AtPRA1.F4, an isoform of Arabidopsis prenylated Rab acceptor1 protein, regulates the exit of post-Golgi proteins from the Golgi apparatus.
Abstract
Prenylated Rab acceptor1 (PRA1) functions in the recruitment of prenylated Rab proteins to their cognate organelles. Arabidopsis (Arabidopsis thaliana) contains a large number of proteins belonging to the AtPRA1 family. However, their physiological roles remain largely unknown. Here, we investigated the physiological role of AtPRA1.F4, a member of the AtPRA1 family. A T-DNA insertion knockdown mutant of AtPRA1.F4, atpra1.f4, was smaller in stature than parent plants and possessed shorter roots, whereas transgenic plants overexpressing HA:AtPRA1.F4 showed enhanced development of secondary roots and root hairs. However, both overexpression and knockdown plants exhibited increased sensitivity to high-salt stress, lower vacuolar Na+/K+-ATPase and plasma membrane ATPase activities, lower and higher pH in the vacuole and apoplast, respectively, and highly vesiculated Golgi apparatus. HA:AtPRA1.F4 localized to the Golgi apparatus and assembled into high-molecular-weight complexes. atpra1.f4 plants displayed a defect in vacuolar trafficking, which was complemented by low but not high levels of HA:AtPRA1.F4. Overexpression of HA:AtPRA1.F4 also inhibited protein trafficking at the Golgi apparatus, albeit differentially depending on the final destination or type of protein: trafficking of vacuolar proteins, plasma membrane proteins, and trans-Golgi network (TGN)-localized SYP61 was strongly inhibited; trafficking of TGN-localized SYP51 was slightly inhibited; and trafficking of secretory proteins and TGN-localized SYP41 was negligibly or not significantly inhibited. Based on these results, we propose that Golgi-localized AtPRA1.F4 is involved in the exit of many but not all types of post-Golgi proteins from the Golgi apparatus. Additionally, an appropriate level of AtPRA1.F4 is crucial for its function at the Golgi apparatus.
Newly synthesized proteins destined for various endomembrane compartments are first cotranslationally targeted to the endoplasmic reticulum (ER) and subsequently transported from the ER to their final destination via vesicle trafficking (Rothman and Wieland, 1996; Schekman and Orci, 1996; Hawes et al., 1999; Kirchhausen, 2000; Lee et al., 2002). These targeting and trafficking processes are mediated by an extremely large number of proteins that ensure specific targeting of proteins to their final destination. Among the large number of proteins involved in protein trafficking, Rab proteins are key players at almost every step of all trafficking pathways involving endomembrane compartments (Novick and Zerial, 1997; Somsel Rodman and Wandinger-Ness, 2000; Waters and Hughson, 2000; Pfeffer, 2001; Segev, 2001; Ueda and Nakano, 2002; Sohn et al., 2003). Rab proteins are members of the small GTPase family, and their roles have been studied extensively at the biochemical, molecular biological, and cell biological levels (Chavrier and Goud, 1999; Pereira-Leal and Seabra, 2000). These studies have shown that Rab proteins undergo cycling between GDP-bound and GTP-bound forms and that they also cycle between membrane-bound and soluble forms. The GDP-bound form remains in the cytosol, whereas the GTP-bound form is bound to organellar membranes. In their target organelles, Rabs are involved in the fusion of incoming vesicles to target organelles or the generation of specific vesicles from organellar membranes (Chavrier and Goud, 1999; Pereira-Leal and Seabra, 2000). All eukaryotic cells, including plant cells, contain a large number of Rab isoforms, which are distributed in various endomembrane compartments (Lazar et al., 1997; Pereira-Leal and Seabra, 2000; Zerial and McBride, 2001; Vernoud et al., 2003; Colicelli, 2004; Saito-Nakano et al., 2005; Zhang et al., 2007).
