Transcriptome analyses of key stages in wheat inflorescence development uncover molecular pathways and candidate genes that may be essential for final seed setting and yield improvement in wheat.
Abstract
Early reproductive development in cereals is crucial for final grain number per spike and hence the yield potential of the crop. To date, however, no systematic analyses of gene expression profiles during this important process have been conducted for common wheat (Triticum aestivum). Here, we studied the transcriptome profiles at four stages of early wheat reproductive development, from spikelet initiation to floral organ differentiation. K-means clustering and stage-specific transcript identification detected dynamically expressed homeologs of important transcription regulators in spikelet and floral meristems that may be involved in spikelet initiation, floret meristem specification, and floral organ patterning, as inferred from their homologs in model plants. Small RNA transcriptome sequencing discovered key microRNAs that were differentially expressed during wheat inflorescence development alongside their target genes, suggesting that miRNA-mediated regulatory mechanisms for floral development may be conserved in cereals and Arabidopsis. Our analysis was further substantiated by the functional characterization of the ARGONAUTE1d (AGO1d) gene, which was initially expressed in stamen primordia and later in the tapetum during anther maturation. In agreement with its stage-specific expression pattern, the loss of function of the predominantly expressed B homeolog of AGO1d in a tetraploid durum wheat mutant resulted in smaller anthers with more infertile pollens than the wild type and a reduced grain number per spike. Together, our work provides a first glimpse of the gene regulatory networks in wheat inflorescence development that may be pivotal for floral and grain development, highlighting potential targets for genetic manipulation to improve future wheat yields.
Grain number per spike is one of the key components of cereal crop yield. It is a multifactorial trait primarily determined in the early stages of reproductive development (Sreenivasulu and Schnurbusch, 2012); therefore, proper spike development is crucial for maximizing yield potential. The development of the spike begins with the transition of the meristem from a vegetative to a reproductive state and the initiation of inflorescence growth and ends when the plant matures. During this process, spike meristem initiation and floral differentiation are two key points that determine final seed set (Sreenivasulu and Schnurbusch, 2012).
To date, most of our understanding of the molecular regulatory pathways for early inflorescence development, especially meristem development, is derived from model plants such as Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). In rice, for example, this process requires genes that maintain inflorescence meristem identity, such as ABERRANT PANICLE ORGANIZATION1 (an F-box protein related to Arabidopsis UNUSUAL FLORAL ORGANS), RICE CENTROLADIALLIS1 (RCN1; related to Arabidopsis TERMINAL FLOWER1), and GRAIN NUMBER1 (a cytokinin oxidase; Nakagawa et al., 2002; Ashikari et al., 2005; Ikeda et al., 2005). MADS-box genes are other major transcription factors (TFs) that are involved in inflorescence meristem development, such as the SEPALLATA (SEP) gene PANICLE PHYTOMER2/OsMADS34, OsMADS14, OsMADS58, and MOSAIC FLORAL ORGANS1/AGAMOUS-LIKE (MFO1/AGL6) that also work in subsequent floral development (Yamaguchi et al., 2006; Ohmori et al., 2009; Gao et al., 2010; Kobayashi et al., 2010, 2012). Recently, a nuclear protein called TAWAWA1 (TAW1) was found to suppress the transition from the inflorescence meristem to the spikelet meristem phase by regulating the expression of the SHORT VEGETATIVE PHASE (SVP)-like MADS-box genes OsMADS22, OsMADS47, and OsMADS55 (Liu et al., 2013b; Yoshida et al., 2013).
On the other hand, APETALA2 (AP2)-like TFs are involved in spikelet determinacy and the transition to the floral meristem, such as BRANCHED SILKLESS1/FRIZZY PANICLE1/BRANCHED FLORETLESS1 (Chuck et al., 2002; Komatsu et al., 2003), INDETERMINATE SPIKELET1 (IDS1; Chuck et al., 1998, 2008), and SISTER OF INDETERMINATE SPIKELET1/SUPERNUMERARY BRACT1 (Chuck et al., 2008; Lee and An, 2012). In wheat (Triticum aestivum), the Q gene, which confers the free-threshing domestication trait, is a close homolog of the maize and rice IDS genes ZmIDS1/OsIDS1 (Zhang et al., 2011), while wheat FRIZZY PANICLE controls the number of spikelets produced (Dobrovolskaya et al., 2015). Moreover, AP2 transcripts are targets of microRNA 172 (miR172), providing another layer of regulation in spike and floret development (Chuck et al., 2007; Nair et al., 2010; Zhu and Helliwell, 2011; Wang et al., 2015). In Arabidopsis, additional miRNAs, such as miR159, miR167, and miR319, interactively target important TFs that function in meristems to regulate floral organogenesis, such as v-myb avian myeloblastosis viral oncogene homolog (MYBs), Teosinte Branched1/Cycloidea/Proliferating cell factors (TCPs), and auxin response factors (ARFs) (Millar and Gubler, 2005; Wu and Poethig, 2006; Rubio-Somoza and Weigel, 2013; Schommer et al., 2014; Hong and Jackson, 2015; Teotia and Tang, 2015). Despite their pivotal roles in meristem activities, only a few miRNAs have been functionally studied in grasses.
It has been shown that the basic genetic framework underlying meristem maintenance and floral organ development (e.g. the “ABCDE” model) is largely conserved between eudicots and monocots (Dornelas et al., 2011); however, the variation in grass inflorescence architecture and floral structure requires modified gene regulatory pathways. For example, the variation in the numbers of key TFs in different species suggests diversification in gene regulatory functions (Dornelas et al., 2011). Many orthologous TFs for floral organ development in Arabidopsis are functionally conserved in rice; for instance, the rice ortholog of the Arabidopsis B-function MADS-box gene AP3 is SUPERWOMAN1/OsMADS16 which, when mutated, changes the lodicule and stamen into palea-like and carpel-like organs, respectively (Nagasawa et al., 2003). However, divergence was also observed. In rice, two MADS-box genes, OsMADS3 and OsMADS58, are considered to be homologs of Arabidopsis AGAMOUS (AG), but they have distinct roles when compared with AG in Arabidopsis because the combined loss for function of these two genes produces abnormal carpel-like organs (Yamaguchi et al., 2006), demonstrating the subfunctionalization of these genes and the divergence in molecular pathways for flower development in the monocots.
Common wheat bears an inflorescence in the form of a spike, with a main axis of two ranks of lateral sessile distichous spikelets directly attaching to the rachis. Such a structure is different to that of rice, which forms secondary branches within its inflorescence. In the Triticeae tribe, which includes important crops like wheat, barley (Hordeum vulgare), and rye (Secale cereale), the number of spikelets per rachis node and the grain number on a spike are key taxonomic traits (Sakuma et al., 2011). Like other grass species, wheat reproductive development begins with spike initiation or spikelet formation, which occurs before the beginning of stem elongation (Waddington et al., 1983; Gardner et al., 1985; Digel et al., 2015). Despite the importance of these development stages (Appleyard et al., 1982; Kitchen and Rasmusson, 1983), little is known about underlying molecular mechanisms.
Recently, large-scale transcriptome analyses were performed on young inflorescences of maize (Zea mays; Eveland et al., 2014), rice (Furutani et al., 2006; Harrop et al., 2016), and barley (Digel et al., 2015), leading to the systematic discovery of regulatory modules of floral development in these crops. To understand the molecular pathways regulating wheat inflorescence development, we performed a comparative transcriptome analysis on shoot apical tissues at the spikelet initiation stage, the floral differentiation stage, and the young spike with immature florets using mRNA-seq and small RNA-seq. Our data provide new insights into the gene regulatory framework for wheat inflorescence development and highlight important candidate genes that may be manipulated to increase wheat grain yield.
RESULTS
Morphology of Early Wheat Inflorescence and RNA-seq Analysis
Wheat inflorescence development involves a series of morphological changes to the shoot apex (Waddington et al., 1983; Gardner et al., 1985; Digel et al., 2015). Stem extension, especially the elongation of the shoot apex, typically indicates the transition from the vegetative phase to the reproductive phase. The young inflorescence then develops ridges composed of bract primordia, followed by the development of spikelet meristems as axillary buds, forming the double ridge. Each spikelet meristem initiates a glume primordium followed by a tiered series of six to eight floral meristems, which have the potential to differentiate into three stamen primordia and one pistil primordium. The subsequent phase of reproductive growth before anthesis involves the differentiation of the floral organs and pollen cell meiosis and occurs concurrently with the elongation of the spike from less than 1 cm to its final length. Spikelet development starts from the middle of each spike and spreads toward both ends.
The early inflorescence development, especially the development of spikelet meristems, floret meristems, and floral organ primordia, is pivotal for subsequent floral development and final seed set (Sreenivasulu and Schnurbusch, 2012). Thus, we collected shoot apices at the double-ridge stage (DR) at the initiation of spike formation and spikelet development (Fig. 1, A and E) when spikelet meristems were the first visible reproductive structures. For Chinese Spring under our growth conditions, plants had 5 to 6 leaves with the appearance of the first internode. The next stage would take about 1 week when we observed the spikes with the appearance of the floret meristems (FMs), glume primordia, and lemma primordia (Fig. 1, B and F). This stage is critical for floret initiation. At the third stage, stamen and pistil primordia emerged from FMs with visible anther primordia for some florets (Fig. 1, C and G). This stage often took 5 d to develop and is important for the differentiation of stamens and pistils and the fertility of the floret (Shitsukawa et al., 2009). We designated this stage as the anther primordia (AM) stage. At the fourth stage, young florets began to differentiate with immature anthers and unelongated pistils (Fig. 1, D and H). At this stage, the pollen mother cells have completed the meiosis to form the tetrads, and therefore the stage was designated as the tetrads stage (TS). These developmental stages roughly corresponded to W2.0 (DR), W3.0 (FM), W4.0 (AM), and W7.5 (TS) on the Waddington scale, respectively (Waddington et al., 1983).
Figure 1.
