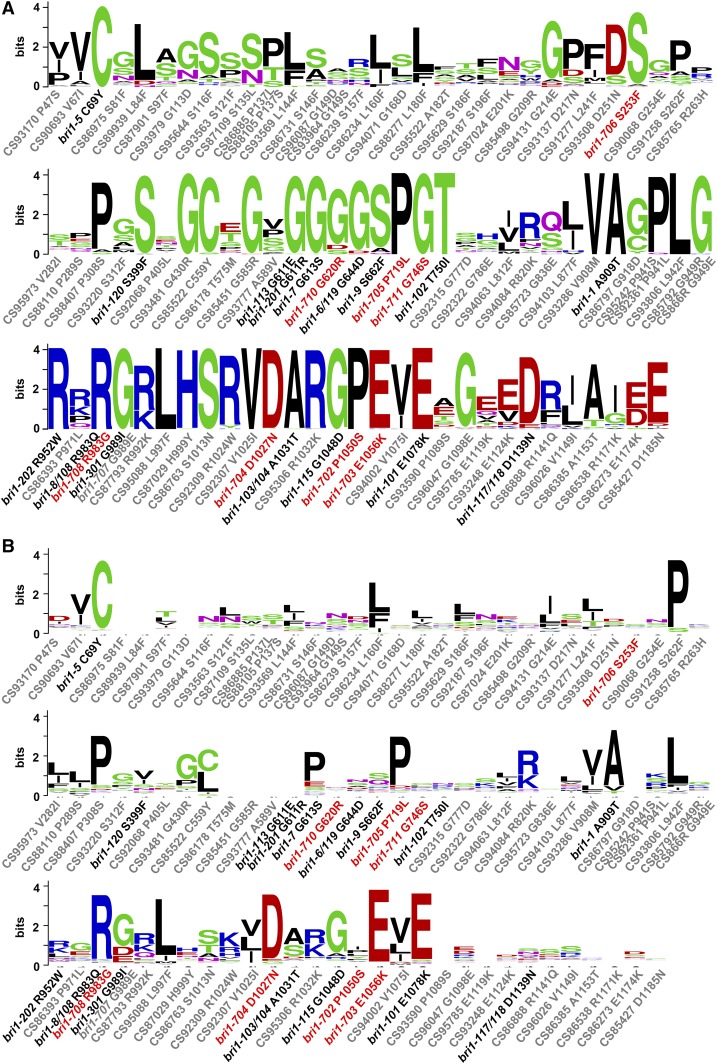
Eighty-three missense mutations of BRI1 were isolated in this study, among which nine point mutations resulted in typical bri1 phenotypes, including one mutation that generated a premature stop codon, and 74 point mutations did not show any significant phenotypes in normal laboratory growing conditions. There were 16 different bri1 point mutants identified in previous reports (premature stop mutants are not included). Altogether, 98 point mutations of BRI1 have been found so far. We were curious whether these 24 bri1 point mutants (16 previously identified plus eight newly identified) with mutant phenotypes all contain point mutations in conserved residues. We thus carried out a sequence alignment among 70 BRI1s or BRLs from 18 sequenced flowering plant species (Fig. 9A). As expected, all of these mutants are mutated at conserved residues, but their phenotypes are not positively related to the degree of conservation. Surprisingly, not all mutations of conserved residues exhibit defective phenotypes (Fig. 9A). As BRI1 is a typical LRR-RLK with multiple functional domains, we also performed sequence analysis among all LRR-RLKs from Arabidopsis (Fig. 9B). These results indicated that not all mutation residues of 24 bri1 mutants are conserved among all LRR-RLKs. These residues may be particularly important for specific BRI1 functions that have been evolutionarily selected. We also found that not all mutations of conserved residues among all LRR-RLKs exhibit defective phenotypes (Fig. 9B).
Figure 9.