Rab protein function depends on specific targeting to its cognate organelle, but it is not fully understood how this process occurs. Rab proteins are posttranslationally modified with prenyl groups at their C termini, and these hydrophobic appendages function as lipid membrane anchors (Seabra et al., 1992; Glomset and Farnsworth, 1994). Prenylated Rab acceptor1 (PRA1), which functions as a GDP dissociation inhibitor displacement factor, is thought to play a role in targeting prenylated Rab proteins to their cognate organelles (Sivars et al., 2003, 2005). The first PRA1 family member was identified in yeast as Ypt1-interacting protein (Yip1; Yang et al., 1998). Numerous Yip-related proteins have since been identified in yeast and mammals, including the Yip1-interacting factor Yif1 (Calero et al., 2002) that is a member of the Yip/PRA1 family (Pfeffer and Aivazian, 2004). In yeast and mammals, Yip/PRA1 family members are reported to facilitate the delivery of Rab GTPases to their cognate organellar membranes by mediating the dissociation of the Rab GDP dissociation inhibitor complex (Sivars et al., 2003).
Similar to animal cells and yeast, plant cells also contain a large number of small membrane proteins that share varying degrees of amino acid sequence homology with animal and yeast Yip/PRA1 proteins. In Arabidopsis (Arabidopsis thaliana), 19 PRA1 homologs (AtPRA1s) have been identified and characterized in terms of their expression and subcellular localization (Alvim Kamei et al., 2008). AtPRA1.B6 localizes to the ER and Golgi apparatus and plays a role in the trafficking of cargo proteins destined for various endomembrane compartments (Jung et al., 2011; Lee et al., 2011). An Arabidopsis PRA1-related protein has been reported to interact with the cauliflower mosaic virus (CaMV) movement protein (Huang et al., 2001). A PRA1 homolog in rice (Oryza sativa), OsPRA1, interacts with OsRab7 and localizes to the prevacuolar compartment (PVC). OsPRA1 is believed to play a role in protein trafficking to the vacuole at the PVC (Heo et al., 2010).
In this study, we investigated the physiological role of the AtPRA1.F4 isoform of AtPRA1. The exact functions of most AtPRA1 isoforms have not been elucidated. This is partly due to the large number of isoforms present and the high degree of sequence homology shared. In this work, to elucidate the physiological role of AtPRA1.F4, we examined the phenotypes of AtPRA1.F4 knockdown mutant (atpra1.f4) plants and transgenic plants overexpressing AtPRA1.F4 tagged at the N terminus with a small hemagglutinin (HA) epitope at both the cellular and whole-plant levels. Compared with wild-type plants, transgenic plants overexpressing HA:AtPRA1.F4 exhibited altered morphology, including increases in the number and length of secondary roots and root hairs. However, both variants showed hypersensitivity to high concentrations of NaCl and KCl as well as alterations in the vacuolar and apoplastic pH. At the cellular level, both HA:AtPRA1.F4-overexpressing and atpra1.f4 knockdown mutant plants displayed highly vesiculated Golgi apparatus and defective vacuolar trafficking.
RESULTS
AtPRA1.F4 Knockdown Mutant Plants Display Growth Defects and Low Seed Yield
The At3g13710 locus encodes AtPRA1.F4 as part of a bicistronic gene together with At3g13700, which encodes an RNA recognition motif-containing protein (Thimmapuram et al., 2005). We identified a mutant (GABI-114H12) with a T-DNA insertion between At3g13710 and At3g13700 (Supplemental Fig. S1, A and B Supplementary Table S1; Supplementary Materials and Methods S1). In this mutant, levels of At3g13710 and At3g13700 transcripts were reduced to 40% and 75%, respectively, compared with the corresponding genes in wild-type plants (Supplemental Fig. S1B). This indicates that the mutation has a more detrimental effect on the expression of AtPRA1.F4; hence, the mutation was named atpra1.f4. We examined the phenotype of atpra1.f4 plants, and growth defects were clearly apparent. When grown in B5 medium or soil, atpra1.f4 plants were smaller than wild-type plants in terms of leaf size and overall height (Fig. 1A). Mutant plants also possessed shorter roots compared with wild-type plants (Fig. 1B), and the seed yield was reduced to 40% (Fig. 1C). However, the germination rate was not affected in atpra1.f4 plants (Fig. 1D).
