The first structure of a plant eIF4E bound to an eIF4G interacting peptide supports a universal mechanism of regulation of translation initiation in higher eukaryotes.
Abstract
The association-dissociation of the cap-binding protein eukaryotic translation initiation factor 4E (eIF4E) with eIF4G is a key control step in eukaryotic translation. The paradigm on the eIF4E-eIF4G interaction states that eIF4G binds to the dorsal surface of eIF4E through a single canonical alpha-helical motif, while metazoan eIF4E-binding proteins (m4E-BPs) advantageously compete against eIF4G via bimodal interactions involving this canonical motif and a second noncanonical motif of the eIF4E surface. Metazoan eIF4Gs share this extended binding interface with m4E-BPs, with significant implications on the understanding of translation regulation and the design of therapeutic molecules. Here we show the high-resolution structure of melon (Cucumis melo) eIF4E in complex with a melon eIF4G peptide and propose the first eIF4E-eIF4G structural model for plants. Our structural data together with functional analyses demonstrate that plant eIF4G binds to eIF4E through both the canonical and noncanonical motifs, similarly to metazoan eIF4E-eIF4G complexes. As in the case of metazoan eIF4E-eIF4G, this may have very important practical implications, as plant eIF4E-eIF4G is also involved in a significant number of plant diseases. In light of our results, a universal eukaryotic bipartite mode of binding to eIF4E is proposed.
The initiation of protein synthesis is a key control and highly regulated step in eukaryotic gene expression (Sonenberg and Hinnebusch, 2009). Translation initiation leads to the assembly of the large (60S) and small (40S) ribosomal subunits into an active 80S ribosome that is able to locate the correct start codon of the mRNA molecule. This is facilitated by the coordinated actions of at least 12 protein initiation factors (Browning, 2004; Jackson et al., 2010; Hinnebusch, 2011; Hinnebusch and Lorsch, 2012; Browning and Bailey-Serres, 2015). In cap-dependent translation, binding of the eukaryotic translation initiation factor 4F (eIF4F) to the 7-methyl guanosine cap (m7G cap) at the 5′ end of mRNA drives the attachment of the mRNA to the ribosomal 43S preinitiation complex. In mammals, eIF4F is built by three proteins: the mRNA 5′ cap binding protein eIF4E, the RNA helicase eIF4A, and the scaffolding protein eIF4G, which contains binding domains for eIF4E, eIF4A, eIF3, and poly(A)-binding protein (PABP; Marcotrigiano et al., 1997; Pestova et al., 2001; Gross et al., 2003). In plants, purified eIF4F heterodimers contain eIF4E and eIF4G (Hinnebusch and Lorsch, 2012); plant eIF4A appears to be loosely associated and is therefore easily lost during purification (Lax et al., 1986). EIF4G associates with eIF3, recruiting the 40S subunit of the ribosome and initiating scanning of the mRNA in the 5′ to 3′ direction (Jackson et al., 2010; Park et al., 2011). EIF4G additionally binds simultaneously to eIF4E and PABP, the latter being able to interact with the mRNA poly(A) tail (Aitken and Lorsch, 2012; Browning and Bailey-Serres, 2015), resulting in a transient circularization of the mRNA.
Higher plants contain an additional isoform of the eIF4F complex, eIFiso4F (Lax et al., 1986; Browning and Bailey-Serres, 2015). Plant eIF4E and eIFiso4E share approximately 50% amino acid identity, while eIFiso4G differs from eIF4G in having a truncated N terminus, but shares similar binding domains for eIF4E, eIF3, and eIF4A. EIF4G interacts with eIF4E through a highly conserved Y(X)4Lϕ amino acid sequence, where X is variable and ϕ is hydrophobic, known as the canonical (C) binding motif (Mader et al., 1995; Marcotrigiano et al., 1999; Gross et al., 2003). The C motif is also conserved in the plant-specific eIFiso4G subunit. Although eIFiso4G can form mixed complexes with eIF4E that retain activity in vitro, dissociation constants showed molecular specificity, suggesting a differential role of the eIFiso4F complex in translation initiation (Gallie and Browning, 2001; Mayberry et al., 2011). Metazoan eIF4E-binding proteins (m4E-BPs) also contain the C motif (Mader et al., 1995; Marcotrigiano et al., 1999) and inhibit translation initiation by competing for the same binding site on the eIF4E surface, thus blocking the assembly of active translation eIF4F complexes (Mader et al., 1995; Matsuo et al., 1997; Marcotrigiano et al., 1999; Gross et al., 2003). In addition to the C motif, m4E-BPs also contain a downstream noncanonical (NC) domain that forms a loop and binds to a highly conserved hydrophobic lateral surface of eIF4E (Mizuno et al., 2008; Gosselin et al., 2011; Paku et al., 2012; Lukhele et al., 2013; Igreja et al., 2014; Peter et al., 2015a, 2015b). An elbow loop downstream of the C motif of m4E-BPs induces the bending of the peptide backbone, thus allowing the NC loop to reach and contact the lateral hydrophobic pocket of eIF4E (Kinkelin et al., 2012; Peter et al., 2015a, 2015b). In plants, only lipoxygenase 2 (LOX2) and the beta subunit of the nascent polypeptide-associated complex (BTF3) have been identified as putative eIF4E interacting proteins (Freire et al., 2000; Freire, 2005), and very little is known about their association with eIF4E.
The NMR structure of the yeast eIF4E-eIF4G393-490 complex has revealed that the C-terminal region downstream of the C motif folds back toward the flexible N-terminal tail of eIF4E, not interacting with the lateral surface of eIF4E (Gross et al., 2003). Since the lateral interaction of the NC motif seemed to occur only in m4E-BPs, it was proposed to be the reason for the known advantage of m4E-BPs over eIF4G in binding eIF4E, leading to translation repression (Paku et al., 2012; Lukhele et al., 2013; Igreja et al., 2014). However, the very recently solved crystal structure of Homo sapiens and Drosophila melanogaster eIF4E-eIF4G peptide complexes has revealed that eIF4G also binds eIF4E through both C and NC motifs, coming into contact with the same dorsal and lateral eIF4E surfaces, respectively, as m4E-BPs (Grüner et al., 2016). Additionally, Grüner et al. (2016) have shown that the known competitive advantage of m4E-BPs over eIF4G is provided by the amino acid composition of the linker and the NC binding motif.
Here we present high-resolution structural data of melon (Cucumis melo) eIF4E (Cm eIF4E) in complex with a Cm eIF4G peptide containing both C and NC possible motifs. Our previous work (Miras et al., 2017b) identified Cm eIF4E residues implicated in binding to the C domain of Cm eIF4G. Amino acid replacements in the corresponding positions resulted in reduction of the Cm eIF4E affinity for Cm eIF4G980–1159 and, concomitantly, reduced cap-independent translation activity of a reporter mRNA carrying the viral Ma5TE cap-independent translation enhancer (CITE; Miras et al., 2017b). The structural analysis shown here indicates that Cm eIF4G, similarly to metazoan eIF4G, binds eIF4E through the C but also through the NC domain. Our in vitro and in vivo functional analyses support the importance of both motifs for a stable eIF4E-eIF4G complex formation. As in the case of metazoan eIF4E-eIF4G interaction, our findings may have important practical implications, because the plant eIF4E-eIF4G complex is involved in a significant number of plant diseases caused by viruses (Truniger and Aranda, 2009; Sanfaçon, 2015). In light of our results, a universal eukaryotic bipartite mode of binding to eIF4E is proposed.
RESULTS
Crystal Structure of Cm eIF4E Bound to eIF4G1003-1092
To gain insight into the binding mode of plant eIF4G to eIF4E, we crystallized and solved the structures of Cm eIF4E in its free form, in the m7GDP cap analog bound state, and in complex with a Cm eIF4G peptide to 2.2 Å, 3.5 Å, and 1.9 Å resolution, respectively. Previous structural studies with other eIF4Es from diverse organisms have shown that the full-length protein is resistant to crystallization. However, N-terminally truncated versions of eIF4E, retaining their cap-binding abilities, have been successfully crystallized (Marcotrigiano et al., 1997, 1999; Matsuo et al., 1997; Monzingo et al., 2007). We expressed an N-terminally truncated Cm eIF4E for crystallization starting at amino acid 51, thus lacking the putative N-terminal flexible region (eIF4E51-235). The Cm eIF4G peptide used for complex formation and crystallization, Cm eIF4G1003-1092, contained the canonical eIF4E binding motif and a downstream extra sequence that included a hydrophobic patch conserved in other eIF4G, eIFiso4G, and m4E-BP proteins (Fig. 1; Supplemental Fig. S1).
Figure 1.
Structure-based sequence alignment of the eIF4E-interacting regions of eIF4G and m4E-BP orthologs. A sequence alignment from superposition of the different eIF4E crystal structures in complex with eIF4E interacting proteins from higher eukaryotes available at the Protein Data Bank (PDB IDs: Cm eIF4G, this work; Hs eIF4G, 5T46; Dm eIF4G, 5T47; Hs 4E-BP1, 4UED; Dm 4E-T, 4UE9; Dm Thor, 4UE8; Dm Mxt, 5ABU) was performed using Chimera (Pettersen et al., 2004). The canonical and NC eIF4E binding motifs (4E-BM) and the elbow loop are framed. The invariant Tyr residue of the canonical motif is typed in bold red and hydrophobic residues conserved in the NC motif are shadowed in blue. Red open circles above the alignment indicate key eIF4G residues within the domains interacting with eIF4E and mutated in this study. The conserved Ser residue in metazoan 4E-BPs is shown in bold blue. The two conserved Ser and Thr residues present in metazoan 4E-BPs implicated in phospho-regulation are colored in orange. Sequences are from Cucumis melo (Cm), Homo sapiens (Hs), and Drosophila melanogaster (Dm).
