A combined phylogenetic and transcriptomic analysis of root hair development genes in seven vascular plant species reveals unexpected diversification in gene structure and expression.
Abstract
The molecular genetic program for root hair development has been studied intensively in Arabidopsis (Arabidopsis thaliana). To understand the extent to which this program might operate in other plants, we conducted a large-scale comparative analysis of root hair development genes from diverse vascular plants, including eudicots, monocots, and a lycophyte. Combining phylogenetics and transcriptomics, we discovered conservation of a core set of root hair genes across all vascular plants, which may derive from an ancient program for unidirectional cell growth coopted for root hair development during vascular plant evolution. Interestingly, we also discovered preferential diversification in the structure and expression of root hair development genes, relative to other root hair- and root-expressed genes, among these species. These differences enabled the definition of sets of genes and gene functions that were acquired or lost in specific lineages during vascular plant evolution. In particular, we found substantial divergence in the structure and expression of genes used for root hair patterning, suggesting that the Arabidopsis transcriptional regulatory mechanism is not shared by other species. To our knowledge, this study provides the first comprehensive view of gene expression in a single plant cell type across multiple species.
A fundamental feature of organismal evolution is the creation and diversification of cell type-specific differentiation programs. These programs are responsible for generating the cellular diversity, and associated division of labor, that is the hallmark of multicellular organisms (Arendt, 2008). However, we still know relatively little about the evolution of the genetic and molecular mechanisms that establish cell differentiation programs and how they differ between species.
The root hair cell is a useful single cell type for experimental studies in plant biology, and its development, physiology, and cell biology have been studied intensively in many plant species (Cormack, 1935; Emons and Ketelaar, 2009; Datta et al., 2011; Qiao and Libault, 2013; Grierson et al., 2014). Root hairs are long tubular extensions of root epidermal cells that greatly increase the root surface area and thereby assist in water and nutrient absorption. The development of root hairs occurs in three basic stages: specification of the root hair cell fate, initiation of a root hair outgrowth, and elongation of the hair via tip growth. Root hairs are found in nearly all vascular plants, including angiosperms, gymnosperms, and lycophytes, and they exhibit similar cellular features, suggesting a common evolutionary origin. However, different plant species are known to vary in their root hair distribution patterns and their root hair morphology (Clowes, 2000; Pemberton et al., 2001; Datta et al., 2011), implying that genetic differences exist in root hair development programs.
Root hairs have been studied intensively in Arabidopsis (Arabidopsis thaliana). In particular, molecular genetic analyses have led to the identification of numerous root hair genes, which provide insight into the mechanisms of Arabidopsis root hair development (Bruex et al., 2012; Gu and Nielsen, 2013; Grierson et al., 2014; Balcerowicz et al., 2015; Salazar-Henao et al., 2016). Root hair-bearing cells in Arabidopsis are specified by a set of early-acting patterning genes that generate a cell position-dependent distribution of root hair cells and nonhair cells via a complex transcriptional regulatory network (Grierson et al., 2014; Salazar-Henao et al., 2016). Once specified, the presumptive root hair cells initiate the outgrowth of the root hair through the action of the ROOT HAIR DEFECTIVE6 (RHD6) gene, which encodes a basic helix-loop-helix (bHLH) transcription factor that induces an extensive root hair gene expression program through the activation of additional regulatory genes (Masucci and Schiefelbein, 1994; Menand et al., 2007; Yi et al., 2010; Bruex et al., 2012). This suite of downstream root hair morphogenesis genes generates the unidirectional expansion (tip growth) of the root hair (Datta et al., 2011; Balcerowicz et al., 2015). These genes encode proteins involved in secretory activities, cell wall synthesis, ion transport, reactive oxygen species regulation, and many other processes (Balcerowicz et al., 2015; Salazar-Henao et al., 2016). The expression profiles of the patterning genes, initiation genes, and morphogenesis genes differ along the longitudinal length of the root tip, which reflects their temporal importance in root hair development (Datta et al., 2011; Grierson et al., 2014).
The wealth of knowledge concerning the genetic control of root hair development in Arabidopsis provides an opportunity to evaluate the similarity in root hair development programs in other plants and thereby address fundamental issues regarding the evolution of cell differentiation mechanisms. Several focused studies have begun to investigate this issue by analyzing individual root hair genes/families in Arabidopsis and selected species to examine their molecular relationships (Kim et al., 2006, 2007; Brady et al., 2007b; Ding et al., 2009; Karas et al., 2009). In general, the results from these studies suggest that root hair developmental genes tend to share similar function in different species, implying conservation in their root hair programs.
In this study, we sought to comprehensively analyze root hair differentiation programs across vascular plants. We first defined the root hair transcriptome and root hair development genes in Arabidopsis and then analyzed the distribution and expression of these genes in six other vascular plant species. In addition, total root hair gene expression was assessed directly by analyzing transcript accumulation in purified root hairs. Although we found that many root hair genes are conserved across these species and, therefore, likely share similar roles, we also discovered significant differences in the structure and/or expression of some root hair development genes. Altogether, this broad analysis of gene expression in a single cell type across multiple species provides new insight into the conservation and diversification of plant cell differentiation programs in vascular plants.
RESULTS
Arabidopsis Root Hair Development Genes
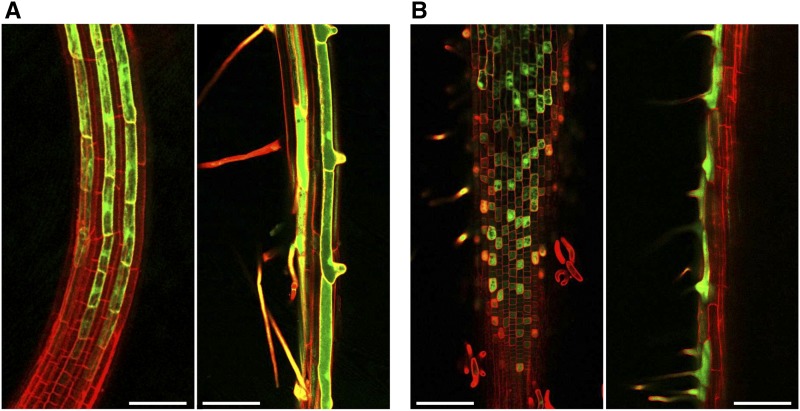
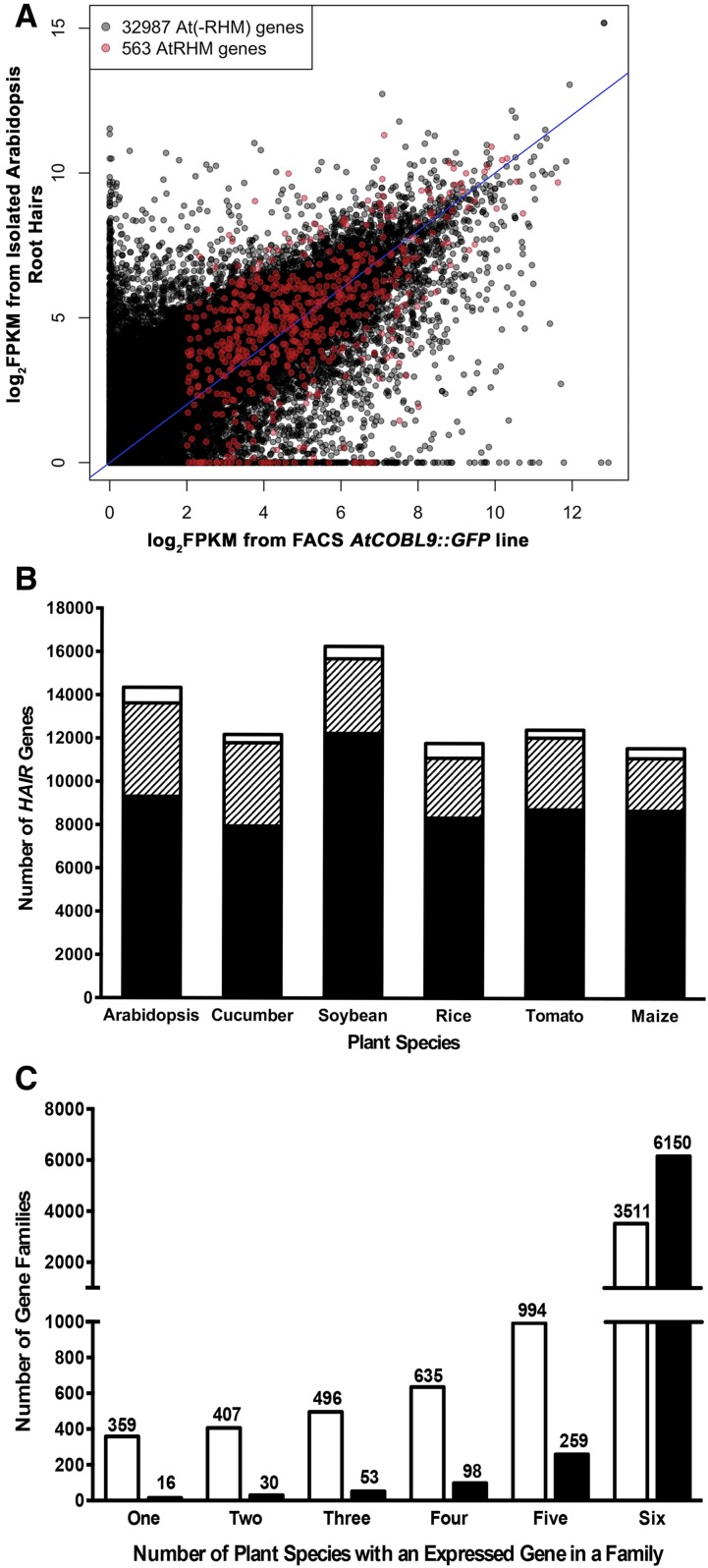
To compare root hair gene activity across plant species, we first defined the genes expressed in differentiating root hair cells of Arabidopsis. A transgenic line containing the GFP and GUS reporters under the control of the COBRA-LIKE9 promoter (AtCOBL9::GFP; Brady et al., 2007b) was used for this purpose, because it specifically accumulates GFP in root hair cells beginning in the elongation zone (prior to hair emergence) through root hair maturation (Fig. 1A). The GFP-expressing cells were isolated by protoplasting and fluorescence-activated cell sorting (FACS) of the AtCOBL9::GFP root tips, and their transcripts were purified and subjected to RNA sequencing (RNA-seq) analysis (using three biological replicates; for details, see “Materials and Methods”). Transcripts from 12,691 genes were identified from among the total of 33,550 Arabidopsis genes (TAIR10) surveyed (mean fragments per kilobase per million mapped reads [FPKM] ≥ 3; >0 FPKM in ≥2 replicates; Supplemental Data Set S1). These 12,691 genes are designated AtRH (Arabidopsis thaliana root hair) genes. As validation, we found that this AtRH gene set includes all 17 of the individual genes reported previously to be root hair specific using nontranscriptome methods, and it possesses 90% to 97% overlap with four previously reported root hair gene data sets defined by transcriptome-based methods (Brady et al., 2007a; Lan et al., 2013; Becker et al., 2014; Li and Lan, 2015; Supplemental Data Set S1; Supplemental Table S1).
