LNK1 and LNK2 act as transcriptional corepressors necessary for expression of the phenylpropanoids biosynthesis gene C4H through recruitment to its promoter via interaction with MYB3.
Abstract
Subgroup 4 of R2R3-MYB transcription factors consists of four members, MYB3, MYB4, MYB7, and MYB32, which possess the conserved EAR repression motif (pdLHLD/LLxiG/S) in their C termini. Here, we show that MYB3 is a newly identified repressor in Arabidopsis (Arabidopsis thaliana) phenylpropanoid biosynthesis. However, the repression mechanism of MYB3 is completely different from MYB4, MYB7, and MYB32. Yeast two-hybrid screening using MYB3 as a bait isolates NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED1 (LNK1) and LNK2, members of a small family of four LNK proteins. The repression activity of MYB3 to cinnamate 4-hydroxylase (C4H) gene expression is directly regulated by corepressors LNK1 and LNK2, which could facilitate binding of MYB3 with C4H promoter. The two conserved Asp residues in both region 1 and 2 domain of LNKs are essential to mediate protein-protein interaction. Importantly, the Extra N-terminal Tail domain plays a negative role in LNK-MYB3 transcription complex-dependent repression of the C4H gene. We conclude that LNK1 and LNK2 act as transcriptional corepressors necessary for expression of the phenylpropanoids biosynthesis gene C4H through recruitment to its promoter via interaction with MYB3.
The phenylpropanoid metabolic pathway is a well-described plant secondary metabolite pathway. Different classes of compounds are produced by the phenylpropanoid pathway and have different functions, such as lignin, which plays a structural role in the secondary cell walls formation (Vanholme et al., 2010), while flavonoids mediate plants against UV radiation and act as a visual signal for attracting pollinators (Mol et al., 1998; Bradshaw and Schemske, 2003). The biosynthesis of these phenylpropanoid compounds are initiated with deamination of Phe by Phe ammonia-lyase (PAL), followed by reactions catalyzed by a P450 enzyme, cinnamate 4-hydroxylase (C4H), to form β-coumaric acid (Winkel-Shirley, 2002; Du et al., 2010). The subsequent reactions catalyzed by a series of enzymes, such as 4-coumarate coenzyme A ligase (4CL), O-methyltransferase (OMT), ferulate-5-hydroxylase (F5H), and cinnamyl alcohol dehydrogenase (CAD; Winkel-Shirley, 2002; Du et al., 2010). During the last few years, many efforts have been made to discover the transcription regulators that regulate the phenylpropanoid compounds biosynthesis in the model plant Arabidopsis (Arabidopsis thaliana), and an increasing amount of evidence reveals that R2R3-MYB type transcription factors (TFs) mainly positively increase anthocyanin biosynthesis (Zhou et al., 2015a). It has been reported that PAP1/MYB75, PAP2/MYB90, MYB113, and MYB114 positively regulate anthocyanin metabolism (Zimmermann et al., 2004; Teng et al., 2005; Stracke et al., 2007; Gonzalez et al., 2008; Qi et al., 2011). However, MYB4, MYB7, MYB32, and MYBL2 repress the phenylpropanoids biosynthesis (Jin et al., 2000; Preston et al., 2004; Dubos et al., 2008, 2010; Fornalé et al., 2014; Zhou et al., 2015b).
The R2R3-MYB TFs are classified into subgroups based on the presence of their C-terminal conserved domain and several studies demonstrate that same subgroup TFs share similar functions (Dubos et al., 2010; Zhou et al., 2015b). R2R3-MYB subgroup 4 TFs consist of four members, MYB3, MYB4, MYB7, and MYB32, which possess the conserved LLsrGIDPxT/SHRxI/L motif and the EAR repression motif (pdLHLD/LLxiG/S) in their C-termini (Jin et al., 2000). In addition, our previous report showed that MYB4, MYB7, and MYB32, but not MYB3, has a putative zinc-finger domain (CX1-2CX7-12CX2C) and a conserved GY/FDFLGL motif in their C termini (Zhou et al., 2015b). Further experiments showed that the GY/FDFLGL motif contributes to the interaction between MYB TFs and the Sensitive to ABA and Drought 2 (SAD2) (Zhou et al., 2015b). SAD2, an importin β-like protein, mediates the transport of MYB4, MYB7, and MYB32 into the nucleus and then increases the repression activity on their target gene (such as C4H) expression (Zhou et al., 2015b). Until now, three of four R2R3-MYB subgroup 4 TFs, MYB4, MYB7, and MYB32, are involved in repression of the phenylpropanoid pathway (Jin et al., 2000; Preston et al., 2004; Dubos et al., 2010; Fornalé et al., 2014; Zhou et al., 2015b). MYB4 is responsive to UV-B irradiation and acts as a repressor in the phenylpropanoid pathway through negatively regulating the expression of the C4H gene (Jin et al., 2000; Zhao et al., 2007). MYB32 has been described as a pollen-specific repressor of lignin biosynthesis (Preston et al., 2004). MYB7 is partly involved in regulation of UV sunscreens through repressing flavonol biosynthesis (Fornalé et al., 2014). Additionally, it has been reported that the R3-MYB-related factor MYBL2 also negatively regulates the expression of anthocyanin genes in Arabidopsis seedlings (Dubos et al., 2008). However, nothing is known about the function of MYB3, the closest MYB4, MYB7, and MYB32 homolog.
Therefore, we investigated the role of MYB3 in the regulation of the phenylpropanoid biosynthesis. Recently, NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED1 (LNK1) and LNK2 were demonstrated to play important roles in Arabidopsis period determination, circadian oscillator, and anthocyanin biosynthesis (Rugnone et al., 2013; Xie et al., 2014; Pérez-García et al., 2015). Here, we report that MYB3, like MYB4, MYB7, and MYB32, is involved in the repression of phenylpropanoid biosynthesis. Through the use of yeast two-hybrid (Y2H) screening, we identified the MYB3-interacting factors, LNK1 and LNK2, known to function in the control of circadian rhythm-associated gene expression as a transcriptional coactivator and anthocyanin biosynthesis-associated gene expression as a transcriptional repressor, respectively (Xie et al., 2014; Pérez-García et al., 2015). We demonstrated that the repression activity of MYB3 is directly regulated by corepressors LNK1 and LNK2, which could facilitate binding of MYB3 with its target gene C4H promoter. We also found that the two conserved Asp residues in both region 1 and 2 domains of LNKs are essential to mediate LNK-MYB3 interactions. Importantly, we provide evidence that the Extra N-terminal Tail (ENT) domain of LNKs plays a negative role in LNK-MYB3 transcription complex-dependent repression of C4H gene. Together, our results suggest that LNKs act as corepressors for the regulation of MYB3-dependent gene expression.
