The miR858-MYB83 regulatory system regulates a substantial number of genes encoding key etiological factors mediating cyst nematode parasitism of Arabidopsis.
Abstract
MicroRNAs (miRNAs) recently have been established as key regulators of transcriptome reprogramming that define cell function and identity. Nevertheless, the molecular functions of the greatest number of miRNA genes remain to be determined. Here, we report cooperative regulatory functions of miR858 and its MYB83 transcription factor target gene in transcriptome reprogramming during Heterodera cyst nematode parasitism of Arabidopsis (Arabidopsis thaliana). Gene expression analyses and promoter-GUS fusion assays documented a role of miR858 in posttranscriptional regulation of MYB83 in the Heterodera schachtii-induced feeding sites, the syncytia. Constitutive overexpression of miR858 interfered with H. schachtii parasitism of Arabidopsis, leading to reduced susceptibility, while reduced miR858 abundance enhanced plant susceptibility. Similarly, MYB83 expression increases were conducive to nematode infection because overexpression of a noncleavable coding sequence of MYB83 significantly increased plant susceptibility, whereas a myb83 mutation rendered the plants less susceptible. In addition, RNA-seq analysis revealed that genes involved in hormone signaling pathways, defense response, glucosinolate biosynthesis, cell wall modification, sugar transport, and transcriptional control are the key etiological factors by which MYB83 facilitates nematode parasitism of Arabidopsis. Furthermore, we discovered that miR858-mediated silencing of MYB83 is tightly regulated through a feedback loop that might contribute to fine-tuning the expression of more than a thousand of MYB83-regulated genes in the H. schachtii-induced syncytium. Together, our results suggest a role of the miR858-MYB83 regulatory system in finely balancing gene expression patterns during H. schachtii parasitism of Arabidopsis to ensure optimal cellular function.
Mature microRNAs (miRNAs) are small 21- to 22-nucleotide-long noncoding RNAs that are processed from transcripts forming a stem-loop secondary structure. miRNAs operate through base pairing with their target genes (Bartel, 2004; Voinnet, 2009). Once they bind to their target sequences, miRNAs can trigger mRNA degradation or translational repression, causing down-regulation of the target genes. With the fast expansion of high-throughput sequencing platforms, genome-wide identification and differential expression analysis of miRNAs have been accomplished in an increasing number of plant species (Zhang et al., 2011; Li et al., 2014). Despite the large number of miRNA genes showing differential expression under various developmental and stress conditions, only few of these miRNAs have been functionally characterized.
Initial functional studies of miRNA genes revealed their involvement in regulating a number of developmental processes (Kidner and Martienssen, 2005; Chen, 2009; Chuck et al., 2009; Weiberg et al., 2014). Nonetheless, the key regulatory roles of miRNAs in mediating plant responses to pathogen infection are now being increasingly recognized (Seo et al., 2013; Staiger et al., 2013; Weiberg et al., 2014; Fei et al., 2016). Recent studies have generated compelling proof for the implication of miRNAs in regulating defense signaling and immune responses during plant interaction with various phytopathogens, including bacteria, fungi, oomycetes, viruses, and nematodes (Seo et al., 2013; Gupta et al., 2014; Yang and Huang, 2014; Hewezi and Baum, 2015). miRNAs can function as negative regulators of plant defenses, leading to increasing plant susceptibility to pathogen infection. For instance, miR844 and miR400 were found to enhance plant susceptibility to Pseudomonas syringae and Botrytis cinerea, respectively, when overexpressed in Arabidopsis (Arabidopsis thaliana; Park et al., 2014; Lee et al., 2015). In barley (Hordeum vulgare), various miR9863 family members have been demonstrated to target distinct alleles of the Mla immune receptors to inhibit immune response signaling in response to infection by the powdery mildew fungus, Blumeria graminis (Liu et al., 2014a). A limited number of miRNA genes have also been shown to modulate plant innate immunity. For instance, overexpression of Arabidopsis miR160a, which targets ARF10, ARF16, and ARF17, activated callose deposition, resulting in enhanced plant resistance to P. syringae (Li et al., 2010). More recently, miR444 has been found to activate plant innate immunity against rice stripe virus in rice. The expression of miR444 was activated upon virus infection, and this activation was accompanied by down-regulation of its target genes OsMADS23, OsMADS27a, and OsMADS57, the repressors of RNA-DEPENDENT RNA POLYMERASE1 (RdRP1), leading to activation of the RdRP1-dependent antiviral silencing pathway (Wang et al., 2016).
Detailed functional studies also revealed that the same miRNA gene can execute diverse functions against different pathogens. For example, overexpression of rice miR398b resulted in increasing plant susceptibility to P. syringae via inhibiting callose deposition (Li et al., 2010). In contrast, miR398b overexpression can also enhance plant resistance against the blast fungus Magnaporthe oryzae via increasing the production of hydrogen peroxide (Li et al., 2014). Similarly, it has been shown that miR863-3p can mutually regulate negative and positive mediators of defense signaling in a pathogen infection stage-specific fashion to fine-tune the timing of defense response (Niu et al., 2016).
Plant-parasitic cyst nematodes are most destructive root parasites, causing substantial yield losses in many crop plants. These obligate parasites form, in the root vascular tissues, a specialized multinucleate feeding site, termed syncytium. The syncytium is a metabolically hyperactive sink-like structure from which the nematodes feed throughout the parasitic stages. Formation of functional syncytia is a sophisticated cellular process that involves an intricate interplay of numerous signaling and developmental pathways, whose regulation remains poorly understood. However, recent studies point at vital regulatory functions of miRNAs in syncytium formation and function. For example, miR396 targeting of growth regulating factor 1 (GRF1) and GRF3 was found to control syncytium initiation and development via regulating numerous hormonal signaling and developmental pathways (Hewezi et al., 2012; Hewezi and Baum, 2012; Liu et al., 2014b). More recently, it has been shown that Heterodera schachtii-induced up-regulation of miR827 posttranscriptionally silences the NITROGEN LIMITATION ADAPTATION gene specifically in the syncytium to permanently attenuate immune responses and enable successful parasitism (Hewezi et al., 2016). Also, miRNAs seem to have functional roles in regulating phytohormone signaling during plant interactions with root-knot nematodes. It has been recently demonstrated that tomato (Solanum lycopersicum) miR319 regulates jasmonic acid level during Meloidogyne incognita infection (Zhao et al., 2015). Another recent study has suggested a role of Arabidopsis miR319 in modulating auxin signaling during Meloidogyne javanica parasitism (Cabrera et al., 2016).
miRNA-mediated post transcriptional control of gene activity is a highly dynamic process that determines not only transcript stability and protein level but also allows plant cells to establish metabolic and physiological readjustment to cope with new functions or fluctuating conditions. In plants, a significant number of miRNA genes target transcription factors (Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004). In turn, miRNA-regulated transcription factors have tremendous potential to achieve such readjustment in cellular metabolism and physiology because of their ability to control numerous downstream targets. In addition, miRNA genes and their targeted transcription factors may correspondingly adjust the expression of each other through feedback regulatory loops, in which the transcription factors directly regulate the expression of their negative regulators, resulting in tight control over gene expression patterns (Meng et al., 2011). Furthermore, a transcription factor and its miRNA regulators may antagonistically regulate common targets, although such mechanisms have not yet been described in plants.
In Arabidopsis, miR858 posttranscriptionally silences the expression of several MYB transcription factors including MYB6, MYB11, MYB12, MYB13, MYB20, MYB42, MYB63, MYB83, and MYB111 (Fahlgren et al., 2007; Addo-Quaye et al., 2008; Sharma et al., 2016). These miR858-targeted MYBs are involved in a variety of cellular processes, including plant responses to drought (MYB20 and 60), the phenylpropanoid pathway (MYB11, 12, and 111), and secondary wall biosynthesis (MYB46 and 83; Cominelli et al., 2005; McCarthy et al., 2009; Oh et al., 2011; Gao et al., 2014; Sharma et al., 2016). Here, we report a novel function of the miR858-MYB83 regulatory system in plant-cyst nematode interaction. Both miR858 and MYB83 were transcriptionally activated in the syncytia of H. schachtii, and modulation of their expression through gain- and loss-of-function approaches altered Arabidopsis response to nematode infection. In line with the function of MYB83 in facilitating nematode infection of Arabidopsis, our transcriptome analysis revealed that MYB83 regulates a substantial number of syncytial genes encoding components essential for syncytium formation and function. Also, our results establish that MYB83 through a feedback loop activates the expression of miR858, thereby stabilizing its own transcript abundance and its downstream regulated genes during the initiation and progression of nematode parasitism.
RESULTS
miR858 Is Expressed in the Syncytium during the Initiation and Progression of Nematode Parasitism
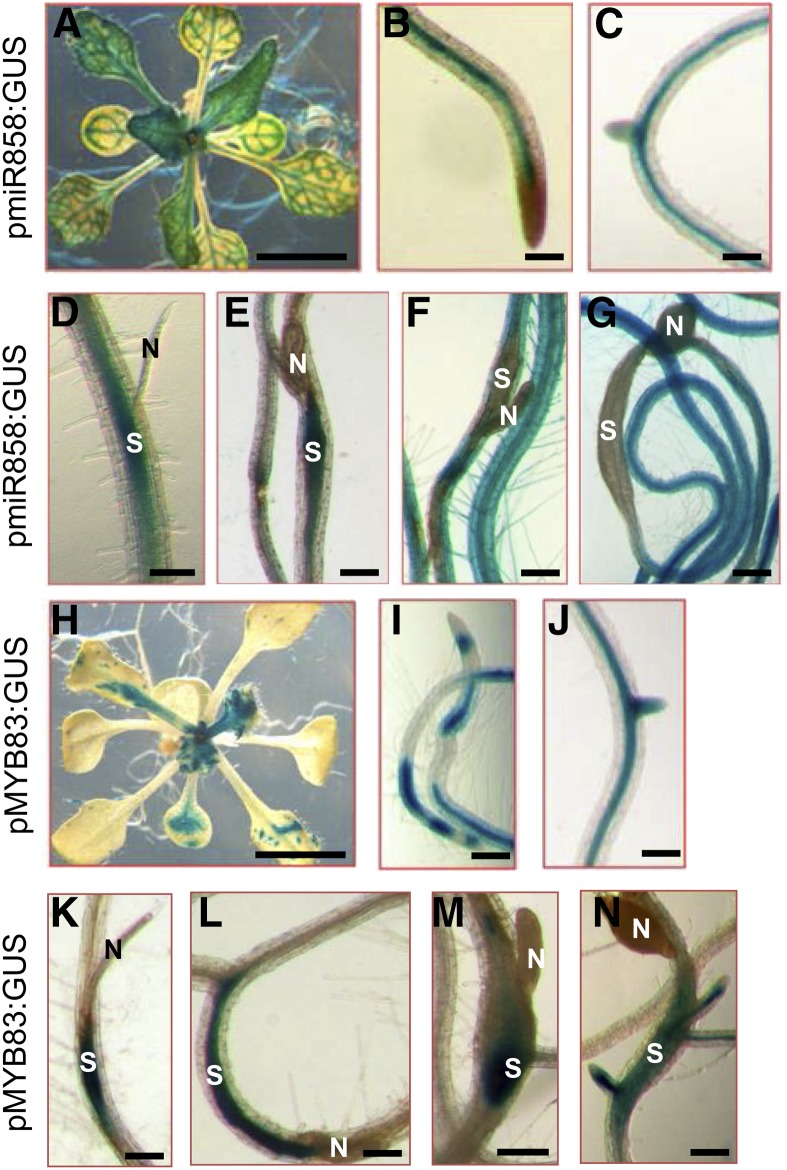
In Arabidopsis, miR858 is encoded by one functional genomic locus (miR858a, AT1G71002) that produces 21-nucleotide mature molecules and targets ten MYB transcription factor mRNAs that contain the miR858 complementary sequences (Sharma et al., 2016). To investigate the functional role of miR858 during the compatible interaction between Arabidopsis and the beet cyst nematode H. schachtii, we first produced transgenic Arabidopsis lines expressing the β-glucuronidase (GUS) reporter gene driven by the miR858 promoter (pmiR858:GUS). GUS activity of four independent transgenic lines (T2 generation) were assayed both under noninfected and H. schachtii-infected conditions. In noninfected 2-week-old-plants, GUS staining was observed in leaf and root vascular tissues (Fig. 1, A–C). Under H. schachtii-infected conditions, GUS activity was observed in the developing syncytium of the second stage nematode juvenile (J2) at 3 d postinfection (dpi) as well as in the syncytium of the early J3 stage at 7 dpi (Fig. 1, D and E). However, at 10 and 14 dpi (late J3 and J4 stages), GUS staining in the syncytium was absent (Fig. 1, F and G). The expression pattern of miR858 in the nematode feeding sites points to a functional role of miR858 in suppressing its target genes during the initiation and progression of nematode parasitism. Therefore, reduced miR858 promoter activity at later stages suggests an uninhibited expression of these target genes at these time points.
