Pistil transcriptomes of Arabidopsis species show a clear overlap between responses to fungal infection and pollination and suggest novel roles for cysteine-rich peptides in fertilization.
Abstract
Independent lines of evidence suggest that members from ancient and polymorphic gene families such as defensins and receptor-like kinases mediate intercellular communication during both the immune response and reproduction. Here, we report a large-scale analysis to investigate the extent of overlap between these processes by comparing differentially expressed genes (DEGs) in the pistil transcriptomes of Arabidopsis thaliana and Arabidopsis halleri during self-pollination and interspecific pollination and during infection with Fusarium graminearum. In both Arabidopsis species, the largest number of DEGs was identified in infected pistils, where genes encoding regulators of cell division and development were most frequently down-regulated. Comparison of DEGs between infection and various pollination conditions showed that up to 79% of down-regulated genes are shared between conditions and include especially defensin-like genes. Interspecific pollination of A. thaliana significantly up-regulated thionins and defensins. The significant overrepresentation of similar groups of DEGs in the transcriptomes of reproductive and immune responses of the pistil makes it a prime system in which to study the consequences of plant-pathogen interactions on fertility and the evolution of intercellular communication in pollination.
In flowering plants (angiosperms), intercellular communication during pollination involves a complex cross talk between the gametophytes and several maternal tissues of the receptive pistil. Diverse self/nonself recognition mechanisms determine whether pollen germination takes place and whether maternal reproductive tissues support compatible pollen tube growth. In Arabidopsis thaliana stigmatic tissues, the transmitting tract and ovules generate a variety of signaling molecules like Exo70A1, S-azadecalins, plantacyanin, as well as LUREs and EC1s, all essential for successful pollen hydration, germination, growth across the transmitting tract, and double fertilization (for review, see Dresselhaus and Franklin-Tong, 2013). In addition to signaling molecules, both stigmatic papilla and the extracellular matrix of the transmitting tract also provide nutrients like carbohydrates, proteins, and glycoproteins, which support pollen tube growth. Several studies have shown that especially members of subgroups of the Cys-rich peptide (CRP) superfamily play key roles during reproduction (Schopfer et al., 1999; Park et al., 2000; Okuda et al., 2009; Amien et al., 2010; Sprunck et al., 2012). Initially, plant CRPs with antimicrobial activity were characterized and named as defensins (DEFs) due to their activity and structural similarity to insect and mammalian defensins. Like their animal counterparts, plant defensins were believed to be involved primarily in the immune response (Terras et al., 1995). Later, it was shown that plant CRPs are strongly overrepresented and highly expressed in both male and female gametophytes (pollen and embryo sacs), respectively (Huang et al., 2015). This indicated that they may be involved in protecting the important generative cells and may have novel roles in reproduction or even multiple functions. In particular, members of the DEF-like (DEFL) group of CRPs have been shown to be involved in self-incompatibility responses (SCR/SP11), species-preferential pollen tube guidance (LUREs), or pollen tube burst (ZmES4). Some still possess antimicrobial activity (for review, see Qu et al., 2015; Bircheneder and Dresselhaus, 2016). Like DEFs/DEFLs, members of other ancient and polymorphic gene families such as receptor-like kinase (RLKs) appear to mediate intercellular communication during both the immune response and reproduction. FERONIA, for example, controls pollen tube burst and acts as a negative regulator of the immune response to bacterial and fungal pathogens (Escobar-Restrepo et al., 2007; Kessler et al., 2010).
These studies point toward the necessity of investigating the convergence of angiosperm fertilization and immunity in a more systematic manner to obtain a deeper insight into the makeup and evolution of the complex network of intercellular signals exchanged by male and female gametophytes and with the maternal tissues during reproduction.
The pistil represents an ideal system in which to investigate and compare the molecular and evolutionary connections of reproduction and immune response, because the nutrient-rich environment regulating germination, invasion, and growth of pollen tubes also promotes fungal colonization. Previously, it has been suggested that the response of the pistil to foreign or incompatible pollen might bear similarities with the reaction to fungal invasion (Sanabria et al., 2008; Dresselhaus and Márton, 2009). To test this hypothesis, we used different Arabidopsis species to study the relationship between the immune response and pollination-associated processes at the transcriptomic level. Specifically, we chose closely related Arabidopsis thaliana (self-compatible) as well as Arabidopsis halleri and Arabidopsis lyrata (both self-incompatible) as our model systems. Notably, A. lyrata and A. halleri also form hybrids in nature (Shimizu-Inatsugi et al., 2009) and are reproductively isolated from Arabidopsis thaliana. Transcriptomic studies in these species are supported by the availability of their completely sequenced genomes as well as curated functional annotations and gene ontologies for A. thaliana. We chose Fusarium graminearum, a well-known flower-infecting fungus and one of the economically most important pathogens causing Fusarium head blight (FHB) in many cereals and cob rot in maize (Zea mays; McMullen et al., 2012). Infection by F. graminearum is facilitated at anthesis by conidia or ascospores attaching to pollen and anthers, from which mycelia colonize the soft tissue of stigmas and ovaries (Miller et al., 2004). Similarly, F. graminearum successfully infects flowers of the model plant species A. thaliana, thus advancing a useful translational model for more rapid studies of host-pathogen interactions (Brewer and Hammond-Kosack, 2015).
We identified candidate genes differentially expressed in either or both processes by comparing differentially expressed genes (DEGs) in pistil transcriptomes of A. thaliana and A. halleri responding either to fungal infection or to self-specific as well as interspecific pollination. Here, we report the identification of genes associated with biological processes commonly as well as specifically regulated during both defense and various pollinations. The data set presented is a valuable resource for in-depth analyses of plant-pathogen interactions and puts forward candidate genes that may be involved in fertilization and the establishment of prezygotic reproductive barriers in the genus Arabidopsis.
RESULTS
Morphological Characteristics of A. thaliana and A. halleri Pistils during Pollination and Infection with F. graminearum
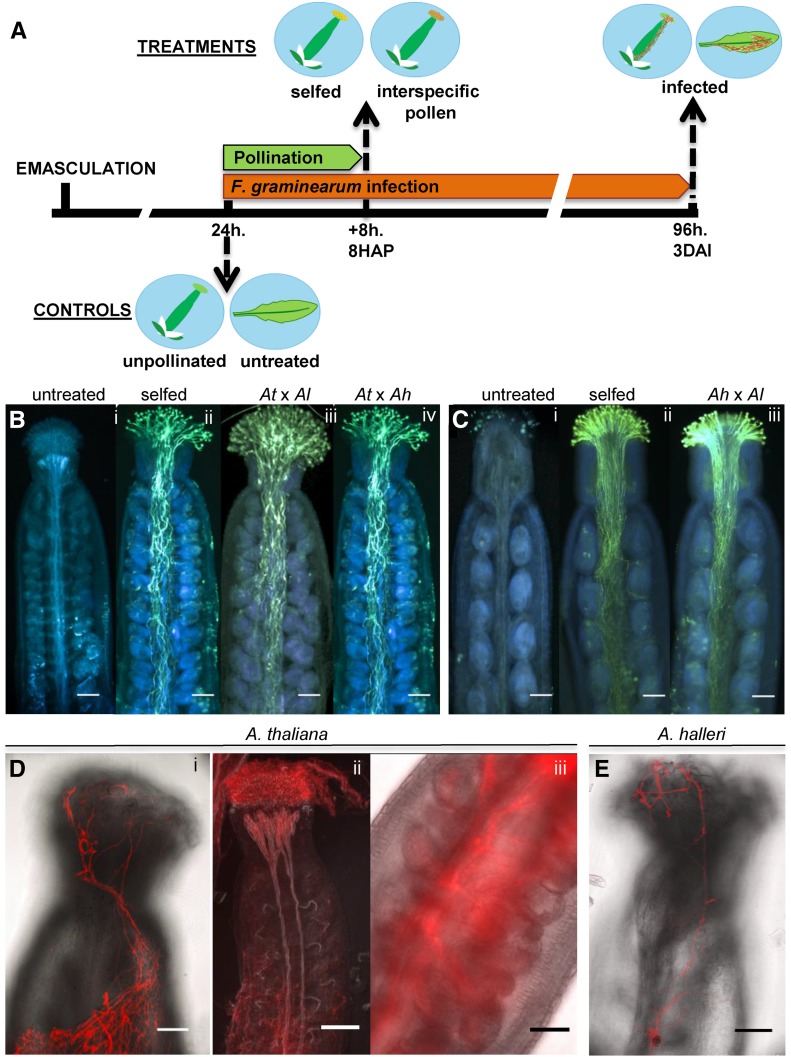
To identify optimal time points for transcriptome profiling, we first carried out a morphological characterization of Arabidopsis pistils during pollination and fungal infection. All experimental and control treatments were performed 24 h after flower emasculation (Fig. 1A). Pistils treated with pollen from the same species (selfed) or with pollen from A. halleri or A. lyrata were collected 8 h after pollination (HAP). In all cases, substantial pollen tube growth was observed, making this time point ideal for sample collection (Fig. 1, B and C). Although pollen tubes reached the abaxial end of the transmitting tract and several of them advanced along the funiculus toward the ovule, seed set was detected only from selfings of A. thaliana and to a lesser extent also from A. halleri. Crosses involving A. halleri or A. lyrata pistils and A. thaliana pollen were not performed because of previously documented unilateral incompatibility of crosses between pistils of self-incompatible and pollen of self-compatible species (Lewis and Crowe, 1958; Müller et al., 2016).
Figure 1.
Study design and morphology of pistils during pollination or infection with F. graminearum. A, Diagram describing the timeline for the treatment and collection of Arabidopsis pistils and leaves employed for transcriptome profiling of pollination or F. graminearum infection. B, Aniline Blue-stained mature emasculated A. thaliana pistils: unpollinated (i) and 8 HAP with own pollen (ii), pollen from A. lyrata (iii), or pollen from A. halleri (iv). C, Mature emasculated A. halleri pistils: unpollinated (i) and 8 HAP with own pollen (ii) and pollen from A. lyrata (iii). D, F. graminearum hyphae stained with wheat germ agglutinin-tetramethylrhodamine (WGA-TMR) 3 DAI on the surface of pistils from A. thaliana (i). At 3 DAI, F. graminearum hyphae proliferate heavily on the stigma and colonize the interior of the pistil using the vasculature (ii). F. graminearum hyphae grow through the transmitting tract (iii). E, F. graminearum hyphae stained with WGA-TMR 3 DAI on the surface of pistils from A. halleri. All scale bars are 50 μm.
A separate group of pistils treated exclusively with F. graminearum was collected 3 d after dipping and spraying emasculated flowers with conidia solution and incubation in a moist chamber under long-day conditions. This inoculation approach ensured effective Fusarium infection and avoided artifactual responses (e.g. through incubation in the dark or leaf infiltration with a syringe).