Sequence analyses of all 98 BRI1 point mutations. A, Sequence analyses among 70 BRI1s or BRLs from 18 sequenced flowering plant species. Mutants showing defective phenotypes all result from mutations of conserved residues. But not all mutations in the conserved residues show defective phenotypes. B, Sequence analyses among all 223 LRR-RLKs of Arabidopsis. Not all mutation sites from mutants with defective phenotypes are conserved, and vice versa. Only the amino acid sites with mutations are shown. New mutants from this study are labeled in red. Previously known mutants are labeled in black. New mutations without significant phenotypes are labeled in gray. The size of the logo is positively correlated to the degree of conservation.
DISCUSSION
The activation loop is a crucial region for protein kinase activity (Oh et al., 2000). Sequence analysis indicated that most residues in the activation loop are extremely conserved in 70 BRI1s/BRLs obtained from 18 sequenced flowering plant species (Supplemental Fig. S4A). Besides, there are several residues in the activation loop of BRI1 that also are conserved in all Arabidopsis LRR-RLKs (Supplemental Fig. S4B). Two new bri1 strong alleles, bri1-703 and bri1-704, contain mutations in the last and first residues of the BRI1 activation loop, respectively, and both of them are extremely conserved not only in BRI1s or BRLs but also in all other Arabidopsis LRR-RLKs. Two previously reported alleles, bri1-103/104 and bri1-115, with mutations on extremely conserved residues among BRI1s or BRLs but less conserved among all other LRR-RLKs, also show strong bri1 phenotypes (Supplemental Fig. S4; Li and Chory, 1997; Friedrichsen et al., 2000). We found that bri1-702, the only weak allele in the activation loop of BRI1, contains a mutation in Pro-1050 that is extremely conserved in BRI1s or BRLs but not conserved in all other LRR-RLKs (Supplemental Fig. S4). However, there also is a mutation found via TILLING in Arg-1032Lys that has the same degree of conservation with bri1-103/104 among BRI1s or BRLs but showed no defective phenotypes, possibly because it is mutated to Lys, which shows a similar degree of conservation with Arg in other LRR-RLKs (Supplemental Fig. S4). Among all Arabidopsis LRR-RLKs, several residues mutated in bri1 alleles are not even conserved. We suggest that these mutation residues may be related to the specificity of the BRI1-mediated signaling pathway (Fig. 9B).
In previous studies, the importance of the BRI1 kinase activity was questioned because bri1-301-KD showed no kinase activity in vitro, either via an autophosphorylation assay or an analysis of its phosphorylation activity toward BAK1, but exhibits a weak bri1 phenotype (Xu et al., 2008). In our studies, the bri1-301-CD fusion protein extracted from E. coli also could not be stained by the phosphoprotein Pro-Q Diamond (Fig. 6, A and B). However, we noticed that, unlike in a bri1 null allele, the BAK1 phosphorylation level in bri1-301 is even higher than that of the wild type as detected by immunoblotting using a phospho-Thr antibody (Fig. 6C). This is possibly caused by higher endogenous levels of BL in bri1-301, as was found in other bri1 mutants (Noguchi et al., 1999). The phosphorylation level of BAK1 in bri1-301 can be induced significantly by BL in vivo (Fig. 6C). This result indicated that bri1-301-CD possesses kinase activity for its substrate BAK1 in vivo. The undetectable autophosphorylation of bri1-301-CD in vitro may be caused by protein misfolding when expressed in E. coli.
In this study, we also have some interesting findings from analyzing new bri1 mutant alleles. For example, bri1-707 (Gly-989Glu, Col-0 accession) contains the same mutation as bri1-301 (Gly-989Ile, Col-0 accession), but with a different amino acid substitution. As a result, bri1-707 shows a wild-type-like phenotype (Fig. 2). Actually, the change of Gly to Glu is predicted to substitute a larger side chain than Gly to Ile because of an additional carboxyl group in Glu. Similarly, the new strong allele bri1-708 (Arg-983Gly, Col-0 accession) also bears a point mutation at the same residue as bri1-8 (Arg-983Gln, WS2 accession) and bri1-108-112 (Arg-983Gln, Col-0 accession). However, the Arg-to-Gly substitution causes a greater phenotypical alteration than the Arg-to-Gln substitution. These results indicated that the mechanisms of point mutation are complicated and may involve structural changes.
