The chloroplast-located DnaJ-like protein HCF222 is indispensable for accumulation of the cytochrome b6f complex and to a lesser extent necessary for wild-type levels of PSI and PSII.
Abstract
To understand the biogenesis of the thylakoid membrane in higher plants and to identify auxiliary proteins required to build up this highly complex membrane system, we have characterized the allelic nuclear mutants high chlorophyll fluorescence222-1 (hcf222-1) and hcf222-2 and isolated the causal gene by map-based cloning. In the ethyl methanesulfonate-induced mutant hcf222-1, the accumulation of the cytochrome b6f (Cytb6f) complex was reduced to 30% compared with the wild type. Other thylakoid membrane complexes accumulated to normal levels. The T-DNA knockout mutant hcf222-2 showed a more severe defect with respect to thylakoid membrane proteins and accumulated only 10% of the Cytb6f complex, accompanied by a reduction in photosystem II, the photosystem II light-harvesting complex, and photosystem I. HCF222 encodes a protein of 99 amino acids in Arabidopsis (Arabidopsis thaliana) that has similarities to the cysteine-rich zinc-binding domain of DnaJ chaperones. The insulin precipitation assay demonstrated that HCF222 has disulfide reductase activity in vitro. The protein is conserved in higher plants and bryophytes but absent in algae and cyanobacteria. Confocal fluorescence microscopy showed that a fraction of HCF222-green fluorescent protein was detectable in the endoplasmic reticulum but that it also could be recognized in chloroplasts. A fusion construct of HCF222 containing a plastid transit peptide targets the protein into chloroplasts and was able to complement the mutational defect. These findings indicate that the chloroplast-targeted HCF222 is indispensable for the maturation and/or assembly of the Cytb6f complex and is very likely involved in thiol-disulfide biochemistry at the thylakoid membrane.
In true leaves of plants, chloroplasts develop from proplastids, which are small, colorless precursors surrounded by the inner and outer envelope, with rudimentary membranes inside. During leaf growth, these plastids differentiate into mature chloroplasts with an elaborate membrane system that is densely packed with the pigment-protein complexes of the photosynthetic machinery. This machinery is composed of PSII, where light-induced water oxidation takes place, the cytochrome b6f (Cytb6f) complex, PSI, and the ATP synthase. Biogenesis of these membrane complexes requires regulation and assistance mainly at posttranscriptional and posttranslational steps of gene expression; consequently, genetic and biochemical approaches identified many auxiliary factors required during these processes (Lyska et al., 2013b).
The Cytb6f complex acts as a plastoquinol-plastocyanin reductase and is the smallest of the four photosynthetic complexes. The native form in higher plants and algae is a symmetric dimer of 220 kD (Mosser et al., 1997; Kurisu et al., 2003). The monomeric complex in higher plants consists of eight subunits: the plastome-encoded PetA (cytochrome f), PetB (cytochrome b6), PetD, PetG, PetL, and PetN, and the nucleus-encoded PetC (Rieske iron-sulfur protein), and PetM. These subunits bound seven prosthetic groups, comprising two b-type hemes and atypically bound heme cn in cytochrome b6, a c-type heme in cytochrome f, a [2Fe-2S] cluster in the Rieske protein, and chlorophyll a and β-carotene (Baniulis et al., 2008). In a subpopulation of the Cytb6f complex, the ferredoxin-NADP+-reductase (FNR) was identified as a stoichiometric component (Zhang et al., 2001). The integrity of the complex depends on all subunits except PetL and FNR (Schöttler et al., 2015). Despite detailed knowledge about the structure and composition of the complex, less is known about protein maturation and the mechanisms involved in its assembly.
A transthylakoid disulfide-reducing pathway, which is homologous to the system I disulfide-reducing pathway in the plasma membrane of bacteria, is required during covalent heme binding to cytochrome f (Kranz et al., 2009). Central components of this pathway are CCDA as a membrane integral protein and the thioredoxin-like HIGH CHLOROPHYLL FLUORESCENCE164 (HCF164)/CCS5, which is exposed to the lumenal site of the thylakoids (Lennartz et al., 2001; Page et al., 2004; Gabilly et al., 2010). It is suggested that electrons provided by stromal thioredoxin are transferred to the lumen via CCDA and HCF164/CCS5 to the oxidized CxxCH motif of the apocytochrome f, thereby generating the necessary sulfhydryls for heme ligation (Motohashi and Hisabori, 2006). Recently, the protein CCS4 was identified in Chlamydomonas reinhardtii that also is an essential component for heme attachment to cytochrome f (and cytochrome c6); its exact role is yet unclear, but there is evidence that it is involved in the disulfide-reducing pathway at the thylakoids (Gabilly et al., 2011). While heme binding in the lumen absolutely requires the transthylakoid disulfide-reducing pathway, covalent attachment of heme cn to Cys-35 of cytochrome b6 requires the CCB (for cofactor-binding, Cytb6f complex, and cytochrome b6) factors 1 to 4 at the stromal site of the membrane (Saint-Marcoux et al., 2009). These membrane integral proteins are suggested to transiently interact with cytochrome b6 during heme delivery to the apocytochrome. Much less is known about incorporation of the iron-sulfur cluster into the Rieske protein. It is suggested that an assembly machinery related to the eubacterial sulfur utilization factor system is involved (Balk and Pilon, 2011); however, its involvement in assembly of the Rieske [2Fe-2S] cluster remains to be shown. Two additional proteins, DAC and HCF153, are required for normal accumulation of the Cytb6f complex; whereas DAC is suggested to play a role during PetD assembly/stabilization (Xiao et al., 2012), the precise function of HCF153 remains to be determined (Lennartz et al., 2006).
Protein maturation is coupled with the folding of proteins into their native conformation and requires, in many cases, the formation of disulfide bridges between thiols of conserved Cys residues. The process is designated as oxidative folding, and diverse studies revealed existence of many disulfide-bonded proteins in each chloroplast compartment except the intermembrane space (Hall et al., 2010; Kieselbach, 2013). Protein precursors are imported into chloroplasts in an unfolded conformation, a state that is favored by the reducing environment of the cytosol. Following import, oxidative protein folding takes place at the final chloroplast compartment: the stroma, the thylakoid lumen, or the thylakoid membrane. Oxidative folding of proteins is studied thoroughly in the endoplasmic reticulum (ER), where disulfide formation is catalyzed by various protein disulfide isomerases (PDIs), which donate disulfides to substrate proteins via a thiol-disulfide exchange reaction (Aller and Meyer, 2013; Onda, 2013). The reduced PDI is reoxidized by a disulfide-generating enzyme, which relays the oxidizing power from molecular oxygen. Only a few plastid proteins are known that have disulfide bond-forming or disulfide isomerase activity (Kieselbach, 2013). One example is LUMEN THIOL OXIDOREDUCTASE1, which catalyzes the formation of a disulfide bond in the extrinsic PsbO subunit of the oxygen-evolving complex of PSII (Feng et al., 2011; Karamoko et al., 2011). The protein consists of a vitamin K epoxide reductase domain with four putative transmembrane helices and a soluble, lumen-exposed thioredoxin-like domain, which interacts with PsbO. The zinc-finger domain of DnaJ proteins characterizes a different group of thylakoid-associated thiol-disulfide oxidoreductases. In DnaJ, this domain contains two CxxCxnCxxC motifs, able to bind two zinc atoms (Shi et al., 2005). The zinc finger is indispensable for thiol-disulfide oxidoreductase activity of the Escherichia coli chaperone DnaJ (Tang and Wang, 2001), and it has the capacity to recognize and bind unfolded or misfolded proteins (Langer et al., 1992; Szabo et al., 1996). Two representative DnaJ-like zinc-finger proteins of the chloroplast are LOW QUANTUM YIELD1 (LQY1) and PHOTOSYSTEM ASSEMBLY2 (PSA2). LQY1 is involved in PSII repair and reassembly (Lu et al., 2011), and PSA2 is essential during PSI biogenesis (Fristedt et al., 2014). As both proteins showed PDI-like activity, a function in the folding of specific target proteins was suggested. Although classical PDIs and PDI-like proteins were found in chloroplasts, their involvement in protein folding was not demonstrated (Kieselbach, 2013). In contrast, a role during light regulation of D1 synthesis was reported for Arabidopsis (Arabidopsis thaliana) PDI6 (Wittenberg et al., 2014), comparable to the PDI RB60 in C. reinhardtii (Alergand et al., 2006).
To understand the regulatory and functional roles of auxiliary proteins during biogenesis of the photosynthetic apparatus and, thus, the thylakoid membrane, we exploited an ethyl methanesulfonate (EMS)-mutagenized Arabidopsis population and isolated mutants with impaired photosynthetic efficiency. In this screen, we identified a mutant of the gene HCF222. To improve the functional analysis of the encoded gene product, we also investigated the T-DNA knockout mutant and characterized the phenotypes of both mutants in detail. Our results show that HCF222 is required predominantly for stable accumulation of the Cytb6f complex but, in addition, it is also necessary to ensure adequate levels of PSI and PSII. HCF222 encodes a small protein with homology to the zinc-finger domain of E. coli DnaJ. The protein is dually located to the ER and the chloroplast. Complementation analysis revealed that the mutational lesion is related to the plastid-located HCF222.
RESULTS
Mutation of the Gene At5g15802 Results in a Seedling-Lethal Photosynthesis Mutant
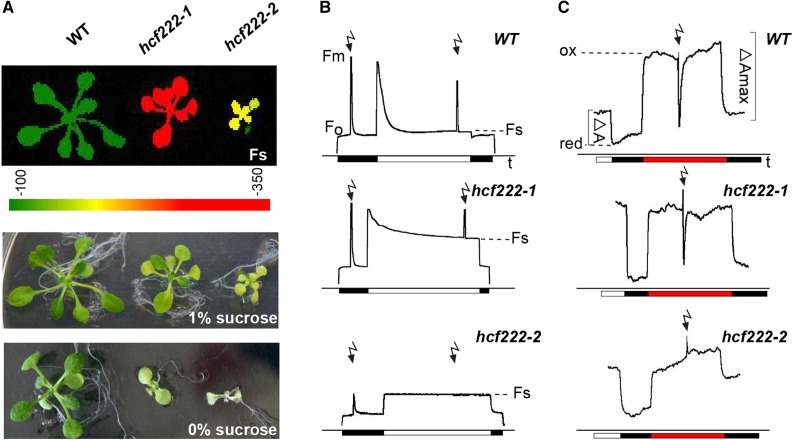
In an EMS-mutagenized population of Arabidopsis (ecotype Columbia-0 [Col-0]), we screened for mutants with an hcf phenotype. This phenotype is indicative for reduced photosynthetic performance of plants and can be the result of impaired thylakoid membrane biogenesis in chloroplasts (Meurer et al., 1996). In this screen, the mutant hcf222-1 was selected. Imaging of chlorophyll a fluorescence with a FluorCam system showed significantly increased steady-state fluorescence of hcf222-1 seedlings compared with the wild type (Fig. 1A). On Suc-supplemented medium, mutant growth was retarded slightly and leaves were paler than in the wild type (Fig. 1A). In contrast, under autotrophic conditions on Suc-free substrate, mutant seedlings showed severe growth retardation and leaves were much paler than in the wild type. After 3 weeks, hcf222-1 seedlings only developed two tiny rosette leaves compared with four to six leaves in the wild type (Fig. 1A). During cultivation on soil, mutant seedlings died after 4 to 5 weeks; thus, seeds can only be obtained from heterozygous plants. For all experiments, the progeny of heterozygous mutants were grown on 1% Suc-supplemented medium and mutants were selected by their hcf phenotype.
Figure 1.
Phenotype and photosynthetic capacity of hcf222 mutants. A, In the top image, steady-state fluorescence (Fs) of mutants was visualized with an imaging fluorometer. The signal intensity of Fs is indicated by the color scale bar below the image. The other images show representative mutant plants grown under autotrophic or heterotrophic conditions. Seedlings were cultivated for 3 weeks on one-half-strength Murashige and Skoog medium, supplied with Suc as indicated. B, Chlorophyll fluorescence induction kinetics were recorded after dark adaption of seedlings for 5 min. Saturating white light pulses (flash symbols) of 5,000 μmol m−2 s−1 were applied to determine Fm and Fm′. Actinic light has an intensity of 120 μmol m−2 s−1. C, P700 absorbance kinetics were measured with a modified PAM fluorometer. After illumination of seedlings with actinic light for 5 min, light was switched off and P700 was oxidized by far-red light of 720 nm and 15 μmol m−2 s−1; saturating white light pulses were like those for chlorophyll fluorescence measurements. Black bar, actinic light off; white bar, actinic light on; red bar, far-red light on. Twenty- to 24-d-old intact seedlings were used for all measurements. One representative curve for at least five different measurements for each mutant is depicted. WT, Wild type.
In order to examine the photosynthetic capacity of the mutant more closely, we measured chlorophyll fluorescence induction kinetics with 3-week-old intact plants. The maximum quantum efficiency of PSII photochemistry was comparable to that of the wild type in hcf222-1, indicating efficient PSII photochemistry (Fig. 1B; Table I). In contrast, fluorescence quenching in continuous actinic light was reduced drastically in the mutant (Fig. 1B), and the parameter qL, which reflects the fraction of oxidized QA in PSII reaction centers in continuous light, reached only 20% of the wild-type value in hcf222-1 (Table I). This behavior can be attributed to impaired electron transport capacities behind PSII, either due to defects in the Cytb6f complex or PSI or even both.