Key developmental stages of early wheat inflorescences. A and E, DR stage with initiating spikelet meristems (SM, indicated by asterisk). B and F, FM stage with the glume primordia (GP) followed by lemma primordia (LP) and floret meristems. C and G, AM stage with stamen and pistil primordia (STP and PP, respectively). STPs are indicated by asterisks. D and H, TS with elongating styles. E to H are magnifications of A to D. A to C and E to G are SEM images. D and H were taken under a stereomicroscope. Bars: A to C, 0.5 mm; D, 1 cm; E, 0.05 mm; F, 0.25 mm; G and H, 0.5 mm.
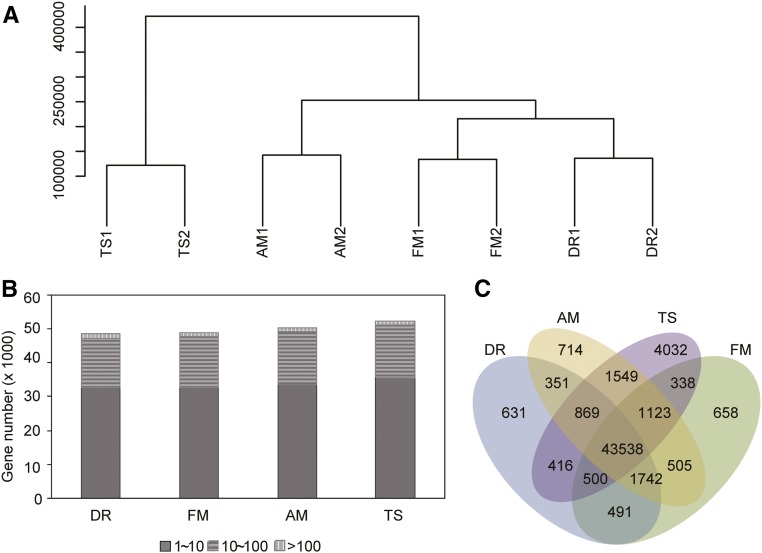
RNAs were isolated from 10 to 50 whole spikes and subjected to next-generation sequencing. RNA-seq reads were mapped to the wheat genomic sequences (Ensemble Triticum_aestivum.IWGSC2.25; Supplemental Table S1). Unique reads were determined according to Mayer et al. (2014) and used to quantify gene models in reads per kilobase per million reads (RPKM). For simplicity, we have defined each homeolog as a gene or used the terms “gene” and “homoeolog” interchangeably. An expressed gene was required to have an average RPKM of ≥ 1 in at least one stage. Thus, ∼50,000 genes or homeologs were expressed in each stage (Fig. 2B). A cluster dendrogram analysis showed that gene expression levels between replicated samples were highly related, with correlation efficiencies of higher than 0.95 (R2 > 0.95; Fig. 2A; Supplemental Figure S1). Among the expressed genes, 43,538 (87%) were expressed in all four stages (Fig. 2C). The numbers of genes expressed in only one stage were 631, 658, and 714 for DR, FM, and AM, respectively, with TS having the highest number of stage-specific genes (4032), indicating relatively homogenous gene expression profiles in meristematic and primordial tissues (DR, FM, AM) when compared with that of more differentiated young florets (TS).
Figure 2.
Gene expression profiles in developing wheat inflorescences. A, Cluster dendrogram of gene expression profiles between biological replicates and among different developmental stages. The y axis measures the degree of variance (see “Materials and Methods”). B, Numbers of genes expressed during each stage with an average number of RPKM of ≥ 1. C, A Venn diagram showing the numbers of expressed genes shared between inflorescences at different developmental stages.
k-means Clustering Analysis of Differentially Expressed Genes
A pairwise differential analysis was conducted on gene expression levels among the four developmental stages using DESeq (Anders and Huber, 2010) R packages. Genes with an adjusted P value of < 0.05 were considered to be differentially expressed (Supplemental Table S2). We found 753 (DR versus FM) and 1082 (FM versus AM) differentially expressed genes (DEGs) between the meristematic tissues at different stages. By contrast, more than 10,000 DEGs were identified between the meristematic tissues and the young florets (Supplemental Figure S2). In total, the pairwise comparison identified 19,060 DEGs that were further studied.
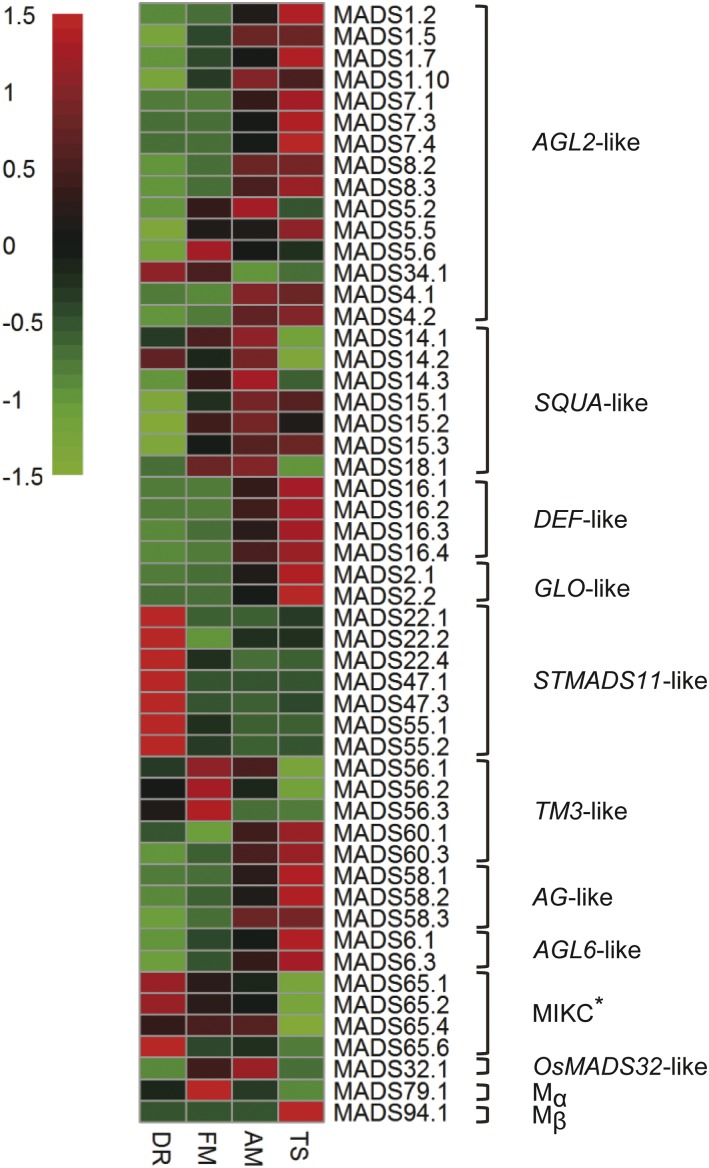
A k-means clustering analysis was performed on all DEGs with k set as 24 (Supplemental Table S3). These clusters displayed various gene expression patterns that may correlate with their functions during wheat inflorescence development. As shown in Supplemental Figure S3, we found that the 4804 genes in clusters 5, 12, and 22 displayed the highest expression level in the DR stage that were down-regulated over the course of inflorescence development, while clusters 2, 16, and 19 conferred 597 genes with expression patterns specific to FM. Only cluster 17 (2961 genes) contained genes most highly expressed in AM, whereas 562 genes of clusters 1 and 2 were predominantly expressed in both FM and AM. The remaining 16 clusters represented 10,525 genes whose expression peaked in TS, after various patterns of expression throughout early development. Gene expression patterns are good indicators of their functions. Homologs of MADS-box genes that play important roles, from reproductive initiation to floret organogenesis, were present in clusters of all classes described above (Supplemental Table S4). As shown in Figure 3, MADS-box genes involved in floral meristem activity (such as SVP/StMADS11-like genes) were highly expressed at DR, while the “ABCDE” genes AGL2-like, DEF-like, GLO-like, and AG-like were up-regulated toward the development of floral organs.
Figure 3.
Differential expression of MADS-box transcription factors during early inflorescence development in wheat. Wheat MADS-box genes are named following the nomenclature of their closest rice homolog (Supplemental Table S4) and were classified according to Arora et al. (2007). The suffix numbers are serial numbers of homeologs similar to the same rice gene (for details, see Supplemental Table S4). Note the expression patterns of MADS-box genes, such as the SVP-like genes (STMADS11-like) are specifically expressed in the DR phase, while the expression of others gradually increased during wheat inflorescence and floral organ development (AGL2, AQUA, DEF, GLO, AG, and AGL6-like). The heatmap was generated using the R package pheatmap, where the numbers represent relative expression levels of genes normalized across the four developmental stages. Clustering was performed using Pearson correlation distance.