Figure 1.
Root hair cell-specific expression of GFP marker lines in Arabidopsis and rice roots. A, GFP accumulation in the AtCOBL9::GFP line in immature root hair cells in the elongation zone (left) and in the differentiation zone (right) of Arabidopsis roots. B, GFP accumulation in the OsEXPA30::GFP line in immature root hair cells in the elongation zone (left) and in the differentiation zone (right) of rice roots. Roots were stained with propidium iodide (red fluorescence). Bars = 100 μm.
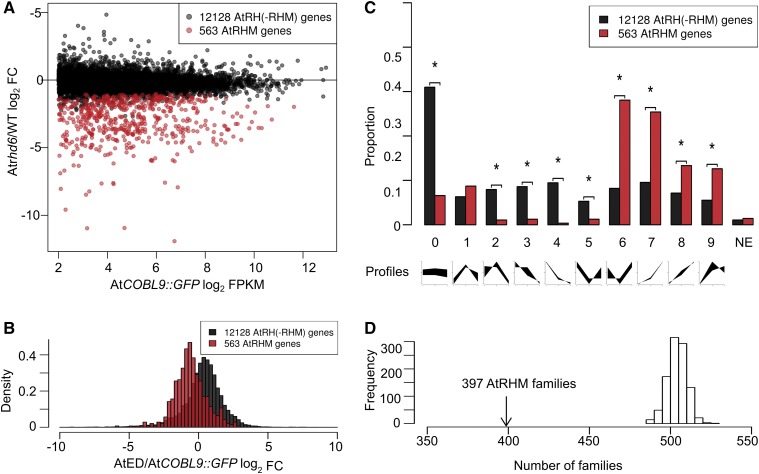
It is likely that many of these AtRH genes are associated with functions common to most/all cells (e.g. housekeeping functions). To identify the subset of AtRH genes closely associated with root hair cell differentiation, we assessed their transcript levels in the hairless rhd6 mutant relative to the wild type. We also included in these lines a WER::GFP marker, which accumulates GFP in the entire developing root epidermis (Lee and Schiefelbein, 1999), to limit transcript acquisition to differentiating epidermal cells. Following protoplasting/FACS and RNA-seq analysis, we compared transcript accumulation in rhd6 WER::GFP versus wild-type WER::GFP roots (three biological replicates per line) and identified 563 AtRH genes that are significantly down-regulated in rhd6 (false discovery rate [FDR] ≤ 0.01, fold change ≥ 2, counts per million from three or more of six replicates ≥ 1; Fig. 2A; Supplemental Data Set S2). We specially designate this subset of 563 AtRH genes to be called AtRHM genes (Arabidopsis thaliana root hair morphogenesis genes) because their expression is RHD6 dependent and, therefore, they are more closely associated with root hair formation than the other AtRH genes. As validation, we found that all six of the root hair genes shown previously to be positively regulated by RHD6 based on nontranscriptome methods are among the AtRHM genes, and 122 of the 126 genes (97%) reported previously to be down-regulated in rhd6 in a microarray study (Bruex et al., 2012) also are present in the AtRHM gene set (Supplemental Data Set S2; Supplemental Table S1). Furthermore, as expected, Gene Ontology (GO) analysis of the AtRHM genes showed significant overrepresentation (FDR < 0.01) of root hair-associated terms, including root hair cell differentiation, unidimensional cell growth, and trichoblast differentiation (Supplemental Table S2).
Figure 2.
Analysis of AtRH and AtRHM genes. A, Distribution of AtRH(-RHM) genes and AtRHM genes, based on transcript levels in the FACS-purified root hair cells of AtCOBL9::GFP and log2 fold change (FC) in transcript levels from FACS-purified cells of rhd6 WER::GFP versus wild-type WER::GFP. Data are means from three biological replicates. B, Distribution of AtRH(-RHM) genes and AtRHM genes, based on log2 fold change in transcript level from FACS-purified root hair cells of AtCOBL9::GFP versus wild-type root elongation zone and differentiation zone segments. C, Distribution of AtRH(-RHM) genes and AtRHM genes, based on relative transcript levels in the meristematic, elongation, and differentiation zones of wild-type roots (i.e. expression profiles). The nine expression profile types (defined by Huang and Schiefelbein [2015]) are indicated in the graphs at bottom (from left to right: meristematic, elongation, and differentiation zones). NE, No expression detected. Asterisks indicate profile types with significantly different proportions between the two groups (P < 0.01, χ2 test, Bonferroni corrected). D, Distribution of the number of gene families resulting from 1,000 random draws of 543 genes from the 12,449 total AtRH genes in the GreenPhyl family database. The observed numbers of gene families (397) that contain the 543 AtRHM genes are indicated.
Given that they are positively regulated by RHD6, the AtRHM genes might exhibit preferential expression in root hair cells, as compared with the remainder of the AtRH genes [which we designate AtRH(-RHM) genes]. To test this, we calculated the ratio of transcript accumulation in root hair cells (FPKM from COBL9::GFP) to transcript accumulation in the entire root elongation zone and differentiation zone (FPKM from published root segment RNA-seq data; Huang and Schiefelbein, 2015) for each of these genes (Supplemental Data Set S1). The distribution of these values differs significantly between the AtRHM and AtRH(-RHM) gene groups (P < 10−15, Wilcoxon rank-sum test), indicating that, as a whole, the AtRHM genes possess a relatively greater degree of preferential root hair expression than the other AtRH genes (Fig. 2B).
We also analyzed the temporal expression profiles of the AtRHM genes relative to the AtRH(-RHM) genes. Because they are associated with root hair formation, AtRHM genes might be expected to exhibit relatively high transcription in the differentiation zone of the root, where root hairs emerge and grow (Grierson et al., 2014). To examine this, we compared each gene’s transcript accumulation in the three major longitudinal root zones (meristematic zone, elongation zone, and differentiation zone) using transcriptome data reported previously from these Arabidopsis root segments (Huang and Schiefelbein, 2015). We found that a majority of the AtRHM genes (79%), but not the AtRH(-RHM) genes (30%), exhibit temporal expression profiles associated with relatively high transcript accumulation in the differentiation zone (expression profile types 6–9; Supplemental Data Set S1), a statistically significant enrichment (P < 0.01 for each of these four profile types, χ2 test; Fig. 2C).
Next, we sought to determine whether the AtRHM genes tend to be related to one another in sequence. To assess this, we analyzed the distribution of the AtRHM genes among Arabidopsis gene families. Using an established plant gene family database (GreenPhyl version 4; Rouard et al., 2011), we found that 543 of the 563 AtRHM genes have been assigned to a total of 397 GreenPhyl-defined gene families (Supplemental Data Set S2). This number of families is substantially less than the numbers obtained from 1,000 random draws of 543 genes from the 12,449 AtRH genes included in the GreenPhyl database (Fig. 2D), indicating that AtRHM genes tend to be related to one another and cluster in families. Consistent with this, we observed several families that contain high proportions of AtRHM genes, including six two-gene families in which both members are AtRHM genes and, in the most extreme case, an 11-member gene family (the PRP3 family) composed entirely of AtRHM genes (Supplemental Data Set S2). This suggests conservation in (RHD6-regulated) root hair gene expression in certain gene lineages.
Rice Root Hair Development Genes
To determine whether the root hair development genes identified in Arabidopsis are similar in other plants, we defined the root hair transcriptome of rice (Oryza sativa). RNA was isolated from protoplasted/FACS-treated roots from a rice transgenic line, OsEXPA30::GFP (Kim et al., 2006), that specifically accumulates GFP in root hair cells in a manner similar to the GFP accumulation in the Arabidopsis COBL9::GFP line (Fig. 1B). Following RNA-seq analysis (three biological replicates), we defined 13,342 genes expressed in these EXPA30::GFP sorted cells (mean FPKM ≥ 3; >0 FPKM in ≥2 replicates, same parameters used for the Arabidopsis COBL9::GFP analysis; Supplemental Data Set S3). These are designated as OsRH genes because they are root hair-expressed genes from rice, and they include all six of the previously reported root hair-specific genes defined by nontranscriptome methods in rice (Supplemental Data Set S3; Supplemental Table S1).
We tested whether the root hair genes in Arabidopsis are related to the root hair genes in rice by analyzing the distribution of the AtRH and non-AtRH genes relative to the OsRH and non-OsRH genes within GreenPhyl-defined gene families. Among families that possess at least one Arabidopsis gene and at least one rice gene, we discovered a statistically significant nonrandom distribution (controlling for family size), indicating preferential associations of AtRH genes with OsRH genes and non-AtRH genes with non-OsRH genes in these families (Table I; Supplemental Data Set S4). This familial association indicates that root hair-expressed genes tend to be conserved in these two plant species.
Table I. P value results of Fisher’s exact test analyzing the familial association between Arabidopsis and rice root hair gene sets.