Figure 1.
Histochemical staining of GUS activity driven by miR858 and MYB83 promoters in transgenic Arabidopsis lines in response to H. schachtii infection. A to C, GUS activity of the pmiR858:GUS plants under noninfected conditions. Shown are GUS activity in the vascular tissues of leaves (A) and roots (B and C) of 2-week-old plants. D to G, GUS activity of the pmiR858:GUS plants in response to H. schachtii infection. Strong GUS activity was observed in the H. schachtii-induced syncytia at 3 (D) and 7 (E) dpi, whereas at 10 and 14 dpi, GUS activity was absent in the syncytia (F and G). H to J, GUS activity of the pMYB83:GUS plants under noninfected conditions. Shown are GUS activity in leaves (H) and vascular root tissues (I and J) of 2-week-old plants. K to N, GUS activity of the pMYB83:GUS plants in response to H. schachtii infection. Strong GUS activity was observed in the H. schachtii-induced syncytia at 3 (K), 7 (L), 10 (M), and 14 (N) dpi. N, Nematode; S, syncytium. Bars = 100 μm, except for A and H, which are 1 cm.
miR858 Posttranscriptionally Regulates MYB83 during H. schachtii Parasitism of Arabidopsis
It has been shown recently that miR858 posttranscriptionally silences the 10 MYB transcription factors MYB6, MYB11, MYB12, MYB13, MYB20, MYB42, MYB48, MYB63, MYB83, and MYB111 that contain the miR858 binding site (Sharma et al., 2016). Recent functional analyses have implicated MYB83 (AT3G08500) in the coordination of secondary wall biosynthesis and cell wall modifications in Arabidopsis (McCarthy et al., 2009; Zhong and Ye, 2012), both of which are fundamental cellular processes impacting syncytium formation and development (Bohlmann and Sobczak, 2014; Hewezi, 2015). Therefore, we directed our focus to elucidating the potential regulatory role of the miR858-MYB83 system in establishing the interactions between Arabidopsis and H. schachtii. We generated pMYB83:GUS transgenic lines to determine if MYB83 shares the temporal expression patterns with miR858 in the syncytium, which would suggest that MYB83 is posttranscriptionally down-regulated by miR858 also in the nematode feeding site. GUS activities were assayed in four independent transgenic lines (T2 generation) both under noninfected and H. schachtii-infected conditions. GUS activity of 2-week-old noninfected plants was observed in leaf and root vascular tissues (Fig. 1, H–J). Under H. schachtii infection, robust GUS staining was detected in the syncytium during the J2, early J3, late J3, and J4 developmental stages at 3, 7, 10, and 14 dpi, respectively (Fig. 1, K–N). The coincident up-regulation of miR858 and MYB83 promoters in the syncytium at 3 and 7 dpi suggests that MYB83 is targeted by miR858 for posttranscriptional regulation during the early syncytium development stage. At later stages, miR858 expression was down-regulated in the syncytium; thus, MYB83 is unlikely to be posttranscriptionally silenced by miR858.
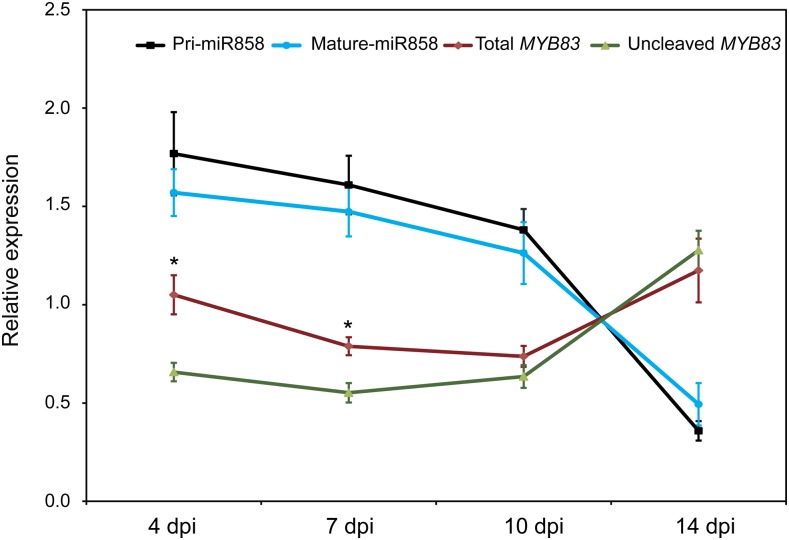
In order to provide additional evidence that miR858 mediates posttranscriptional regulation of MYB83 during H. schachtii infection of Arabidopsis, we used quantitative real-time RT-PCR (qPCR) to quantify the levels of miR858 primary transcripts (pri-miR858), mature miR858, as well as total and uncleaved transcript levels of MYB83 in the roots of wild-type (Col-0) Arabidopsis plants inoculated with H. schachtii at 4, 7, 10, and 14 dpi, relative to the corresponding noninoculated controls. The relative levels of uncleaved MYB83 transcripts were determined using a primer pair flanking the miR858 binding site, whereas the relative levels of total (cleaved and uncleaved) MYB83 transcripts were determined using a primer pair located downstream of the miR858 binding site as previously described by Hewezi et al. (2016). Gene expression data from three biologically independent replicates revealed up-regulation of both primary and mature miR858 in infected roots at 4 and 7 dpi compared with noninfected roots (Fig. 2), a result that is consistent with the increased activity of the miR858 promoter in the syncytium at the same time points. Meanwhile, the levels of total MYB83 transcripts were significantly higher (P < 0.05) than the level of uncleaved transcripts in infected root tissues compared with noninfected roots at both 4 and 7 dpi (Fig. 2). At 10 dpi, the expression levels of both primary and mature miR858 were slightly up-regulated, and this up-regulation was accompanied with small insignificant (P = 0.26) reduction in the level of uncleaved MYB83 transcripts compared with the total transcript level (Fig. 2). At 14 dpi, the expression of primary and mature miR858 was sharply decreased in the infected roots. At the same time, total and uncleaved MYB83 transcripts accumulated at similar levels (Fig. 2). These temporal expression patterns, which show that the abundance of uncleaved MYB83 transcripts are inversely correlated with the expression level of miR858, indicate that MYB83 is subjected to posttranscriptional regulation by miR858 following H. schachtii infection. Given our promoter data, this regulation likely is at work in the syncytium.
Figure 2.
miR858 posttranscriptionally down-regulates MYB83 during H. schachtii parasitism of Arabidopsis. The abundance of primary and mature miR858 as well as total and uncleaved transcript levels of MYB83 were measured using qPCR in the roots of wild-type Col-0 plants at 4, 7, 10, and 14 d after H. schachtii infection, relative to noninfected control plants. The total transcript levels of MYB83 were inversely correlated with the expression levels of primary and mature miR858 at the four time points. In addition, the levels of uncleaved MYB83 transcripts were lower than the level of total transcripts at 4 and 7 dpi, indicative of a posttranscriptional down-regulation of MYB83 by miR858 at these two time points. Data were obtained from three biological samples and represented as mean ± se. Normalization of the expression levels of miR858 and MYB83 was carried out using U6 and Actin8 as internal reference genes, respectively. Statistically significant differences between the levels of total and uncleaved MYB83 transcripts were determined using t tests (*P < 0.05).
Overexpression of miR858 Confers Enhanced Resistance to H. schachtii
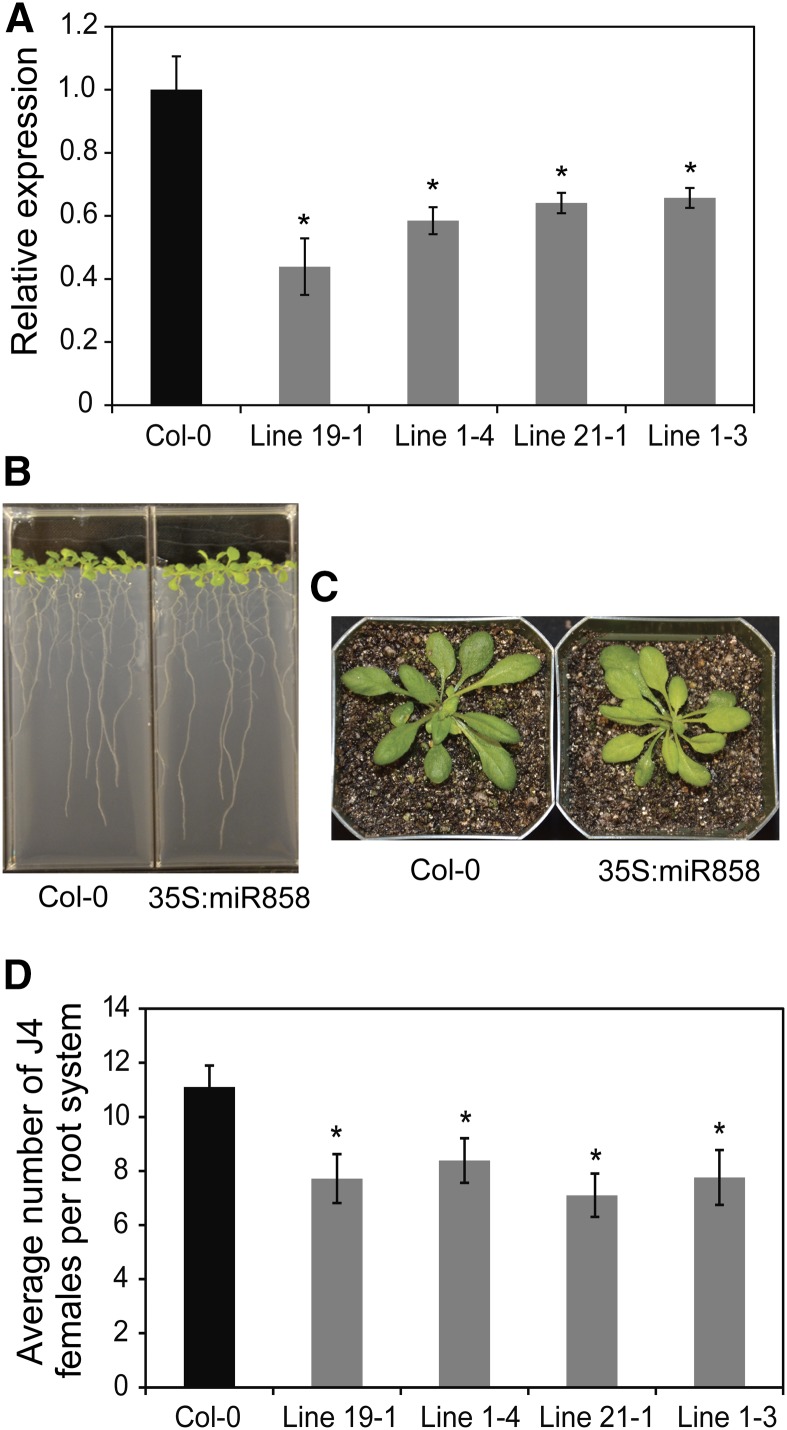
Promoter and qPCR analyses of miR858 expression during nematode infection revealed two distinct patterns (i.e. up-regulation during J2 and early J3 stages and down-regulation during late J3 and J4 stages). Thus, we investigated whether constitutive overexpression of miR858 would modulate Arabidopsis susceptibility to H. schachtii. To this end, we produced transgenic Arabidopsis lines overexpressing the primary miR858 sequence under the control of 35S promoter (35S:miR858). Four independent nonsegregating T2 overexpression lines (1-3, 1-4, 19-1, and 21-1) showing between 6- and 18-fold increases in miR858 expression levels relative to the Col-0 plants were selected (Supplemental Fig. S1A). MYB83 transcript levels were significantly decreased in these lines relative to Col-0 plants (Fig. 3A), which is consistent with posttranscriptional degradation of MYB83 transcripts by miR858. The root lengths of miR858 overexpression lines were comparable to Col-0 plants, with the exception that line 21-1 showed a slight increase of about 5% (Fig. 3B; Supplemental Fig. S2A). No other developmental defects were observed when these lines were grown under standard growth conditions (Fig. 3, B and C), confirming the results recently obtained by Sharma et al. (2016). The four miR858 overexpression lines were assayed for H. schachtii susceptibility. These lines displayed statistically significant decreases in susceptibility levels with 30 to 42% reduction in J4 female nematode counts compared to Col-0 plants (Fig. 3D). These results indicate that constitutive overexpression of miR858 interferes with H. schachtii parasitism of Arabidopsis.