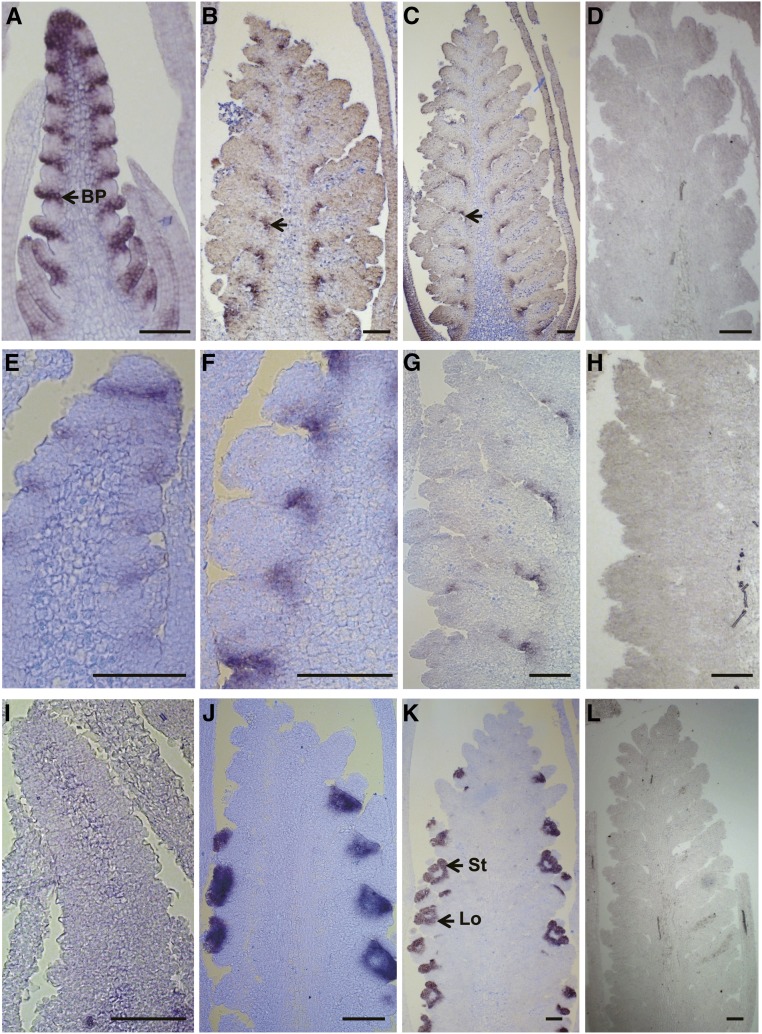
We then performed quantitative reverse transcription (qRT)-PCR analyses to verify our RNA-seq data. Genes with various expression patterns were selected and detected using homeolog-specific primers and RNAs extracted from independent samples. As shown in Supplemental Figure S4, 14 out of 15 investigated homeologs (except for TaSPL14-7D; SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE14) displayed similar expression patterns in the qRT-PCR assays to those quantified by RNA-seq. Gene expression patterns were further validated by in situ hybridization. Figure 4 shows the expression patterns of three genes, homologs of rice SPL14, BOWMAN-BIRK TYPE BRAN TYPSIN INHIBITOR (BBTI), and MADS7, which displayed the highest expression levels in DR, FM, and AM, respectively. In particular, the SPL14 homolog was highly expressed in the leaf and bract primordia at the DR stage, and its expression decreased with the development of the wheat inflorescence (Fig. 4, A–D), suggesting a role of this gene in spikelet initiation. The homolog of BBTI was highly expressed during FM, with expression localized to the basal portions of the spikelet and the floret meristems (Fig. 4, E–H). OsMADS7 is a SEP-like MADS-box gene, and the expression of its wheat homolog was initially detected at the floret meristem, where it was specifically expressed in the stamen and lodicule primordia during the development of the floral organs (Fig. 4, I–L), pointing to a similar role as its rice homolog in specifying floral determinacy and organ identities (Cui et al., 2010). Results of these assays also support the accuracy of our RNA-seq data.
Figure 4.
In situ hybridization analysis of genes preferentially expressed at key stages of wheat inflorescence development. In situ localization of homologs of rice OsSPL14 (A–D), OsBBTI (E–H), and OsMADS7 (I–L): DR stage (A, E, and I), FM stage (B, F, and J), AM stage (C, G, and K), and sense probe (D, H, and L). The wheat homolog of OsSPL14 was highly expressed in leaf and bract primordia in the DR stage and decreased with the development of wheat inflorescence (arrows in A–C). The wheat homolog of OsBBTI was highly expressed during FM at the bases of the floret and spikelet meristems (E–H). The expression of the OsMADS7 wheat homolog was initially detected in the floret meristems and later was specifically expressed in the stamen and lodicule primordia during the development of floral organs. BP, Bract primordium; St, stamen; Lo, lodicule. Bars: 200 μm.
Stage-Specific Genes Responsible for Spikelet and Floret Meristem Development
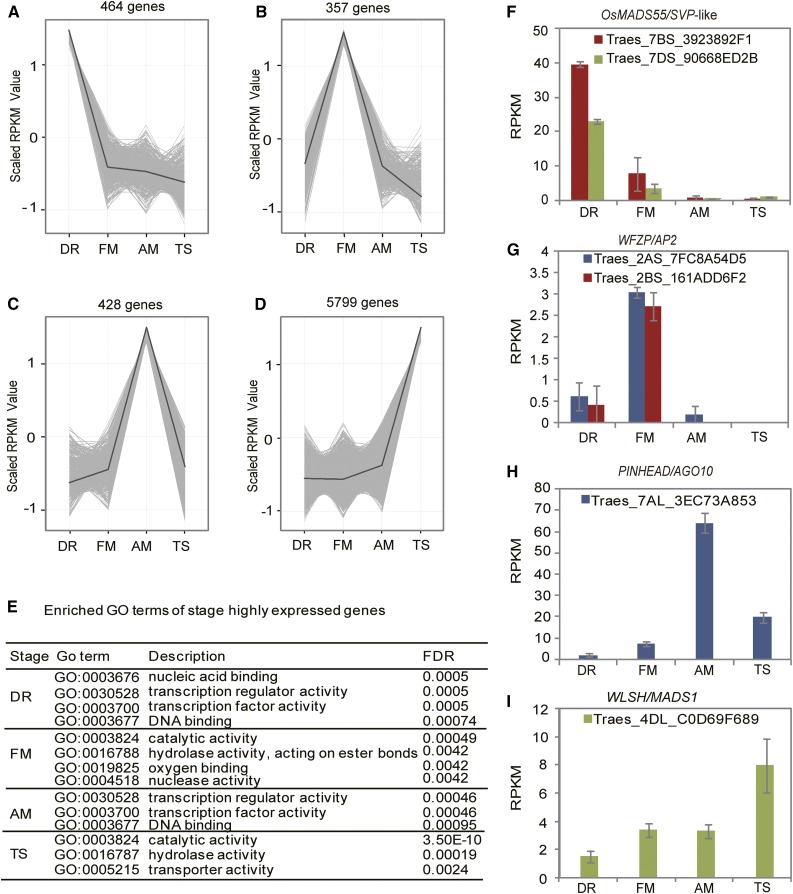
To identify genes that are closely associated with a particular developmental stage, we manually screened the expression patterns of DEGs. We defined DEGs with expression levels in one stage that were at least 2-fold or more than those in the remaining three stages as “stage-specific” genes (Supplemental Dataset 1). Among these genes, some showed “strict” stage specificity with nearly no expression in three stages (RPKM < 1) but expressed at one stage with a much higher expression level. There were other genes that were expressed at one stage with expression level much higher (>2-fold change) than those in the other three that were also expressed. In other words, these genes were expressed at all four stages but were particularly up-regulated at one stage. Under such conditions, 464 genes were considered to be specifically expressed at the DR stage (Supplemental Dataset 1). A Gene Ontology (GO) enrichment analysis showed that these DR-specific genes were enriched for “nucleic acid binding,” “transcription regulator activity,” “transcription factor activity,” and “DNA binding” functions (Fig. 5). Among these genes were homologs of AP2 TF AINTEGUMENTA/PLETHORA2 and OsMADS22, OsMADS47, and OsMADS55, hallmark meristematic genes (Figs. 3 and 5). These genes are involved in panicle branching, flowering time determination, and floral meristem specification in Arabidopsis and rice (Liu et al., 2013b; Teo et al., 2014). Meanwhile, homologs of the SPL7, SPL14, and SPL17 genes that are involved in rice reproductive shoot architecture and grain number determination (Xie et al., 2006; Jiao et al., 2010; Miura et al., 2010; Wang et al., 2015; Liu et al., 2016) also displayed DR-specific expression patterns, suggesting similar roles for these genes in wheat. Such a notion was supported by the in situ hybridization assay, where TaSPL14 was found to be highly expressed in the wheat leaf and bract primordia, and subsequently repressed along the development of the young spike (Fig. 4, A–C). Additional DR-specific genes were homologs of CLAVATA1 (CLV1), RCN4 (homologous to TERMINAL FLOWER1; Traes_2AL_9EEAEBACF, Traes_2BL_20095A876), and FLORICAULA/LEAFY (Traes_2AL_83D0D0C3F) that are well known to function in meristems. Characteristic hormone-related genes present at this stage were those involved in auxin, gibberellin (GA), and salicylic acid (SA) signaling, as shown by MapMan classification (Supplemental Table S5).
Figure 5.
Expression patterns of stage-specific genes during early wheat inflorescence development. A to D, Expression patterns of genes specific to each developmental stage, with the average expression trend highlighted. The y axis represents centered and scaled expression levels of genes across the four developmental stages by the R function scale. E, Enriched GO terms among stage-specific genes. F to I, Examples of expression patterns of stage-specific genes with rice homologs that are functionally characterized in rice inflorescence development. Blue, red, and green colors indicate the A, B, and D subgenomes, respectively. Each point of the RPKM represents the average of two biological replicates. Error bars indicate sd.
A total of 357 homeologs that may be involved in floral meristem initiation were classified as being FM specific. A GO analysis showed that this stage was enriched in genes with “catalytic activity,” “hydrolase activity,” “oxygen binding,” and “nuclease activity” (Fig. 5). Homologs of functionally characterized TFs from the model species Arabidopsis and rice were scarcely present at this stage. Despite this, two genes were worthy to mention that were homologous to the rice MADS-box gene: OsMADS50 and the Arabidopsis gene SUPPRESSOR OF OVEREXPRESSION OF CO1, one of the key genes involved in flowering time determination, floret meristem specification, and floral organ patterning (Lee et al., 2004). In addition, two Arabidopsis PINHEAD gene homologs were highly expressed in the floret meristem tissue (Supplemental Dataset 1). PINHEAD/ZWILLE encodes AGO10, a small RNA-binding protein that plays a unique role in maintaining the shoot apical meristem by sequestering the functions of miR165/166 (Moussian et al., 1998; Liu et al., 2009; Zhu et al., 2011; Zhang and Zhang, 2012). Hormone-related genes at this stage were featured by those involved in jasmonic acid (JA)- and SA-related functions (Supplemental Table S5).