Table I. Chlorophyll fluorescence parameters measured in leaves of wild-type, hcf222 mutant, and complemented plants (introduced fusion construct is indicated in subscript).
Mean values for at least three plants are shown. Whole seedlings were measured with the following light intensities: 500-ms saturating light pulses, 5,000 μmol m−2 s−1; and 4 min of actinic light, 120 μmol m−2 s−1. Fv/Fm, Maximum quantum yield of PSII; ΦPSII, effective quantum yield of PSII; qL, fraction of open PSII centers; NPQ, nonphotochemical quenching. Student’s t test significance levels are as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
| Plant | Fv/Fm | ΦPSII | qL | NPQ |
|---|---|---|---|---|
| Wild-type Col-0 | 0.87 ± 0.02 | 0.68 ± 0.07 | 0.55 ± 0.08 | 0.54 ± 0.2 |
| hcf222-1 | 0.80 ± 0.06 | 0.28 ± 0.15* | 0.11 ± 0,05*** | 0.10 ± 0.02** |
| hcf222-2 | 0.65 ± 0.06** | 0.008 ± 0.01*** | 0.004 ± 0.007*** | 0.008 ± 0.01** |
| Complemented plants | ||||
| hcf222-1cHCF222-HA | 0.86 ± 0.02 | 0.74 ± 0.03 | 0.51 ± 0.08 | 0.34 ± 0.13 |
| hcf222-2cHCF222-HA | 0.85 ± 0.009 | 0.64 ± 0.03 | 0.45 ± 0.1 | 0.30 ± 0.25 |
| hcf222-2cHCF222-GFP | 0.85 ± 0.04 | 0.71 ± 0.06 | 0.42 ± 0.05* | 0.43 ± 0.17 |
| hcf222-2CTP-cHCF222-GFP | 0.86 ± 0.01 | 0.65 ± 0.02 | 0.44 ± 0.01 | 0.30 ± 0.03 |
To differentiate between these alternatives, we studied the redox activity of the PSI reaction center by measuring the absorbance kinetics of P700 at 820 to 870 nm (Fig. 1C). In wild-type plants, P700 was reduced in the dark and became completely oxidized under illumination with far-red light, reflected by an increased absorbance. A similar absorbance increase was observed for hcf222-1, suggesting the integrity of PSI reaction centers. A saturating light pulse was able to reduce P700 in the wild type and in hcf222-1. However, in continuous actinic light, P700 was completely oxidized in the mutant, whereas the redox state of wild-type P700 equilibrates to a steady-state level, corresponding to a partly oxidized state (Fig. 1C). This behavior indicated the disruption of electron transport between PSII and PSI, thus pointing to a defect in the Cytb6f complex.
To confirm that the observed phenotype of hcf222-1 is caused by a single nuclear mutation, we analyzed the segregation ratio of hcf mutants and the wild type in the progeny of heterozygous plants. In a representative analysis with 527 individuals, we found the expected mendelian 1:3 (mutants:wild type) ratio indicating a recessive, monogenic, nuclear mutation (Table II).
Table II. Segregation ratio of mutant lines hcf222-1 and hcf222-2.
Segregation of the hcf mutant phenotype was determined on Suc-supplemented medium. Segregation of antibiotic resistance was examined separately on sulfadiazine-containing medium. Numbers represent individual seedlings determined, and the percentage is given in parentheses. GK-038B09BC, Backcrossed T-DNA mutant.
| Mutant | hcf Phenotype | Wild Type | Resistant Seedlings | Sensitive Seedlings |
|---|---|---|---|---|
| hcf222-1 | 119 (24%) | 384 (76%) | – | – |
| hcf222-2 (GK-038B09BC) | 175 (25%) | 517 (75%) | 373 (79%) | 100 (21%) |
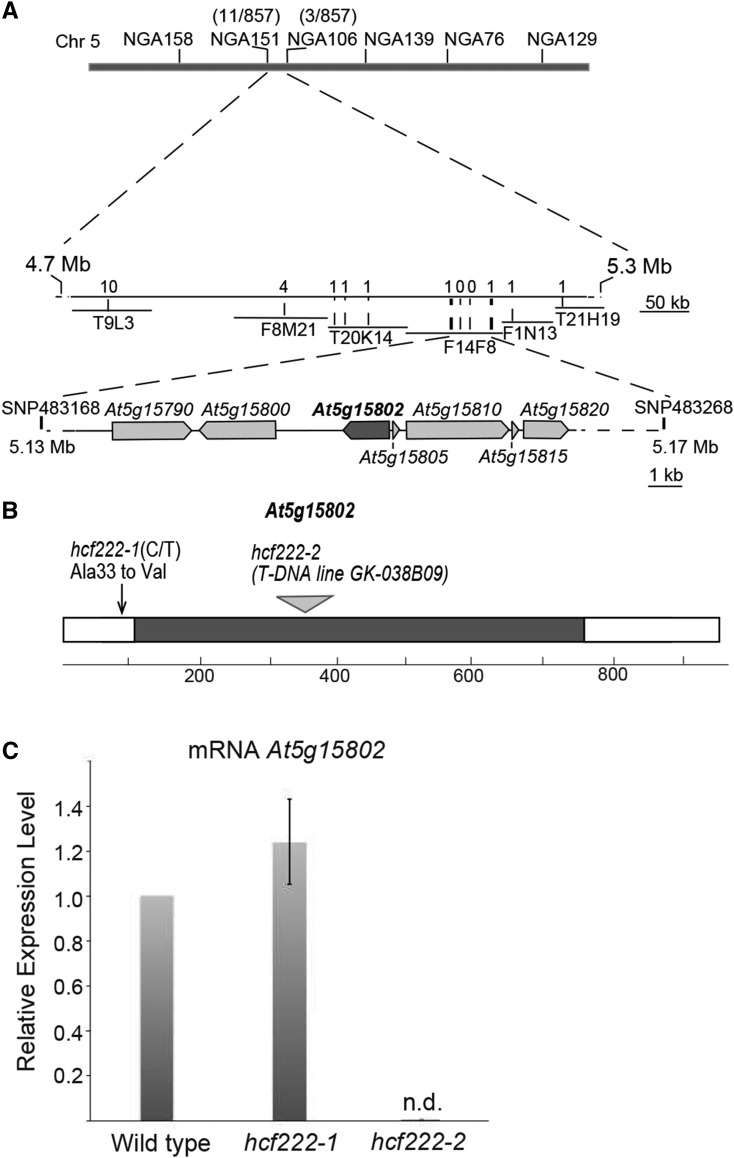
In order to map the hcf222 gene, heterozygous mutants were crossed to the ecotype Landsberg erecta (Ler), and a mapping population of 857 homozygous F2 mutant plants was genotyped using simple sequence length polymorphisms and cleaved-amplified polymorphic sequences as genetic markers. The mutated gene was mapped to a 35.8-kb fragment on the upper arm of chromosome 5 (Fig. 2A). Of the remaining 10 genes in this chromosomal region, all 10 were amplified in the mutant background and sequenced. Comparison of mutant and wild-type genomic sequences revealed a C-to-T transition in the first exon of the gene At5g15802 (nucleotide 98 of the reading frame), leading to an amino acid exchange from Ala-33 to Val in the mutant allele (Fig. 2B).
Figure 2.
Map-based cloning of HCF222 and gene expression in the mutants. A, Mapping of the hcf222-1 locus. The gene was mapped on the upper arm of chromosome 5 between the single-nucleotide polymorphism markers 483168 and 483268. Genes located in the region around the gene HCF222/At5g15802 are depicted. B, Gene structure of HCF222 and mutant alleles. The coding sequence of HCF222 comprises 943 nucleotides consisting of two exons separated by an intron at nucleotide positions 114 to 756. Sequence analysis of the mutant allele hcf222-1 revealed a point mutation at nucleotide 98 in exon 1. This mutation modifies codon GCT into GTT, causing an amino acid exchange from the original Ala to Val. In hcf222-2 (GABI-Kat line 038B09), the T-DNA insertion was detected in the intron between nucleotides 347 and 348. C, Determination of HCF222 mRNA levels in the wild type and the two allelic mutants by real-time PCR. n.d., Not detectable. Expression levels of HCF222 were determined in three biological replicates, and means with sd are depicted.
Knockout of HCF222 Results in More Severe Deficiencies of Mutant Seedlings
The importance of the identified gene At5g15802 for growth and photosynthesis of Arabidopsis seedlings was further examined using the T-DNA insertion line GK-038B09 from the GABI-Kat collection (Kleinboelting et al., 2012), which possesses an insertion in the intron of At5g15802 between nucleotides 347 and 348 (Fig. 2B). As more than one T-DNA insertion was assumed for this mutant, a backcross with the wild type (Col-0) was performed to obtain a progeny containing a single insertion. Heterozygous F2 plants from this backcrossed line (GK-038B09BC) segregated pale green hcf mutants according to the typical mendelian ratio (Table II), revealing that the mutant phenotype results from inactivation of a single, recessive nuclear gene. A proportion of 21% sulfadiazine-sensitive seedlings in the F2 generation showed that only one T-DNA insertion exists in the mutant genome (Table II). In the progeny of the backcrossed T-DNA line, we found mutants with a more severe phenotype than that of the EMS mutant. Under heterotrophic growth conditions, mutants stayed very tiny and their leaves bleached much stronger than observed for hcf222-1 mutants (Fig. 1A). On Suc-free medium, seedlings appeared transparent and grew even slower (Fig. 1A).
Due to the minimal chlorophyll content of the mutant hcf222-2, the steady-state fluorescence of leaves was increased only slightly compared with the wild type (Fig. 1A). Recording of fluorescence induction kinetics revealed a reduced maximum quantum efficiency of PSII photochemistry by 25%. Furthermore, there was more or less no fluorescence quenching in the light; thus, qL tends to 0 (Fig. 1B; Table I). The P700 absorbance resembled that of hcf222-1; however, a saturating light pulse in the far-red background was unable to reduce the reaction center of PSI (Fig. 1C). Altogether, these results indicate a severe defect in the Cytb6f complex as well as impaired PSII photochemistry in the T-DNA mutant.
To verify that the identified gene is responsible for the phenotypic alterations in the two mutants hcf222-1 and GK-038B09BC, we introduced the wild-type cDNA of At5g15802 into heterozygous plants by Agrobacterium tumefaciens-mediated transformation. To this end, we used the Gateway-based vector pAUL1 (Lyska et al., 2013a), which confers a 35S promoter and the coding region for a C-terminal 27-amino acid-long triple hemagglutinin (HA) epitope tag to the cDNA. In the T2 progeny of transformed heterozygous plants from both mutants, we were able to identify seedlings that were homozygous for the hcf222-1 locus (five independent transformed plants) or the GK-038B09 T-DNA insertion (four independent transformed plants) but that grew like the wild type under autotrophic conditions (Supplemental Fig. S1). Measurement of chlorophyll a fluorescence revealed wild-type kinetics for the transformed lines hcf222-1cHCF222-HA and hcf222-2cHCF222-HA (Table I); consequently, the complemented phenotypes confirmed the identity of the gene HCF222 as the causal mutant gene in hcf222-1 and hcf222-2 and revealed that the At5g15802 cDNA encodes a functional protein.
To examine the expression level of At5g15802 in the two mutants, real-time PCR was used to quantify the corresponding mRNA levels. In the EMS mutant, the corresponding transcript reached the wild-type level, whereas the T-DNA line lacked this mRNA (Fig. 2C). This demonstrates that the inserted T-DNA leads to a knockout of the gene At5g15802, corresponding to the more severe phenotype of this mutant allele.