Figure 3.
Overexpression of miR858 confers enhanced resistance to H. schachtii. A, Constitutive overexpression of miR858 in four independent transgenic Arabidopsis lines resulted in significant decreases in MYB83 expression levels. The expression levels of MYB83 were determined in the roots of 2-week-old transgenic lines relative to the wild-type Col-0 plants using qPCR. Shown are average expression levels obtained from three biological samples ± se. Statistically significant differences were determined using t tests (*P < 0.01). B and C, Root (C) and shoot (B) phenotypes of 3-week-old transgenic Arabidopsis plants overexpressing miR858. D, Nematode infection assays of the miR858 overexpression lines showing reduced susceptibility to H. schachtii compared with the wild-type Col-0 plants. Shown are average numbers of J4 females per root system ± se (n = 20) at 3 weeks postinoculation. Statistically significant differences from wild-type Col-0 plants were determined using t tests (*P < 0.05).
Overexpression of a Mimic Sequence for miR858 Augments Plant Susceptibility to H. schachtii
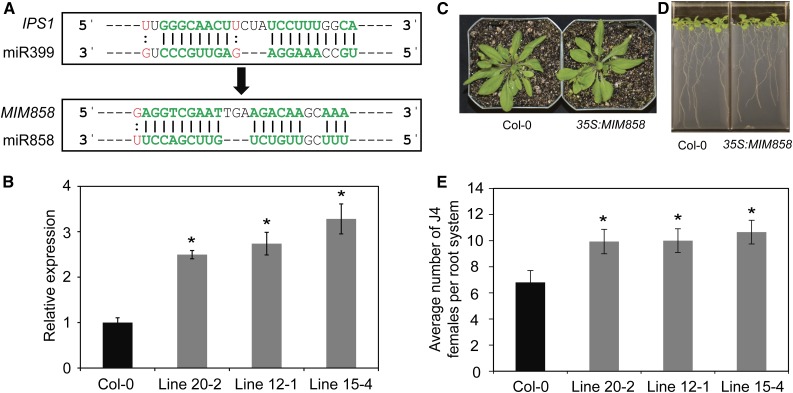
The implication of miR858 in modulating plant response to nematode infection was further examined by generating transgenic Arabidopsis plants with reduced miR858 expression. This was accomplished by expressing a mimic sequence for miR858 in its mature form (MIM858; Fig. 4A). The artificial noncleavable binding site for the mature miR858 contained a three-nucleotide bulge (TGA) that does not interfere with miR858 binding but would prevent transcript cleavage and hence sequester miR858 activity. Three independent transgenic lines showing between 2.7- and 6.8-fold reduction in the mature miR858 expression levels were selected (Supplemental Fig. S1B). qPCR quantification revealed that miR858 down-regulation in the MIM858 lines was correlated with significant increases in MYB83 expression levels (Fig. 4B), a finding that confirms the efficiency of our target mimicry construct in sequestering the activity of miR858. Other than minor reductions in root lengths of MIM858 plants, no noticeable morphological differences between MIM858 lines and Col-0 plants were detected (Fig. 4, C and D; Supplemental Fig. S2B). Interestingly, when the susceptibility of the MIM858 lines to H. schachtii was determined, these lines were significantly more susceptible than the wild-type plants, showing up to 56% increase in the number of J4 nematodes (Fig. 4E). Together, these data confirm that increased expression of miR858 is accountable for the reduced susceptibility phenotype seen in miR858 overexpression plants.
Figure 4.
Constitutive down-regulation of miR858 increased plant susceptibility to H. schachtii. A, Approach for creating a mimic binding site for miR858 (MIM858). The miR399 mimic sequence in the IPS1 was replaced by an artificial noncleavable binding site for the mature miR858. The artificial binding site contained a three-nucleotide bulge (TGA) that would prevent transcript cleavage and hence sequester miR858 activity. B, Constitutive overexpression of MIM858 in three independent transgenic Arabidopsis lines resulted in significant up-regulation of MYB83. The expression levels of MYB83 were quantified in the roots of 2-week-old transgenic lines relative to the wild-type Col-0 plants using qPCR. Shown are average expression levels obtained from three biological samples ± se. Statistically significant differences were determined using t tests (*P < 0.01). C and D, Shoot (C) and root (D) phenotypes of 3-week-old transgenic Arabidopsis plants overexpression MIM858. E, Nematode infection assays of the MIM858 overexpression lines showing increased susceptibility to H. schachtii compared with the wild-type Col-0 plants. Shown are average numbers of J4 females per root system ± se (n = 20) at 3 weeks postinoculation. Statistically significant differences from wild-type Col-0 plants were determined using t tests (*P < 0.05).
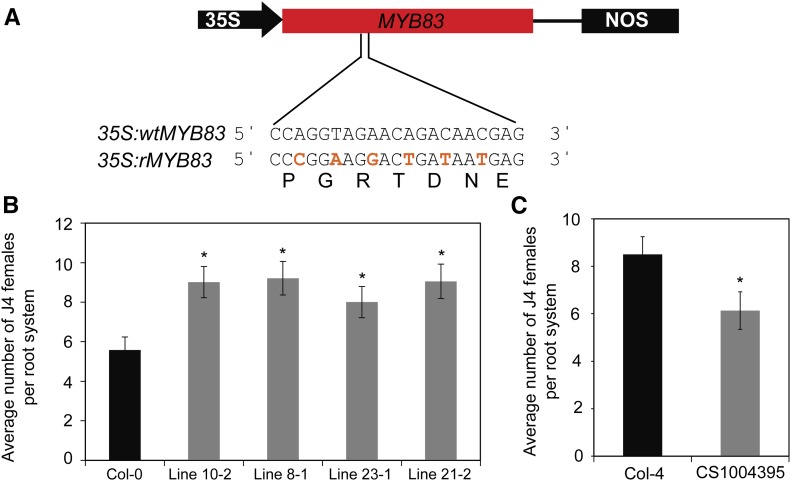
Ectopic Overexpression of a Nondegradable Coding Sequence of MYB83 Enhances Arabidopsis Susceptibility to H. schachtii
The gene expression analyses and nematode susceptibility assays of miR858 and MIM858 overexpression lines mentioned above indicate that inhibition of miR858 activity facilitates H. schachtii infection, most likely through the up-regulation of its MYB target genes. If miR858 modulates plant susceptibility mainly through posttranscriptional regulation of MYB83, manipulation of MYB83 expression should also influence plant susceptibility to H. schachtii but in the opposite direction. To investigate this assumption, we generated transgenic Arabidopsis lines overexpressing a miR858-resistant variant of MYB83 under the control of 35S promoter (35S:rMYB83). The noncleavable variant rMYB83 was constructed by creating six-nucleotide mismatches in the miR858 target site without altering the encoded protein sequences (Fig. 5A). Four nonsegregating T2 lines showing between 16- and 31-fold MYB83 mRNA up-regulation were selected and phenotypically analyzed (Supplemental Fig. S1C). These lines were indistinguishable from the nontransgenic plants in term of root and shoot morphology and development (Supplemental Fig. S2C). Interestingly, when these lines were used in H. schachtii infection assays, all lines exhibited statistically significant increases in susceptibility compared with Col-0 plants (Fig. 5B). In addition, a T-DNA insertional mutant of MYB83 (CS1004395; Supplemental Fig. S3) was identified, and no obvious morphological defects in roots or shoots were observed. In contrast to the rMYB83 overexpression lines, the myb83 mutant showed reduced nematode susceptibility relative to the wild-type Col-4 plants (Fig. 5C). Taken together, these results link the activity of the MYB83 to the function of miR858 in modulating plant responses to H. schachtii infection.
Figure 5.
Constitutive overexpression of miR858-resistant variant of MYB83 increased plant susceptibility to H. schachtii. A, Schematic representation showing the generation of a miR858-resistant variant of MYB83 (rMYB83) by introducing synonymous mutations to the miR858 binding site in the MYB83 coding sequence. B and C, Nematode infection assays of rMYB83 overexpression lines and a MYB83 mutant line. Three independent transgenic lines overexpressing 35S:rMYB83 construct showed increased susceptibility to H. schachtii compared with the wild-type Col-0 plants (B). In contrast, the myb83 knockout mutant line CS1004395 showed reduced susceptibility compared with the wild-type Col-4 plants (C). Shown are average numbers of J4 females per root system ± se (n = 20) at 3 weeks postinoculation. Statistically significant differences from the corresponding wild-type plants were determined using t tests (*P < 0.05).