We then examined the 428 genes that were specifically expressed in AM. A GO enrichment analysis revealed genes with transcription regulator activities (Fig. 5; Supplemental Dataset 1). Seven homeologs were found to be homologous to AP2 TFs. One homeolog was similar to OsMADS1/LEAFY HULL STERILE1, a SEP-like gene that has been found to specify spikelet meristem identity in rice (Tanaka et al., 2013). Again, homologs of CLV1, RCN4, and PINHEAD were present at this stage. Interestingly, a number of homeologs were similar to genes encoding PAZ domain-containing proteins, several of which were annotated as AGO1d (Supplemental Dataset 1). AGO1d is one of the four AGO1-like proteins (a–d) in grasses that are essential for the miRNA-mediated regulation of development and stress responses (Wu et al., 2009; Rogers and Chen, 2013; Zhang et al., 2015). The AM-specific expression of AGO1d may suggest a role of this gene in anther development. This stage was characterized by increased expression of genes for cytokinin (CK) metabolism, in addition to those that were clearly annotated as being involved in ethylene, JA, abscisic acid, and auxin metabolism or signaling (Supplemental Table S5).
Genes Involved in Wheat Floral Organogenesis
In contrast to the three meristematic and primordial stages, substantially more genes (5799) were specifically expressed in TS, a stage for floral organ differentiation. These genes were enriched in biochemical functions, such as catalytic, hydrolytic, and transporter activities (Fig. 5E; Supplemental Dataset 1). Although transcription regulator activity was not enriched by the GO analysis, numerous homologs of TFs were present that play important functions during floral development in model plants, including multiple homeologs similar to TCPs, SPLs, and MADS-box TFs. Interestingly, a different set of SPL genes, homologs of SPL8, SPL10, SPL16, and SPL18, were specifically expressed in TS, indicating stage specific functions of SPL genes during wheat inflorescence development. Numerous hormone-related genes were expressed, including those associated with brassinosteroid signaling, which were not detected in the other stages (Supplemental Table S5).
MiRNAs in Early Wheat Inflorescence Development
MiRNAs are a class of noncoding RNAs that posttranscriptionally regulate target gene expression levels. MiRNAs have been shown to play pivotal roles in Arabidopsis during the transition from vegetative to reproductive development and the subsequent floral patterning (Chen, 2004; Nag et al., 2009; Luo et al., 2013). To assess the roles of miRNAs in wheat inflorescence development, we sequenced the small RNA pools at key inflorescence developmental stages.
A miRNA was considered to be expressed if three or more copies were detected in at least one developmental stage. We used 525 mature miRNAs from Brachypodium distachyon as queries and searched our small RNA dataset for sequences with maximum 2 nucleotide differences. A total of 166 small RNAs were found with sequence similarity to Brachypodium miRNAs that fell in 21 families (Supplemental Dataset 2). Among these, 39 from 17 families were identical to those of Brachypodium and had higher copy numbers than related variants. Differential analysis using the DESeq program showed that only one miRNA, miR156b-5p (MIMAT0020679), was differentially expressed between stages DR and FM (Supplemental Table S6). By contrast, five miRNAs (miR160a-5p, miR167c-5p, miR319a-5p, miR319b-3p, and miR319c-5p) were differentially expressed between stages FM and AM. Similar to protein-coding genes, a much larger number of miRNAs (24) were differentially expressed between the TS and AM stages. A clustering analysis showed that miRNAs also displayed stage-specific expression patterns (Supplemental Figure S5), indicating that miRNAs, like protein-coding genes, are widely involved in wheat inflorescence development.
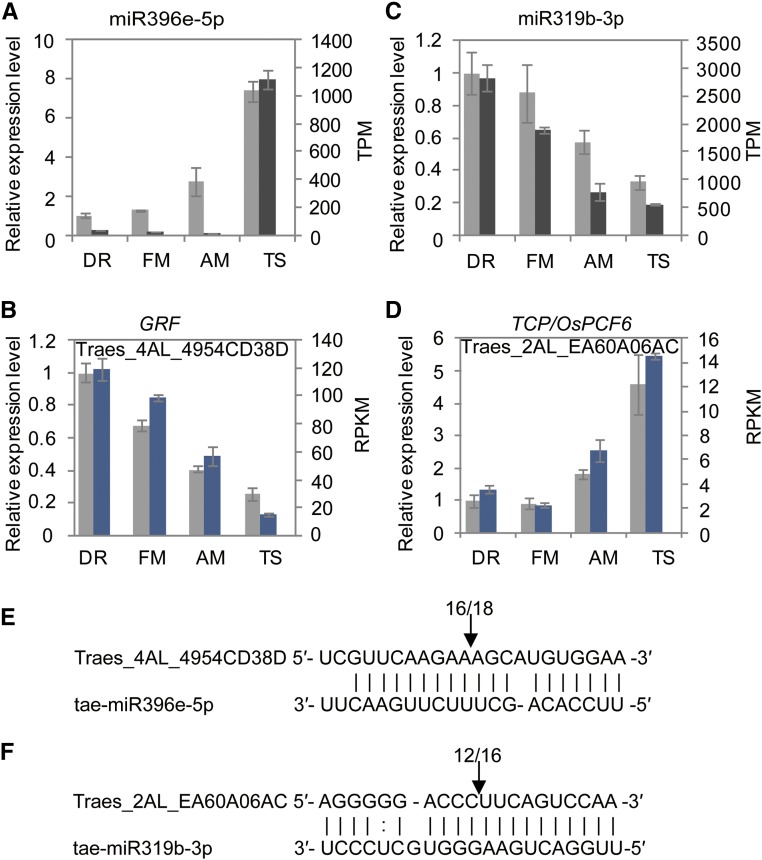
Interestingly, several miRNAs were exceptionally highly expressed in wheat inflorescence, up to hundreds to thousands of transcripts per million (TPM; Supplemental Table S6). For example, miR159-3p.1 had a copy number of >6000 TPM at the DR stage, which was reduced to ∼1600 TPM when the inflorescence reached the TS stage. Additional most abundant miRNAs were members of the miR166, miR319, and miR396 families, while miR160, miR167, and miR171 were modestly expressed, with TPMs between 50 and several hundred. The remaining miRNAs showed a much lower expression level. These highly expressed miRNAs were of particular interest because, in Arabidopsis, miR159, miR167, and miR319 and their targets form a regulatory circuit during floral organ patterning (Rubio-Somoza and Weigel, 2013). Moreover, the miR319-TCP module directly regulates miR396 expression, which in turn represses a number of growth-regulating factors (GRFs), while GRFs may repress meristem cell-promoting genes in actively proliferating cells (Schommer et al., 2014; Rodriguez et al., 2015). In wheat, the high expression levels of miR159b and miR319b were sharply down-regulated from DR to TS, while miR396e remained low in the first three stages and was significantly increased (>50-fold) in TS (Fig. 6; Supplemental Fig. S7). At least one member of the miR167 family, miR167a, was down-regulated (>10-fold) during wheat inflorescence development (Supplemental Dataset 2). The expression patterns of these miRNAs were readily confirmed by additional loop PCRs (Fig. 6; Supplemental Fig. S7). These data suggest similar functions of wheat miRNAs to those of Arabidopsis.
Figure 6.
Expression patterns of the highly expressed miRNAs and their predicted target genes in wheat inflorescence meristems. A to D, The expression level of miRNAs and their targets based on TPM/RPKM (black and blue) and qPCR (gray) data. The 18s rRNA was used as an endogenous reference to normalize the qPCR results. Each point of the TPM/RPKM represents the average of two biological replicates. Two biological replicates with similar pattern were performed for qPCR. Each point represents the average of three technical replicates. Error bars indicate sd. E and F, MiRNAs cleavage sites in targets mRNAs determined by RNA ligase-mediated 5′-RACE. The frequency of 5′-RACE clones corresponding to the site are indicated by numbers above arrows.
We then investigated whether the targets of these highly abundant miRNAs were accordingly regulated during wheat inflorescence development. As shown in Supplemental Figure S6, for each of these miRNAs, a number of targets were predicted and their expressions were validated by qRT-PCRs (Fig. 6; Supplemental Fig. S7). MiR319, for example, was predicted to target three transcripts, two from homeolog group 2 (2AL and 2BL) and the third from 5BL (Supplemental Fig. S6). Sequence comparison showed that the target sites of these three transcripts were perfectly complementary to the first 11 nucleotides of miR319b, which is critical for efficient miRNA cleavage (Mallory et al., 2004; Schwab et al., 2005). A database search found that these wheat homeologs were homologous to rice PROLIFERATING CELL FACTOR6 and Arabidopsis TCP2 (Supplemental Fig. S6; Martín-Trillo and Cubas, 2010). Similarly, the predicted targets of miR167 were similar to the rice gene models annotated as auxin response factors, such as rice ARF6 (ARFY_ORYSJ), while, as expected, the predicted targets of miR396 were highly similar to rice GRF9 (GRF9_ORYSJ). Moreover, previous work has shown that wheat miR159 is able to cleave MYB-like gene transcripts derived from 3AL and 3DL (Wang et al., 2012), which were identical to the targets predicted here. We performed separate RNA ligase-mediated rapid amplification of 5′-cDNA ends (5′-RACE) assays and found that the wheat miR319b, miR396e, miR159b, and miR167a were indeed all capable of cleaving the predicted targets (Fig. 6; Supplemental Fig. S7). The coordinated expression of enriched miRNAs and their targets imply conserved regulatory mechanisms in plant inflorescence development.
