Arabidopsis acyl-CoA-binding protein ACBP1 forms an endoplasmic reticulum-associated complex with STEROL C4-METHYL OXIDASE1-1 and negatively modulates sterol synthesis during embryogenesis.
Abstract
Fatty acids (FAs) and sterols are primary metabolites that exert interrelated functions as structural and signaling lipids. Despite their common syntheses from acetyl-coenzyme A, homeostatic cross talk remains enigmatic. Six Arabidopsis (Arabidopsis thaliana) acyl-coenzyme A-binding proteins (ACBPs) are involved in FA metabolism. ACBP1 interacts with PHOSPHOLIPASE Dα1 and regulates phospholipid composition. Here, its specific role in the negative modulation of sterol synthesis during embryogenesis is reported. ACBP1, likely in a liganded state, interacts with STEROL C4-METHYL OXIDASE1-1 (SMO1-1), a rate-limiting enzyme in the sterol pathway. Proembryo abortion in the double mutant indicated that the ACBP1-SMO1-1 interaction is synthetic lethal, corroborating with their strong promoter activities in developing ovules. Gas chromatography-mass spectrometry revealed quantitative and compositional changes in FAs and sterols upon overexpression or mutation of ACBP1 and/or SMO1-1. Aberrant levels of these metabolites may account for the downstream defect in lipid signaling. GLABRA2 (GL2), encoding a phospholipid/sterol-binding homeodomain transcription factor, was up-regulated in developing seeds of acbp1, smo1-1, and ACBP1+/−smo1-1 in comparison with the wild type. Consistent with the corresponding transcriptional alteration of GL2 targets, high-oil, low-mucilage phenotypes of gl2 were phenocopied in ACBP1+/−smo1-1. Thus, ACBP1 appears to modulate the metabolism of two important lipid classes (FAs and sterols) influencing cellular signaling.
Sterols are structural components that control eukaryotic membrane fluidity and permeability (Schaller, 2004). Unlike cholesterol, which is a principal animal sterol, there exist more than 100 plant sterols (i.e. phytosterols), prevalently as sitosterol, campesterol, and stigmasterol (Hartmann, 1998). Sterols are primary metabolites synthesized from cycloartenol, the first stable tetracyclic derivative from acetyl-CoA via the isoprenoid pathway (Bach, 1995). Functional sterols are produced after the removal of two C4 methyl groups from cycloartenol, which is catalyzed successively by a multienzyme complex containing STEROL C4-METHYL OXIDASE (SMO) in animals and fungi (Li and Kaplan, 1996). In plants, demethylation occurs nonconsecutively under the catalysis of nonhomologous SMO1 and SMO2, which are encoded by three and two loci in Arabidopsis (Arabidopsis thaliana), respectively (Darnet and Rahier, 2004). Five of the six steps in the early sterol pathway have been investigated using Arabidopsis mutants that exhibited embryonic and vascular patterning defects (Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000, 2002; Souter et al., 2002; Willemsen et al., 2003; Kim et al., 2005; Men et al., 2008). SMO1 catalyzes the remaining one that has not been studied by reverse genetics but shown to be rate limiting, representing a potential target for enhancing flux toward higher phytosterol accumulation (Lange et al., 2015). Its catalytic product also is known to regulate polar auxin transport (Mialoundama et al., 2013). Studies on sterol biosynthetic mutants have indicated the existence of other sterol-derived signals that shape plant development, although many remain unknown (Carland et al., 2010; Qian et al., 2013; Zhang et al., 2016).
Downstream protein participants in sterol-mediated signaling are better understood. Sterols are bound by the steroidogenic acute regulatory protein (StAR)-related lipid transfer (START) domain (Ponting and Aravind, 1999), which facilitates StAR-mediated cholesterol transport in animal mitochondria (Stocco, 2001). In plants, this domain is distributed mostly within the homeodomain (HD)-Leu zipper transcription factor (TF) family (Schrick et al., 2004), of which the unidentified ligands are deemed important developmental and stress regulators (Yu et al., 2008). The Arabidopsis genome encodes 21 HD-START TFs, most of which regulate cell fate determination and patterning (Schrick et al., 2014). Among them, GLABRA2 (GL2) is the sole known member that negatively regulates seed oil biosynthesis and promotes seed coat mucilage formation (Shen et al., 2006; Shi et al., 2012). Whether the START domain is essential for GL2 function had remained unclear until recent findings that its truncation or site-directed mutagenesis affected the complementation of Arabidopsis gl2 phenotypes (Schrick et al., 2014). The START domain of GL2 was shown recently to bind phospholipids (PLs) in addition to sterols (Schrick et al., 2014), corroborating earlier evidence that GL2 modulates PL signaling in root hair patterning (Ohashi et al., 2003). While PLs and sterols functionally converge on the same downstream target (START domain) in signal transduction, their close metabolic relationship also is inferred from a reaction that esterifies them into fatty acyl sterol esters (Schaller, 2004; Bouvier-Navé et al., 2010).
Acetyl-CoA also fuels the synthesis of fatty acids (FAs), which are used for acyl-lipid (e.g. PLs and triacylglycerols) assembly after their activation into CoA esters (Block et al., 1983). In the cytosol, these esters are maintained homeostatically by nonenzymic proteins such as acyl-CoA-binding proteins (ACBPs; Xiao and Chye, 2011). Apart from cytosolic members, there exist other plant ACBPs that also are conserved to bind acyl-CoA esters and PLs (Lung and Chye, 2016a). The six Arabidopsis ACBPs are grouped into four classes by size and domain architecture (Meng et al., 2011), which are linked differently to plant development and stress responses (Xiao and Chye, 2011; Du et al., 2016; Lung and Chye, 2016b). ACBP1 and its homolog, ACBP2, are targeted to the endoplasmic reticulum (ER) and the plasma membrane (Chye, 1998; Chye et al., 1999; Li and Chye, 2003). Both proteins are highly expressed during seed development, and the acbp1acbp2 double mutant is embryo lethal (Chen et al., 2010). While ACBP1 binds very-long-chain acyl-CoA esters and contributes to stem cuticle synthesis (Xue et al., 2014), its affinity to phosphatidylcholine (PC) and phosphatidic acid (PA) is pertinent to PL signal transduction (Du et al., 2010, 2013). ACBP1 interacts with PHOSPHOLIPASE Dα1 (PLDα1) at the plasma membrane to regulate the generation of PA, a messenger for abscisic acid signaling (Du et al., 2013). acbp1 is more freezing tolerant, as its rosettes contain a lower level of PLDα1-derived PA, a damaging agent of biomembranes during cold stress (Du et al., 2010). In siliques, acbp1 exhibits an aberrant PL composition (Chen et al., 2010).
Other than its roles in PL metabolism and signaling, ACBP1 is reported here to negatively modulate sterol synthesis during embryogenesis via protein-protein interaction (PPI) with another partner, SMO1-1. Reverse genetics studies revealed a relationship between FA homeostasis and sterol production as linked by ACBP1. The aberrant FA and sterol compositions of mutant siliques may account for the downstream defect in lipid-mediated signaling that involves GL2.
RESULTS
ACBP1 and SMO1-1 Are ER-Localized Protein Partners
Yeast two-hybrid (Y2H) screens of an Arabidopsis cDNA library using ACBP1 as bait had identified a partial in-frame sequence of SMO1-1 (Tse, 2005). This PPI was confirmed by Y2H analysis using the soluble region (i.e. residues 41–338) of ACBP1 and the open reading frame (ORF) encoding SMO1-1 (Fig. 1A). SMO1-1 did not bind ACBP132–216 lacking the C-terminal ANK domain (Fig. 1A), a PPI structure (Vo et al., 2015). The absence of blue colonies using ANK alone (i.e. ACBP1217–338) further suggested that both ACB and ANK domains are essential for PPI (Fig. 1A). To study if a functional ACB domain is required, the conserved Tyr at 171 was substituted with Ala, because an equivalent substitution had interrupted 16:0-CoA binding to recombinant ACBP2 (Chye et al., 2000). In Y2H screens, this mutation weakened PPI, implying that a liganded form of ACBP1 may facilitate SMO1-1 interaction (Fig. 1A).
Figure 1.
PPI of ACBP1 with SMO1-1. A, Y2H assays. The soluble domain of ACBP1, its derivative without the ankyrin repeat (ANK) or acyl-CoA-binding (ACB) domains, and its mutated (Y171A) versions were cloned into the bait vector. The full-length SMO1-1 sequence was cloned into the prey vector. Cotransformants of bait and prey constructs were verified on double dropout (DDO) plates. The appearance of blue colonies on triple dropout selection plates (TDO/X/A) indicates PPI. The pGADT7-T construct was cotransformed with pGBKT7-53 and pGBKT7-Lam as positive and negative controls, respectively. B, Localization of SMO1-1:EGFP in transgenic Arabidopsis. Leaf epidermal cells of 3-week-old plants and root cells of 1-week-old seedlings were imaged by confocal laser scanning microscopy. Signals were detected at the perinuclear ER (white arrowheads), ER bodies (open arrowheads), and tubular ER network throughout the cells. Signals were colocalized at the membrane of ER-derived vesicles (red arrowheads) using the ER-Tracker in root hair cells (bottom). Bars = 20 µm. C, Colocalization of SMO1-1:EGFP with DsRed:ACBP1. Root and hypocotyl cells of 1-week-old transgenic Arabidopsis seedlings and agroinfiltrated tobacco leaf epidermal cells were imaged by confocal laser scanning microscopy. Signals were colocalized at the plasma membrane, membrane of ER-derived vesicles (red arrowheads), ER cisternae (arrows), and perinuclear ER (white arrowheads). Bars = 20 µm. D, Subcellular fractionation of SMO1-1:HA:StrepII and ACBP1. Proteins (20 µg per lane) from total crude extracts (T), nuclei (N), membranes (M), large particles including mitochondria, plastids, and peroxisomes (LP), and soluble fractions (S) were prepared from aboveground tissues of 6-week-old Arabidopsis and analyzed by western-blot analysis and Coomassie Blue-stained gels. Total proteins from an empty vector line (VC) served as a negative control. Arrowheads indicate the positions of the expected bands (37 kD for SMO1-1:HA:StrepII and 38 kD for ACBP1). E, Strep-Tactin pull-down assays. Membrane proteins were isolated from transgenic Arabidopsis of SMO1-1:HA:StrepII and the vector control, solubilized, and incubated with Strep-Tactin beads. Eluents were analyzed by western-blot analysis using anti-ACBP1 and anti-HA antibodies. F, Coimmunoprecipitation of SMO1-ACBP1 complexes. Membrane proteins were prepared as described in E and incubated with anti-ACBP1 antibodies that had been covalently coupled to Affi-Gel 10 beads. Eluents were analyzed by western-blot analysis using anti-ACBP1 and anti-HA antibodies.
To investigate the PPI site, SMO1-1 was expressed in fusion with an enhanced GFP (EGFP) in transgenic Arabidopsis. SMO1-1:EGFP was detected at the perinuclear ER in guard cells and ER bodies in root cells, apart from the tubular ER network, by confocal laser scanning microscopy (Fig. 1B, top). In root hair cells, signals were visualized at the plasma membrane and colocalized at the membrane of ER-derived vesicles with the ER marker (Fig. 1B, bottom). In Arabidopsis seedlings, SMO1-1:EGFP was colocalized with DsRed:ACBP1 at the membrane of ER-derived vesicles in root cells and the ER cisternae in hypocotyl cells, in addition to the plasma membrane of both cell types (Fig. 1C). In agroinfiltrated tobacco (Nicotiana tabacum) leaf epidermal cells, the signals were colocalized at the plasma membrane, reticular ER network, and perinuclear ER (Fig. 1C, bottom). These observations coincided well with the predicted location of SMO1-1 (Supplemental Fig. S1A) and the reports of ACBP1 (Chye, 1998; Li and Chye, 2003) at the ER, the site of sterol biosynthesis (Benveniste, 2004).