We identified an extracellular subtle mutant, bri1-705, which has an amino acid substitution close to the BRI1-BAK1 interaction surface. Unlike other weak alleles, such as bri1-5, bri1-702, and bri1-301, overexpression of mBAK1 in bri1-705 failed to generate a dominant negative phenotype (Fig. 7). This result suggested that the mutation in bri1-705 may affect the interaction between the extracellular portions of BRI1 and BAK1. This is further supported by the MD simulation results, indicating that the mutation in bri1-705 results in significant local conformational changes, causing the disruption of three pairs of hydrogen bonds formed between BRI1-Asn-705 and BAK1-His-61, BRI1-Asn-705 and BL, and BAK1-His-61 and BL. Alterations of this hydrogen bonding network could potentially destabilize the pairwise interactions among BRI1, BAK1, and BL and lead to decreased binding affinity of the BRI1-BAK1 extracellular portions. Our data, therefore, provide additional genetic and structural evidence for the crucial role of BL-dependent BRI1-BAK1 heterodimerization in the early events of BR signaling.
During our study, we identified another surprising bri1 mutant, bri1-706. bri1-706 only shows a subtle morphological phenotype compared with other weak alleles isolated so far (Fig. 2). However, root inhibition and hypocotyl promotion assays showed that bri1-706 is greatly insensitive to treatment with BL (Fig. 4A; Supplemental Fig. S5A). When pBRI1::BRI1 was introduced in bri1-706, both the phenotype and insensitivity to BL were completely restored (Fig. 3B; Supplemental Fig. S5B). Endo H analyses indicated that bri1-706 proteins are partially retained in ER in 10-d-old seedlings, in which roots and shoots of the seedlings showed no significant differences (Supplemental Figs. S5, C and D, and S6). But CPD down-regulation and unphosphorylated BES1 accumulation in response to exogenous BL in bri1-706 are similar to those of wild-type seedlings (Supplemental Fig. S5, E–G). To our knowledge, this is the first bri1 mutant showing that phenotypic severity is not positively correlated with the degree of insensitivity to exogenous BL application. These data suggest that the specific mutation found in bri1-706 may have destroyed the interaction of bri1-706 with an unknown protein that may regulate a branch of the BR signaling pathway. Detailed molecular mechanisms regarding how bri1-706 affects root growth sensitivity to BL need to be investigated further in the near future.
We identified a strong allele, bri1-709, in which a point mutation results in a premature stop codon within the juxtamembrane region (Supplemental Fig. S7A). Interestingly, a specific band that is smaller than that in Col-0 can be detected by an immunoblot assay using total proteins from bri1-709 plants with an anti-BRI1 antibody (Supplemental Fig. S7B). This result suggests that the early stop codon of bri1-709 did not disrupt the protein transport of BRI1 and that a partial BRI1 protein without the kinase domain and the C terminus is expressed in bri1-709. This mutant can be used for future genetic and biochemical analyses.
In previous studies, two extracellular weak alleles, bri1-5 and bri1-9, were found to be retained in the ER, because the mutational BRI1 proteins interact with ER chaperones and fail to pass the ER quality control system (Jin et al., 2007, 2009; Hong et al., 2008). In order to test whether BRI1 is retained in the ER for our newly identified extracellular mutation alleles, we performed an Endo H analysis (Supplemental Fig. S6). The results indicated that BRI1 is not retained on ER in bri1-710 and bri1-711 and is partially retained on ER in bri1-705 and bri1-706. In bri1-705 and bri1-706, there is still a considerable amount of BRI1 that is resistant to Endo H cleavage, suggesting that partially retained bri1-705 or bri1-706 on ER may not be the main reason for their defective phenotypes.