The surface of infected pistils of both species collected 3 d after infection (DAI) showed profuse hyphal growth (Fig. 1, Di and E), particularly on and inside the stigma as well as inside the vasculature and transmitting tract (Fig. 1D, ii and iii). Pistils collected 24 h after emasculation, but otherwise untreated, were used for comparison with pistil treatments. However, this approach does not address all possible effects, including inoculation stress and the age differences of some samples. Samples of untreated and infected cauline leaves were included to identify pistil-specific differential gene expression by excluding genes up- or down-regulated in both pistils and leaves during Fusarium infection. As documented in pistils, Fusarium hyphae also grew on the surface of inoculated leaves of both species (Supplemental Fig. S1); however, proliferation was weaker, possibly hindered by trichomes.
Global Comparison of Pistil Transcriptomes between A. thaliana and A. halleri
Total RNA from pistils infected with Fusarium or pollinated (selfed or interspecific) was employed to compare the dynamics of gene expression with untreated pistils. RNA sequencing yielded reads with a mean quality score of 36 or greater for over 94% of the reads in all biological replicates (Table I; Supplemental Table S1), indicating that the base call accuracy of sequencing was well above 99.9% (Ewing and Green, 1998). After quality control and trimming, reads were mapped to the corresponding Columbia-0 TAIR10 version of the A. thaliana genome or to version 1.1 of the A. halleri genome. Table I summarizes the most important aspects of RNA sequencing (RNA-seq), mapping, and differential gene expression analysis. The reproducibility of RNA-seq results was confirmed with quantitative PCR (qPCR) assays on a set of 14 candidate DEFLs, as shown in Supplemental Figures S2 and S3. Correlation analysis showed a value of r2 = 0.83 between the log2 fold changes detected by RNA-seq and qPCR.
Table I. Characteristics of the transcriptomes sequenced.
| Sample | Total Reads | Mean Quality Score | Mapped Reads | Genes Expresseda | Up-Regulatedb | Down-Regulatedb |
|---|---|---|---|---|---|---|
| A. thaliana mature pistils unpollinated | 109,362,994 | 36.51 | 89,537,597 | 69.79 | ||
| A. thaliana pistils selfed 8 HAP | 101,942,362 | 36.52 | 96,071,451 | 69.06 | 81 (0.28) | 260 (0.90) |
| A. thaliana pistils × A. lyrata pollen 8 HAP | 100,332,498 | 36.84 | 95,899,345 | 68.89 | 181 (0.63) | 296 (1.03) |
| A. thaliana pistils × A. halleri pollen 8 HAP | 103,676,026 | 36.9 | 98,639,610 | 69.28 | 211 (0.73) | 297 (1.03) |
| A. thaliana pistils 3 DAI with Fusarium | 97,213,704 | 36.59 | 70,661,826 | 65.38 | 1,928 (6.73) | 2,007 (7.01) |
| A. thaliana leaf untreated | 80,329,562 | 35.59 | 74,144,556 | 58.16 | ||
| A. thaliana leaf 3 DAI with Fusarium | 95,285,902 | 36.88 | 91,697,050 | 57.79 | 606 (2.11) | 785 (2.74) |
| A. halleri mature pistils unpollinated | 106,156,558 | 36.88 | 75,012,556 | 73.92 | ||
| A. halleri pistils selfed 8 HAP | 93,961,972 | 36.71 | 67,574,404 | 72.59 | 173 (0.69) | 211 (0.84) |
| A. halleri pistils × A. lyrata pollen 8 HAP | 121,254,382 | 36.44 | 87,023,215 | 73.3 | 184 (0.73) | 197 (0.78) |
| A. halleri pistil 3 DAI with Fusarium | 83,771,668 | 36.68 | 56,172,094 | 66.81 | 1,724 (6.89) | 1,994 (7.97) |
| A. halleri leaf untreated | 127,526,910 | 36.42 | 89,168,588 | 65.78 | ||
| A. halleri leaf 3 DAI with Fusarium | 93,279,706 | 36.76 | 66,208,233 | 65.35 | 1,349 (5.39) | 517 (2.06) |
As a percentage of the total number of genes included in the reference genomes: A. thaliana TAIR10, 28,642 (including mitochondrial genes); A. halleri V1, 25,008 (including 68 chloroplast and mitochondrial genes). Genes expressed are those with RPKM ≥ 1.
For each species, nonpollinated pistils or untreated leaves were the points of reference for calculating differential expression. Differential expression is log2 fold change ≥ 2 for up-regulation or ≤ −2 for down-regulation, FDR-corrected P ≤ 0.0005. Values in parentheses are the number of genes differentially expressed, as a percentage of the total number of genes in each species.
Of the total number of reads obtained from A. thaliana samples, 90% or more mapped to the reference genome, indicating the reliability of the sampling. In most of the samples from A. halleri, about 70% of the reads obtained could be mapped. In both species, the lowest proportion of mapped reads corresponds to samples obtained from pistils infected with F. graminearum, indicating that a portion of unmapped reads corresponded to the fungal transcriptome (Table I). On average, A. thaliana expressed 18,754 genes in pistils (65% of the total annotated genes), while the expression of about 17,411 genes (70%) was detected in A. halleri.
To maintain similar levels of variation between the biological replicates investigated, principal component analysis and box plots were employed to select the two most similar of three samples initially obtained for each experimental and control condition (Supplemental Table S1). Differential gene expression analysis was based on read counts from pollinated and infected pistils compared with those obtained from unpollinated pistils of A. thaliana and A. halleri, respectively. DEGs were those with a false discovery rate (FDR)-corrected P < 0.0005 and an expression log2 fold change ≥ 2 (up-regulation) or ≤ −2 (down-regulation) and that are expressed with an RPKM (reads per kilobase per million mapped reads) ≥ 1. The lists of DEGs in each comparison as well as their corresponding fold change and RPKM values are provided in Supplemental Data S1. Generally, in both A. thaliana and A. halleri, differential expression analysis detected a significantly higher number of down-regulated than up-regulated genes (Table I). While pistil infection resulted in transcriptional changes ranging from 6.7% to 8% of the total annotated genes, pistil pollination resulted in differential expression of at most 1% (Table I). In comparison with infected pistils, leaf infection resulted in an even smaller proportion, ranging from 2% to 5% DEGs (Table I).
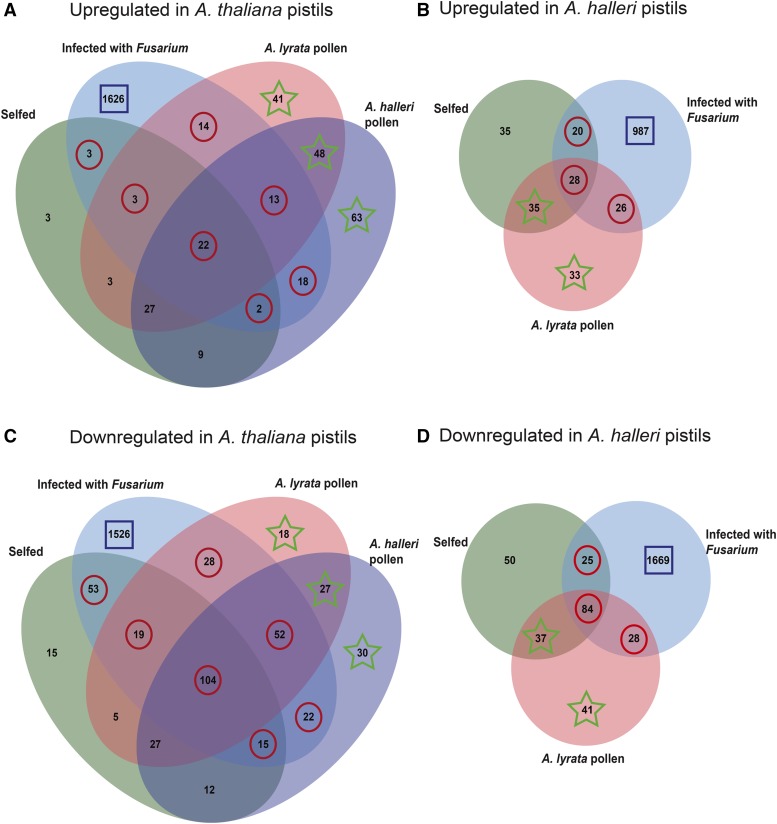
To determine genes that are significantly represented in response to pollination and fungal infection, we focused on three major groups of DEGs: (1) those detected only in Fusarium-infected pistils, (2) those identified in both infected and pollinated pistils, and (3) those found only in pistils treated with interspecific pollen (the three groups are indicated with squares, circles, or stars in Fig. 2). To increase the representation of pistil-specific genes in our analysis, we excluded all DEGs present in both pistils and Fusarium-infected leaves (indicated with black circles in Supplemental Fig. S4).
Figure 2.
Venn diagrams identify and indicate the number of DEGs unique or common to pollination and Fusarium infection. The comparison of pistil transcriptomes was based on Gene Ontology (GO) enrichment tests performed on three major groups of DEGs. Highlighted with squares are the numbers of DEGs exclusive to pistils infected with F. graminearum. Circles indicate DEGs identified in response to both Fusarium infection and pollination. Stars highlight the number of DEGs present exclusively in response to interspecific pollination. Note that, in the case of A. halleri, this last group also included DEGs identified both in selfing and in response to A. lyrata pollen.
Differential Gene Expression Exclusive to Fusarium-Infected Pistils of Arabidopsis
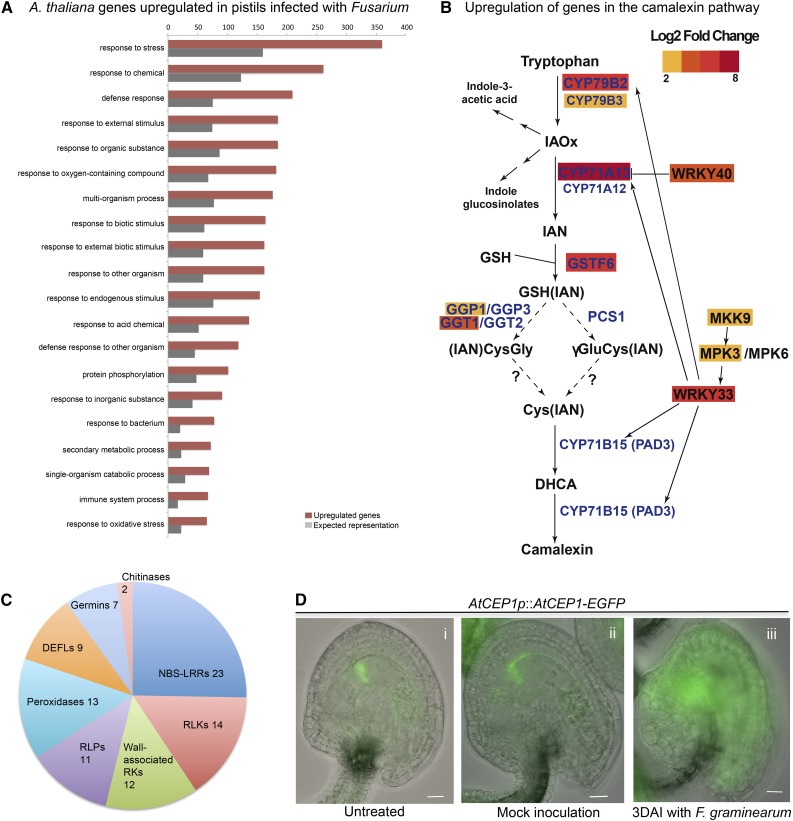
In both Arabidopsis species analyzed, the largest proportion of DEGs were those expressed exclusively in pistils 3 DAI with F. graminearum (indicated with squares in Fig. 2). GO analysis of DEGs up-regulated in infected pistils shows an overrepresentation of those involved in the immune response to Fusarium as well as the associated hypersensitive response (Fig. 3A). Among the diverse aspects of the immune response represented in the infected pistil transcriptome are detoxification, production of antifungal substances, response to hormone signaling, pathogen detection, and killing and programmed cell death (PCD).