RESULTS
MYB3 Represses Phenylpropanoids Biosynthesis
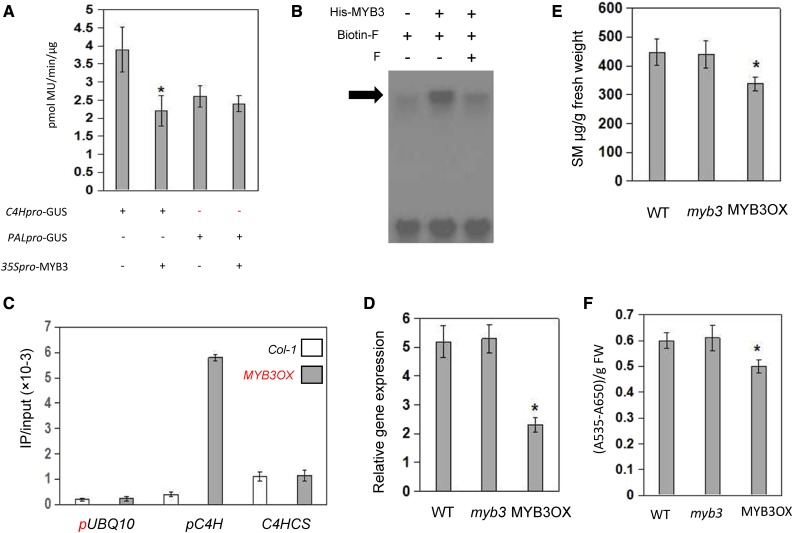
MYB4, MYB7, and MYB32 have been described as transcriptional repressors of the phenylpropanoid pathway (Jin et al., 2000; Preston et al., 2004; Dubos et al., 2010; Fornalé et al., 2014; Zhou et al., 2015b). Thus, we hypothesized that MYB3 may also repress the phenylpropanoids biosynthesis. As a first step to test this hypothesis, the effect of MYB3 on C4H gene expression was tested by protoplast transactivation assays. Cotransformation of Arabidopsis protoplasts with a C4Hpro-GUS reporter construct (Zhou et al., 2015b) and an effector CaMV35S-MYB3 resulted in significant repression of around 2-fold, while MYB3 did not affect the GUS activity of PALpro-GUS reporter, as expected (Fig. 1A). To test whether the directly interaction between MYB3 protein and C4H promoter, electrophoresis mobility shift assay (EMSA) and chromatin immunoprecipitation assay (ChIP) were performed. As shown in Figure 1, B and C, MYB3 was able to bind to the C4H promoter fragment. As a negative control, MYB3 did not bind to the reference gene UBQ10 promoter and C4H CDS fragments (Fig. 1C). These results indicate that MYB3 directly represses the C4H gene expression. Furthermore, to functionally test the role of MYB3 on phenylpropanoids metabolism in planta, we analyzed the MYB3-HA overexpressing and myb3 mutant Arabidopsis plants (Supplemental Fig. S1). A myb3 T-DNA insertion line did not express the MYB3 transcript and was also morphologically indistinguishable from wild-type plants (Supplemental Fig. S1). qRT-PCR analysis performed with 2-week-old seedlings showed that the C4H gene was highly repressed in MYB3-HA-overexpressing plants compared with myb3 and wild-type plants (Fig. 1D). Additionally, overexpression of MYB3 also affected the key enzyme genes, such as CHS, 4CL1, and 4CL3, which were unaffected in the myb3 mutant plants (Supplemental Fig. S2). The transcript levels of PAL2, OMT, F5H, and CAD1 were not regulated by MYB3 factor (Supplemental Fig. S2). Therefore, we want to functionally test the role of MYB3 on the sinapoyl malate (SM) and anthocyanin accumulation (Jin et al., 2000; Zhou et al., 2015b). As shown in Figure 1, E and F, the level of SM and anthocyanin were significantly reduced in plants overexpressing MYB3-HA compared with myb3 and wild-type plants. It has been reported that the accumulation of SM in plants are more tolerant of UV-B (Jin et al., 2000). Our results showed that overexpression of MYB3 enhanced the sensitivity to UV-B (Supplemental Fig. S3).
Figure 1.
MYB3 represses phenylpropanoids biosynthesis. A, Transactivation assays in Arabidopsis protoplasts that were cotransfected with plasmids carrying C4Hpro-GUS and overexpression vectors containing 35S:MYB3, as indicated. Values are means ± sd of three biological repeats. Asterisks indicate statistically significant differences compared with C4Hpro-GUS (P < 0.05, Student’s t test). B, EMSA of a probe including the MYB3 binding site AATAGTT with His-MYB3 purified from Escherichia coli BL21 (DE3). The arrow indicates the protein probe complex. C, Direct binding of MYB3 with the promoter of C4H. ChIP assays were conducted by real-time PCR after normalizing with the input DNA. The fragment of C4H coding sequence and the reference gene UBQ10 promoter were used as a negative control. D to F, Measurement of C4H gene expression levels (D), SM (E), and anthocyanin (F) contents in indicated genotypes by LC/MS and quantitative RT-PCR, respectively. Values are means ± sd of three biological repeats of three independent transgenic lines. Asterisks indicate statistically significant differences compared with wild-type and myb3 plants (P < 0.05, Student’s t test). FW, Fresh weight.
Identification of LNKs That Interact with MYB3
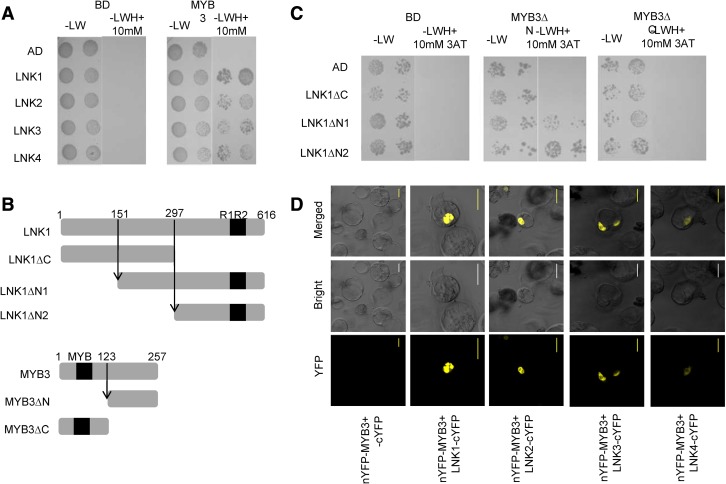
To identify proteins that interact with MYB3, Y2H screenings were performed using full-length MYB3 fused to the GAL4 DNA-binding domain (BD) in the vector pAS2.1 as a bait. Screening of 5.4 × 105 yeast transformants obtained with Arabidopsis cDNA library generated from ecotype Col-0 seedlings in the vectors pACT2 resulting in nine colonies that were able to grow on medium lacking His. Recovered prey plasmids were retransformed and five plasmids conferred growth on selective medium. From these candidate MYB3 interactors, only one cDNA sequence was in frame with the GAL4 activation domain (AD) and encoded the C-terminal 294 amino acids of the protein named LNK1 (At5g64170). LNK1 is one of a small family of four LNK proteins, which has been shown to interact with Myb-like REVEILLE TFs (RVE4 and RVE8) and regulate some circadian rhythm and anthocyanin biosynthesis-associated genes expression as a transcriptional coregulator (Rugnone et al., 2013; Xie et al., 2014; Pérez-García et al., 2015). To test whether all four LNKs interact with MYB3, Y2H assays were performed. These results showed that all four LNKs interacted with MYB3 (Fig. 2A). To investigate the functional domains mediating the interaction between MYB3 and LNKs, we tested the interaction of threeLNK1 deletion derivatives (LNK1∆N1, LNK1∆N2, and LNK1∆C) with two MYB3 deletion derivatives (MYB3∆N and MYB3∆C; Fig. 2B). As shown in Figure 2C, the interaction observed between LNKs and MYB3 requires the C-terminal domain of both LNKs and MYB3. However, LNKs did not interact with another subgroup 4 MYB TF MYB4, MYB7, and MYB32 (Supplemental Fig. S4).