The test was performed in nine groups with the family size fixed (a combination of one to three Arabidopsis genes and one to three rice genes per family). The design table for each test is given below. AtRH families = families with at least one AtRH gene, non-AtRH families = families with no AtRH gene, OsRH families = families with at least one OsRH gene, non-OsRH families = families with no OsRH gene, AtRHM families = families with at least one AtRHM gene, and AtRH(-RHM) families = families that do not contain an AtRHM gene and contain at least one AtRH gene. P values were corrected by the Bonferroni method to control for multiple testing error rate.
| Test Design Table | AtRH Families | Non-AtRH Families | |
|---|---|---|---|
| OsRH families | |||
| Non-OsRH families | |||
| Rice gene(s) per family | |||
| Arabidopsis gene(s) per family | 1 | 2 | 3 |
| 1 | 1.6699E-152 | 5.66047E-34 | 1.125E-09 |
| 2 | 1.85754E-25 | 6.73908E-17 | 6.9186E-10 |
| 3 | 7.94086E-06 | 1.15231E-05 | 0.006749827 |
| Test Design Table | AtRH(-RHM) Families | Non-AtRH Families | |
|---|---|---|---|
| OsRH families | |||
| Non-OsRH families | |||
| Rice gene(s) per family | |||
| Arabidopsis gene(s) per family | 1 | 2 | 3 |
| 1 | 3.2434E-154 | 1.68647E-34 | 1.1001E-09 |
| 2 | 3.9281E-25 | 6.8512E-17 | 1.37369E-09 |
| 3 | 3.58292E-06 | 1.45424E-05 | 0.006405968 |
| Test Design Table | AtRHM Families | Non-AtRH families | |
|---|---|---|---|
| OsRH families | |||
| Non-OsRH families | |||
| Rice gene(s) per family | |||
| Arabidopsis gene(s) per family | 1 | 2 | 3 |
| 1 | 1 | 1 | 1 |
| 2 | 0.000578923 | 0.239459721 | 0.016056802 |
| 3 | 1 | 0.072914349 | 0.905572755 |
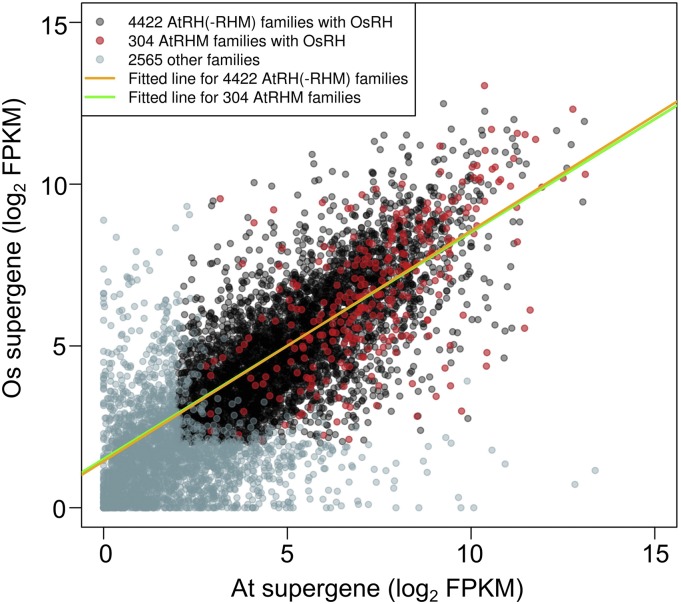
Next, we compared the relative levels of root hair expression from Arabidopsis and rice genes present in the same family. To avoid complications associated with differences in gene number per family between these species, we calculated the total root hair transcript accumulation for all Arabidopsis genes (by summing FPKM values from the AtCOBL9::GFP data set) and for all rice genes (by summing FPKM values from the OsEXPA30::GFP data set) from each individual family (Supplemental Data Set S5). These aggregate genes are referred to as supergenes. A comparison of the transcript levels for Arabidopsis and rice supergenes from the same families reveals a strong positive correlation (Pearson’s r = 0.77; Fig. 3), indicating similar total root hair expression for Arabidopsis and rice genes in the same family.
Figure 3.
Comparison of Arabidopsis and rice supergene expression from common families. The distribution of GreenPhyl-defined gene families is based on combined transcript levels (FPKM, log2 scaled) for all Arabidopsis genes (from FACS-purified AtCOBL9::GFP) and for all rice genes (from FACS-purified OsEXPA30::GFP) from each of the 7,291 families that possess at least one Arabidopsis gene and one rice gene. Pearson’s correlation coefficient is r = 0.77 for the total 7,291 families. Least-square fitted lines were generated for the 304 AtRHM families containing one or more OsRH gene (red dots, green line; adjusted r2 = 0.39) and the 4,422 AtRH(-RHM) families containing one or more OsRH gene (black dots, orange line; adjusted r2 = 0.51).
We also analyzed the possibility that the developmental profile of gene expression is conserved for those AtRH and OsRH genes present in the same families. Using Fisher’s exact test, we discovered a significant preferential familial association of profile types between AtRH and OsRH genes (P < 0.01, corrected value; Supplemental Table S3). Thus, in addition to possessing sequence similarity, Arabidopsis and rice root hair genes from the same families also tend to exhibit similar transcript levels and developmental patterns of gene expression.
In our next series of experiments, we compared the rice root hair (OsRH) genes with the subset of Arabidopsis root hair genes associated with root hair morphogenesis (i.e. the AtRHM genes). As above, we first assessed the association between the AtRHM genes and OsRH genes within gene families. Surprisingly, unlike the strong familial association of AtRH genes with OsRH genes (and non-AtRH genes with non-OsRH genes), the AtRHM genes do not associate strongly with OsRH genes within families (Table I). This suggests that, as a group, the AtRHM genes exhibit less similarity to rice root hair-expressed genes than do the other AtRH genes.
To extend this, we constructed phylogenetic trees containing the Arabidopsis and rice genes from each AtRHM GreenPhyl family and identified well-supported subfamilies within these that possess at least one Arabidopsis gene and one rice gene (see “Materials and Methods”; Supplemental Fig. S1). Consistent with our family-level results, we discovered that subfamilies containing AtRH(-RHM) genes preferentially included OsRH rather than non-OsRH rice genes (P < 0.01, Fisher’s exact test) but subfamilies containing AtRHM genes did not exhibit a statistically significant preference for OsRH genes over non-OsRH genes (P = 0.45, Fisher’s exact test).
These results suggest greater diversification of the genes in the AtRHM families, relative to the AtRH(-RHM) families, between Arabidopsis and rice. If so, we might expect that a greater fraction of the AtRHM families would lack a rice gene member entirely, relative to the AtRH families. Indeed, controlling for family size (one to three Arabidopsis genes per family), 25.4% of the AtRHM-containing GreenPhyl gene families lacked a rice gene, whereas only 12% of the AtRH(-RHM)-containing gene families lacked a rice gene member (Supplemental Table S4).
We also compared the expression levels of the AtRHM and AtRH(-RHM) supergenes versus OsRH supergenes from the same family to determine whether transcript levels also have diversified preferentially in the AtRHM families. We discovered a significant difference (P < 0.01, Student’s t test, Bonferroni corrected) between mean transcript levels from AtRHM and OsRH supergenes from the same family but not between AtRH(-RHM) and OsRH supergenes from the same family (Supplemental Fig. S2). Furthermore, as shown in Figure 3, the adjusted r2 value for AtRHM versus OsRH is smaller than AtRH versus OsRH (0.39 versus 0.51), showing that AtRHM versus OsRH exhibits more variation (greater scatter in the plot) that cannot be explained by the regression model.
We also analyzed the degree of similarity in gene expression profiles for families containing AtRHM and OsRH genes. In contrast to the results from this test using the entire set of AtRH genes, we did not find a significant association of Arabidopsis and rice genes possessing the same expression profile within these AtRHM families (Supplemental Table S3).
Together, these findings indicate that AtRHM-related genes in rice are less conserved in structure and expression compared with AtRH(-RHM)-related genes, suggesting preferential divergence in the root hair developmental program used by Arabidopsis and rice.
Root Hair Development Gene Relatives in Other Plant Species
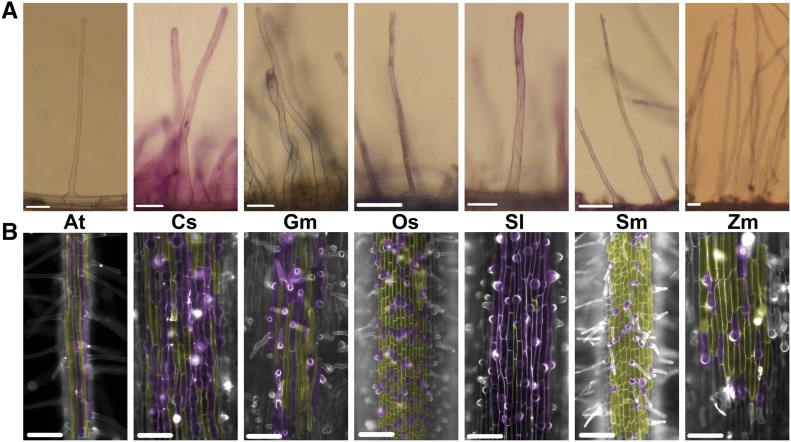
To determine whether rice is unique among vascular plants in its divergent AtRHM-related genes, we analyzed relatives of these root hair genes in four additional angiosperm species (soybean [Glycine max], tomato [Solanum lycopersicum], maize [Zea mays], and cucumber [Cucumis sativus]) and in a lycophyte species (Selaginella moellendorffii; Fig. 4A). First, we analyzed the composition of AtRHM and AtRH(-RHM) gene families to determine whether these additional species possess related genes. Consistent with our results with rice, we found that a greater fraction of the AtRHM families lacked genes from these species as compared with the AtRH(-RHM) families (approximately 2-fold difference for each species; Supplemental Table S4). Thus, the preferential divergence of AtRHM-related genes does not appear to be unique to rice.
Figure 4.
Root hairs in diverse vascular plants. A, Photographs of individual root hairs from Arabidopsis (At), cucumber (Cs), soybean (Gm), rice (Os), tomato (Sl), Selaginella (Sm), and maize (Zm). Bars = 50 μm. B, Root epidermis from Arabidopsis, cucumber, soybean, rice, tomato, Selaginella, and maize roots stained with fluorescent dye (Fluorescent Brightener 28 or propidium iodide). The root hair cells were pseudocolored in purple, and the nonhair cells were pseudocolored in yellow. Only the Arabidopsis root possesses the longitudinal file-specific (type 3) pattern of root hair cells; the other species exhibit a random distribution of root hair cells (type 1). Bars = 100 μm.