Figure 3.
The transcriptome of Fusarium-infected pistils is enriched in up-regulated genes associated with the immune response. A, GO term overrepresentation of genes up-regulated only in A. thaliana pistils infected with F. graminearum. The top 20 terms with enrichment of 2-fold or greater and Bonferroni-corrected P ≤ 0.05 are considered. Complete overrepresentation analyses are provided in Supplemental Data S2 (A. thaliana) and Supplemental Data S3 (A. halleri). B, In infected pistils, genes of the secondary metabolism are up-regulated encoding proteins involved in the synthesis of antimicrobial substances like camalexin. This pathway highlights the relationship between key regulators of camalexin biosynthesis, which are up-regulated 2 log2 fold or greater in A. thaliana pistils 3 DAI with F. graminearum. This diagram was adapted from Ahuja et al. (2012). C, Number and diversity of up-regulated genes encoding pathogenesis-related proteins as well as other proteins involved in the immune response. D, Localization pattern of the PCD marker AtCEP1-EGFP in the ovule as indicated. Scale bars are 50 μm.
Deoxynivalenol (DON) is the most prevalent and economically important Fusarium toxin because it provokes acute and chronic disease symptoms in mammals that consume contaminated grains (Bennett and Klich, 2003). In the transcriptome of infected pistils, we observed the up-regulation of the key detoxification gene DON-Glucosyltransferase1 (AT2G36800; Poppenberger et al., 2003; Desmond et al., 2008). Other UDP-glycosyltransferases also were up-regulated in pistils of both species 3 DAI (Supplemental Data S2 and S3), suggesting that the enzymes encoded by this gene family have an important role in the response to Fusarium infection. Furthermore, we detected a significant increase in the expression of several genes regulating the production of secondary metabolites with antifungal properties, specifically those encoding CYP79B2, CYP79B3, CYP71A13, GSTF6, and GGT1 (Fig. 3B; Supplemental Data S2), which are involved in the synthesis of Trp-derived indole glucosinolates and camalexin, the major phytoalexin of A. thaliana (Bak and Feyereisen, 2001; Smolen and Bender, 2002; Naur et al., 2003; Clay et al., 2009). Camalexin is probably increased further by the up-regulated gene products MKK9 and MPK3, which transcriptionally activate and phosphorylate WRKY33, an important positive regulator of this pathway (Fig. 3B; Xu et al., 2008). In this context, the simultaneous up-regulation of 16 candidate genes encoding glutathione S-transferases indicates that indole glucosinolates might be further metabolized into additional secondary metabolites with antimicrobial activity (Supplemental Data S2).
Salicylic acid (SA), jasmonic acid (JA), and ethylene play major roles in regulating plant defense responses against diverse pathogens. GO analysis suggests that, in infected pistils of A. thaliana, almost as many candidate genes responsive to JA and SA signaling were up-regulated (Supplemental Data S2; some candidates are shared between lists). Furthermore, in infected pistils of both species, we observed the up-regulation of LIPOXYGENASE1, a key enzyme in the synthesis of JA, as well as the ethylene- and jasmonate-responsive plant defensin genes PDF1.2b, PDF1.2c, and PDF1.3 and the A. halleri homolog of PDF1.2a. On the other hand, three known markers of SA signaling were significantly up-regulated in A. thaliana infected pistils: NONEXPRESSOR OF PR1 and PHYTOALEXIN DEFICIENT4 as well as several genes encoding PR proteins.
In the transcriptome of infected pistils, we further detected significant up-regulation of pattern-recognition receptor-like kinases, which recognize and initiate pattern-triggered immunity. The signal-specific activation of plant pattern-recognition receptors by microorganism-associated molecular patterns leads to the up-regulation of genes encoding PR proteins, like peroxidases, DEFLs, germins, and chitinases (Fig. 3C). Genes encoding NBS-LRR proteins, cytoplasmic immune receptors, are the largest family of receptors significantly up-regulated (Fig. 3C; Supplemental Data S2).
To determine whether the immune response to Fusarium triggered PCD in pistils 3 DAI, we performed a cross-comparison between DEGs in Fusarium-infected pistils and predictive markers for PCD induced by biotic stress (Olvera-Carrillo et al., 2015). This analysis identified a group of 47 and 24 up-regulated genes from diverse gene families in A. thaliana and A. halleri, respectively (Supplemental Data S2 and S3). Among the up-regulated markers of PCD are AtCEP1, AtCEP2, and CONSTITUTIVELY ACTIVATED CELL DEATH1 (Morita-Yamamuro et al., 2005; Helm et al., 2008). To investigate whether AtCEP1 protease activity also is induced after Fusarium infection, we studied AtCEP1-EGFP expression and its influence on tissue morphology in ovules of A. thaliana 3 DAI with F. graminearum. Specifically, fluorescent signals of AtCEP1-EGFP were distributed over the entire ovules of infected pistils (Fig. 3Diii). In untreated and mock-infected pistils, the signal was restricted to the nucellus cells surrounding the chalazal pole of the central cell (Fig. 3D, i and ii), as documented previously (Zhou et al., 2016). Associated with a widespread expression pattern of AtCEP1-EGFP during fungal infection, we observed the disintegration of outer ovule integuments and the collapse of tissues (Fig. 3Diii). In summary, we observed signs of PCD triggered by fungal infection at both the transcriptional and morphological levels.
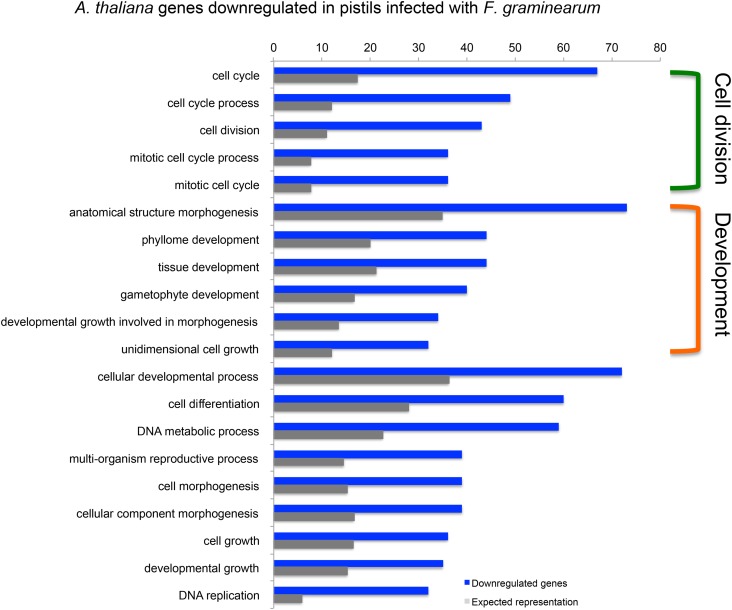
At least 634 genes (between 35% and 40%) down-regulated during pistil Fusarium infection are shared between A. thaliana and A. halleri. In this group, genes associated with GO terms involving cell division and growth are overrepresented (Fig. 4). Table II summarizes the complex immune response by listing genes annotated with four or more of the 12 GO terms shared by the down-regulated genes of A. thaliana and A. halleri. These include genes encoding kinesins, chromatin assembly factors, the transcription factor CUC2, as well as the LRR receptor-like kinases STRUBBELIG and ERECTA. While almost half of the genes listed are involved in cell division, there is also a large group of candidates additionally associated with growth; thus, a clear-cut grouping of the processes affected and the genes involved was not possible.
Figure 4.
GO terms describing processes associated with cell division and development prevail among the top 20 terms overrepresented in genes down-regulated in A. thaliana infected pistils. Terms in common between the GO term enrichment of A. thaliana and A. halleri are indicated with brackets. Only terms with enrichment of 2-fold or greater and Bonferroni-corrected P ≤ 0.05 are considered. The complete overrepresentation analyses as well as the annotation of genes differentially expressed are provided in Supplemental Data S2 (A. thaliana) and Supplemental Data S3 (A. halleri).
Table II. Genes down-regulated by F. graminearum and annotated with four or more overrepresented cell division or development GO terms in both A. thaliana and A. halleri.
Each gene belongs to either or both of two major groups of overrepresented GO terms. X, Cell division GO terms: mitotic cell cycle process (GO:1903047), mitotic cell cycle (GO:0000278), cell cycle process (GO:0022402), cell division (GO:0051301), cell cycle (GO:0007049). Y, Development GO terms: regulation of anatomical structure morphogenesis (GO:0022603), unidimensional cell growth (GO:0009826), developmental growth involved in morphogenesis (GO:0060560), gametophyte development (GO:0048229), phyllome development (GO:0048827), anatomical structure morphogenesis (GO:0009653), tissue development (GO:0009888).