The solved structures of Cm eIF4E (Fig. 2), in their free state and in complex with a cap analog m7GDP, are similar to those observed for other eIF4Es from diverse organisms (Marcotrigiano et al., 1997; Gross et al., 2003; Monzingo et al., 2007; Paku et al., 2012; Papadopoulos et al., 2014; Peter et al., 2015a; 2015b). The Cm eIF4E crystals contain four independent copies in the asymmetric unit, all adopting a crescent-shaped conformation formed by a strongly bent beta sheet composed of eight antiparallel β-strands. The convex surface is decorated by three α-helices that form the dorsal surface (Fig. 2A). Major differences are confined to the cap-binding pocket, located on the concave, ventral surface of eIF4E. In all the structures described so far, this pocket is built by two conserved Trp residues located in loops L1 and L3, respectively. In Cm eIF4E and Cm eIF4E-m7GDP structures only loop L3 is visible (Fig. 2A) and adopts a conformation that closely resembles those described for other eIF4E structures in complex with a cap analog (Marcotrigiano et al., 1997, 1999; Matsuo et al., 1997; Monzingo et al., 2007). The orientation adopted by residue W128 is fully compatible with cap binding in all structures, though, in our structure, the m7GDP cap analog is observed in only two of the four independent molecules in the asymmetric unit interacting with W128 located in loop L3 (Fig. 2A). Binding is additionally stabilized through interactions between the phosphates in the cap and charged residues R114, R178, and K183. In contrast, loop L1 was never observed in Cm eIF4E and Cm eIF4E-m7GDP structures except for molecule C, in which it adopts a fixed conformation positioning the loop far away from the binding pocket due to intermolecular contacts with a neighboring subunit within the asymmetric unit. When Cm eIF4E is bound to the Cm eIF4G peptide (Fig. 2, B–G), the formation of a disulfide bond between Cys residues 133 and 171 is observed. This introduces a structural rearrangement in loop L3, which promotes the formation of a helix 310 (from residue P131 to residue A134) and pulls W128 out of the cap-binding cavity (Fig. 2D), thus rendering a putatively “auto-inhibited” conformation, unable to bind the cap. Even though the two Sϒ atoms are also in proximity (4.1 Å) in the free Cm eIF4E structure, no disulfide bridge was observed, and any attempts to crystallize a Cm eIF4F complex without the disulfide bond failed.
Figure 2.
Structure and molecular details of Cm eIF4E unbound or bound to Cm eIF4G1003-1092. A, Ribbon diagram of Cm eIF4E51-235 in complex with the m7GDP cap analog. The m7GDP is located in the cap-binding pocket. Residue W128, in direct interaction with the cap, is marked. B and C, Structure of the Cm eIF4E-eIF4G1003-1092 complex shown as a cartoon and surface rendering, respectively. Cm eIF4G1003-1092 (orange) contains the C and NC eIF4E binding motifs (4E-BM). D, Structural superposition of Cm eIF4E (in gray as cartoon) and Cm eIF4E-eIF4G1003-1092 (in blue as cartoon) binding pocket structures. The formation of a disulfide bridge (shown as sticks in yellow in the structure) promotes the formation of a helix 310 in loop L3 and pulls W128 from m7GDP-interacting position 1 to an apparently “auto-inhibited” position 2 (both labeled in the figure). E, Close-up views of the interaction of the Cm eIF4G C motif (orange) with the dorsal surface of Cm eIF4E (blue) in two orientations. F, Close-up view of the Cm eIF4G linker region (orange) and interactions with Cm eIF4E (blue). G, Close-up view of the NC interaction loop of Cm eIF4G (orange) showing all interactions with the lateral surface of Cm eIF4E (blue). Selected interface residues are shown as blue sticks for Cm eIF4E and as orange sticks for Cm eIF4G. In E to G, residues implicated in the interactions between Cm eIF4E and Cm eIF4G1003-1092 are shown as sticks.
EIF4G Exploits Both the C and NC Domains for Binding to eIF4E
The initial difference maps of the Cm eIF4E-eIF4G1003-1092 complex clearly showed the presence of an extra continuous electron density beyond the C domain extending to the lateral hydrophobic surface of Cm eIF4E (Supplemental Fig. S2). Taking the well-characterized C domain as an anchoring point, we were able to fit 37 amino acid residues of Cm eIF4G (from K1057 to N1093) within this density (Fig. 2, B and C). The structure revealed an N-terminal α-helix and a loop connected by a linker region (Fig. 2, E–G). Each of these three structural elements binds to a defined Cm eIF4E surface: (1) The N-terminal α-helix is formed by the C domain and interacts with the dorsal surface of Cm eIF4E (Fig. 2E); (2) an elbow after the canonical α-helix that bends the peptide backbone by approximately 90°, orienting the linker region downward to engage the lateral surface of Cm eIF4E (Fig. 2F); and (3) the loop contacts a hydrophobic pocket on the lateral surface of Cm eIF4E, indicating the existence of a NC binding motif (Fig. 2, E–G). Importantly, this second motif has been identified in m4E-BPs from D. melanogaster and in vertebrates (Kinkelin et al., 2012; Peter et al., 2015a, 2015b), and very recently in metazoan eIF4G (Grüner et al., 2016). This binding mode is structurally analogous to that observed in metazoan eIF4G and m4E-BPs (Supplemental Fig. S3) and is clearly distinct from the bracelet-like structure formed in the Sc eIF4E-eIF4G393-490 complex (Supplemental Fig. S3C).
As expected, the C motif of Cm eIF4G presents an α-helical fold that extends from residue Y1059 to A1068 (Fig. 2E), which resembles all the canonical peptides in complex with eIF4E solved so far (Gross et al., 2003; Mizuno et al., 2008; Umenaga et al., 2011; Kinkelin et al., 2012; Paku et al., 2012). Thus, the interactions established between the α-helical region containing the consensus sequence Y(X)4Lϕ and eIF4E are highly conserved; the hydroxyl group of the side chain of Y10594G is hydrogen-bonded to the carboxyl oxygen of P614E within the conserved H60-P61-L62 motif in eIF4E and additionally establishes lateral hydrophobic interactions with F10634G. L10644G is buried in a hydrophobic pocket built by Cm eIF4E residues V95, W99, L157, and I160. The next residue defined by the consensus sequence L10654G makes contacts with W994E and L1574E. In addition, the side chain of residue R10614G contacts Y1544E, while F10674G contacts the lateral chain of H604E and W994E (Fig. 2E). All these residues are well conserved in all eukaryotic eIF4Es and participate in other eIF4Gs and m4E-BPs interactions through the C motif (Supplemental Fig. S4; Marcotrigiano et al., 1999; Kinkelin et al., 2012; Peter et al., 2015a, 2015b).
The characteristic elbow loop, present in m4E-BPs and metazoan eIF4Gs, which begins immediately after the R/Q residues at position 9 of the canonical motifs (Hs Q621 and Dm R630) and contains a half helical turn beginning at the tip of the elbow (Hs F624-S626 and Dm K633-S635), is not observed in our structure. Instead, the α-helix containing the C domain presents an extra turn-reaching residue L10724G, which is stabilized through main-chain interactions between residues A10684G and F10714G and the side chain of residue N1034E (Fig. 2F). Hydrophobic interactions are also established between the side chains of residues A10684G and F10714G and residues W994E and E964E. This extra helical turn slightly bends the helix toward the lateral surface of Cm eIF4E and thus allows the NC domain of Cm eIF4G to contact the hydrophobic lateral core of Cm eIF4E (Figs. 2G and 3A). An intramolecular network of hydrophobic interactions is built by Cm eIF4G residues L1074, F1078, V1080, I1084, and M1088. I894E, a conserved hydrophobic residue in eukarya, is central in the interaction with Cm eIF4G and provides a hydrophobic core around which hydrophobic residues F10784G and I10844G are arranged (Fig. 3A). On the beta-turn of Cm eIF4G, L1087 contacts the main-chain conserved residues F704E and I1054E and engages the hydrophobic pocket also with L1114E and L1174E. M10884G makes side chain contacts with I1054E and with the carbonyl oxygen of H1064E. In the N terminus of the NC motif, F10784G contacts V1014E of α-helix α1 and Y904E of β-strand β1. In addition to the hydrophobic interactions, S10894G and T10924G show hydrogen bonds with N1044E located on helix α1 (Fig. 2G). The last C-terminal extension is stabilized mainly through hydrogen bonds.
Figure 3.
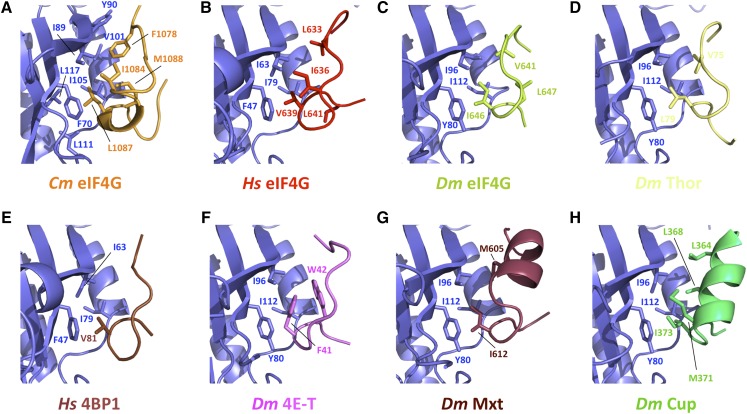
Structures of the eIF4E lateral hydrophobic pocket interacting with eIF4G and m4E-BP peptides. Hydrophobicity of residues in all eIF4E-interacting domains is conserved. Close-up views of eIF4E hydrophobic patch and residues involved in the binding of Cm eIF4E and eIF4G1003-1092 (A), Hs eIF4E and eIF4G592-653 (B; PDB ID: 5T46; Grüner et al., 2016), Dm eIF4E and eIF4G601-660 (C; PDB ID: 5T47; Grüner et al., 2016), Dm eIF4E and m4E-BP Thor50-83 (D; PDB ID: 4UE8; Peter et al., 2015a), Hs eIF4E and m4E-BP 4E-BP150-83 (E; PBD ID: 4UED; Peter et al., 2015a), Dm eIF4E and m4E-BP 4E-T9-44 (F; PBD ID: 4UE9; Peter et al., 2015a), Dm eIF4E and m4E-BP Mxt577-640 (G; PBD ID: 5ABV; Peter et al, 2015b), and Dm eIF4E and m4E-BP Cup325-376 (H; PBD ID: 4AXG; Kinkelin et al., 2012).