Next, we analyzed the overall degree of conservation of AtRHM-related genes and gene expression among these seven vascular plant species. For each of the 397 GreenPhyl-defined AtRHM gene families, we assigned each species a similarity score based on whether the species possesses a gene in that family and the degree to which its gene(s) matches the root expression profile of the AtRHM gene (see “Materials and Methods”; Supplemental Methods S1). The comparative analysis of these similarity scores yielded a species-wise hierarchical clustering with a tree topology that mirrored the evolutionary relationships between the species (Fig. 5; Supplemental Data Set S6), indicating that changes in the gene family structure and expression are positively correlated with the divergence time from common ancestors. The family-wise groupings, generated by hard cutoffs of the similarity scores, produced distinct clusters of gene families with common across-species AtRHM relationships (Fig. 5; see “Materials and Methods”).
Figure 5.
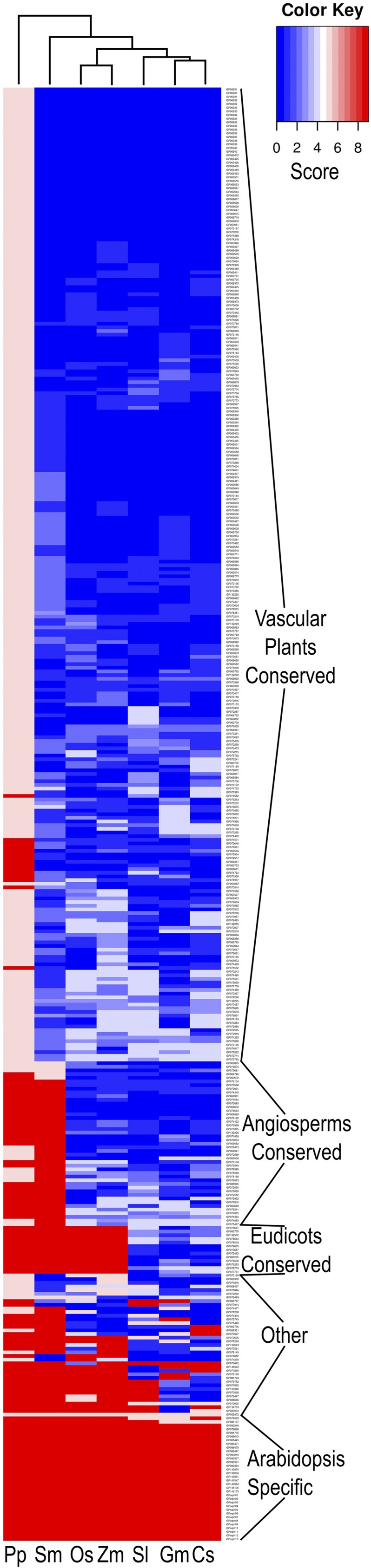
Conservation of Arabidopsis root hair morphogenesis genes in other plants. A differential matrix heat map was generated for the AtRHM-containing GreenPhyl gene families. Each species (from left to right: P. patens, Selaginella, rice, maize, tomato, soybean, and cucumber; ordered by hierarchical clustering) was scored for its degree of conservation (blue = highest and red = lowest) based on the presence/absence of a gene and its expression profile, relative to the AtRHM gene in each family. Major categories of AtRHM genes are indicated; a detailed list of these is given in Supplemental Data Set S6.
The largest cluster of gene families, designated vascular plant conserved, includes 266 AtRHM families that possess a root-expressed gene from each of the plant species tested, indicating that these are the most ancient families and likely contain genes with common root hair functions shared by all vascular plants (Fig. 5). This cluster includes many of the well-characterized Arabidopsis root hair genes (e.g. EXPA7, IRT2, AHA7, RHD2, LRX1, COW1, MRH1, MRH6, IRE, and PIP5K3; Supplemental Data Set S6) and includes a disproportionate share (93%) of the AtRHM genes encoding secretory pathway activities. It is noteworthy that the degree of conservation of the AtRHM root developmental expression profile varies among these families (Fig. 5), suggesting that the regulation or developmental role of these genes has diverged in some of the families.
A second cluster of gene families, angiosperm conserved, possesses root-expressed AtRHM-related genes from all six angiosperms, but Selaginella either lacks a related gene or lacks root expression of its gene (Fig. 5), suggesting that these root hair gene functions arose after the lycophyte-euphyllophyte split. These AtRHM genes encode a relatively high proportion (40%) of putative regulatory proteins (e.g. AP2-, GATA-, and WRKY-related transcription factors and various protein kinases; Supplemental Data Set S6) that may have evolved to provide angiosperms new mechanisms to control root hair growth.
A cluster designated eudicot conserved includes 13 families of AtRHM-related genes that possess root-expressed members exclusively from the four eudicot species tested. Another cluster, Arabidopsis specific, includes 34 families that do not possess a root-expressed AtRHM-related gene from any of the other six species tested. These two clusters are dominated (10 of 13 and 22 of 34) by genes encoding unknown or uncharacterized proteins (Supplemental Data Set S6), which may contribute to novel species- or lineage-specific root hair features. The Arabidopsis-specific cluster also contains six families encoding cell wall-related proteins, including an arabinogalactan protein (AGP3) and several Pro-rich family proteins.
A final cluster of gene families, designated other, contains unusual distributions of AtRHM-related genes among the species, consistent with relatively rare lineage-specific gene loss/gain (Fig. 5). For instance, the family containing the Arabidopsis FERRIC REDUCTION OXIDASE4 (FRO4) and FRO5 genes include root-expressed genes from all vascular plant species tested except rice and maize. This implies the loss of this root hair-related gene activity during monocot evolution, perhaps associated with distinct strategies used by grasses for iron acquisition (Jain et al., 2014).
We also analyzed these GreenPhyl-defined AtRHM families for the presence of related genes from the moss Physcomitrella patens. Interestingly, although moss lacks roots and root hairs, we found that most of the AtRHM gene families (277 of 397) contain a P. patens gene (Fig. 5; Supplemental Data Sets S4 and S6), implying that these root hair developmental genes evolved from genes possessing a related function in a root-hairless ancestor of vascular plants. Conversely, we find that 20 of the AtRHM families lack a P. patens gene member yet possess a root-expressed gene from Selaginella and at least one angiosperm, which defines families likely to have arisen during vascular plant evolution coincident with the evolution of root hairs (Supplemental Data Set S6).
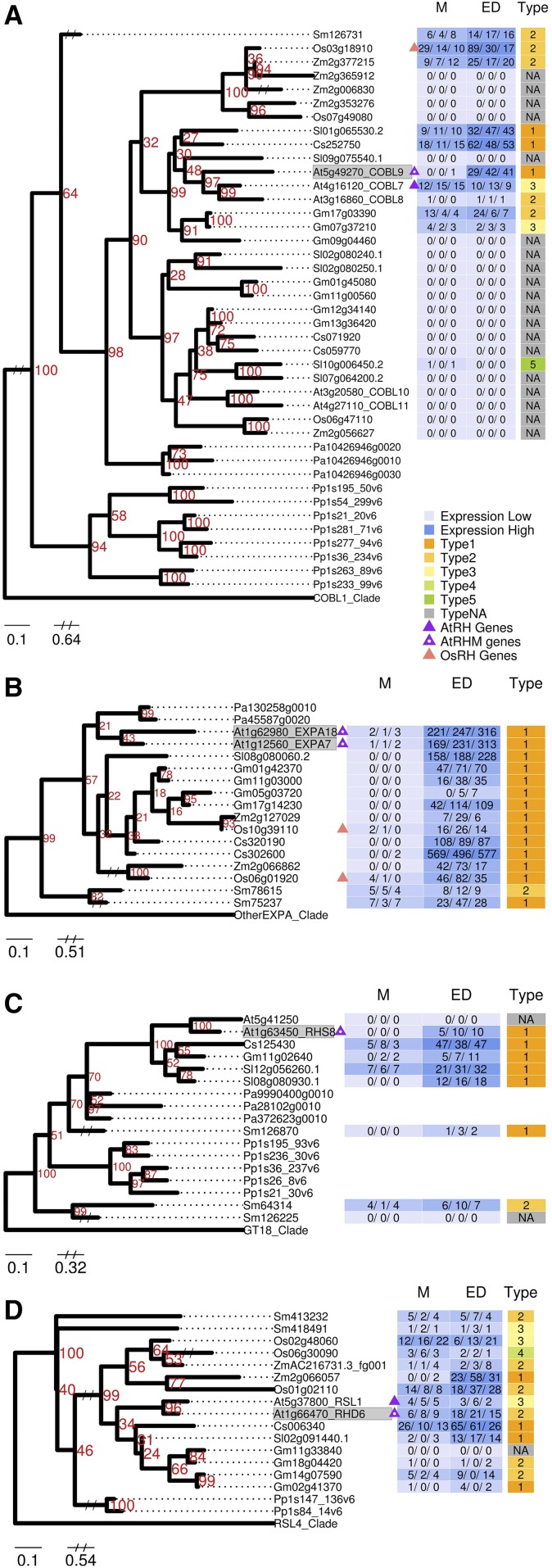
To rigorously analyze specific families containing AtRHM genes with a demonstrated role in root hair development, we constructed maximum likelihood phylogenetic trees using the protein sequences from relatives of the 18 AtRHM genes that, when mutated, exhibit an abnormal root hair phenotype (Supplemental Table S5). For this analysis, we included related genes from the seven vascular plant species and their associated root developmental zone transcript data as well as related genes from P. patens and Norway spruce (Picia abies). A total of 15 trees were generated from these 18 AtRHM genes (Fig. 6; Supplemental Fig. S3), and overall, the gene relationships largely mirrored the results obtained from the similarity score clustering analysis described above. For example, in the COBL9 family (one of the vascular plant conserved families from Fig. 5), each species possesses a gene with strong similarity in sequence and transcript accumulation to the Arabidopsis COBL9 (Fig. 6A). In a few trees, we observed variation in the structure or expression of AtRHM-related genes in certain species. The EXPA7 gene tree contains a well-supported clade including root-expressed EXPA7/EXPA18-related genes from all vascular plant species, but not from P. patens (Fig. 6B), which suggests that this expansin subgroup may have evolved (in part) for use in root hair development. Consistent with this, the two rice genes from this clade (Os10g39110 and Os06g01920) have previously been shown to participate in root hair differentiation (Kim et al., 2006; ZhiMing et al., 2011). In another case, the RHS8 gene is part of a well-supported clade that lacks rice and maize members but contains root-expressed genes from all other species tested (Fig. 6C), which suggests a monocot lineage-specific loss of this AtRHM gene/function. Altogether, these results indicate that the AtRHM-related gene divergence uncovered in the similarity score matrix analysis (Fig. 5) likely represents an underestimate of the actual variation in AtRHM-related gene function across vascular plants. More generally, these results demonstrate the utility of a combined phylogenetic and transcriptomic approach, enabling a high-resolution view of the likely evolutionary and functional relationships between genes in large families.