Accumulation of the Cytb6f Complex Is Drastically Impaired in hcf222 Mutants
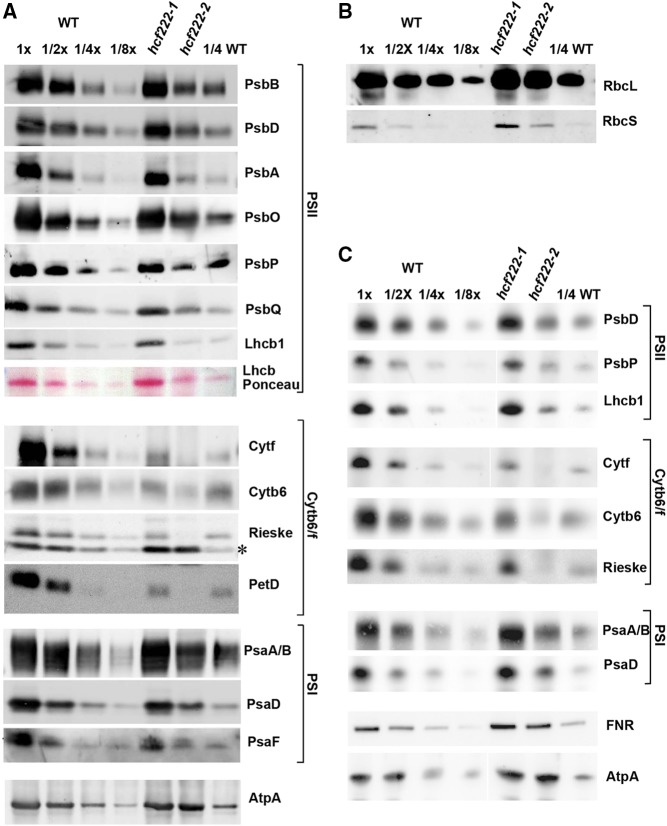
To address the question of whether the defect in photosynthetic electron transport of the mutants is based on reduced accumulation of thylakoid membrane protein complexes, levels of the major plastome- and nucleus-encoded components of PSII, the Cytb6f complex, PSI, and the ATP synthase were determined by immunoblot analyses (Fig. 3A). In both mutants, the most pronounced reduction was found for the levels of all four large Cytb6f subunits. However, the mutants differed in the degree of protein reduction. Whereas the EMS mutant hcf222-1 accumulated around 30% of the plastome-encoded cytochrome f, cytochrome b6, and PetD as well as nucleus-encoded Rieske protein, these subunits were around or even below the 10% level in the T-DNA mutant hcf222-2. Subunits of the core complex of PSII (plastid encoded: PsbB, PsbD, and PsbA) and PSI (plastid encoded: PsaA/B; nucleus encoded: PsaD and PsaF) reached wild-type levels in hcf222-1 but only ranged between 25% and 50% in the T-DNA line hcf222-2. Notably, nucleus-encoded subunits of the oxygen-evolving complex of PSII also showed diminished levels in the mutant hcf222-2 (PsbO, 50%; PsbP, 25%; and PsbQ, 30%; Fig. 3A). Ponceau Red staining of separated membrane proteins revealed a significant reduction of the LHCB complex in hcf222-2 (Fig. 3A). A specific antibody against LHCB1 detected wild-type amounts of this protein in hcf222-1, but only about 25% was detected in hcf222-2. The α-subunit of ATP synthase reached wild-type amounts or even somewhat higher levels in both mutants.
Figure 3.
Accumulation of chloroplast proteins in hcf222 mutants. A, Levels of chloroplast membrane proteins from seedlings cultivated for 20 d under moderate light conditions (40–60 μmol m−2 s−1). Whole-cell membranes from wild-type (WT) and mutant seedlings were separated by SDS-PAGE and transferred to nitrocellulose membranes. The dilution series contains 30 µg of protein (1x) or the indicated dilutions. Mutants contain 30 µg of membrane proteins. Antibodies used for detection are indicated on the right; the asterisk indicates an unspecific signal detected by the Rieske antibody. B, Levels of the large and small subunits of Rubisco (RbcL and RbcS) detected in whole-cell proteins of hcf222 mutants. C, Accumulation of thylakoid membrane proteins in hcf222 mutant seedlings grown under dim light conditions (5–10 μmol m−2 s−1). Plants were cultivated for 24 d. Whole-cell proteins were loaded on the gel. The dilution series in B and C contain 40 µg of protein (1x) or the indicated dilutions. Mutants contain 40 µg of whole-cell protein. Even if images were cut between lanes, all bands of a protein were detected simultaneously on one blot. Representative results of at least three biological replicates are depicted for each antibody.
The observed reduction of nearly all tested photosynthetic proteins in hcf222-2 might be the consequence of a generally impaired protein synthesis rate in chloroplasts of this mutant. Besides membrane proteins, also soluble plastid proteins should be affected by this defect. Thus, we measured the amount of the large subunit of Rubisco as a soluble representative in whole cell extracts of the wild type and mutants (Fig. 3B). Normal accumulation of this abundant protein demonstrated that the reduction of the different thylakoid membrane proteins in hcf222-2 is not the consequence of a generally decreased synthesis rate of plastid proteins. Similar amounts of protein also were detected for the Rubisco small subunit in the wild type and mutants (Fig. 3B).
Due to the strong deficiencies for the Cytb6f complex in hcf222-2, secondary effects based on damage through applied growth light cannot be excluded. To examine the extent of this effect, mutant seedlings were cultivated under dimmed light with the intensity of 5 to 10 µmol m−2 s−1. In order to get a more accurate quantitation of thylakoid membrane proteins in the T-DNA mutant, selected proteins were determined by immunoblot analyses of whole-cell proteins from both mutants. The obtained data showed that membrane proteins of PSII, PSI, the Cytb6f complex, and the LHCB complex are still reduced significantly in hcf222-2 (Fig. 3C); however, some of these proteins accumulate to slightly higher levels (30% LHCB1, 70% PsaD, and 40% PsbP) under dimmed light. The most severe reduction was again observed for the large subunits of the Cytb6f complex, which accumulated to around 10% (Fig. 3C). This indicated only minimal protein damage through applied light; thus, reduced protein levels of hcf222-2 are primarily the result of the mutation. In addition, this experiment showed that quantification of the protein levels in hcf222-2 is comparable in membrane and whole-cell protein fractions. Besides the integral membrane proteins and lumen-exposed, peripheral membrane proteins, we also tested the accumulation of FNR, which is exposed to the stroma. Like the α-subunit of the ATP synthase, FNR accumulated to wild-type levels in both mutants, indicating that the protein is unaffected by the strong reduction of the Cytb6f complex.
Taken together, our results imply that the phenotype of the hcf222-1 mutant is attributable primarily to equally reduced steady-state levels of the large subunits of the Cytb6f complex. In the knockout mutant hcf222-2, levels of Cytb6f subunits were much more strongly reduced; moreover, the accumulation of PSII, LHCB, and PSI subunits was also significantly impaired.
Impaired Protein Accumulation in hcf222 Mutants Is Caused by a Posttranslational Defect
To address the question of whether the reduced protein levels are caused by reduced mRNA amounts, RNA gel-blot hybridization using probes against transcripts of the large Cytb6f proteins was performed with whole-leaf mRNA from hcf222-1. Compared with the wild type, we found no reduction or modified processing pattern for petA, petB, petD, and petC mRNAs in the mutant (Supplemental Fig. S2A). For the T-DNA mutant hcf222-2, we analyzed levels of several plastid- and nucleus-encoded mRNAs of thylakoid proteins by quantitative real-time PCR (Supplemental Fig. S2B). For the transcripts petA, petB, petD, and petC, differences between the wild type and hcf222-2 were within the normal fluctuation range. The same was observed for transcripts of PSI proteins (psaA, psaB, and psaD), the α-subunit of the ATP synthase (atpA), and the rbcL mRNA. The three PSII transcripts tested (psbA, psbB, and psbD) were reduced slightly, ranging from 60% to 80% of the wild-type levels. The amounts of the related proteins were around 30%; thus, it can be assumed that mRNA levels are not limiting for the synthesis of the corresponding proteins.
As RNAs for the large Cytb6f subunits accumulate normally, reduced protein levels might be the consequence of reduced translation. Thus, we fractionated ribosome-bound leaf RNA of the wild type and mutants on Suc density gradients to analyze ribosomal loading of plastid-encoded RNAs for Cytb6f subunits. In both mutants, gel-blot hybridization with gene-specific probes revealed a distribution pattern for petB/D, petA, petG/L, petN, and the nucleus-encoded petC RNA that resembles the distribution in the wild type (Supplemental Fig. S3). Thus, we conclude that the low amounts of Cytb6f proteins in the two allelic mutants are not due to reduced mRNA levels or to reduced translational efficiency but are very likely a failure of a specific posttranslational modification that, in turn, leads to increased degradation of the affected proteins.
The stability of proteins is determined, among other factors, by the correct association of their nonproteinaceous cofactors. Therefore, we examined covalently attached hemes in cytochrome f and b6 by the peroxidation of luminol, which results in chemiluminescence emission. In several heme attachment mutants of C. reinhardtii and Arabidopsis, heme was undetectable but apocytochromes accumulated to low amounts (Lennartz et al., 2001; Kuras et al., 2007; Lyska et al., 2007). In addition, the migration of apocytochrome b6 is increased slightly compared with the holoform, leading to a band shift in heme cn-binding mutants (Lezhneva et al., 2008).
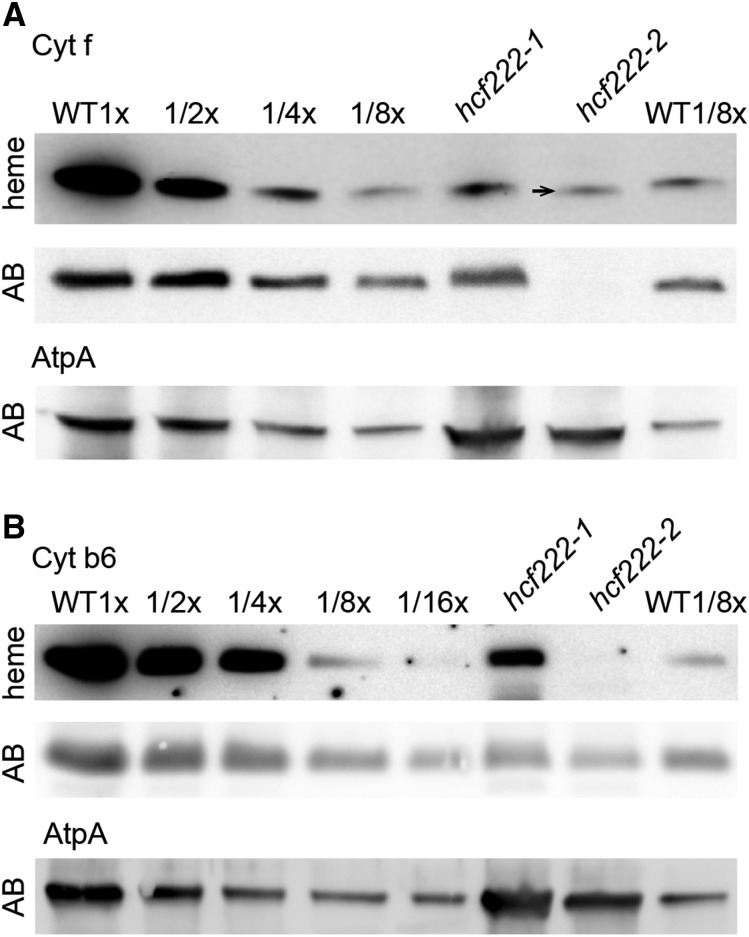
Analysis of peroxidase activity in protein extracts of hcf222-1 revealed that the heme amount in cytochrome f corresponded to the level of the accumulating protein, indicating that remaining protein amounts represent holocytochrome f (Fig. 4A). In hcf222-2, anti-cytochrome f detected no protein band; hence, protein levels were significantly below 10%. In contrast, heme staining revealed about 10% heme. A closer look showed that the detected band was shifted slightly downward in all our heme-staining experiments (Fig. 4A; data not shown). Therefore, it is unlikely that this band represents holocytochrome f. It is possible that this is mitochondrial cytochrome c1, whose size is very similar to that of cytochrome f (Meyer et al., 2008) and that might be up-regulated in the mutant hcf222-2. For cytochrome b6 of hcf222-1, heme and protein levels reached around 20% (Fig. 4B). In the mutant hcf222-2, heme cn was below the detection limit and cytochrome b6 protein levels were around 6% (Fig. 4B). Although a direct comparison of heme and protein levels was not feasible for this mutant, a shift of the protein band, as can be expected for accumulating apocytochrome b6, was not visible.
Figure 4.
Comparison of heme and protein levels in the wild type (WT) and hcf222 mutants. Covalently bound heme groups were detected by their peroxidase activity using luminol as a substrate. A, Detection of heme in cytochrome f. The arrow marks the slight shift of the heme signal in hcf222-2. Corresponding levels of the cytochrome f protein were detected with an antibody (panel AB). B, Heme detection in cytochrome b6 and the corresponding protein accumulation (gel AB). The AtpA subunit of the ATP synthase was used as a loading control. A total of 40 µg of protein (1x) or the indicated dilutions of the wild type was applied to the gel. The mutant lanes contained 40 µg of protein.
Taken together, these results indicate that covalent heme attachment to cytochrome f and cytochrome b6 was not impaired in hcf222 mutants.
HCF222 Encodes a Small Cys-Rich Zinc-Finger Motif Protein
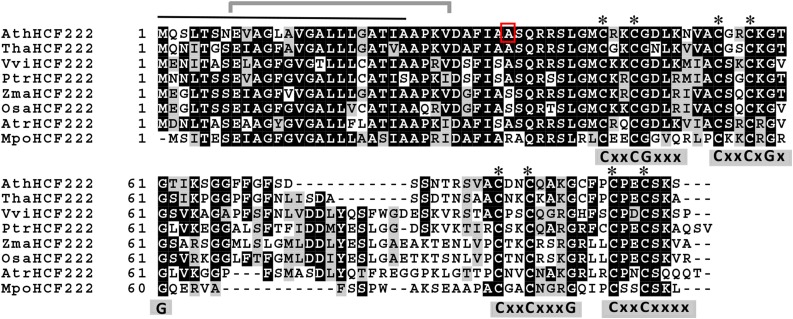
The reading frame of HCF222 encodes a small protein of 99 amino acids that is characterized by eight conserved Cys residues arranged in two CxxCx7CxxC clusters (where x represents any amino acid; Fig. 5). The two Cys clusters show homology with the zinc-finger domain of E. coli chaperone DnaJ proteins. In this motif, two zinc atoms are coordinated between the Cys residues of four CxxC motifs, forming a loop structure (Shi et al., 2005). In DnaJ proteins, the sequence CxxC is often followed by two conserved Gly residues (CxxCxGxG). In HCF222 only, in one of the four CxxC motifs, conserved Gly residues were found. Besides the Cys-rich zinc-finger motif, HCF222 contains no further known domains.