Figure 2.
MYB3 interacts with a small family of four LNK proteins in yeast and in planta. A to C, LNK1, LNK2, LNK3, LNK4, and LNK1 deletion derivatives (LNK1∆N1 and LNK1∆N2) interact with MYB3 and MYB3∆N in yeast. B, Schematic representation of the deletion derivatives of both LNK1 and MYB3. Yeast cells expressing LNK1, LNK2, LNK3, LNK4, LNK1∆N1, LNK1∆N2, and LNK1∆C proteins fused to the GAL4 AD and MYB3, MYB3∆N, and MYB3∆C fused to the GAL4 BD were spotted on SD/-LW medium to select for the plasmids and on SD/-LWH medium containing 10 mm 3-amino-1,2,4-triazole (3AT) to select for transcriptional activation of the His-3 gene. Growth was monitored after 5 d. Yeast cells transformed with empty plasmids pAS2.1 and pACT2, expressing GAL4 BD and AD, respectively, were used as controls in A and C. D, BiFC assays in planta. YFP fluorescence images alone or merged with bright-field images of Arabidopsis cell suspension protoplasts cotransfected with constructs encoding the indicated fusion proteins with YFP at the C terminus or the N terminus. Bar = 20 µm.
To confirm the interaction of MYB3 with LNKs in planta, a bimolecular fluorescence complementation (BiFC) assay was performed. The constructs were transiently coexpressed in all possible combinations of YN and YC fusion proteins in Arabidopsis suspension cell protoplasts. As shown in Figure 2D, strong YFP signals were observed in the nucleus of Arabidopsis protoplasts upon coexpression of LNK-cYFP with nYFP-MYB3. No YFP fluorescence was detected upon coexpression of single plasmids and any combination of empty YFP vectors. These results demonstrated that MYB3 can interact with LNKs in the nucleus of plant cells.
Two Conserved Asp Residues of LNKs Are Required for Protein Interactions
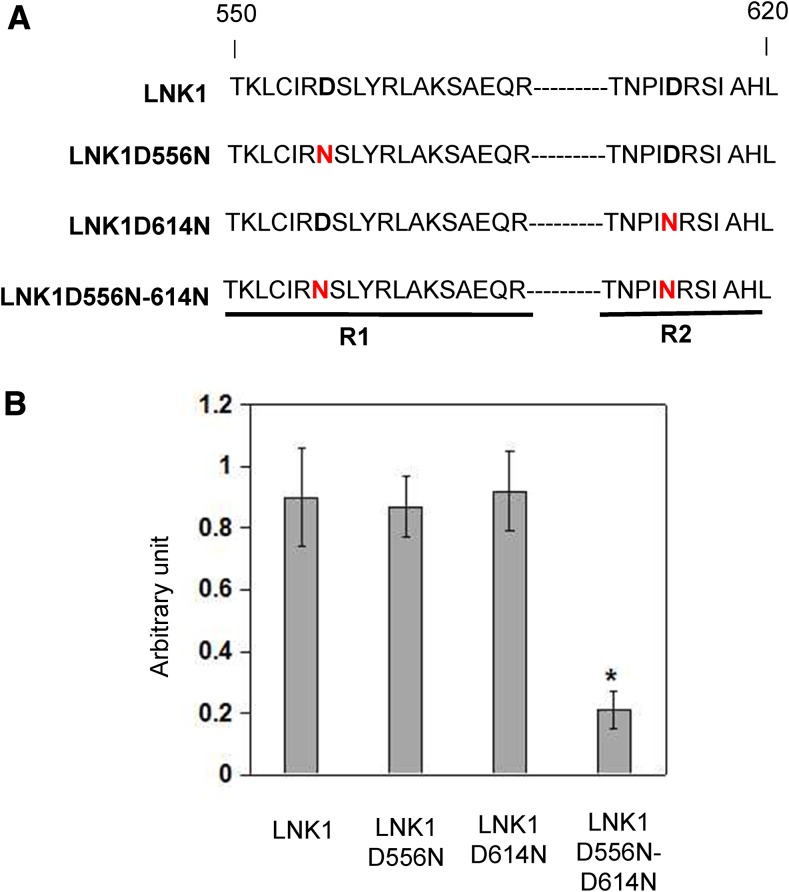
The two plant-specific conserved regions (R1 and R2) of C termini of LNK family members have previously been shown to interact with the Myb-like TF RVE4 (Xie et al., 2014). Mutation of two conserved residues in either R1 or R2 region (Arg-555Asp-556 to GlyGly or Asp-614Arg-615 to GlyGly) of LNK1 significantly diminished the interaction between LNK1 and RVE4 (Xie et al., 2014). Several studies demonstrated the conserved Asp residue contributes to the protein-protein interaction, such as the Asp residue in the JAZ interaction domain of MYC2 and the SID motif of MYB4 (Goossens et al., 2015; Zhou et al., 2015b). To obtain insight into the role of the Asp residue in LNK1 interaction, we tested the interaction of Asp-556 to Asn (D556N), Asp-614 to Asn (D614N), or Asp-556-Asp-614 to Asn-Asn (D556N-D614N) mutated LNK1 with MYB3 protein (Fig. 3A). Immunoblot analysis revealed that LNK1 and LNK1 derivatives were present at similar levels in yeast (Supplemental Fig. S5), indicating that LNK1 protein stability was not affected by this mutation. Quantitative Y2H assays showed that the interaction of LNK1D556N or LNK1D614N with MYB3 was not significantly diminished (Fig. 3B). However, LNK1D556N-D614N greatly reduces the ability of LNK1 to interact with MYB3 (Fig. 3B). Taken together, these results suggest that the two conserved Asp residues in R1 and R2 of LNKs’ C termini are important for interaction of MYB3 with LNKs.
Figure 3.
Two conserved Asp residues of LNKs are required for protein interactions. A, Schematic representation of mutation of two conserved residues in either R1 or R2 region of LNK1. B, LNK1D556N, LNK1D614N, and LNK1D556N-D614N interact with MYB3 in quantitative Y2H assays. A liquid culture β-galactosidase assay was performed on the transformed yeasts. The activity of β-galactosidase was measured in arbitrary units. Values are means ± sd of three biological repeats. Asterisks indicate statistically significant differences compared with MYB3, MYB3D556N, and MYB3D614N (P < 0.05, Student’s t test).