Despite the apparent lack of sequence conservation in the NC motif of eIF4Gs and m4E-BPs, the structure-based amino acid sequence alignment indicates that this patch of hydrophobic residues within this domain is mostly conserved between plants, vertebrates, and insects (Fig. 1; Supplemental Fig. S1). In fact, the spatial arrangement of residues F10784G, I10844G, and L10874G in our structure closely relates to those observed in H. sapiens and D. melanogaster complexes (Fig. 3). More specifically, residues L6334G, I6364G and V6394G and V6414G, I6464G and L6474G in the human and D. melanogaster eIF4F complexes occupy equivalent positions (Fig. 3, B and C; Grüner et al., 2016). This cluster of hydrophobic interactions is also observed, although to a lesser extent, in other m4E-BPs (Fig. 3, D–H); the highly conserved eIF4E hydrophobic pocket interacts with side chains of Thor V75 and L79, 4BP1 V81, 4E-T F41 and W42, Mxt M605, and I373 or Cup M371 and I373 (Fig. 3, D–H; Peter et al., 2015a, 2015b). Thus, binding to the lateral hydrophobic surface of eIF4E appears to be a conserved feature of m4E-BPs, eukaryotic eIF4Gs, and plant-specific eIFiso4G isoforms.
Functional Validation of the Cm eIF4E-eIF4G1003-1092 Structure
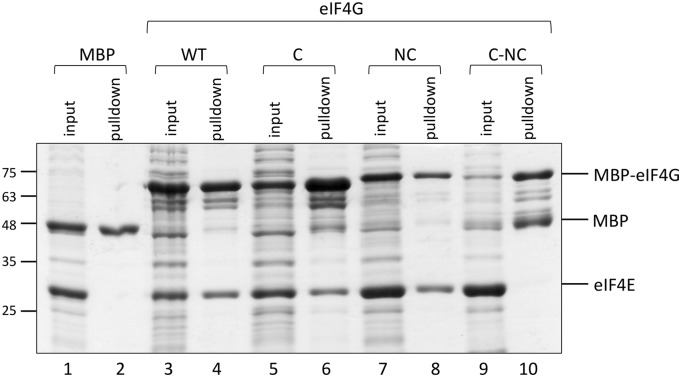
To validate our structural results, we substituted key residues in both proteins and performed in vitro pull-down assays (Fig. 4). We tested recombinant fragments of wild-type Cm eIF4G1003-1092 (eIF4G-WT) and mutated versions of it for binding to full-length Cm eIF4E. Mutations were introduced in the C (eIF4G-C), the NC (eIF4G-NC), and both (eIF4G-C-NC) Cm eIF4E-binding motifs. In the well-conserved C motif Y(X)4Lϕ of eIF4G-C, three residues, Y1059, L1064, and L1065, were substituted by Ala, while the hydrophobic residues F1078, I1084, and L1087 of the proposed NC motif in eIF4G-NC were substituted by Asp residues. All eIF4G fragments were expressed in Escherichia coli as N-terminal fusions of the maltose binding protein (MBP), and purified Cm eIF4E was added to the Cm eIF4G1003-1092 (wild type and mutants) cleared lysates in a fixed concentration. This purely qualitative pull-down analysis showed that substitutions in either the C or the NC binding-motifs did not disrupt the association of Cm eIF4G1003-1092 with Cm eIF4E (Fig. 4, lanes 3–8), while no binding to Cm eIF4E was observed when amino acid substitutions in the C and NC domains (eIF4G-C-NC) were combined (Fig. 4, lanes 9–10). To confirm these results, we compared the binding efficiencies of each mutant in pull-down assays using increasing concentrations (0.5–3 µm) of Cm eIF4E with a fixed concentration of Cm eIF4G1003-1092 (Fig. 5). In agreement with the results above, EIF4G-WT and eIF4G-NC seemed to display similar affinities for Cm eIF4E, binding Cm eIF4E at the lowest concentration (Fig. 5, A and D; MBP control in Fig. 5B), while the eIF4G-C interaction with Cm eIF4E was clearly impaired (Fig. 5C). Again, no binding was observed for the eIF4G-C-NC mutant, even if Cm eIF4E was added in excess (Fig. 5E). We quantified band intensities of the pulled-down Cm eIF4E and plotted them versus the Cm eIF4E input (Fig. 5F). When a hyperbolic binding curve was fitted to the resulting concentration data, approximate dissociation constants (Kd) of 2.15, 2.67, and 1.90 µm were obtained for eIF4G-WT, eIF4G-C, and eIF4G-NC, respectively. Therefore, in our assay, each of the binding sites seemed to be sufficient for a similarly productive interaction, and only simultaneous mutations affecting the two binding sites resulted in binding disruption. It should be pointed out that there are important differences in the Kd values estimated here and the value estimated for the wheat (Triticum aestivum) full-length eIF4E-eIF4G complex (Mayberry et al., 2011), and these may result from the nature of the Cm eIF4G peptide versus the wheat full-length protein; still, our data are fully useful for the relative comparison of the Cm eIF4G peptides analyzed here.
Figure 4.
In vitro interaction of Cm eIF4E and Cm eIF4G1003-1092. MBP pull-down assay showing the interaction of Cm eIF4E (wild type, full length) and MBP-Cm eIF4G1003-1092 (either wild type or mutated in C, NC, or C-NC). Bacterial lysates expressing MBP-tagged Cm eIF4G1003-1092 (wild type and mutants) were incubated with purified Cm eIF4E 1 μm in a volume of 300 μL. Of these, 15 μL (5%; input) was analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining. Bound proteins were eluted in 60 μL, and 10 μL (16%; pulldown) was loaded on the gel. MBP served as a negative control (lanes 1 and 2).
Figure 5.
Binding affinity of Cm eIF4E and Cm eIF4G1003-1092. MBP pull-down assays showing the interaction of purified Cm eIF4E (wild type, full length; increasing concentrations, 0.5–3 μm) and MBP-tagged Cm eIF4G1003-1092 (wild type or mutant in bacterial lysates at a fixed concentration). A, Pull-down with Cm eIF4G1003-1092-WT. B, Pull-down with MBP used as a negative control. C to E, Pull-down with MBP-eIF4G1003-1092 mutants in C, NC, and C-NC domains. EIF4G lysates (wild type and mutants) were incubated with purified Cm eIF4E at concentrations of 0.5, 1, 2, and 3 μm (corresponding to lanes 1, 2, 3, and 4 of each gel, respectively) in a volume of 300 μL. The input samples (lanes 1 to 4) and pull-down fractions (lanes 5 to 8) were analyzed in a 12% SDS-PAGE followed by Coomassie Brilliant Blue staining, loading in gels 5% and 16% of the total volumes, respectively. F, Graph representing the specific binding of Cm eIF4E with Cm eIF4G1003-1092 (wild type and mutants), plotted as a function of Cm eIF4E concentration. Data points in the binding curves represent means calculated from data points of two different experiments as in A, C, and D. Each data was fitted using a one-site binding model.
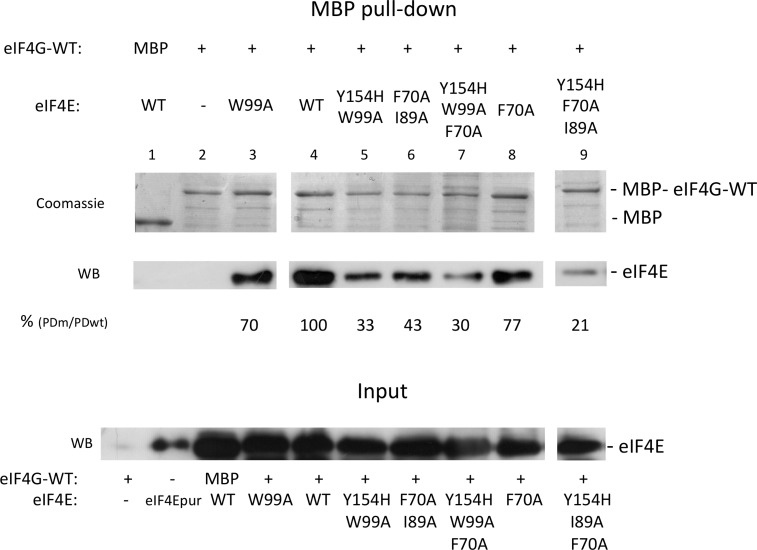
To further validate these results, we examined the effect of substitutions in Cm eIF4E residues proposed to be involved in Cm eIF4G binding, including F70A, I89A, W99A, and Y154H, even as double and triple mutants Y154H-W99A, F70A-I89A, Y154H-W99A-F70A, and Y154H-I89A-F70A. Lysates of MBP-tagged eIF4G-WT, and wild-type or mutant Cm eIF4Es were incubated and pulled-down using the MBP-tag. In this case, Cm eIF4E was visualized by western-blot analysis. Substitutions W99A and Y154H-W99A on the dorsal surface of Cm eIF4E reduced its Cm eIF4G1003-1092-binding capacity to 70% and 33%, respectively, compared to eIF4E-WT (Fig. 6, lanes 3 and 5). On the other hand, the substitution of the hydrophobic residues of the lateral pocket of Cm eIF4E, F70A, and double mutant F70A-I89A reduced its Cm eIF4G1003-1092-binding efficiency to 77% and 43% compared to eIF4E-WT, respectively (Fig. 6, lanes 6 and 8). Interestingly, binding was affected to similar extents for single and double mutants in dorsal and lateral surfaces. When substitutions in both surfaces were combined, binding was strongly impaired, resulting in 30% and 21% of the eIF4E-WT efficiency (Fig. 6, lanes 7 and 9), respectively. These results suggest that Cm eIF4G needs to interact with both the dorsal and lateral surfaces of Cm eIF4E for efficient binding.
Figure 6.