Figure 6.
Representative maximum likelihood phylogenetic trees of AtRHM gene families. A, COBL9. B, EXPA7. C, RHS8. D, RHD6. For each tree, the defining Arabidopsis AtRHM gene(s) associated with a mutant phenotype is shaded in gray. Gene expression FPKM values are shown for each replicate and converted to a heat map with high expression in darker blue and low expression in light blue. Expression profile types generated from the fold change between two developmental zones are shown in different colors as indicated in the key. Triangles indicate AtRHM genes (purple with white dot), AtRH genes (solid purple), and OsRH genes (pink). Numbers in red indicate support levels from 1,000 bootstrap. Gene identifiers are abbreviated (Arabidopsis, At; cucumber, Cs; soybean, Gm; rice, Os; tomato, Sl; Selaginella, Sm; maize, Zm; P. patens, Pp; Norway spruce, Pa). M, Meristematic zone; ED, combined elongation plus differentiation zones; Type, expression profile types. The complete set of 14 AtRHM family trees is presented in Supplemental Figure S3.
We also generated a maximum likelihood tree for the RHD6-related genes from these species. We find that RHD6 is included in a well-supported clade that contains the partially functionally redundant Arabidopsis RSL1 gene (Menand et al., 2007) as well as root-expressed genes from each of the other species examined (Fig. 6D), consistent with a previous study showing broad conservation of the RHD/RSL gene sequence (Pires et al., 2013). It is notable that each of these species possesses an RHD6-related gene with transcript accumulation in the meristematic region of the root, similar to Arabidopsis RHD6 (Fig. 6D), implying that each of these species might use an RHD6 homolog to regulate early root hair cell differentiation.
Root Hair-Expressed Genes
To analyze root hair gene expression more broadly, including genes expressed in mature root hairs, we defined transcript accumulation in purified root hair preparations. Using a previously established method (Lee et al., 2008), root hairs were isolated from seedling roots and rapidly frozen in liquid nitrogen, and RNA was purified and subjected to RNA-seq analysis (three biological replicates each; see “Materials and Methods”). Since this isolation method is not restricted to plant species with available root hair GFP marker lines, it allowed for a comprehensive analysis of root hair gene expression across species. However, this method does not capture differentiating cells prior to hair emergence, so it is not likely to identify root hair genes that are expressed primarily at early developmental stages (e.g. genes regulating root hair cell fate).
We first analyzed transcripts from isolated Arabidopsis root hairs using this method and discovered 14,919 expressed genes (mean FPKM ≥ 3; >0 FPKM in ≥2 replicates; Supplemental Data Set S7), which we designate AtHAIR genes. These include 11,041 (87%) of the 12,691 AtRH genes and 501 (89%) of the 563 AtRHM genes (Supplemental Data Sets S1 and S2), indicating substantial overlap between the gene transcripts defined by our two root hair cell isolation methods (FACS of root hair-specific GFP expression and direct root hair purification). Consistent with this, we also observed an overall positive correlation in transcript levels obtained from these two root hair cell isolation methods (Pearson’s r = 0.78; Fig. 7A). Furthermore, we discovered that the 1,650 AtRH genes and the 62 AtRHM genes that are not present in the AtHAIR gene set are significantly underrepresented for temporal expression profiles associated with transcript accumulation in the differentiation zone (i.e. expression profile types 6–9; P < 0.001 for each comparison, χ2 test; Supplemental Data Set S1). This indicates that, as expected, transcriptomes generated from the direct purification of root hairs are less likely to define root hair genes preferentially expressed at early developmental stages.
Figure 7.
Gene expression from isolated root hairs. A, Scatterplot comparing transcript accumulation from isolated Arabidopsis root hairs and from the FACS-purified Arabidopsis COBL9::GFP line. B, Bar graph displaying the number of root hair-expressed (HAIR) genes from each of six species, subdivided by genes present in conserved families (black bars), nonconserved families (hatched bars), and species-specific families (white bars). C, Bar graph showing the number of families that contain a root hair-expressed (HAIR) gene (white bars) or a root-expressed gene (black bars) from Arabidopsis, rice, cucumber, soybean, tomato, and/or maize among the 17,042 total GreenPhyl families that possess at least one gene from each species.
Next, we generated transcript data from isolated root hairs from rice, tomato, soybean, cucumber, and maize, which yielded 12,229 OsHAIR genes, 12,970 SlHAIR genes, 16,652 GmHAIR genes, 12,622 CsHAIR genes, and 14,471 ZmHAIR genes, respectively (mean FPKM ≥ 3; >0 FPKM in ≥2 replicates; Supplemental Data Sets S8–S12). By comparing the distribution of these HAIR genes among GreenPhyl-defined gene families, we were able to identify root hair genes that are shared or are unique in these species. First, we discovered that the majority (65%–75%) of the HAIR genes from each species are members of gene families that include a HAIR gene from all six species (Fig. 7B; Supplemental Data Set S4). These 3,511 conserved root hair gene families likely contain genes important for root hair cell formation, metabolism, and/or function in all angiosperms. Similarly, 79% of the 1,363 Arabidopsis root hair proteins identified in a previous proteome study (Petricka et al., 2012) are encoded by members of these conserved families (Supplemental Data Set S4). In addition, we found that 3,026 (86%) of these 3,511 families also possess a root-expressed gene from Selaginella (Supplemental Data Set S4), implying that these are shared by all vascular plants, and, therefore, provide an estimate of the minimum gene set necessary for the root hair cell type. Interestingly, 2,882 (95%) of these 3,026 families also possess a related gene from the rootless moss P. patens, implying an ancient origin for the majority of these conserved root hair-expressed genes.
In addition to the 3,511 conserved families that possess a HAIR gene from all six species, we discovered that a large number of families (4,177) possess HAIR genes from a subset (two to five) of these six species (Supplemental Data Set S4). These nonconserved HAIR gene families likely result from diversification in the root hair gene expression program across these species, due to lineage-specific gain or loss of genes and/or root hair expression. Indeed, we found that the proportion of families sharing HAIR genes from different species is largely related to their phylogenetic relationships. For example, the largest fraction (32%) of the 1,174 families containing HAIR genes from exactly two species contained rice and maize HAIR genes, presumably representing monocot-specific root hair genes (Supplemental Data Set S4). Notably, the overall proportions of these nonconserved HAIR gene families is much greater than the equivalent nonconserved root-expressed gene families identified previously from these same species (controlling for family size; Fig. 7C; Huang and Schiefelbein, 2015). Thus, like the preferential divergence of AtRHM-related genes (described above), these results suggest a relatively greater degree of diversification of HAIR genes during the evolution of these species.
Finally, the comparison of these HAIR gene data sets enabled us to define 2,623 species-specific root hair gene families (i.e. families that possess a root hair-expressed gene from only one of the species; Supplemental Data Set S4). These range in number from 278 to 595 per species and likely reflect root hair functions unique to each species. Using GO analysis of the Arabidopsis-specific HAIR genes, we find an array of enriched terms (including several ubiquitin-related and DNA-associated terms) among this gene set, implying that the evolution of diverse gene functions is responsible for these (Supplemental Table S6). Notably, we find that these species-specific HAIR gene families include a substantial fraction (14%) of families that contain related genes (but not HAIR genes) from all five of the other species as well as a large fraction (52%) of families that contain no genes from the other five species. These results indicate that modifications in gene expression as well as gene structure were responsible for the evolution of these species-specific HAIR genes.
Root Hair Patterning Gene Relatives in Diverse Plants
In addition to analyzing root hair-expressed genes, we also sought to determine whether early-acting genes responsible for patterning root hair cells might be conserved across vascular plant species. Arabidopsis is unique among the plants analyzed in this study because its root hair pattern is position dependent, with root hair cells limited to longitudinal cell files in particular locations (type 3), whereas the other six species produce root hair and nonhair epidermal cells in a random distribution (type 1; Fig. 4B; Clowes, 2000; Pemberton et al., 2001; Balcerowicz et al., 2015; Salazar-Henao et al., 2016). We generated maximum likelihood trees and analyzed root gene expression for relatives of 12 Arabidopsis patterning genes (present in seven gene families; Supplemental Table S7; Supplemental Fig. S4). In Arabidopsis, each of these genes acts early in root epidermis development (beginning in the meristematic zone) and ultimately regulates RHD6 transcription to specify the root hair cell pattern (Grierson et al., 2014; Balcerowicz et al., 2015; Salazar-Henao et al., 2016).
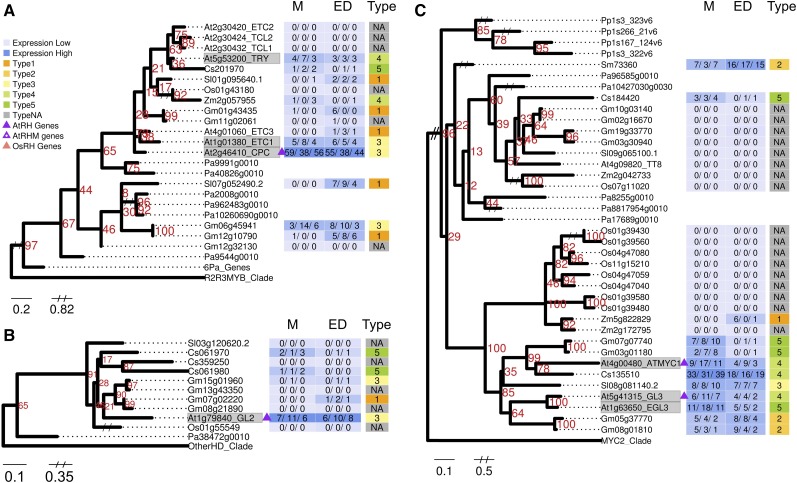
The CPC/TRY/ETC1 patterning genes encode small one-repeat MYB proteins (Wada et al., 1997; Kirik et al., 2004), and we found that they are all present in a clade that includes genes from all euphyllophytes, but only cucumber and soybean genes share similar consistent meristem zone transcript accumulation (Fig. 8A). The GL2 gene encodes an HD-Zip transcription factor that promotes the nonhair fate (Masucci et al., 1996), and it occupies a well-supported clade containing root-expressed genes from cucumber and soybean only (Fig. 8B). The GL3, EGL3, and MYC1 genes encode partially redundant bHLH proteins (Bernhardt et al., 2003; Bruex et al., 2012), and our maximum likelihood tree shows that they reside in a subgroup that contains meristem zone-expressed genes from eudicots only (Fig. 8C). Similarly, we found conservation of gene structure and root expression in eudicots only for our maximum likelihood tree containing the TTG2 gene (Supplemental Fig. S4), which encodes a WRKY transcription factor (Johnson et al., 2002). These four trees are similar in showing conservation among (some) eudicots only, suggesting functional divergence for these patterning genes during eudicot evolution or, possibly, the loss of gene/function in the monocot lineage.