To further detect the ACBP1-SMO1-1 complex, Arabidopsis was transformed with 35S::SMO1-1:HA:StrepII for Strep-Tactin pull-down and coimmunoprecipitation assays. These plants also served as overexpressors (OEs) in subsequent studies. First, subcellular fractionation showed that SMO1-1:HA:StrepII was enriched in the membrane fraction and less abundant in the fraction of large particles containing mitochondria, plastids, and peroxisomes (Fig. 1D). The trace amount of nucleus-associated signals (Fig. 1D) could be attributed to the perinuclear localization arising from the connection of nuclear envelopes with ER membranes (Fig. 1, B and C). A similar subcellular distribution of ACBP1 was observed (Fig. 1D, right), consistent with a previous observation (Chye, 1998). Subsequently, SMO1-1:HA:StrepII enriched in the membrane fraction was solubilized and captured on Strep-Tactin beads for pull-down assays. Western-blot analysis of coeluted proteins using ACBP1 antibodies produced a cross-reacting band from SMO1-1:HA:StrepII but not the vector control (Fig. 1E). Reciprocally, ACBP1 antibodies coimmunoprecipitated SMO1-1:HA:StrepII from the solubilized membrane fraction (Fig. 1F). Taken together, it is concluded that both ACB and ANK domains of ACBP1 are essential for its association with SMO1-1 at the ER, which is likely facilitated if ACBP1 is in a liganded state.
SMO1-1 Is Expressed in Floral Organs and Developing Seeds
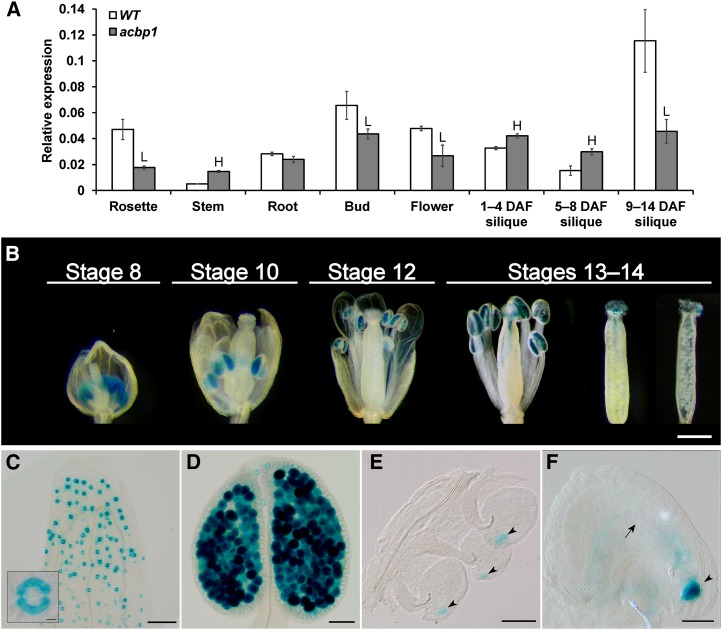
Quantitative real-time (qRT)-PCR detected SMO1-1 transcripts in rosettes, stems, roots, floral buds, flowers, and siliques (Fig. 2A). Its expression was up- or down-regulated in all aboveground tissue samples of acbp1, indicating the functional relevance of SMO1-1 with ACBP1 (Fig. 2A). Microarray data showed that SMO1-1 and ACBP1 were expressed in seeds and dissected embryos from zygotic to maturation stages throughout embryogenesis (Supplemental Fig. S2, B and C). Promoter-driven GUS assays had indicated ACBP1 expression in embryos and floral parts, including stigmas, styles, ovaries, and sepals (Du et al., 2013). SMO1-1pro:GUS lines were generated for comparative studies. Signals were detected in pollen at all floral stages and in pollen tubes at anthesis (Fig. 2B). Microscopically, SMO1-1pro:GUS was observed in guard cells of sepals and anthers (Fig. 2, C and D) and the micropylar end of fertilized ovules at stage 13 (Fig. 2E). At stage 14, it was found in the egg apparatus and, to a lesser extent, in elongated embryo sacs (Fig. 2F). At the quadrant embryo stage, SMO1-1pro:GUS was visualized in the basal suspensor cells and chalazal endosperm (Supplemental Fig. S2A). At maturation, it was expressed in cotyledons of embryos (Supplemental Fig. S2, B and C). ACBP1pro:GUS in fertilized ovules and developing seeds appeared less spatially restricted (Supplemental Fig. S2, D–F), consistent with its immunolocalization pattern (Chye et al., 1999). Hence, microarray data mining, qRT-PCR, and GUS assays suggested the coexpression of SMO1-1 and ACBP1 in reproductive tissues, where functional significance was addressed subsequently by reverse genetics.
Figure 2.
Spatial expression pattern of SMO1-1 in Arabidopsis. A, Relative expression levels of SMO1-1 in various tissues of 6-week-old acbp1 and the wild type (WT). Transcript levels were measured by qRT-PCR and normalized against ACTIN2. Each bar represents the mean of three replicates ± sd. H and L indicate statistically significant (P < 0.05) elevation and reduction, respectively, in comparison with the wild type by Student’s t test. B, SMO1-1pro:GUS expression pattern at different stages of floral development. GUS signals were detected in 8-week-old transgenic Arabidopsis. Intact and open pistils at stages 13 and 14 are shown. Bar = 1 mm. C to F, Whole-mount floral parts of SMO1-1pro:GUS, including sepals (inset, guard cells; C), anthers (D), ovaries at stage 13 (E), and ovules at stage 14 (F). Arrowheads and the arrow indicate egg apparatus and a central cell, respectively. Bars = 100 µm (C), 10 µm (inset in C), 50 µm (D and E), and 20 µm (F).
ACBP1 and SMO1-1 Mutation Led to Reproductive Phenotypes
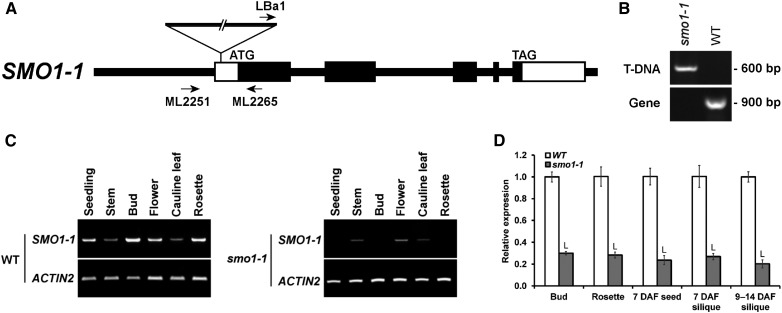
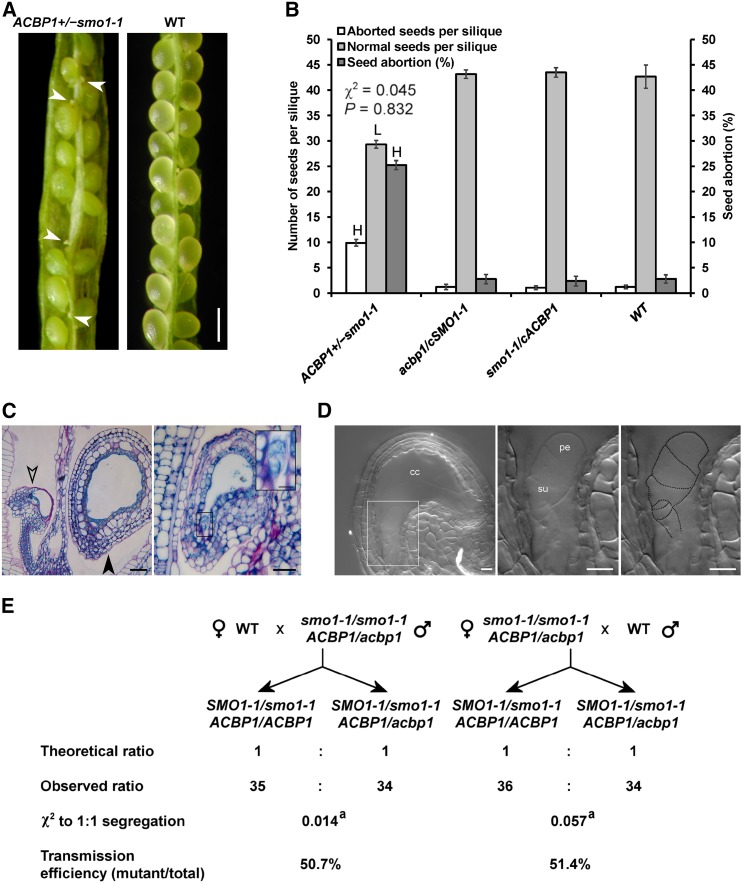
A T-DNA insertion line was first characterized to be a knockdown mutant of smo1-1 by reverse transcription (RT)-PCR (Fig. 3, A–C). qRT-PCR revealed that its SMO1-1 levels in various tissues were 70% to 80% lower than in the wild type (Fig. 3D). This mutant was crossed subsequently with the acbp1 knockout mutant. After genotyping more than 100 F2 and F3 progeny, ACBP1+/−smo1-1 (i.e. hemizygous for acbp1 and homozygous for smo1-1) and acbp1SMO1-1+/− were obtained without encountering double homozygotes. The locations of ACBP1 and SMO1-1 on different chromosomes (chromosomes 5 and 4, respectively) ruled out a linkage relationship, implying a synthetic-lethal interaction of the two loci. The double mutant was propagated as ACBP1+/−smo1-1 (since acbp1 was Basta resistant and smo1-1 lacked antibiotic resistance), of which selfed progeny gave a Basta-resistant to Basta-sensitive ratio of 2.19:1, deviating from the 3:1 Mendelian ratio (Supplemental Table S1). As controls, selfed progeny of double hemizygotes and those of ACBP1+/−smo1-1 complemented with 35S:SMO1-1 (cSMO1-1) exhibited a 3:1 ratio (Supplemental Table S1). In mature green siliques of selfed ACBP1+/−smo1-1, high frequencies of aborted and senesced ovules were observed (Fig. 4A). The 25% abortion rate was higher than in the wild type and complemented lines (2.4%–2.8%) and close to the 1:3 Mendelian ratio (Fig. 4B). These results implied fatal arrest postfertilization rather than gametophytic sterility, which would have yielded a 1:1 ratio. In selfed ACBP1+/−smo1-1 at 3 to 4 d after flowering (DAF), some fertilized ovules had shrunken while others were normal (Fig. 4C). Aborted acbp1smo1-1 proembryos displayed distorted-shaped suspensors (Fig. 4D). To further discriminate embryo lethality from gametophytic defects, ACBP1+/−smo1-1 was crossed reciprocally with the wild type. Irrespective of whether ACBP1+/−smo1-1 was a pollen donor or a maternal line, ACBP1smo1-1 and acbp1smo1-1 gametes were carried forward at efficiencies not different (P > 0.05) from the hypothesized 1:1 ratio (Fig. 4E). Taken together, the data indicated that acbp1smo1-1 was synthetic lethal at the proembryo stage.
Figure 3.
Characterization of the smo1-1 mutant. A, Genomic structure of the SMO1-1 gene, indicating the locations of the T-DNA insertion in smo1-1 and the primers for genotyping. Black and white boxes represent coding and untranslated regions of exons, respectively. B, PCR genotyping of smo1-1. The primer combination LBa1/ML2265 was diagnostic for the presence of T-DNA, and ML2251/ML2265 was diagnostic for the wild-type (WT) gene. C, RT-PCR showing knockdown mutation in smo1-1 as measured using 2-week-old seedlings and various organs of 6-week-old plants. ACTIN2 served as a loading control. D, The relative expression levels of SMO1-1 in smo1-1 were compared with those in the wild type (defined value of 1) using 6-week-old plants by qRT-PCR. Transcript levels were normalized against ACTIN2. Each bar represents the mean of three replicates ± sd. L indicates statistically significant (P < 0.05) reduction in comparison with the wild type by Student’s t test.
Figure 4.