Recovery of Cm eIF4E (wild type and mutants) in pull-down assays with MBP-Cm eIF4G1003-1092. Pull-down assays showing the interaction of wild-type and mutant Cm eIF4E with MBP-Cm eIF4G-WT. A fixed volume of a lysate from bacteria expressing Cm eIF4G1003-1092-WT with a N-terminal MBP tag was incubated with Cm eIF4E bacterial lysates (wild type and mutants) in a volume of 300 μL. Middle, Pulled-down wild-type and mutant Cm eIF4Es as determined by Western blot (WB) using a Cm eIF4E-specific antibody. Top, The same gel was stained with Coomassie Brilliant Blue to compare the amount of eluted MBP-tagged Cm eIF4G-WT in each experiment (Coomassie). Bottom, The amount of Cm eIF4E present in the input visualized by western blot. Bands from WB were quantified and a binding was estimated using the band intensity ratio between sample Cm eIF4E-WT (input, lane 5) and Cm eIF4E-WT (MBP pull-down, lane 4) as 100% reference value. The interacting factors in each experiment are shown below the gels: pull-down proteins, either MBP or MBP-Cm eIF4G, and interacting Cm eIF4E proteins (wild type or mutants). Input and pull-down correspond to a 1% of the incubation volume and 5% of the elution volumes, respectively.
Testing Substitutions in Cm eIF4E-Binding Residues in a Translation Efficiency Assay
Recently, we developed a translation efficiency assay to test Cm eIF4E mutants in vivo (Miras et al., 2017b). This assay is based on the RNA element called Ma5TE located in the 3′-UTR of Melon necrotic spot virus (MNSV, family Tombusviridae), which requires and has a strong specificity for Cm eIF4E alleles to efficiently drive cap-independent translation of the messenger RNAs bearing it (Nieto et al., 2006; Truniger et al., 2008; Rodríguez-Hernández et al., 2012). Briefly, this assay is based on the capacity of Cm eIF4E-WT to complement the Ma5TE-controlled translation of RNAs when it is transiently expressed in melons that carry the Cm eIF4EH228L allele in homozygosity, which does not support Ma5TE-driven translation (Nieto et al., 2006; Miras et al., 2017b). Thus, we transiently expressed Cm eIF4E mutants in eIF4EH228L melons and studied their ability to complement Ma5TE-controlled translation of reporter RNAs consisting of the luciferase gene flanked by the 5′-and 3′-UTRs of MNSV.
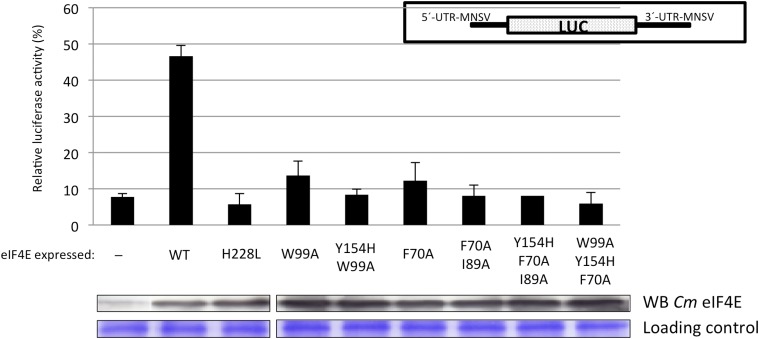
Recapitulating previous results (Miras et al., 2017b), our data showed that transient expression of Cm eIF4E-WT increased Ma5TE-driven translation 6-fold with respect to translation in its absence (Fig. 7, (-) and wild type), reaching nearly 50% of the translation activity of an eIF4E-independent reporter RNA used to normalize the data (see “Materials and Methods” for details); in contrast, transient overexpression of Cm eIF4EH228L did not support translation (Fig. 7, H228L). We then studied the effect on translation of the substitutions that reduced Cm eIF4G-binding in vitro. Single and double substitutions in residues of the dorsal surface of Cm eIF4E, in contact with the C domain of Cm eIF4G, resulted in a drastic reduction of the Ma5TE-driven translation efficiency (Fig. 7, W99A and Y154H-W99A; Miras et al., 2017b). In contrast to in vitro binding assays, substitutions in the lateral surface of Cm eIF4E, involved in interactions with the NC binding domain in Cm eIF4G, also reduced translation efficiency (Fig. 7, F70A and F70A-I89A). Similarly, Cm eIF4E-mutants with substitutions in residues at both surfaces were unable to complement translation in this assay (Fig. 7, Y154H-F70A-I89A and Y154H-W99A-F70A). These results strongly support the importance of the Cm eIF4E residues implicated in binding to Cm eIF4G through the NC domain for eIF4F-dependent translation. The interaction of both Cm eIF4E surfaces with Cm eIF4G is required for efficient translation.
Figure 7.
Effect of substitutions in Cm eIF4E on cap-independent translation efficiency. Relative luciferase activity (%) obtained with a reporter RNA that depends on Cm eIF4E for cap-independent translation (see inset in the right upper part of figure) melon protoplasts. Protoplasts carried Cm eIF4EH228L, which is unable to efficiently complement cap-independent translation of the reporter RNA. Protoplasts were transiently transformed to express different Cm eIF4E variants (wild type, which is able to complement cap-independent translation of the reporter; Miras et al., 2017b; and mutants). For normalization of measurements in different protoplast preparations, luciferase activity obtained with a Cm eIF4E independently translated reporter RNA construct 5′-UTR-luc-3′-UTR was set to 100% for each protoplast preparation. The transiently expressed Cm eIF4E mutants are indicated below each bar: -, silencing suppressor P19 alone; WT, wild type; engineered Cm eIF4E mutations, H228L, W99A and Y154H-W99A (in residues in the dorsal surface proposed to be involved in canonical Cm eIF4G interaction; Miras et al., 2017b), and F70A and F70A-I89A (in residues in the lateral surface proposed to be involved in NC Cm eIF4G interaction); and mutations in both surfaces Y154H-F70A-I89A and W99A-Y154H-F70A. Error bars are ±sd. Panels below the graph show expression of each Cm eIF4E mutant in melon cotyledons visualized by western blot using antibodies against Cm eIF4E. Bottom, Loading as visualized by Coomassie Brilliant Blue staining.
For the mutant proteins tested here, we studied their ability to complement the growth of the eIF4E-deficient yeast strain JO55. This Saccharomyces cerevisiae strain lacks its endogenous eIF4E gene and requires complementation with an external eIF4E to survive, for example the human eIF4E expressed from the pGAL-eIF4E-URA3 plasmid in a Gal-dependent manner (Altmann et al., 1989). This assay is a good indicator of all the eIF4E functions that affect yeast growth, including mRNA translation (Altmann et al., 1989; Charron et al., 2008; German-Retana et al., 2008; Ashby et al., 2011), and we have used it previously to test the effect of single Cm eIF4E mutations (Miras et al., 2017b). Almost all Cm eIF4E mutants analyzed here complemented yeast growth, confirming their functionality in this organism (Supplemental Fig. S6), with the exception of the triple mutant Y154H-F70A-I89A (Supplemental Fig. S6) harboring substitutions in two residues involved in NC and one residue in canonical binding to Cm eIF4G.
DISCUSSION
Architecture of the Cap Binding Site
All the structural information available to date shows that the eIF4E binding pocket is built by two principal regions when bound to either the m7GDP or m7GTP cap analogs; a conserved Trp pair located in the loop region and a cluster of Arg/Lys residues interacting with the phosphates in the cap. NMR cap-free eIF4E structures show that loops L1 and L3 are quite flexible and adopt multiple conformations in solution (Volpon et al., 2006). However, they get ordered upon binding the cap through stacking interactions with these two Trp residues each located in each loop (Marcotrigiano et al., 1997, 1999; Matsuo et al., 1997; Monzingo et al., 2007). As mentioned before, in our structure only loop L3 is visible and adopts a conformation, either in its m7GDP bound or unbound state, that closely resembles those described for other eIF4E structures in complex with a cap analog and thus positioning residue W128 in a conformation fully compatible with cap binding. A defined conformation for both loops has been observed in the crystal structure of the human eIF4E in its unbound state, with the four eIF4E molecules in the asymmetric unit arranged as dimers and the static position of both loops due to intermolecular contacts between the concave surface of both monomers in each dimer (Siddiqui et al., 2012). Whether the static conformation observed for loop L3 in our structure is driven by such contacts is difficult to infer, since there are four independent Cm eIF4E molecules in the asymmetric unit that result in a different local environment for each loop. In contrast, loop L1 was never observed in our Cm eIF4E and Cm eIF4E-m7GDP structures except for molecule C, in which it adopts a fixed conformation. However, this appeared to be a conformational selection triggered by crystal packing rather than a functionally relevant state in terms of cap binding, since it positions the loop far away from the binding pocket. It should be noted that after structure refinement, local distribution of B factors in this region indicated that the binding pocket was not fully occupied in the two Cm eIF4E molecules in which we observed extra density, so occupancy for m7GDP was finally adjusted to a value of 0.6 to match local B factor distribution. It is plausible then, that the local concentration of m7GDP in the binding pocket is too low to drive the transition of loop L1 to its cap-binding conformation, while loop L3 is already in a competent conformation to interact with the guanine base of the cap. Structural studies in solution should be performed in the future to analyze the flexibility of these loops in Cm eIF4E.
A Universal Bimodal Binding Mode of eIF4E to eIF4G in Higher Eukaryotes?
This work provides the first description of the structure of a plant eIF4G peptide in complex with eIF4E and shows that it binds to both dorsal and lateral surfaces of Cm eIF4E interacting through the C and NC domains, similarly to metazoan eIF4Gs and 4E-BPs (Kinkelin et al., 2012; Peter et al., 2015a, 2015b; Grüner et al., 2016). Using in vitro pull-down assays with proteins mutated in these domains, we showed that motifs in both domains were required for efficient complex formation. Also, our functional analysis using an in vivo cap-independent eIF4E-dependent translation system (Miras et al., 2017b) supports the importance of this bimodal type of binding.