Figure 8.
Representative maximum likelihood phylogenetic trees of Arabidopsis root hair patterning genes. A, CPC/TRY/ETC1. B, GL2. C, GL3/EGL3/MYC1. For each tree, the defining Arabidopsis root hair patterning gene(s) is shaded in gray. Gene expression FPKM values are shown for each replicate and converted to a heat map with high expression in darker blue and low expression in light blue. Expression profile types generated from the fold change between two developmental zones are shown in different colors as indicated in the key. Triangles indicate AtRHM genes (purple with white dot), AtRH genes (solid purple), and OsRH genes (pink). Numbers in red indicate support levels from 1,000 bootstrap. Gene identifiers are abbreviated (Arabidopsis, At; cucumber, Cs; soybean, Gm; rice, Os; tomato, Sl; Selaginella, Sm; maize, Zm; P. patens, Pp; Norway spruce, Pa). M, Meristematic zone; ED, combined elongation plus differentiation zones; Type, expression profile types. The complete set of seven patterning gene family trees is presented in Supplemental Figure S4.
Two other patterning genes exhibit broader potential conservation. The TTG1 gene, encoding a WD protein required to repress root hair specification (Galway et al., 1994), is in a clade with similarly root-expressed genes from all vascular plants tested (Supplemental Fig. S4). The SCM (aka SUB) gene encodes a receptor-like kinase that influences the positional expression of the other patterning genes (Kwak et al., 2005; Kwak and Schiefelbein, 2007), and we find root-expressed SCM-related genes in each of the vascular plant species tested (Supplemental Fig. S4). However, SCM’s preferential meristematic transcript accumulation is only shared by SCM-related genes from eudicots.
The R2R3 MYB transcription factors WER and MYB23 are partially redundant and negatively regulate root hair differentiation (Lee and Schiefelbein, 1999; Kang et al., 2009). Our maximum likelihood analysis places these two MYB genes in a clade (previously defined as MYB subgroup 15; Stracke et al., 2001) that also includes the trichome development gene GLABROUS1 (GL1; Oppenheimer et al., 1991) but does not include related genes from any of the other plant species tested.
The substantial divergence in the structure and expression of these patterning genes across vascular plant species strongly suggests that these are not generally used to specify root hair cells in all vascular plants. Altogether, our analysis of root hair genes from these species provides a broad outline of the evolution of genes controlling root hair development in vascular plants (Fig. 9).
Figure 9.
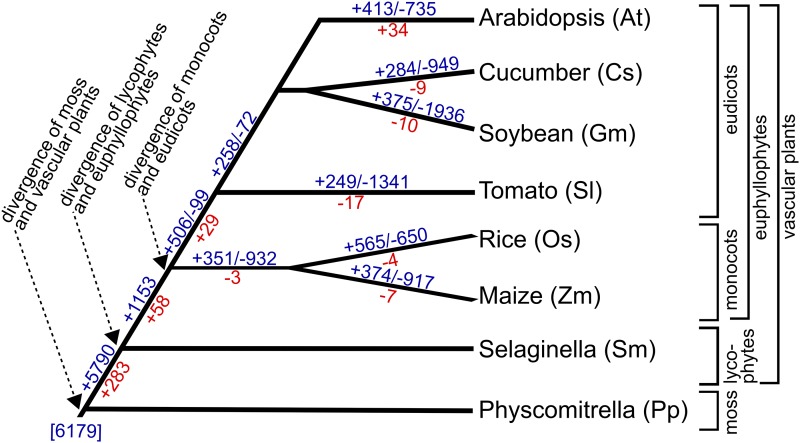
Summary of the evolutionary history of root hair gene families. The tree indicates the distribution and putative origin of root hair gene expression in the 10,311 gene families (blue numbers) possessing one or more root hair-expressed genes identified in this study. Positive numbers refer to putative lineage-specific gain of families containing root hair-expressed genes, and negative numbers refer to putative lineage-specific loss of families containing root hair-expressed genes. The numbers in red specifically indicate the evolutionary history of the 404 key Arabidopsis root hair gene families (397 AtRHM + seven patterning gene families). Genes related to 6,179 of these families also are present in the root hair-less plant P. patens, indicated at the base of the tree (in brackets). For discussion, see text.
DISCUSSION
This large-scale study combined phylogenetic and transcriptome analyses to define and compare root hair genes from seven diverse vascular plant species, including eudicots, monocots, and a lycophyte. To our knowledge, this provides the first comprehensive view of gene expression in a single plant cell type across multiple species. A major finding was that most root hair genes are similar in structure and expression in all species tested, suggesting that a core program for root hair development and function is conserved across the vascular plants. Indeed, we were able to define ∼3,000 such conserved root hair gene families, which provides an initial estimate for the minimal gene set necessary for the root hair cell type.
Furthermore, we found that nearly all of the vascular plant-conserved root hair-expressed genes (2,882 of 3,026) and putative root hair development genes (251 of 266) possess close relatives in the rootless moss P. patens, implying that the core root hair program did not evolve de novo in the vascular plant lineage but likely was coopted from a preexisting program in a land plant ancestor. An attractive possibility is that this ancient program was responsible for the unidirectional cellular growth (tip growth) of exploratory or invasive cell types in the ancestral species and was recruited for tip-growing root hair cells during vascular plant evolution. Related to this, we found that a disproportionate share (93%) of the AtRHM genes encoding predicted secretory pathway proteins (likely involved in tip growth) are among this vascular plant-conserved group. Furthermore, cellular and physiological mechanisms employed by tip-growing cells are similar across different groups of organisms, including fungi, bryophytes, and vascular plants, consistent with the possibility of an evolutionarily ancient underlying program (Geitmann and Emons, 2000; Jones and Dolan, 2012; Rounds and Bezanilla, 2013; Sanati Nezhad and Geitmann, 2013).
In addition to identifying conserved root hair genes, we also discovered significant diversification in the genetic program associated with root hair development among the vascular plants. We initially discovered this by comparing root hair-expressed genes from Arabidopsis and rice. Specifically, we found that the Arabidopsis root hair development (AtRHM) genes exhibit significantly greater divergence in their structure and expression from their rice relatives (within the same gene families) compared with non-AtRHM root hair-expressed genes. This was unexpected, because we had previously found that root-expressed genes are generally conserved between Arabidopsis and rice (Huang and Schiefelbein, 2015). The underlying reason for the preferential divergence of root hair development genes is unclear. It may be that, as a single cell type, the root hair may be relatively less constrained in its developmental options, due to minimal coordination with neighboring cells. Another possibility is that, as a cell that extends from the plant body into the rhizosphere, the root hair may evolve and utilize multiple developmental strategies to effectively interact with and adapt to a varying environment. In support of this, root hair growth in many species is known to be strongly influenced by nutrient availability (Perry et al., 2007; Nestler et al., 2016; Salazar-Henao et al., 2016). Alternatively, the strong selection for high yield imposed on most of these species during their domestication may be responsible for the high degree of root hair gene divergence. Future studies that include closely related wild species will help to resolve this issue.
Overall, approximately one-third of the root hair-expressed genes from each species, as well as one-third of the Arabidopsis root hair development genes, differ substantially in structure or expression in one or more of the other six vascular plant species tested. Considering that the conserved root hair genes may define a core root hair growth program (as discussed above), then these diverged genes may be responsible for regulating and/or modifying this core program in ways appropriate for particular species or lineages. The proportion of diverged genes within species largely followed phylogenetic lines, with Selaginella exhibiting the greatest differences in gene structure and expression (Fig. 9). It is notable that, among the AtRHM gene families lacking a Selaginella gene, those encoding putative regulatory proteins were highly represented, suggesting that new mechanisms of root hair developmental control evolved following the divergence of lycophytes and euphyllophytes. Among the Arabidopsis-specific AtRHM genes, those encoding proteins with unknown or uncharacterized functions were overrepresented, which may prove fruitful for further study to understand the evolution of novel cell type developmental activities or characteristics.
The analysis of Arabidopsis genes controlling root hair patterning was of particular interest in this study, because Arabidopsis differs from the other analyzed species by producing a particular pattern of root hair cells (dependent on cell position; type 3) rather than a random distribution of root hair cells (type 1) in the root epidermis (Clowes, 2000; Pemberton et al., 2001). Consistent with this, we detected greater divergence in gene structure and expression within the seven families of Arabidopsis genes involved in patterning compared with families containing AtRHM genes. In particular, five of these seven families possess a clade that includes the Arabidopsis patterning genes but lack a related root-expressed gene from one or more of the other angiosperm species. These results strongly suggest a linkage between the structure/expression of these patterning genes and the evolution of the type 3 root hair pattern in Arabidopsis. Furthermore, this implies that the type 1 root hair distribution mechanism relies on other, as yet unknown, cell fate regulators. In this respect, it is notable that all of these species possess and express an RHD6-related bHLH gene similar to Arabidopsis RHD6 (Figs. 6D and 8). Indeed, it has been shown previously that RHD6 homologs are widespread and function similarly in divergent species (Menand et al., 2007; Pires et al., 2013), suggesting that RHD6 acts as the critical regulator of root hair initiation in all vascular plants. Given that the Arabidopsis root hair-patterning genes specify cell fate via the transcriptional regulation of RHD6 (Grierson et al., 2014; Balcerowicz et al., 2015), type 1 species may similarly achieve their root hair cell distribution by regulating their RHD6 homologs, but employing a different mechanism to do so.