Early embryo abortion in self-pollinated ACBP1+/−smo1-1. A, Representative image of an open silique from a self-pollinated F3 plant showing developing seeds and aborted ovules (as indicated by arrowheads). Bar = 500 µm. B, Seed counts using eight siliques per plant. Each bar represents the mean calculated from four plants ± sd. H and L indicate statistically significant (P < 0.01) elevation and reduction, respectively, in comparison with the wild type (WT) by Student’s t test. A χ2 test showed no significant (P > 0.05) deviation from the hypothesized 3:1 segregation ratio. C, Histological examination of the aborted ovules from self-pollinated ACBP1+/−smo1-1. Siliques at 3 to 4 DAF were sectioned, stained, and imaged by light microscopy. Open and closed arrowheads indicate an aborted ovule and a developing seed, respectively. Bars = 20 µm and 5 µm (inset). D, Nomarski image of the aborted ovule. A representative aborted ovule from acbp1smo1-1 showed a distorted appearance of suspensor cells (left), as magnified in the middle and outlined at right. cc, Central cell; pe, early proembryo; su, suspensor. Bars = 10 µm. E, Genetic transmission of acbp1smo1-1 gametophytes after reciprocal crossing with the wild type. a, Statistically insignificant (P > 0.05) deviation from the hypothesized 1:1 segregation ratio.
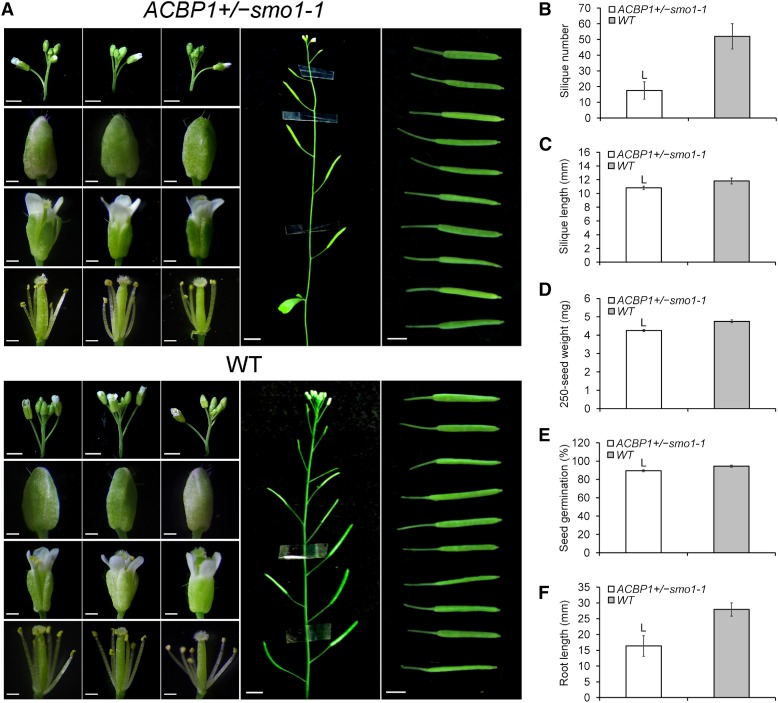
Besides embryogenesis, developmental phenotypes of ACBP1+/−smo1-1 were examined. Macroscopic examination of inflorescences showed fewer floral buds than in the wild type, while the size and morphology of their floral buds and open flowers were normal (Fig. 5A). ACBP1+/−smo1-1 showed a decline in silique number (−66%) and length (−8%) and a lighter seed weight (−10%), accompanied by the reduced germination rate (89.5%), when compared with the wild type (94.5%; Fig. 5, B–E). The lower seed quality also compromised postgerminative growth, as reflected by the shorter (−41%) primary root length of seedlings (Fig. 5F).
Figure 5.
Phenotypic aberrance of ACBP1+/−smo1-1. A, Representative images showing various organs of 6-week-old plants, including inflorescences (bars = 2 mm), floral buds (bars = 0.25 mm), open flowers with/without sepals and petals removed (bars = 0.5 mm), floral stems (bars = 1 cm), and mature green siliques (bars = 0.4 cm). B to F, Comparison of ACBP1+/−smo1-1 with the wild type (WT) in terms of average silique number per plant (B), average silique length (C), average 250-seed weight (D), germination rate using 250 dry after-ripened seeds sown on Murashige and Skoog (MS) plates for 7 d (E), and primary root length of 8-d-old seedlings (F). Each bar represents the mean obtained from seven plants ± sd (except n = 35 in C and n = 48 in F). L indicates statistically significant (P < 0.01) reduction in comparison with the wild type by Student’s t test.
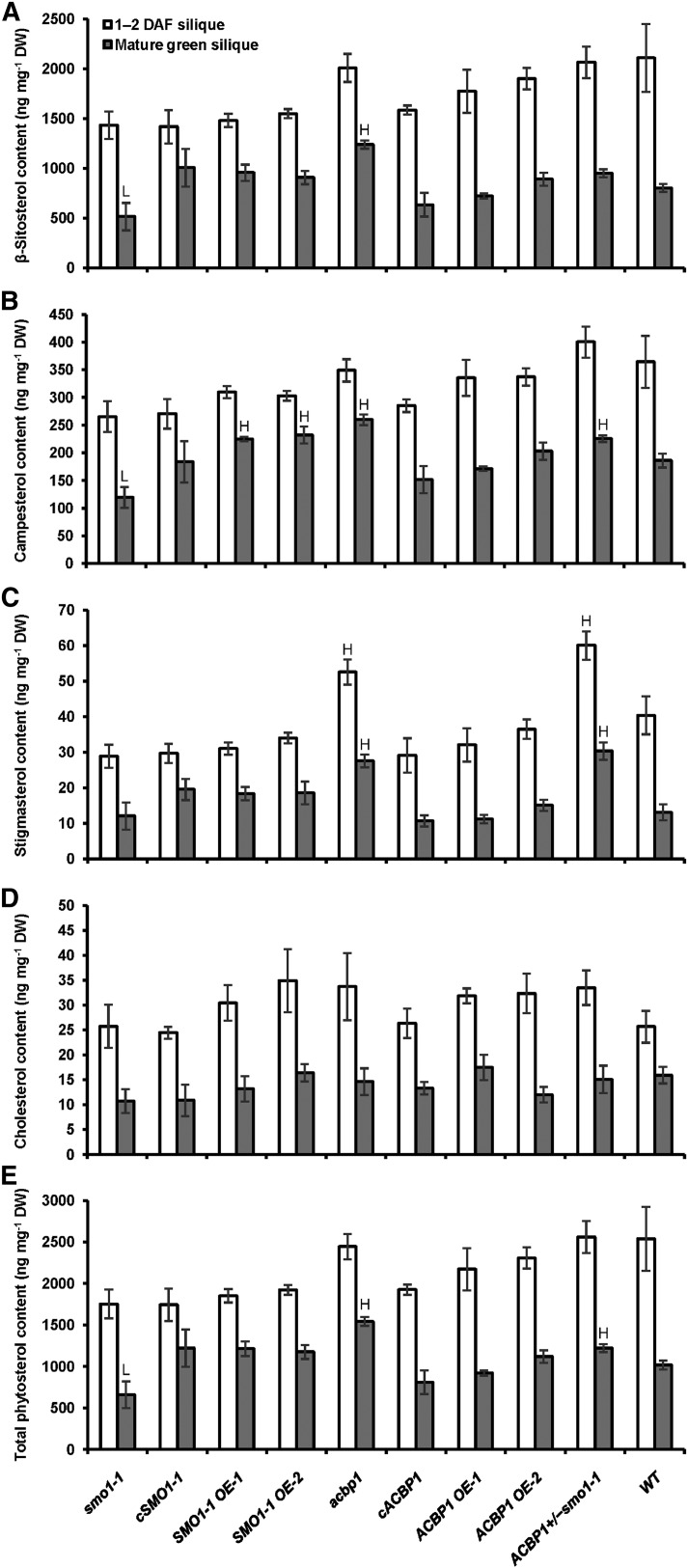
Sterols and FAs Were Altered in Mutant and OE Lines of ACBP1 and SMO1-1
To justify the phenotypic aberrance of ACBP1+/−smo1-1 by a shift in FA-sterol homeostasis, sterols were analyzed in siliques by gas chromatography-mass spectrometry (GC-MS). The total content and composition of major sterols in the mutant, complemented, and OE lines of SMO1-1 and ACBP1 were compared with those of the wild type. In mature green siliques, the level of β-sitosterol, a predominant species in Arabidopsis, was reduced in smo1-1 (−36%) but elevated in acbp1 (+54%), while their opposing effects were negated in ACBP1+/−smo1-1 (Fig. 6A). Campesterol was less abundant in smo1-1 (−36%) but enriched in SMO1-1 OEs (+21% to +25%), acbp1 (+40%), and ACBP1+/−smo1-1 (+21%; Fig. 6B). Stigmasterol was elevated in mature green siliques of acbp1 (+110%) and ACBP1+/−smo1-1 (+140%) as well as in 1- to 2-DAF siliques of acbp1 (+30%) and ACBP1+/−smo1-1 (+48%; Fig. 6C), whereas their cholesterol levels remained unchanged (Fig. 6D). In fact, SMO1-1 does not act on cycloartenol (a cholesterol precursor), but its C24 methylated toward sitosterol, campesterol, and stigmasterol production (Diener et al., 2000). Overall, the total sterol content in mature green siliques was higher in acbp1 (+51%) and ACBP1+/−smo1-1 (+20%) and lower in smo1-1 (−35%; Fig. 6E).
Figure 6.
Major sterol content of mutant, complemented, and OE lines of SMO1-1 and ACBP1. Phytosterols were extracted from young (1–2 DAF) and mature green siliques of 6- to 7-week-old Arabidopsis and analyzed by GC-MS. Each bar represents the mean of four replicates ± sd. H and L indicate statistically significant (P < 0.01) elevation and reduction, respectively, in comparison with the wild type by Student’s t test. A, β-Sitosterol. B, Campesterol. C, Stigmasterol. D, Cholesterol. E, Total phytosterols. DW, Dry weight.
FA methyl esters from mutants and OEs also were compared with the wild type by GC-MS. The total FA level was higher (+24%) in mature green siliques of acbp1, while the 1- to 2-DAF and mature green siliques of smo1-1 (+27% and +11%) and ACBP1+/−smo1-1 (+20% and +9%) also accumulated more FAs (Table I). With respect to acyl composition, no change was observed with FA species derived from the ER-localized eukaryotic pathway, including 18:2-, 18:3-, 20:0-, 20:1-, 20:2-, 20:3-, 22:0-, and 22:1-FAs (Table I). In 1- to 2-DAF siliques, mutation in SMO1-1 and/or ACBP1 increased 16:3-FA at the expense of 18:0-FA, whereas 18:1Δ9-FA was elevated in smo1-1 and ACBP1 OEs (Table I). In mature green siliques, the 35S:ACBP1 transgene enhanced 18:0-FA, while plastidial 16:1- and 16:3-FAs decreased in smo1-1 but increased in SMO1-1 OEs at the expense of 18:0-FA (Table I). Similar changes were not detected in ACBP1+/−smo1-1, possibly due to the negating effect of ACBP1 mutation that boosted 16:1- and 16:3-FAs (Table I). Taken together, the homeostasis of sterols and FAs was affected quantitatively and compositionally upon manipulating SMO1-1 and/or ACBP1 expression.
Table I. Fatty acid content and composition of 1- to 2-DAF and mature green siliques from mutant, complemented, and OE lines of SMO1-1 and ACBP1.