The eIF4G residues belonging to the C motif are generally conserved in eukaryotic eIF4Gs (Miras et al., 2017b). In contrast, the residues of the NC motif identified here are not conserved, but the hydrophobicity of the lateral eIF4E core and of the NC motif are conserved in eukaryotic eIF4G and m4E-BPs (Fig. 1; Supplemental Fig. S4). Moreover, this patch of hydrophobic residues within the NC domain is also conserved in the plant-specific eIFiso4G, suggesting a similar bimodal interaction in the isoform complex eIFiso4F (Supplemental Fig. S1). Thus, our structure of the Cm eIF4E-eIF4G1003-1092 complex very much resembles those of metazoan complexes, where the canonical helix is bound to the dorsal surface of eIF4E while binding extends to the lateral interface (Supplemental Fig. S3, A and B; Grüner et al., 2016). Melon and metazoan complexes are structurally similar, but differ from that of yeast (Supplemental Fig. S3C; Gross et al., 2003; Grüner et al., 2016). This difference could be due to the lack of sequence conservation. However, the yeast eIF4E-eIF4G393-490 structure was determined by NMR in the presence of a zwitterionic detergent that may interfere with hydrophobic bonds (Matsuo et al., 1997; Gross et al., 2003; Grüner et al., 2016). Therefore, our data together with those of Grüner and coworkers (Grüner et al., 2016) strongly support a universal bimodal binding mode of eIF4E to eIF4G in higher eukaryotes.
Peculiarities of the Plant eIF4E-eIF4G1003-1092 Complex
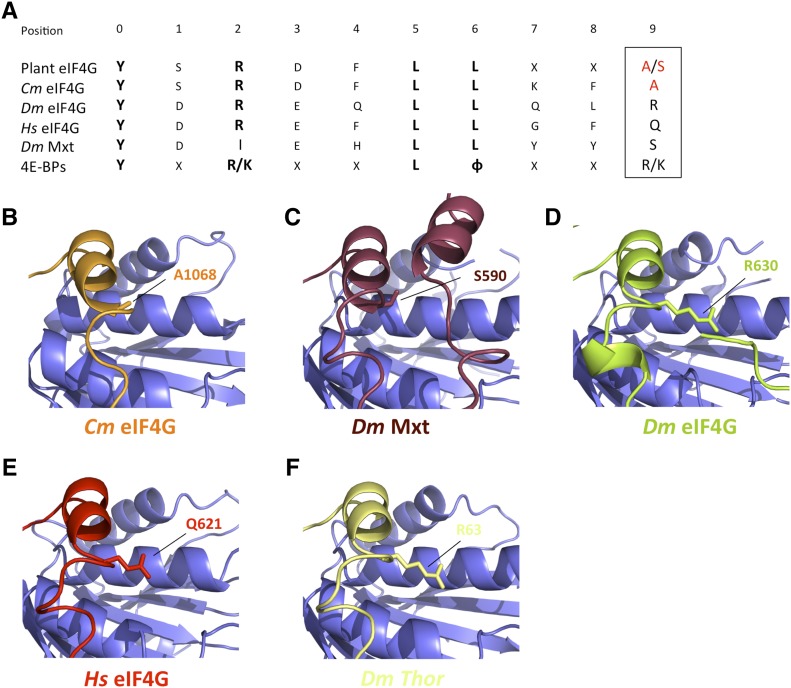
A search for structural neighbors within the database of the Dali server revealed that Drosophila Mextli protein (Dm Mxt) shares its topology with Cm eIF4G at its maximum score (Z = 2.4), even higher than Dm and Hs eIF4G (Z = 1.9 and Z = 1.6, respectively). In plants, the eIF4G C motif can be extended to the consensus sequence [YSRDFLLX2(A/S)] (Patrick and Browning, 2012; Fig. 8A), in which Cm eIF4G position 10 (underlined) is occupied by an Ala (A1068). Interestingly, while metazoan eIF4Gs and most of the m4E-BPs contain long aliphatic residues (K/Q/R) at this position, in Dm Mxt, shorter residues (Ile or Ser) are present, which allows for a tripartite binding mode for Dm Mxt (Peter et al., 2015b); an auxiliary helical motif can be set on the dorsal surface of eIF4E, replacing the interactions and covering the surface occupied by the side chains of the aliphatic residues present in the majority of m4E-BPs and metazoan eIF4Gs (Fig. 8, B–F). Thus, it may be hypothesized that plant eIF4Gs may use this tripartite binding mode, making the complexes more stable and granting them with distinct functional properties.
Figure 8.
Structural comparison of Cm eIF4G, metazoan eIF4Gs, and 4E-BP linker regions. A, Extended consensus sequence for the plant eIF4G canonical eIF4E binding domain from all available databank sequences (Patrick and Browning, 2012) and Dm Mxt and consensus m4E-BPs. Invariant residues are typed in bold, and plant-specific Ala or Ser residues in position 9 (boxed) are red colored. B to F, Close-up views of the canonical helices and the linker region of Cm eIF4G (B), Dm Mxt (C), Dm eIF4G (D), Hs eIF4G (E), and Dm Thor (as m4E-BP model; F) bound to the dorsal surface of eIF4E (PDB IDs: 5T46, 5T47, 5ABV, 4UE8; Peter et al., 2015a, 2015b; Grüner et al., 2016). Residues of the canonical helix in position 9 are shown as sticks.
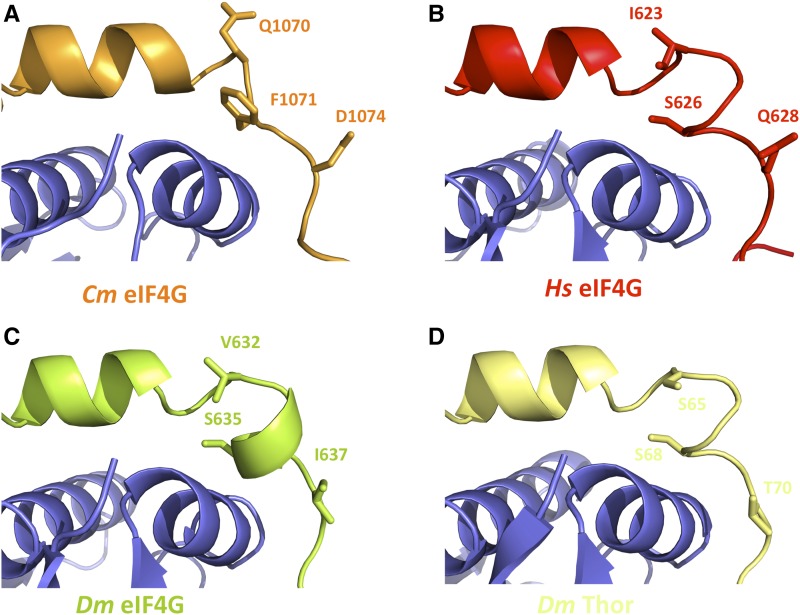
As mentioned before, the characteristic half-helical turn beginning at the tip of the elbow loop (Hs F624-S626 and Dm K633-S635) present in all the metazoan 4E-BPs and eIF4Gs structures solved so far was not observed in the Cm eIF4E-eIF4G1003-1092 complex (Supplemental Fig. S3). The contact established between the side chains of the highly conserved S/T residues located at the end of the elbow loop in metazoan eIF4Gs and 4E-BPs and carbonyl oxygens of the preceding residues fixing the backbone were lost in our structure (Peter et al., 2015a). Instead, the side chain of Cm F10714G occupied the position of the S/T residue present in its metazoan counterparts, introducing a two-amino acid gap (Figs. 2 and 9A), and explaining why the half-helical turn is lost in the Cm eIF4E-eIF4G1003-1092 structure. Interestingly, two conserved S/T residues have been located at both ends of the canonical elbow loop in all the m4E-BP crystal structures solved so far (Fig. 9, B–D; Peter et al., 2015a; Grüner et al., 2016). The phosphorylation of these linker residues, not present in metazoan eIF4Gs (Fig. 1), is sufficient to reduce the strength of m4E-BPs as translational repressors and the mechanism behind this appears to be a phosphorylation-induced disorder-to-order transition that drastically reduces the affinity of m4E-BPs for eIF4E (Bah et al., 2015). In plants, only LOX2 and BTF3 have been identified as putative eIF4E interacting proteins (Freire et al., 2000, 2005). While BTF3 does not contain an eIF4E-recognizable binding motif, LOX2 possesses a sequence that resembles a C domain, but it is poorly conserved. In contrast to m4E-BPs, there is no apparent hydrophobic patch that would correspond to a NC interacting region (Supplemental Fig. S5). Moreover, as in the case of Cm eIF4G, the S or T residues are not conserved downstream of the C domain from LOX2. Thus, the lack of a “metazoan-like” elbow loop and the fact that the characteristic phosphorylable S/T residue pairs in metazoans seems not to be conserved in plants suggest that a different mechanism may control the assembly and stability of the plant eIF4E-eIF4G complex.
Figure 9.
Structural comparison of the elbow loops of Cm eIF4G, metazoan eIF4Gs, and 4E-BP. A to D, Close-up views of the canonical helices and elbow loops of Cm eIF4G (A), Hs eIF4G (B), Dm eIF4G (C), and Dm Thor (as a m4E-BP model; D) bound to the dorsal surface of eIF4E (PDB IDs: 5T46, 5T47, 4UE8; Peter et al., 2015a; Grüner et al., 2016). Selected residues mediating phosphorylation and their analogs are shown as sticks.
Another peculiarity seems to be the formation of a disulfide bridge in the Cm eIF4E-eIF4G1003-1092 complex. As expected, the Cm eIF4E structure has much in common with that of mammalian and other plant eIF4Es (Marcotrigiano et al., 1999; Monzingo et al., 2007; Ashby et al., 2011). Interestingly, the main difference between Cm eIF4E either in its free or cap-bound state with Cm eIF4E bound to Cm eIF4G peptide is the formation of a disulfide bridge in the Cm eIF4E-eIF4G1003-1092 complex, which seems to block the ability of Cm eIF4E to bind to the cap (Fig. 2D). The formation of a disulfide bridge has been previously observed in the wheat eIF4E crystal structure, where cocrystallization with the cap was only possible using an eIF4E mutant in which one of the C residues was replaced to avoid disulfide bridge formation (Monzingo et al., 2007). Despite that the two cysteines involved are strictly conserved in plant eIF4Es, suggesting that they may have some important biological role, affinity-binding assays under both reducing and oxidizing conditions showed that in both cases wheat eIF4E retained the ability to bind the cap (Monzingo et al., 2007). However, further studies using mass spectrometry and a Lys-specific chemical probe indicated structural changes upon altering the redox state, supporting the hypothesis of a redox sensor or switch for eIF4E (O’Brien et al., 2013). Whether the oxidation state of eIF4E may modulate cap binding in a redox-sensing manner in plants and regulate the level of translation in response to the redox state of the cell remains uncertain.