| GO Groups | Gene Identifier | Log2 FCa | Gene Name and Symbol |
|---|---|---|---|
| X | AT4G26760 | −2.36 | 65-kD microtubule-associated protein2; MAP65-2 |
| X | AT2G32590 | −2.63 | Condensin complex subunit2; CAPH |
| X | AT2G21800 | −2.33 | Crossover junction endonuclease EME1A |
| X | AT5G49160 | −2.44 | DNA (cytosine-5)-methyltransferase1; DMT1 |
| X | AT5G63920 | −2.58 | DNA topoisomerase3-α; TOP3A |
| X | AT1G53140 | −2.82 | Dynamin-related protein5A; DRP5A |
| X | AT5G37630 | −2.41 | Embryo-defective protein2656; EMB2656 |
| X | AT5G13840 | −2.61 | FIZZY-RELATED3; FZR3 |
| X | AT3G53760 | −2.04 | γ-Tubulin complex component4; GCP4 |
| X | AT4G05190 | −3.56 | Kinesin5; ATK5 |
| X | AT4G14150 | −3.02 | Kinesin-like protein KIN12A |
| X | AT5G67270 | −2.81 | Microtubule-associated protein RP/EB family member 1C; EB1C |
| X | AT2G40550 | −2.52 | Minichromosome maintenance complex-binding protein; ETG1 |
| X | AT1G49910 | −2.61 | Mitotic checkpoint protein BUB3.2 |
| X | AT2G20635 | −2.32 | Mitotic checkpoint Ser/Thr protein kinase BUB1 |
| X | AT2G35190 | −2.7 | Novel plant SNARE11; NPSN11 |
| X | AT3G19050 | −3.17 | Phragmoplast-orienting kinesin2; POK2 |
| X | AT3G17360 | −3.27 | Phragmoplast-orienting kinesin1; POK1 |
| X | AT4G32830 | −3.48 | Ser/Thr protein kinase Aurora1; AUR1 |
| X | AT2G25880 | −3.45 | Ser/Thr protein kinase Aurora2; AUR2 |
| X | AT2G45490 | −2.68 | Ser/Thr protein kinase Aurora3; AUR3 |
| X | AT5G52950 | −2.67 | Uncharacterized protein; At5g52950 |
| X | AT1G02970 | −2.84 | Wee1-like protein kinase; WEE1 |
| Y | AT1G68090 | −4.62 | Annexin D5; ANN5 |
| Y | AT1G04020 | −2.59 | BRCA1-associated RING domain protein1; BARD1 |
| Y | AT1G65470 | −2.25 | Chromatin assembly factor1 subunit FAS1 |
| Y | AT5G64630 | −2.21 | Chromatin assembly factor1 subunit FAS2 |
| Y | AT5G53950 | −3.18 | CUP-SHAPED COTYLEDON2; NAC098 |
| Y | AT5G07180 | −2.97 | LRR receptor-like Ser/Thr protein kinase ERL2 |
| Y | AT3G18000 | −2.44 | Phosphoethanolamine N-methyltransferase1; NMT1 |
| Y | AT2G19690 | −2.53 | Phospholipase A2-β; PLA2-β |
| Y | AT1G79860 | −13.48 | Rop guanine nucleotide exchange factor12; ROPGEF12 |
| Y | AT1G11130 | −2.16 | STRUBBELIG; SUB |
| XY | AT5G51600 | −2.08 | 65-kD microtubule-associated protein3; MAP65-3 |
| XY | AT1G78770 | −2.36 | Anaphase-promoting complex subunit 6; APC6 |
| XY | AT5G60880 | −3.05 | BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE; BASL |
| XY | AT3G57860 | −2.62 | GIGAS CELL1; GIG1 |
| XY | AT5G38110 | −2.21 | Histone chaperone ASF1B |
| XY | AT1G18370 | −3.89 | Kinesin-like protein NACK1; NACK1 |
| XY | AT5G07280 | −2.03 | Leu-rich repeat receptor protein kinase EMS1/EXS |
| XY | AT2G26330 | −2.01 | LRR receptor-like Ser/Thr protein kinase ERECTA |
| XY | AT5G56580 | −3.23 | MEK6; MKK6 |
| XY | AT2G33560 | −4.08 | Mitotic spindle checkpoint protein BUBR1 |
| XY | AT3G25980 | −2.83 | Mitotic spindle checkpoint protein MAD2; MAD2 |
| XY | AT2G42260 | −2.04 | Protein POLYCHOME; PYM |
| XY | AT1G50240 | −2.59 | Ser/Thr protein kinase TIO |
| XY | AT3G59550 | −3.19 | Sister chromatid cohesion1 protein3; SYN3 |
| XY | AT3G18730 | −2.31 | TONSOKU; TSK |
| XY | AT5G46700 | −2.98 | TORNADO 2; TRN2 |
Relative to mature emasculated pistils, FC = log2 fold change ≥ 2 for up-regulation or ≤ −2 for down-regulation, FDR-corrected P ≤ 0.0005.
DEFLs Form a Large, Evolutionarily Conserved Group of Genes Down-Regulated during Both Pollination and Fusarium Infection
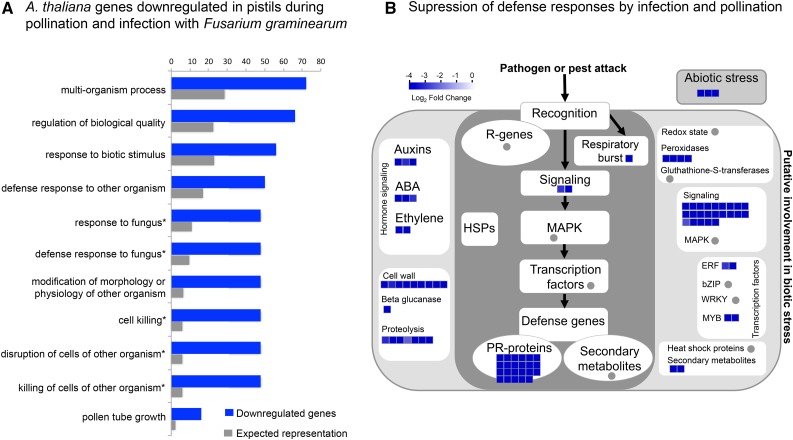
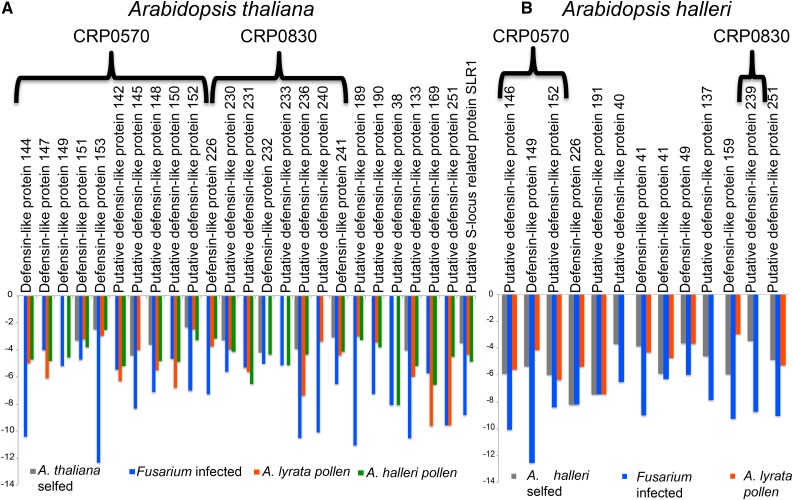
The second major group of DEGs was formed by those down-regulated during both fungal infection and pollination (numbers circled in Fig. 2). In both species, down-regulated genes are up to 79% of those differentially expressed during pollination and in response to fungal infection (Fig. 2, C and D). Interestingly, GO term overrepresentation analysis among down-regulated genes shows that a significant number are involved in defense responses to fungus or cell killing (Fig. 5A), are classified as pathogenesis-related proteins (Fig. 5B), and most encode DEFLs (Fig. 6; Supplemental Data S2 and S3). In A. thaliana, 75% of these DEFLs are closely related gene duplicates and form part of CRP subgroups CRP0570 and CRP0830. Whereas members of CRP0570 are annotated as low-molecular-weight Cys-rich peptides (LCRs), those in subgroup CRP0830 encode pollen coat proteins related to the S-locus Cys-rich protein SCR and the S-locus-related1-binding protein SLR1-BP (Fig. 6A). In the same conditions, A. halleri pistils reproduce the pattern of expression observed in A. thaliana pistils during pollination and in fungal infection. In A. halleri, several of the down-regulated DEFLs also belong to subgroups CRP0570 and CRP0830 (Fig. 6B). However, we noticed that several additional A. halleri members of these subgroups are down-regulated only in Fusarium-infected pistils (Supplemental Fig. S5).
Figure 5.
Among the genes down-regulated both in infected pistils and during pollination, those involved in pathogen killing prevail. A, Results of GO term overrepresentation analyses of genes down-regulated during both Fusarium infection and pollination. GO terms indicated with asterisks are those overrepresented in both A. thaliana and A. halleri lists of down-regulated genes. B, MapMan diagram illustrates the different aspects of biotic stress represented among down-regulated genes in A. thaliana pistils during both Fusarium infection and pollination. Color intensity reflects log2 fold change ≥ 2. Mapman figure re-drawn from original to improve image quality.
Figure 6.
Conserved down-regulation of DEFL protein genes during both Fusarium infection and pollination. A, Genes encoding DEFLs are the largest group of down-regulated genes during both pollination and fungal infection. In A. thaliana, about 75% of these belong to subgroups CRP0570 and CRP0830. B, In A. halleri, a similar pattern was observed.
Genes Involved in the Response to Stress Are Up-Regulated during Both Pollination and Pathogen Infection
In A. thaliana and A. halleri, up-regulated genes are at most 35% of those differentially expressed both during pollination and in the course of fungal infection (Fig. 2, A and B). Among these genes, the GO terms response to stimulus and response to stress are most significantly overrepresented (Supplemental Data S2 and S3). Genes associated with these terms encode proteins deployed to counteract xenobiotic toxins and the damaging effects of reactive oxygen species, mostly glutathione S-transferases, members of the cytochrome P450 gene family, camalexin synthase, heat shock proteins, peroxidases, chitinase, the LRR-RLK IOS1, a well-known regulator of pattern-triggered immunity (Hok et al., 2011, 2014), as well as the transcription factor JUNGBRUNNEN1, an important regulator of cellular hydrogen peroxide levels and reactive oxygen species-induced stress responses (Wu et al., 2012).
Genes Associated with Pathogen Detection and Killing Are Up-Regulated in Arabidopsis Pistils Exposed to Foreign Pollen
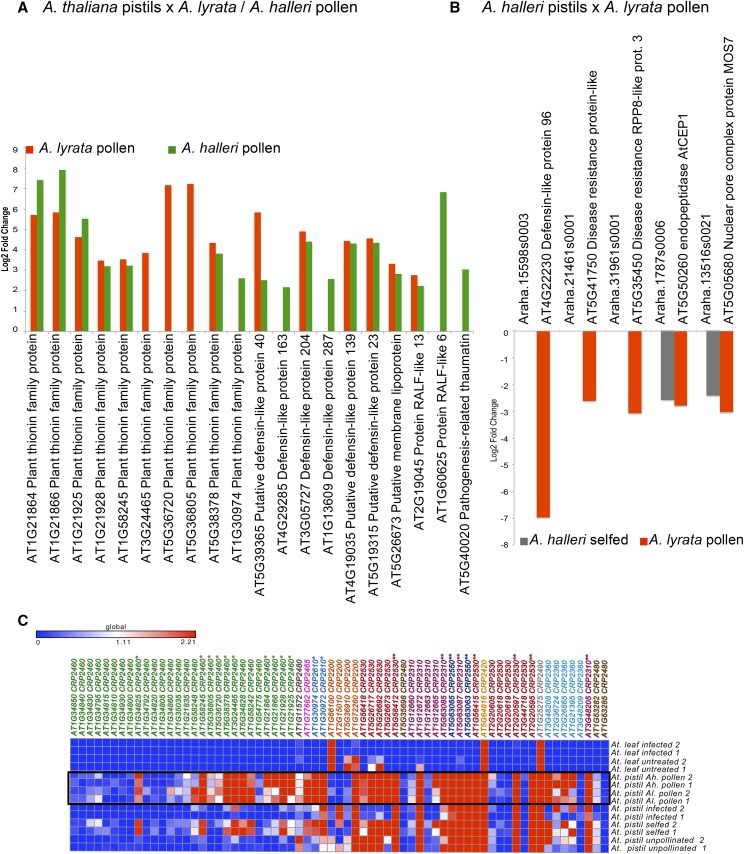
Genes differentially expressed only in response to pollen from other Arabidopsis species form the third major group detected in our analysis (indicated with stars in Fig. 2). The 152 differentially up-regulated genes expressed in A. thaliana pistils pollinated with A. lyrata or A. halleri pollen were significantly enriched with genes encoding extracellular proteins, mostly associated with plant defense-like thionins, DEFLs, and thaumatin but also others like RALFs (Fig. 7A; Supplemental Data S2). In contrast, pollination of A. halleri pistils with A. lyrata pollen did not result in the up-regulation of those gene families (Supplemental Data S3). In fact, we detected the down-regulation of several genes most similar to A. thaliana NBS-LRR disease resistance genes, a DEFL, the PCD protease AtCEP1, and the gene encoding MOS7, a regulator of the immune response (Cheng et al., 2009; Fig. 7B).