Figure 1.
atpra1.f4 mutant plants show alterations in development. A, atpra1.f4 plants in soil adopt a smaller stature. B, Shorter root length of atpra1.f4 plants. The root length was measured from wild-type (WT) and atpra1.f4 plants grown on one-half-strength Murashige and Skoog (MS) plates for the indicated time periods. Error bars represent sd (n = 6). Bar = 1 cm. C, Seed yield of atpra1.f4 plants. D, Germination rate of atpra1.f4 plants. Asterisks indicate statistically significant differences at P ≤0.001 (***) between WT and atpra1.f4.
Transgenic Plants Overexpressing HA:AtPRA1.F4 Display Alterations in Secondary Root and Root Hair Development and Salt Stress Responses
To further elucidate the physiological role of AtPRA1.F4, we generated transgenic plants overexpressing HA-tagged AtPRA1.F4 (HA:AtPRA1.F4) under the control of the strong CaMV 35S promoter or an empty vector as a control. HA:AtPRA1.F4 expression was examined by western-blot analysis using anti-HA antibody. HA:AtPRA1.F4-overexpressing (AtPRA1.4 OX) plants produced a strong band at the expected size of 22 kD that was absent in control plants (Supplemental Fig. S2A). HA:AtPRA1.F4 OX plants also produced multiple discrete bands and a smearing in the high-Mr region (Supplemental Fig. S2B; Supplementary Materials and Methods 1). To further investigate this apparent high-Mr complex formation, HA:AtPRA1.F4 was immunoprecipitated with anti-HA antibody, and immunoprecipitated proteins were analyzed by western blotting. Immunoprecipitates clearly produced multiple bands corresponding to HA:AtPRA1.F4 monomer, dimer, trimer, and tetramer, even after denaturation with SDS, indicating that HA:AtPRA1.F4 forms highly stable high-Mr complexes, most likely via homomeric interactions.
We examined the phenotype of HA:AtPRA1.F4 OX plants at various stages of plant growth and development. Overall, HA:AtPRA1.F4 OX plants did not show any significant morphological alterations compared with control plants, except for significant increases in the number and length of lateral roots and root hairs (Supplemental Fig. S3, A–D). HA:AtPRA1.F4 OX plants also showed a slight delay (2−3 d) in flowering (Supplemental Fig. S3E). The phenotypes of HA:AtPRA1.F4 OX and atpra1.f4 plants were examined further by testing their responses to salt stress. Both types of plants exhibited higher hypersensitivity to 100 mm NaCl and 100 mm KCl compared with control plants harboring the empty vector pBIB (Fig. 2), indicating that both high and low levels of AtPRA1.F4 expression are detrimental for plant stress responses to high salt concentrations.
Figure 2.
HA:AtPRA1.F4 OX and atpra1.f4 mutant plants show hypersensitivity to high concentrations of NaCl and KCl. A to C, Effects of HA:AtPRA1.F4 overexpression and AtPRA1.F4 knockdown mutation on the responses to high concentrations of NaCl and KCl. Plants were grown on one-half-strength MS plates supplemented with or without 100 mm NaCl or 100 mm KCl. Images were taken 29 d after planting. D, Fresh weight of whole plants from 29-d-old plants grown on one-half-strength MS plates supplemented with 100 mm KCl. Three independent experiments were performed on 51 plants. Error bars represent sd (n = 51). Asterisks indicate significant differences at P < 0.01 (**) or P < 0.001 (***) as calculated using Student’s t test. WT, Wild type.
Both HA:AtPRA1.F4 OX and atpra1.f4 Plants Have Lower Vacuolar Na+/K+-ATPase and Plasma Membrane ATPase Activities
To elucidate the mechanisms underlying the hypersensitivity of atpra1.f4 and HA:AtPRA1.F4 OX plants to high concentrations of NaCl and KCl, we first measured the activity of vacuolar Na+/K+-ATPase, which is responsible for the sequestration of excess Na+ and K+ ions from the cytosol into the vacuole. Vacuolar Na+/K+-ATPase activity was measured in vacuoles isolated from control, HA:AtPRA1.F4 OX, and atpra1.f4 plants, and it was significantly lower in both HA:AtPRA1.F4 OX and atpra1.f4 plants than in control plants (Fig. 3A). This finding is consistent with the fact that both HA:AtPRA1.F4 OX and atpra1.f4 plants were hypersensitive to high concentrations of NaCl and KCl (Fig. 2).
Figure 3.