Figure 5.
Alignment of HCF222 orthologs from seed plants and a bryophyte. Multiple amino acid sequence alignment of Arabidopsis HCF222 (AtHCF222) and homologs from Tarenaya hassleriana (Tha), Vitis vinifera (Vvi), Zea mays (Zma), Oryza sativa (Osa), Populus trichocarpa (Ptr), Amborella trichopoda (Atr), and the bryophyte M. polymorpha (Mpo). Identical amino acids are marked white on black; similar amino acids are black on gray. The predicted signal peptide of HCF222 is marked by a black line on top, the gray bracket indicates the predicted membrane-spanning helix, and the red frame indicates the mutated amino acid (A→V) in the mutant allele hcf222-1. Asterisks mark conserved Cys residues, and, in addition, the four CxxC motifs and conserved Gly residues are shown below the sequence. The amino acid sequence alignment was generated using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and Boxshade 3.21 (http://www.ch.embnet.org/software/BOX_form.html).
Phylogenetic analysis showed orthologs of HCF222 in land plants, but no related proteins were found in algae or cyanobacteria or in other taxa. The ortholog of HCF222 from Populus trichocarpa was assigned to a protein family designated as glutaredoxin-like proteins (Navrot et al., 2006). The designation glutaredoxin-like is merely based upon the existence of the CxxC motifs but no other similarities. In Arabidopsis, the protein family contains 15 members. The alignment of the amino acid sequence of Arabidopsis HCF222 with sequences from dicots, monocots, the primitive angiosperm Amborella trichopoda, and the liverwort Marchantia polymorpha revealed conservation for the regions comprising the CxxC motifs (Fig. 5). The highest similarity, of around 80% to 70%, was found between the N-terminal half of HCF222 (the first 42 amino acids) and its orthologs (Fig. 5; Navrot et al., 2006). Based on targeting predictions summarized in the SUBA3 database of Arabidopsis proteins (Tanz et al., 2013), the N terminus of HCF222 was identified to contain a signal peptide that directs the protein into the secretory pathway. The TargetP 1.1 program (Emanuelsson et al., 2007) predicts the possible cleavage site between amino acids 24 and 25. The predicted cellular targets of the protein were inconsistent, varying between ER, Golgi, and the extracellular space, and also plastid localization was suggested, although with low confidence.
Transient and Stably Expressed HCF222-GFP Is Transported to the ER and the Chloroplast
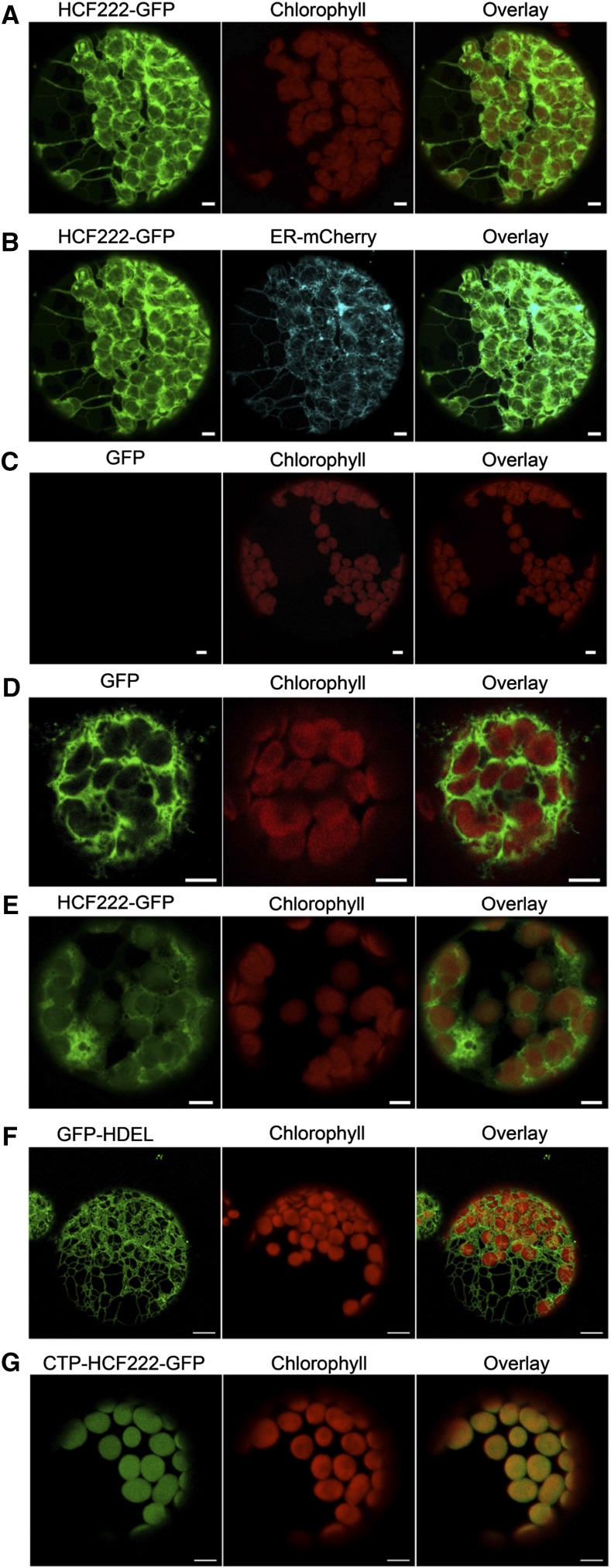
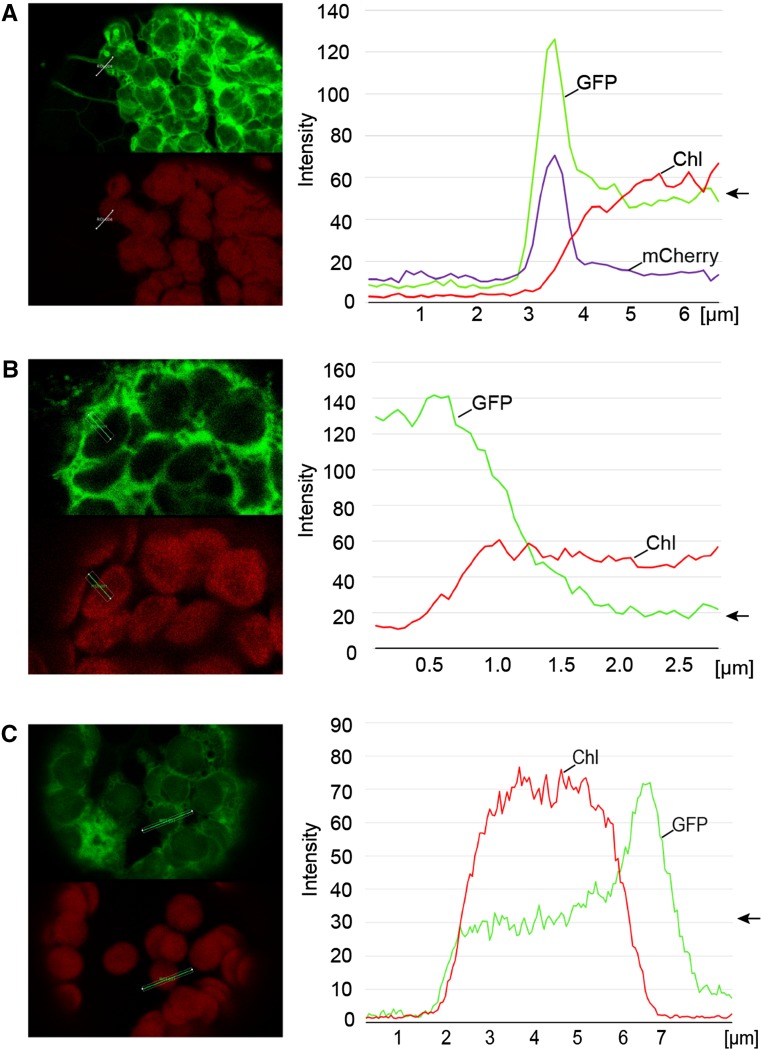
To examine the localization of HCF222 in the cell, we analyzed the transient expression of HCF222 fused with GFP in Nicotiana benthamiana mesophyll cells. The full-length cDNA of HCF222 was cloned into the Gateway destination vector pGWB5, which provided a 35S promoter and a C-terminal GFP (Nakagawa et al., 2007). After infiltration of N. benthamiana leaves with transformed Agrobacterium tumefaciens, protoplasts were isolated and analyzed by confocal laser scanning microscopy. Transient expression of HCF222-GFP for 72 h resulted in a fluorescence signal that labeled a net-like structure throughout the whole mesophyll protoplast (Fig. 6A). The pattern of HCF222-GFP fluorescence suggested that the fusion protein is partly located to the ER. Thus, we used the ER marker protein ER-mCherry (ER-rk) to label this cell compartment (Nelson et al., 2007). Figure 6B shows a clear overlap between ER-mCherry and HCF222-GFP fluorescence. In addition to the ER, GFP fluorescence also was detectable in chloroplasts; however, this fluorescence had a lower intensity but was visible in each chloroplast (Fig. 6A). To verify that the detected fluorescence in chloroplasts was caused by HCF222-GFP alone and not overlaid by chlorophyll autofluorescence, we recorded the fluorescence of wild-type protoplasts under the same microscope settings as for HCF222-GFP detection. Wild-type protoplasts showed no fluorescence emission in the GFP channel (Fig. 6C), confirming that the signal in chloroplasts of transformed leaves is caused by HCF222-GFP. To present a more quantitative picture of the detected HCF222-GFP fluorescence, we generated a fluorescence profile of Figure 6A reflecting signals in the ER and a chloroplast (Fig. 7A). The GFP profile of the first 4 µm revealed a peak of the mCherry profile, because the profile line passes through the ER. Inside the chloroplast, reflected by increased chlorophyll autofluorescence, mCherry fluorescence declined sharply to the basic level. The GFP profile line also declined, but only to an intermediate level, which is significantly above the basic level and which arises from the HCF222-GFP located inside the chloroplast (Fig. 7A). In contrast to the fusion protein, GFP without a target peptide showed fluorescence in the cytosol and no GFP signals inside the chloroplasts (Fig. 6D). Accordingly, the profile of GFP declined to a low level inside this cell compartment (Fig. 7B).
Figure 6.
Localization of HCF222 in the cell by confocal fluorescence microscopy. A, Analysis of N. benthamiana protoplasts transformed with HCF222-GFP. B, Visualization of the compartment marker ER-mCherry in relation to HCF222-GFP fluorescence (same protoplast as in A). C, Wild-type protoplast as a control, to verify that autofluorescence of chloroplasts did not influence GFP fluorescence (same microscope settings as in A and B). D, Control with cytosolic GFP. E, Visualization of HCF222-GFP fluorescence in Arabidopsis protoplasts that stably expressed the fusion protein in the mutant background of hcf222-2. F, Stably expressed compartment marker ER-GFP in the Arabidopsis line ER-gk (CS16251). G, Stable expression of CTP-HCF222-GFP in the mutant background of hcf222-2. Bars = 5 µm.
Figure 7.
Quantification of fluorescence emission in chloroplasts and ER. A, Analysis of N. benthamiana protoplasts transformed with HCF222-GFP and the corresponding fluorescence profile. B, Control with cytosolic GFP. C, Fluorescence profile of HCF222-GFP fluorescence in Arabidopsis protoplasts that stably expressed the fusion protein in the mutant background of hcf222-2. The arrows mark the levels of GFP fluorescence inside the chloroplasts. All profiles were directed from the cytosol/vacuole into the chloroplast. Fluorescence profiles were generated with the profile tool of the software package LAS X (Leica Microsystems). Chl, Chlorophyll autofluorescence.
To confirm the localization of HCF222, we repeated our studies with protoplasts from hcf222-2 mutants that stably expressed the HCF222-GFP fusion protein. The mutant phenotype of these transformed plants was restored, because all photosynthetic parameters were comparable to the wild type (Table I, plants designated hcf222-2cHCF222-GFP). This indicates that HCF222-GFP is functional and targeted correctly. GFP fluorescence in protoplasts from these lines resembled the distribution found in N. benthamiana protoplasts. In each chloroplast, GFP fluorescence was detectable, and partly net-like structures were visible in the cytosol (Fig. 6E). The fluorescence profile revealed a perfect correspondence between GFP and chlorophyll fluorescence on the left side of the selected chloroplast, whereas the GFP profile increased at the right margin, flanked by the ER (Fig. 7C). For comparison, we analyzed protoplasts from an Arabidopsis ER-GFP marker line, ER-gk (Nelson et al., 2007), which showed the typical interconnected ER membrane network but no fluorescence in the chloroplast compartment (Fig. 6F).