CONCLUSION
In this study, we analyzed 83 BRI1 point mutations generated by TILLING, among which nine mutants showed typical bri1 mutant phenotypes. bri1-702 is a weak allele. bri1-705, bri1-706, bri1-710, and bri1-711 are subtle alleles. bri1-703, bri1-704, bri1-708, and bri1-709 are strong alleles, similar to bri1-701 (Fig. 1; Table I). bri1-702, the first weak allele in the activation loop of BRI1 identified so far, exhibited not only reduced autophosphorylation activity of BRI1 in vitro but also reduced sensitivity of the BAK1 phosphorylation response to exogenously applied BL in vivo (Fig. 6). bri1-705, a new subtle allele in the extracellular portion, contains a BRI1 mutation that may disrupt the formation of hydrogen bonds among BRI1, BL, and BAK1 (Fig. 8). Surprisingly, bri1-706, a subtle morphological mutant with a mutation in the extracellular portion of BRI1, showed insensitive root growth inhibition to exogenously applied BL, which is similar to the null bri1 mutants (Figs. 2A and 4A). The identification of unexpected characteristics of bri1-706 suggests that there are still many unknown aspects in the early events of the BR signaling pathway waiting to be elucidated in the future.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Mutant Detection
Original Arabidopsis (Arabidopsis thaliana) bri1 TILLING seeds obtained from the Arabidopsis Biological Resource Center are all in the Col-0 accession, which contains an er mutation. After er and additional mutations were segregated out via backcrossing for at least three generations, each homozygous bri1 was isolated and used for phenotypic determination and detailed analyses. Other known bri1 mutants used in this study include bri1-5 (WS2 background), bri1-9 (Col-0 background), bri1-301 (Col-0 background), and bri1-701 (SALK_116202). All plants were grown in a greenhouse under a long-day lighting condition (16 h of light and 8 h of dark) or under darkness at 22°C. Seedlings were grown on one-half-strength MS agar plates or in liquid medium supplemented with 1% (w/v) Suc.
The dCAPS approach was used to confirm the point mutations (Neff et al., 1998). dCAPS Finder 2.0 (http://helix.wustl.edu/dcaps/dcaps.html) was used to design mismatched PCR primers to generate or disrupt a restriction enzyme recognition site relative to the mutation (Neff et al., 2002). All primers and enzymes used in this study for genotyping bri1 TILLING mutations are listed in Supplemental Table S1.
Root Inhibition and Hypocotyl Growth Assays
Seeds were surface sterilized with 75% (w/v) ethanol for 30 s, followed by 1% NaClO for an additional 7 min, and washed with desterilized water five times. The seeds were then placed on one-half-strength MS plates with 0.8% (w/v) agar, 1% (w/v) Suc, and different concentrations of 24-epibrassinolide (Sigma) or BRZ (Santa Cruz Biotechnology). The seeds were vernalized at 4°C for 3 d and transferred to different growth conditions as indicated in the figure legends. Before being placed in the dark, plates were exposed to white light for 6 h to induce germination. All measurements were carried out using ImageJ (http://rsb.info.nih.gov/ij/).
Vector Construction and Transgenic Plant Generation
Gateway technology (Invitrogen, Life Technologies) was used in cloning all genes into expression vectors in this study. The full-length coding sequence fragments of BRI1, BRI1-CD, and BAK1 were amplified and cloned into a pDONR/Zeo entry clone vector. Site-directed mutagenesis was carried out to generate mutated entry clones of BRI1, BRI1-CD, and BAK1 based on the method described previously (Gou et al., 2012). All the cloned genes were sequence verified. Genes in the entry clone vectors were then cloned into various destination vectors for different purposes, such as pBRI1-GWR-Flag, pBIB-BASTA-35s-GWR-Flag, and pBIB-HYG-35s-GWR-GFP. All expression constructs were transferred into appropriate plant materials by the floral dip method (Clough and Bent, 1998).