MYB3 Repressor Activity Is Enhanced by LNK1 and LNK2
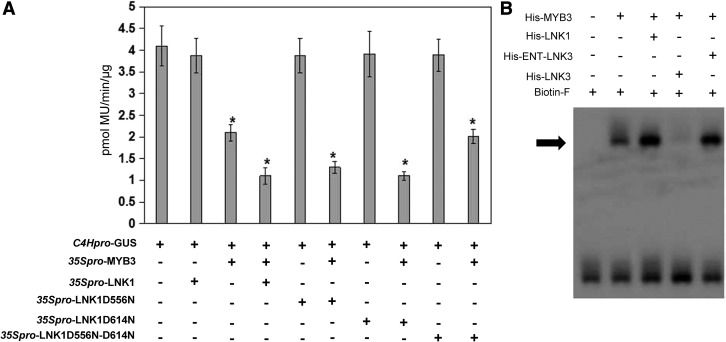
In order to elucidate the functional significance of the interaction between LNKs and MYB3, transactivation assays were performed. Effector constructs for MYB3, LNK1, LNK2, LNK3, LNK4, or LNK1 derivatives were expressed under the control of the CaMV 35S promoter. Arabidopsis cell suspension protoplasts were transiently cotransformed with a C4Hpro-GUS reporter (Zhou et al., 2015b) and effectors. As shown in Figure 4A and Supplemental Figure S6, we noticed that all four LNKs and LNK1 derivatives failed to affect the C4Hpro-GUS activity. Therefore, we asked whether LNKs were coregulators necessary for the repression of C4H transcription by MYB3. The MYB3 protein repressed the expression of C4Hpro-GUS about 2-fold, whereas coexpression of LNK1, LNK2, LNK1D556N, or LNK1D614N with MYB3 greatly reduced the expression of C4Hpro-GUS about 4-fold (Fig. 4A), indicating that LNK1 and LNK2, but not LNK3 and LNK4, act as corepressors to enhance MYB3 repression activity (Supplemental Fig. S6).
Figure 4.
MYB3 repressor activity is enhanced by LNK1 and LNK2. A, Transactivation assays in Arabidopsis protoplasts that were cotransfected with plasmids carrying C4Hpro-GUS and overexpression vectors containing 35S:MYB3, 35S:LNK1, 35S:LNK1D556N, 35S:LNK1D614N, or 35S:LNK1D556N-D614N, as indicated. Values are means ± sd of three biological repeats. Asterisks indicate statistically significant differences compared with C4Hpro-GUS, 35S:MYB3 or 35S:LNK1 (P < 0.05, Student’s t test). B, EMSA of a probe including the MYB3 binding site AATAGTT with His-MYB3, His-LNK1, His-ENT-LNK3, and His-LNK3 purified from E. coli BL21 (DE3). The arrow indicates the protein probe complex.
The data presented above indicate that LNK1 and LNK2 serve as transcriptional corepressors for C4H transcription, which suggests that LNK1 and LNK2 should be recruited to the C4H promoter. We then investigated whether LNK1 affected the interaction between MYB3 and C4H promoter using competitive EMSA. As shown in Figure 4B, the interactions between MYB3 and C4H promoter was inhibited by the addition of His-LNK3, but not His-LNK1 and His-ENT-LNK3. Taken together, these results reveal that LNK1 probably facilitate binding of MYB3 with C4H promoter.
The N-Terminal Domain of LNK1 Is Required for MYB3/LNK1 Complex-Dependent Repression of C4H
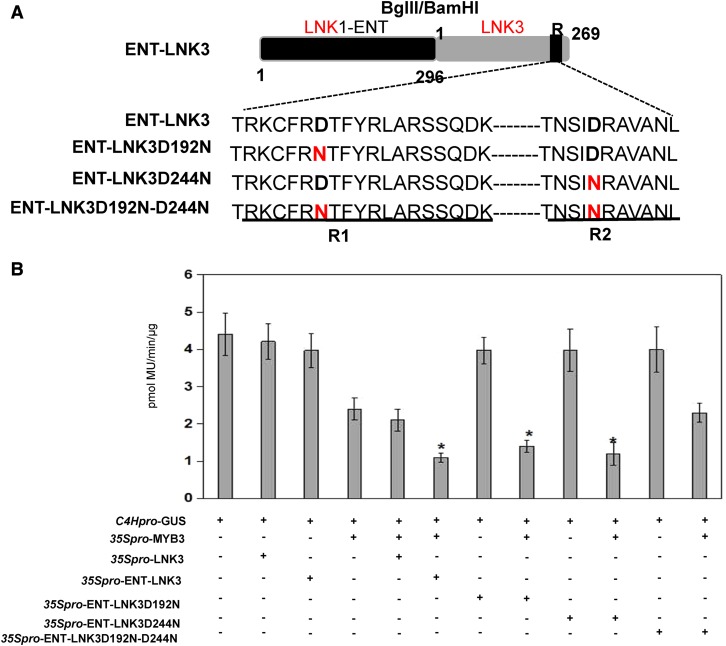
As noted above, both LNK3 and LNK4 did not enhance the MYB3 repression activity, suggesting that both LNKs lack the transcription regulation domain. The amino acid sequence alignment of four LNK proteins shows that LNK1 and LNK2 possess the ENT domain that is around 300 amino acids long compared to LNK3 and LNK4 (Supplemental Fig. S7). Therefore, we hypothesized that the ENT domain is essential for LNK transcription activity. To test this hypothesis, we generated a chimeric protein ENT-LNK3 that is the ENT domain of LNK1 fused to the N termini of LNK3, and ENT-LNK3 derivatives (Fig. 5A). As shown in Figure 5B, cotransformation of ENT-LNK3, ENT-LNK3D192N, or ENT-LNK3D244N and MYB3 significantly enhanced the ability of MYB3 to repress the expression of C4Hpro-GUS in Arabidopsis cell suspension protoplast. As shown in Figure 4B, LNK3 inhibited MYB3 binding to C4H promoter, but chimeric protein ENT-LNK3 facilitated the MYB3 binding activity. These results indicate that ENT-LNK3, like LNK1, can be as a corepressor to enhance MYB3 repression activity. Taken together, we conclude the ENT domain of LNKs is required for MYB3/LNK complex-dependent repression of C4H.
Figure 5.
ENT domain of LNK1 promotes the activity of MYB3/LNK1 complex-dependent repression of C4H. A, Schematic representation of mutation of two conserved residues in either the R1 or R2 region of LNK3 and chimeric protein of ENT-LNK3. B, Transactivation assays in Arabidopsis protoplasts that were cotransfected with plasmids carrying C4Hpro-GUS and overexpression vectors containing 35S:MYB3, 35S:LNK3, 35S:ENT-LNK3, 35S:ENT-LNK3D192N, 35S:ENT-LNK3D244N, or 35S:ENT-LNK3D192N-D244N, as indicated. Values are means ± sd of three biological repeats. Asterisks indicate statistically significant differences compared with C4Hpro-GUS, 35S:MYB3, 35S:LNK3, or 35S:ENT-LNK3 (P < 0.05, Student’s t test).