Altogether, our analysis revealed that HCF222 is dually located to the ER and the chloroplast. Under the conditions we examined in N. benthamiana and Arabidopsis, only a minor fraction of the fusion protein is located to chloroplasts. The observed import into the ER compartment corresponds to the predicted N-terminal signal peptide of 24 amino acids in HCF222.
Chloroplast-Targeted HCF222 Can Complement the Mutant Phenotype
As HCF222 is dually targeted, the question arises of which of the two locations is relevant for the observed mutational lesion. To be able to define the role of the chloroplast-located HCF222, we stably transformed the mutant hcf222-2 with a full-length HCF222 fusion protein that was provided with the chloroplast transit peptide (CTP) of the RbcS protein. This protein version should be able to restore the functional lesion in the chloroplast. We selected nine independent transformants that had a homozygous mutant background and were able to grow under photoautotrophic conditions (Supplemental Fig. S1). Analyses of chlorophyll fluorescence kinetics showed wild-type behavior for these transformed plants with respect to all fluorescence parameters tested (Table I, plants designated hcf222-2CTP-cHCF222-GFP). Detection of GFP fluorescence in leaf protoplasts confirmed the localization of the fusion protein CTP-HCF222-GFP inside chloroplasts (Fig. 6G). No further GFP signals outside the organelles could be visualized.
The successful complementation strongly suggests that chloroplast targeting of HCF222 is sufficient to restore the mutational lesion, indicating that HCF222 is acting in this cell compartment during thylakoid membrane biogenesis. However, we cannot exclude that spurious amounts of HCF222 enter the ER or remain in the cytosol and contribute to complementation of the mutant phenotype.
HCF222 Is Associated with the Membrane Fraction and Has Disulfide Reductase Activity in Vitro
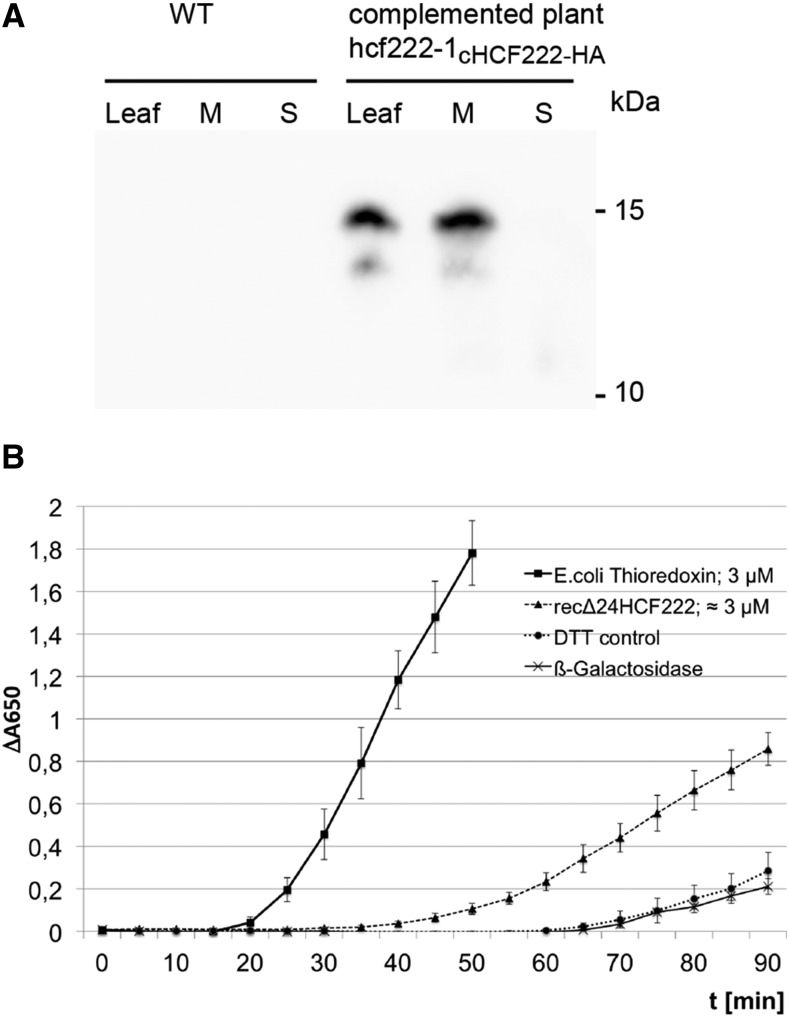
To obtain more information about HCF222, we isolated protein extracts from 3-week-old seedlings and detected HCF222-HA using an antibody against the C-terminal epitope tag. The immunoblot revealed a protein band around 15 kD exclusively in whole-cell protein extracts of the complemented line hcf222-1-cHCF222-HA and not in the wild type, indicating the specificity of the antibody to HCF222-HA (Fig. 8A). The segregation distance of the protein correlates roughly with the size of the full-length protein (10 kD) plus 3 kD for the triple HA tag. The nature of a faint band migrating below this major signal could be the result of a partial loss of the HA tag, as found for HA-tagged PsbH protein in a stably transformed Arabidopsis mutant (Levey et al., 2014). When whole-cell proteins were separated into membrane and soluble proteins, HCF222 was found in the whole-cell membrane fraction and no fusion protein was detectable in the soluble fraction (Fig. 8A). We then tried to detect HCF222-HA in intact chloroplasts isolated from 6- to 8-week-old seedlings and separated on a Percoll gradient, but immunodetection with the anti-HA antibody gave no reproducible results because protein levels were at or below the detection limit (data not shown). To get a first insight into the membrane-binding capacity of HCF222, we isolated whole-cell membrane extracts from 3-week-old seedlings and treated the membranes with different salts and detergent (Supplemental Fig. S4). Removal of HCF222-HA from the membrane could only be achieved by detergent treatment, whereas sodium bromide had no effect and sodium thiocyanate extracted minor amounts of the protein. This suggests that HCF222 contains at least one hydrophobic helix, which anchors the protein in the membrane. The only hydrophobic stretch of the protein is located in the N terminus as part of the signal peptide (Fig. 5). The program TopPred 1.10 (Claros and von Heijne, 1994) predicted this helix from amino acids 8 to 28 as a certain transmembrane helix (score 1,575).
Figure 8.
Characterization of HCF222. A, Immunodetection of HCF222-HA in leaf extracts. After SDS-PAGE, the protein pattern of whole-leaf extracts (Leaf) and whole-leaf membrane (M) and soluble fraction (S) from wild-type (WT) and complemented plants was transferred to nitrocellulose, and the fusion protein was detected with anti-HA antibody. B, Measurement of disulfide reductase activity of recombinant HCF222-6xHis by the reductive insulin assay. The reduction of disulfide bonds in insulin was monitored by an increase in A650, which results from insulin precipitation. The concentration of recombinant HCF222-6xHis was estimated after SDS-PAGE and Coomassie Blue staining in comparison with a dilution series of lysozyme. DTT without enzyme served as a control. In addition, recombinant β-galactosidase-6xHis from similarly purified E. coli extracts served as a negative control to estimate contaminants from E. coli. Values represent means of three independent experiments, each with three replicates; β-galactosidase activity is shown as means of three replicates from one purification. Means with sd are depicted.
HCF222 consists almost entirely of the zinc-finger domain of DnaJ proteins, as found for the chloroplast proteins LQY1 and PSA2. Based on this Cys-rich motif, thiol disulfide oxidoreductase activity was measured for each of these proteins (Lu et al., 2011; Fristedt et al., 2014). Therefore, we used the reductive insulin assay to determine a corresponding catalytic activity of HCF222. The assay is based on the reduction of two interchain disulfide bonds between the A and B chains of insulin by DTT (Holmgren, 1979). The free B chain precipitates, and turbidity formation can be measured at 650 nm. Thioredoxin from E. coli accelerates disulfide reduction about 20-fold compared with the noncatalyzed reaction (Holmgren, 1979). Recombinant HCF222 lacking the N-terminal 24 amino acids was produced in E. coli and purified by affinity chromatography on Ni-NTA columns via the attached C-terminal 6× His tag. As shown in Figure 8B, recombinant Δ24HCF222 significantly accelerated insulin reduction compared with the control reactions that contained either recombinant β-galactosidase, purified by the same procedure as HCF222, or DTT only. E. coli thioredoxin also catalyzed insulin reduction; however, the precipitation rates revealed that this enzyme is roughly 3 to 4 times more efficient than HCF222. This result demonstrated that purified, recombinant HCF222 has disulfide reductase activity, suggesting that HCF222 is required for dithiol-disulfide transactions in specific chloroplast-located target proteins.
DISCUSSION
HCF222 encodes a small protein of 99 amino acids in Arabidopsis that has similarities to the Cys-rich zinc-binding domain of DnaJ chaperones. HCF222 is conserved in higher plants and the liverwort M. polymorpha but is absent in algae and cyanobacteria. Confocal fluorescence microscopy and complementation analysis showed that HCF222 is a chloroplast protein. However, the protein also can be detected in the ER corresponding to the signal peptide at the N terminus. The examination of two allelic Arabidopsis mutants revealed that HCF222 is required predominantly for the accumulation of the Cytb6f complex but also necessary, although to a lesser extent, for wild-type levels of PSI and PSII.
HCF222 Is Required for the Formation of a Functional Thylakoid Membrane and Acts at the Posttranslational Level
Knockout of the gene HCF222 as well as a point mutation, leading to an amino acid exchange in the protein, resulted in mutants that are unable to survive under autotrophic growth conditions (Fig. 1). This severe phenotype is caused by drastically reduced photosynthetic capacity in both mutants, which is restricted mainly by a block in intersystem electron transport (Fig. 1). Correspondingly, in the EMS mutant hcf222-1, all four large subunits of the Cytb6f complex were reduced equally to around 25% to 30%, whereas other photosynthetic membrane complexes accumulated normally. The knockout line showed only traces (∼10%) of the Cytb6f complex subunits; in addition, components of PSII, PSI, and the LHCB complex were reduced substantially (Fig. 3). This pleiotropic defect is obviously not due to secondary effects of the mutation caused by excess light energy, as mutants grown under a very low-light regime (5–10 µmol m−2 s−1) showed comparable deficiencies (Fig. 3). Furthermore, other Cytb6f mutants, like prfB3 (Stoppel et al., 2011), hcf208 (Lyska et al., 2007), and dac (Xiao et al., 2012), exhibit weak photoinhibitory effects, and these are only related to the reaction center of PSI. Hence, our results strongly suggest that HCF222 is indispensable for the accumulation of the Cytb6f complex; in addition, it is required for the normal accumulation of proteins from PSII, PSI, and the LHCB complex. In contrast to this, the ATP synthase accumulated in both mutants to high levels (Fig. 3). This also was observed in Cytb6f mutants of N. benthamiana and could be a compensatory effect, which possibly counteracts reduced ATP production (Schwenkert et al., 2007).
The strong reduction of the large Cytb6f complex subunits in the two mutants is not the consequence of reduced levels or altered processing of the corresponding mRNAs (Supplemental Fig. S2). The analysis of the polysomal association of all plastid-encoded Cytb6f transcripts in the mutants revealed that ribosomal loading resembles the wild type (Supplemental Fig. S3); hence, all subunits of the complex are synthesized in the mutants. Together, our findings suggest that the factor HCF222 is required for posttranslational processes during maturation or the assembly of photosynthetic membrane complexes, which applies in particular to the Cytb6f complex.
HCF222 Belongs to a Group of DnaJ-Like Zinc-Finger Proteins That Play Defined Roles in Thylakoid Membrane Biogenesis
HCF222 is a member of a protein family consisting of one domain with four conserved CxxC motifs, which is typical for the Cys-rich domain found in DnaJ proteins (InterPro IDIPR001305; Fig. 5). In DnaJ of E. coli, these CxxC motifs are required for the coordinated binding of zinc, forming the so-called zinc finger, which is necessary for the recognition, binding, and stabilization of unfolded proteins (Banecki et al., 1996; Szabo et al., 1996). Moreover, it contains the disulfide isomerase activity of the chaperone (de Crouy-Chanel et al., 1995). In orthologs of HCF222 from phylogenetically diverse species, the eight Cys residues are well conserved (Fig. 5), indicating that the function of HCF222 is related to the CxxC motifs. The disulfide reductase activity assay revealed that HCF222 has the ability to reduce disulfide bridges in a protein substrate (Fig. 8B). Altogether, these observations point out that the factor might act as a thiol-disulfide oxidoreductase of specific target proteins in vivo. In contrast to DnaJ, HCF222 lacks the classical J-domain and the G/F module, through which the protein binds its chaperone partner Hsp70. Thus, it is unlikely that HCF222 operates together with this heat shock protein.
HCF222 possesses a highly conserved N terminus, of which the first 24 amino acids were predicted to be a signal peptide. According to this target peptide, fluorescence microscopic investigations showed that the GFP-tagged HCF222 was located in the ER, but, in addition, we also detected the protein in the chloroplast, although in lower quantities (Figs. 6 and 7). The localization in chloroplasts is supported by a recent semiquantitative proteome study, where HCF222 was detected for the first time in this cell compartment (Tomizioli et al., 2014). Furthermore, our successful complementation of the knockout mutant with a chloroplast-directed HCF222 strongly suggests that the main site of action of this protein is located to the chloroplast (Table II; Fig. 6G; Supplemental Fig. S1), where it is involved in essential processes during thylakoid biogenesis.