Figure 7.
CRPs up-regulated in A. thaliana pistils during interspecific pollination with A. lyrata or A. halleri. A, In A. thaliana, most of these genes encode members of PR protein families like thionins and DEFLs. B, In A. halleri, PR genes showed an opposite regulation. C, Up-regulated thionins belong mostly to CRP2460 and CRP2610 subgroups (indicated with asterisks).
Mapping the patterns of expression of thionin genes on their molecular phylogeny showed that those genes up-regulated in interspecific pollination were closely related and belong to three well-supported clades with duplications specific to A. thaliana (Fig. 7C; Supplemental Fig. S6). Most of the thionins belong to the CRP2460 and CRP2410 subgroups, which, like other CRP subgroups, are composed of genes forming several smaller clades.
A closer look to the annotation of genes up-regulated in response to foreign pollen identified RLP24, encoding a receptor-like protein with an extracellular LRR domain (AT2G33020), and NOI1, an RPM1-interacting4 family protein (AT5G63270). Both candidates belong to families usually associated with the detection of pathogens or their elimination (Supplemental Data S2). Similarly, in A. halleri, genes encoding proteins homologous to the receptors EFR and FLS2 were up-regulated. In A. thaliana, these LRR receptor-like Ser/Thr protein kinases are key in the identification of pathogen molecules and the initiation of the intracellular signaling cascade that triggers the immune response (Greeff et al., 2012). In summary, the up-regulation of genes during interspecific pollination encoding proteins involved in the recognition of pathogens and defense responses indicates evolutionary connections between the reaction to pathogens and foreign pollen that are relevant to the establishment of prezygotic reproductive barriers.
DISCUSSION
CRP Group Genes Are Differentially Regulated in Pistils Invaded by Fusarium or Foreign Pollen Tubes
The superfamily of CRPs includes well-known effectors of the immune response in plants, like DEFLs, thionins, lipid transfer proteins, and snakins (Epple et al., 1997; Silverstein et al., 2007). While many CRPs also are involved in other functions, like stem cell regulation, cell proliferation, and epidermal patterning, in the last 10 years, a growing number of studies describe the involvement of CRPs (mostly DEFLs) in key processes during plant reproduction ranging from self-incompatibility, pollen tube guidance and burst, to sperm cell activation and seed development (for review, see Bircheneder and Dresselhaus, 2016). In this study, we report two novel patterns of differential gene expression of specific CRP genes: (1) the up-regulation of genes encoding thionins, DEFLs, and RALFs in pistils of A. thaliana exposed to foreign pollen, and (2) the down-regulation of other DEFLs during defense and fertilization.
The observation that A. thaliana is reproductively isolated from A. halleri and A. lyrata suggests that the up-regulation of genes associated with immunity in response to interspecific pollen (e.g. IOS1, RLP, NOI1, thionins, DEFLs, and RALFs) may represent part of the active process of pollen tube rejection. Thionins are plant-specific CRPs toxic to bacteria, fungi, and yeast, and their expression can be induced by phytopathogenic fungi (Terras et al., 1993; Vignutelli et al., 1998; Iwai et al., 2002). In A. thaliana, nearly 70 genes encoding thionins have been annotated; most of them are expressed in the inflorescence and siliques at relatively low levels, while a handful are expressed all over the plant (Supplemental Fig. S7). Most of the up-regulated thionins in pistils pollinated with foreign pollen are closely related and specifically expressed in flowers. The expression of some thionins in pollen, pollen tubes, developing seeds, ovules, and individual cells of the female gametophyte has been reported before in A. thaliana (Jones-Rhoades et al., 2007; Wuest et al., 2010; Leydon et al., 2013; Huang et al., 2015), but their specific roles in reproduction have not been investigated. Other groups of up-regulated genes encoding secreted peptides that could feasibly contribute to interspecific pollen tube rejection include RALF-like CRP peptides. In this context, AtRALFL6 and AtRALFL13 might contribute to the inhibition of pollen tube growth, as has been reported in vitro for AtRALF4 and SlPRALF from Solanum lycopersicum (Covey et al., 2010; Morato do Canto et al., 2014).
This hypothesis is further supported by the finding that, in A. halleri pistils treated with pollen from A. lyrata, neither thionins nor other genes associated with immunity are up-regulated. Instead, in A. halleri pistils selfed or pollinated with A. lyrata pollen, we observed the down-regulation of NBS-LRRs, a DEFL, the PCD protease AtCEP1, as well as MOS7, a component of the nuclear pores required for innate immunity (Fig. 7B). Because A. halleri and A. lyrata form natural hybrids (Shimizu-Inatsugi et al., 2009), this response suggests that A. halleri pistils actively recognized their own and A. lyrata pollen as compatible, and this resulted in the down-regulation of genes that might potentially inhibit pollen tube growth.
Most of the A. thaliana and A. halleri genes down-regulated during Fusarium infection and in response to pollination belong to two large groups of CRPs homologous to those that, in Brassica spp., encode the SCR male component of self-incompatibility, SCR-like genes (SCRLs), and DEF-related pollen coat proteins (PCPs; Vanoosthuyse et al., 2001). The roles of both SCRLs and LCRs, putative homologs of Brassica spp. PCP genes (Broekaert et al., 1995), in self-compatible A. thaliana are not clear, but their down-regulation suggests that they might fulfill similar roles, such as protecting pollen and other reproductive structures from pathogen attack. Like other DEFLs, these LCR and SCRL genes potentially encode antifungal peptides (Thevissen et al., 2004; Sagaram et al., 2011). We hypothesize that their basal expression in unpollinated pistils and in pollen is part of the innate defense mechanisms protecting flower organs and gametophytes and, thus, is down-regulated by Fusarium infection (Chen et al., 2009). During pollination, these genes are likely down-regulated, because their products could cause nonspecific pollen tube growth arrest and burst, as has been shown for the gametophyte-specific DEFL ZmES4 (Amien et al., 2010).
Immunity and Pollen Defense Responses Appear to Be Initiated by Similar Recognition Mechanisms
It is already known that recognition mechanisms in self-incompatibility systems and host-pathogen interactions share common characteristics regarding their morphological, genetic, and signaling pathways (Sanabria et al., 2008; Dresselhaus and Márton, 2009). Pistils are an ideal experimental model in which to systematically investigate the genetic basis of these two processes and in which to identify candidate genes mediating intraspecific and interspecific pollen recognition. Our comparative meta-analysis showed that many DEGs are similarly regulated in response to fungal infection as well as interspecific and intraspecific pollination.
In both plant species, we identified a set of DEGs encoding proteins involved in immunity, like IOS1 and NOI1. These candidates are particularly interesting because they could interact with nonself molecules derived from pathogens or foreign pollen in the reception of interspecific pollen and further relay signals that result in pollen tube rejection and/or activation of the hypersensitive response. Evidence favoring a double role for IOS1 is that it contains a malectin-like extracellular domain like FERONIA (FER), a CrRLK1 LRR-RLK involved in both pollen tube reception and infection by powdery mildew fungus (Escobar-Restrepo et al., 2007; Kessler et al., 2010; Hok et al., 2011, 2014). However, the role of IOS1 in pollen tube growth and/or fertilization still needs to be established. The up-regulation of NOI1 in pistils pollinated with A. lyrata and A. halleri pollen caught our attention because it encodes a RIN4 family member, well-known key regulators of defense. According to the guard hypothesis, the R protein RPM1 is activated by RIN4 phosphorylation and initiates effector-triggered immunity (Eschen-Lippold et al., 2016). The up-regulation of NOI1 during interspecific pollen-pistil interactions suggests that this intracellular protein also might participate in the signaling pathway triggering pollen tube rejection.
The Transcriptomics of Arabidopsis Pistil Infection Provides Valuable Insights into the Genetic Basis of Plant-Fusarium Interactions
The thin-walled and nutrient-rich structures of the stigma and the transmitting tract facilitate fungal infection and provide direct access to ovules and developing seeds. Because of these characteristics, the Arabidopsis-Fusarium graminearum flower phytopathogenic model provides direct insight into the mechanisms that enable fungal infection and contribute to the reduction of seed set and yield decrease (Urban et al., 2002; Poppenberger et al., 2003; Brewer and Hammond-Kosack, 2015). Our transcriptome analysis of Fusarium-infected pistils provides a valuable resource for developing A. thaliana as a translational phytopathogenic model for FHB, because it reflects key biological processes also documented in the immune response of barley (Hordeum vulgare) and wheat (Triticum aestivum) to F. graminearum. As observed in the transcriptome of Fusarium-infected pistils, the infection of barley inflorescences and leaves up-regulates genes involved in the production of PR proteins, the detoxification of DON, as well as genes encoding cytochrome P450 monooxygenases, glutathione S-transferases, ABC transporters, glycosyltransferases, and peroxidases (Boddu et al., 2006; Chen et al., 2009; Bischof et al., 2011).
Phytoalexins are important secondary metabolites with antimicrobial activity, synthesized as a result of biotic stress (for review, see Ahuja et al., 2012). The transcriptomic profile of infected Arabidopsis pistils detected the up-regulation of genes encoding phytoalexin synthesis enzymes of the Brassicaceae-specific camalexin pathway (Su et al., 2011; Brewer and Hammond-Kosack, 2015) and other enzymes of the Trp secondary metabolic pathways important for the synthesis of additional antifungal substances (Bak and Feyereisen, 2001; Bednarek et al., 2009). The results of our analysis allow comparison of phytoalexin production pathways in pistils with those triggered by infection in leaves and roots, where most studies have been carried out.