Loss of Function of AGO1d in Tetraploid Wheat Reduced Floral Fertility
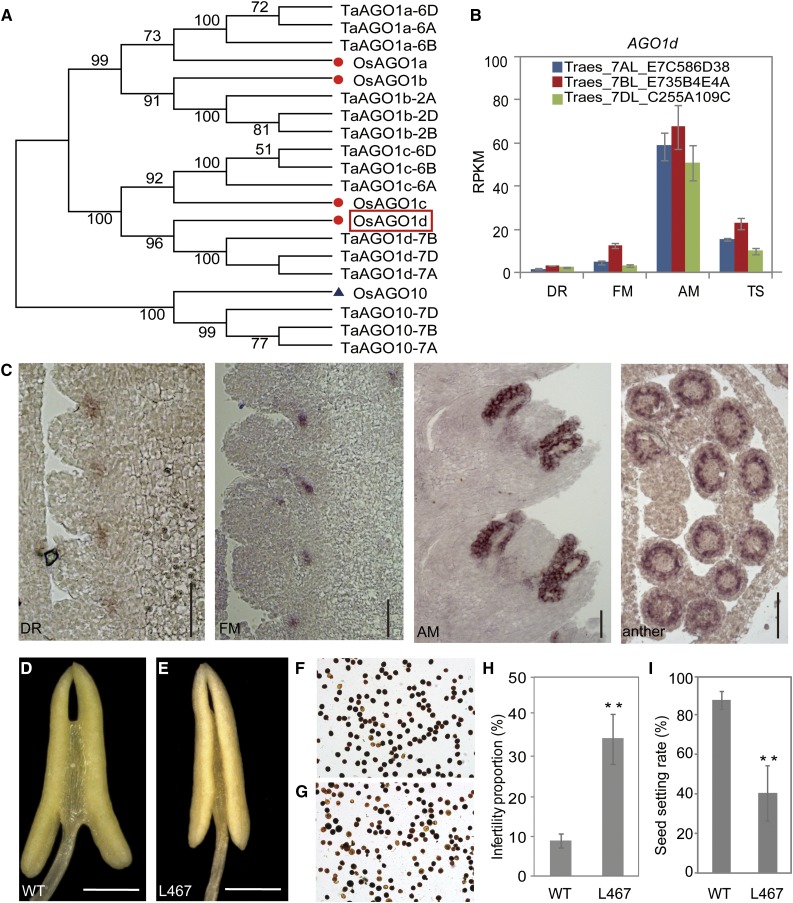
In rice, four AGO1 proteins, AGO1a–d, are known to play both redundant and specialized functions in their small RNA loading capability, especially miRNAs (Wu et al., 2009). One of them, AGO1d, caught our attention because it exhibited an AM-specific expression pattern (Supplemental Dataset 1). By sifting through the wheat draft genomes, we were able to find four AGO1 genes in wheat, each with three homeologs (Fig. 7A). Despite this, only the homeologs of AGO1b and AGO1d were all expressed at similar levels in the developing wheat inflorescence (Fig. 7B; Supplemental Fig. S8B), while only one and two homeologs were expressed for AGO1a and AGO1c, respectively (Supplemental Fig. S8, A and C). Homeologs of the AGO1a–c genes were all down-regulated during wheat inflorescence development, except for AGO1d. An in situ hybridization assay showed that, in the DR and FM stages, AGO1d was expressed in the abaxial side of spikelet meristems, close to the rachis (Fig. 7C). By the late AM stage, the expression signals of AGO1d were mostly detected at the anther primordia. Later, a transverse section of maturing anthers showed that AGO1d was mainly expressed at locations corresponding to the tapetum, suggesting a role of this gene in anther development.
Figure 7.
Functional characterization of the wheat AGO1d gene in anther and pollen development. A, A neighbor-joining tree of wheat and rice AGO1 homologs using AGO10 homologs as an outgroup. Red dots indicate rice AGO1a–d proteins, and the protein in red box is the rice AGO1d. B, Expression patterns of wheat AGO1d homeologs during early inflorescence development. C, In situ hybridization of homologs of AGO1d during the DR, FM, late AM (spike at 1 cm long) stages and anther transverse sections (spike at 2 cm long) in common wheat cv Chinese Spring. Bars, 100 µm. D to E, Anther of wild-type (cv Kronos (D) and the EMS mutant line L467 (E) of tetraploid durum wheat plants. L467 contains a premature stop codon at the predominantly expressed AGO1d B homeolog (see Supplemental Fig. S8 for more information). Bars, 500 µm. F and G, Pollen grains of wild-type (F) and L467 (G) tetraploid wheat lines stained by I2-KI. H, Infertility rates in the wild-type and mutant lines. At least 400 pollen grains were counted. Error bars represent sd (**P < 0.01, Student’s t test). I, Seed setting rates of the wild-type and L467 lines. Error bars represent sd (n = 17; **P < 0.01, Student’s t test).
To study the functions of AGO1d, we turned to tetraploid durum wheat, Triticum turgidum ssp. durum (AABB), which is closely related to common wheat as a putative tetraploid progenitor. EMS mutant lines derived from the desert durum variety “Kronos” are available (Uauy et al., 2009), and their mutation sites have been identified using exome sequencing (Henry et al., 2014; Krasileva et al., 2017). We first determined that AGO1d had a similar expression pattern in both durum wheat and common wheat. As shown by an in situ hybridization (Supplemental Fig. S8, D and E), AGO1d was expressed in the stamen primordia of durum wheat, similar to that in common wheat, suggesting comparable functions of this gene in tetraploid and hexaploid wheat. A BLAST search against the mutant sequence dataset found that the mutant line L467 carried a nonsense mutation on the AGO1d B homeolog (the predominantly expressed homeolog), which generated a premature stop codon (Supplemental Fig. S8F). A real-time PCR assay showed that the expression of the B homeolog was also significantly reduced in the mutant plants (Supplemental Fig. S8J). Morphological observation revealed that the loss of AGO1d-B function resulted in significantly shorter spikes than those of the wild type (Supplemental Figure S8, G–I). Anthers of the mutant plants were smaller, with a higher fraction of infertile pollen grains (Fig. 7, D–H), yielding much fewer seeds per spike relative to the wild type (Fig. 7I). Together, these data support our hypothesis that AGO1d plays an important role in wheat anther and pollen development. Furthermore, our results validated our approach in identifying stage-specific genes during wheat inflorescence development that may have an effect on wheat grain yield.
DISCUSSION
For cereal plants, inflorescence architecture affects both the number and the size of seeds, which are central demographic parameters in wild species and critical economic traits in grain production that determine crop yields (Kellogg et al., 2013). Inflorescence development, especially the spikelet and floret initiation stages, is critical for the production and survival of the florets, and thus the maximal yield potential. Despite its importance, molecular studies of wheat inflorescences have been limited until recently due to the lack of quality reference genome sequences (Jia et al., 2013; Ling et al., 2013; Mayer et al., 2014). To date, only a few genes have been found to be associated with spike development in wheat (Simons et al., 2006; Boden et al., 2015; Dobrovolskaya et al., 2015). A systematic study of genes and their regulatory networks for wheat spike development is urgently needed. The transcriptome profiling of several key developmental stages of wheat inflorescence should provide novel insights into the molecular mechanisms for wheat inflorescence development.
Functional Modifications of Key Regulatory Genes for Wheat Spikelet Development
The indeterminate nature of wheat spikelet meristems enables more than eight florets to be formed within one spikelet, while only three to five grains are harvested per spikelet in most cultivars, indicating a dynamic regulation for spikelet and floral development in wheat. Gene expression profiles revealed that key regulatory genes for spikelet meristem development may be conserved between wheat and rice. In rice, for example, the SVP-like MADS-box genes OsMADS22, OsMADS47, and OsMADS55 regulate inflorescence branching and repress spikelet meristem identity (Liu et al., 2013b). OsMADS22 and OsMADS55 are activated by the G1-like transcriptional activator G1-like5 (TAW1; Yoshida et al., 2013) and were repressed in expression between primary branch meristems and the shoot apical meristem (Harrop et al., 2016). We found that wheat SVP-like MADS-box genes and the homolog of TAW1 were highly expressed during spikelet meristem initiation in the DR stage and were down-regulated over the course of inflorescence development. These conserved expression patterns may suggest a largely similar mode of action for these key regulatory genes in the two grass species. This is especially true for genes that are involved in meristem maintenance such as the CLV1 homologs (e.g. Traes_4AS_04E4A0063) that are highly expressed throughout the stages containing meristematic/primordial tissues, indicating that major functions of these key genes are conserved across the plant kingdom (Clark et al., 1997; Suzaki et al., 2004; Kobayashi et al., 2012; Liu et al., 2014b).
Nevertheless, modifications in orthologous gene expression patterns may be necessary due to the morphological differences in inflorescences of different cereal crops. The rice SNB gene, for instance, is a member of the IDS1 subgroup of the AP2 family. The mutation of SNB delays the transition from spikelet meristem to floret meristem, causing the development of multiple rudimentary glumes in the alternative phyllotaxy, as well as aberrant floral shape (Lee et al., 2007). In wheat, the SNB homologs were highly expressed at the DR stage and were down-regulated significantly in developing floral organs, suggesting similar roles in spikelet meristem regulation but possibly different functions in floral organ development. Similarly, the wheat SPL14 homeologs were specifically expressed at the DR stage in bract primordia, but not at the spikelet meristems. In rice, SPL14 is predominantly expressed in the shoot apex, particularly the bract primordia and the primary and secondary branches, consistent with its role in panicle architecture determination (Jiao et al., 2010; Miura et al., 2010; Luo et al., 2012). Several additional SPL genes also displayed DR-specific expression patterns, such as homologs of SPL7 and SPL17, which are important for rice grain number and tiller numbers (Liu et al., 2016). The RICE FLORICAULA/LEAFY/ABERRANT PANICLE ORGANIZATION2 gene differs from its Arabidopsis ortholog, FLO/LFY, because it is required for inflorescence meristem identity specification (Rao et al., 2008; Ikeda-Kawakatsu et al., 2012). In wheat, the LFY homolog (Traes_2AL_83D0D0C3F) was highly expressed in DR and rapidly down-regulated during inflorescence development; however, unlike in rice, the wheat LFY homolog seemed not to be expressed in developing floral organs. Thus, the differences in inflorescence structures may be accounted for expression pattern changes of the conserved genes in different crops.
Molecular Regulation of Floral Organ Patterning in Wheat
The transition of the wheat inflorescence from meristematic status to floral organ differentiation is accompanied by the down-regulation of meristem activity genes and a significant up-regulation of a large number of genes associated with organ development. Like in other plants, wheat homologs of various MADS-box genes may play central roles during this process. Homologs of rice OsMADS6, OsMADS7, and OsMADS8 were expressed early in the FM stage and increased toward later stages of floral development. In addition to the transcriptional regulatory activities of various TFs, phytohormones were also involved in this process, with elevated expression of auxin-, GA-, and SA-related genes at the initial stages of wheat inflorescence development and JA-, abscisic acid-, cytokinin-, and brassinosteroid-related genes kicking in at the later stages of floral development.