Among the seven families containing Arabidopsis patterning genes, the WER/MYB23 family was unique in possessing its patterning genes in an Arabidopsis-specific subgroup (previously defined as MYB subgroup 15; Stracke et al., 2001). Thus, it is tempting to speculate that the evolution of the WER/MYB23 genes was the critical factor in the origin of the Arabidopsis type 3 pattern. However, a recent extensive analysis of MYB genes in multiple species showed that subgroup 15 includes members from several type 1 eudicots (Du et al., 2015), complicating the potential linkage between this subgroup and the type 3 pattern. Interestingly, the patterning of epidermal hairs (trichomes) on the leaf surface of Arabidopsis also relies on a member of this MYB subgroup 15, the GL1 gene (Larkin et al., 1993), implying shared evolution of these patterning mechanisms. This study provides a foundation for further analyses of the evolutionary events responsible for the origin of these cell type patterns.
MATERIALS AND METHODS
Plant Materials and RNA Isolation
For the analysis of root hair gene expression, seeds of the Arabidopsis (Arabidopsis thaliana) COBL9::GFP line (Brady et al., 2007b) and the rice (Oryza sativa) EXPA30::GFP line (Kim et al., 2006) were grown on agarose-solidified nutrient medium under constant light as described previously (Schiefelbein and Somerville, 1990). The growing tips of the seedling primary roots were cut, and FACS/protoplasting was performed as described (Bruex et al., 2012). Total RNA was extracted from frozen samples using the Qiagen RNeasy Plant Mini Kit. Library construction was carried out by the University of Michigan Sequencing Core using the Illumina TruSeq Kit followed by sequencing on the Illumina HiSeq 2000 System.
Root hairs were physically isolated from seedling roots using a previously published protocol (Lee et al., 2008). Briefly, seeds were surface sterilized and incubated on Murashige and Skoog medium under continuous light. Seedling roots were harvested and either held with tweezers while submerged and agitated in liquid nitrogen (for Arabidopsis, tomato [Solanum lycopersicum], and soybean [Glycine max]) or placed in liquid nitrogen and stirred with glass rods (for rice, cucumber [Cucumis sativus], and maize [Zea mays]). Root hairs were then purified by filtration through a 250-μm-mesh membrane. RNA was extracted using the RNeasy Plant Mini Kit (Qiagen), and its purity was verified by analyzing the expression of cell-specific root genes, including homologs of the Arabidopsis ACTIN8, EXPANSIN7, SCARECROW, SHORTROOT, and PLETHORA1 genes. cDNA libraries were prepared using the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech).
Microscopy
Young seedlings of Arabidopsis and rice (4–5 d after plating) were stained with propidium iodide for 1 min, and the roots were examined with a Leica SP5 laser scanning confocal microscope. The excitation wavelength was 488 nm for the detection of GFP signals and 561 nm for the propidium iodide.
Young seedlings of all vascular plants were stained with Toluidine Blue for 5 to 10 s, and the roots and root hairs were examined with a Leica Laborlux S microscope or a Wild M420 Makroskop.
For the analysis of root hair distribution, the root epidermis of each species was examined with an Olympus IX81 after the root was stained with Fluorescent Brightener 28 for 30 to 60 s or propidium iodide for 1 min. The root hair cells were pseudocolored in purple, and the nonhair cells were pseudocolored in yellow.
RNA-seq Analysis
Sequencing reads were processed and analyzed as described previously (Huang and Schiefelbein, 2015). In brief, the first 15 bp of each 50-bp-long read was trimmed before mapping to a reference genome using TopHat (version 2.0.3; Kim et al., 2013) with default settings (–segment length 17). Gene expression was calculated using Cufflinks2 (version 2.1.1; Trapnell et al., 2013) with multiread correction (-u -G). Reads generated from rice samples were processed using an updated version of TopHat (version 2.0.9) with the other steps unchanged. Reference genomes and the annotation of Arabidopsis and rice were both downloaded from the Ensembl Plant database (version 19; http://plants.ensembl.org/index.html).
Gene Differential Expression Analysis
The number of raw counts mapped to each gene was quantified by HTSeq (version 0.6.1; Anders et al., 2015) with the following setting (-m intersection-strict -s no -f bam) and analyzed using edgeR (Robinson et al., 2010) for differential expression analysis. First, genes with expression lower than the cutoff (counts per million > 1 for at least three out of six samples) were filtered out. Second, raw counts were normalized using the default trimmed mean of M-values method, and the variation was modeled using a tag-wise dispersion. Next, the calculated P values were corrected for multiple testing by the method of Benjamini and Hochberg (1995). Significant differentially expressed genes were identified using a cutoff of fold change ≥ 2 and an FDR q value ≤ 0.01. The log2-scaled gene expression value was added to 1 before the log2 transformation.
Gene Family Information
The composition of gene families in the seven plant species was obtained from GreenPhyl (version 4; Rouard et al., 2011) and is presented in Supplemental Data Set S2.
For the analysis of family size, a set of 543 genes was randomly drawn from the total 12,449 AtRH genes (excluding genes that are not included in the GreenPhyl database), and the number of families of these 543 genes was recorded. The process was repeated 1,000 times, and the distribution of the number of families was plotted as a histogram.
Heat Map Construction
The distance matrix used to evaluate the expression similarity is included as Supplemental Methods S1. For each GreenPhyl-defined AtRHM family, the expression difference between each AtRHM gene and every gene from the other (non-Arabidopsis) species was calculated, and the minimal value for each species was used as the similarity score. The heat map was generated by the gplots package (https://cran.r-project.org/web/packages/gplots/index.html). The angiosperm data are based on a comparison of gene expression from three developmental zones (i.e. 10 profile types), and the Selaginella data are based on two zone comparisons (i.e. five profile types).
The groups of families with similar species distributions were defined as follows: vascular plant conserved, score = 0 to 4 in all vascular plants; angiosperm conserved, score = 0 to 4 in all angiosperms and score = 5 or 9 in Selaginella; eudicot conserved, score = 0 to 4 in all eudicots and score = 5 or 9 in maize, rice, and Selaginella; Arabidopsis specific, score = 5 or 9 in all vascular plants.
Subfamily Analysis of GreenPhyl Arabidopsis-Rice Families
To analyze Arabidopsis-rice subfamilies of the AtRHM GreenPhyl-defined families, Arabidopsis and rice protein sequences were obtained from each of the 304 GreenPhyl families that possess at least one AtRHM gene and one OsRH gene. Sequences from each family were aligned by MAFFT (version 6.864b; –genafpair–ep 0–maxiterate 1000; Katoh and Standley, 2013) if the tree included fewer than 200 genes; otherwise, an automatic parameter was used to align sequences (-auto). Phylogenetic trees were reconstructed using FastTree (version 2.1; -gamma; Price et al., 2009). Trees were rooted between two vascular clades, if applicable, or at the midpoint of the total tree and plotted by the ete2 package in Python (Huerta-Cepas et al., 2010). Well-supported (greater than 0.85) subfamilies containing at least one Arabidopsis gene and one rice gene were identified, and the distribution of AtRHM, AtRH(-RHM), non-AtRH, OsRH, and non-OsRH genes was analyzed within these subfamilies.
Statistical Analyses
All statistical analyses and graph plotting were performed in the R statistical computing environment (https://www.R-project.org) unless mentioned otherwise.
The built-in R function fisher.test was used to calculate the P value for Fisher’s exact test. The background total was GreenPhyl-defined families with a specific family size. A total of nine combinations of different family sizes were tested (permutations drawn from one to three Arabidopsis genes and one to three rice genes). All families that met the size requirement were divided into four groups for the test: expression in both species; expression in Arabidopsis only; expression in rice only; and no expression in either. The alternative hypothesis was that the observed data had greater association than expected from the null.
The Fisher’s exact test for the association of the temporal expression profiles between AtRH and OsRH genes followed the previous analysis (Huang and Schiefelbein, 2015), with the background total to be the families with exactly one AtRH gene and one OsRH gene, exactly one AtRH gene and two OsRH genes, and exactly two AtRH genes and one OsRH gene with the expression profile types 1 to 9. The AtRHM families were used for the association between AtRHM and OsRH genes.
Supergene Expression Analysis
Supergene expression was calculated as described previously (Huang and Schiefelbein, 2015). In brief, the FPKM expression values were summed for genes from the same family in a given species. For this analysis, only the expression values in the root hair cells were processed.
GO Term Enrichment Test
GO term enrichment analysis was performed by DAVID (http://david.abcc.ncifcrf.gov/) on the 563 AtRHM genes versus the background total of 33,550 Arabidopsis genes in the genome. Significantly enriched terms with Benjamini and Hochberg (1995) corrected P ≤ 0.01 are included in Supplemental Table S2.
Phylogenetic Analysis
Phylogenetic trees were reconstructed using maximum likelihood or an approximate maximum likelihood similar to previously published methods (Huang and Schiefelbein, 2015). Briefly, homologous sequences were identified using BLAST (version 2.2.26+; Camacho et al., 2009) and then clustered into groups. Groups with more than 200 members were aligned using MAFFT (version 6.864b; Katoh and Standley, 2013; -auto option), whereas smaller groups were aligned using MAFFT (-genafpair-ep 0-maxiterate 1000 option). Next, large family alignment (≥100) was sent to FastTree (version 2.1; Price et al., 2009; version 2.1.9; -gamma) for the approximate maximum likelihood tree reconstruction. A well-supported clade (a monophyletic clade with at least one gene from each species, unless the gene was included in another well-supported clade) with local support ≥ 0.85 and its neighboring well-supported clade (or the closest Arabidopsis gene with highest BLASTp score as outgroup) were realigned using MAFFT (-genafpair-ep 0-maxiterate 1,000 option). Alignment was trimmed using trimAl (version 1.2rev59; Capella-Gutiérrez et al., 2009) with the -automated 1 option. Finally, trees were reconstructed using RAxML (version 7.7.8; Stamatakis, 2006; -m PROTGAMMAJTTF -f a -N 1000). Trees were rooted between two well-supported clades or after the divergence of the Arabidopsis outgroup. The heat map aligned with the tree was generated using the ggtree package (https://www.bioconductor.org/packages/release/bioc/html/ggtree.html).
Accession Numbers
Sequence data from this article can be found in the Gene Expression Omnibus under accession number GSE85516.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. FastTree phylogenetic analysis of 304 GreenPhyl-defined families that contain at least one AtRHM gene and one OsRH gene.
Supplemental Figure S2. Box plot of the absolute expression difference between Arabidopsis and rice supergenes.
Supplemental Figure S3. Maximum likelihood phylogenetic trees for the AtRHM genes associated with a root hair mutant phenotype.