The significant down-regulation of genes relevant to cell division and development in Fusarium-infected pistils provides an informative group of candidates in which to elucidate the basis of reduced seed set resulting from FHB as well as growth inhibition triggered by immunity (Smedegaard-Petersen and Tolstrup, 1985; Tian et al., 2003). Initially, it was proposed that the growth-immunity tradeoff reflects competition for available energy resources and nutrients that, when limited, cannot be allocated to both processes simultaneously. Recent studies suggest that this tradeoff is better explained by the conflictive activation of hormone signaling pathways shared by the immune response and growth processes (for review, see Eichmann and Schäfer, 2015). The identification of over 40 genes participating in the generalized, evolutionarily conserved down-regulation of cell division and development now provides a relevant group of candidates in which to investigate the hormonal and molecular basis of the growth-immunity tradeoff in the pistil. In addition to hormone signaling pathways, other factors like mycotoxins also could contribute to the observed down-regulation of genes involved in cell division in Fusarium-infected pistils. DON, the best characterized F. graminearum toxin, has mitotoxic effects on dividing plant cells, leading to chromosomal aberrations and other abnormalities that interfere with mitosis and development (Packa, 1998; Packa and Sliwinska, 2005; Masuda et al., 2007).
CONCLUSION
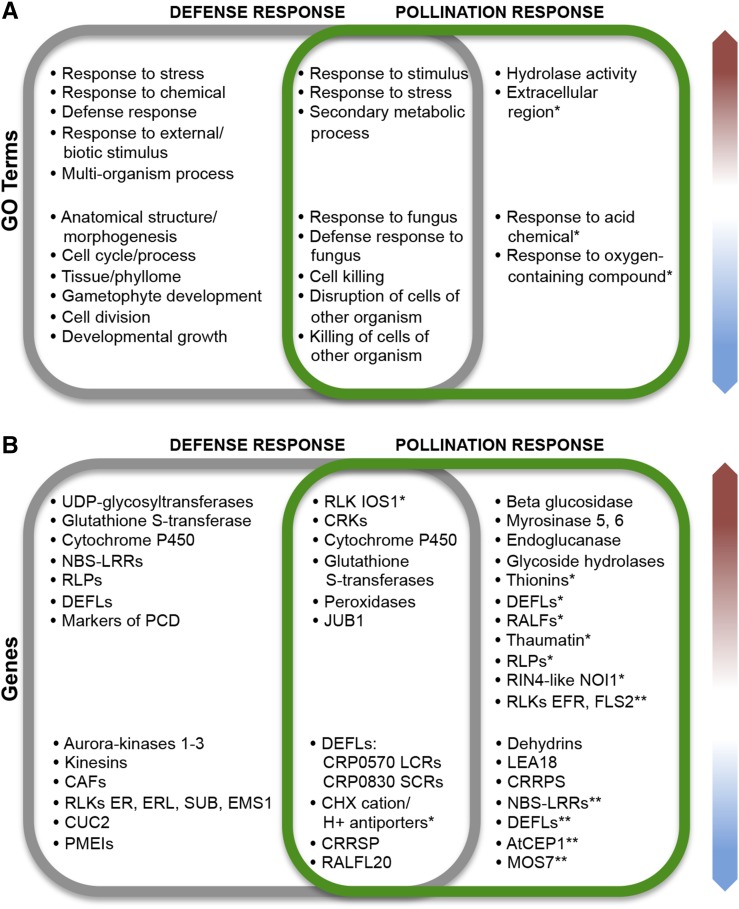
Similarities between reproductive and immune responses in the pistil make it an attractive and informative system in which to approach the effects of plant-pathogen interactions in plant fertility and the evolution of cross talk pathways between gametophytes (for summary, see Fig. 8). Our analysis showed that A. thaliana and A. halleri display similar transcriptomic responses to Fusarium infection. Similarly, at the intersection of infection-pollination, both species showed significant down-regulation of a conserved group of DEFLs. However, A. thaliana and A. halleri diverge in their transcriptional response to foreign pollen, possibly due to their distinct reproductive compatibility with A. lyrata. Overall, our transcriptomic analysis identified distinct groups of CRPs and RLKs usually associated with immune responses, like those encoding thionins and DEFLs or RLKs like IOS1, FLS2, and EFR, which, given their patterns of expression, also might participate in the reception of interspecific pollen (Fig. 8). This suggests that, during the evolution of reproductive structures, flowering plants likely adapted existing signaling pathways from the immune system to recognize and react to their own and foreign pollen grains/tubes. This hypothesis is supported by the pattern of gene expression shared by pistils during both infection and pollination. However, RLKs, DEFLs, RALFs, and NBS-LRRs form functionally dynamic gene families probably even more ancient than the plant immune response, which have been repeatedly coopted to mediate communication in different biological processes.
Figure 8.
Summary comparing the main aspects of pistil immune and pollination responses in A. thaliana and A. halleri. A, Top overrepresented GO terms shared by DEGs from both plant species (red arrow indicates up-regulated and blue arrow indicates down-regulated genes). B, Genes or gene families differentially expressed under the same conditions in A. thaliana and A. halleri pistils. Asterisks mark GO terms and genes differentially expressed only in A. thaliana (*) or only in A. halleri (**).
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of Arabidopsis thaliana ecotype Columbia were sown on soil and kept for 3 d at 4°C in the dark to promote seed stratification. Seedlings were grown in long-day conditions (16 h of light at 21°C and 8 h of darkness at 18°C, in 60% humidity) to induce flowering. Seeds of Arabidopsis halleri ecotype DE-1-Heidelberg were surface sterilized (washing stepwise in 70% ethanol (v/v), 50% bleach (v/v), and sterile distilled water) and grown on a Murashige and Skoog plate during stratification at 4°C for 10 d, after which they were moved into long-day conditions. Germinated plants were transferred to soil after 14 d. A. halleri plants with healthy rosettes were transferred to a vernalization chamber for 10 weeks at 4°C before they were moved to long-day conditions to induce flowering. Seeds of Arabidopsis lyrata MN47 were surface sterilized and kept in sterile water at 4°C for about 3 weeks in a horizontally placed Falcon vial. Seedlings were transferred to soil and grown under long-day conditions for 2 months. Next, plants with healthy rosettes were vernalized at 4°C for 10 weeks, after which they were moved back to long-day conditions to induce flowering.
Pollination Experiments
Flowers of Arabidopsis species at developmental stage 12 (Smyth et al., 1990) were emasculated 24 h before pollination. Unpollinated pistils of Arabidopsis species were collected 24 h after emasculation and quickly frozen in liquid nitrogen. Pistils of emasculated flowers of A. thaliana or A. halleri were selfed or pollinated with pollen from other Arabidopsis species, as illustrated in Figure 1. For each condition tested, three biological replicates, each consisting of about 60 pistils, were collected 8 HAP. Material was immediately frozen in liquid nitrogen and stored at −80°C before further processing.
Aniline Blue Staining
Aniline Blue staining of pistils was performed after Coimbra et al. (2010) as follows. Pollinated pistils were fixed with absolute ethanol:glacial acetic acid (3:1, v/v) for 1 to 3 h, washed three times for 5 min with sterile distilled water, and bleached overnight with 8 n NaOH. Samples were washed three times with water for 1 h and thereafter stained with Aniline Blue (0.1% Aniline Blue [w/v] dissolved in 100 mL of 0.1M K3PO4) for 30 to 60 min and observed with a fluorescence microscope at 350 to 400 nm.
Fusarium graminearum Culturing and Infection
We employed F. graminearum strain SG005/Fg005, a single-spore isolate obtained in 2008 from spring barley (Hordeum vulgare) grain harvested in Freising, Germany (Linkmeyer, 2012). This strain was propagated in induction medium (1 g L−1 yeast extract, 15 g L−1 carboxyl methyl cellulose, 1 g L−1 NH4NO3, 1 g L−1 KH2PO4, and 0.5 g L−1 MgSO40∙7H2O) at 28°C for 10 to 14 d and then filtered through a sterile Spandex bandage. The filtrate was centrifuged at 5,000 rpm for 10 min at 4°C, and the supernatant was discarded. Inoculation medium for infection consists of conidia resuspended in sterile distilled water containing 1% Tween to a final concentration of 8 to 9 × 105 spores mL−1. As described above, flowers of Arabidopsis species were emasculated 24 h before inoculation. Flowers were dipped into conidial suspension for 20 min followed by spraying of conidial solution onto pistils. Plants were covered with a plastic bag sprayed with water to maintain relative humidity and favor the development of Fusarium infection for 72 h under long-day conditions. Following incubation, pistils and cauline leaves were collected, immediately frozen in liquid nitrogen, and stored at −80°C before further processing.
Fusarium infection was visualized in leaves and pistils by WGA-TMR staining after Hoefle et al. (2011). In brief, leaves and pistils were fixed in a solution of 750 mL of ethanol, 250 mL of chloroform, and 1.5 g of trichloroacetic acid for 2 d, then washed with distilled water, incubated for 20 min in 1× phosphate-buffered saline buffer (8 g of NaCl, 2.8 g of Na2HPO4∙2H2O, 0.24 g of KH2PO4, and 0.2 g of KCl in 1 L of water, pH 7.4), and transferred to 5 mL of WGA-TMR staining solution (5 mL of 1× phosphate-buffered saline, 50 µg µL−1 BSA, and 50 µL of WGA-TMR). After vacuum infiltration for 20 min, leaves and pistils were incubated in the staining solution for at least 24 h in the dark. Before staining, some samples were cleared with chloral hydrate as follows. Pistils were fixed overnight in ethanol:acetic acid (9:1, v/v) and then washed stepwise for 30 to 60 min with 90% (v/v) and 70% ethanol (v/v). Finally, samples where cleared overnight in a solution of chloral hydrate (2.5 g of chloral hydrate to 1 mL of 30% glycerol v/v).
Leaves and pistils were viewed using a confocal laser scanning microscope (LSM 510). Samples were excited by a 561-nm laser line, and emission was detected at 571 to 610 nm.
RNA Extraction and Preparation of cDNA Libraries
Total RNA was extracted from all tissues using the RNeasy Mini Plant Kit (Qiagen), cleaned from any residual genomic DNA, and tested for integrity and concentration with a Bioanalyzer 2100 using an RNA 6000 Nano assay (Agilent Technologies). All cDNA libraries were prepared from 500 ng of total RNA using the TruSeq RNA sample preparation kit (Illumina) at the Center for Fluorescent Bioanalytics (University of Regensburg). Libraries were quantified using the KAPA SYBR FAST ABI Prism Library Quantification Kit (Kapa Biosystems). Equimolar amounts of each library were pooled, and the pools were used for cluster generation using cBot (TruSeq PE Cluster Kit version 3). Sequencing runs were performed on a HiSeq 1000 instrument using the indexed 2 × 100 cycles paired-end protocol and TruSeq SBS version 3 reagents. Image analyses and base calling, which yield .bcl files, were converted into .fastq files with CASAVA 1.8.2 software. Libraries were multiplexed to obtain 50 to 60 million reads with a mean quality score of at least 37 per biological replicate.
RNA-seq and Transcriptomic Analyses of Differential Gene Expression
Read quality was assessed with FastQC (Babraham 2011) and trimmed in the first and last 15 residues of every read. Trimmed reads were mapped with CLC Genomics Workbench 7 (Qiagen) to the corresponding Columbia-0 TAIR10 version of the A. thaliana genome or to version 1.1 of the A. halleri genome downloaded from Phytozome (phytozome.jgi.doe.gov) as follows: mapping only to gene regions (A. thaliana) or to genic and intergenic regions (A. halleri), 10 maximum number of hits for a read, both strands, count paired reads as two, expression value as total counts, no global alignment, similarity fraction = 0.8, length fraction = 0.8, mismatch cost = 2, insertion cost = 3, and deletion cost = 3.