HA:AtPRA1.F4 OX and atpra1.f4 plants have lower vacuolar Na+/K+-ATPase and plasma membrane (PM) ATPase activities. A, Effects of HA:AtPRA1.F4 overexpression and AtPRA1.F4 knockdown mutation on vacuolar Na+/K+-ATPase activity. Vacuolar membrane proteins were prepared from isolated vacuoles and used to measure Na+/K+-ATPase activity as described in “Materials and Methods.” Control plants harbor the empty pBIB vector. Asterisks indicate statistically significant differences at P ≤ 0.001 (***) between pBIB and HA:AtPRA1.F4 OX plants and at P ≤ 0.05 (*) between wild-type (WT) and atpra1.f4 plants. Error bars represent sd (n = 3). B, Effects of HA:AtPRA1.F4 overexpression and AtPRA1.F4 knockdown mutation on apoplastic pH. Apoplastic fluid samples were prepared and pH was measured as described in “Materials and Methods.” Error bars represent sd (n = 3). Asterisks indicate statistically significant differences at P ≤ 0.01 (**) between pBIB and HA:AtPRA1.F4 OX plants and between wild-type and atpra1.f4 plants. C, Effects of HA:AtPRA1.F4 overexpression and AtPRA1.F4 knockdown mutation on rhizosphere pH. Ten-day-old seedlings grown on one-half-strength MS plates were incubated in one-half-strength liquid MS medium supplemented with 0.005% Bromocresol Purple for 2 d, and the pH of the incubation medium was measured as described in “Materials and Methods.” Error bars represent sd (n = 3). Asterisks indicate statistically significant differences at P ≤ 0.1 (*) between pBIB and HA:AtPRA1.F4 OX plants and at P ≤ 0.001 (***) between wild-type and atpra1.f4 plants. D, Effects of HA:AtPRA1.F4 overexpression and AtPRA1.F4 knockdown mutation on PM ATPase activity. Inside-out vesicles of PM factions were used to measure PM ATPase activity. Asterisks indicate statistically significant differences at P ≤ 0.001 (***) between pBIB and HA:AtPRA1.F4 OX plants and at P ≤ 0.05 (*) between wild-type and atpra1.f4 plants. Error bars represent sd (n = 3).
Another important mechanism used to increase salt tolerance involves pumping Na+ and K+ ions out of the cytosol and into the apoplast. PM H+-ATPases play a crucial role in this mechanism. Therefore, we examined whether HA:AtPRA1.F4 OX and atpra1.f4 plants displayed detectable alterations in the pH of the apoplast and rhizosphere (Fig. 3, B and C). The pH in both apoplast and rhizosphere was much higher in HA:AtPRA1.F4 OX and atpra1.f4 plants than in control plants, indicating that both plants have lower PM H+-ATPase activity. To confirm this result, we prepared PM fractions from control, atpra1.f4, and HA:AtPRA1.F4 OX plants and measured ATPase activity (Fig. 3D). The ATPase activity was significantly lower in both atpra1.f4 and HA:AtPRA1.F4 OX plants than in control plants, consistent with their higher apoplastic and rhizosphere pH values. These results suggest that PM H+-ATPase activity is lower in both atpra1.f4 and HA:AtPRA1.F4 OX plants, which, in turn, results in a failure to pump excess N+ and K+ ions out of the cytosol and into the apoplast, thereby leading to hypersensitive responses to high concentrations of NaCl and KCl.
HA:AtPRA1.F4 Localizes to the Golgi Apparatus
PRA1 family members function in protein trafficking (Liu et al., 2006; Heo et al., 2010; Lee et al., 2011). Therefore, we hypothesized that the altered phenotypes observed in HA:AtPRA1.F4 OX plants could be related to a defect in protein trafficking. To test this hypothesis, we first examined the subcellular localization of HA:AtPRA1.F4 in transgenic plants by performing immunohistochemical analysis with anti-HA antibody. Previous studies showed that AtPRA1 family members localize to various endomembrane compartments when expressed as GFP fusions (Alvim Kamei et al., 2008). The HA-tagged form of a close homolog of AtPRA1.F4, namely AtPRA1.B6, localizes to the ER and Golgi apparatus (Jung et al., 2011; Lee et al., 2011). Our results showed that HA:AtPRA1.F4 produced a punctate staining pattern in root tissues of HA:AtPRA1.F4 OX plants (Fig. 4A).