With respect to the zinc-finger domain, the size, and membrane binding, HCF222 is related to the chloroplast proteins LQY1 and PSA2. LQY1 is bound to thylakoid membranes and involved in PSII repair and reassembly (Lu et al., 2011; Jin et al., 2014). The protein is able to bind zinc and has disulfide isomerase activity. Its molecular function is very likely accompanied by interaction with the PSII core proteins CP47 and CP43 (Lu et al., 2011). Recently, PSA2 was identified that is specifically required for PSI accumulation (Fristedt et al., 2014). The lumen-located, membrane-associated PSA2 has disulfide reductase activity, and it is suggested that this factor mediates dithiol-disulfide transactions required for PSI assembly. PSA2 can be detected in a PSI-related subcomplex containing PsaG, and coimmunoprecipitation showed an association with PsaG, defining this subunit as a putative target protein of PSA2 (Fristedt et al., 2014). The cotyledon-specific biogenesis factor CYO1/SCO2 also is related to the DnaJ-like zinc-finger proteins but harbors a modified motif (Shimada et al., 2007; Albrecht et al., 2008). Disulfide isomerase activity was measured with different assays (Shimada et al., 2007), and interaction studies using the split-ubiquitin system showed an interaction of CYO1/SCO2 with core complex subunits of PSII (CP43 and CP47) and PSI (PsaA/B; Muranaka et al., 2012), suggesting that the factor is necessary to accelerate the folding of these thylakoid membrane proteins by repeatedly breaking and creating disulfide bonds (Muranaka et al., 2012). Thus, it can be assumed that small DnaJ-type zinc-finger proteins, including HCF222, form a group of proteins that is required for dithiol-disulfide transactions in chloroplasts, maybe particularly at the thylakoid membrane.
HCF222 Possibly Catalyzes Protein Maturation at the Thylakoids
Considering the observed phenotype of the allelic hcf222 mutants, it can be assumed that HCF222 is engaged in the thiol-disulfide biochemistry of specific Cytb6f proteins. Three large subunits of the complex contain Cys residues, which are either involved in cofactor binding (cytochrome f and b6) or in disulfide bond formation (Rieske protein). To analyze covalent binding of hemes in the two cytochromes, we examined their peroxidase activity in the mutants (Fig. 4). For cytochrome attachment mutants, it has been shown that immature apoproteins are highly instable (Howe and Merchant, 1992; Kuras et al., 1995); however, small amounts of apocytochromes can be detected in the mutants of C. reinhardtii and Arabidopsis (Kuras et al., 2007; Lyska et al., 2007). We could not detect the apoproteins of cytochrome f or cytochrome b6 in the two mutants, showing that HCF222 is not required for the attachment of covalent hemes to these two cytochromes.
A putative target of HCF222 could be the Rieske protein. Crystal structure analysis showed that the [2Fe-2S] cluster-binding domains of chloroplast and mitochondrial Rieske proteins are virtually identical (Carrell et al., 1997). The stability and the redox properties of this [2Fe-2S] cluster depend on a disulfide bridge in this domain between Cys-164 and Cys-180, as shown for the mitochondrial Rieske subunit (Merbitz-Zahradnik et al., 2003; Leggate and Hirst, 2005). Due to the conserved structure, a similar important role can be expected for the disulfide bridge in the Rieske protein from chloroplasts. It is conceivable that a specific disulfide isomerase catalyzes the correct formation of this disulfide. Whether HCF222 can fulfill this role will be examined by means of interaction studies and in vitro disulfide assays with this enzyme.
Further deficiencies in PSII and PSI in the knockout mutant suggest that HCF222 also targets proteins from the other photosynthetic complexes and/or auxiliary factors required for their assembly. With respect to this, it should be noted that many lumenal proteins are targeted by thioredoxin affinity chromatography, indicating that they are putative candidates for thiol-disulfide biochemistry (Hall et al., 2010). Identified proteins of this study were the structural proteins PsbO, PsbP, and PsbN as well as the peptidyl-prolyl-isomerases CYP38 and FKBP20-2. The impact of HCF222 on the accumulation of the LHCB complex is yet unclear, but this reduction corresponds to the phenotype of the mutant cyo1/sco2, which also shows pleiotropic reduction of the photosynthetic complexes, including the LHCB complex (Shimada et al., 2007). CYO1/SCO2 also interacts with LHCB1, and it was suggested that this interaction is related to alternative transport and membrane insertion of LHCB proteins in cotyledons (Tanz et al., 2012).
Providing that HCF222 posttranslationally modifies thylakoid membrane proteins, it should be located in or at this membrane system. In the proteome study of Tomizioli et al. (2014), who analyzed thylakoid subfractions from Arabidopsis chloroplasts, HCF222 was detected exclusively in the fraction of the stromal thylakoid membrane system. Our salt and detergent treatment of the cell membrane fraction also identified HCF222 as a membrane-bound protein (Supplemental Fig. S4). The only hydrophobic part of HCF222 is located in the N terminus. The first 42 amino acids are highly conserved (80%–70% identity) in all HCF222 orthologs investigated (Fig. 5). Therefore, we assume that the N terminus is part of the mature protein and necessary to anchor HCF222 in the thylakoid membrane via the hydrophobic helix from amino acids 8 to 28 (Fig. 5). An important function of the N terminus is further reflected by the dramatic alteration of the hcf222-1 phenotype, which is based on an exchange of amino acid 34 from Ala to Val.
The immunological detection of HCF222 in Arabidopsis leaf extracts succeeded differently. Although we detected HCF222-HA in the leaf membrane fraction of 3-week-old seedlings, we were unable to detect HCF222-HA in chloroplasts from 6- to 8-week-old plants. A possible explanation could be that this is due to the differential expression of HCF222 in young and mature plants or leaves. The expression profile of HCF222 in the TRAVA database (Klepikova et al., 2016), which comprises RNA sequencing data for different stages and tissues of Arabidopsis, supports this assumption (Supplemental Fig. S5). HCF222 expression is highest in leaf tissue, but this applies only to young and developing leaves, as in 1-d-old seedlings or during the first 2 weeks after germination. In flowering plants, levels of HCF222 mRNA declined by a factor of 5 relative to the maximum expression in apical meristems of 1-d-old seedlings. This is consistent with the proteome study, where HCF222 was measured with only a few counts in chloroplasts from 4- to 5-week-old seedlings (Tomizioli et al., 2014). Together, these observations suggest that HCF222 shows highest expression levels in young leaf stages. This expression behavior is typically found for protein factors involved in the biogenesis of the thylakoid membrane, like the PSII assembly factors HCF136 (Meurer et al., 1998) and PAM68 (Armbruster et al., 2010) or the PSI biogenesis factors PSA2 and Y3IP1 (Fristedt et al., 2014). Therefore, we assume that HCF222, like these factors, is especially required during early processes of thylakoid membrane development.
HCF222 Is Localized to the ER and the Chloroplast
With respect to our localization data, one can conclude that HCF222 is dually located to the ER and the chloroplast compartment (Fig. 6). A similar dual trafficking was observed for the PDI RB60 from C. reinhardtii (Levitan et al., 2005). This protein contains an N-terminal 50-amino acid targeting sequence sufficient for import into chloroplasts and the ER. In vitro import studies with microsomes and chloroplasts showed that RB60 engages both the cotranslational SRP pathway for ER import and posttranslational translocation via the TIC/TOC machinery (Levitan et al., 2005). RB60 contains a KDEL retention signal at the C terminus, indicating that the protein persists in the ER. A retention signal for soluble ER proteins is lacking in HCF222; also, motifs for ER membrane retention, like a di-Lys motif, are not obvious, suggesting that the factor is not a resident ER protein. However, confocal fluorescence microscopy did not show that HCF222-GFP is located to other endomembrane compartments. Further investigations are necessary to solve the questions concerning the final localization and the function of HCF222 in the endomembrane system.
As HCF222 possesses a signal peptide at its N terminus, the question arises of how the protein is recognized and imported into chloroplasts. Studies of chloroplast proteomes from Arabidopsis revealed several proteins that lack transit peptides (Kleffmann et al., 2004; Zybailov et al., 2008). For some selected noncanonical chloroplast proteins, it was shown by in vitro import experiments that they are substrates of the TOC/TIC machinery (Armbruster et al., 2009). Thus, it is possible that HCF222 is imported into chloroplasts by the classical TOC/TIC pathway. However, we cannot exclude that the factor uses an alternative route, which involves the ER and the secretory pathway. This so-called endomembrane system-mediated chloroplast import pathway was shown only for Arabidopsis α-carbonic anhydrase, rice (Oryza sativa) nucleotide pyrophosphatase/phosphodiesterase, and two rice α-amylases, which contain an N-terminal signal peptide for cotranslational import into the ER (Asatsuma et al., 2005; Villarejo et al., 2005; Nanjo et al., 2006; Kitajima et al., 2009). It was suggested that Golgi-mediated vesicles target these proteins to chloroplasts, where they fuse with the outer envelope membrane (Kitajima et al., 2009). This transport through the secretory system is accompanied by the addition of N- and O-linked glycans to carbonic anhydrase and α-amylases (Kitajima et al., 2009; Burén et al., 2011). It will be an important next step to define such modifications of purified HCF222 by mass spectrometry.
With respect to the import of HCF222 into chloroplasts, it is interesting that the genomes of the grasses maize (Zea mays) and sorghum (Sorghum bicolor) contain a gene that encodes HCF222 with a chloroplast transit peptide. The maize ortholog of HCF222 is encoded by the gene GRMZM2G145527 (http://maizegdb.org.). Due to alternative transcription start sites and different splicing events, two transcripts were found for this gene, one encoding HCF222 with a signal peptide (GRMZM2G145527-P02) and a variant that contains an open reading frame for a chloroplast transit peptide in addition to the full-length sequence (GRMZM2G145527-P01; Supplemental Fig. S6). In sorghum, at least two genes exist, one encoding HCF222 with a signal peptide (Sb01g032530) and the other with a chloroplast-targeting peptide in addition to the signal peptide sequence (Sorbi_009g237800; Supplemental Fig. S6; http://www.plantgdb.org/SbGDB/). It is unclear so far, due to limited sequence data from grasses, if the existence of two protein variants is unique to maize and sorghum; however, in rice (subsp. japonica), a second gene or an mRNA variant that encodes HCF222 with a plastid-targeting sequence was not reported in the databases. The relevance of HCF222 variants in maize and sorghum has to be proven; however, they provide a further hint that the chloroplast is one target compartment of HCF222 in the two grasses. Whether the acquisition of a transit peptide is related to a modified chloroplast-targeting efficiency of HCF222 will be addressed in the future.
Small DnaJ-like zinc-finger proteins with disulfide isomerase activity are not frequent in chloroplasts, but for the known thylakoid-bound representatives CYO1/SCO2, LQY1, and PSA2, the involvement in thiol-disulfide biochemistry in this membrane or in the thylakoid lumen is very likely. We showed that the zinc-finger protein HCF222 is a fourth relative of this group of chloroplast proteins. The factor is indispensable for a posttranslational step during biogenesis of the Cytb6f complex. Determination of the precise role of HCF222, also with respect to PSII and PSI, will be the aim of future studies. Furthermore, it will be interesting to define by which of the known pathways HCF222 enters the chloroplast.
MATERIALS AND METHODS
Growth Conditions
The mutant hcf222-1 was selected from the M2 progeny of an EMS-treated Arabidopsis (Arabidopsis thaliana) population (ecotype Col-0) due to its high chlorophyll fluorescence phenotype (Meurer et al., 1996). The mutants selected in this screen were named with the abbreviation hcf and a serial number starting with 100. In order to map the hcf222-1 gene, heterozygous mutants were crossed to the ecotype Ler, and selected F2 mutant plants were genotyped using simple sequence length polymorphisms and cleaved-amplified polymorphic sequences as genetic markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994). The mutated gene was mapped to chromosome 5 between the markers SNP483168 (primers SNP483168-F/SNP483168-R; Supplemental Table S1; polymorphism HpyF3I site in Col-00) and SNP483268 (primers SNP483268-F/SNP483268-R; Supplemental Table S1; polymorphism Eam1104I site in Ler). Of the remaining 10 genes in this chromosomal region, all 10 were amplified in the mutant background and sequenced to identify mutations in their coding regions. The T-DNA mutant line GK_038B09 was generated in the context of the GABI-Kat program (Kleinboelting et al., 2012) and obtained from the Arabidopsis Biological Resource Center (N403573).
For mutant selection, surface-sterilized seeds were sown on petri dishes containing 0.3% (w/v) solidified gelrite supplemented with one-half-strength Murashige and Skoog nutrients (Murashige and Skoog, 1962) and 1% (w/v) Suc. During the day, seedlings were illuminated at a photon flux density (PFD) of 40 to 60 μmol m−2 s−1 with a 16-h photoperiod and 23°C. To minimize secondary effects of light, mutants were cultivated under dim light with an intensity of 8 to 10 μmol m−2 s−1. Alternatively, plants were cultivated on soil in a growth chamber operating at a 16-h-light/8-h-dark period with a constant temperature of 23°C. For seed collection, the Arasystem (Betatech; http://www.gelifesciences.com) was used.