Boldface values indicate statistically significant (P < 0.01) difference from the wild type by Student’s t test: H, higher than wild type; L, lower than wild type. ND, Not detected. Fatty acids were extracted from siliques of 6- to 7-week-old Arabidopsis. Each value represents the mean of four replicates ± sd.
| Genotype | Silique Stage | Total | Relative Composition |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16:0 | 16:1 | 16:3 | 16:0-OH | 18:0 | 18:1Δ9 | 18:1Δ11 | 18:2 | 18:3 | 20:0 | 20:1Δ11 | 20:1Δ13 | 20:2 | 20:3 | 22:0 | 22:1 | |||
| % dry wt | mol % | |||||||||||||||||
| smo1-1 | 1–2 DAF | 5.06 ± 0.45H | 21.12 ± 0.62 | 1.61 ± 0.04 | 4.63 ± 0.09H | 0.39 ± 0.05 | 2.34 ± 0.10L | 2.69 ± 0.09H | 0.30 ± 0.00 | 27.42 ± 0.49 | 32.62 ± 0.46 | 0.95 ± 0.04 | 2.12 ± 0.70 | ND | 2.88 ± 1.25 | ND | 0.92 ± 0.12 | ND |
| Mature green | 7.50 ± 0.25H | 15.84 ± 0.44 | 0.93 ± 0.05L | 2.39 ± 0.04L | 0.25 ± 0.01 | 3.60 ± 0.11 | 9.47 ± 0.49 | 1.96 ± 0.05 | 30.44 ± 1.42 | 21.14 ± 0.60 | 1.61 ± 0.10 | 6.88 ± 0.63 | 1.63 ± 0.17 | 2.25 ± 0.15 | 0.22 ± 0.07 | 0.85 ± 0.09 | 0.54 ± 0.06 | |
| cSMO1-1 | 1–2 DAF | 4.30 ± 0.13 | 21.40 ± 0.15 | 1.41 ± 0.05 | 3.75 ± 0.12 | 0.47 ± 0.06 | 2.57 ± 0.09 | 2.59 ± 0.02 | 0.28 ± 0.02 | 28.46 ± 0.53 | 29.38 ± 0.23 | 0.92 ± 0.06 | 2.54 ± 0.15 | ND | 5.23 ± 0.80 | ND | 0.99 ± 0.05 | ND |
| Mature green | 6.60 ± 0.13 | 16.74 ± 0.31 | 1.17 ± 0.10 | 2.71 ± 0.09H | 0.25 ± 0.03 | 3.47 ± 0.06 | 9.06 ± 0.40 | 1.96 ± 0.15 | 29.87 ± 0.50 | 21.07 ± 0.32 | 1.56 ± 0.05 | 6.80 ± 0.16 | 1.52 ± 0.18 | 2.14 ± 0.57 | 0.27 ± 0.09 | 0.85 ± 0.13 | 0.56 ± 0.10 | |
| SMO1-1 OE-1 | 1–2 DAF | 4.44 ± 0.18 | 21.32 ± 0.28 | 1.48 ± 0.07 | 4.05 ± 0.21 | 0.33 ± 0.02 | 2.48 ± 0.14 | 2.48 ± 0.03 | 0.32 ± 0.02 | 28.79 ± 1.21 | 31.30 ± 1.16 | 0.98 ± 0.03 | 2.15 ± 0.09 | ND | 3.35 ± 0.64 | ND | 0.98 ± 0.07 | ND |
| Mature green | 6.83 ± 0.13 | 16.17 ± 0.08 | 1.30 ± 0.03H | 2.65 ± 0.08H | 0.24 ± 0.03 | 3.36 ± 0.07L | 8.97 ± 0.34 | 1.92 ± 0.07 | 29.85 ± 0.26 | 22.03 ± 0.13 | 1.59 ± 0.05 | 6.75 ± 0.09 | 1.55 ± 0.02 | 1.95 ± 0.11 | 0.23 ± 0.01 | 0.87 ± 0.03 | 0.57 ± 0.04 | |
| SMO1-1 OE-2 | 1–2 DAF | 4.09 ± 0.17 | 23.12 ± 0.81 | 1.56 ± 0.09 | 3.72 ± 0.18 | 0.34 ± 0.03 | 2.58 ± 0.13 | 2.60 ± 0.12 | 0.32 ± 0.03 | 29.73 ± 0.85 | 29.60 ± 0.47 | 1.00 ± 0.02 | 2.35 ± 0.23 | ND | 2.11 ± 1.43 | ND | 0.98 ± 0.04 | ND |
| Mature green | 6.58 ± 0.54 | 16.20 ± 0.52 | 1.29 ± 0.07H | 2.67 ± 0.05H | 0.28 ± 0.03 | 3.14 ± 0.18L | 9.41 ± 0.20 | 2.00 ± 0.02 | 30.68 ± 0.09 | 20.65 ± 0.22 | 1.58 ± 0.04 | 6.75 ± 0.37 | 1.53 ± 0.07 | 2.11 ± 0.15 | 0.30 ± 0.08 | 0.87 ± 0.04 | 0.57 ± 0.04 | |
| acbp1 | 1–2 DAF | 4.50 ± 0.44 | 20.94 ± 0.52 | 1.43 ± 0.17 | 4.43 ± 0.11H | 0.42 ± 0.10 | 2.42 ± 0.13L | 2.39 ± 0.10 | 0.28 ± 0.03 | 27.38 ± 1.66 | 31.72 ± 1.02 | 0.97 ± 0.07 | 2.49 ± 0.51 | ND | 4.18 ± 1.48 | ND | 0.96 ± 0.09 | ND |
| Mature green | 8.37 ± 0.50H | 14.38 ± 0.12L | 1.24 ± 0.06H | 2.78 ± 0.13H | 0.26 ± 0.01 | 3.55 ± 0.08 | 9.88 ± 0.19 | 1.97 ± 0.12 | 30.66 ± 0.32 | 21.08 ± 0.30 | 1.65 ± 0.11 | 7.25 ± 0.77 | 1.54 ± 0.07 | 1.96 ± 0.27 | 0.31 ± 0.05 | 0.93 ± 0.06 | 0.56 ± 0.11 | |
| cACBP1 | 1–2 DAF | 4.06 ± 0.32 | 21.93 ± 0.58 | 1.42 ± 0.04 | 3.66 ± 0.13 | 0.48 ± 0.12 | 2.80 ± 0.13 | 2.67 ± 0.14 | 0.29 ± 0.02 | 27.42 ± 1.02 | 30.53 ± 1.30 | 1.01 ± 0.06 | 2.13 ± 0.63 | ND | 4.68 ± 1.66 | ND | 0.99 ± 0.11 | ND |
| Mature green | 6.90 ± 0.28 | 16.28 ± 0.69 | 1.21 ± 0.08 | 2.55 ± 0.06 | 0.29 ± 0.02 | 3.90 ± 0.08H | 9.49 ± 0.04 | 1.99 ± 0.05 | 30.39 ± 0.30 | 20.86 ± 0.63 | 1.67 ± 0.04 | 6.31 ± 0.23 | 1.41 ± 0.08 | 2.05 ± 0.35 | 0.24 ± 0.03 | 0.88 ± 0.05 | 0.48 ± 0.03 | |
| ACBP1 OE-1 | 1–2 DAF | 4.12 ± 0.12 | 21.81 ± 0.24 | 1.46 ± 0.17 | 3.85 ± 0.03 | 0.36 ± 0.06 | 2.68 ± 0.03 | 2.69 ± 0.05H | 0.29 ± 0.02 | 28.17 ± 0.46 | 32.07 ± 0.54 | 0.96 ± 0.01 | 2.10 ± 0.39 | ND | 2.63 ± 0.90 | ND | 0.93 ± 0.04 | ND |
| Mature green | 6.58 ± 0.08 | 15.78 ± 0.21 | 1.14 ± 0.04 | 2.34 ± 0.08 | 0.25 ± 0.02 | 3.71 ± 0.03H | 9.17 ± 0.10 | 1.94 ± 0.04 | 30.34 ± 0.54 | 21.59 ± 0.50 | 1.64 ± 0.01 | 6.74 ± 0.13 | 1.40 ± 0.08 | 2.23 ± 0.18 | 0.27 ± 0.04 | 0.93 ± 0.04 | 0.52 ± 0.00 | |
| ACBP1 OE-2 | 1–2 DAF | 3.83 ± 0.30 | 22.25 ± 0.13 | 1.58 ± 0.10 | 4.13 ± 0.04 | 0.38 ± 0.02 | 2.74 ± 0.02 | 2.78 ± 0.07H | 0.31 ± 0.02 | 27.21 ± 0.26 | 31.83 ± 0.40 | 1.04 ± 0.04 | 2.21 ± 0.32 | ND | 2.51 ± 0.24 | ND | 1.02 ± 0.02 | ND |
| Mature green | 6.78 ± 0.06 | 16.09 ± 0.11 | 1.19 ± 0.03 | 2.45 ± 0.18 | 0.29 ± 0.05 | 4.17 ± 0.01H | 9.33 ± 0.18 | 1.95 ± 0.06 | 30.08 ± 0.62 | 21.40 ± 0.55 | 1.58 ± 0.04 | 6.37 ± 0.17 | 1.50 ± 0.23 | 2.05 ± 0.12 | 0.20 ± 0.05 | 0.79 ± 0.04 | 0.56 ± 0.03 | |
| ACBP1+/−smo1-1 | 1–2 DAF | 4.78 ± 0.27H | 22.00 ± 0.85 | 1.30 ± 0.13 | 4.45 ± 0.06H | 0.32 ± 0.04 | 2.10 ± 0.12L | 2.35 ± 0.15 | 0.30 ± 0.02 | 27.49 ± 0.62 | 32.03 ± 0.37 | 1.02 ± 0.10 | 2.48 ± 0.25 | ND | 3.15 ± 1.07 | ND | 1.01 ± 0.01 | ND |
| Mature green | 7.34 ± 0.06H | 15.10 ± 0.40L | 1.14 ± 0.15 | 2.46 ± 0.17 | 0.26 ± 0.03 | 3.54 ± 0.04 | 9.45 ± 0.05 | 1.91 ± 0.09 | 30.45 ± 0.27 | 21.87 ± 0.26 | 1.66 ± 0.13 | 6.82 ± 0.85 | 1.57 ± 0.07 | 2.02 ± 0.13 | 0.25 ± 0.06 | 0.89 ± 0.03 | 0.61 ± 0.08 | |
| Wild type | 1–2 DAF | 3.97 ± 0.32 | 21.65 ± 0.36 | 1.44 ± 0.10 | 3.86 ± 0.26 | 0.48 ± 0.13 | 2.80 ± 0.10 | 2.42 ± 0.09 | 0.27 ± 0.02 | 27.94 ± 0.57 | 29.73 ± 1.58 | 1.02 ± 0.10 | 2.45 ± 0.66 | ND | 4.94 ± 1.89 | ND | 0.99 ± 0.04 | ND |
| Mature green | 6.75 ± 0.26 | 16.46 ± 0.38 | 1.08 ± 0.06 | 2.48 ± 0.02 | 0.27 ± 0.02 | 3.52 ± 0.04 | 9.39 ± 0.28 | 1.95 ± 0.04 | 30.18 ± 0.40 | 21.38 ± 0.85 | 1.58 ± 0.04 | 6.48 ± 0.34 | 1.47 ± 0.06 | 2.15 ± 0.44 | 0.23 ± 0.02 | 0.86 ± 0.05 | 0.51 ± 0.05 | |
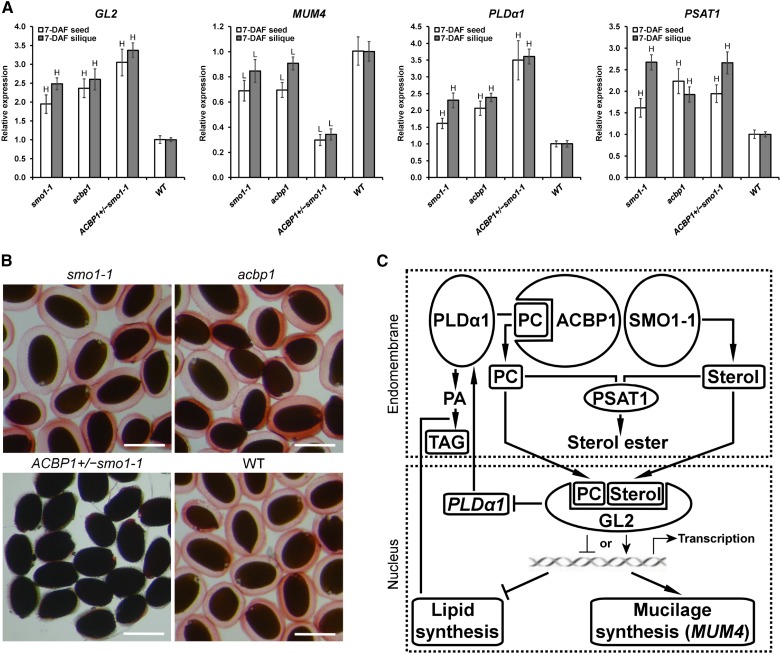
ACBP1 and SMO1-1 Mutation Affected GL2 Function
The higher FA accumulation in mature green siliques of smo1-1, acbp1, and ACBP1+/−smo1-1 (Table I) coincided with the high-oil phenotype of gl2 seeds (Shen et al., 2006; Shi et al., 2012). Hence, it was investigated if the aberrant sterol and FA compositions affected the activity of PL/sterol-binding GL2, a downstream participant in lipid signaling. qRT-PCR revealed a significant (P < 0.01) up-regulation of GL2 in developing seeds and siliques of smo1-1, acbp1, and ACBP1+/−smo1-1 over the wild type (Fig. 7A). The expression of a GL2 target, MUCILAGE MODIFIED4 (MUM4; Shi et al., 2012), was reduced in smo1-1 and acbp1 and more drastically in ACBP1+/−smo1-1 (Fig. 7A). Consistently, ACBP1+/−smo1-1 seeds failed to accumulate mucilage, as indicated by Ruthenium Red staining (Fig. 7B). In agreement with the direct suppression of the PLDα1 promoter by GL2 (Liu et al., 2014), PLDα1 levels were higher in smo1-1, acbp1, and ACBP1+/−smo1-1 (Fig. 7A). In these genotypes, up-regulation of PHOSPHOLIPID:STEROL ACYLTRANSFERASE1 (PSAT1) further indicated a change in PL and sterol metabolism (Fig. 7A).