The AM stage, with the emergence of the stamen meristems and their subsequent differentiation, is one of the most critical phases in wheat inflorescence development. It is a key period that may affect pollen maturation and hence the fertility of florets and the final grain numbers (Guo and Schnurbusch, 2015). At this stage, we found a number of genes that encode proteins working for with small RNA functions, such as AGO1d, that is specifically expressed at the tapetum, suggesting the wide involvement of small RNAs (both miRNAs and siRNAs) in wheat floral development. Such an observation may indicate that AGO1d have been subfunctionalized for anther development. The expression pattern was conserved in tetraploid wheat, and its function can be inferred from AGO1d EMS mutant plants. The functions of the wheat AGO1d homolog deserve further investigation.
The Roles of MiRNAs in Wheat Inflorescence Development
MiRNAs are extensively involved in meristem functions during plant morphogenesis, especially the establishment of the reproductive phase and organ patterning (Zhang and Zhang, 2012; Spanudakis and Jackson, 2014; Hong and Jackson, 2015). Arabidopsis miRNAs, such as miR159, miR167, and miR319, are involved in reproductive processes such as flowering and floral patterning (Smoczynska and Szweykowska-Kulinska, 2016), but not much has been studied in grass species. In wheat, miR159, miR167, and miR319 were exceptionally abundantly expressed in inflorescence meristems, with the sharp increase of miR396 expression later during floret maturation. Predicted wheat target genes were conserved as those in Arabidopsis and rice and expressed in cooperative manners. The capability of these miRNAs to cleave their targets was also confirmed in wheat. More detail should be explored regarding the role of small RNAs in wheat inflorescence, spikelet, and floral development.
Besides AGO1d and AGO10, additional genes involved in small RNA functions were found among DEGs during wheat inflorescence development, including homologs to rice SHOOTLESS2, SHOOTLESS4/SHOOT ORGANIZATION2, and SHOOT ORGANIZATION1 (encoding RNA-dependent RNA polymerase 6, AGO7, and DICER-like 4, respectively; Satoh et al., 2003; Nagasaki et al., 2007), as well as the homolog of rice WAVY LEAF1, an ortholog of Arabidopsis HUA ENHANCER1, which regulates shoot development by maintaining the accumulation of miRNAs and transacting small interfering RNAs (Abe et al., 2010). Thus, despite the modification in inflorescence morphology, the principle modes of action underlying mechanisms mediated by miRNAs seem to be conserved in wheat, rice, and Arabidopsis inflorescence development.
CONCLUSION
An optimal growth condition for inflorescence development is critical to achieve a spike with more spikelets, higher floret survival rate, and increased final grain number. With the progress of wheat genome sequencing and the availability of an improved genome sequence, more questions can be pursued regarding gene regulatory mechanisms for spike development in this hexaploid species, such as how the interaction between different homeologs and the overall expression level of all homeologs of key regulatory genes contribute to the final yield. Together with other technical advances, such as laser capture microdissection for precision tissue collection, genome editing by CRISPR/Cas9, and increased transformation efficiency, we will be able to better dissect the regulatory pathways for wheat inflorescence development. Such work should provide valuable candidate genes that can be genetically targeted for yield improvement in wheat.
MATERIALS AND METHODS
Plant Growth and Tissue Collection
Plants of common wheat cv Chinese Spring (CS) were grown in the experimental station in Beijing (39.97°N, 116.34°E) in the springs of 2014 and 2015. The developmental stages of the young inflorescences were determined using a stereomicroscope (S8 APO, Leica Microsystems). Four stages of tissues were collected: the double ridge/spikelet meristem stage (DR), the floret meristem stage (FM), the anther primordia stage (AM), and the young floret stage, when the styles have just emerged from the pistils (TS). The inflorescences in DR stage were collected when wheat plants had five leaves and the first node of stem was visible. The second stage was marked by the emergence of the second node and floret meristems (hence FM stage) about 1 week later. After 5 more days, the third stem internode began to extend, and anther primordia were formed and the plant were ready to enter the booting stage. This stage was designated as the AM stage. In the following 2 weeks, floral organs experienced differentiation and development with the completion of pollen mother cells and the formation of tetrads. The stage was designated as TS. Plants at this stage conferred a flag leaf about 3 cm above the penultimate leaf.
Whole wheat plants were collected in the morning and spikes were hand dissected, immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction. For each stage, 10 to 50 spikes were pooled for each of the two biological replicates. For in situ hybridization and mutant analysis experiments, tissues were collected from CS plants and tetraploid wheat (Triticum turgidum ssp. durum) plants grown in a greenhouse under 22°C/20°C day/night, 16 h/8 h light/dark, and 50% relative humidity conditions.
RNA Isolation and Sequencing Library Construction
Total RNA was extracted with TRIzol reagent (Invitrogen). The digital gene expression libraries were generated using a NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biosystems), following the manufacturer’s recommendations. Small RNA libraries were prepared as described previously (Liu et al., 2014a).
RNA Sequencing and Data Analysis
Sequencing was performed on a HiSEquation 2000 (Illumina). Approximately 10 to 17 million 49-bp single-end reads were generated for each sample. Reference genome sequences and annotations were downloaded from the CS genome website (Ensemble Triticum_aestivum.IWGSC2.25: ftp://ftp.ensemblgenomes.org/pub/plants/release-25/fasta/triticum_aestivum/dna/). Reads were aligned to the genome using TopHat (v.2.09; Trapnell et al., 2009; Kim et al., 2013) with the following parameters: read-mismatches = 2; segment mismatches = 1; max-multihits = 20; r = 0. Unique reads were determined according to Mayer et al. (2014) to distinguish between wheat subgenomes. HTSeq v.0.6.1 (www-huber.embl.de/users/anders/HTSeq/) was used to count the read numbers mapped to the gene models. For all comparisons, read counts were normalized to the aligned RPKM (Mortazavi et al., 2008) to obtain the relative levels of expression. Differential expression analyses between different developmental stages were performed using the DESeq (Anders and Huber, 2010) R packages. In brief, differential gene expression analysis between stages (each with two biological replicates) was implemented in the DESeq program based on the assumption that the number of reads in each sample assigned to each gene could be modeled by a negative binomial distribution. The mean and variance were estimated from the read counts and the differential expression between different conditions was determined under the null hypothesis: Total read counts in one condition were equal to those in the other condition. Therefore, just one test was performed between two conditions (Anders and Huber, 2010). For each gene, an adjusted P value was computed by DESeq, and those with an adjusted P value of < 0.05 were considered differentially expressed.
The sample dendrogram was constructed using the R function flashClust. The Pearson correlations between biological replicates were calculated using the R function cor, based on the RPKM values. The Venn diagram was made using the vennDiagram function in R, using a gene list for each stage derived by combining the genes from all genotypes. AgriGO (http://bioinfo.cau.edu.cn/agriGO/; Du et al., 2010) and MapMan (http://mapman.gabipd.org/; Sreenivasulu et al., 2008) were used for the GO analysis and the identification of pathways of stage-specific genes, respectively. GO terms with a corrected false discovery rate of <0.05 were considered to be significantly enriched. Clustering was performed in R using the k-means function, where k = 24 within the cluster package by Euclidean distance. Heat maps were drawn using the Pheatmap package in R and were clustered using Pearson correlation distance.
Small RNA Sequencing and Bioinformatics Analysis
Small RNA sequencing was performed on the same tissues used for RNA sequencing, with two biological replicates. About 8 to 18 million reads were obtained for each library. Reads were removed if they were of low quality; contained 5′ primer contaminants, more than a 10% N content, or a poly A sequence; did not contain the insert fragments or the 3′ primer; or were shorter than 18 nucleotides. The remaining clean reads were aligned to the genome (ftp://ensemblgenomes.org/pub/release-30/plants/fasta/triticum_aestivum/) using Bowtie (http://bowtie-bio.sourceforge.net/index.shtml; Langmead et al., 2009; Supplemental Table S7). The mapped reads were also compared to Rfam (v.11, http://rfam.janelia.org/) with BLAST or Bowtie to remove rRNA-, scRNA-, snoRNA-, snRNA-, and tRNA-associated reads. Custom PERL scripts were then used to compare the mapped reads with 525 mature miRNAs from Brachypodium distachyon downloaded from miRBase (Release21, http://www.mirbase.org/; Kozomara and Griffiths-Jones, 2011, 2014). Sequences with a maximum of 2-nucleotide differences in length compared with the Brachypodium miRNAs were collected.
Differential expression analyses between the samples were performed using DESeq (Anders and Huber, 2010) R packages. Small RNAs with an adjusted P value of ≤ 0.05 and a log2FoldChange absolute value of ≥ 1 were considered to be differentially expressed. MiRNA target prediction was conducted using psRobot (http://omicslab.genetics.ac.cn/psRobot/) and TargetFinder (Bo and Wang, 2005), and common targets were identified.
qRT-PCR Expression Analysis of Genes and MiRNAs
Total RNA was extracted with TRIzol reagent (Invitrogen). Reverse transcription was performed with 1 μg RNA using a PrimeScript RT reagent Kit with gDNA Eraser (Takara Bio Inc.) using an oligo(dT)18 primer. Real-time PCR was performed using SYBR Premix Ex Taq II (Takara Bio) on an ABI 7300 Real-Time PCR system (Applied Biosystems). Wheat GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an endogenous reference gene (forward primer: 5′-TTAGACTTGCGAAGCCAGCA-3′; reverse primer: 5′-AAATGCCCTTGAGGTTTCCC-3′) to normalize the data. Three technical and two biological replicates were performed to produce the average expression levels relative to the reference, using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
For miRNA, real-time PCR was performed using specific stem-loop primers for the reverse transcription, as described previously (Chen et al., 2005). The forward and reverse primers for PCR were gene specific and universal (universal primer: 5′-CAGTGCAGGGTCCGAGGTA-3′), respectively. The 18s rRNA was used as an endogenous reference gene (forward primer: 5′-AGGCCTTCACCAAGTATGCTCTG-3′; reverse primer: 5′-TGGGCGATAACACGGACAACAGTA-3′). All other primer sequence information is listed in Supplemental Dataset 3.