Complementation of the Mutants
For a first complementation analysis of the hcf222 mutant phenotype, a construct consisting of the At5g15802 open reading frame and the 5′ untranslated region was created in the Gateway vector system pAUL1 (Lyska et al., 2013a). The HCF222 cDNA was amplified by PCR with the primers 5-5802att-B1 and 5-5802att-B2 (Supplemental Table S1) using wild-type cDNA as a template. To create the entry clone pENTRY-HCF222, a BP Clonase reaction (Invitrogen) between the HCF222 PCR product and the donor vector pDONR221 was performed according to the Gateway manual. pENTRY-HCF222 was subjected to sequence analysis. By performing an LR Clonase reaction (Invitrogen) with this entry clone, HCF222 was introduced into various binary T-DNA destination vectors. The created expression clones were transferred to heterozygous hcf222-1 and hcf222-2 plants by Agrobacterium tumefaciens-mediated transformation according to the floral dip method (Clough and Bent, 1998). To determine the genotype of transformed plants and their progeny, a single-nucleotide polymorphism marker was created (SNP15802) based on the loss of an Fnu4HI site in the mutant allele (primers 5-5802-4.1F/5-5802-4R (Supplemental Table S1). To create the HCF222-GFP fusion constructs, the destination vector pGWB5 (Nakagawa et al., 2007) was used. For construction of the synthetic fusion product RbcS-TP-cHCF222-GFP, the rbcS-TP cDNA was amplified by PCR with the primers rbcS-att-B1 and R-rbcS(BamHI) (Supplemental Table S1). The cDNA of HCF222 was amplified with the primers F-5-5802(BamHI) and 5-5802att-B2 (Supplemental Table S1). Both PCR products were fused via their BamHI sites, inserted into the donor vector pDONR221 (Thermo) by Gateway cloning, and finally introduced into pGWB5.
Spectroscopic Measurements
Chlorophyll fluorescence and P700 absorbance measurements were performed as described (Meurer et al., 1996) with the following modifications. Saturating light pulses had a length of 500 ms and a PFD of 5,000 μmol m−2 s−1, and actinic light had a PFD of 120 μmol m−2 s−1. Plants grown under dimmed light (5–10 µmol m−2 s−1) were measured with actinic light of 30 μmol m−2 s−1. The parameter qL was determined according to Kramer et al. (2004) as qL = (Fm′ − F′/Fm′)(Fo/F′). The steady-state fluorescence of 3-week-old seedlings was visualized with a FluorCam 800MF (Photon Systems Instruments) controlled by the software package FluorCam 6.
Protein Extraction, Immunoblot Analysis, and Heme Staining
To isolate whole-leaf membrane proteins, leaves from wild-type and mutant plants were ground in liquid nitrogen and the cell powder was homogenized in ice-cold lysis buffer (50 mm Tris-HCl, pH 8, 10 mm EDTA, 2 mm EGTA, and 10 mm DTT). The protein extract was filtered through Miracloth and centrifuged for 20 min at 4°C and 16,000g. For total leaf protein extracts, the amount of lysis buffer was reduced to 4 mL g−1 leaf fresh weight and filtered to glass wool placed in the tip of a syringe. Protein extracts were separated by SDS-PAGE (Schägger and von Jagow, 1987) and subsequently used for immunoblotting as described (Levey et al., 2014). To analyze the strength of the membrane association of HCF222, whole-leaf membrane fractions were resuspended in 50 mm HEPES/KOH, pH 7.5, or HEPES buffer supplemented with 2 m NaBr, 2 m NaSCN, or 1% (w/v) octyl-β-d-glucopyranoside. Membranes were centrifuged for 20 min at 4°C and 24,000g. Supernatants were precipitated with 15% (w/v) TCA. Resuspended samples were separated by SDS-PAGE and analyzed with specific antisera. Antibodies used for the immunodetection of specific thylakoid membrane proteins were as described (Oswald et al., 1990; Meurer et al., 1996) or purchased from Agrisera (anti-PetB [AS03034], anti-LHCB1 [AS09522], anti-RbcL [AS03037], and anti-RbcS [AS07259]). To detect covalently bound heme in cytochrome f and b6, leaf membrane proteins were isolated in lysis buffer as described above. Subsequently, protein extracts were separated directly by SDS-PAGE and blotted to nitrocellulose membranes. The dry membrane was incubated with the high-sensitivity enhanced chemiluminescence substrate Super Signal West Femto (Thermo) and exposed for 2 to 10 min in the image analyzer LAS 4000 (Fujifilm).
RNA Isolation, Gel-Blot Analysis, and Quantitative Real-Time PCR
Isolation of total leaf RNA, electrophoresis, gel-blot analysis, and hybridization were performed according to Westhoff et al. (1991). The specific RNA probes used for hybridization are listed by Meurer et al. (1996). For real-time PCR, RNA was extracted from Arabidopsis rosette leaves using the Plant RNA MiniPrep Kit (Zymo Research) and reverse transcribed with the QuantiTect Reverse Transcription Kit (Qiagen). Quantitative PCR was carried out on a 7500 Fast Real-Time PCR system with SYBR Green PCR master mix (Applied Biosystems). Primers used to quantify HCF222 transcript levels and various transcripts of photosynthetic genes are listed in Supplemental Table S1. Transcript levels of actin were used as a reference. Polysomal fractions of RNA were isolated as described (Barkan, 1993). For the detection of RNAs, we used the protocol cited above. Probes against petG, petL, and petN were described (Lennartz et al., 2006).
Expression of Recombinant Protein and Enzymatic Analysis of Δ24HCF222
A cDNA fragment of HCF222 comprising amino acids 25 to 99 was amplified by the PCR primers P2-HCF222-F and P2-HCF222-R (Supplemental Table S1). This cDNA encoded a truncated protein lacking the predicted 24-amino acid-long signal peptide of HCF222 (Δ24HCF222). The cleaved DNA fragment (BamHI and XhoI underlined in the primer sequence) was cloned into the expression vector pET-21a(+) (Novagen) and transformed into Escherichia coli Rosetta 2 (DE3) (Novagen). The protein was expressed under standard conditions (37°C, induction with 0.5 mm isopropyl-β-d-thiogalactopyranoside, 3 h of expression). A C-terminal 6× His tag enabled the purification of the protein by metal chelate affinity chromatography on Ni-NTA resin under native conditions according to the QIAexpressionist protocol (Qiagen). For the purification of preparative protein amounts, His GraviTrap columns (GE Healthcare) were used. Isolated amounts of HCF222 were determined by comparison with a dilution series of lysozyme after SDS-PAGE. To measure the disulfide reductase activity of HCF222, the insulin precipitation assay was conducted (Holmgren, 1979). The reaction contained 100 mm sodium phosphate buffer (pH 7), 1 mm EDTA, 0.17 mm insulin (Sigma-Aldrich), 0.35 mm DTT, and either 3 µm E. coli thioredoxin, ∼3 µm Δ24HCF222, or an equivalent volume of β-galactosidase-6xHis (∼1 µm), which was isolated from E. coli extracts under the same conditions as Δ24HCF222. The assay was started by the addition of DTT, and precipitation of the insulin B chain was measured spectrophotometrically at 650 nm. β-Galactosidase was used as a control to monitor the possible impact of contaminating proteins from E. coli during enzyme measurements. Disulfide reduction also was measured under conditions without enzyme (DTT control).
Transient Expression of Fusion Proteins in Nicotiana benthamiana
Transformation of N. benthamiana and preparation of protoplasts from infiltrated leaf discs were performed as described (Breuers et al., 2012). N. benthamiana plants were 4 to 5 weeks old, but only youngest leaves were used for infiltration. Images of GFP or mCherry fluorescence were taken 48 to 72 h after infiltration.
Confocal Laser Scanning Microscopy and ER Staining
Protoplasts were analyzed using a Leica TCS SP8 STED 3X confocal laser scanning microscope (Leica Microsystems). GFP and chlorophyll were excited with the 488-nm laser line of an argon laser. Emission of GFP was detected between 491 and 533 nm and chlorophyll autofluorescence between 620 and 670 nm. Under these settings, discrimination between GFP and chlorophyll fluorescence was possible, and faint GFP fluorescence of chloroplasts could be collected with high sensitivity. The ER marker mCherry HDEL (Nelson et al., 2007) was excited at 561 nm, and emission was collected at 575 to 615 nm. Images were taken using a 40× water-immersion objective and 12-bit resolution. All images were produced with a pinhole of 1 Airy unit to ensure perfect confocality. Image processing was performed with the software package LAS X (Leica Microsystems).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative, the Maize Genetics and Genomics, or the GenBank/EMBL databases under the following accession numbers: Arabidopsis thaliana HCF222 (At5g15802) Tarenaya hassleriana HCF222 (XP_010549962.1), Vitis vinifera HCF222 (XP_003634726.1), Zea mays GRMZM2G145527-P02 (AFW79256), Z. mays GRMZM2G145527-P01 (AFW79255), Populus trichocarpa HCF222 (XP_006373394.1), Oryza sativa japonica group HCF222 (XP_015629048.1), Sorghum bicolor HCF222 SORBIDRAFT_01g032530 (XP_002465131.1), S. bicolor HCF222 SORBI_009G237800 (KXG22558), Amborella trichopoda HCF222 (XP_006846238.1), and Marchantia polymorpha HCF222 (OAE31695).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Complementation analysis of hcf222 mutants.
Supplemental Figure S2. Accumulation of RNAs for photosynthetic proteins in the wild type and hcf222 mutants.
Supplemental Figure S3. Polysome association of plastid- and nucleus-encoded Cytb6f RNAs.
Supplemental Figure S4. Membrane binding of HCF222.
Supplemental Figure S5. Expression of HCF222 in different tissues and developmental stages of Arabidopsis.
Supplemental Figure S6. HCF222 homologs from maize and sorghum.
Supplemental Table S1. Sequences of primers used for PCR.
Acknowledgments
We thank the Center for Advanced Imaging (Heinrich Heine University Düsseldorf) for technical assistance with confocal microscopy, the GABI-Kat and the Arabidopsis Biological Resource Center for providing seeds of the T-DNA line N403573, and Peter Jahns for support with the FluorCam system.
Glossary
- Cytb6f
cytochrome b6f
- ER
endoplasmic reticulum
- EMS
ethyl methanesulfonate
- Col-0
Columbia-0
- PFD
photon flux density
Footnotes
This work was supported by the German Science Foundation (grant no. SFB 1208).
Articles can be viewed without a subscription.