RNA Extraction and Quantitative Real-Time PCR
Seven-day-old seedlings of Col-0 and bri1 mutants grown on one-half-strength MS plates were collected and then grown with or without 1 μm 24-epibrassinolide in liquid medium for 4 h. Total RNAs were extracted using an RNA simple total RNA kit (Tiangen) and reverse transcribed to total cDNAs utilizing Moloney murine leukemia virus reverse transcriptase (Invitrogen, Life Technologies). Quantitative real-time PCR was performed by the StepOnePlus Real-Time PCR System (Applied Biosystems) utilizing SYBR Green reagent (Takara). Detailed experimental procedures were the same as reported previously (Zhao et al., 2016).
Membrane Protein Isolation, Immunoprecipitation, Western Blotting, and Phosphoprotein Staining
Ten-day-old seedlings of 35S::BAK1-GFP, 35S::BRI1-FLAG, Col-0 35S::BAK1-GFP, bri1-702 35S::BAK1-GFP, bri1-301 35S::BAK1-GFP, or bri1-701 35S::BAK1-GFP were treated with or without 100 nm 24-epibrassinolide for 1.5 h and then ground to a fine powder in liquid N2. The membrane protein isolation procedure was the same as reported previously (Li et al., 2002). The soluble protein samples were immunoprecipitated utilizing an anti-GFP antibody (Invitrogen) combined with Protein A/G Plus-Sefinose Resin (BBI). The immunoprecipitated proteins were separated by 10% SDS-PAGE and tested by western analyses with anti-GFP and anti-phospho-Thr antibodies.
The same plant materials used for quantitative real-time PCR also were used in the detection of BES1 phosphorylation status. Total proteins were separated by 12% SDS-PAGE and detected via a specific anti-BES1 antibody (made by J.L.’s laboratory). The transgenic plants in Figure 7 were identified by detecting their GFP tag utilizing an anti-GFP antibody (Invitrogen). The Endo H analyses shown in Supplemental Figures S5 and S6 were performed as described previously (Hong et al., 2008). The total proteins of wild-type and mutant plants in Supplemental Figure S7 were separated by 8% SDS-PAGE and detected via a specific anti-BRI1 antibody (Agrisera; A177246). Horseradish peroxidase-linked anti-rabbit or anti-mouse antibody was used as a secondary antibody, and the signal was detected by Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer) in all western-blot experiments.
For the phosphoprotein staining assay, the cytoplasmic domain sequences of wild-type BRI1 and mutated BRI1 (residues 815–1,196) were cloned into the expression vector pMAL-cRI-MBP-GWR. The recombinant proteins were expressed in Escherichia coli and induced by 0.1 mm isopropylthio-β-galactoside for 6 h at 18°C. The cell lysates were boiled and centrifuged. The supernatant was separated by 10% SDS-PAGE and stained sequentially with Pro-Q Diamond (Invitrogen) and Coomassie Blue Silver as described previously (Taylor et al., 2013).