Figure 7.
Knockdown of SMO1-1 and/or ACBP1 affected GL2 function in seeds. A, Relative expression levels of GL2, MUM4, PLDα1, and PSAT1 in various genotypes against the wild type (WT; defined value of 1) using 6-week-old plants by qRT-PCR. Transcript levels were normalized against ACTIN2. Each bar represents the mean of three (7-DAF silique) or four (7-DAF seed) replicates ± sd. H and L indicate statistically significant (P < 0.01) elevation and reduction, respectively, in comparison with the wild type by Student’s t test. B, Seed coat mucilage staining. Mature seeds were hydrated and stained with Ruthenium Red. Bars = 0.5 mm. C, A mechanistic model illustrating the proposed effect of ACBP1 on GL2 function. At endomembranes, ACBP1 interacts with PLDα1 and SMO1-1 to regulate PC and PA metabolism (Du et al., 2013) and sterol synthesis (this study), respectively. The homeostatic levels of PLs and sterols are maintained by converting excessive molecules into sterol esters under the catalysis of PSAT1. In the nucleus, GL2 binds PC and sterols (Ponting and Aravind, 1999; Schrick et al., 2014) that are crucial for its transcriptional regulatory activities, including the activation of MUM4 expression for mucilage synthesis (Shi et al., 2012) and the inhibition of PLDα1 expression (Liu et al., 2014) and lipid synthesis (Shen et al., 2006). This signaling pathway could be affected upon ACBP1 and/or SMO1-1 knockdown, as reflected by changes in mRNA expression of the proteins involved and levels of lipid metabolites.
DISCUSSION
ACBP1 Cooperates with SMO1-1 during Early Embryogenesis
Previously, knockouts of ACBP1 and its homolog ACBP2 arrested embryo development, possibly due to an aberrant PL composition (Chen et al., 2010). In this study, a discrete role of ACBP1 in modulating sterol synthesis via PPI with SMO1-1 is reported. The importance of their interaction is reflected by the severe proembryo abortion phenotype of acbp1smo1-1 (Fig. 4). acbp1smo1-1 differed from all other early sterol biosynthetic mutants with embryonic abnormalities detected at or beyond the globular stage, and their lethality (if any) occurred postembryonically (Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000, 2002; Souter et al., 2002; Willemsen et al., 2003; Kim et al., 2005; Men et al., 2008). Coincidentally, smo2-1smo2-2 is the only embryo-lethal genotype among all mutants in the entire phytosterol pathway characterized thus far (Zhang et al., 2016), although SMO1 and SMO2 are catalytically independent (Darnet and Rahier, 2004). Unlike the arrest of acbp1smo1-1 at the preglobular stage (Fig. 4D), smo2-1smo2-2 embryos were aborted at globular to heart-like stages (Zhang et al., 2016). Consistent with specific SMO1-1pro:GUS signals at the micropylar end in the egg apparatus (Fig. 2, E and F) and basal cells (Supplemental Fig. S2A), acbp1smo1-1 proembryos were aborted with collapsed suspensors (Fig. 4D), likely from loss in cell turgidity (Almagro et al., 2008). As sterols regulate membrane fluidity and osmotic permeability (Schaller, 2004), the distortion of suspensor cells could arise from malfunctioning membranes with nonphysiological sterol content, reminiscent of the increased pathogen susceptibility upon knockout of STEROL METHYLTRANSFERASE2 (SMT2; Wang et al., 2012). Suspensor cell vacuolation could be affected by abnormal sterol-lipid distribution, analogous to the vacuolar fragmentation in yeast lacking oxysterol-binding proteins (Beh and Rine, 2004). Besides, the sterol-mediated formation of microdomains to which sterol-dependent proteins bind may be affected by an imbalance in sterol homeostasis (Hartmann, 1998). Recently, the smt1 proteome revealed changes in the microdomain association of many transporters and enzymes, including those in FA and acyl-lipid synthesis, sterol glycosylation, and vesicle trafficking (Zauber et al., 2014). Similar changes may account for the severe acbp1smo1-1 phenotype (Fig. 4).
Furthermore, in several sterol biosynthetic mutants, cell signaling is compromised by altered membrane sterol composition, which affects endocytosis and polar auxin transport (Men et al., 2008; Zhang et al., 2016). Phospholipases as lipid signal generators rely on microdomain association (Gardiner and Marc, 2013). For instance, phospholipase C was less active in smt1 when it became less abundant in the sterol-associated membrane fraction (Zauber et al., 2014). ACBP1 cooperates with PLDα1 and PLDδ in the freezing stress response (Du et al., 2010) and partners with PLDα1 in abscisic acid signaling (Du et al., 2013). Considering its interactions with PLDα1 (Du et al., 2013) and SMO1-1 (Fig. 1), which generate PL (Wang, 2001) and sterol (Mialoundama et al., 2013) signals, respectively, ACBP1 is positioned to modulate cross talk between the two lipogenic signaling pathways. The magnitude of PL or sterol change may not be drastic enough to affect embryogenesis in single mutants. In acbp1smo1-1, the cumulative variation of both lipidic signals may result in proembryo lethality when the dual signaling pathways converge on shared downstream targets, as represented by HD-START TFs (Schrick et al., 2004).
ACBP1 Links FA Homeostasis to Sterol Production
To provide insight into the biochemical significance of the ACBP1-SMO1-1 interaction, sterols (Fig. 6) and FAs (Table I) were analyzed in mutant, complemented, and OE lines. Resembling other sterol biosynthetic mutants (Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000, 2002; Kim et al., 2005; Men et al., 2008; Carland et al., 2010; Qian et al., 2013; Zhang et al., 2016) and SMO1-silenced tomato (Solanum lycopersicum) and Nicotiana benthamiana (Sonawane et al., 2016), smo1-1 accumulated abnormal levels of end-product sterols (Fig. 6). The altered sterol profile of acbp1 suggested a role for ACBP1 in sterol metabolism, and changes in ACBP1+/−smo1-1 further revealed semidominant effects of the ACBP1 mutation (Fig. 6). Differences in the sterol composition of ACBP1+/−smo1-1 in comparison with acbp1 and smo1-1 (Fig. 6) explained its phenotypic aberrance in plant development not seen in single mutants (Fig. 5). For instance, the poor silique development of ACBP1+/−smo1-1 (Fig. 5) was consistent with its shifted ratio of campesterol to sitosterol (Fig. 6), a crucial factor for Arabidopsis growth (Schaeffer et al., 2001). Among all genotypes tested, ACBP1+/−smo1-1 siliques also contained the highest level of stigmasterol (Fig. 6), a cell proliferation modulator (Hartmann, 1998). In mature green siliques, down-regulation of SMO1-1 reduced the total sterol content, whereas ACBP1 mutation produced an opposing effect and complemented the change in smo1-1, demonstrating that ACBP1 inhibits SMO1-1 function (Fig. 6). The inhibitory effect of ACBP1 likely occurs in its liganded form, because site-directed mutagenesis at its ACB domain weakened the interaction with SMO1-1 (Fig. 1A). Accordingly, the ligand-binding status of ACBP1 may be a cue that reflects the cellular levels of FA esters and determines the sterol synthetic rate, indicating a link between FA homeostasis and sterol production in plants. This PPI-mediated regulation contrasts with mammalian systems, in which ACBPs regulate cholesterol homeostasis at the transcriptional level in conjunction with sterol regulatory element-binding protein1 (Neess et al., 2006; Oikari et al., 2008), which modulates StAR gene expression (Yokoyama et al., 1993).
ACBPs are analogous to some ATP-binding cassette (ABC) transporters that mobilize acyl-CoA esters and PLs (Du et al., 2016). Recently, ABCG9, ABCG11, and AGCG14 were postulated to mediate the long-distance transport of sterols or sterol conjugates in the phloem sap (Le Hir et al., 2013), besides their roles in cuticle formation (Hwang et al., 2016). Given the functional resemblance of ACBP1 in stem cuticle synthesis and its high expression in the vasculature (Du et al., 2013; Xue et al., 2014), it is pertinent to study whether these plasma membrane-localized proteins cooperate in sterol transport. Similar to other sterol biosynthetic mutants (Zauber et al., 2014), abcg9, abcg11, and abcg14 exhibited FA compositional changes (Le Hir et al., 2013), supporting an interrelationship of sterol and FA metabolism. In this study, FA profiles (particularly 16:0- and 18:0-FAs) were significantly altered in mutants and OEs of SMO1-1 and ACBP1 (Table I). As 16:0- and 18:0-FAs were the two major species elevated in Arabidopsis OEs of acyl-CoA:sterol acyltransferase with substrate specificity for 16:0- and 18:0-CoA esters (Chen et al., 2007), our results suggest a regulatory role for ACBP1 in sterol acylation via its association with these acyl-CoA species. Deviations of 16:1- and 16:3-FA levels in mutants and OEs of SMO1-1 and ACBP1 also revealed a compositional change in plastidial lipids (Table I). This observation is consistent with a previous analysis of acbp1 siliques indicating acyl compositional variations in galactolipids and phosphatidylglycerol, which are crucial thylakoid membrane components for embryo and cotyledon development (Chen et al., 2010). Although ACBP1 does not function in plastids, the interorganellar transport of lipids is possible via ER-plastid contact sites (Hurlock et al., 2014). By analogy, the ER-localized ABCA9 facilitates the plastid-ER transport of FAs (Kim et al., 2013). A functional ER-plastid relationship also is evident in the contribution of plastid-derived isopentenyl diphosphate via the methylerythritol phosphate pathway to subsequent sterol biosynthesis at the ER (Laule et al., 2003).
The Potential Role of the ACBP1-SMO1-1 Interaction in Lipid Signaling
Among several transcriptional regulators of seed oil synthesis, GL2 is the only known TF that binds lipids and sterols via its START domain (Schrick et al., 2014). Similar to the boosted FA levels (9%–24%) in mature green siliques of acbp1, smo1-1, and ACBP1+/−smo1-1 (Table I), Arabidopsis gl2 seeds accumulated 8% more oil than the wild type (Shen et al., 2006; Shi et al., 2012). Resembling gl2 (Shi et al., 2012), a loss in seed coat mucilage was observed in ACBP1+/−smo1-1 but not in acbp1 and smo1-1 single mutants (Fig. 7B), indicating a semidominant mutation of ACBP1 when SMO1-1 was knocked down simultaneously. Possibly, a cumulative change in ACBP1 and SMO1-1 protein levels was sufficient to affect lipid signaling, in agreement with the ectopic root hair formation in hemizygous gl2 mutants in a semidominant fashion (Masucci et al., 1996). Considering the concomitant transcriptional regulation of GL2 and its downstream targets (Fig. 7A), the high-oil, low-mucilage phenotype of ACBP1+/−smo1-1 may be attributed to a reduction in GL2 activity when concentrations of its ligands (PL and sterols) are suboptimal. The up-regulation of GL2 at the mRNA level may be a compensatory response to its loss of transactivating activity at the protein level, given that GL2 expression is regulated transcriptionally via a positive feedback loop involving other TFs (Khosla et al., 2014). In our proposed model (Fig. 7C), ACBP1 may sense the metabolic status of a cell by its ability to bind acyl-CoA esters and PLs (Chye, 1998; Chen et al., 2010; Du et al., 2010, 2013). It may then modulate the synthesis of PC and sterol signals via PPI with PLDα1 (Du et al., 2013) and SMO1-1 (Fig. 1), respectively.