References
- Albrecht V, Ingenfeld A, Apel K (2008) Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Mol Biol 66: 599–608 [DOI] [PubMed] [Google Scholar]
- Alergand T, Peled-Zehavi H, Katz Y, Danon A (2006) The chloroplast protein disulfide isomerase RB60 reacts with a regulatory disulfide of the RNA-binding protein RB47. Plant Cell Physiol 47: 540–548 [DOI] [PubMed] [Google Scholar]
- Aller I, Meyer AJ (2013) The oxidative protein folding machinery in plant cells. Protoplasma 250: 799–816 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Hertle A, Makarenko E, Zühlke J, Pribil M, Dietzmann A, Schliebner I, Aseeva E, Fenino E, Scharfenberg M, et al. (2009) Chloroplast proteins without cleavable transit peptides: rare exceptions or a major constituent of the chloroplast proteome? Mol Plant 2: 1325–1335 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Zühlke J, Rengstl B, Kreller R, Makarenko E, Rühle T, Schünemann D, Jahns P, Weisshaar B, Nickelsen J, et al. (2010) The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asatsuma S, Sawada C, Itoh K, Okito M, Kitajima A, Mitsui T (2005) Involvement of alpha-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol 46: 858–869 [DOI] [PubMed] [Google Scholar]
- Balk J, Pilon M (2011) Ancient and essential: the assembly of iron-sulfur clusters in plants. Trends Plant Sci 16: 218–226 [DOI] [PubMed] [Google Scholar]
- Banecki B, Liberek K, Wall D, Wawrzynów A, Georgopoulos C, Bertoli E, Tanfani F, Zylicz M (1996) Structure-function analysis of the zinc finger region of the DnaJ molecular chaperone. J Biol Chem 271: 14840–14848 [DOI] [PubMed] [Google Scholar]
- Baniulis D, Yamashita E, Zhang H, Hasan SS, Cramer WA (2008) Structure-function of the cytochrome b6f complex. Photochem Photobiol 84: 1349–1358 [DOI] [PubMed] [Google Scholar]
- Barkan A. (1993) Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Breuers FK, Bräutigam A, Geimer S, Welzel UY, Stefano G, Renna L, Brandizzi F, Weber AP (2012) Dynamic remodeling of the plastid envelope membranes: a tool for chloroplast envelope in vivo localizations. Front Plant Sci 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burén S, Ortega-Villasante C, Blanco-Rivero A, Martínez-Bernardini A, Shutova T, Shevela D, Messinger J, Bako L, Villarejo A, Samuelsson G (2011) Importance of post-translational modifications for functionality of a chloroplast-localized carbonic anhydrase (CAH1) in Arabidopsis thaliana. PLoS ONE 6: e21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell CJ, Zhang H, Cramer WA, Smith JL (1997) Biological identity and diversity in photosynthesis and respiration: structure of the lumen-side domain of the chloroplast Rieske protein. Structure 5: 1613–1625 [DOI] [PubMed] [Google Scholar]
- Claros MG, von Heijne G (1994) TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci 10: 685–686 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Crouy-Chanel A, Kohiyama M, Richarme G (1995) A novel function of Escherichia coli chaperone DnaJ: protein-disulfide isomerase. J Biol Chem 270: 22669–22672 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Feng WK, Wang L, Lu Y, Wang XY (2011) A protein oxidase catalysing disulfide bond formation is localized to the chloroplast thylakoids. FEBS J 278: 3419–3430 [DOI] [PubMed] [Google Scholar]
- Fristedt R, Williams-Carrier R, Merchant SS, Barkan A (2014) A thylakoid membrane protein harboring a DnaJ-type zinc finger domain is required for photosystem I accumulation in plants. J Biol Chem 289: 30657–30667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabilly ST, Dreyfuss BW, Karamoko M, Corvest V, Kropat J, Page MD, Merchant SS, Hamel PP (2010) CCS5, a thioredoxin-like protein involved in the assembly of plastid c-type cytochromes. J Biol Chem 285: 29738–29749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabilly ST, Kropat J, Karamoko M, Page MD, Nakamoto SS, Merchant SS, Hamel PP (2011) A novel component of the disulfide-reducing pathway required for cytochrome c assembly in plastids. Genetics 187: 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M, Mata-Cabana A, Akerlund HE, Florencio FJ, Schröder WP, Lindahl M, Kieselbach T (2010) Thioredoxin targets of the plant chloroplast lumen and their implications for plastid function. Proteomics 10: 987–1001 [DOI] [PubMed] [Google Scholar]
- Holmgren A. (1979) Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem 254: 9627–9632 [PubMed] [Google Scholar]
- Howe G, Merchant S (1992) The biosynthesis of membrane and soluble plastidic c-type cytochromes of Chlamydomonas reinhardtii is dependent on multiple common gene products. EMBO J 11: 2789–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Liu B, Luo L, Feng D, Wang P, Liu J, Da Q, He Y, Qi K, Wang J, et al. (2014) HYPERSENSITIVE TO HIGH LIGHT1 interacts with LOW QUANTUM YIELD OF PHOTOSYSTEM II1 and functions in protection of photosystem II from photodamage in Arabidopsis. Plant Cell 26: 1213–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamoko M, Cline S, Redding K, Ruiz N, Hamel PP (2011) Lumen Thiol Oxidoreductase1, a disulfide bond-forming catalyst, is required for the assembly of photosystem II in Arabidopsis. Plant Cell 23: 4462–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieselbach T. (2013) Oxidative folding in chloroplasts. Antioxid Redox Signal 19: 72–82 [DOI] [PubMed] [Google Scholar]
- Kitajima A, Asatsuma S, Okada H, Hamada Y, Kaneko K, Nanjo Y, Kawagoe Y, Toyooka K, Matsuoka K, Takeuchi M, et al. (2009) The rice α-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell 21: 2844–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjölander K, Gruissem W, Baginsky S (2004) The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol 14: 354–362 [DOI] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2012) GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA (2016) A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J 88: 1058–1070 [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Kramer DM, Johnson G, Kiirats O, Edwards GE (2004) New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth Res 79: 209. [DOI] [PubMed] [Google Scholar]
- Kranz RG, Richard-Fogal C, Taylor JS, Frawley ER (2009) Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol Mol Biol Rev 73: 510–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras R, Büschlen S, Wollman FA (1995) Maturation of pre-apocytochrome f in vivo: a site-directed mutagenesis study in Chlamydomonas reinhardtii. J Biol Chem 270: 27797–27803 [DOI] [PubMed] [Google Scholar]
- Kuras R, Saint-Marcoux D, Wollman FA, de Vitry C (2007) A specific c-type cytochrome maturation system is required for oxygenic photosynthesis. Proc Natl Acad Sci USA 104: 9906–9910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu G, Zhang H, Smith JL, Cramer WA (2003) Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science 302: 1009–1014 [DOI] [PubMed] [Google Scholar]
- Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU (1992) Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356: 683–689 [DOI] [PubMed] [Google Scholar]
- Leggate EJ, Hirst J (2005) Roles of the disulfide bond and adjacent residues in determining the reduction potentials and stabilities of respiratory-type Rieske clusters. Biochemistry 44: 7048–7058 [DOI] [PubMed] [Google Scholar]
- Lennartz K, Bossmann S, Westhoff P, Bechtold N, Meierhoff K (2006) HCF153, a novel nuclear-encoded factor necessary during a post-translational step in biogenesis of the cytochrome bf complex. Plant J 45: 101–112 [DOI] [PubMed] [Google Scholar]
- Lennartz K, Plücken H, Seidler A, Westhoff P, Bechtold N, Meierhoff K (2001) HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b(6)f complex in Arabidopsis. Plant Cell 13: 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey T, Westhoff P, Meierhoff K (2014) Expression of a nuclear-encoded psbH gene complements the plastidic RNA processing defect in the PSII mutant hcf107 in Arabidopsis thaliana. Plant J 80: 292–304 [DOI] [PubMed] [Google Scholar]
- Levitan A, Trebitsh T, Kiss V, Pereg Y, Dangoor I, Danon A (2005) Dual targeting of the protein disulfide isomerase RB60 to the chloroplast and the endoplasmic reticulum. Proc Natl Acad Sci USA 102: 6225–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezhneva L, Kuras R, Ephritikhine G, de Vitry C (2008) A novel pathway of cytochrome c biogenesis is involved in the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J Biol Chem 283: 24608–24616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Hall DA, Last RL (2011) A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23: 1861–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyska D, Engelmann K, Meierhoff K, Westhoff P (2013a) pAUL: a Gateway-based vector system for adaptive expression and flexible tagging of proteins in Arabidopsis. PLoS ONE 8: e53787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyska D, Meierhoff K, Westhoff P (2013b) How to build functional thylakoid membranes: from plastid transcription to protein complex assembly. Planta 237: 413–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyska D, Paradies S, Meierhoff K, Westhoff P (2007) HCF208, a homolog of Chlamydomonas CCB2, is required for accumulation of native cytochrome b6 in Arabidopsis thaliana. Plant Cell Physiol 48: 1737–1746 [DOI] [PubMed] [Google Scholar]
- Merbitz-Zahradnik T, Zwicker K, Nett JH, Link TA, Trumpower BL (2003) Elimination of the disulfide bridge in the Rieske iron-sulfur protein allows assembly of the [2Fe-2S] cluster into the Rieske protein but damages the ubiquinol oxidation site in the cytochrome bc1 complex. Biochemistry 42: 13637–13645 [DOI] [PubMed] [Google Scholar]
- Meurer J, Meierhoff K, Westhoff P (1996) Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198: 385–396 [DOI] [PubMed] [Google Scholar]
- Meurer J, Plücken H, Kowallik KV, Westhoff P (1998) A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J 17: 5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EH, Taylor NL, Millar AH (2008) Resolving and identifying protein components of plant mitochondrial respiratory complexes using three dimensions of gel electrophoresis. J Proteome Res 7: 786–794 [DOI] [PubMed] [Google Scholar]
- Mosser G, Breyton C, Olofsson A, Popot JL, Rigaud JL (1997) Projection map of cytochrome b6 f complex at 8 A resolution. J Biol Chem 272: 20263–20268 [DOI] [PubMed] [Google Scholar]
- Motohashi K, Hisabori T (2006) HCF164 receives reducing equivalents from stromal thioredoxin across the thylakoid membrane and mediates reduction of target proteins in the thylakoid lumen. J Biol Chem 281: 35039–35047 [DOI] [PubMed] [Google Scholar]
- Muranaka A, Watanabe S, Sakamoto A, Shimada H (2012) Arabidopsis cotyledon chloroplast biogenesis factor CYO1 uses glutathione as an electron donor and interacts with PSI (A1 and A2) and PSII (CP43 and CP47) subunits. J Plant Physiol 169: 1212–1215 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nanjo Y, Oka H, Ikarashi N, Kaneko K, Kitajima A, Mitsui T, Muñoz FJ, Rodríguez-López M, Baroja-Fernández E, Pozueta-Romero J (2006) Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-Golgi to the chloroplast through the secretory pathway. Plant Cell 18: 2582–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navrot N, Gelhaye E, Jacquot JP, Rouhier N (2006) Identification of a new family of plant proteins loosely related to glutaredoxins with four CxxC motives. Photosynth Res 89: 71–79 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Onda Y. (2013) Oxidative protein-folding systems in plant cells. Int J Cell Biol 2013: 585431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald A, Streubel M, Ljungberg U, Hermans J, Eskins K, Westhoff P (1990) Differential biogenesis of photosystem-II in mesophyll and bundle-sheath cells of ‘malic’ enzyme NADP(+)-type C4 plants. A comparative protein and RNA analysis. Eur J Biochem 190: 185–194 [DOI] [PubMed] [Google Scholar]
- Page MLD, Hamel PP, Gabilly ST, Zegzouti H, Perea JV, Alonso JM, Ecker JR, Theg SM, Christensen SK, Merchant S (2004) A homolog of prokaryotic thiol disulfide transporter CcdA is required for the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J Biol Chem 279: 32474–32482 [DOI] [PubMed] [Google Scholar]
- Saint-Marcoux D, Wollman FA, de Vitry C (2009) Biogenesis of cytochrome b6 in photosynthetic membranes. J Cell Biol 185: 1195–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Schöttler MA, Tóth SZ, Boulouis A, Kahlau S (2015) Photosynthetic complex stoichiometry dynamics in higher plants: biogenesis, function, and turnover of ATP synthase and the cytochrome b6f complex. J Exp Bot 66: 2373–2400 [DOI] [PubMed] [Google Scholar]
- Schwenkert S, Legen J, Takami T, Shikanai T, Herrmann RG, Meurer J (2007) Role of the low-molecular-weight subunits PetL, PetG, and PetN in assembly, stability, and dimerization of the cytochrome b6f complex in tobacco. Plant Physiol 144: 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YY, Tang W, Hao SF, Wang CC (2005) Contributions of cysteine residues in Zn2 to zinc fingers and thiol-disulfide oxidoreductase activities of chaperone DnaJ. Biochemistry 44: 1683–1689 [DOI] [PubMed] [Google Scholar]
- Shimada H, Mochizuki M, Ogura K, Froehlich JE, Osteryoung KW, Shirano Y, Shibata D, Masuda S, Mori K, Takamiya K (2007) Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. Plant Cell 19: 3157–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppel R, Lezhneva L, Schwenkert S, Torabi S, Felder S, Meierhoff K, Westhoff P, Meurer J (2011) Recruitment of a ribosomal release factor for light- and stress-dependent regulation of petB transcript stability in Arabidopsis chloroplasts. Plant Cell 23: 2680–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Korszun R, Hartl FU, Flanagan J (1996) A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J 15: 408–417 [PMC free article] [PubMed] [Google Scholar]
- Tang W, Wang CC (2001) Zinc fingers and thiol-disulfide oxidoreductase activities of chaperone DnaJ. Biochemistry 40: 14985–14994 [DOI] [PubMed] [Google Scholar]
- Tanz SK, Castleden I, Hooper CM, Vacher M, Small I, Millar HA (2013) SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res 41: D1185–D1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanz SK, Kilian J, Johnsson C, Apel K, Small I, Harter K, Wanke D, Pogson B, Albrecht V (2012) The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LHCB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings. Plant J 69: 743–754 [DOI] [PubMed] [Google Scholar]
- Tomizioli M, Lazar C, Brugière S, Burger T, Salvi D, Gatto L, Moyet L, Breckels LM, Hesse AM, Lilley KS, et al. (2014) Deciphering thylakoid sub-compartments using a mass spectrometry-based approach. Mol Cell Proteomics 13: 2147–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarejo A, Burén S, Larsson S, Déjardin A, Monné M, Rudhe C, Karlsson J, Jansson S, Lerouge P, Rolland N, et al. (2005) Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat Cell Biol 7: 1224–1231 [DOI] [PubMed] [Google Scholar]
- Westhoff P, Offermann-Steinhard K, Höfer M, Eskins K, Oswald A, Streubel M (1991) Differential accumulation of plastid transcripts encoding photosystem II components in the mesophyll and bundle-sheath cells of monocotyledonous NADP-malic enzyme-type C4 plants. Planta 184: 377–388 [DOI] [PubMed] [Google Scholar]
- Wittenberg G, Levitan A, Klein T, Dangoor I, Keren N, Danon A (2014) Knockdown of the Arabidopsis thaliana chloroplast protein disulfide isomerase 6 results in reduced levels of photoinhibition and increased D1 synthesis in high light. Plant J 78: 1003–1013 [DOI] [PubMed] [Google Scholar]
- Xiao J, Li J, Ouyang M, Yun T, He B, Ji D, Ma J, Chi W, Lu C, Zhang L (2012) DAC is involved in the accumulation of the cytochrome b6/f complex in Arabidopsis. Plant Physiol 160: 1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Whitelegge JP, Cramer WA (2001) Ferredoxin:NADP+ oxidoreductase is a subunit of the chloroplast cytochrome b6f complex. J Biol Chem 276: 38159–38165 [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]