Implications in Plant Disease: EIF4G and Potyviral VPg Possibly Share an eIF4E-Binding Surface
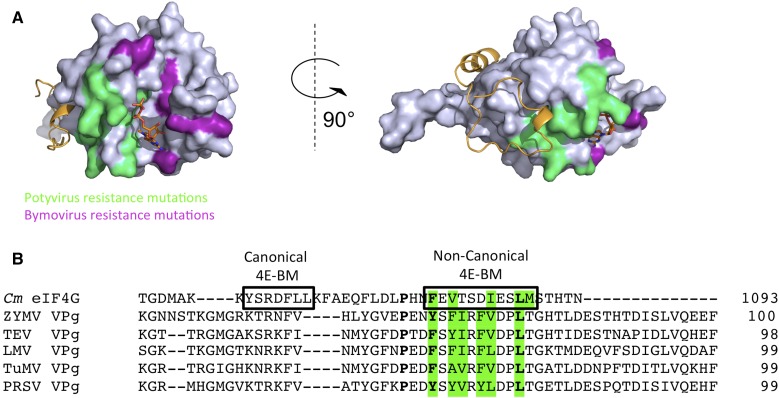
Plants harboring mutations in eIF4E lose their susceptibility to a number of plant viruses; thus specific eIF4E alleles confer recessive resistance to viruses (Truniger and Aranda, 2009; Sanfaçon, 2015; Dinkova et al., 2016). One example is the Cm eIF4E-mediated resistance of melon to MNSV (Tombusviridae) that results from a single amino acid substitution in this protein (Nieto et al., 2006). The resulting Cm eIF4E variant is still functional for translation of plant mRNAs, but it does not promote translation of MNSV RNAs (Truniger et al., 2008). We show here that mutations in Cm eIF4E affecting its interaction with Cm eIF4G reduce the eIF4E-dependent translation of MNSV RNAs, confirming that the Cm eIF4E-eIF4G complex is required for MNSV translation (Miras et al., 2017b). However, we cannot rule out that these mutations could reduce its ability to bind the 3′-CITE, even though the Cm eIF4E residues mutated here are not in the vicinity of the residue H228 involved in 3′-CITE binding (Miras et al., 2017b). But nearly all of the eIF4E-dependent viruses belong to the family Potyviridae (Sanfaçon, 2015). In many of these cases, the resistance phenotype arises from disruption of the direct interaction between the potyviral genome-linked protein (VPg) and eIF4E (Léonard et al., 2004; Yeam et al., 2007; Charron et al., 2008; Miras et al., 2017a). Overall, two regions of eIF4E have been mapped to be involved in the interaction with VPg, one near the cap-binding pocket and another in the lateral surface of the protein. This latter surface interacts with the eIF4G NC motif (Fig. 10A), but a C motif could not be identified in VPgs, which on the other hand are recalcitrant to crystallization (Rantalainen et al., 2008). Despite the lack of sequence conservation, an alignment of Cm eIF4G and VPgs identified three conserved amino acids in a hydrophobic region that may correspond to a putative NC motif (Fig. 10B). Therefore, eIF4E and VPgs may bind through NC motifs. At least one amino acid substitution in the lateral surface of tomato eIF4E, G107R, disrupted in planta its interaction with VPg from Tobacco etch virus (Yeam et al., 2007). Moreover, analysis of mutations in the lateral surface of pea (Pisum sativum) and lettuce (Lactuca sativa) eIF4Es revealed that the mutant proteins could not complement viral infection of two potyviruses, Pea seed-borne mosaic virus and Lettuce mosaic virus (German-Retana et al., 2008; Ashby et al., 2011). Interestingly, several studies have suggested that potyviral VPg is able to inhibit cap-dependent translation (Khan et al., 2008; Miyoshi et al., 2008; Eskelin et al., 2011). Thus, it can be hypothesized that VPg competes with eIF4G for binding to the lateral surface of eIF4E through its NC binding motif, impairing eIF4E-eIF4G interaction and hijacking eIF4E for viral genome translation. However, it should be pointed out that apart from translation, other roles have been proposed for the VPg-eIF4E interaction, including viral cell-to-cell movement (Gao et al., 2004).
Figure 10.
Plant virus resistance mutations modeled on the melon eIF4E-eIF4G1003-1092 complex. A, The residues implicated in plant potyvirus and bymovirus resistance are shaded in green and magenta, respectively (Gao et al., 2004; Kang et al., 2005; Kanyuka et al., 2005; Ruffel et al., 2005; Stein et al., 2005; Nicaise et al., 2007; German-Retana et al., 2008; Ashby et al., 2011). The Cm eIF4E molecule is shown as a surface rendering and Cm eIF4G1003-1092 is shown as a cartoon representation, both colored in gray and orange, respectively. The m7GDP cap-analog is located in the cap-binding pocket of Cm eIF4E. B, Sequence alignment of Cm eIF4G (residues 1052–1093) and potyviral VPgs (ZYMV NP477522.1; TEV NP734204.1; LMV KF285932.1; TuMV NP734219.1; PRSV AEC04846.1). C and NC motifs are boxed in the Cm eIF4G sequence. Conserved residues are typed in bold and hydrophobic residues located in the putative NC motif are green-colored.
In conclusion, we have identified and characterized key structural principles determining plant eIF4E-eIF4G interaction. The identification of extended and conserved interacting motifs supports a universal binding mode in eukaryotes and offers targets for the design and edition of eIF4E genes to regulate translation and block viral infection.
MATERIALS AND METHODS
Plasmids
The plasmids expressing Cm eIF4G-WT and Cm eIF4E were previously described (Miras et al., 2017b). Briefly, Cm eIF4G (residues 1003–1092) and Cm eIF4E (residues 51–235) were amplified by PCR from plasmids pTOPO-Cm-4G and pET15b-4EVed (Nieto et al., 2006) and cloned by LIC technology into the p2CT (which provides an N-terminal MPB tag followed by a TEV protease cleavage site) and p2AT expression vectors, respectively (Macrolab, UC Berkeley), yielding p2CT-eIF4G-p10 and p2AT-eIF4E-WT (Miras et al., 2017b). For expression of the Cm eIF4E-eIF4G1003-1092 complex, a dicistronic expression vector was generated by cloning into plasmid p2D (Macrolab), yielding p2D-4Fp10. Cm eIF4E was cloned into cassette 1 using BamHI/XbaI restriction sites, and Cm eIF4G1003-1092-WT was cloned into cassette 3 using SbfI/AscI restriction sites. All the Cm eIF4E and Cm eIF4G mutants were generated by site-directed mutagenesis using plasmids pET15b-4EVed (H228) and p2CT-eIF4G-p10, respectively, and the oligonucleotide primers shown in Supplemental Table S1. For transient Cm eIF4E expression in the complementation experiments, the Cm eIF4E constructs were cloned into the binary vector pBIN61 using XbaI/XmaI restriction sites. All mutants were confirmed by DNA sequencing. Luc construct with the 5 ´- and 3 ´-UTR from MNSV-Mα5 and MNSV-264 flanking the firefly luciferase gene has been previously described (Truniger et al., 2008). Reporter RNA was obtained by in vitro transcription (RiboMAX Large Scale RNA production, Promega).
Protein Expression and Purification
All proteins were expressed in Escherichia coli Rosetta (DE3) pLysS cells (Novagen) that were grown in Luria-Bertani medium at 37°C to an OD600 of 0.6. At this point, isopropylthio-β-galactoside was added to a final concentration of 0.4 mm to induce protein expression O/N at 20°C. Cells were resuspended in nickel buffer A (25 mm HEPES, pH 7.5, 400 mm NaCl, 20 mm imidazole, 10% glycerol, 1 mm dithiothreitol) and supplemented with DNaseI (Roche) and protease inhibitor cocktail (Roche) and lysed by sonication. Expressed Cm eIF4E-eIF4G1003-1092 complex was eluted from a HisTrap HP column (GE Healthcare) using nickel buffer B (same as nickel buffer A but with 400 mM imidazole). After buffer exchange, the eluted protein was quantified by spectrometry at OD280. The His6 and MBP tags were removed after cleavage with TEV protease at a 1:20 mass ratio and overnight incubation at 4°C. Finally, the protein was loaded onto the same HisTrap HP column and eluted with Nickel buffer B followed by loading onto a size-exclusion Superdex 200 16/60 column (GE Healthcare). Cm eIF4E51-235 was expressed from p2A-eIF4E-WT as described above, purified using a HisTrap HP column (GE Healthcare), and passed through a final size exclusion chromatography column (Superdex 200 16/60, GE Healthcare). Purification of full-length Cm eIF4E was carried out as previously described (Nieto et al., 2006). All proteins were concentrated to a final concentration of 10 mg/mL, and buffers were exchanged to the final buffer 25 mm HEPES, pH 7.5, 200 mm NaCl, and 10% glycerol.
Crystallization, Data Collection, and Processing
Crystals were obtained using the hanging drop vapor diffusion method. Briefly, 1 μL of protein solution (10 mg/mL) was mixed with an equal volume of reservoir solution and incubated at 20°C until crystals suitable for x-ray diffraction were obtained. Cm eIF4E crystals were grown using 1.0 m sodium/potassium tartrate in 100 mm MES, pH 6.0, as the precipitant, and Cm eIF4E-m7GDP complex was obtained by incubating native crystals O/N in the presence of 1:1.1 excess ligand. Cm eIF4E-eIF4G1003-1092 complex was crystallized using 1.4 to 1.6 m ammonium sulfate, 100 mm sodium acetate, pH 4.5, as the reservoir solution. Small needles were obtained when plates were incubated at 20°C. Suitable crystals for x-ray data collection were obtained using microseeding techniques. Data were collected using synchrotron radiation at ALBA-CELLS after flash freezing under liquid nitrogen using 20% glycerol as the cryo-protectant. Diffraction images from one single crystal for each complex were processed using XDS (Emsley and Cowtan, 2004) and SCALA (Collaborative, 1994).