Figure 4.
HA:AtPRA1.F4 localizes to the Golgi apparatus. A to C, Localization of HA:AtPRA1.F4 in HA:AtPRA1.F4 OX plants. Root tissues of HA:AtPRA1.F4 OX plants were fixed and immunostained with anti-HA antibody (A) followed by fluorescein isothiocyanate (FITC)- or tetramethylrhodamine (TRITC)-labeled anti-rat IgG or anti-γ-COP antibody (B) or anti-AtRabF2a antibody (C) followed by TRITC- or FITC-labeled anti-rabbit IgG. To quantify the colocalization in B and C, 30 independent cells were analyzed using the Pearson and Spearman correlation coefficient colocalization plug-in of ImageJ software. The results are shown as scatterplots (right-most images). SE, Stable expression in transgenic plants. Bars = 20 μm. D to F, AtPRA1.F4 localization in protoplasts. HA:AtPRA1.F4 was transformed into protoplasts prepared from transgenic plants expressing ST:GFP or RFP:AtRabF2b, and HA:AtPRA1.F4 localization was examined by immunostaining with anti-HA antibody. ST:GFP and RFP:AtRabF2b were observed directly. TE, Transient expression in protoplasts; WT, wild type. Bars = 10 μm.
Next, we compared the localization of HA:AtPRA1.F4 with that of γ-COP and AtRabF2a. Endogenous γ-COP and AtRabF2a were detected by immunohistochemistry using anti-γ-COP and anti-AtRabF2a antibodies, respectively. The results showed that γ-COP, a component of the COPI coatomer complex, localized to the Golgi apparatus and AtRabF2a localized to the PVC (Lee et al., 2004). HA:AtPRA1.F4 closely overlapped with γ-COP in the punctate foci in roots (Fig. 4B) but did not overlap with AtRabF2a (Fig. 4C), indicating that HA:AtPRA1.F4 localized to the Golgi apparatus. This result is not in agreement with the findings of a previous study showing that AtPRA1.F4 labeled with a C-terminal GFP localized to vesicular structures often associated with a network of ER strands (Alvim Kamei et al., 2008). Certain PRA1 proteins contain a targeting signal at their C terminus (Abdul-Ghani et al., 2001; Liang et al., 2004; Jung et al., 2011). This suggests that the C-terminal GFP moiety may have disrupted the proper localization of AtPRA1.F4 in the previous study.
To obtain supporting evidence for HA:AtPRA1.F4 localization, protoplasts from transgenic plants expressing ST:GFP or RFP:AtRABF2b were transformed with HA:AtPRA1.F4, and HA:AtPRA1.F4 localization was compared with that of ST:GFP or RFP:AtRABF2b. HA:AtPRA1.F4 was detected by immunostaining with anti-HA antibody, whereas ST:GFP and RFP:AtRABF2b were observed directly. ST:GFP, a chimeric protein consisting of full-length rat sialyltransferase (ST) and GFP, localized to the trans-Golgi, whereas RFP:AtRABF2b localized primarily to the PVC, with a minor portion localized at the Golgi apparatus (Kim et al., 2001; Sohn et al., 2003; Kotzer et al., 2004). HA:AtPRA1.F4 closely overlapped with ST:GFP in the punctate foci in roots (Fig. 4E) and also closely overlapped with RFP:AtRABF2b in a small proportion of punctate foci in roots (Fig. 4F). These combined results confirm that HA:AtPRA1.F4 localizes to the Golgi apparatus.
The degree of overlap was analyzed using Pearson (rp) and Spearman (rs) correlation coefficients (French et al., 2008), which were as follows: rp = 0.88 and rs = 0.83 for HA:AtPRA1.F4 and γ-COP; rp = 0.82 and rs = 0.7 for HA:AtPRA1.F4 and ST:GFP; rp = 0.35 and rs = 0.19 for HA:AtPRA1.F4 and RabF2a; and rp = 0.56 and rs = 0.3 for HA:AtPRA1.F4 and RFP:AtRABF2b. Therefore, HA:AtPRA1.F4 closely colocalized with anti-γ-COP and ST:GFP but not with AtRabF2a and RFP:AtRABF2b. These results confirm that HA:AtPRA1.F4 localizes to the Golgi apparatus, in particular to the trans-Golgi.