RNA-Seq Analysis of miR858 and rMYB83 Overexpression Plants
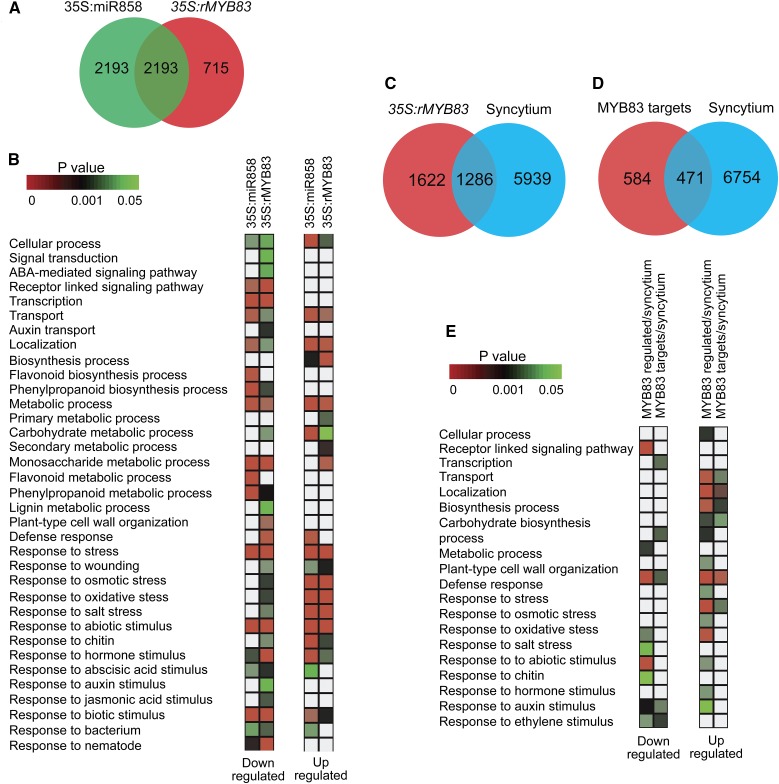
The robust effects of MYB83 overexpression and knockout mutant lines on nematode susceptibility suggest that this transcription factor may control downstream target genes encoding proteins necessary for syncytium formation/function. Therefore, we performed RNA-seq analysis on root tissues isolated from the 35S:miR858 (line 1-4), 35S:rMYB83 (line 8-1), and wild-type Col-0 plants in order to identify the downstream targets that may be directly or indirectly controlled by MYB83. Three biological samples of root tissues were collected from each plant line (2 weeks old) for mRNA isolation and library preparation. Differentially expressed genes (DEGs) were determined using adjusted P values < 0.05. We identified 4,386 and 2,908 DEGs in P35S:miR858 and 35S:rMYB83, respectively, compared with Col-0 plants. Out of the 4,386 DEGs identified in the 35S:miR858 plants, 2,082 genes were up-regulated and 2,304 genes were down-regulated (Supplemental Data Set 1). In the 35S:rMYB83 plants, 1,249 genes were up-regulated and 1,659 genes were down-regulated (Supplemental Data Set 2). Comparison of the DEGs in these two transgenic lines revealed that 2,193 genes were common to both sets (Fig. 6A; Supplemental Data Set 3). The fact that this overlapping gene list represents more than 50% of the DEGs identified in the 35S:miR858 plants, indicates that MYB83 is the main target of miR858 in roots.
Figure 6.
Functional classification and GO enrichment analyses of differentially expressed genes identified in 35S:miR858 and 35S:rMYB83 lines. A, Venn diagram displaying the number and overlap of the DEGs identified in miR858 and rMYB83 overexpression lines. B, GO classification and enrichment analyses of the DEGs identified in 35S:miR858 and 35S:rMYB83 lines. Enrichment analyses of up-regulated and down-regulated genes were performed separately. C, Venn diagram shown the overlap between MYB83-regulated genes and syncytium DEGs. D, Venn diagram shown the overlap between MYB83 putative targets and syncytium DEGs. E, GO classification and enrichment analyses of MYB83-regulated genes and its putative direct targets overlapping with syncytium DEGs. Enrichment analyses of up-regulated and down-regulated genes were performed separately. Enrichment analysis was performed using Fisher’s exact test and Bonferroni multitest correction with a significance cutoff P < 0.05.
Gene Ontology (GO) classification and enrichment analyses of the DEGs in the 35S:miR858 and 35S:rMYB83 plants were performed. Thirty-five GO biological process terms, which are mainly associated with metabolic processes and response to biotic and abiotic stimuli, were identified (Fig. 6B). While several GO terms were enriched among the up-regulated or down-regulated genes in both lines, enriched GO terms specific to each line were also seen. For example, GO terms corresponding to defense response, and responses to bacterium and abscisic acid stimulus were enriched uniquely among the up-regulated genes in 35S:miR858 plants (Fig. 6B). Similarly, GO terms corresponding to carbohydrate metabolic processes as well as secondary metabolic process were significantly overrepresented uniquely among the up-regulated genes in 35S:rMYB83 plants (Fig. 6B). The same observation is equally evident among the down-regulated genes. For example, enrichment of GO terms corresponding to flavonoid biosynthesis and metabolic processes were identified only among the down-regulated genes in 35S:miR858 plants (Fig. 6B). GO terms corresponding to signal transduction, auxin transport, lignin metabolic process, cell wall organization, and responses to wounding, osmotic stress, oxidative stress, salt stress, chitin, auxin stimulus, and jasmonic acid stimulus were significantly enriched exclusively among the down-regulated genes in the 35S:rMYB83 plants (Fig. 6B).
Identification of Putative Direct Targets of MYB83
Recently, the ACC(A/T)A(A/C)(T/C) consensus sequence was identified as the MYB83 cis-binding element in Arabidopsis (Zhong and Ye, 2012). Therefore, we scanned the promoters of the DEGs identified in the 35S:rMYB83 plants (2,908 genes) for the presence of this cis-element within 1.5 kb upstream of the transcription start site (TSS) to identify putative direct targets of MYB83. The number of cis-elements identified in these DEGs ranged between 0 and 15 elements, with the large majority containing between 0 and 2 elements (Supplemental Fig. S4A). Also, we determined the average number of this cis-element in the promoters of 2,908 randomly selected genes to be 1.02 elements. Thus, DEGs with at least three MYB83 cis-binding elements in the promoters of the MYB83-regulated genes were considered as putative direct targets of MYB83 (P value 2.69E-65, Fisher’s exact test). As a result, 1,055 of the MYB83-regulated genes were identified as bona fide direct target candidates (Supplemental Data Set 4). The cis-elements are equally distributed across the gene promoters (Supplemental Fig. S4B). However, 77% (815/1055) of these putative direct targets contain at least one cis-element within 500 bp of the TSS (Supplemental Fig. S4C). GO enrichment analysis revealed that genes involved in transport, primary metabolic processes, secondary metabolic processes, particularly glucosinolate, and responses to biotic and abiotic stresses were significantly enriched among the direct targets that were positively regulated in the 35S:rMYB83 plants. GO categories associated with transcription, metabolic process, defense response, and responses to stress, nematode, hormone, and biotic stimuli were significantly enriched among the putative direct target genes that were negatively regulated in the 35S:rMYB83 plants (Supplemental Fig. S5).
MYB83 Regulates Key Cellular Processes in the Syncytium of H. schachtii
Consistent with the function of MYB83 in promoting plant susceptibility to H. schachtii, we found a significant overlap between the MYB83-regulated genes and the syncytium DEGs previously identified by Szakasits et al. (2009). Out of the 2,908 MYB83-regulated genes 1,286 overlapped with the 7,725 syncytium DEGs (Fig. 6C; Supplemental Data Set 5). This significant overlap (44.2%, χ2 = 224.729, P value = 1.909E-48) indicates that 16.6% of the syncytium transcriptome is regulated by MYB83. Also, we compared the identified 1,055 direct target genes of MYB83 with the syncytium DEGs to determine the extent to which MYB83 directly regulates gene expression in the syncytium. A common set of 471 genes (44.6%, χ2 = 79.765, P value = 3.452E-17) was identified, implying that 6% of syncytium genes are under direct control of MYB83 (Fig. 6D; Supplemental Data Set 6). Of these 471 genes, 216 (46%) were up-regulated in of MYB83 overexpression plants and 255 (54%) were down-regulated, suggesting that MYB83 has a dual transactivation and transrepression function. GO term analysis of the 1,286 MYB83-regulated genes overlapping with the syncytium DEGs revealed an enrichment of genes involved in receptor-mediated signaling pathways, transport, metabolic processes, cell wall organization, and responses to stress, chitin, and bacterium, together with responses to biotic, abiotic, hormone, auxin, and ethylene stimuli (Fig. 6E). When this analysis was conducted to include only the 471 genes predicted as putative direct targets of MYB83 in the syncytium, GO terms associated with transcription, transport, metabolic process, responses to stress, oxidative stress, bacterium, and biotic stimulus were significantly enriched (Fig. 6E).
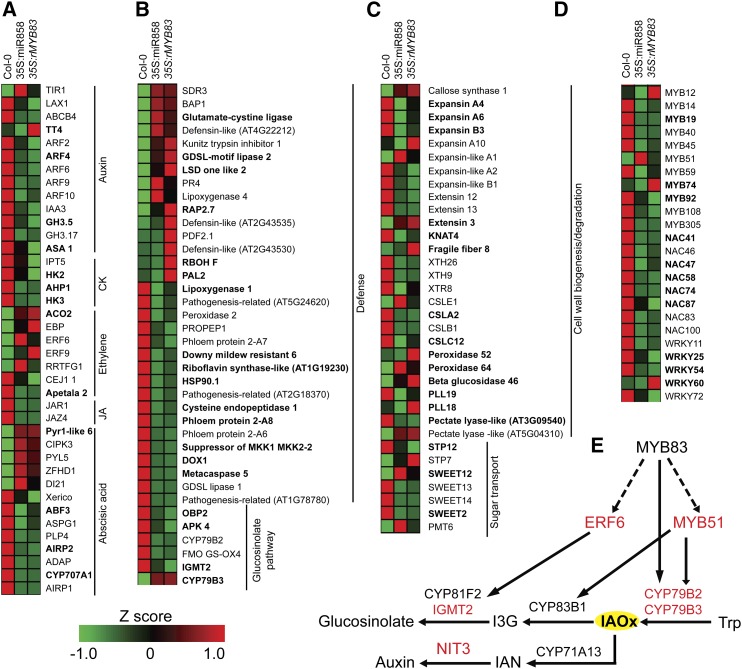
In addition to GO analysis, careful examination of the known functions of MYB83-regulated genes overlapping with the syncytium DEGs enabled more detailed insights into the functional role of MYB83 during H. schachtii infection. As shown in Figure 7, genes involved in hormone signaling pathways (Fig. 7A), defense response and glucosinolate biosynthesis (Fig. 7, B and E), cell wall modification and sugar transport (Fig. 7C), and transcriptional control (Fig. 7D) seem to be the key etiological factors of MYB83 in facilitating nematode parasitism of Arabidopsis.
Figure 7.
Differential expression patterns of a set of MYB83-regulated genes involved in key biological processes associated with nematode parasitism. The RKPM values of the selected MYB83-regulated genes overlapping with the syncytium DEGs were row-wise normalized using Z score and used to construct the heat maps. Shown are genes involved in hormone signaling pathways (A), defense response (B), cell wall modification and sugar transport (C), and transcriptional control (D). Putative direct targets of MYB83 are highlighted in bold. Not that MYB83-regulated genes were identified under noninfected conditions and were compared with the syncytium DEGs previously identified by Szakasits et al. (2009) at 5 and 15 dpi. E, Schematic depicting the glucosinolate biosynthesis pathway in which 6 MYB83-regulated genes are highlighted in red.