Cm eIF4E-eIF4G1003-1092 Structure Solution and Refinement
Initial phases were determined by molecular replacement with the MOLREP program (Collaborative, 1994) using the crystal structure of wheat (Triticum aestivum) eIF4E as a search model (Monzingo et al., 2007; PDB ID: 2IDR). Rigid body refinement was conducted using REFMAC5 (Murshudov et al., 1997). Manual rebuilding was performed using COOT (Emsley and Cowtan, 2004) and refinement using Refmac5 and PHENIX (Brünger et al., 1998; Adams et al., 2002). Statistics for both data collection and refinement are summarized in Supplemental Table S2.
Protein Pull-Down Assays
Pull-down experiments were performed following other authors (Igreja et al., 2014; Peter et al., 2015a, 2015b). In Figure 4, Cm eIF4G1003-1092-WT and mutants were expressed with an N-terminal MBP tag in E. coli Rosetta (DE3) pLysS cells as described above. The bacterial cells were resuspended in 2 mL of lysis buffer and lysed by sonication. A fixed concentration of purified Cm eIF4E (1 μm) was added to the MBP-tagged eIF4G1003-1092 (wild type and mutants) expressing cleared bacterial lysates, adjusted to 300 μL with lysis buffer (input fraction), and incubated with 30 μL of amylose resin (New England Biolabs) for 1 h at 4°C. The beads were washed three times with lysis buffer and eluted with 60 μL of lysis buffer containing 25 mm maltose (bound fraction). To obtain roughly similar amounts of pulled-down Cm eIF4E, Cm eIF4G1003-1092 (wild type, C, NC, and C-NC) cleared lysates were incubated with purified Cm eIF4E in a ratio of 3:1, 5:1, 2:1, and 4:1, respectively. Similarly, a cleared lysate of MBP-expressing bacteria was incubated with purified Cm eIF4E as negative control. Proteins were analyzed by a 12% SDS-PAGE followed by Coomassie Brilliant Blue staining. In Figure 6, MBP-tagged Cm eIF4G-WT and Cm eIF4E (wild type and mutants) were expressed in E. coli Rosetta (DE3) pLysS cells as described above and cleared lysates were used. Pull-down assays were performed using a 3:1 ratio of Cm eIF4G-WT and Cm eIF4E lysates (wild type and mutants). Eluted proteins were visualized by Coomassie Brilliant Blue Blue staining and western blot (eIF4E) using a rabbit polyclonal antibody against Cm eIF4E (Miras et al., 2017b).
Binding Efficiency
For binding affinity experiments shown in Figure 5, cleared bacterial lysates expressing MBP-tagged wild-type and mutant Cm eIF4G-peptides (C, NC, and C-NC) were adjusted to the same recombinant protein concentration using lysis buffer. Serial concentrations of 0.5, 1, 2, and 3 μm of purified Cm eIF4E were added to each Cm eIF4G lysate in a total volume of 300 μL and incubated with amylose resin for 1 h at 4°C. Protein elution and determination of eluted proteins were done as before. Band intensity of pulled-down Cm eIF4Es was quantified with Quantity One software (Bio-Rad) and plotted versus total Cm eIF4E input. Apparent dissociation constants were calculated using a one-site binding model (GraphPad Prism).
Translation Complementation by Transiently Expressed Cm eIF4E
Cm eIF4EH228 and mutant proteins were transiently expressed from binary plasmids in cotyledons of melon (Cucumis melo) accession C46, which is homozygous for Cm eIF4EH228L, as previously described (Nieto et al., 2006). At 3 to 4 d postagroinfiltration, protoplasts were prepared from infiltrated tissues. In vivo translation assays were performed as previously described (Truniger et al., 2008) by electroporating separately with two 5′-UTR-luc-3′-UTR RNA constructs, differing in the UTRs flanking the luciferase gene that were either from isolate MNSV-Mα5 (eIF4E-dependent) or from the eIF4E-independent isolate MNSV-264. Translation of MNSV-264 can take place in protoplasts from eIF4E knocked-down lines, so it can function in the absence of eIF4E (Rodríguez-Hernández et al., 2012) and therefore serves for normalization of the different protoplast preparations. After 5 to 6 h incubation in the dark at 25°C, protoplasts were lysed in 1x Passive lysis buffer (Promega). Firefly luciferase activities were measured with the Luciferase assay system (Promega). For each protoplast preparation, the translation efficiency determined for the construct of MNSV-264 was set to 100% and translation of MSNV-Mα5 was compared to it. The expression levels of Cm eIF4E were analyzed in protein extracts (extraction buffer: 0.1 m Tris HCl, pH 9.0, 0.1 m NaCl, 5 m urea, 10 mm EDTA, 0.1 m β-mercaptoethanol) of infiltrated cotyledons and visualized by western blot using a rabbit polyclonal antibody against Cm eIF4E (Miras et al., 2017b).
Yeast Complementation
For the analysis of the complementation capacity of the Cm eIF4E mutants in JO55 yeast, the cDNAs encoding each of the Cm eIF4E variants were introduced into the SpeI/BamHI restriction sites of the Trp-selectable yeast-E. coli shuttle vector p424-GDP/TRP1 (Mumberg et al., 1995) and used to transform JO55 selected on Gal-containing minimal medium in the absence of Ura and Trp. After transformation, yeast cells were grown at 30°C in liquid Gal/Raf-Ura-Trp medium until OD600 = 1, washed with sterile water, and serially diluted in 10-fold steps until reaching 1,000-fold. Drops (5 mL) of dilutions were placed on both control Gal/Raf-Ura-Trp solid medium and nitrogen base medium containing 2% Glc in the absence of Ura/Trp. JO55 transformed with an empty p424-GPD/TRP1 vector was used as the negative control. Positive controls included JO55 transformed with Arabidopsis thaliana eIF4E gene present in vector p424-GPD/TRP1:At-eIF4E (Charron et al., 2008).
Accession Numbers
Coordinates for the structures described in this study have been deposited in the Protein Data Bank under the following accession numbers: 5ME5, Cm eIF4E:eIF4G1003-1092; 5ME6, Cm eIF4E:m7GDP; and 5ME7, Cm eIF4E.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The canonical and NC eIF4E-binding domain in eIF4G and eIFiso4G.
Supplemental Figure S2. Fo-Fc difference map obtained after molecular replacement using the C. melo eIF4E crystal structure as the search model.
Supplemental Figure S3. Structural comparisons of the melon, metazoan, and fungal eIF4G peptides bound to eIF4E.
Supplemental Figure S4. Sequence alignment of eIF4E orthologous proteins.
Supplemental Figure S5. Sequence alignment of plant eIF4Gs (top) and LOX2 proteins (bottom).
Supplemental Figure S6. Complementation of translation by Cm eIF4E mutants in eIF4E-deficient yeast.
Acknowledgments
We thank Mari Carmen Montesinos and Blanca Gosalvez for their excellent technical assistance. Mario Fon edited the manuscript.
Glossary
- MNSV
Melon necrotic spot virus
Footnotes
Work in Murcia was financially supported by grants AGL2015-65838 and PCIN-2013-043 (MINECO, Spain-FEDER). Work in Barcelona was supported by grants BIO2014-54588-P (MINECO, Spain-FEDER) and Maria de Maeztu Unit of Excellence MDM-2014-0435. X-ray data were collected at ALBA-CELLS (beamline XALOC), Cerdanyola del Valles, Barcelona, Spain, with the collaboration of ALBA staff and at ESRF (Grenoble, France). Financial support was also provided by ALBA and ESRF.