Variations in the levels of expression between the biological replicates of each condition were assessed via box plots and principal component analysis. To maintain similar levels of variation between biological replicates, only the two most similar samples of each experimental and control condition were further considered for the analysis of differential gene expression (Supplemental Table S1). Differential gene expression was investigated with the exact test for two-group comparisons from edgeR (Robinson et al., 2010) implemented in the CLC Workbench. In this analysis, read counts obtained from pollinated or infected pistil treatments described previously were compared with the corresponding samples of untreated pistils of A. thaliana or A. halleri. We considered genes as DEGs that had an FDR-corrected P < 0.0005 and an expression fold change ≥ 2 (up-regulation) or ≤ −2 (down-regulation) and that were expressed with RPKM ≥ 1.
Analysis of GO Terms
Venn diagrams were employed to identify unique and shared elements in the lists of DEGs in each experimental condition (Bardou et al., 2014). First, genes differentially expressed in both pistils and leaves were identified and excluded. The resulting data set of pistil-exclusive genes was further analyzed to define three major groups of genes exclusively up- and down-regulated in infected pistils, pistils pollinated with foreign pollen, or differentially expressed both in pistils during infection with F. graminearum and during pollination (selfing and interspecific). These groups of DEGs were tested for significant overrepresentation of GO terms based on their annotation in the Panther classification system of the Gene Ontology Reference Genome Project (version 11.0; pantherdb.org). Overrepresentation tests were performed based on the reference list of A. thaliana and the complete annotation data sets of GO terms describing biological processes. Overrepresentation tests for A. halleri DEGs were based on GO annotation of best hits in the A. thaliana genome. All analyses were Bonferroni corrected for multiple testing.
qPCR Assays
cDNA was synthesized from total RNA samples originally employed for transcriptome sequencing. This assay involved two biological replicates, each represented by two cDNA pools that provided the template for two technical replicate reactions. The synthesis of cDNA was performed with the SuperScript III RT kit (Invitrogen), starting from 1 µg of total RNA and using poly-T primer AB05 as described previously (Mondragón-Palomino and Theissen, 2011). All primers were designed with the programs Primer Premier 6 and Beacon Designer 8 from Premier Biosoft and ordered from the supplier Biomers. Amplification efficiencies were calculated based on standard curves as described previously (Mondragón-Palomino and Theissen, 2011). Primer sequences with their corresponding melting temperatures and amplification efficiencies are provided in Supplemental Table S2. Standard 20-µL qPCR with KAPA SYBR FAST qPCR MasterMix Universal (Peqlab) was performed in a Mastercycler ep realplex gradient S (Eppendorf). Each assay included the following controls: nontemplate, -RT control, and positive. The specificity of reactions was assessed via melting curves and amplicon size on agarose gels. Resulting Cq (cycle of quantification) values were imported into qbase+ (Biogazelle). Quality control settings required that Cq values from technical replicates differ by less than 0.5 cycles, and the specific amplification efficiencies were determined by standard curves. Normalization was based on two internal reference genes, AT2G19270 and AT1G10310. These genes were chosen because they are not differentially expressed in any of the conditions investigated. The resulting calibrated normalized relative quantities were then exported into Excel and log2 transformed for further analysis and calculation of fold change values.
Molecular Phylogeny
The amino acid sequences for annotated thionins of A. thaliana, A. halleri, A. lyrata, and Capsella grandiflora were downloaded from Phytozome version 10.3 and aligned with the MUSCLE program implemented in SeaView 4.5.3. (Gouy et al., 2010). The maximum likelihood gene tree was estimated with the program PhyML using the GTR model, starting with the topology of an optimized BioNJ tree. Branch support was calculated with aLRT (SH-like), and invariable sites and across-site rate variation were optimized. Tree-searching operations took the best result from Nearest-Neighbor-Interchange and Subtree-Pruning-and-Regrafting.
Accession Numbers
Data will be available from the National Center for Biotechnology Information Sequence Read Archive under project number PRJNA384934. All other materials are available from the authors upon request.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Proliferation of F. graminearum on leaves.
Supplemental Figure S2. Correlation between RNA-seq and qPCR fold change measurements of 14 DEFL genes.
Supplemental Figure S3. Comparison of log2 fold change values obtained in the differential expression analysis of RNA-seq and qPCR of 14 DEFLs from A. thaliana.
Supplemental Figure S4. Venn diagrams for identifying DEGs in both pistils and leaves of A. thaliana or A. halleri.
Supplemental Figure S5. Additional members of DEFL subgroups CRP0570 and CRP0830 are down-regulated in A. halleri Fusarium-infected pistils.
Supplemental Figure S6. Maximum likelihood tree of Arabidopsis thionins.
Supplemental Figure S7. Genevestigator RNA-seq expression in the anatomy of A. thaliana.
Supplemental Table S1. Characteristics of the samples employed for RNA-seq analysis.
Supplemental Table S2. Primers employed in the qPCR assay.
Supplemental Data S1. DEGs in A. thaliana and A. halleri pistils and leaves.
Supplemental Data S2. GO term overrepresentation and annotation of genes differentially expressed in A. thaliana pistils.
Supplemental Data S3. GO term overrepresentation and annotation of genes differentially expressed in A. halleri pistils.
Acknowledgments
We thank Nina Lantzouni and Claus Schwechheimer for introduction and access to the CLC Genomics Workbench; Andrea Linkmeyer and Ralph Hückelhoven for providing F. graminearum strain SG005 and Lena Müller for providing seeds from A. halleri; Carolin Delker for providing seeds of A. lyrata; Kriss Spalvins for drawing the pistil diagrams; Benedikt Müller and Mihaela Márton for helpful consultation on confocal microscopy; and Alice Cheung and Kevin Begcy for discussions of the article. The U.S. Department of Energy Joint Genome Institute produced the genome data of A. halleri employed to map our RNA-seq reads.
Glossary
- FHB
Fusarium head blight
- DEGs
differentially expressed genes
- HAP
hours after pollination
- DAI
days after infection
- RNA-seq
RNA sequencing
- qPCR
quantitative PCR
- FDR
false discovery rate
- PCD
programmed cell death
- DON
deoxynivalenol
- SA
salicylic acid
- JA
jasmonic acid
- GO
Gene Ontology
- WGA-TMR
wheat germ agglutinin-tetramethylrhodamine
Footnotes
This work was supported by the German Research Council through the collaborative research project EVOREP (grant no. DR 334/8-1 to T.D.) and by the Bavarian Program for the Realization of Equal Opportunities for Women in Research (to M.M.-P.).
Articles can be viewed without a subscription.
References
- Ahuja I, Kissen R, Bones AM (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17: 73–90 [DOI] [PubMed] [Google Scholar]
- Amien S, Kliwer I, Márton ML, Debener T, Geiger D, Becker D, Dresselhaus T (2010) Defensin-like ZmES4 mediates pollen tube burst in maize via opening of the potassium channel KZM1. PLoS Biol 8: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babraham B. (2011) FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Institute, Cambridge, UK [Google Scholar]
- Bak S, Feyereisen R (2001) The involvement of two p450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol 127: 108–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C (2014) jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 15: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P, Piślewska-Bednarek M, Svatoš A, Schneider B, Doubský J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bennett JW, Klich M (2003) Mycotoxins. Clin Microbiol Rev 16: 497–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bircheneder S, Dresselhaus T (2016) Why cellular communication during plant reproduction is particularly mediated by CRP signalling. J Exp Bot 67: 4849–4861 [DOI] [PubMed] [Google Scholar]
- Bischof M, Eichmann R, Hückelhoven R (2011) Pathogenesis-associated transcriptional patterns in Triticeae. J Plant Physiol 168: 9–19 [DOI] [PubMed] [Google Scholar]
- Boddu J, Cho S, Kruger WM, Muehlbauer GJ (2006) Transcriptome analysis of the barley-Fusarium graminearum interaction. Mol Plant Microbe Interact 19: 407–417 [DOI] [PubMed] [Google Scholar]
- Brewer HC, Hammond-Kosack KE (2015) Host to a stranger: Arabidopsis and Fusarium ear blight. Trends Plant Sci 20: 651–663 [DOI] [PubMed] [Google Scholar]
- Broekaert WF, Terras FR, Cammue BP, Osborn RW (1995) Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol 108: 1353–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Steed A, Travella S, Keller B, Nicholson P (2009) Fusarium graminearum exploits ethylene signalling to colonize dicotyledonous and monocotyledonous plants. New Phytol 182: 975–983 [DOI] [PubMed] [Google Scholar]
- Cheng YT, Germain H, Wiermer M, Bi D, Xu F, García AV, Wirthmueller L, Després C, Parker JE, Zhang Y, et al. (2009) Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21: 2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coimbra S, Costa M, Mendes MA, Pereira AM, Pinto J, Pereira LG (2010) Early germination of Arabidopsis pollen in a double null mutant for the arabinogalactan protein genes AGP6 and AGP11. Sex Plant Reprod 23: 199–205 [DOI] [PubMed] [Google Scholar]
- Covey PA, Subbaiah CC, Parsons RL, Pearce G, Lay FT, Anderson MA, Ryan CA, Bedinger PA (2010) A pollen-specific RALF from tomato that regulates pollen tube elongation. Plant Physiol 153: 703–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond OJ, Manners JM, Stephens AE, Maclean DJ, Schenk PM, Gardiner DM, Munn AL, Kazan K (2008) The Fusarium mycotoxin deoxynivalenol elicits hydrogen peroxide production, programmed cell death and defence responses in wheat. Mol Plant Pathol 9: 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T, Franklin-Tong N (2013) Male-female crosstalk during pollen germination, tube growth and guidance, and double fertilization. Mol Plant 6: 1018–1036 [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Márton ML (2009) Micropylar pollen tube guidance and burst: adapted from defense mechanisms? Curr Opin Plant Biol 12: 773–780 [DOI] [PubMed] [Google Scholar]
- Eichmann R, Schäfer P (2015) Growth versus immunity: a redirection of the cell cycle? Curr Opin Plant Biol 26: 106–112 [DOI] [PubMed] [Google Scholar]