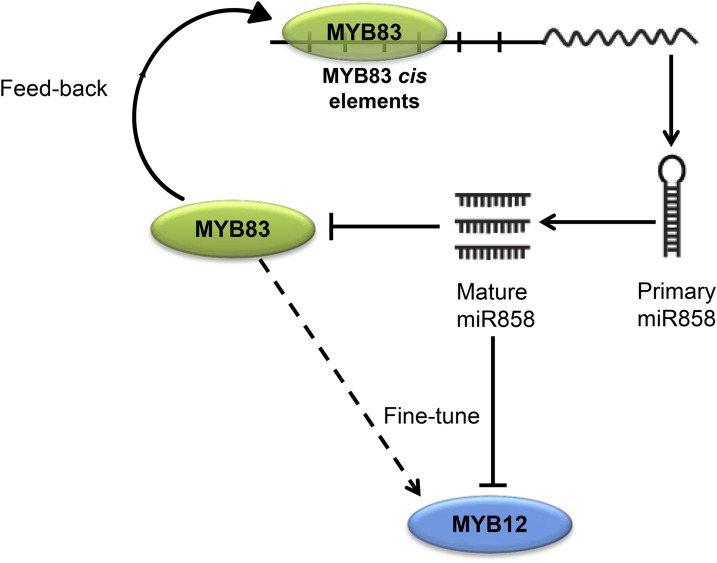
miR858 and MYB83 Constitute a Feedback Regulatory Loop That Involves MYB12
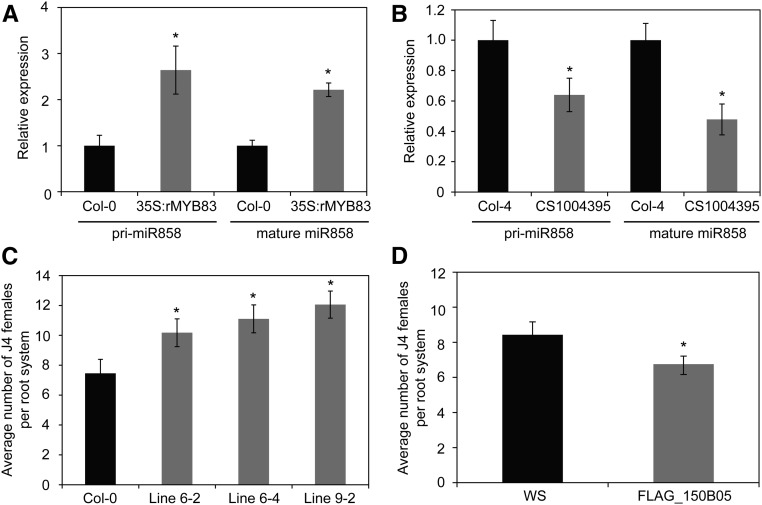
We next examined the promoter of miR858, 2 kb upstream of the TSS, for the presence of MYB83 cis-binding element. Interestingly, seven cis-binding elements were identified, emphasizing the possibility that MYB83 regulates the expression of miR858. To investigate this possibility, we quantified the expression of primary and mature miR858 in the 35S:rMYB83 plants as well as the myb83 knockout mutant line (CS1004395) using qPCR. Data from three biological samples indicated about 2-fold up-regulation of both primary and mature miR858 in the transgenic plants overexpressing MYB83 (Fig. 8A) compared to Col-0 plants. In contrast, both primary and mature miR858 transcripts were down-regulated in the myb83 mutant compared with the wild-type Col-4 (Fig. 8B). Taken together, these results imply that MYB83 positively regulates the expression of its negative regulator through a feedback regulatory loop to maintain proper level of its transcripts.
Figure 8.
miR858/MYB83 regulatory loop involves MYB12. A and B, MYB83 positively regulates the expression of miR858. The expression levels of primary and mature miR858 transcripts were quantified in the roots of 2-week-old rMYB83 overexpression plants (A) as well as the myb83 mutant line CS1004395 (B) relative to the wild-type Col-0 or Col-4 plants, respectively using qPCR. Shown are relative expression values obtained from three biological samples ± se. Statistically significant differences were determined using t tests (*P < 0.05). C and D, MYB12 phenocopied the effects of MYB83 on plant susceptibility to H. schachtii. Three independent transgenic lines overexpressing the 35S:rMYB12 construct increased susceptibility to H. schachtii compared with the wild-type Col-0 plants (C), whereas the myb12 mutant line FLAG_150B05 showed reduced susceptibility compared with the wild-type Wassilewskija plants (D). Shown are average numbers of J4 females per root system ± se (n = 20) at 3 weeks postinoculation. Statistically significant differences from the corresponding wild-type plants were determined using t tests (*P < 0.05).
We then examined our RNA-seq data set to find out if any of the confirmed targets of miR858 were inversely regulated in the MYB83 overexpression plants and hence constitute part of the regulatory loop. Interestingly, we identified MYB12, a confirmed target of miR858, among the MYB83 positively regulated genes. This finding guided us to test whether MYB12 is an integral part of the miR858/MYB83 regulatory circuit impacting plant response to nematode infection. To this end, we generated transgenic plants overexpressing a miR858-resistant variant of MYB12 (rMYB12) driven by the 35S promoter (Supplemental Fig. S6). A nematode infection assay of three independent overexpression lines displayed significant increases in plant susceptibility to nematode infection compared with Col-0 plants (Fig. 8C). In addition, a T-DNA insertional mutant of MYB12 (FLAG_150B05; Supplemental Fig. S7) was identified, and no obvious morphological defects in roots or shoots were detected. Contrary to rMYB12 overexpression lines the myb12 mutant exhibited reduced susceptibility compared with the wild-type Wassilewskija plants (Fig. 8D). Together, these results suggest that MYB12 may constitute part of miR858/MYB83 regulatory loop regulating plant response to nematode infection.
DISCUSSION
Arabidopsis miR858 has been shown to regulate various growth and plant developmental processes (Guan et al., 2014; Jia et al., 2015; Sharma et al., 2016). However, a regulatory function of miR858 in plant-pathogen interactions has not been reported. Here, we report a crucial regulatory role of miR858 during H. schachtii parasitism of Arabidopsis. In response to H. schachtii infection, miR858 exhibited a biphasic expression pattern, including strong activation in the developing syncytia at 3 and 7 dpi and a subsequent down-regulation in the mature syncytia at 10 and 14 dpi. This pattern of miR858 expression suggests different functions during the two distinct stages of syncytium formation and maintenance. As a result, constitutive overexpression of miR858 resulted in significant decreases in nematode infection. In contrast, inactivation of miR858 by overexpressing an artificial target mimic sequence produced the opposite phenotype of enhanced susceptibility. It may be important to mention that no complementary sequences for miR858 were identified in Heterodera spp. when we scanned all available transcripts in the databases. Thus, it is unlikely that miR858 triggered host-induced gene silencing during nematode feeding on the transgenic lines overexpressing miR858.
The influence of miR858 expression changes on plant responses to H. schachtii seems to be mediated through posttranscriptional regulation of its MYB transcription factor genes, specifically MYB83. The MYB83 promoter was predominantly active in the syncytium during all nematode parasitic stages. Posttranscriptional silencing of MYB83 by miR858 was evident at 4 and 7 dpi as shown by low levels of uncleaved MYB83 transcripts compared with the total transcript levels. MYB83 expression increase seems to be conducive to H. schachtii infection of Arabidopsis because rMYB83 overexpression enhanced plant susceptibility, whereas a myb83 mutation rendered the plants less susceptible.
The regulatory relationship between miR858 and MYB83 seems to be established through a feedback regulatory loop. Our finding that the MYB83 binding motif occurs repeatedly in the miR858 promoter led us to examine a possible role of MYB83 in regulating the expression of miR858. The transcript abundance of pri-miR858 and mature miR858 was considerably increased in the rMYB83 overexpressing plants but decreased in the myb83 mutant, indicating that MYB83 participates in a feedback loop with its negative regulator to stabilize its own transcript abundance. A reciprocal feedback loop controlling the expression of miR396 and its target transcription factors GRF1 and GRF3 has been demonstrated to coordinate transcriptional events required for proper syncytium formation and function (Hewezi and Baum, 2012; Hewezi et al., 2012; Liu et al., 2014b). In addition, a number of miRNAs and their transacting targets were found to be intricately connected through feedback circuits in different growth and developmental contexts, where robust and adaptable transcriptional responses were established (Xie et al., 2003; Gutierrez et al., 2009; Wu et al., 2009; Marin et al., 2010; Yant et al., 2010; Merelo et al., 2016). Our data suggest that the miR858/MYB83 regulatory circuit may involve MYB12, a confirmed target of miR858, whose transcript abundance was positively regulated by MYB83. Thus, miR858 appears to fine-tune the function of MYB83 at various levels. MYB12 is also of functional importance for nematode parasitism since constitutive changes in its expression levels through overexpression and T-DNA insertional mutant lines altered plant response to nematode infection.
The miR858/MYB83 regulatory loop may enable controlling precise expression levels of genes involved in critical cellular processes required for syncytium differentiation without turning gene expression on and off to prevent syncytium degeneration and collapse. Our finding that 1,286 genes of the 2,193 MYB83-regulated genes were among the previously identified syncytium DEGs (Szakasits et al., 2009) may reflect a key regulatory function of MYB83 in reprogramming syncytium transcriptomes. It may be worth mentioning that other than significant differences in the numbers of female nematodes, we did not observe aberrant phenotype for nematode and syncytium development between the transgenic lines and wild-type controls. The transcriptome reprogramming mediated by the coordinated function of miR858 and MYB83 could be facilitating the formation of functional syncytium. We therefore focused our discussion on the potential importance of these genes for syncytium formation and nematode parasitism. Levels and signaling of phytohormones play fundamental roles in determining syncytium cell fate reprogramming and differentiation (Grunewald et al., 2009b; Gheysen and Mitchum, 2011; Goverse and Bird, 2011; Cabrera et al., 2015; Kammerhofer et al., 2015). In particular, auxin signaling has been shown to be rapidly activated upon nematode infection leading to syncytium differentiation and development (Goverse et al., 2000; Karczmarek et al., 2004; Grunewald et al., 2009a; Absmanner et al., 2013; Hewezi et al., 2014). Several genes encoding numerous functions of the auxin signal transduction cascade, including the auxin receptor TIR1, the auxin response factors 2, 4, 6, 9, and 10, the auxin influx carrier LAX1, and the auxin efflux transporter ABCB4 were among MYB83-regulated genes in the syncytium (Fig. 7A). Additional key genes involved in the auxin response (SHY2, ARGOS, and PLDP2) and auxin homeostasis (GH3.17) were also regulated by MYB83 in the syncytium (Fig. 7A). It has been recently reported that cytokinin signaling is critical for syncytium development and successful H. schachtii parasitism of Arabidopsis (Shanks et al., 2016; Siddique et al., 2015). MYB83-regulated genes overlapping with syncytium DEGs included various components of cytokinin signaling pathway, namely, the cytokinin synthase IPT5, the His kinase receptors AHK2 and AHK4, and the His phosphotransfer protein AHP (Fig. 7A). Together, these results indicate that MYB83 regulates auxin and cytokinin responses at various levels of biosynthesis, signal transduction, and downstream responses.
Also, ethylene has been shown to play contrasting dual functions during various nematode parasitic stages (Wubben et al., 2001; Kammerhofer et al., 2015). Notably, numerous ethylene response factors (ERFs), which control the downstream signaling of ethylene response, were also identified among the MYB83-indcued genes overlapping with the syncytium DEGs (Fig. 7A). This included ERF6, ERF9, and ERF72, which play key regulatory functions in biotic stress responses (Ogawa et al., 2005; Camehl and Oelmüller, 2010; Moffat et al., 2012; Maruyama et al., 2013; Meng et al., 2013; Chen et al., 2014; Xu et al., 2016), as well as ERF109, which regulates the accumulation of reactive oxygen species following biotic and abiotic stresses stimuli (Matsuo et al., 2015). Remarkably, a substantial number of genes associated with gibberellin, jasmonic acid, and abscisic acid signal transduction networks were directly or indirectly regulated by MYB83 in the syncytium (Fig. 7A). While the function of jasmonic acid and abscisic acid signaling in directing plant response to cyst nematodes is not fully understood (Kammerhofer et al., 2015), it has been recently demonstrated that these pathways regulate defense responses and basal immunity against sedentary and migratory nematodes (Nahar et al., 2011, 2012; Ozalvo et al., 2014).