LNK1/2-MYB3 Complex Represses Phenylpropanoids Biosynthesis
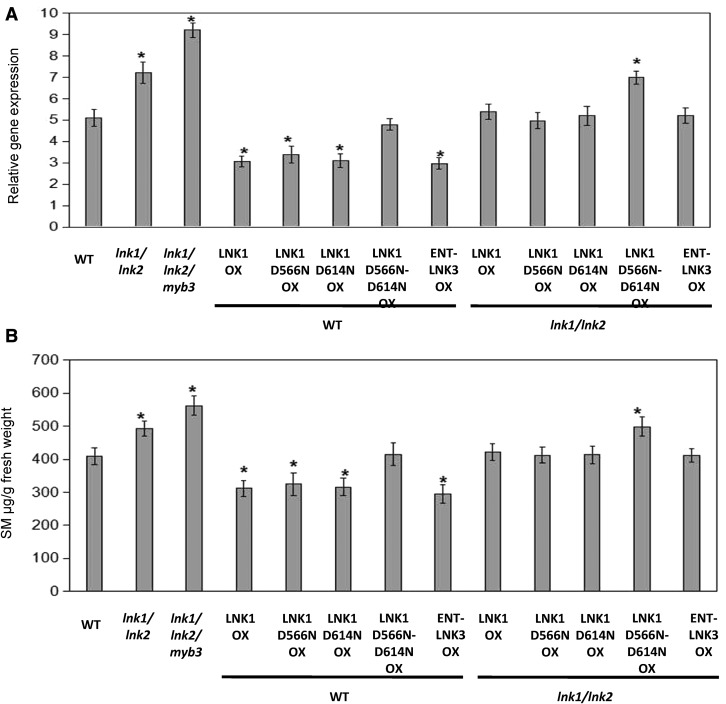
As noted above, overexpression of MYB3 represses phenylpropanoids biosynthesis. Our results from transactivation assays indicated that LNK1 or LNK2 acts as corepressor to enhance MYB3 repression activity. Therefore, we wanted to functionally test the role of these proteins in plants. First, we generated double-mutant plants homozygous for T-DNA insertion alleles of lnk1 and lnk2 to circumvent possible redundant roles of these two proteins. Then, the triple mutant lnk1/lnk2/myb3 plants were generated from a cross between lnk1/lnk2 and myb3 plants. qRT-PCR analysis confirmed that mutant seedlings failed to accumulate LNK1, LNK2, and MYB3 transcripts (Supplemental Fig. S8). The double mutant lnk1/lnk2 was morphologically indistinguishable from wild-type plants, which is agreement with the previous reports (Xie et al., 2014; Pérez-García et al., 2015). Additionally, the triple mutant lnk1/lnk2/myb3 plants were also morphologically indistinguishable from wild-type plants. To further investigate whether the two conserved Asp residues in R1 and R2 of LNK C termini and ENT motif affect the activity of LNKs, Arabidopsis lnk1/lnk2 and wild-type plants constitutively overexpressing LNK1-HA, LNK1D556N-HA, LNK1D614N-HA, LNK1D556N/D614N-HA, and ENT-LNK3-HA were generated. qRT-PCR analysis of 2-week-old seedlings showed that the C4H gene expression level was significantly higher in lnk1/lnk2 plants and LNK1D556N/D614N- HA/lnk1/lnk2 plants compared to wild-type plants LNK1-HA/lnk1/lnk2 plants, respectively (Fig. 6A). In Arabidopsis wild-type background, the C4H expression level was reduced in MYB3-HA, LNK1-HA, LNK1D556N-HA, LNK1D614N-HA, and ENT-LNK3-HA-overexpressing plants compared with plants overexpressing LNK1D556N/D614N-HA and wild-type plants. Interestingly, the C4H gene expression level was the highest in lnk1/lnk2/myb3 plants compared with wild-type and lnk1/lnk2 plants (Fig. 6A). It has been reported that the REV8 directly regulated anthocyanin biosynthesis-related gene UGT79B1 (At5g54060, anthocyanin 3-O-glucoside 2′-O-xylosyltransferase) is significantly up-regulated in lnk1/lnk2 plants (Pérez-García et al., 2015). Here, we found that the UGT79B1 gene was not changed in MYB3-HA-overexpressing plants compared with wild-type plants, indicating that MYB3 does not regulate the UGT79B1 gene expression (Supplemental Fig. S9A). Additionally, the UGT79B1 expression level was down-regulated in LNK1-HA, LNK1D556N-HA, LNK1D614N-HA, and ENT-LNK3-HA-overexpressing plants, while significantly up-regulated in lnk1/lnk2 and LNK1D556N/D614N-HA/lnk1/lnk2 plants, compared with plants overexpressing LNK1D556N/D614N-HA and wild-type plants, respectively (Supplemental Fig. S9A). These results are in agreement with data showing that LNK1 and LNK2 act as transcriptional repressors of RVE8-activated UGT79B1 gene expression (Pérez-García et al., 2015). Taken together, our observation suggests that LNK1 and LNK2 act as corepressors of MYB3 to modulate C4H gene expression in plants, and this regulation depends on the two conserved Asp residues in R1 and R2 of LNKs’ C-termini and the ENT domain of N termini.
Figure 6.
LNK1 and LNK2 repress phenylpropanoids biosynthesis. A and B, Measurement of C4H gene expression (A) and SM (B) levels in indicated genotypes by LC/MS and qRT-PCR, respectively. Values are means ± sd of three biological repeats. Asterisks indicate statistically significant differences compared with wild-type and lnk1/lnk2 plants (P < 0.05, Student’s t test).
To determine whether modulation of LNK1 and LNK1 derivatives activity affects the MYB3-regulated phenylpropanoid metabolism, the content of SM and anthocyanin were analyzed for all genotypes. Liquid chromatography/mass spectrometry (LC/MS) assays showed that SM accumulated at a significantly higher level in lnk1/lnk2 plants and LNK1D556N/D614N-HA/lnk1/lnk2 plants than wild-type and myb3 plants. The level of SM was significantly reduced in plants overexpressing MYB3-HA, LNK1-HA, LNK1D556N-HA, LNK1D614N-HA, and ENT-LNK3-HA compared with plants overexpressing LNK1D556N/D614N-HA and wild-type plants (Fig. 6B). As shown in Supplemental Figure S9B, decreased anthocyanin accumulation was observed in MYB3-HA, LNK1-HA, LNK1D556N-HA, LNK1D614N-HA, and ENT-LNK3-HA-overexpressing plants, whereas the anthocyanin content was significantly higher in lnk1/lnk2 plants and LNK1D556N/D614N-HA/lnk1/lnk2 plants. The contents of both SM and anthocyanin were the highest in lnk1/lnk2/myb3 plants compared to all other plants (Fig. 6; Supplemental Fig. S9), as expected. We did not observe the change of the anthocyanin concentration in the LNK1D556N/D614N-HA-overexpression plants. These results are consistent with the transcriptional changes observed in these plants and with the negative role for MYB3 and the corepressors of LNK1 and LNK2 in the control of the phenylpropanoid pathway.
DISCUSSION
R2R3-MYB TFs possess an N-terminal DNA-binding domain and a C-terminal regulation domain. Based on the highly varied C-terminal domains, R2R3-MYB TFs have been divided into subgroups. The function is broadly conserved for the R2R3-MYB TFs of the same subgroup in different angiosperms. Subgroup 4 R2R3-MYB TFs, MYB3, MYB4, MYB7, and MYB32 are considered as transcriptional repressors due to the presence of the EAR repression motif (pdLHLD/LLxiG/S) in their C termini (Dubos et al., 2010). MYB4 is responsive to UV-B irradiation and controls sinapate ester biosynthesis through negatively regulating the expression of the C4H gene (Jin et al., 2000), while MYB32 regulates pollen wall composition due to be a pollen-specific repressor of lignin biosynthesis (Preston et al., 2004). Additionally, MYB7 represses flavonol biosynthesis and partly involved in regulation of UV sunscreens (Fornalé et al., 2014). Here, we show that MYB3, like MYB4, negatively regulates the expression of the C4H gene and represses the sinapate ester biosynthesis. However, the regulatory activity of four members of subgroup 4 R2R3-MYB TFs is different. MYB4, MYB7, and MYB32 possess a conserved GY/FDFLGL motif in their C termini, which can be recognized by an importin β-like protein SAD2, and then transports the SAD2-MYBs complex into the nucleus (Zhou et al., 2015b). Here, we demonstrated that the repression activity of MYB3 is directly regulated by LNKs, and this interaction depends on the varied C-terminal domain. However, the R3-MYB factor MYBL2 repressed flavonoid biosynthesis through directly inhibiting the activity of MYB-BHLH-WDR complexes (Dubos et al., 2008). Therefore, the same subgroup R2R3-MYB TFs share similar functions but have different regulation mechanisms.