RNA Ligase-Mediated 5′-RACE Assays
To verify the target cleavage capability of selected miRNAs, the RNA ligase-mediated rapid amplification of 5′-cDNA ends (5′-RACE) was performed using the FirstChoice RLM-RACE Kit (Invitrogen), according to the manufacturer’s instructions. In brief, RNAs were ligated to the 5′-RACE RNA adapter, and reverse transcription reactions were then performed with the ligated RNA. Nested PCRs were performed using universal and gene-specific outer and inner sense primers (Supplemental Dataset 3). PCR products were cloned into the pGEM-T Easy vector (Promega Corporation) and sequenced.
In Situ Hybridization
RNA in situ hybridization was carried out as described previously (Liu et al., 2013a). Fresh young spikes were fixed in formalin-acetic acid-alcohol overnight at 4°C, dehydrated through a standard ethanol series, embedded in Paraplast Plus tissue-embedding medium (Sigma-Aldrich), and sectioned at 8 μm using a microtome (RM2235, Leica Microsystems). The gene-specific regions were amplified to generate sense and antisense RNA probes. Digoxigenin-labeled RNA probes were synthesized using a DIG northern Starter Kit (Roche), according to the manufacturer’s instructions. Primer sequences used for probe synthesis are listed in Supplemental Dataset 3.
Microscopy and Scanning Electron Microscopy (SEM)
Photomicrographs of young spikes were taken using a stereomicroscope (S8 APO, Leica Microsystems) equipped with a digital camera (A640, Canon Inc.). For SEM, young spikes from each stage were fixed in 2% glutaraldehyde in 0.2 m phosphate buffer (pH 7.2), dehydrated through a standard ethanol series, and critical point dried using CO2. The dried tissues were coated with platinum and observed and photographed using a scanning electron microscope (SEM SU8010, Hitachi, Ltd.).
Phylogenetic Analysis
Sequence alignment was performed using ClustalX (http://www.clustal.org/), and a phylogenetic tree was constructed with the Mega5 program (http://www.megasoftware.net/) using AGO1 protein sequences manually collected from the newly released genome sequence (v0.4: https://urgi.versailles.inra.fr/download/iwgsc/IWGSC-WGA_Sequences/v0.4/). A BLAST search was performed using 400-amino acid segments of the rice AGO protein sequences (residues approximately 260–650; Supplemental Table S8). Sequences were manually screened and tested with a phylogenetic analysis.
Mutant Analysis
EMS mutant desert durum wheat cultivar Kronos lines were kindly provided by Dr. Jorge Dubcovsky at the University of California, Davis. DNA was extracted using a Plant Genomic DNA Kit (Tiangen Biotech [Beijing] Co., Ltd.). Regions containing mutation sites were amplified from wild-type and mutant plants (forward primer: 5′-GTGCTTTGTCTCAACCCTT-3′; reverse primer: 5′-TCAACGCATCAGCCAGTACA-3′) and sequenced to identify single nucleotide polymorphisms.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers PRJNA383677 and PRJNA383333 for RNA-seq and small RNA data respectively.
SUPPLEMENTAL MATERIALS
The following supplemental materials are available.
Supplemental Figure S1. Correlation of gene expression profiles between biological replicates and among tissues of different stages during the early development of wheat inflorescences.
Supplemental Figure S2. Numbers of DEGs between the specified wheat inflorescence development stages.
Supplemental Figure S3. Expression patterns of genes in the 24 k-means clusters.
Supplemental Figure S4. Quantitative RT-PCR validation and RPKM values of the homeologs of genes preferentially expressed during wheat inflorescence development.
Supplemental Figure S5. Clustering analysis of miRNA expression levels during wheat inflorescence development.
Supplemental Figure S6. Multiple sequence alignments of miRNAs and their predicted targets.
Supplemental Figure S7. Expression patterns of miR167a, miR159b-3p.1, and their predicted target genes during wheat inflorescence development.
Supplemental Figure S8. Characterization of wheat AGO1 genes and the function of AGO1d.
Supplemental Table S1. RNA-seq data statistics and genome mapping rates.
Supplemental Table S2. Differential analysis results based on pairwise comparisons.
Supplemental Table S3. List of differentially expressed genes in the 24 k-means clusters.
Supplemental Table S4. List of wheat homeologs similar to rice MADS-box genes.
Supplemental Table S5. List of stage-specific transcription factors and hormone-related genes classified by MapMan.
Supplemental Table S6. Differential analysis results of miRNA based on pairwise comparisons.
Supplemental Table S7. Small RNA sequencing statistics and genome mapping rates.
Supplemental Table S8. AGO1 sequences used for phylogenetic analysis.
Supplemental Dataset 1. List of stage-specific genes.
Supplemental Dataset 2. Expression of known miRNAs and their variants in wheat inflorescence development.
Supplemental Dataset 3. Primers used for qRT-PCRs, in situ hybridizations, and RLM-5′-RACE.
Acknowledgments
We thank Professor Jorge Dubcovsky at University of California Davis for making the Kronos EMS mutant lines of durum wheat available.
Glossary
- TF
transcription factor
- DR
double-ridge stage
- FM
floret meristem stage
- AM
anther primordia stage
- TS
tetrads stage
- RPKM
reads per kilobase per million reads
- DEG
differentially expressed gene
Footnotes
This work is supported in part by National Science Foundation of China (#31571665) and the National Key R&D Plan Project (#2016YFD0102002) from Ministry of Science and Technology of China to A.L, and the National Key Program on Transgenic Research from the Ministry of Agriculture (#2016ZX08009-001) and the CAAS Innovation Program to L.M.
Articles can be viewed without a subscription.
References
- Abe M, Yoshikawa T, Nosaka M, Sakakibara H, Sato Y, Nagato Y, Itoh J (2010) WAVY LEAF1, an ortholog of Arabidopsis HEN1, regulates shoot development by maintaining MicroRNA and trans-acting small interfering RNA accumulation in rice. Plant Physiol 154: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleyard M, Kirby EJM, Fellowes G (1982) Relationships between the duration of phases in the pre-anthesis life cycle of spring barley. Aust J Agric Res 33: 917–925 [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S (2007) MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745 [DOI] [PubMed] [Google Scholar]
- Bo X, Wang S (2005) TargetFinder: A software for antisense oligonucleotide target site selection based on MAST and secondary structures of target mRNA. Bioinformatics 21: 1401–1402 [DOI] [PubMed] [Google Scholar]
- Boden SA, Cavanagh C, Cullis BR, Ramm K, Greenwood J, Jean Finnegan E, Trevaskis B, Swain SM (2015) Ppd-1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nat Plants 1: 14016. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. (2005) Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley RB, Hake S (1998) The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev 12: 1145–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Hake S (2008) Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135: 3013–3019 [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Irish E, Sakai H, Hake S (2007) The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet 39: 1517–1521 [DOI] [PubMed] [Google Scholar]
- Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ (2002) The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298: 1238–1241 [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 [DOI] [PubMed] [Google Scholar]
- Cui R, Han J, Zhao S, Su K, Wu F, Du X, Xu Q, Chong K, Theissen G, Meng Z (2010) Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J 61: 767–781 [DOI] [PubMed] [Google Scholar]
- Digel B, Pankin A, von Korff M (2015) Global transcriptome profiling of developing leaf and shoot apices reveals distinct genetic and environmental control of floral transition and inflorescence development in barley. Plant Cell 27: 2318–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolskaya O, Pont C, Sibout R, Martinek P, Badaeva E, Murat F, Chosson A, Watanabe N, Prat E, Gautier N, et al. (2015) FRIZZY PANICLE drives supernumerary spikelets in bread wheat. Plant Physiol 167: 189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornelas MC, Patreze CM, Angenent GC, Immink RGH (2011) MADS: The missing link between identity and growth? Trends Plant Sci 16: 89–97 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveland AL, Goldshmidt A, Pautler M, Morohashi K, Liseron-Monfils C, Lewis MW, Kumari S, Hiraga S, Yang F, Unger-Wallace E, et al. (2014) Regulatory modules controlling maize inflorescence architecture. Genome Res 24: 431–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani I, Sukegawa S, Kyozuka J (2006) Genome-wide analysis of spatial and temporal gene expression in rice panicle development. Plant J 46: 503–511 [DOI] [PubMed] [Google Scholar]
- Gao X, Liang W, Yin C, Ji S, Wang H, Su X, Guo C, Kong H, Xue H, Zhang D (2010) The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol 153: 728–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JS, Hess WM, Trione EJ (1985) Development of the young wheat spike: A sem study of Chinese Spring wheat. Am J Bot 72: 548–559 [Google Scholar]
- Guo Z, Schnurbusch T (2015) Variation of floret fertility in hexaploid wheat revealed by tiller removal. J Exp Bot 66: 5945–5958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrop TWR, Ud Din I, Gregis V, Osnato M, Jouannic S, Adam H, Kater MM (2016) Gene expression profiling of reproductive meristem types in early rice inflorescences by laser microdissection. Plant J 86: 75–88 [DOI] [PubMed] [Google Scholar]
- Henry IM, Nagalakshmi U, Lieberman MC, Ngo KJ, Krasileva KV, Vasquez-Gross H, Akhunova A, Akhunov E, Dubcovsky J, Tai TH, et al. (2014) Efficient genome-wide detection and cataloging of EMS-induced mutations using exome capture and next-generation sequencing. Plant Cell 26: 1382–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Jackson S (2015) Floral induction and flower formation—the role and potential applications of miRNAs. Plant Biotechnol J 13: 282–292 [DOI] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K, Maekawa M, Izawa T, Itoh J, Nagato Y (2012) ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J 69: 168–180 [DOI] [PubMed] [Google Scholar]
- Ikeda K, Nagasawa N, Nagato Y (2005) ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev Biol 282: 349–360 [DOI] [PubMed] [Google Scholar]
- Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, International Wheat Genome Sequencing Consortium, et al. (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496: 91–95 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42: 541–544 [DOI] [PubMed] [Google Scholar]