- Epple P, Apel K, Bohlmann H (1997) Overexpression of an endogenous thionin enhances resistance of Arabidopsis against Fusarium oxysporum. Plant Cell 9: 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschen-Lippold L, Jiang X, Elmore JM, Mackey DM, Shan L, Coaker GL, Scheel D, Lee J (2016) Bacterial AvrRpt2-like cysteine proteases block activation of the Arabidopsis mitogen-activated protein kinases, MPK4 and MPK11. Plant Physiol 171: 2223–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U (2007) The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317: 656–660 [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194 [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224 [DOI] [PubMed] [Google Scholar]
- Greeff C, Roux M, Mundy J, Petersen M (2012) Receptor-like kinase complexes in plant innate immunity. Front Plant Sci 3: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Schmid M, Hierl G, Terneus K, Tan L, Lottspeich F, Kieliszewski MJ, Gietl C (2008) KDEL-tailed cysteine endopeptidases involved in programmed cell death, intercalation of new cells, and dismantling of extensin scaffolds. Am J Bot 95: 1049–1062 [DOI] [PubMed] [Google Scholar]
- Hoefle C, Huesmann C, Schultheiss H, Börnke F, Hensel G, Kumlehn J, Hückelhoven R (2011) A barley ROP GTPase ACTIVATING PROTEIN associates with microtubules and regulates entry of the barley powdery mildew fungus into leaf epidermal cells. Plant Cell 23: 2422–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hok S, Allasia V, Andrio E, Naessens E, Ribes E, Panabières F, Attard A, Ris N, Clément M, Barlet X, et al. (2014) The receptor kinase IMPAIRED OOMYCETE SUSCEPTIBILITY1 attenuates abscisic acid responses in Arabidopsis. Plant Physiol 166: 1506–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hok S, Danchin EG, Allasia V, Panabières F, Attard A, Keller H (2011) An Arabidopsis (malectin-like) leucine-rich repeat receptor-like kinase contributes to downy mildew disease. Plant Cell Environ 34: 1944–1957 [DOI] [PubMed] [Google Scholar]
- Huang Q, Dresselhaus T, Gu H, Qu LJ (2015) Active role of small peptides in Arabidopsis reproduction: expression evidence. J Integr Plant Biol 57: 518–521 [DOI] [PubMed] [Google Scholar]
- Iwai T, Kaku H, Honkura R, Nakamura S, Ochiai H, Sasaki T, Ohashi Y (2002) Enhanced resistance to seed-transmitted bacterial diseases in transgenic rice plants overproducing an oat cell-wall-bound thionin. Mol Plant Microbe Interact 15: 515–521 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Borevitz JO, Preuss D (2007) Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genet 3: 1848–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U (2010) Conserved molecular components for pollen tube reception and fungal invasion. Science 330: 968–971 [DOI] [PubMed] [Google Scholar]
- Lewis D, Crowe LK (1958) Unilateral interspecific incompatibility in flowering plants. Heredity 12: 233–256 [Google Scholar]
- Leydon AR, Beale KM, Woroniecka K, Castner E, Chen J, Horgan C, Palanivelu R, Johnson MA (2013) Three MYB transcription factors control pollen tube differentiation required for sperm release. Curr Biol 23: 1209–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkmeyer A. (2012) Fusarium head blight of barley: epidemiology and host-pathogen interaction. PhD thesis. Technical University of Munich, Munich, Germany
- Masuda D, Ishida M, Yamaguchi K, Yamaguchi I, Kimura M, Nishiuchi T (2007) Phytotoxic effects of trichothecenes on the growth and morphology of Arabidopsis thaliana. J Exp Bot 58: 1617–1626 [DOI] [PubMed] [Google Scholar]
- McMullen M, Bergstrom G, De Wolf E (2012) A unified effort to fight an enemy of wheat and barley: Fusarium head blight. Plant Dis 96: 1712–1728 [DOI] [PubMed] [Google Scholar]
- Miller SS, Chabot D, Ouellet T (2004) Use of a Fusarium graminearum strain transformed with green fluorescent protein to study infection in wheat (Triticum aestivum). Can J Plant Pathol 26: 453–463 [Google Scholar]
- Mondragón-Palomino M, Theissen G (2011) Conserved differential expression of paralogous DEFICIENS- and GLOBOSA-like MADS-box genes in the flowers of Orchidaceae: refining the ‘orchid code.’ Plant J 66: 1008–1019 [DOI] [PubMed] [Google Scholar]
- Morato do Canto A, Ceciliato PH, Ribeiro B, Ortiz Morea FA, Franco Garcia AA, Silva-Filho MC, Moura DS (2014) Biological activity of nine recombinant AtRALF peptides: implications for their perception and function in Arabidopsis. Plant Physiol Biochem 75: 45–54 [DOI] [PubMed] [Google Scholar]
- Morita-Yamamuro C, Tsutsui T, Sato M, Yoshioka H, Tamaoki M, Ogawa D, Matsuura H, Yoshihara T, Ikeda A, Uyeda I, et al. (2005) The Arabidopsis gene CAD1 controls programmed cell death in the plant immune system and encodes a protein containing a MACPF domain. Plant Cell Physiol 46: 902–912 [DOI] [PubMed] [Google Scholar]
- Müller LM, Lindner H, Pires ND, Gagliardini V, Grossniklaus U (2016) A subunit of the oligosaccharyltransferase complex is required for interspecific gametophyte recognition in Arabidopsis. Nat Commun 7: 10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naur P, Petersen BL, Mikkelsen MD, Bak S, Rasmussen H, Olsen CE, Halkier BA (2003) CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol 133: 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S, Tsutsui H, Shiina K, Sprunck S, Takeuchi H, Yui R, Kasahara RD, Hamamura Y, Mizukami A, Susaki D, et al. (2009) Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361 [DOI] [PubMed] [Google Scholar]
- Olvera-Carrillo Y, Van Bel M, Van Hautegem T, Fendrych M, Huysmans M, Simaskova M, van Durme M, Buscaill P, Rivas S, Coll N, et al. (2015) A conserved core of programmed cell death indicator genes discriminates developmentally and environmentally induced programmed cell death in plants. Plant Physiol 169: 2684–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packa D. (1998) Potential genotoxicity of Fusarium mycotoxins in Vicia and Pisum cytogenetic tests. J Appl Genet 2: 171–192 [Google Scholar]
- Packa D, Sliwinska E (2005) Trichothecene fusarial toxins perturb the cell cycle in meristematic cells of Secale cereale L., Triticum aestivum L. and Vicia faba L. Caryologia 58: 86–93 [Google Scholar]
- Park SY, Jauh GY, Mollet JC, Eckard KJ, Nothnagel EA, Walling LL, Lord EM (2000) A lipid transfer-like protein is necessary for lily pollen tube adhesion to an in vitro stylar matrix. Plant Cell 12: 151–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B, Berthiller F, Lucyshyn D, Sieberer T, Schuhmacher R, Krska R, Kuchler K, Glössl J, Luschnig C, Adam G (2003) Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J Biol Chem 278: 47905–47914 [DOI] [PubMed] [Google Scholar]
- Qu LJ, Li L, Lan Z, Dresselhaus T (2015) Peptide signalling during the pollen tube journey and double fertilization. J Exp Bot 66: 5139–5150 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagaram US, Pandurangi R, Kaur J, Smith TJ, Shah DM (2011) Structure-activity determinants in antifungal plant defensins MsDef1 and MtDef4 with different modes of action against Fusarium graminearum. PLoS ONE 6: e18550–e18513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanabria N, Goring D, Nürnberger T, Dubery I (2008) Self/nonself perception and recognition mechanisms in plants: a comparison of self-incompatibility and innate immunity. New Phytol 178: 503–514 [DOI] [PubMed] [Google Scholar]
- Schopfer CR, Nasrallah ME, Nasrallah JB (1999) The male determinant of self-incompatibility in Brassica. Science 286: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Shimizu-Inatsugi R, Lihová J, Iwanaga H, Kudoh H, Marhold K, Savolainen O, Watanabe K, Yakubov VV, Shimizu KK (2009) The allopolyploid Arabidopsis kamchatica originated from multiple individuals of Arabidopsis lyrata and Arabidopsis halleri. Mol Ecol 18: 4024–4048 [DOI] [PubMed] [Google Scholar]
- Silverstein KA, Moskal WA Jr, Wu HC, Underwood BA, Graham MA, Town CD, VandenBosch KA (2007) Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J 51: 262–280 [DOI] [PubMed] [Google Scholar]
- Smedegaard-Petersen V, Tolstrup K (1985) The limiting effect of disease resistance on yield. Annu Rev Phytopathol 23: 475–490 [Google Scholar]
- Smolen G, Bender J (2002) Arabidopsis cytochrome P450 cyp83B1 mutations activate the tryptophan biosynthetic pathway. Genetics 160: 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprunck S, Rademacher S, Vogler F, Gheyselinck J, Grossniklaus U, Dresselhaus T (2012) Egg cell-secreted EC1 triggers sperm cell activation during double fertilization. Science 338: 1093–1097 [DOI] [PubMed] [Google Scholar]
- Su T, Xu J, Li Y, Lei L, Zhao L, Yang H, Feng J, Liu G, Ren D (2011) Glutathione-indole-3-acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell 23: 364–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terras FR, Eggermont K, Kovaleva V, Raikhel NV, Osborn RW, Kester A, Rees SB, Torrekens S, Van Leuven F, Vanderleyden J, et al. (1995) Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell 7: 573–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terras FR, Torrekens S, Van Leuven F, Osborn RW, Vanderleyden J, Cammue BP, Broekaert WF (1993) A new family of basic cysteine-rich plant antifungal proteins from Brassicaceae species. FEBS Lett 316: 233–240 [DOI] [PubMed] [Google Scholar]
- Thevissen K, Warnecke DC, François IE, Leipelt M, Heinz E, Ott C, Zähringer U, Thomma BP, Ferket KK, Cammue BP (2004) Defensins from insects and plants interact with fungal glucosylceramides. J Biol Chem 279: 3900–3905 [DOI] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77 [DOI] [PubMed] [Google Scholar]
- Urban M, Daniels S, Mott E, Hammond-Kosack K (2002) Arabidopsis is susceptible to the cereal ear blight fungal pathogens Fusarium graminearum and Fusarium culmorum. Plant J 32: 961–973 [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V, Miege C, Dumas C, Cock JM (2001) Two large Arabidopsis thaliana gene families are homologous to the Brassica gene superfamily that encodes pollen coat proteins and the male component of the self-incompatibility response. Plant Mol Biol 46: 17–34 [DOI] [PubMed] [Google Scholar]
- Vignutelli A, Wasternack C, Apel K, Bohlmann H (1998) Systemic and local induction of an Arabidopsis thionin gene by wounding and pathogens. Plant J 14: 285–295 [DOI] [PubMed] [Google Scholar]
- Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor MI, Asensi-Fabado MA, Munné-Bosch S, Antonio C, Tohge T, et al. (2012) JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24: 482–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenführer J, von Mering C, Grossniklaus U (2010) Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol 20: 506–512 [DOI] [PubMed] [Google Scholar]
- Xu J, Li Y, Wang Y, Liu H, Lei L, Yang H, Liu G, Ren D (2008) Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J Biol Chem 283: 26996–27006 [DOI] [PubMed] [Google Scholar]
- Zhou LZ, Höwing T, Müller B, Hammes UZ, Gietl C, Dresselhaus T (2016) Expression analysis of KDEL-CysEPs programmed cell death markers during reproduction in Arabidopsis. Plant Reprod 29: 265–272 [DOI] [PubMed] [Google Scholar]