The regulation of gene expression needs interlocked transcriptional feedback loops with both activating and repressive components. In the Arabidopsis circadian clock, many interlocked transcriptional components have been identified. For example, transcription of the morning phased CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) gene is repressed by the sequential binding of PSEUDORESPONSE REGULATOR (PRR) family protein to the CCA1 and LHY promoters, and this repression requires a corepressor, encoded by members of the TOPLESS/TOPLESS-RELATED gene family (Nakamichi et al., 2010; Wang et al., 2013). LNK1 and LNK2 have been demonstrated to be as transcriptional coactivators via protein-protein interactions to enhance the transcription activity of RVE8 and RVE4 in the control of evening-expressed clock genes (Xie et al., 2014). However, it has been reported recently that LNK1 and LNK2 act as repressors of the expression of anthocyanin structural genes (such as UGT79B1), a repressive role that counteracts RVE8-activating function (Pérez-García et al., 2015). These studies thus unravel a dual sign for the regulatory activity of RVE8-LNK interaction. Here, we identified MYB3 as a new binding partner of LNK1 and LNK2. Our results showed that LNK1 and LNK2 serve as transcriptional corepressors interacting with MYB3 and facilitating binding of MYB3 with C4H promoter. Additionally, we found that LNK1 alone does not directly activate the C4H promoter (Fig. 4A), which contradicts the reported data showing LNK1 is recruited to elements of PRR5 promoter and affects their activity (Xie et al., 2014). Therefore, LNKs probably have the different regulatory functions in different biological processes.
LNKs are plant-specific family proteins and consist of two conserved regions (R1 and R2). Both R1 and R2 domains contribute to the interaction between LNKs and multiple clock TFs (Xie et al., 2014). Mutation of two residues in either R1 or R2 domain (Arg-555Asp-556 to GlyGly or Asp-614Arg-615 to GlyGly) of LNK1 reduces the ability of LNK1 to interact with RVE4 (Xie et al., 2014). It has been widely accepted that the conserved Asp residue plays an important role in mediating protein-protein interaction. For example, the conserved Asp residue in the JAZ interaction domain of MYC2 and MYC3 plays a key role in binding to most JAZ proteins (Fernández-Calvo et al., 2011; Frerigmann et al., 2014; Goossens et al., 2015). Our recent publication showed that the conserved Asp residue mutation of SID motif of MYB4, MYB7, and MYB32 greatly reduces the interaction with SAD2 and is not transported into the nucleus (Zhou et al., 2015b). Here, we mutated the conserved Asp residue in either the R1 or R2 domain of LNK1, which does not affect the interaction between LNK1 and MYB3. However, two conserved Asp residues mutation from both R1 and R2 domains of LNK1 completely abolishes the interaction with MYB3. Thus, we postulate that the conserved Asp residue mutation of LNK1 greatly reduces the protein-protein interaction, probably by perturbing the LNK1 protein structure and thereby indirectly compromising the MYB3 binding domain.
Many plant nucleotide-binding Leu-rich repeat proteins possess an N-terminal domain that plays distinct roles in regulating downstream signaling pathways (Collier and Moffett, 2009). For example, the extended N-terminal domain of tomato (Solanum lycopersicum) Prf interacts with Pto kinase that directly interacts with AvrPtoB, which mediates downstream resistance response (Gutierrez et al., 2010). Tomato Mi-1.1 and 1.2 also contain an extra N-terminal domain play dual regulatory roles in signaling host cell death (Lukasik-Shreepaathy et al., 2012). Here, we also found that LNK1 and LNK2 possess an ENT domain around 300 amino acids long that is essential for LNK transcription activity. Therefore, we postulate that LNK1 and LNK2 serve as coregulators connecting the components of the transcription complex to regulate the downstream signaling pathway. It would be interesting to learn in the future whether the multilayered regulatory mechanism exists for LNKs protein. Further investigation of the ENT domain and the identification of LNKs’ ENT domain interaction partners could provide the potential for discovering new links into a range of plant signaling pathways.
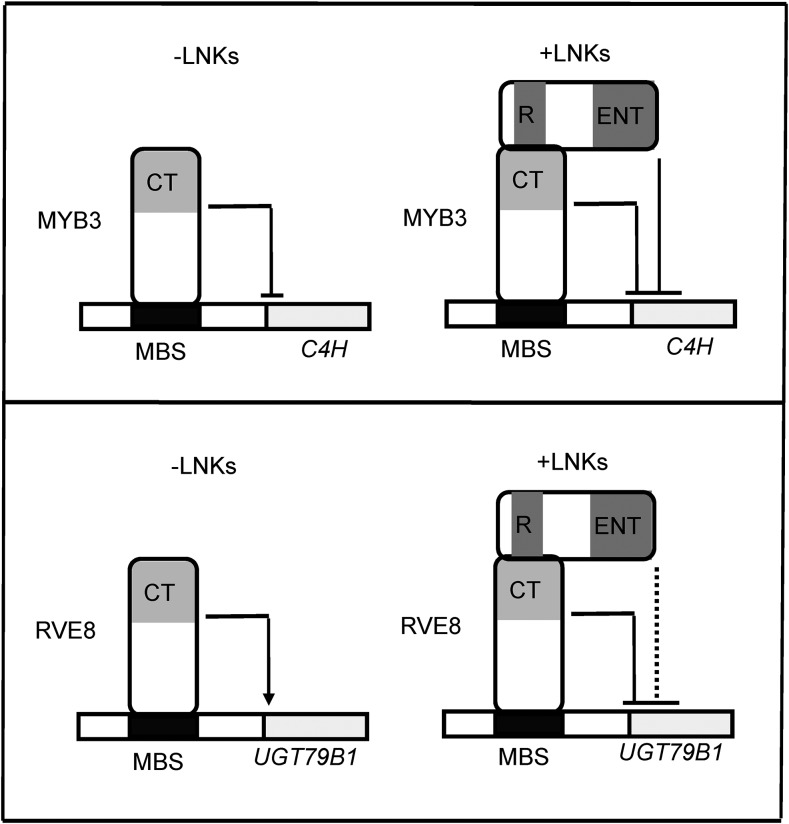
In summary, the subgroup 4 R2R3-MYB TF MYB3 is a novel identified repressor in the phenylpropanoid biosynthesis. The repression activity of MYB3 to C4H gene expression is directly regulated by corepressors LNK1 and LNK2, which also act as corepressors of the expression of anthocyanin structural gene UGT79B1 activated by MYB TF RVE8. The two conserved Asp residues in both R1 and R2 domains of LNKs are essential to mediate protein-protein interaction. Importantly, the ENT domain became a negative regulator in LNK-MYB3 transcription complex-dependent repression of C4H gene expression. Therefore, our data can be combined into a coherent model (Fig. 7) and may ultimately be useful for the design of artificial TFs for plant metabolic engineering.
Figure 7.