Interestingly, we noted that genes encoding functions that mediate the interplay between various hormone signaling pathways were also regulated by MYB83 (Fig. 7A). This included, for example, ERF109 and ANTHRANILATE SYNTHASE ALPHA SUBUNIT1, which mediate the interplay between jasmonic acid and auxin biosynthesis and transport in roots (Sun et al., 2009; Cai et al., 2014), and the acyl acid amido synthetase GH3.5, which regulates the homeostasis and responses of salicylic acid and auxin following pathogen infection (Zhang et al., 2007; Westfall et al., 2016). Thus, miR858/MYB83-mediated precise regulation of transcript levels of various phytohormone signaling genes may allow infected root cells to properly differentiate and develop into functional syncytia in a stage-specific fashion, taking into consideration that the levels of these phytohormones are anticipated to vary throughout various stages of syncytium initiation, formation, and maintenance. It is plausible also that MYB83 may integrate signals from these hormone pathways to fine-tune the biosynthesis of defense components. In this context, pathogenesis-related (PR) genes, whose expression is linked to the signaling pathways of salicylic acid (thaumatin-like) and jasmonic acid (PR4 and PDF2.1), were among the identified MYB83-regulated genes in the syncytium (Fig. 7B). PDF2.1 was recently confirmed to be strongly expressed in the syncytium using reporter lines (Siddique et al., 2011). Interestingly, two genes encoding the cytochrome P450 enzymes CYP79B2 and CYP79B3, which are involved in the conversion of Trp to indole-3-acetaldoxime (Hull et al., 2000; Mikkelsen et al., 2000), were oppositely regulated by MYB83 (Fig. 7, B and E). The fact that indole-3-acetaldoxime is the metabolic branch node bringing about the biosynthesis of auxin and indole glucosinolate (Bak et al., 2001) suggests a role of MYB83 in regulating the balance between auxin homeostasis and glucosinolate biosynthesis. In support with this suggestion, several syncytium DEGs that are involved in the biosynthesis of glucosinolate were among the identified MYB83-regulated genes, from which four were considered as direct target gene candidates (Fig. 7E).
Several transcription factors of MYB, NAC, and WRKY families were among the MYB83-regulated genes in the syncytium (Fig. 7D), suggesting a role of MYB83 in forming a complex and highly interconnected regulatory network in the syncytium. Of the MYB transcription factors, MYB108, which regulates wound-induced cell death in an abscisic acid-dependent manner (Cui et al., 2013), and MYB51, a key regulator of indole glucosinolate biosynthesis (Frerigmann and Gigolashvili, 2014) were found. Additional MYB transcription factors included MYB12 and MYB59 that are involved in phenylpropanoid biosynthesis and cell cycle progression, respectively (Mehrtens et al., 2005; Mu et al., 2009). Thus, cross-regulation among certain MYB transcription factors in the syncytium may constitute a subregulatory network that contributes to the establishment of a syncytium-specific transcriptional program. Of the WRKY transcription factors regulated by MYB83, WRKY72 was previously reported to contribute to basal resistance against the root-knot nematode M. incognita and the oomycete Hyaloperonospora arabidopsidis (Bhattarai et al., 2010). Also, WRKY60 and WRKY11, the negative regulators of defense response (Journot-Catalino et al., 2006; Xu et al., 2006), were found to be regulated by MYB83 in opposite direction, implying a role of MYB83 in the control of defense response and inhibition of autoimmunity.
Positive and negative regulators of plant immunity are frequently dysregulated upon cyst nematode infection (Szakasits et al., 2009; Kandoth et al., 2011). Notably, master regulators of plant immunity were identified among the MYB83-regulated genes in the syncytium (Fig. 7B). This included KUNITZ TRYPSIN INHIBITOR1 and alpha-dioxygenase1, which encode functions that antagonize oxidative stress and cell death during pathogen infection (De León et al., 2002; Li et al., 2008). Other regulators of plant immunity included, for example, PROPEP1, the precursor of Pep1, which stimulates the transcription of the plant defensin gene PDF1.2 (Huffaker et al., 2006), the SUPPRESSOR OF MKK1 MKK2 2, an immune receptor that is involved in triggering defense responses against bacteria (Zhang et al., 2012), and BON ASSOCIATION PROTEIN1, a general suppressor of defense responses and programmed cell death (Yang et al., 2006, 2007).
Further inspection of the MYB83-regulated genes in the syncytium provided additional insights into the function of MYB83 during nematode parasitism. Interestingly, a number of genes encoding transmembrane sugar transport proteins were positively or negatively regulated by MYB83, including SWEET2, 12, 13, and 14, the sugar transporter protein 7 and 12, and the MONOSACCHARIDE TRANSPORTER6 (Fig. 7C). These sugar transporters may function in sugar remobilization to the syncytium during nematode feeding and development (Hofmann et al., 2009). As shown in Figure 7C, MYB83-regulated genes in the syncytium also included several expansins and genes coding for enzymes that participate in cell wall biogenesis and modification, comprising cellulose synthases, β-glucosidases, pectate lyases, and peroxidases; some of them were previously shown to modulate plant-nematode interactions (Wieczorek et al., 2006; Jin et al., 2011; Bohlmann and Sobczak, 2014; Wieczorek et al., 2014). Collectively, these data suggest a functional role of MYB83 in a variety of cellular processes associated with nematode infection.
Finally, we propose a model for miR858-MYB83 interaction during H. schachtii parasitism of Arabidopsis (Fig. 9). H. schachtii-induced activation of miR858 during the initiation and progression of nematode parasitism posttranscriptionally silences MYB83. MYB83 in turn positively regulates the expression of miR858, which contains several MYB83 cis-binding elements in its promoter. This feedback regulatory circuit may function as a homeostatic control mechanism to ensure proper expression levels of more than a thousand of MYB83-regulated genes in the H. schachtii-induced syncytium. The miR858/MYB83 regulatory system may also involve MYB12, which was oppositely regulated by miR858 and MYB83, providing additional layer of tight control over unidentified MYB12-regulated genes in the syncytium.
Figure 9.
Model for miR858-MYB83 interaction. Our results indicate that miR858 and MYB83 expression are connected through a feedback circuit in which miR858 regulates the expression of MYB83 and responds to its expression levels. This regulatory mechanism ensures proper expression levels of more than a thousand MYB83-regulated genes in the H. schachtii-induced syncytium. This fine-tuning mechanism appears to include MYB12, which was oppositely regulated by miR858 and MYB83, providing additional layer of tight control over gene expression.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All transgenic Arabidopsis (Arabidopsis thaliana) lines were generated in the Col-0 background. The myb83 T-DNA insertional mutant (CS1004395) in the Col-4 background was obtained from the Arabidopsis Biological Resource Center. The myb12 T-DNA mutant (FLAG_150B05) in the Wassilewskija background was obtained from the Genomic Resource Center, INRA-Versailles, France. Plants were grown at 24°C under light conditions of 16 h light and 8 h dark.
Nematode Infection Assay
Seeds of the transgenic and mutant lines along with the wild-type controls (Col-0, Col-4, or Wassilewskija) were sterilized using a 2.8% bleach solution for 5 min followed by four washes with sterilized double-distilled water. The sterilized seeds were then randomly distributed in 12-well culture plates (BD Biosciences) containing modified Knop’s medium solidified with 0.8% Daishin agar (Brunschwig Chemie) with each line being replicated 20 times. The plates were placed in a growth chamber at 24°C with 16-h-light/8-h-dark conditions. Freshly hatched J2 Heterodera schachtii nematodes were surface-sterilized using a fresh solution of 0.01% mercuric chloride for 5 min followed by four washes with sterilized double-distilled water. The J2 nematodes were then suspended in a 0.1% agarose solution and used to inoculate 10-d-old seedlings with ∼250 nematodes per seedling. The nematode susceptibility of the lines was determined 3 weeks after inoculation by counting the number of female nematodes per plant using a dissecting microscope. Statistically significant differences between the lines and the corresponding wild-type control were determined using t test on SAS with a P value cutoff of 0.05. Nematode infection assays were repeated at least two times and similar results were obtained.
Histochemical Analysis of GUS Activity
GUS activity of the pmiR858:GUS and pMYB83:GUS transgenic plants was determined by staining the plants at various time points post H. schachtii infection according to Jefferson et al. (1987). All tissues were stained for ∼6 h with the exception that pmiR858:GUS infected plants at 10 and 14 dpi were stained overnight to confirm the complete absence of the promoter activity in the syncytium at these two time points. At least 50 syncytia at each time point were examined, and the staining patterns were common to at least 90% of the examined syncytia in four independent transgenic lines. The images of both infected and noninfected plants were taken using a Zeiss digital camera and then analyzed with the Zeiss Axio Vision SE64 software (version 4.8).
Vector Construction and Production of Transgenic Plants
The binary vector of miR858 overexpression was constructed by amplifying the miR858 precursor (200 bp) from Col-0 genomic DNA using a primer pair containing BamHI and SacI restriction sites as overhangs. The amplified fragment was digested, gel-purified, and then ligated into the binary vector pBI121 under the control of 35S promoter. The wild-type MYB83 coding sequence was amplified from first-strand cDNA and the noncleavable MYB83 variant was constructed by creating 10 mismatches in the miR858-binding sites without altering the amino acid sequences. The modified MYB83 sequence was then cloned in the pBI121 binary vector under 35S promoter using XbaI and SacI restriction sites. The MIM858 overexpression was generated as recently described by Hewezi et al. (2016). Briefly, the 22-nucleotide miR399-complementary region in the Arabidopsis IPS1 gene, a noncoding phosphate starvation-induced transcript, was substituted with a mimic sequence for the mature miR858 sequences. The miR858 mimic sequence contained a three-nucleotide bulge (TGA) between the nucleotide numbers 10 and 11 of the binding region and two additional mismatches at the nucleotides numbers 1 and 10 of the binding site. The modified IPS1 genes containing the miR858 mimic sequence was cloned in the pBI121 vector under the control of 35S promoter using SacI and BamHI restriction sites.
The miR858 promoter (2,513 bp upstream of the miR858 TSS) was amplified from Col-0 genomic DNA using a primer pair containing BamHI and SacI restriction sites. Similarly, the MYB83 promoter (1,970 bp upstream of the translation start codon) was PCR amplified using a primer pair containing BamHI and SalI restriction sites. The PCR-amplified products were digested, gel-purified, and finally ligated to the binary vector pBI101 in the corresponding restriction sites to drive GUS gene expression. All constructs were confirmed by sequencing and introduced into Agrobacterium tumefaciens strain C58 by the freeze-thaw method. The bacteria were used to transform Arabidopsis wild-type Col-0 plants by the floral dip method (Clough and Bent, 1998). Transgenic T1 lines were identified by screening the seeds on Murashige and Skoog agar medium supplemented with 50 mg/L kanamycin. Transgene expression in various transgenic lines was quantified using qPCR. The primers used for binary plasmid construction are included in Supplemental Table S1.