MD Simulation Analysis
The initial complex coordinates of BRI1-BAK1-BL were retrieved from the Protein Data Bank (http://www.rcsb.org) under accession code 4M7E (Sun et al., 2013). MD simulations were carried out using the AMBER 10 software package (Case et al., 2005) to check the stability of BRI1 and BAK1 in complex with BL. We used the AMBER ff12SB (Hornak et al., 2006) force field for the protein and water and the general AMBER force field (Wang et al., 2004) for the BL molecule. Atom types and AM1-BCC atomic charges were generated for BL using the Antechamber module. The LEaP module of the AMBER 10 software package was used to prepare the protein-ligand complex. The complex was solvated using the explicit TIP3P (Jorgensen et al., 1983) water model, which extended a minimum 10-Å distance from the box surface to any atom of the solute. Two consecutive steps of minimization were carried out. All bond lengths were constrained using the SHAKE algorithm (Ryckaert et al., 1977), and the equations of motion were integrated with a 2-fs time step using the Verlet leapfrog algorithm. To eliminate possible bumps between the solute and the solvent, the entire system was minimized in two steps. First, the complex was restrained with a harmonic potential with a force constant k = 50 kcal mol−1 Å−2. The water molecules and counter ions were optimized using the steepest descent method of 1,000 steps, followed by the conjugate gradient method for 2,000 steps. Second, the entire system was optimized using the first step method without any constraints. Subsequently, the entire system was heated gradually in the NVT ensemble from 0 to 300 K over 400 ps, with a weak restraint (k = 5 kcal mol−1 Å−2) for the complex (Wu et al., 2016). The final coordinates, obtained after the temperature equilibration simulation, were used for a 120-ns MD simulation, during which the temperature was kept by the Langevin thermostat with a collision frequency of 2 ps−1. A constant pressure periodic boundary was used to maintain the pressure of the system using an isotropic position scaling algorithm with a relaxation time of 2 ps. After MD simulation, distances between two atoms were calculated using the cpptraj program, and the dictionary of secondary structure of protein program developed by Kabsch and Sander (1983) was used to assign the secondary structures of the BRI1-BAK1-BL complex. All the structure figures were prepared using PyMol (www.pymol.org).
Sequence Analysis
For sequence analysis, the sequences of 70 BRI1s or BRLs from 18 flowering plants and 223 LRR-RLK sequences from Arabidopsis were downloaded and analyzed in MEGA software (http://www.megasoftware.net/). The final alignment data were generated to a logo (Fig. 9; Supplemental Fig. S4) on the WebLogo Web site (http://weblogo.berkeley.edu/logo.cgi).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Eighty-three total mutations of BRI1 identified in this study and 22 previously reported bri1 mutants.
Supplemental Figure S2. Overexpression of mBAK1 in bri1-5 resulted in a dominant negative phenotype.
Supplemental Figure S3. Monitoring of the equilibration of the MD trajectories of the wild-type complex and the BRI1-Pro-719Leu complex.
Supplemental Figure S4. Sequence analyses of the BRI1 activation loop.
Supplemental Figure S5. Endo H cleavage of BRI1 and responses of BES1 phosphorylation and CPD expression to BL treatment showed no significant differences between shoots and roots of bri1-706 seedlings.
Supplemental Figure S6. Endo H analysis indicated that BRI1 is not retained in the ER in bri1-710 and bri1-711 and is partially retained in the ER in bri1-705 and bri1-706.
Supplemental Figure S7. In bri1-709, the mutation results in a premature stop codon.
Supplemental Table S1. List of primers and enzymes used in this study for genotyping bri1 TILLING mutations.
Acknowledgments
We thank J.L. laboratory member Yao Xiao for generating the BES1 antibody and Steven Henikoff and his team for their hard work on the Arabidopsis TILLING project.
Glossary
- BR
brassinosteroid
- BL
brassinolide
- ER
endoplasmic reticulum
- EMS
ethyl methanesulfonate
- TILLING
targeted induced local lesions in genomes
- dCAPS
derived cleaved-amplified polymorphic sequence
- Col-0
Columbia-0
- BRZ
brassinazole
- MD
molecular dynamic
- WS2
Wassilewskija-2
- MS
Murashige and Skoog
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31530005 and 31470380 to J.L.).