Simplified transcriptional networks of LNKs as corepressors in Arabidopsis phenylpropanoids biosynthesis. LNKs serve as transcriptional corepressors recruited to the promoters of target genes C4H and UGT79B1 (Pérez-García et al., 2015), via protein-protein interaction with the DNA binding MYB TFs MYB3 and RVE8. Both regions 1 and 2 (R1 and R2) of C termini of LNKs contribute to the interaction with the C termini of MYB3. The ENT domain of LNKs is required for its repression activity. Arrows indicate activation, T-shaped lines indicate inhibition, and broken lines indicate that this regulation was not reported to date. CT, C termini; MBS, MYB binding site; UGT79B1, anthocyanin 3-O-glucoside.
MATERIALS AND METHODS
Yeast Two-Hybrid Assays
Full-length MYB3 cloned in pAS2.1 was used as bait for yeast two-hybrid screening. Using the Stratagene cDNA synthesis kit, amplified cDNA libraries representing 2 × 106 primary transformants were prepared from an equal mixture of RNA from stems, leaves, roots, and flowers of mature ecotype Col-0 Arabidopsis (Arabidopsis thaliana) plants. The cDNA in this library had been fused to the GAL4 activation domain (GAL4 AD) into pACT2 plasmid library (James et al., 1996). Cotransformation of bait and cDNA library was performed into yeast strain PJ64-4A according to a modified yeast transformation protocol (James et al., 1996). Transformants were plated on minimal synthetic defined (SD)-Glc medium containing 10 mm 3-amino-1,2,4-triazole and lacking Trp, Leu, and His (-LWH).The full-length genes LNK1, LNK2, LNK3, and LNK4, the deletion derivatives LNK1∆N1, LNK1∆N2, LNK1∆C, MYB3∆N, and MYB3∆C, and the mutated LNK1 genes LNK1D556N, LNK1D614N, and LNK1D556N-D614N were cloned into pACT2 or pAS2.1. The point mutations were constructed using the GeneTailor site-directed mutagenesis system (Life Technologies). All constructed plasmids were sequenced. All primer sequences are presented in Supplemental Table S1. Interaction assays were performed by cotransformation of bait and prey plasmids into yeast strain PJ69-4A according to the protocol as previously described (Zhou et al., 2015b). The β-galactosidase assay was performed as described in the Yeast Protocols Handbook (Clontech).
Arabidopsis Protoplast Transactivation Assays
Full-length MYB3-HA, LNK1-HA, LNK2-HA, LNK3-HA, and LNK4-HA, or mutatedLNK1D556N-HA, LNK1D614N-HA, LNK1D556N/D614N-HA, and chimeric ENT-LNK3-HA (HA tag, influenza virus hemagglutinin tag) were PCR amplified cloned into effector plasmid pRT101 under the CaMV 35S promoter (Töpfer et al., 1987). The promoter fragments of C4H-fused GUS cloned in reporter plasmid GusXX was described previously (Zhou et al., 2015b). All primer sequences are presented in Supplemental Table S1. Arabidopsis protoplast suspensions were cotransformed with a reporter plasmid combined with effector plasmids by polyethylene glycol-mediated transfection as previously described (Schirawski et al., 2000). GUS activity assays were performed as described (van der Fits and Memelink, 1997).
Plant Materials and Growth Conditions
The T-DNA insertion lines myb3 (CS478221), lnk1-3 (SALK_024353), lnk2-5 (CS807006), lnk3-2 (SALK_085551C), and lnk4-2 (CS120858) were obtained from the ABRC and had been described previously (Xie et al., 2014; Pérez-García et al., 2015). Pollen from homozygous lnk1-3 plants were used to pollinate emasculated homozygous lnk2-5 flowers to generate lnk1/lnk2 double homozygous plants. The triple mutant lnk1/lnk2/myb3 plants were generated from a cross between lnk1/lnk2 and myb3 plants. For construction of transgenic lines constitutively overexpressing MYB3-HA, LNK1-HA, LNK1D556N-HA, LNK1D614N-HA, LNK1D556N/D614N-HA, and ENT-LNK3-HA, the CaMV 35S cassettes containing the open reading frames of these genes were digested from pRT101 using SphI and cloned into pCAMBIA1300 digested using SphI. The binary vectors pCAMBIA1300 and empty vector were introduced into Agrobacterium tumefaciens strain LBA4404. A. tumefaciens was then used to transform Arabidopsis homozygous lnk1/lnk2 plants and ecotype Col-0 by floral dip (Clough and Bent, 1998). Transgenic Arabidopsis plants were identified using the methods that were described by our previous report (Zhou et al., 2015b).
BiFC
The N-terminal (YN) or C-terminal (YC) fragments of the yellow fluorescent protein (nYFP or cYFP) were fused either N-terminally or C-terminally with MYB3, LNK1, LNK2, LNK3, and LNK4 and cloned in pRTL2-YNEE (nYFP-) or pRTL2-HAYC (-cYFP). All primer sequences are presented in Supplemental Table S1. The constructs were transiently coexpressed in all possible combinations of nYFP and cYFP fusion proteins in Arabidopsis cell suspension by polyethylene glycol-mediated transfection as previously described (Schirawski et al., 2000). Images of triplicate transfected protoplasts were acquired with a Leica DM IRBE confocal laser scanning microscope as described previously (Zhou et al., 2015b). Microscopy images were analyzed using the ImageJ software (Abràmoff et al., 2004).
qRT-PCR Analyses
Total RNA extraction and reverse transcription (Revert Aid first-strand cDNA synthesis kit; Fermentas) were described in the manufacturer’s instructions and our previous publication (Zhou et al., 2010). qRT-PCR was performed as previously described (Zhou et al., 2010). The PCR primers for C4H, CHS, 4CL1, 4CL3, PAL, F5H, OMT, CAD, UGT79B1, and reference gene UBQ10 were as previously described (Jin et al., 2000; Fornalé et al., 2014; Xie et al., 2014; Pérez-García et al., 2015).
ChIP Assay
Detached leaves from 4-week-old Col-0 and MYB3OX (35S::MYB3-HA) plants were incubated in darkness for 24 h. HA antibody (Roche) was used for immunoprecipitation. ChIP assays were quantified by real-time PCR after normalizing with the input DNA. qPCR was performed using primers flanking the MYB-binding sequence of C4H promoter. Primers used for qRT-PCR presented in Supplemental Table S1. The coding sequence region of C4H gene and the reference gene UBQ10 promoter were used as a negative control. ACTIN2 was used as a reference gene. ChIP assays were performed as previously described (Saleh et al., 2008).
Protein Purification and Western Blot
Overexpression of HA-tagged genes LNK1-HA, LNK1D556N-HA, LNK1D614N-HA, LNK1D556N/D614N-HA, and ENT-LNK3-HA in Arabidopsis seedlings were ground in protein extraction buffer (25 mm sodium phosphate buffer, pH 7.5, 1 mm EDTA, 7 mm β-mercaptoethanol, 1% Triton X-100, and 10% glycerol). Proteins were separated on 10% (w/v) SDS-polyacrylamide gels, transferred to Protran nitrocellulose by semidry blotting, and detected with anti-HA antibody (Roche). The detailed extraction method and western blot were described in our previous publication (Zhou et al., 2015b).
EMSA
To produce His-tagged proteins, MYB3, LNK1, LNK3, and ENT-LNK3 were amplified and cloned in pASK-IBA45plus. All primer sequences are presented in Supplemental Table S1. These constructs were transformed into Escherichia coli strain BL21 (DE3) pLysS selecting on 50 µg/mL chloramphenicol and 200 µg/mL carbenicillin, and protein expression was induced by 0.2 µg/mL anhydrotetracycline. The soluble His fusion proteins were extracted and immobilized onto Ni-NTA agarose beads (Qiagen). The 200-bp probe sequence including MYB3 binding site is presented in Supplemental Table S1. EMSAs were performed using biotin-labeled double-stranded probes and the Light Shift Chemiluminescent EMSA Kit according to the manufacturer’s instructions (Thermo Fisher).