Supplemental Figure S4. Maximum likelihood phylogenetic trees for Arabidopsis genes required for root hair pattern formation.
Supplemental Table S1. Root hair genes identified by other studies.
Supplemental Table S2. GO-enriched terms for the 563 AtRHM genes versus the total 33,550 Arabidopsis genes.
Supplemental Table S3. Fisher’s exact test of association between AtRH/AtRHM and OsRH gene expression profiles.
Supplemental Table S4. Frequencies of the AtRHM/AtRH(-RHM) gene families lacking a member from other species.
Supplemental Table S5. AtRHM genes associated with a root hair mutant phenotype.
Supplemental Table S6. GO-enriched terms for the 1,349 Arabidopsis genes from 584 families versus the total 33,550 Arabidopsis genes.
Supplemental Table S7. List of Arabidopsis root hair patterning genes.
Supplemental Methods S1. Distance matrix for evaluation of the similarity of gene expression profiles between species.
Supplemental Data Set S1. List of 12,691 AtRH genes and their transcript accumulation characteristics.
Supplemental Data Set S2. List of 563 AtRHM genes and their transcript accumulation characteristics.
Supplemental Data Set S3. List of 13,342 OsRH genes and their transcript accumulation in rice root hair cells.
Supplemental Data Set S4. List of 18,110 GreenPhyl-defined gene families and distribution of the AtRH, AtRHM, OsRH, and HAIR genes within these families.
Supplemental Data Set S5. Supergene expression data for the 7,291 GreenPhyl families that contain at least one Arabidopsis gene and one rice gene.
Supplemental Data Set S6. Classification of AtRHM families according to similarity in sequence and expression with genes in other plant species.
Supplemental Data Set S7. List of 14,919 AtHAIR genes and their transcript accumulation.
Supplemental Data Set S8. List of 12,229 OsHAIR genes and their transcript accumulation.
Supplemental Data Set S9. List of 12,970 SlHAIR genes and their transcript accumulation.
Supplemental Data Set S10. List of 16,652 GmHAIR genes and their transcript accumulation.
Supplemental Data Set S11. List of 12,622 CsHAIR genes and their transcript accumulation.
Supplemental Data Set S12. List of 14,471 ZmHAIR genes and their transcript accumulation.
Acknowledgments
We thank Dr. Hyung-Taeg Cho for providing the OsEXPA30::GFP-expressing rice transgenic line and the University of Michigan Sequencing Core and Bioinformatics Core for assistance in the acquisition and analysis of sequence data.
Glossary
- bHLH
basic helix-loop-helix
- RNA-seq
RNA sequencing
- FACS
fluorescence-activated cell sorting
- FPKM
fragments per kilobase per million mapped reads
- FDR
false discovery rate
- GO
Gene Ontology
Footnotes
This work was supported by the National Science Foundation (grant no. IOS-1444400) and the University of Michigan Rackham Graduate School.
Articles can be viewed without a subscription.
References
- Anders S, Pyl PT, Huber W (2015) HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D. (2008) The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet 9: 868–882 [DOI] [PubMed] [Google Scholar]
- Balcerowicz D, Schoenaers S, Vissenberg K (2015) Cell fate determination and the switch from diffuse growth to planar polarity in Arabidopsis root epidermal cells. Front Plant Sci 6: 1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JD, Takeda S, Borges F, Dolan L, Feijó JA (2014) Transcriptional profiling of Arabidopsis root hairs and pollen defines an apical cell growth signature. BMC Plant Biol 14: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57: 289–300 [Google Scholar]
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J (2003) The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007a) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brady SM, Song S, Dhugga KS, Rafalski JA, Benfey PN (2007b) Combining expression and comparative evolutionary analysis: the COBRA gene family. Plant Physiol 143: 172–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, Kang YH, Bernhardt C, Xia Y, Zheng X, Wang JY, Lee MM, Benfey P, et al. (2012) A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowes FAL. (2000) Pattern in root meristem development in angiosperms. New Phytol 146: 83–94 [Google Scholar]
- Cormack RGH. (1935) Investigations on the development of root hairs. New Phytol 34: 30–54 [Google Scholar]
- Datta S, Kim CM, Pernas M (2011) Root hairs: development, growth and evolution at the plant-soil interface. Plant Soil 346: 1–14 [Google Scholar]
- Ding W, Yu Z, Tong Y, Huang W, Chen H, Wu P (2009) A transcription factor with a bHLH domain regulates root hair development in rice. Cell Res 19: 1309–1311 [DOI] [PubMed] [Google Scholar]
- Du H, Liang Z, Zhao S, Nan MG, Tran LS, Lu K, Huang YB, Li JN (2015) The evolutionary history of R2R3-MYB proteins across 50 eukaryotes: new insights into subfamily classification and expansion. Sci Rep 5: 11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emons AMC, Ketelaar T (2009) Root Hairs. Springer-Verlag, Heidelberg, Germany [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW (1994) The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol 166: 740–754 [DOI] [PubMed] [Google Scholar]
- Geitmann A, Emons AM (2000) The cytoskeleton in plant and fungal cell tip growth. J Microsc 198: 218–245 [DOI] [PubMed] [Google Scholar]
- Grierson C, Nielsen E, Ketelaarc T, Schiefelbein J (2014) Root hairs. The Arabidopsis Book 12: e0172 doi/10.1199/tab.0172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Nielsen E (2013) Targeting and regulation of cell wall synthesis during tip growth in plants. J Integr Plant Biol 55: 835–846 [DOI] [PubMed] [Google Scholar]
- Huang L, Schiefelbein J (2015) Conserved gene expression programs in developing roots from diverse plants. Plant Cell 27: 2119–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Dopazo J, Gabaldón T (2010) ETE: a Python Environment for Tree Exploration. BMC Bioinformatics 11: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Wilson GT, Connolly EL (2014) The diverse roles of FRO family metalloreductases in iron and copper homeostasis. Front Plant Sci 5: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones VA, Dolan L (2012) The evolution of root hairs and rhizoids. Ann Bot (Lond) 110: 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YH, Kirik V, Hulskamp M, Nam KH, Hagely K, Lee MM, Schiefelbein J (2009) The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 21: 1080–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas B, Amyot L, Johansen C, Sato S, Tabata S, Kawaguchi M, Szczyglowski K (2009) Conservation of lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiol 151: 1175–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CM, Park SH, Je BI, Park SH, Park SJ, Piao HL, Eun MY, Dolan L, Han CD (2007) OsCSLD1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiol 143: 1220–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho HT (2006) Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18: 2958–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Simon M, Huelskamp M, Schiefelbein J (2004) The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol 268: 506–513 [DOI] [PubMed] [Google Scholar]
- Kwak SH, Schiefelbein J (2007) The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Dev Biol 302: 118–131 [DOI] [PubMed] [Google Scholar]
- Kwak SH, Shen R, Schiefelbein J (2005) Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307: 1111–1113 [DOI] [PubMed] [Google Scholar]
- Lan P, Li W, Lin WD, Santi S, Schmidt W (2013) Mapping gene activity of Arabidopsis root hairs. Genome Biol 14: R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin JC, Oppenheimer DG, Pollock S, Marks MD (1993) Arabidopsis GLABROUS1 gene requires downstream sequences for function. Plant Cell 5: 1739–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J (1999) WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483 [DOI] [PubMed] [Google Scholar]
- Lee Y, Bak G, Choi Y, Chuang WI, Cho HT, Lee Y (2008) Roles of phosphatidylinositol 3-kinase in root hair growth. Plant Physiol 147: 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Lan P (2015) Re-analysis of RNA-seq transcriptome data reveals new aspects of gene activity in Arabidopsis root hairs. Front Plant Sci 6: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW (1996) The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW (1994) The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol 106: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L (2007) An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316: 1477–1480 [DOI] [PubMed] [Google Scholar]
- Nestler J, Keyes SD, Wissuwa M (2016) Root hair formation in rice (Oryza sativa L.) differs between root types and is altered in artificial growth conditions. J Exp Bot 67: 3699–3708 [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493 [DOI] [PubMed] [Google Scholar]
- Pemberton LMS, Tsai SL, Lovell PH, Harris PJ (2001) Epidermal patterning in seedling roots of eudicotyledons. Ann Bot (Lond) 87: 649–654 [Google Scholar]
- Perry P, Linke B, Schmidt W (2007) Reprogramming of root epidermal cells in response to nutrient deficiency. Biochem Soc Trans 35: 161–163 [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Schauer MA, Megraw M, Breakfield NW, Thompson JW, Georgiev S, Soderblom EJ, Ohler U, Moseley MA, Grossniklaus U, et al. (2012) The protein expression landscape of the Arabidopsis root. Proc Natl Acad Sci USA 109: 6811–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires ND, Yi K, Breuninger H, Catarino B, Menand B, Dolan L (2013) Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc Natl Acad Sci USA 110: 9571–9576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Z, Libault M (2013) Unleashing the potential of the root hair cell as a single plant cell type model in root systems biology. Front Plant Sci 4: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouard M, Guignon V, Aluome C, Laporte MA, Droc G, Walde C, Zmasek CM, Périn C, Conte MG (2011) GreenPhylDB v2.0: comparative and functional genomics in plants. Nucleic Acids Res 39: D1095–D1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounds CM, Bezanilla M (2013) Growth mechanisms in tip-growing plant cells. Annu Rev Plant Biol 64: 243–265 [DOI] [PubMed] [Google Scholar]
- Salazar-Henao JE, Vélez-Bermúdez IC, Schmidt W (2016) The regulation and plasticity of root hair patterning and morphogenesis. Development 143: 1848–1858 [DOI] [PubMed] [Google Scholar]
- Sanati Nezhad A, Geitmann A (2013) The cellular mechanics of an invasive lifestyle. J Exp Bot 64: 4709–4728 [DOI] [PubMed] [Google Scholar]
- Schiefelbein JW, Somerville C (1990) Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K (1997) Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Yi K, Menand B, Bell E, Dolan L (2010) A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat Genet 42: 264–267 [DOI] [PubMed] [Google Scholar]
- ZhiMing Yu, Bo K, XiaoWei H, ShaoLei L, YouHuang B, WoNa D, Ming C, Hyung-Taeg C, Ping W (2011) Root hair-specific expansins modulate root hair elongation in rice. Plant J 66: 725–734 [DOI] [PubMed] [Google Scholar]