References
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Durbak A, Wierzba M, Schmitz RJ, Aguirre A, Michel R, Rowe S, Fujioka S, Tax FE (2010) Intragenic suppression of a trafficking-defective brassinosteroid receptor mutant in Arabidopsis. Genetics 185: 1283–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell EM, Lin WC, Husbands AY, Yu L, Jaganatha V, Jablonska B, Mangeon A, Neff MM, Girke T, Springer PS (2012) Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc Natl Acad Sci USA 109: 21146–21151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Cheatham TE III, Darden T, Gohlke H, Luo R, Merz KM Jr, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26: 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiwanon J, Wang ZY (2015) Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr Biol 25: 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zhu W, Chen Y, Ito S, Asami T, Wang X (2014) Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. eLife 3: e02525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse SD. (1996) Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J 10: 1–8 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC (1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451 [DOI] [PubMed] [Google Scholar]
- Domagalska MA, Schomburg FM, Amasino RM, Vierstra RD, Nagy F, Davis SJ (2007) Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development 134: 2841–2850 [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J (2000) Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 123: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Liu JS, Fan M, Bai MY, Wenkel S, Springer PS, Barton MK, Wang ZY (2012) Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc Natl Acad Sci USA 109: 21152–21157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-García S, Russinova E, Caño-Delgado AI (2011) Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859 [DOI] [PubMed] [Google Scholar]
- Gou X, Yin H, He K, Du J, Yi J, Xu S, Lin H, Clouse SD, Li J (2012) Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet 8: e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudesblat GE, Russinova E (2011) Plants grow on brassinosteroids. Curr Opin Plant Biol 14: 530–537 [DOI] [PubMed] [Google Scholar]
- Gudesblat GE, Schneider-Pizoń J, Betti C, Mayerhofer J, Vanhoutte I, van Dongen W, Boeren S, Zhiponova M, de Vries S, Jonak C, et al. (2012) SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat Cell Biol 14: 548–554 [DOI] [PubMed] [Google Scholar]
- Guo Z, Fujioka S, Blancaflor EB, Miao S, Gou X, Li J (2010) TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S (2011) Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Jin H, Tzfira T, Li J (2008) Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 3418–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Kajiura H, Su W, Jin H, Kimura A, Fujiyama K, Li J (2012) Evolutionarily conserved glycan signal to degrade aberrant brassinosteroid receptors in Arabidopsis. Proc Natl Acad Sci USA 109: 11437–11442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65: 712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J (2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Hong Z, Su W, Li J (2009) A plant-specific calreticulin is a key retention factor for a defective brassinosteroid receptor in the endoplasmic reticulum. Proc Natl Acad Sci USA 106: 13612–13617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Yan Z, Nam KH, Li J (2007) Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol Cell 26: 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey R, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79: 926–935 [Google Scholar]
- Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22: 2577–2637 [DOI] [PubMed] [Google Scholar]
- Kim TW, Michniewicz M, Bergmann DC, Wang ZY (2012) Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Choe S (2005) Brassinosteroid biosynthesis and dwarf mutants. J Plant Biol 48: 1–15 [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Lease KA, Tax FE, Walker JC (2001a) BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 5916–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J (2001b) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 127: 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang C, Wang D, Su W, Liu L, Wang M, Li J (2015) EBS7 is a plant-specific component of a highly conserved endoplasmic reticulum-associated degradation system in Arabidopsis. Proc Natl Acad Sci USA 112: 12205–12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S (2000) Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiol 123: 439–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-García S, Vert G, Yin Y, Caño-Delgado A, Cheong H, Chory J (2004) Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE (1998) dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J 14: 387–392 [DOI] [PubMed] [Google Scholar]