The fate of PC and sterol signals also may be influenced by PSAT1 activity, given its transcriptional regulation in acbp1 and smo1-1 mutants (Fig. 7A). By altering the cellular levels of these lipid signals, ACBP1 may modulate the transcriptional regulatory activity of downstream targets, including GL2. Schrick et al. (2014) reported that deletion of the START domain or site-directed mutagenesis of its lipid-binding residues abolished GL2 activity. By analogy, varying its ligand concentrations may result in similar effects, as shown here. In fact, the involvement of GL2 in PL- and steroid-mediated signaling has been proposed previously (Ohashi et al., 2003; Kuppusamy et al., 2009). Besides GL2, other HD-START TFs are promising subjects for future studies. The proembryo abortion of acbp1smo1-1 (Fig. 4) may be linked to some HD-START TFs. For instance, ARABIDOPSIS THALIANA MERISTEM LAYER1 and PROTODERMAL FACTOR2 have been ascribed a role in embryogenesis, as double mutants were arrested at the globular stage (Ogawa et al., 2015). Triple mutants lacking CORONA, PHABULOSA, and PHAVOLUTA also displayed ovule abnormalities (Kelley et al., 2009). HOMEODOMAIN GLABROUS9 is another candidate, given its specific expression in the embryo sacs (Nakamura et al., 2006), coinciding with the SMO1-1pro:GUS expression pattern (Fig. 2, E and F). Sterol intermediates and derivatives per se could act as signals for embryonic and postembryonic development (Carland et al., 2010; Qian et al., 2013; Zhang et al., 2016), including the catalytic product of SMO1 (Mialoundama et al., 2013). It remains to be explored whether the ACBP1-SMO1-1 interaction regulates the generation of an as yet unidentified signal that binds to any HD-START protein(s) during embryogenesis.
Potential Applications for Plant Oil Modification
In the interest of promoting phytosterol intake for lowering blood cholesterol (Moreau et al., 2002) and cancer therapies (Woyengo et al., 2009), higher phytosterol content in seeds is a nutraceutically valuable trait in plant seed oil production. In this study, the phytosterol increase in mature green siliques of acbp1 and ACBP1+/−smo1-1 (Fig. 6) opens up a new strategy for boosting phytosterol production in seed crops. It is plausible to identify and deplete a SMO1-interacting equivalent of ACBP1 in crop species to eliminate its inhibitory effect on sterol biosynthesis, as is the case in Arabidopsis (Fig. 6). It may present a promising approach to push the flux toward end-product sterol production, because SMO1 catalyzes one of the rate-limiting steps in sterol biosynthesis (Lange et al., 2015). If the carbon partitioning of photoassimilates in developing seeds shifts toward a higher accumulation of lipids (sterols and FAs), as observed in acbp1 and ACBP1+/−smo1-1 (Table I; Fig. 6), the genetically modified crops will exhibit a high-oil phenotype of great agronomic significance. Nonetheless, considering the versatile roles of ACBP1 in other aspects of plant development and stress responses, the drawbacks of ACBP1 depletion in Arabidopsis, such as defective stem cuticle formation (Xue et al., 2014) and higher sensitivity to heavy metal stress (Xiao et al., 2008), should be taken into consideration. The negative impacts of ACBP1/SMO1-1 cosilencing on reproductive development also present some concerns (Fig. 5). Whether these issues can be solved by seed-specific silencing awaits further investigations.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used in this study. Seeds were stratified at 4°C for 2 d and germinated on MS plates with 2% (w/v) Suc (unless specified otherwise) at 22°C under a 16-h-light/8-h-dark cycle. Potted plants were grown with a day/night regime of 16/8 h and 23°C/21°C. The acbp1 (Basta-resistant) mutant, cACBP1, and ACBP1 OEs have been described previously (Xiao et al., 2008). The smo1-1 mutant (SALK_021394) was selected from the SALK collection of TAIR, and its T-DNA was identified by PCR using the left border primer LBa1 and the gene-specific primer ML2265 (Fig. 3A). By sequencing the PCR product, the T-DNA insert was mapped to the 5′ untranslated region at 102 bp upstream of the translation start site (Fig. 3A). The homozygous mutant was confirmed by PCR using the primer pair ML2251/ML2265 (Fig. 3A). All primers used in this study are listed in Supplemental Table S2. Transgenic lines were generated by Agrobacterium tumefaciens-mediated transformation of wild-type Arabidopsis by the floral dip method (Clough and Bent, 1998).
Generation of OE and Complemented Lines
The full-length SMO1-1 ORF was PCR amplified using the primer pair ML2804/ML2805. The 900-bp product was blunt-end ligated into the SmaI sites on pXCS-HAStrep (Witte et al., 2004) to generate plasmid pAT830 for Arabidopsis transformation. The SMO1-1 OE plants were screened on MS plates containing 20 mg L−1 Basta and PCR genotyped using the primer pair 35SB/ML2805. The 35S promoter-driven expression of HA/StrepII-tagged protein was confirmed by western-blot analysis in homozygous T3 plants, which were used for subsequent studies. Similarly, plasmid pAT830 was introduced into smo1-1 to generate cSMO1-1 complemented lines.
Genetic Crosses
Homozygous smo1-1 and acbp1 were genetically crossed. With no double homozygote identified after genotyping more than 100 F2 progeny, the double mutant was maintained as ACBP1+/−smo1-1. From the F3 progeny of these plants, the absence of the acbp1 homozygote was further confirmed by PCR and Basta selection. To complement the embryo lethality of acbp1smo1-1 by overexpression of ACBP1 or SMO1-1 ORFs, ACBP1+/−smo1-1 was crossed with cACBP1 or cSMO1-1, respectively. Homozygous lines were identified from F2 progeny by genotyping. The seed abortion rate of each line was scored in the F3 population in comparison with the wild type using a dissecting microscope.
Generation of SMO1-1:EGFP and DsRed:ACBP1 Lines
The full-length SMO1-1 ORF was PCR amplified using the primer pair ML2237/ML2238. The 900-bp product was cloned in frame into the BamHI site on pBI121-EGFP (Shi et al., 2005) to generate plasmid pAT735 for Arabidopsis transformation. Homozygous T3 plants were identified by kanamycin selection and PCR genotyping. The DsRed:ACBP2 fragment was PCR amplified from plasmid pAT226 (Li and Chye, 2004) using the primer pair ML2433/ML2434. The 1.9-kb product was digested with SalI and HindIII and cloned into XhoI and HindIII sites on pXCS-HAStrep (Witte et al., 2004) to generate plasmid pAT763. The full-length ACBP1 ORF was PCR amplified using the primer pair ML2435/ML2436 and blunt-end ligated into SmaI sites on plasmid pAT763 (replacing ACBP2) to generate plasmid pAT764, which was introduced into SMO1-1:EGFP lines. After confirmation by Basta selection and PCR genotyping, homozygous T3 plants were used for confocal laser scanning microscopy.
Confocal Laser Scanning Microscopy
For transient expression of EGFP and DsRed fusion proteins, plasmids pAT735 and pAT764 were introduced into A. tumefaciens strain GV3101, which was used for agroinfiltration of 4-week-old tobacco (Nicotiana tabacum var SR1) according to Yang et al. (2000). Agroinfiltrated tobacco leaves and transgenic Arabidopsis were examined with a Zeiss LSM 710 NLO confocal laser scanning microscope following Li and Chye (2004). For colocalization with an ER marker, 1-week-old seedlings were vacuum infiltrated with the ER-Tracker Red dye (1 µm; Invitrogen catalog no. E34250) for 30 min prior to two 5-min washes in distilled, deionized water. The ER-Tracker signals were excited at 594 nm, and emission was detected at 615 nm. Representative images are shown after observation with consistent results from at least 20 cells.
Y2H Analysis
Y2H analysis was performed using the Matchmaker Gold Y2H System (Clontech) according to the manufacturer’s instructions. The bait vector pGBKT7 containing the coding region of ACBP141–338 (plasmid pAT248) constructed previously (Du et al., 2013) was used. The coding regions of ACBP132–216 and ACBP1217–338 were PCR amplified using primer pairs ML1791/ML1792 and ML2208/ML2209, respectively. The products were cloned into pGEM-T Easy Vector (Promega) to generate plasmids pAT660 and pAT710, respectively, from which EcoRI-BamHI fragments were excised and cloned into similar sites on pGBKT7 to generate plasmids pAT661 and pAT713, respectively. A 1.1-kb EcoRI-EcoRI fragment containing the Y171A-mutated coding region of ACBP141–338 was excised from plasmid pAT259 (Leung et al., 2006) and cloned into the EcoRI site on pGBKT7 to generate plasmid pAT760. The full-length SMO1-1 ORF was PCR amplified using the primer pair ML1794/ML1795. The 900-bp product was cloned into pGEM-T Easy Vector (Promega) to generate plasmid pAT656, from which an EcoRI-XhoI fragment was excised and cloned into similar sites on the prey vector pGADT7 to generate plasmid pAT658. The bait and prey constructs were introduced into Y2HGold cells (Clontech) according to the manufacturer’s instructions, using cotransformation of the supplied pGADT7-T with the supplied pGBKT7-52 and pGBKT7-Lam as positive and negative controls, respectively. Successful cotransformants were identified from synthetic dropout medium/−Leu/−Trp DDO plates after a 3-d incubation at 30°C and resuspended in 100 µL of sterile 0.9% (w/v) NaCl. Five-microliter cell suspensions, diluted to OD600 = 0.1, were spotted on DDO and triple dropout (synthetic dropout medium/−His/−Leu/−Trp containing 40 µg mL–1 X-α-Gal and 125 ng mL–1 aureobasidin A) plates, which were incubated at 30°C for 3 d before photography.
Subcellular Fractionation
Silique-bearing aboveground tissues of 6-week-old Arabidopsis were subjected to subcellular fractionation according to Smith et al. (1988). Proteins were resolved by 12% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Pall) for western-blot analysis. The blots were cross-reacted with rabbit polyclonal anti-ACBP1 (raised against recombinant ACBP1 protein; 1:4,000) or anti-HA (1:5,000; Immunoway) antibodies at 4°C overnight and horseradish peroxidase-conjugated anti-rabbit secondary antibodies (1:20,000; Sigma-Aldrich) at room temperature for 2 h. Signals were developed using the Amersham ECL Prime Detection Reagent (GE Healthcare) followed by autoradiography.
Strep-Tactin Pull-Down and Coimmunoprecipitation Assays
Membrane fractions were isolated from 0.5 g fresh weight of silique-bearing plants according to Smith et al. (1988). All subsequent steps were performed at 4°C. Each membrane pellet was solubilized in 1 mL of binding buffer containing 100 mm Tris-HCl (pH 8), 150 mm NaCl, 1 mm EDTA, 0.5% (v/v) Triton X-100, 1% (v/v) protease inhibitor cocktail (Sigma-Aldrich catalog no. P9599), and 1 mm phenylmethylsulfonyl fluoride. The samples were clarified by centrifugation at 20,000g for 30 min. For Strep-Tactin pull-down assays, the solubilized membrane proteins were loaded onto the Strep-Tactin MacroPrep matrix (IBA catalog no. 2-1505-002). The target proteins were bound, washed, and eluted according to the manufacturer’s instructions prior to western-blot analysis using anti-ACBP1 and anti-HA antibodies as described above. For coimmunoprecipitation assays, anti-ACBP1 antibodies were covalently coupled to Affi-Gel 10 beads (Bio-Rad catalog no. 153-6046) according to the manufacturer’s instructions. The 100-µL beads were incubated with the solubilized membrane proteins for 30 min on a rotary shaker and washed with 1 mL of binding buffer five times. The proteins were eluted with SDS sample buffer and subjected to western-blot analysis using anti-ACBP1 and anti-HA antibodies as described above.