RNA Isolation and Quantitative Real-Time RT-PCR Analysis
To assess the expression level of miR858 (both mature and primary transcripts), total RNA was extracted from 20 mg root tissues using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Total RNA including miRNAs was then polyadenylated and reverse transcribed using the Mir-X miRNA First-Strand Synthesis Kit (Clontech). Approximately 50 ng of the synthesized cDNA was used as a template for qPCR reaction. qPCR was carried out using SYBR Advantage qPCR Premix (Clontech). The mature miR858 sequence appended with two adenines on the 3′ end was used as forward primer sequence to ensure correct binding of the primer to the poly(T) region of the mature miR858 cDNA and preclude potential binding to the miR858 precursor. The primary transcript of miR858 was quantified using a forward primer specific to miRNA precursor and the universal reverse primer mRQ 3′ (provided with the kit). U6 small nuclear RNA was used as an internal control for miRNA gene expression normalization. The PCR reactions were performed in QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) using the following program: 95°C for 3 min followed by 40 cycles of 95°C for 30 s and 60°C for 30 s. The PCR products were then exposed to a temperature ramp to generate the dissociation curves and determine amplification specificity. The dissociation program was 95°C for 15 s and 50°C for 15 s, followed by a slow gradient from 50°C to 95°C. For the quantification of MYB83 expression levels, total RNA was isolated from 20 mg root tissues according to Verwoerd et al. (1989). The isolated total RNA was treated with DNase I (Invitrogen) and ∼25 ng was used in qPCR reactions using Verso SYBR green One-Step qRT-PCR Rox mix (Thermo Scientific) following the manufacturer’s protocol. The PCR amplification products were then subjected to a temperature ramp to create the dissociation curves using the following program: 95°C for 15 s and 60°C for 75 s, followed by a slow gradient from 60°C to 95°C. Primers used for qPCR quantification assays are included in Supplemental Table S1.
RNA-Seq Library Preparation and Data Analysis
P35S:miR858 (line 1-4), P35S:rMYB83 (line 8-1), and Col-0 were grown in Murashige and Skoog plates, and three biological samples of root tissues were collected of 2-week-old plants. mRNA was isolated from 20 mg grounded root tissue using magnetic mRNA isolation kit (NEB) following the manufacturer’s protocol. Approximately 250 ng of mRNA was used for RNA-seq library preparation using NEBnext mRNA library prep master mix (NEB) following the manufacturer’s protocol. The nine RNA-seq libraries were multiplexed and sequenced using HiSeq 2500 system with 100-bp single-end reads. Quality of the sequenced data was assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Low-quality reads were trimmed using Trimmomatic (Bolger et al., 2014). After trimming, uniquely mapped read was aligned to the Arabidopsis reference genome (TAIR10) using TopHat v2.0.14 (Trapnell et al., 2009). Number of reads assigned to individual genes were counted using HTSeq (Anders et al., 2015). DEGs were determined using the R package DESeq2 (Love et al., 2014) using an adjusted P value cutoff of 0.05. GO terms enrichment analysis of the DEGs was performed using agriGO database (Du et al., 2010) with Fisher’s exact test and Bonferroni multitest adjustment with a significance cutoff P value of 0.05.
Accession Numbers
Sequence data of Arabidopsis genes described in this study can be found in The Arabidopsis Information Resource database under the following accession numbers: miR858 (AT1G71002), MYB83 (AT3G08500), MYB12 (AT2G47460), and Actin8 (AT1G49240). The RNA-seq data described in this manuscript were submitted to the National Center for Biotechnology Information, Gene Expression Omnibus under accession number GSE95198.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Gene expression levels of miR858, MIM858, and rMYB83 in transgenic lines.
Supplemental Figure S2. Root lengths of miR858, MIM858, and rMYB83 overexpression lines.
Supplemental Figure S3. Characterization of the MYB83 T-DNA mutant line (CS1004395).
Supplemental Figure S4. Enrichment of MYB83 cis-binding element in the MYB83-regulated genes.
Supplemental Figure S5. Gene Ontology classification and enrichment analyses of the putative direct targets of MYB83.
Supplemental Figure S6. Gene expression levels of MYB12 in overexpression lines.
Supplemental Figure S7. Characterization of the MYB12 T-DNA mutant line (FLAG_150B05).
Supplemental Table S1. List of the primer sequences used in the current study.
Supplemental Data Set 1. List of differentially expressed genes identified in miR858 overexpression plants.
Supplemental Data Set 2. List of differentially expressed genes identified in MYB83 overexpression plants.
Supplemental Data Set 3. List of differentially expressed genes common to miR858 and MYB83 overexpression lines.
Supplemental Data Set 4. List of the identified putative direct targets of MYB83
Supplemental Data Set 5. List of MYB83-regulated genes overlapping with syncytium differentially expressed genes.
Supplemental Data Set 6. List of the putative direct targets of MYB83 overlapping with syncytium differentially expressed genes.
Acknowledgments
We thank several undergraduate students in the Hewezi laboratory and Sujata Agarwal at the UTIA Genomics Hub Laboratory for technical assistance.
Glossary
- miRNA
microRNA
- dpi
days postinfection
- DEGs
differentially expressed genes
- GO
Gene Ontology
- TSS
transcription start site
Footnotes
Articles can be viewed without a subscription.
This work was supported by a grant from the National Science Foundation (award no. IOS-1145053 to T.H. and T.J.B.) and by funds from the University of Tennessee, Institute of Agriculture to Hewezi Laboratory.
References
- Absmanner B, Stadler R, Hammes UZ (2013) Phloem development in nematode-induced feeding sites: the implications of auxin and cytokinin. Front Plant Sci 4: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18: 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R (2001) CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bhattarai KK, Atamian HS, Kaloshian I, Eulgem T (2010) WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J 63: 229–240 [DOI] [PubMed] [Google Scholar]
- Bohlmann H, Sobczak M (2014) The plant cell wall in the feeding sites of cyst nematodes. Front Plant Sci 5: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, Wuyts J, Rouzé P, Van de Peer Y (2004) Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc Natl Acad Sci USA 101: 11511–11516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera J, Barcala M, García A, Rio-Machín A, Medina C, Jaubert-Possamai S, Favery B, Maizel A, Ruiz-Ferrer V, Fenoll C, Escobar C (2016) Differentially expressed small RNAs in Arabidopsis galls formed by Meloidogyne javanica: a functional role for miR390 and its TAS3-derived tasiRNAs. New Phytol 209: 1625–1640 [DOI] [PubMed] [Google Scholar]
- Cabrera J, Diaz-Manzano FE, Fenoll C, Escobar C (2015) Developmental pathways mediated by hormones in nematode feeding sites. In C Escobar, C Fenoll, eds, Advances in Botanical Research, Vol 73 Elsevier, Boston, pp 167–188 [Google Scholar]
- Cai X-T, Xu P, Zhao P-X, Liu R, Yu L-H, Xiang C-B (2014) Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat Commun 5: 5833. [DOI] [PubMed] [Google Scholar]
- Camehl I, Oelmüller R (2010) Do ethylene response factorS9 and -14 repress PR gene expression in the interaction between Piriformospora indica and Arabidopsis? Plant Signal Behav 5: 932–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Wong CL, Muzzi F, Vlaardingerbroek I, Kidd BN, Schenk PM (2014) Root defense analysis against Fusarium oxysporum reveals new regulators to confer resistance. Sci Rep 4: 5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Candela H, Hake S (2009) Big impacts by small RNAs in plant development. Curr Opin Plant Biol 12: 81–86 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15: 1196–1200 [DOI] [PubMed] [Google Scholar]
- Cui F, Brosché M, Sipari N, Tang S, Overmyer K (2013) Regulation of ABA dependent wound induced spreading cell death by MYB108. New Phytol 200: 634–640 [DOI] [PubMed] [Google Scholar]
- De León IP, Sanz A, Hamberg M, Castresana C (2002) Involvement of the Arabidopsis α-DOX1 fatty acid dioxygenase in protection against oxidative stress and cell death. Plant J 29: 61–62 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Zhang Y, Xia R, Meyers BC (2016) Small RNAs add zing to the zig-zag-zig model of plant defenses. Mol Plant Microbe Interact 29: 165–169 [DOI] [PubMed] [Google Scholar]
- Frerigmann H, Gigolashvili T (2014) MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol Plant 7: 814–828 [DOI] [PubMed] [Google Scholar]
- Gao S, Zhang YL, Yang L, Song JB, Yang ZM (2014) AtMYB20 is negatively involved in plant adaptive response to drought stress. Plant Soil 376: 433–443 [Google Scholar]
- Gheysen G, Mitchum MG (2011) How nematodes manipulate plant development pathways for infection. Curr Opin Plant Biol 14: 415–421 [DOI] [PubMed] [Google Scholar]
- Goverse A, Bird D (2011) The role of plant hormones in nematode feeding cell formation. In Jones J, Gheysen G, Fenoll C, eds, Genomics and Molecular Genetics of Plant-Nematode Interactions. Springer, The Netherlands, pp 325–347 [Google Scholar]
- Goverse A, Overmars H, Engelbertink J, Schots A, Bakker J, Helder J (2000) Both induction and morphogenesis of cyst nematode feeding cells are mediated by auxin. Mol Plant Microbe Interact 13: 1121–1129 [DOI] [PubMed] [Google Scholar]
- Grunewald W, Cannoot B, Friml J, Gheysen G (2009a) Parasitic nematodes modulate PIN-mediated auxin transport to facilitate infection. PLoS Pathog 5: e1000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, van Noorden G, Van Isterdael G, Beeckman T, Gheysen G, Mathesius U (2009b) Manipulation of auxin transport in plant roots during Rhizobium symbiosis and nematode parasitism. Plant Cell 21: 2553–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Pang M, Nah G, Shi X, Ye W, Stelly DM, Chen ZJ (2014) miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat Commun 5: 3050. [DOI] [PubMed] [Google Scholar]
- Gupta OP, Sharma P, Gupta RK, Sharma I (2014) Current status on role of miRNAs during plant–fungus interaction. Physiol Mol Plant Pathol 85: 1–7 [Google Scholar]
- Gutierrez L, Bussell JD, Pacurar DI, Schwambach J, Pacurar M, Bellini C (2009) Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T. (2015) Cellular signaling pathways and posttranslational modifications mediated by nematode effector proteins. Plant Physiol 169: 1018–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T, Baum TJ (2012) Complex feedback regulations govern the expression of miRNA396 and its GRF target genes. Plant Signal Behav 7: 749–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T, Baum TJ (2015) Gene silencing in nematode feeding sites. In Escobar C, Fenoll C, eds, Advances in Botanical Research, Vol 73 Elsevier, Boston, pp 221–239 [Google Scholar]
- Hewezi T, Maier TR, Nettleton D, Baum TJ (2012) The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol 159: 321–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T, Piya S, Qi M, Balasubramaniam M, Rice JH, Baum TJ (2016) Arabidopsis miR827 mediates post-transcriptional gene silencing of its ubiquitin E3 ligase target gene in the syncytium of the cyst nematode Heterodera schachtii to enhance susceptibility. Plant J 88: 179–192 [DOI] [PubMed] [Google Scholar]
- Hewezi T, Piya S, Richard G, Rice JH (2014) Spatial and temporal expression patterns of auxin response transcription factors in the syncytium induced by the beet cyst nematode Heterodera schachtii in Arabidopsis. Mol Plant Pathol 15: 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J, Hess PH, Szakasits D, Blöchl A, Wieczorek K, Daxböck-Horvath S, Bohlmann H, van Bel AJ, Grundler FM (2009) Diversity and activity of sugar transporters in nematode-induced root syncytia. J Exp Bot 60: 3085–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci USA 103: 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL (2000) Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci USA 97: 2379–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Shen J, Liu H, Li F, Ding N, Gao C, Pattanaik S, Patra B, Li R, Yuan L (2015) Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta 242: 283–293 [DOI] [PubMed] [Google Scholar]