- Neff MM, Turk E, Kalishman M (2002) Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18: 613–615 [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE (1999) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MH, Ray WK, Huber SC, Asara JM, Gage DA, Clouse SD (2000) Recombinant brassinosteroid insensitive 1 receptor-like kinase autophosphorylates on serine and threonine residues and phosphorylates a conserved peptide motif in vitro. Plant Physiol 124: 751–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert J, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23: 327–341 [Google Scholar]
- Santiago J, Henzler C, Hothorn M (2013) Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341: 889–892 [DOI] [PubMed] [Google Scholar]
- Shang Y, Lee MM, Li J, Nam KH (2011) Characterization of cp3 reveals a new bri1 allele, bri1-120, and the importance of the LRR domain of BRI1 mediating BR signaling. BMC Plant Biol 11: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J, Han Z, Kim TW, Wang J, Cheng W, Chang J, Shi S, Wang J, Yang M, Wang ZY, et al. (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber CM, McCourt P (2001) A role for brassinosteroids in germination in Arabidopsis. Plant Physiol 125: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Liu Y, Xia Y, Hong Z, Li J (2011) Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis. Proc Natl Acad Sci USA 108: 870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Liu Y, Xia Y, Hong Z, Li J (2012) The Arabidopsis homolog of the mammalian OS-9 protein plays a key role in the endoplasmic reticulum-associated degradation of misfolded receptor-like kinases. Mol Plant 5: 929–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Han Z, Tang J, Hu Z, Chai C, Zhou B, Chai J (2013) Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res 23: 1326–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I, Seitz K, Bennewitz S, Walker JC (2013) A simple in vitro method to measure autophosphorylation of protein kinases. Plant Methods 9: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, Johnson JE, Burtner C, Odden AR, Young K, Taylor NE, et al. (2003) Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res 13: 524–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Nemhauser JL, Geldner N, Hong F, Chory J (2005) Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol 21: 177–201 [DOI] [PubMed] [Google Scholar]
- Vilarrasa-Blasi J, González-García MP, Frigola D, Fàbregas N, Alexiou KG, López-Bigas N, Rivas S, Jauneau A, Lohmann JU, Benfey PN, et al. (2014) Regulation of plant stem cell quiescence by a brassinosteroid signaling module. Dev Cell 30: 36–47 [DOI] [PubMed] [Google Scholar]
- Wang J, Jiang J, Wang J, Chen L, Fan SL, Wu JW, Wang X, Wang ZX (2014) Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1. Cell Res 24: 1328–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general Amber force field. J Comput Chem 25: 1157–1174 [DOI] [PubMed] [Google Scholar]
- Wang X, Chory J (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313: 1118–1122 [DOI] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Wang X, Li X, Meisenhelder J, Hunter T, Yoshida S, Asami T, Chory J (2005) Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell 8: 855–865 [DOI] [PubMed] [Google Scholar]
- Wang Y, Sun S, Zhu W, Jia K, Yang H, Wang X (2013) Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev Cell 27: 681–688 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Bai MY, Oh E, Zhu JY (2012) Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet 46: 701–724 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Wei Z, Li J (2016) Brassinosteroids regulate root growth, development, and symbiosis. Mol Plant 9: 86–100 [DOI] [PubMed] [Google Scholar]
- Wu D, Tao X, Chen ZP, Han JT, Jia WJ, Zhu N, Li X, Wang Z, He YX (2016) The environmental endocrine disruptor p-nitrophenol interacts with FKBP51, a positive regulator of androgen receptor and inhibits androgen receptor signaling in human cells. J Hazard Mater 307: 193–201 [DOI] [PubMed] [Google Scholar]
- Xu W, Huang J, Li B, Li J, Wang Y (2008) Is kinase activity essential for biological functions of BRI1? Cell Res 18: 472–478 [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, et al. (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65: 634–646 [DOI] [PubMed] [Google Scholar]
- Yuan T, Fujioka S, Takatsuto S, Matsumoto S, Gou X, He K, Russell SD, Li J (2007) BEN1, a gene encoding a dihydroflavonol 4-reductase (DFR)-like protein, regulates the levels of brassinosteroids in Arabidopsis thaliana. Plant J 51: 220–233 [DOI] [PubMed] [Google Scholar]
- Zhang C, Xu Y, Guo S, Zhu J, Huan Q, Liu H, Wang L, Luo G, Wang X, Chong K (2012) Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet 8: e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Lv M, Feng Z, Campbell T, Liscum E, Li J (2016) TWISTED DWARF 1 associates with BRASSINOSTEROID-INSENSITIVE 1 to regulate early events of the brassinosteroid signaling pathway. Mol Plant 9: 582–592 [DOI] [PubMed] [Google Scholar]
- Zhou A, Wang H, Walker JC, Li J (2004) BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J 40: 399–409 [DOI] [PubMed] [Google Scholar]
- Zhu JY, Sae-Seaw J, Wang ZY (2013) Brassinosteroid signalling. Development 140: 1615–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]