RT-PCR and qRT-PCR
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen) and reverse transcribed using the SuperScript VILO cDNA Synthesis Kit (Invitrogen) primed with oligo(dT) according to the manufacturer’s instructions. For RT-PCR, SMO1-1 was amplified using the primer pair ML2184/ML1799 under the following conditions: 95°C for 2 min, then 32 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, with a final step of 72°C for 7 min. ACTIN2 was amplified using the primer pair ML1124/ML1125 with the following conditions: 95°C for 2 min, then 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 10 s, with a final step of 72°C for 7 min. The cycle numbers were within the predetermined linear amplification range. qRT-PCR was conducted using the FastStart Universal SYBR Green Master (Roche) with the StepOnePlus Real-Time PCR System (Applied Biosystems) with the following conditions: 95°C for 10 min, then 40 cycles of 95°C for 15 s and 60°C for 30 s. Primer pairs for qRT-PCR were ML1124/ML1125 (ACTIN2), ML2252/ML2253 (SMO1-1), ML2918/ML2919 (PSAT1), ML2922/ML2923 (PLDα1), ML2926/ML2927 (GL2), and ML2934/ML2935 (MUM4).
GUS Staining
The 3-kb 5′ flanking region of SMO1-1 was PCR amplified using the primer pair ML2316/ML2317 and cloned into pGEM-T Easy Vector (Promega) to generate plasmid pAT750, from which a BamHI-BamHI fragment was excised and cloned into the BamHI site on pBI101.3 (Clontech) to generate the SMO1-1pro:GUS construct pAT799 for Arabidopsis transformation. After confirmation by kanamycin selection and PCR genotyping, homozygous T3 plants were used. Arabidopsis ACBP1pro:GUS plants were generated by Du et al. (2013). GUS staining was performed according to Kim et al. (2006). Specimens were fixed in 90% (v/v) acetone for 20 min and vacuum infiltrated in a substrate solution containing 50 mm sodium phosphate (pH 7), 0.2% (v/v) Triton X-100, 10 mm EDTA, 2 mm potassium ferricyanide, 2 mm potassium ferrocyanide, and 1 mg mL–1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide for 30 min. The specimens were incubated at 37°C for 6 to 12 h until the development of blue signals and cleared in 70% (v/v) ethanol for photography. Floral stages were numbered according to Smyth et al. (1990). The GUS-stained ovules and developing seeds were further cleared in Herr’s solution as described previously (Chen et al., 2010) prior to differential interference contrast (DIC) microscopy.
Seed Coat Mucilage Staining
Dry seeds were shaken in water for 1 h and stained with an aqueous solution of 0.01% (w/v) Ruthenium Red according to Western et al. (2001). The stained seeds were washed briefly in water and observed with a dissecting microscope.
Light Microscopy
Siliques were vacuum infiltrated in a fixative solution containing 50 mm sodium phosphate (pH 7), 2.5% (v/v) glutaraldehyde, and 1.6% (v/v) paraformaldehyde at room temperature for 1 h and further fixed at 4°C overnight. After dehydration in an ethanol series, the specimens were cleared in Herr’s solution as described previously (Chen et al., 2010). For DIC microscopy, whole-mount tissues were mounted in Hoyer’s medium (Stangeland and Salehian, 2002) and observed with a Nikon Eclipse 80i microscope using DIC optics. For histochemical examination, the specimens were embedded and sectioned in Technovit 7100 (Electron Microscopy Sciences). Sections were stained with the periodic acid-Schiff’s reaction for total insoluble carbohydrates and Amido Black 10B for protein and then imaged with a Leitz photomicroscope using a Nikon DS-Fi2 digital camera according to Hsiao et al. (2015).
Postgerminative Root Growth Studies
Seeds were germinated for 3 d on MS plates with 1% (w/v) Suc. Germinated seeds (12 per plate; n = 4) were transferred to fresh identical plates and grown for 5 d prior to primary root length measurement. Given that the progeny from ACBP1+/−smo1-1 could be segregated into ACBP1+/−smo1-1 and smo1-1 (null) genotypes, the presence of the acbp1 T-DNA in each seedling was determined by PCR using the primer pair ML179/SLB1 following Chen et al. (2010).
GC-MS Analysis of Sterols and FA Methyl Esters
Sterols were extracted according to standard procedures (Liao et al., 2014; Henry et al., 2015) with some modifications. Briefly, 10 mg of freeze-dried siliques at 1 to 2 DAF and 15 mg of freeze-dried mature green siliques were incubated in 4 mL of 2:1 (v/v) chloroform:methanol at 70°C for 1 h, dried under N2 gas, and saponified in 2 mL of 6% (w/v) KOH in methanol at 80°C for 1 h. The samples were cooled down and mixed with 1 mL of distilled, deionized water and then 1 mL of hexane, after which the solvent phase was collected by brief centrifugation. After three rounds of extraction, the solvent phases were combined and dried under N2 gas, followed by derivatization in 100 µL of N,O-bis(trimethylsilyl)trifluoroacetamide at 80°C for 30 min. The extracts were spiked with 40 µg of lupenyl-3,28-diacetate as an internal standard. Sterol masses were determined using an Agilent GC-MS device (5973 inert mass spectrometer combined with 6890N gas chromatograph) equipped with an HP-5MS capillary column as described previously (Liao et al., 2014). For FA quantification and profiling, 4 mg of freeze-dried siliques were subjected to acid-catalyzed transmethylation according to Li-Beisson et al. (2013). The extracts were spiked with 5 µg of C19:0-FA as an internal standard and analyzed by the same GC-MS device equipped with an Agilent J&W DB-WAX capillary column (30 m × 0.25 mm × 0.25 µm) using a 1:40 split-mode injection. The temperature was set at 150°C for 3 min, then increased to 240°C at 10°C min−1, and held at 240°C for 5 min.
Accession Numbers
The sequence data used in this study can be retrieved from TAIR: AT4G12110 (SMO1-1), AT5G53470 (ACBP1), AT1G79840 (GL2), AT1G53500 (MUM4), AT3G15730 (PLDα1), AT1G04010 (PSAT1), and AT3G18780 (ACTIN2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Computational analysis for subcellular localization of SMO1-1 and its expression patterns during seed development.
Supplemental Figure S2. Spatial expression patterns of SMO1-1pro:GUS and ACBP1pro:GUS in transgenic Arabidopsis during seed development.
Supplemental Table S1. Basta-resistant to Basta-sensitive ratio of progeny from self-pollinated F3 plants.
Supplemental Table S2. Sequences of the primers used in this study.
Acknowledgments
We thank Dr. Claus-Peter Witte (Freie Universität Berlin) for providing the pXCS-HAStrep vector.
Glossary
- TF
transcription factor
- PL
phospholipid
- FA
fatty acid
- ER
endoplasmic reticulum
- PC
phosphatidylcholine
- PA
phosphatidic acid
- PPI
protein-protein interaction
- Y2H
yeast two-hybrid
- ORF
open reading frame
- ANK
ankyrin repeat
- ACB
acyl-CoA-binding
- OEs
overexpressors
- qRT
quantitative real-time
- RT
reverse transcription
- DAF
days after flowering
- GC-MS
gas chromatography-mass spectrometry
- OE
overexpression
- MS
Murashige and Skoog
- DDO
double dropout
- DIC
differential interference contrast
Footnotes
This work was supported by the Wilson and Amelia Wong Endowment Fund (to M.-L.C.) and the Research Grants Council of the Hong Kong Special Administrative Region, China (grant no. HKU765813M to M.-L.C.), by a postdoctoral fellowship and Committee on Research and Conference Grants award from the University of Hong Kong (Small Project Funding grant no. 201309176052 to S.-C.L.), and by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (to E.C.Y.).
Articles can be viewed without a subscription.
References
- Almagro A, Lin SH, Tsay YF (2008) Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell 20: 3289–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach TJ. (1995) Some new aspects of isoprenoid biosynthesis in plants: a review. Lipids 30: 191–202 [DOI] [PubMed] [Google Scholar]
- Beh CT, Rine J (2004) A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J Cell Sci 117: 2983–2996 [DOI] [PubMed] [Google Scholar]
- Benveniste P. (2004) Biosynthesis and accumulation of sterols. Annu Rev Plant Biol 55: 429–457 [DOI] [PubMed] [Google Scholar]
- Block MA, Dorne AJ, Joyard J, Douce R (1983) The acyl-CoA synthetase and acyl-CoA thioesterase are located on the outer and inner membrane of the chloroplast envelope, respectively. FEBS Lett 153: 377–381 [Google Scholar]
- Bouvier-Navé P, Berna A, Noiriel A, Compagnon V, Carlsson AS, Banas A, Stymne S, Schaller H (2010) Involvement of the phospholipid sterol acyltransferase1 in plant sterol homeostasis and leaf senescence. Plant Physiol 152: 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland F, Fujioka S, Nelson T (2010) The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol 153: 741–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Steinhauer L, Hammerlindl J, Keller W, Zou J (2007) Biosynthesis of phytosterol esters: identification of a sterol O-acyltransferase in Arabidopsis. Plant Physiol 145: 974–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QF, Xiao S, Qi W, Mishra G, Ma J, Wang M, Chye ML (2010) The Arabidopsis acbp1acbp2 double mutant lacking acyl-CoA-binding proteins ACBP1 and ACBP2 is embryo lethal. New Phytol 186: 843–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chye ML. (1998) Arabidopsis cDNA encoding a membrane-associated protein with an acyl-CoA binding domain. Plant Mol Biol 38: 827–838 [DOI] [PubMed] [Google Scholar]
- Chye ML, Huang BQ, Zee SY (1999) Isolation of a gene encoding Arabidopsis membrane-associated acyl-CoA binding protein and immunolocalization of its gene product. Plant J 18: 205–214 [DOI] [PubMed] [Google Scholar]
- Chye ML, Li HY, Yung MH (2000) Single amino acid substitutions at the acyl-CoA-binding domain interrupt 14[C]palmitoyl-CoA binding of ACBP2, an Arabidopsis acyl-CoA-binding protein with ankyrin repeats. Plant Mol Biol 44: 711–721 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Darnet S, Rahier A (2004) Plant sterol biosynthesis: identification of two distinct families of sterol 4α-methyl oxidases. Biochem J 378: 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W, Whoriskey WJ, Nes WD, Fink GR (2000) STEROL METHYLTRANSFERASE 1 controls the level of cholesterol in plants. Plant Cell 12: 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZY, Arias T, Meng W, Chye ML (2016) Plant acyl-CoA-binding proteins: an emerging family involved in plant development and stress responses. Prog Lipid Res 63: 165–181 [DOI] [PubMed] [Google Scholar]
- Du ZY, Chen MX, Chen QF, Xiao S, Chye ML (2013) Arabidopsis acyl-CoA-binding protein ACBP1 participates in the regulation of seed germination and seedling development. Plant J 74: 294–309 [DOI] [PubMed] [Google Scholar]
- Du ZY, Xiao S, Chen QF, Chye ML (2010) Depletion of the membrane-associated acyl-coenzyme A-binding protein ACBP1 enhances the ability of cold acclimation in Arabidopsis. Plant Physiol 152: 1585–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner J, Marc J (2013) Phospholipases may play multiple roles in anisotropic plant cell growth. Protoplasma 250: 391–395 [DOI] [PubMed] [Google Scholar]
- Hartmann MA. (1998) Plant sterols and the membrane environment. Trends Plant Sci 3: 170–175 [Google Scholar]
- Henry LK, Gutensohn M, Thomas ST, Noel JP, Dudareva N (2015) Orthologs of the archaeal isopentenyl phosphate kinase regulate terpenoid production in plants. Proc Natl Acad Sci USA 112: 10050–10055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao AS, Yeung EC, Ye ZW, Chye ML (2015) The Arabidopsis cytosolic acyl-CoA-binding proteins play combinatory roles in pollen development. Plant Cell Physiol 56: 322–333 [DOI] [PubMed] [Google Scholar]
- Hurlock AK, Roston RL, Wang K, Benning C (2014) Lipid trafficking in plant cells. Traffic 15: 915–932 [DOI] [PubMed] [Google Scholar]
- Hwang JU, Song WY, Hong D, Ko D, Yamaoka Y, Jang S, Yim S, Lee E, Khare D, Kim K, et al. (2016) Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle. Mol Plant 9: 338–355 [DOI] [PubMed] [Google Scholar]