Measurement of Sinapoyl Malate and Anthocyanin
Arabidopsis seedlings were grown in solid acidified Murashige and Skoog medium at pH 5.4 by addition of HCl. Then, 2-week-old leaves were frozen and ground in liquid nitrogen. Total phenolics were extracted in dry ice-cold methanol. The anthocyanin content was measured as described by Swain and Hillis (1959) and presented as (A535 − A650)/g fresh weight. The sinapoyl malate content was analyzed by LC/MS as described previously (Tamagnone et al., 1998). The experiment was repeated three biological times.
Statistical Analysis
Data were analyzed using Student’s t test. A P value < 0.05 was considered to be significant.
Accession Numbers
Sequence data for the genes described in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MYB3 (At1g22640), LNK1(At5g64170), LNK2(At3g54500), LNK3(At3g12320), LNK4(At5g06980), C4H (At2g30490), UGT79B1 (At5g54060), and UBQ10 (At4g05320).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Description and Identification of myb3 Mutant and MYB3-Overexpressing Plants.
Supplemental Figure S2. Measurement of the Transcripts of Phenylpropanoids Biosynthesis Genes in Wild-Type, myb3, and MYB3OX Plants by Quantitative RT-PCR.
Supplemental Figure S3. Effect of Overexpression of MYB3 on the Tolerance of Arabidopsis Seedlings to UV-B.
Supplemental Figure S4. MYB4, MYB7, and MYB32 Do Not Interact with a Small Family of Four LNK Proteins in Yeast.
Supplemental Figure S5. The LNK1D556N, LNK1D614N, and LNK1D556N-D614N Mutations Do Not Affect LNK1 Protein Stability in Transformed Yeasts.
Supplemental Figure S6. MYB3 Repressor Activity Is Not Enhanced by LNK3 and LNK4.
Supplemental Figure S7. Amino Acid Sequence Alignment of a Small Family of Four LNK Proteins.
Supplemental Figure S8. Identification of lnk1/lnk2 Mutant, lnk1/lnk2/myb3 Mutant, and LNK1-HA, LNK1D556N-HA, LNK1D614N-HA, LNK1D556N/D614N-HA, and ENT-LNK3-HA-Overexpressing Plants.
Supplemental Figure S9. LNK1, LNK2, and MYB3 Repress Anthocyanin Accumulation.
Supplemental Table S1. Primers Used in This Study.
Glossary
- TF
transcription factor
- Y2H
yeast two-hybrid
- EMSA
electrophoresis mobility shift assay
- ChIP
chromatin immunoprecipitation assay
- SM
sinapoyl malate
- BiFC
bimolecular fluorescence complementation
- LC/MS
Liquid chromatography/mass spectrometry
References
- Abràmoff MD, Magalhães PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Bradshaw HD, Schemske DW (2003) Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 426: 176–178 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collier SM, Moffett P (2009) NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci 14: 521–529 [DOI] [PubMed] [Google Scholar]
- Du H, Huang Y, Tang Y (2010) Genetic and metabolic engineering of isoflavonoid biosynthesis. Appl Microbiol Biotechnol 86: 1293–1312 [DOI] [PubMed] [Google Scholar]
- Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55: 940–953 [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornalé S, Lopez E, Salazar-Henao JE, Fernández-Nohales P, Rigau J, Caparros-Ruiz D (2014) AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol 55: 507–516 [DOI] [PubMed] [Google Scholar]
- Frerigmann H, Berger B, Gigolashvili T (2014) bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol 166: 349–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53: 814–827 [DOI] [PubMed] [Google Scholar]
- Goossens J, Swinnen G, Vanden Bossche R, Pauwels L, Goossens A (2015) Change of a conserved amino acid in the MYC2 and MYC3 transcription factors leads to release of JAZ repression and increased activity. New Phytol 206: 1229–1237 [DOI] [PubMed] [Google Scholar]
- Gutierrez JR, Balmuth AL, Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, Jones AM, Rathjen JP (2010) Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J 61: 507–518 [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik-Shreepaathy E, Slootweg E, Richter H, Goverse A, Cornelissen BJ, Takken FL (2012) Dual regulatory roles of the extended N terminus for activation of the tomato MI-1.2 resistance protein. Mol Plant Microbe Interact 25: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3: 212–217 [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua N-H, Sakakibara H (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-García P, Ma Y, Yanovsky MJ, Mas P (2015) Time-dependent sequestration of RVE8 by LNK proteins shapes the diurnal oscillation of anthocyanin biosynthesis. Proc Natl Acad Sci USA 112: 5249–5253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW (2004) AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J 40: 979–995 [DOI] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugnone ML, Faigón Soverna A, Sanchez SE, Schlaen RG, Hernando CE, Seymour DK, Mancini E, Chernomoretz A, Weigel D, Más P, Yanovsky MJ (2013) LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc Natl Acad Sci USA 110: 12120–12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Schirawski J, Planchais S, Haenni AL (2000) An improved protocol for the preparation of protoplasts from an established Arabidopsis thaliana cell suspension culture and infection with RNA of turnip yellow mosaic tymovirus: a simple and reliable method. J Virol Methods 86: 85–94 [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain T, Hillis WE (1959) The phenolic constituents of Prunus domestica. I: the quantitative analysis of phenolic constituents. J Agric Food Chem 10: 63–68 [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C (1998) The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Töpfer R, Matzeit V, Gronenborn B, Schell J, Steinbiss HH (1987) A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res 15: 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Memelink J (1997) Comparison of the activities of CaMV 35S and FMV 34S promoter derivatives in Catharanthus roseus cells transiently and stably transformed by particle bombardment. Plant Mol Biol 33: 943–946 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kim J, Somers DE (2013) Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci USA 110: 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Xie Q, Wang P, Liu X, Yuan L, Wang L, Zhang C, Li Y, Xing H, Zhi L, Yue Z, et al. (2014) LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 26: 2843–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang W, Zhao Y, Gong X, Guo L, Zhu G, Wang X, Gong Z, Schumaker KS, Guo Y (2007) SAD2, an importin -like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 19: 3805–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Wei L, Sun Z, Gao L, Meng Y, Tang Y, Wu Y (2015a) Production and transcriptional regulation of proanthocyanidin biosynthesis in forage legumes. Appl Microbiol Biotechnol 99: 3797–3806 [DOI] [PubMed] [Google Scholar]
- Zhou M, Sun Z, Wang C, Zhang X, Tang Y, Zhu X, Shao J, Wu Y (2015b) Changing a conserved amino acid in R2R3-MYB transcription repressors results in cytoplasmic accumulation and abolishes their repressive activity in Arabidopsis. Plant J 84: 395–403 [DOI] [PubMed] [Google Scholar]
- Zhou ML, Zhu XM, Shao JR, Wu YM, Tang YX (2010) Transcriptional response of the catharanthine biosynthesis pathway to methyl jasmonate/nitric oxide elicitation in Catharanthus roseus hairy root culture. Appl Microbiol Biotechnol 88: 737–750 [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40: 22–34 [DOI] [PubMed] [Google Scholar]