- Kellogg EA, Camara PEAS, Rudall PJ, Ladd P, Malcomber ST, Whipple CJ, Doust AN (2013) Early inflorescence development in the grasses (Poaceae). Front Plant Sci 4: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen BM, Rasmusson DC (1983) Duration and inheritance of leaf initiation, spike initiation, and spike growth in barley. Crop Sci 23: 939–943 [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J (2010) PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol 51: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Yasuno N, Sato Y, Yoda M, Yamazaki R, Kimizu M, Yoshida H, Nagamura Y, Kyozuka J (2012) Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24: 1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J (2003) FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130: 3841–3850 [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S (2011) miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S (2014) miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva KV, Vasquez-Gross HA, Howell T, Bailey P, Paraiso F, Clissold L, Simmonds J, Ramirez-Gonzalez RH, Wang X, Borrill P, et al. (2017) Uncovering hidden variation in polyploid wheat. Proc Natl Acad Sci USA 114: E913–E921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D-Y, An G (2012) Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 69: 445–461 [DOI] [PubMed] [Google Scholar]
- Lee S, Kim J, Han J-J, Han M-J, An G (2004) Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J 38: 754–764 [DOI] [PubMed] [Google Scholar]
- Lee D-Y, Lee J, Moon S, Park SY, An G (2007) The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J 49: 64–78 [DOI] [PubMed] [Google Scholar]
- Ling H-Q, Zhao S, Liu D, Wang J, Sun H, Zhang C, Fan H, Li D, Dong L, Tao Y, et al. (2013) Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496: 87–90 [DOI] [PubMed] [Google Scholar]
- Liu Y, Cui S, Wu F, Yan S, Lin X, Du X, Chong K, Schilling S, Theißen G, Meng Z (2013a) Functional conservation of MIKC*-Type MADS box genes in Arabidopsis and rice pollen maturation. Plant Cell 25: 1288–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Harberd NP, Fu X (2016) SQUAMOSA promoter binding protein-like transcription factors: targets for improving cereal grain yield. Mol Plant 9: 765–767 [DOI] [PubMed] [Google Scholar]
- Liu H, Qin C, Chen Z, Zuo T, Yang X, Zhou H, Xu M, Cao S, Shen Y, Lin H, et al. (2014a) Identification of miRNAs and their target genes in developing maize ears by combined small RNA and degradome sequencing. BMC Genomics 15: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Teo ZW, Bi Y, Song S, Xi W, Yang X, Yin Z, Yu H (2013b) A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev Cell 24: 612–622 [DOI] [PubMed] [Google Scholar]
- Liu D, Wang D, Qin Z, Zhang D, Yin L, Wu L, Colasanti J, Li A, Mao L (2014b) The SEPALLATA MADS-box protein SLMBP21 forms protein complexes with JOINTLESS and MACROCALYX as a transcription activator for development of the tomato flower abscission zone. Plant J 77: 284–296 [DOI] [PubMed] [Google Scholar]
- Liu Q, Yao X, Pi L, Wang H, Cui X, Huang H (2009) The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J 58: 27–40 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Δ Δ C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luo Y, Guo Z, Li L (2013) Evolutionary conservation of microRNA regulatory programs in plant flower development. Dev Biol 380: 133–144 [DOI] [PubMed] [Google Scholar]
- Luo L, Li W, Miura K, Ashikari M, Kyozuka J (2012) Control of tiller growth of rice by OsSPL14 and Strigolactones, which work in two independent pathways. Plant Cell Physiol 53: 1793–1801 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J 23: 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P (2010) TCP genes: A family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Mayer KFX, Rogers J, Doležel J, Pozniak C, Eversole K, Feuillet C, Gill B, Friebe B, Lukaszewski AJ, International Wheat Genome Sequencing Consortium (IWGSC), et al. (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 1251788. [DOI] [PubMed] [Google Scholar]
- Millar AA, Gubler F (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17: 705–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, Song X-J, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42: 545–549 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jürgens G, Laux T (1998) Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J 17: 1799–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, King S, Jack T (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106: 22534–22539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H, Itoh J, Hayashi K, Hibara K, Satoh-Nagasawa N, Nosaka M, Mukouhata M, Ashikari M, Kitano H, Matsuoka M, et al. (2007) The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc Natl Acad Sci USA 104: 14867–14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y (2003) SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718 [DOI] [PubMed] [Google Scholar]
- Nair SK, Wang N, Turuspekov Y, Pourkheirandish M, Sinsuwongwat S, Chen G, Sameri M, Tagiri A, Honda I, Watanabe Y, et al. (2010) Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc Natl Acad Sci USA 107: 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Shimamoto K, Kyozuka J (2002) Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J 29: 743–750 [DOI] [PubMed] [Google Scholar]
- Ohmori S, Kimizu M, Sugita M, Miyao A, Hirochika H, Uchida E, Nagato Y, Yoshida H (2009) MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21: 3008–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao NN, Prasad K, Kumar PR, Vijayraghavan U (2008) Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc Natl Acad Sci USA 105: 3646–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RE, Ercoli MF, Debernardi JM, Breakfield NW, Mecchia MA, Sabatini M, Cools T, De Veylder L, Benfey PN, Palatnik JF (2015) MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. Plant Cell 27: 3354–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I, Weigel D (2013) Coordination of flower maturation by a regulatory circuit of three microRNAs. PLoS Genet 9: e1003374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma S, Salomon B, Komatsuda T (2011) The domestication syndrome genes responsible for the major changes in plant form in the Triticeae crops. Plant Cell Physiol 52: 738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh N, Itoh J, Nagato Y (2003) The SHOOTLESS2 and SHOOTLESS1 genes are involved in both initiation and maintenance of the shoot apical meristem through regulating the number of indeterminate cells. Genetics 164: 335–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C, Debernardi JM, Bresso EG, Rodriguez RE, Palatnik JF (2014) Repression of cell proliferation by miR319-regulated TCP4. Mol Plant 7: 1533–1544 [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Shitsukawa N, Kinjo H, Takumi S, Murai K (2009) Heterochronic development of the floret meristem determines grain number per spikelet in diploid, tetraploid and hexaploid wheats. Ann Bot (Lond) 104: 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons KJ, Fellers JP, Trick HN, Zhang Z, Tai Y-S, Gill BS, Faris JD (2006) Molecular characterization of the major wheat domestication gene Q. Genetics 172: 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoczynska A, Szweykowska-Kulinska Z (2016) MicroRNA-mediated regulation of flower development in grasses. Acta Biochim Pol 63: 687–692 [DOI] [PubMed] [Google Scholar]
- Spanudakis E, Jackson S (2014) The role of microRNAs in the control of flowering time. J Exp Bot 65: 365–380 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Schnurbusch T (2012) A genetic playground for enhancing grain number in cereals. Trends Plant Sci 17: 91–101 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Usadel B, Winter A, Radchuk V, Scholz U, Stein N, Weschke W, Strickert M, Close TJ, Stitt M, et al. (2008) Barley grain maturation and germination: Metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol 146: 1738–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Sato M, Ashikari M, Miyoshi M, Nagato Y, Hirano H-Y (2004) The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131: 5649–5657 [DOI] [PubMed] [Google Scholar]
- Tanaka W, Pautler M, Jackson D, Hirano H-Y (2013) Grass meristems II: Inflorescence architecture, flower development and meristem fate. Plant Cell Physiol 54: 313–324 [DOI] [PubMed] [Google Scholar]
- Teo ZWN, Song S, Wang Y-Q, Liu J, Yu H (2014) New insights into the regulation of inflorescence architecture. Trends Plant Sci 19: 158–165 [DOI] [PubMed] [Google Scholar]
- Teotia S, Tang G (2015) To bloom or not to bloom: Role of microRNAs in plant flowering. Mol Plant 8: 359–377 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy C, Paraiso F, Colasuonno P, Tran RK, Tsai H, Berardi S, Comai L, Dubcovsky J (2009) A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol 9: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington SR, Cartwright PM, Wall PC (1983) A quantitative scale of spike initial and pistil development in barley and wheat. Ann Bot 51: 119–130 [Google Scholar]
- Wang Y, Sun F, Cao H, Peng H, Ni Z, Sun Q, Yao Y (2012) TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS One 7: e48445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sun S, Jin J, Fu D, Yang X, Weng X, Xu C, Li X, Xiao J, Zhang Q (2015) Coordinated regulation of vegetative and reproductive branching in rice. Proc Natl Acad Sci USA 112: 15504–15509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhang Q, Zhou H, Ni F, Wu X, Qi Y (2009) Rice MicroRNA effector complexes and targets. Plant Cell 21: 3421–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Wu C, Xiong L (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol 142: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Lee DY, Miyao A, Hirochika H, An G, Hirano H-Y (2006) Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Sasao M, Yasuno N, Takagi K, Daimon Y, Chen R, Yamazaki R, Tokunaga H, Kitaguchi Y, Sato Y, et al. (2013) TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc Natl Acad Sci USA 110: 767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Belcram H, Gornicki P, Charles M, Just J, Huneau C, Magdelenat G, Couloux A, Samain S, Gill BS, et al. (2011) Duplication and partitioning in evolution and function of homoeologous Q loci governing domestication characters in polyploid wheat. Proc Natl Acad Sci USA 108: 18737–18742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Xia R, Meyers BC, Walbot V (2015) Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr Opin Plant Biol 27: 84–90 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhang X (2012) Argonautes compete for miR165/166 to regulate shoot apical meristem development. Curr Opin Plant Biol 15: 652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Helliwell CA (2011) Regulation of flowering time and floral patterning by miR172. J Exp Bot 62: 487–495 [DOI] [PubMed] [Google Scholar]
- Zhu H, Hu F, Wang R, Zhou X, Sze S-H, Liou LW, Barefoot A, Dickman M, Zhang X (2011) Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145: 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]