- Jang JC, Fujioka S, Tasaka M, Seto H, Takatsuto S, Ishii A, Aida M, Yoshida S, Sheen J (2000) A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev 14: 1485–1497 [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Skinner DJ, Gasser CS (2009) Roles of polarity determinants in ovule development. Plant J 57: 1054–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla A, Paper JM, Boehler AP, Bradley AM, Neumann TR, Schrick K (2014) HD-Zip proteins GL2 and HDG11 have redundant functions in Arabidopsis trichomes, and GL2 activates a positive feedback loop via MYB23. Plant Cell 26: 2184–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HB, Schaller H, Goh CH, Kwon M, Choe S, An CS, Durst F, Feldmann KA, Feyereisen R (2005) Arabidopsis cyp51 mutant shows postembryonic seedling lethality associated with lack of membrane integrity. Plant Physiol 138: 2033–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Franceschi VR, Davin LB, Lewis NG (2006) β-Glucuronidase as reporter gene: advantages and limitations. Methods Mol Biol 323: 263–273 [DOI] [PubMed] [Google Scholar]
- Kim S, Yamaoka Y, Ono H, Kim H, Shim D, Maeshima M, Martinoia E, Cahoon EB, Nishida I, Lee Y (2013) AtABCA9 transporter supplies fatty acids for lipid synthesis to the endoplasmic reticulum. Proc Natl Acad Sci USA 110: 773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy KT, Chen AY, Nemhauser JL (2009) Steroids are required for epidermal cell fate establishment in Arabidopsis roots. Proc Natl Acad Sci USA 106: 8073–8076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange I, Poirier BC, Herron BK, Lange BM (2015) Comprehensive assessment of transcriptional regulation facilitates metabolic engineering of isoprenoid accumulation in Arabidopsis. Plant Physiol 169: 1595–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laule O, Fürholz A, Chang HS, Zhu T, Wang X, Heifetz PB, Gruissem W, Lange M (2003) Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6866–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir R, Sorin C, Chakraborti D, Moritz T, Schaller H, Tellier F, Robert S, Morin H, Bako L, Bellini C (2013) ABCG9, ABCG11 and ABCG14 ABC transporters are required for vascular development in Arabidopsis. Plant J 76: 811–824 [DOI] [PubMed] [Google Scholar]
- Leung KC, Li HY, Xiao S, Tse MH, Chye ML (2006) Arabidopsis ACBP3 is an extracellularly targeted acyl-CoA-binding protein. Planta 223: 871–881 [DOI] [PubMed] [Google Scholar]
- Li HY, Chye ML (2003) Membrane localization of Arabidopsis acyl-CoA binding protein ACBP2. Plant Mol Biol 51: 483–492 [DOI] [PubMed] [Google Scholar]
- Li HY, Chye ML (2004) Arabidopsis acyl-CoA-binding protein ACBP2 interacts with an ethylene-responsive element-binding protein, AtEBP, via its ankyrin repeats. Plant Mol Biol 54: 233–243 [DOI] [PubMed] [Google Scholar]
- Li L, Kaplan J (1996) Characterization of yeast methyl sterol oxidase (ERG25) and identification of a human homologue. J Biol Chem 271: 16927–16933 [DOI] [PubMed] [Google Scholar]
- Liao P, Wang H, Wang M, Hsiao AS, Bach TJ, Chye ML (2014) Transgenic tobacco overexpressing Brassica juncea HMG-CoA synthase 1 shows increased plant growth, pod size and seed yield. PLoS ONE 9: e98264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2013) Acyl-lipid metabolism. The Arabidopsis Book 11: e0161, doi/10.1199/tab.0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YF, Li QT, Lu X, Song QX, Lam SM, Zhang WK, Ma B, Lin Q, Man WQ, Du WG, et al. (2014) Soybean GmMYB73 promotes lipid accumulation in transgenic plants. BMC Plant Biol 14: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung SC, Chye ML (2016a) The binding versatility of plant acyl-CoA-binding proteins and their significance in lipid metabolism. Biochim Biophys Acta 1861: 1409–1421 [DOI] [PubMed] [Google Scholar]
- Lung SC, Chye ML (2016b) Deciphering the roles of acyl-CoA-binding proteins in plant cells. Protoplasma 253: 1177–1195 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW (1996) The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Men S, Boutté Y, Ikeda Y, Li X, Palme K, Stierhof YD, Hartmann MA, Moritz T, Grebe M (2008) Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat Cell Biol 10: 237–244 [DOI] [PubMed] [Google Scholar]
- Meng W, Su YCF, Saunders RMK, Chye ML (2011) The rice acyl-CoA-binding protein gene family: phylogeny, expression and functional analysis. New Phytol 189: 1170–1184 [DOI] [PubMed] [Google Scholar]
- Mialoundama AS, Jadid N, Brunel J, Di Pascoli T, Heintz D, Erhardt M, Mutterer J, Bergdoll M, Ayoub D, Van Dorsselaer A, et al. (2013) Arabidopsis ERG28 tethers the sterol C4-demethylation complex to prevent accumulation of a biosynthetic intermediate that interferes with polar auxin transport. Plant Cell 25: 4879–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau RA, Whitaker BD, Hicks KB (2002) Phytosterols, phytostanols, and their conjugates in foods: structural diversity, quantitative analysis, and health-promoting uses. Prog Lipid Res 41: 457–500 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, Takahashi T (2006) Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol 141: 1363–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neess D, Kiilerich P, Sandberg MB, Helledie T, Nielsen R, Mandrup S (2006) ACBP: a PPAR and SREBP modulated housekeeping gene. Mol Cell Biochem 284: 149–157 [DOI] [PubMed] [Google Scholar]
- Ogawa E, Yamada Y, Sezaki N, Kosaka S, Kondo H, Kamata N, Abe M, Komeda Y, Takahashi T (2015) ATML1 and PDF2 play a redundant and essential role in Arabidopsis embryo development. Plant Cell Physiol 56: 1183–1192 [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Oka A, Rodrigues-Pousada R, Possenti M, Ruberti I, Morelli G, Aoyama T (2003) Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300: 1427–1430 [DOI] [PubMed] [Google Scholar]
- Oikari S, Ahtialansaari T, Heinonen MV, Mauriala T, Auriola S, Kiehne K, Fölsch UR, Jänne J, Alhonen L, Herzig KH (2008) Downregulation of PPARs and SREBP by acyl-CoA-binding protein overexpression in transgenic rats. Pflugers Arch 456: 369–377 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L (1999) START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci 24: 130–132 [DOI] [PubMed] [Google Scholar]
- Qian P, Han B, Forestier E, Hu Z, Gao N, Lu W, Schaller H, Li J, Hou S (2013) Sterols are required for cell-fate commitment and maintenance of the stomatal lineage in Arabidopsis. Plant J 74: 1029–1044 [DOI] [PubMed] [Google Scholar]
- Schaeffer A, Bronner R, Benveniste P, Schaller H (2001) The ratio of campesterol to sitosterol that modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2;1. Plant J 25: 605–615 [DOI] [PubMed] [Google Scholar]
- Schaller H. (2004) New aspects of sterol biosynthesis in growth and development of higher plants. Plant Physiol Biochem 42: 465–476 [DOI] [PubMed] [Google Scholar]
- Schrick K, Bruno M, Khosla A, Cox PN, Marlatt SA, Roque RA, Nguyen HC, He C, Snyder MP, Singh D, et al. (2014) Shared functions of plant and mammalian StAR-related lipid transfer (START) domains in modulating transcription factor activity. BMC Biol 12: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, Dangl J, Schmidt J, Jürgens G (2000) FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev 14: 1471–1484 [PMC free article] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Martin G, Bellini C, Kuhnt C, Schmidt J, Jürgens G (2002) Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant J 31: 61–73 [DOI] [PubMed] [Google Scholar]
- Schrick K, Nguyen D, Karlowski WM, Mayer KF (2004) START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol 5: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Sinkevicius KW, Selinger DA, Tarczynski MC (2006) The homeobox gene GLABRA2 affects seed oil content in Arabidopsis. Plant Mol Biol 60: 377–387 [DOI] [PubMed] [Google Scholar]
- Shi DQ, Liu J, Xiang YH, Ye D, Sundaresan V, Yang WC (2005) SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17: 2340–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Katavic V, Yu Y, Kunst L, Haughn G (2012) Arabidopsis glabra2 mutant seeds deficient in mucilage biosynthesis produce more oil. Plant J 69: 37–46 [DOI] [PubMed] [Google Scholar]
- Smith JA, Krauss MR, Borkird C, Sung ZR (1988) A nuclear protein associated with cell divisions in plants. Planta 174: 462–472 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane PD, Pollier J, Panda S, Szymanski J, Massalha H, Yona M, Unger T, Malitsky S, Arendt P, Pauwels L, et al. (2016) Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat Plants 3: 16205. [DOI] [PubMed] [Google Scholar]
- Souter M, Topping J, Pullen M, Friml J, Palme K, Hackett R, Grierson D, Lindsey K (2002) hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14: 1017–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangeland B, Salehian Z (2002) An improved clearing method for GUS assay in Arabidopsis endosperm and seeds. Plant Mol Biol Rep 20: 107–114 [Google Scholar]
- Stocco DM. (2001) StAR protein and the regulation of steroid hormone biosynthesis. Annu Rev Physiol 63: 193–213 [DOI] [PubMed] [Google Scholar]
- Tse MH. (2005) Investigations on recombinant Arabidopsis acyl-coenzyme A binding protein 1. M.Phil. thesis. University of Hong Kong, Hong Kong, China [Google Scholar]
- Vo KTX, Kim CY, Chandran AKN, Jung KH, An G, Jeon JS (2015) Molecular insights into the function of ankyrin proteins in plants. J Plant Biol 58: 271–284 [Google Scholar]
- Wang K, Senthil-Kumar M, Ryu CM, Kang L, Mysore KS (2012) Phytosterols play a key role in plant innate immunity against bacterial pathogens by regulating nutrient efflux into the apoplast. Plant Physiol 158: 1789–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. (2001) Plant phospholipases. Annu Rev Plant Physiol Plant Mol Biol 52: 211–231 [DOI] [PubMed] [Google Scholar]
- Western TL, Burn J, Tan WL, Skinner DJ, Martin-McCaffrey L, Moffatt BA, Haughn GW (2001) Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol 127: 998–1011 [PMC free article] [PubMed] [Google Scholar]
- Willemsen V, Friml J, Grebe M, van den Toorn A, Palme K, Scheres B (2003) Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15: 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte CP, Noël LD, Gielbert J, Parker JE, Romeis T (2004) Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol Biol 55: 135–147 [DOI] [PubMed] [Google Scholar]
- Woyengo TA, Ramprasath VR, Jones PJH (2009) Anticancer effects of phytosterols. Eur J Clin Nutr 63: 813–820 [DOI] [PubMed] [Google Scholar]
- Xiao S, Chye ML (2011) New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Prog Lipid Res 50: 141–151 [DOI] [PubMed] [Google Scholar]
- Xiao S, Gao W, Chen QF, Ramalingam S, Chye ML (2008) Overexpression of membrane-associated acyl-CoA-binding protein ACBP1 enhances lead tolerance in Arabidopsis. Plant J 54: 141–151 [DOI] [PubMed] [Google Scholar]
- Xue Y, Xiao S, Kim J, Lung SC, Chen L, Tanner JA, Suh MC, Chye ML (2014) Arabidopsis membrane-associated acyl-CoA-binding protein ACBP1 is involved in stem cuticle formation. J Exp Bot 65: 5473–5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li R, Qi M (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22: 543–551 [DOI] [PubMed] [Google Scholar]
- Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, Goldstein JL, Brown MS (1993) SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell 75: 187–197 [PubMed] [Google Scholar]
- Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB (2008) Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauber H, Burgos A, Garapati P, Schulze WX (2014) Plasma membrane lipid-protein interactions affect signaling processes in sterol-biosynthesis mutants in Arabidopsis thaliana. Front Plant Sci 5: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sun S, Nie X, Boutté Y, Grison M, Li P, Kuang S, Men S (2016) Sterol methyl oxidases affect embryo development via auxin-associated mechanisms. Plant Physiol 171: 468–482 [DOI] [PMC free article] [PubMed] [Google Scholar]