Articles can be viewed without a subscription.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Aitken CE, Lorsch JR (2012) A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 19: 568–576 [DOI] [PubMed] [Google Scholar]
- Altmann M, Müller PP, Pelletier J, Sonenberg N, Trachsel H (1989) A mammalian translation initiation factor can substitute for its yeast homologue in vivo. J Biol Chem 264: 12145–12147 [PubMed] [Google Scholar]
- Ashby JA, Stevenson CEM, Jarvis GE, Lawson DM, Maule AJ (2011) Structure-based mutational analysis of eIF4E in relation to sbm1 resistance to pea seed-borne mosaic virus in pea. PLoS One 6: e15873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, Sonenberg N, Kay LE, Forman-Kay JD (2015) Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 519: 106–109 [DOI] [PubMed] [Google Scholar]
- Browning KS. (2004) Plant translation initiation factors: it is not easy to be green. Biochem Soc Trans 32: 589–591 [DOI] [PubMed] [Google Scholar]
- Browning KS, Bailey-Serres J (2015) Mechanism of cytoplasmic mRNA translation. Arabidopsis Book 13: e0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Rice LM (1998) Recent developments for the efficient crystallographic refinement of macromolecular structures. Curr Opin Struct Biol 8: 606–611 [DOI] [PubMed] [Google Scholar]
- Charron C, Nicolaï M, Gallois JL, Robaglia C, Moury B, Palloix A, Caranta C (2008) Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J 54: 56–68 [DOI] [PubMed] [Google Scholar]
- Collaborative CP; Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Dinkova TD, Martinez-Castilla L, Cruz-Espíndola MA (2016) The diversification of eIF4E family members in plants and their role in the plant-virus interaction. In Hernández G, Jagus R, eds, Evolution of the Protein Synthesis Machinery and Its Regulation. Springer International Publishing, Cham, Switzerland, pp 187–205 [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Eskelin K, Hafrén A, Rantalainen KI, Mäkinen K (2011) Potyviral VPg enhances viral RNA translation and inhibits reporter mRNA translation in planta. J Virol 85: 9210–9221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire MA. (2005) Translation initiation factor (iso) 4E interacts with BTF3, the β subunit of the nascent polypeptide-associated complex. Gene 345: 271–277 [DOI] [PubMed] [Google Scholar]
- Freire MA, Tourneur C, Granier F, Camonis J, El Amrani A, Browning KS, Robaglia C (2000) Plant lipoxygenase 2 is a translation initiation factor-4E-binding protein. Plant Mol Biol 44: 129–140 [DOI] [PubMed] [Google Scholar]
- Gallie DR, Browning KS (2001) eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J Biol Chem 276: 36951–36960 [DOI] [PubMed] [Google Scholar]
- Gao Z, Johansen E, Eyers S, Thomas CL, Noel Ellis TH, Maule AJ (2004) The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J 40: 376–385 [DOI] [PubMed] [Google Scholar]
- German-Retana S, Walter J, Doublet B, Roudet-Tavert G, Nicaise V, Lecampion C, Houvenaghel MC, Robaglia C, Michon T, Le Gall O (2008) Mutational analysis of plant cap-binding protein eIF4E reveals key amino acids involved in biochemical functions and potyvirus infection. J Virol 82: 7601–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin P, Oulhen N, Jam M, Ronzca J, Cormier P, Czjzek M, Cosson B (2011) The translational repressor 4E-BP called to order by eIF4E: new structural insights by SAXS. Nucleic Acids Res 39: 3496–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JEG, Wagner G (2003) Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115: 739–750 [DOI] [PubMed] [Google Scholar]
- Grüner S, Peter D, Weber R, Wohlbold L, Chung M-Y, Weichenrieder O, Valkov E, Igreja C, Izaurralde E (2016) The structures of eIF4E-eIF4G complexes reveal an extended interface to regulate translation initiation. Mol Cell 64: 467–479 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. (2011) Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75: 434–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Lorsch JR (2012) The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igreja C, Peter D, Weiler C, Izaurralde E (2014) 4E-BPs require non-canonical 4E-binding motifs and a lateral surface of eIF4E to repress translation. Nat Commun 5: 4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CUT, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BC, Yeam I, Frantz JD, Murphy JF, Jahn MM (2005) The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J 42: 392–405 [DOI] [PubMed] [Google Scholar]
- Kanyuka K, Druka A, Caldwell DG, Tymon A, McCallum N, Waugh R, Adams MJ (2005) Evidence that the recessive bymovirus resistance locus rym4 in barley corresponds to the eukaryotic translation initiation factor 4E gene. Mol Plant Pathol 6: 449–458 [DOI] [PubMed] [Google Scholar]
- Khan MA, Miyoshi H, Gallie DR, Goss DJ (2008) Potyvirus genome-linked protein, VPg, directly affects wheat germ in vitro translation: interactions with translation initiation factors eIF4F and eIFiso4F. J Biol Chem 283: 1340–1349 [DOI] [PubMed] [Google Scholar]
- Kinkelin K, Veith K, Grünwald M, Bono F (2012) Crystal structure of a minimal eIF4E-Cup complex reveals a general mechanism of eIF4E regulation in translational repression. RNA 18: 1624–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax SR, Lauer SJ, Browning KS, Ravel JM (1986) Purification and properties of protein synthesis initiation and elongation factors from wheat germ. Methods Enzymol 118: 109–128 [DOI] [PubMed] [Google Scholar]
- Léonard S, Viel C, Beauchemin C, Daigneault N, Fortin MG, Laliberté J-F (2004) Interaction of VPg-Pro of turnip mosaic virus with the translation initiation factor 4E and the poly(A)-binding protein in planta. J Gen Virol 85: 1055–1063 [DOI] [PubMed] [Google Scholar]
- Lukhele S, Bah A, Lin H, Sonenberg N, Forman-Kay JD (2013) Interaction of the eukaryotic initiation factor 4E with 4E-BP2 at a dynamic bipartite interface. Structure 21: 2186–2196 [DOI] [PubMed] [Google Scholar]
- Mader S, Lee H, Pause A, Sonenberg N (1995) The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol 15: 4990–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK (1997) Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89: 951–961 [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK (1999) Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell 3: 707–716 [DOI] [PubMed] [Google Scholar]
- Matsuo H, Li H, McGuire AM, Fletcher CM, Gingras A-C, Sonenberg N, Wagner G (1997) Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol 4: 717–724 [DOI] [PubMed] [Google Scholar]
- Mayberry LK, Allen ML, Nitka KR, Campbell L, Murphy PA, Browning KS (2011) Plant cap-binding complexes eukaryotic initiation factors eIF4F and eIFISO4F: molecular specificity of subunit binding. J Biol Chem 286: 42566–42574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras M, Miller WA, Truniger V, Aranda MA (2017a) Non-canonical translation in plant RNA viruses. Front Plant Sci 8: 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras M, Truniger V, Querol-Audi J, Aranda MA (2017b) Analysis of the interacting partners eIF4F and 3′-CITE required for Melon necrotic spot virus cap-independent translation. Mol Plant Pathol 18: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Okade H, Muto S, Suehiro N, Nakashima H, Tomoo K, Natsuaki T (2008) Turnip mosaic virus VPg interacts with Arabidopsis thaliana eIF(iso)4E and inhibits in vitro translation. Biochimie 90: 1427–1434 [DOI] [PubMed] [Google Scholar]
- Mizuno A, In Y, Fujita Y, Abiko F, Miyagawa H, Kitamura K, Tomoo K, Ishida T (2008) Importance of C-terminal flexible region of 4E-binding protein in binding with eukaryotic initiation factor 4E. FEBS Lett 582: 3439–3444 [DOI] [PubMed] [Google Scholar]
- Monzingo AF, Dhaliwal S, Dutt-Chaudhuri A, Lyon A, Sadow JH, Hoffman DW, Robertus JD, Browning KS (2007) The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond. Plant Physiol 143: 1504–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nicaise V, Gallois JL, Chafiai F, Allen LM, Schurdi-Levraud V, Browning KS, Candresse T, Caranta C, Le Gall O, German-Retana S (2007) Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana. FEBS Lett 581: 1041–1046 [DOI] [PubMed] [Google Scholar]
- Nieto C, Morales M, Orjeda G, Clepet C, Monfort A, Sturbois B, Puigdomènech P, Pitrat M, Caboche M, Dogimont C, et al. (2006) An eIF4E allele confers resistance to an uncapped and non-polyadenylated RNA virus in melon. Plant J 48: 452–462 [DOI] [PubMed] [Google Scholar]
- O’Brien JP, Mayberry LK, Murphy PA, Browning KS, Brodbelt JS (2013) Evaluating the conformation and binding interface of cap-binding proteins and complexes via ultraviolet photodissociation mass spectrometry. J Proteome Res 12: 5867–5877 [DOI] [PubMed] [Google Scholar]
- Paku KS, Umenaga Y, Usui T, Fukuyo A, Mizuno A, In Y, Ishida T, Tomoo K (2012) A conserved motif within the flexible C-terminus of the translational regulator 4E-BP is required for tight binding to the mRNA cap-binding protein eIF4E. Biochem J 441: 237–245 [DOI] [PubMed] [Google Scholar]
- Papadopoulos E, Jenni S, Kabha E, Takrouri KJ, Yi T, Salvi N, Luna RE, Gavathiotis E, Mahalingam P, Arthanari H, et al. (2014) Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G. Proc Natl Acad Sci USA 111: E3187–E3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EH, Walker SE, Lee JM, Rothenburg S, Lorsch JR, Hinnebusch AG (2011) Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1•PABP mRNPs in vivo. EMBO J 30: 302–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick RM, Browning KS (2012) The eIF4F and eIFiso4F complexes of plants: an evolutionary perspective. Comp Funct Genomics 2012: 287814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CUT (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA 98: 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter D, Igreja C, Weber R, Wohlbold L, Weiler C, Ebertsch L, Weichenrieder O, Izaurralde E (2015a) Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol Cell 57: 1074–1087 [DOI] [PubMed] [Google Scholar]
- Peter D, Weber R, Köne C, Chung M-Y, Ebertsch L, Truffault V, Weichenrieder O, Igreja C, Izaurralde E (2015b) Mextli proteins use both canonical bipartite and novel tripartite binding modes to form eIF4E complexes that display differential sensitivity to 4E-BP regulation. Genes Dev 29: 1835–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Rantalainen KI, Uversky VN, Permi P, Kalkkinen N, Dunker AK, Mäkinen K (2008) Potato virus A genome-linked protein VPg is an intrinsically disordered molten globule-like protein with a hydrophobic core. Virology 377: 280–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Hernández AM, Gosalvez B, Sempere RN, Burgos L, Aranda MA, Truniger V (2012) Melon RNA interference (RNAi) lines silenced for Cm-eIF4E show broad virus resistance. Mol Plant Pathol 13: 755–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Gallois JL, Lesage ML, Caranta C (2005) The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol Genet Genomics 274: 346–353 [DOI] [PubMed] [Google Scholar]
- Sanfaçon H. (2015) Plant translation factors and virus resistance. Viruses 7: 3392–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui N, Tempel W, Nedyalkova L, Volpon L, Wernimont AK, Osborne MJ, Park H-W, Borden KLB (2012) Structural insights into the allosteric effects of 4EBP1 on the eukaryotic translation initiation factor eIF4E. J Mol Biol 415: 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein N, Perovic D, Kumlehn J, Pellio B, Stracke S, Streng S, Ordon F, Graner A (2005) The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J 42: 912–922 [DOI] [PubMed] [Google Scholar]
- Truniger V, Aranda MA (2009) Recessive resistance to plant viruses. Adv Virus Res 75: 119–159 [DOI] [PubMed] [Google Scholar]
- Truniger V, Nieto C, González-Ibeas D, Aranda M (2008) Mechanism of plant eIF4E-mediated resistance against a Carmovirus (Tombusviridae): cap-independent translation of a viral RNA controlled in cis by an (a)virulence determinant. Plant J 56: 716–727 [DOI] [PubMed] [Google Scholar]
- Umenaga Y, Paku KS, In Y, Ishida T, Tomoo K (2011) Identification and function of the second eIF4E-binding region in N-terminal domain of eIF4G: comparison with eIF4E-binding protein. Biochem Biophys Res Commun 414: 462–467 [DOI] [PubMed] [Google Scholar]
- Volpon L, Osborne MJ, Topisirovic I, Siddiqui N, Borden KL (2006) Cap-free structure of eIF4E suggests a basis for conformational regulation by its ligands. EMBO J 25: 5138–5149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeam I, Cavatorta JR, Ripoll DR, Kang BC, Jahn MM (2007) Functional dissection of naturally occurring amino acid substitutions in eIF4E that confers recessive potyvirus resistance in plants. Plant Cell 19: 2913–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]