- Jin J, Hewezi T, Baum TJ (2011) Arabidopsis peroxidase AtPRX53 influences cell elongation and susceptibility to Heterodera schachtii. Plant Signal Behav 6: 1778–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Journot-Catalino N, Somssich IE, Roby D, Kroj T (2006) The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerhofer N, Radakovic Z, Regis JM, Dobrev P, Vankova R, Grundler FM, Siddique S, Hofmann J, Wieczorek K (2015) Role of stress-related hormones in plant defence during early infection of the cyst nematode Heterodera schachtii in Arabidopsis. New Phytol 207: 778–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth PK, Ithal N, Recknor J, Maier T, Nettleton D, Baum TJ, Mitchum MG (2011) The Soybean Rhg1 locus for resistance to the soybean cyst nematode Heterodera glycines regulates the expression of a large number of stress- and defense-related genes in degenerating feeding cells. Plant Physiol 155: 1960–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczmarek A, Overmars H, Helder J, Goverse A (2004) Feeding cell development by cyst and root-knot nematodes involves a similar early, local and transient activation of a specific auxin-inducible promoter element. Mol Plant Pathol 5: 343–346 [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA (2005) The developmental role of microRNA in plants. Curr Opin Plant Biol 8: 38–44 [DOI] [PubMed] [Google Scholar]
- Lee HJ, Park YJ, Kwak KJ, Kim D, Park JH, Lim JY, Shin C, Yang KY, Kang H (2015) MicroRNA844-guided downregulation of Cytidinephosphate Diacylglycerol Synthase3 (CDS3) mRNA affects the response of Arabidopsis thaliana to bacteria and fungi. Mol Plant Microbe Interact 28: 892–900 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET (2008) Kunitz trypsin inhibitor: an antagonist of cell death triggered by phytopathogens and fumonisin b1 in Arabidopsis. Mol Plant 1: 482–495 [DOI] [PubMed] [Google Scholar]
- Li Y, Lu YG, Shi Y, Wu L, Xu YJ, Huang F, Guo XY, Zhang Y, Fan J, Zhao JQ, et al. (2014) Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol 164: 1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou JM (2010) Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol 152: 2222–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cheng X, Liu D, Xu W, Wise R, Shen QH (2014a) The miR9863 family regulates distinct Mla alleles in barley to attenuate NLR receptor-triggered disease resistance and cell-death signaling. PLoS Genet 10: e1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rice JH, Chen N, Baum TJ, Hewezi T (2014b) Synchronization of developmental processes and defense signaling by growth regulating transcription factors. PLoS One 9: e98477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A (2010) miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama Y, Yamoto N, Suzuki Y, Chiba Y, Yamazaki K, Sato T, Yamaguchi J (2013) The Arabidopsis transcriptional repressor ERF9 participates in resistance against necrotrophic fungi. Plant Sci 213: 79–87 [DOI] [PubMed] [Google Scholar]
- Matsuo M, Johnson JM, Hieno A, Tokizawa M, Nomoto M, Tada Y, Godfrey R, Obokata J, Sherameti I, Yamamoto YY, Böhmer FD, Oelmüller R (2015) High REDOX RESPONSIVE TRANSCRIPTION FACTOR1 levels result in accumulation of reactive oxygen species in Arabidopsis thaliana shoots and roots. Mol Plant 8: 1253–1273 [DOI] [PubMed] [Google Scholar]
- McCarthy RL, Zhong R, Ye ZH (2009) MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol 50: 1950–1964 [DOI] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138: 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Xu J, He Y, Yang KY, Mordorski B, Liu Y, Zhang S (2013) Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25: 1126–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Shao C, Chen M (2011) Toward microRNA-mediated gene regulatory networks in plants. Brief Bioinform 12: 645–659 [DOI] [PubMed] [Google Scholar]
- Merelo P, Ram H, Pia Caggiano M, Ohno C, Ott F, Straub D, Graeff M, Cho SK, Yang SW, Wenkel S, Heisler MG (2016) Regulation of MIR165/166 by class II and class III homeodomain leucine zipper proteins establishes leaf polarity. Proc Natl Acad Sci USA 113: 11973–11978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA (2000) Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem 275: 33712–33717 [DOI] [PubMed] [Google Scholar]
- Moffat CS, Ingle RA, Wathugala DL, Saunders NJ, Knight H, Knight MR (2012) ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS One 7: e35995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu R-L, Cao Y-R, Liu Y-F, Lei G, Zou H-F, Liao Y, Wang H-W, Zhang W-K, Ma B, Du J-Z, et al. (2009) An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res 19: 1291–1304 [DOI] [PubMed] [Google Scholar]
- Nahar K, Kyndt T, De Vleesschauwer D, Höfte M, Gheysen G (2011) The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol 157: 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar K, Kyndt T, Nzogela YB, Gheysen G (2012) Abscisic acid interacts antagonistically with classical defense pathways in rice-migratory nematode interaction. New Phytol 196: 901–913 [DOI] [PubMed] [Google Scholar]
- Niu D, Lii YE, Chellappan P, Lei L, Peralta K, Jiang C, Guo J, Coaker G, Jin H (2016) miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nat Commun 7: 11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Pan L, Kawai-Yamada M, Yu LH, Yamamura S, Koyama T, Kitajima S, Ohme-Takagi M, Sato F, Uchimiya H (2005) Functional analysis of Arabidopsis ethylene-responsive element binding protein conferring resistance to Bax and abiotic stress-induced plant cell death. Plant Physiol 138: 1436–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JE, Kwon Y, Kim JH, Noh H, Hong SW, Lee H (2011) A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Mol Biol 77: 91–103 [DOI] [PubMed] [Google Scholar]
- Ozalvo R, Cabrera J, Escobar C, Christensen SA, Borrego EJ, Kolomiets MV, Castresana C, Iberkleid I, Brown Horowitz S (2014) Two closely related members of Arabidopsis 13-lipoxygenases (13-LOXs), LOX3 and LOX4, reveal distinct functions in response to plant-parasitic nematode infection. Mol Plant Pathol 15: 319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Lee HJ, Kwak KJ, Lee K, Hong SW, Kang H (2014) MicroRNA400-guided cleavage of Pentatricopeptide repeat protein mRNAs Renders Arabidopsis thaliana more susceptible to pathogenic bacteria and fungi. Plant Cell Physiol 55: 1660–1668 [DOI] [PubMed] [Google Scholar]
- Seo JK, Wu J, Lii Y, Li Y, Jin H (2013) Contribution of small RNA pathway components in plant immunity. Mol Plant Microbe Interact 26: 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks CM, Rice JH, Zubo Y, Schaller GE, Hewezi T, Kieber JJ (2016) The role of cytokinin during infection of Arabidopsis thaliana by the cyst nematode Heterodera schachtii. Mol Plant Microbe Interact 29: 57–68 [DOI] [PubMed] [Google Scholar]
- Sharma D, Tiwari M, Pandey A, Bhatia C, Sharma A, Trivedi PK (2016) MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development in Arabidopsis. Plant Physiol 171: 944–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique S, Radakovic ZS, De La Torre CM, Chronis D, Novák O, Ramireddy E, Holbein J, Matera C, Hütten M, Gutbrod P, et al. (2015) A parasitic nematode releases cytokinin that controls cell division and orchestrates feeding site formation in host plants. Proc Natl Acad Sci USA 112: 12669–12674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique S, Wieczorek K, Szakasits D, Kreil DP, Bohlmann H (2011) The promoter of a plant defensin gene directs specific expression in nematode-induced syncytia in Arabidopsis roots. Plant Physiol Biochem 49: 1100–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D, Korneli C, Lummer M, Navarro L (2013) Emerging role for RNA-based regulation in plant immunity. New Phytol 197: 394–404 [DOI] [PubMed] [Google Scholar]
- Sun J, Xu Y, Ye S, Jiang H, Chen Q, Liu F, Zhou W, Chen R, Li X, Tietz O, et al. (2009) Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21: 1495–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakasits D, Heinen P, Wieczorek K, Hofmann J, Wagner F, Kreil DP, Sykacek P, Grundler FM, Bohlmann H (2009) The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant J 57: 771–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang H, Jiao X, Kong X, Hamera S, Wu Y, Chen X, Fang R, Yan Y (2016) A signaling cascade from miR444 to RDR1 in rice antiviral RNA silencing pathway. Plant Physiol 170: 2365–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A, Wang M, Bellinger M, Jin H (2014) Small RNAs: a new paradigm in plant-microbe interactions. Annu Rev Phytopathol 52: 495–516 [DOI] [PubMed] [Google Scholar]
- Westfall CS, Sherp AM, Zubieta C, Alvarez S, Schraft E, Marcellin R, Ramirez L, Jez JM (2016) Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. Proc Natl Acad Sci USA 113: 13917–13922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek K, Elashry A, Quentin M, Grundler FM, Favery B, Seifert GJ, Bohlmann H (2014) A distinct role of pectate lyases in the formation of feeding structures induced by cyst and root-knot nematodes. Mol Plant Microbe Interact 27: 901–912 [DOI] [PubMed] [Google Scholar]
- Wieczorek K, Golecki B, Gerdes L, Heinen P, Szakasits D, Durachko DM, Cosgrove DJ, Kreil DP, Puzio PS, Bohlmann H, Grundler FM (2006) Expansins are involved in the formation of nematode-induced syncytia in roots of Arabidopsis thaliana. Plant J 48: 98–112 [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wubben MJE II, Su H, Rodermel SR, Baum TJ (2001) Susceptibility to the sugar beet cyst nematode is modulated by ethylene signal transduction in Arabidopsis thaliana. Mol Plant Microbe Interact 14: 1206–1212 [DOI] [PubMed] [Google Scholar]
- Xie Z, Kasschau KD, Carrington JC (2003) Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol 13: 784–789 [DOI] [PubMed] [Google Scholar]
- Xu J, Meng J, Meng X, Zhao Y, Liu J, Sun T, Liu Y, Wang Q, Zhang S (2016) Pathogen-responsive MPK3 and MPK6 reprogram the biosynthesis of indole glucosinolates and their derivatives in Arabidopsis immunity. Plant Cell 28: 1144–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Li Y, Hua J (2006) The C2 domain protein BAP1 negatively regulates defense responses in Arabidopsis. Plant J 48: 238–248 [DOI] [PubMed] [Google Scholar]
- Yang H, Yang S, Li Y, Hua J (2007) The Arabidopsis BAP1 and BAP2 genes are general inhibitors of programmed cell death. Plant Physiol 145: 135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Huang H (2014) Roles of small RNAs in plant disease resistance. J Integr Plant Biol 56: 962–970 [DOI] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M (2010) Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Gao S, Zhou X, Chellappan P, Chen Z, Zhou X, Zhang X, Fromuth N, Coutino G, Coffey M, Jin H (2011) Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol 75: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li Q, Li Z, Staswick PE, Wang M, Zhu Y, He Z (2007) Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol 145: 450–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y (2012) Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263 [DOI] [PubMed] [Google Scholar]
- Zhao W, Li Z, Fan J, Hu C, Yang R, Qi X, Chen H, Zhao F, Wang S (2015) Identification of jasmonic acid-associated microRNAs and characterization of the regulatory roles of the miR319/TCP4 module under root-knot nematode stress in tomato. J Exp Bot 66: 4653–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (2012) MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol 53: 368–380 [DOI] [PubMed] [Google Scholar]