HA:AtPRA1.F4 Overexpression Does Not Affect the Targeting of Golgi-Localized Proteins
The Golgi localization of HA:AtPRA1.F4 suggests a role in protein trafficking that involves the Golgi apparatus. The observed reduction in vacuolar Na+/K+-ATPase and PM ATPase activities, and the increased apoplastic pH in HA:AtPRA1.F4 OX plants, suggest the possibility that high levels of HA:AtPRA1.F4 expression cause a defect in protein trafficking to the tonoplast or PM. Previous studies showed that high levels of AtPRA1.B6 inhibit protein trafficking at the ER. Therefore, we tested whether high levels of HA:AtPRA1.F4 expression inhibit anterograde trafficking of proteins to various endomembrane compartments. First, we introduced ST:GFP, which localizes to the trans-Golgi (Wee et al., 1998; Kim et al., 2001), into protoplasts together with HA:AtPRA1.F4 or R6 and examined ST:GFP localization to the trans-Golgi. ST:GFP produced a punctate staining pattern in protoplasts overexpressing HA:AtPRA1.F4, suggesting that HA:AtPRA1.F4 overexpression does not inhibit the targeting of ST:GFP to the trans-Golgi. To further confirm its localization at the biochemical level, we examined its glycosylation pattern. N-Glycans are modified to a complex type at the trans-Golgi and, thereby, were rendered resistant to endoglycosidase H (Endo H) activity. ST:GFP was cotransformed into protoplasts together with HA:AtPRA1.F4 or the empty R6 vector as a control. Subsequently, protein extracts were prepared from protoplasts, treated with Endo H, and analyzed by western blotting using anti-GFP antibody. In both samples, Endo H treatment did not change the localization/migration pattern of ST:GFP (Fig. 5), indicating that ST:GFP in both samples contains Endo H-resistant N-glycans. These results confirm that HA:AtPRA1.F4 overexpression does not affect ST:GFP localization to the trans-Golgi.
Figure 5.
HA:AtPRA1.F4 overexpression does not inhibit ST:GFP trafficking to the Golgi apparatus. A, Lack of an effect of HA:AtPRA1.F4 on the glycosylation pattern of ST:GFP. ST:GFP was cotransformed with HA:AtPRA1.F4 (10 μg) or R6 (empty vector) into protoplasts prepared from wild-type plants. Protein extracts from protoplasts were treated with Endo H and analyzed by western blotting using anti-GFP and anti-HA antibodies. Gly, Glycosylated form; un-gly, unglycosylated form. B, Trafficking efficiency of ST:GFP to the Golgi apparatus. Trafficking efficiency was determined from the ratio of the Endo H-resistant form versus the total amount of expressed ST:GFP. TE, Transient expression in protoplasts. Error bars represent sd (n = 3).
Both High and Low Levels of AtPRA1.F4 Inhibit Vacuolar Trafficking at the Golgi Apparatus
Next, we examined the role of AtPRA1.F4 in protein trafficking. We used Spo:GFP, a chimeric protein consisting of the N-terminal region of sporamin, a storage protein of sweet potato (Ipomoea batatas), and GFP (Kim et al., 2001) as a reporter protein for vacuolar trafficking. Spo:GFP was cotransformed into wild-type and atpra1.f4 protoplasts, and protein extracts of protoplasts were analyzed by western blotting using anti-GFP antibody. The ratio of processed forms to the total amount of expressed protein was used to estimate the trafficking efficiency, and this was decreased significantly in atpra1.f4 protoplasts compared with wild-type protoplasts (Fig. 6A). Next, we examined whether the defect in vacuolar trafficking could be complemented by coexpression of AtPRA1.F4 in atpra1.f4 protoplasts. Spo:GFP was cotransformed with different amounts (0.1−5 μg) of HA:AtPRA1.F4 in atpra1.f4 protoplasts, and a low amount (0.1 μg) rescued the defect in vacuolar trafficking of Spo:GFP, confirming that the defect is caused by low levels of AtPRA1.F4 in atpra1.f4 plants. However, a higher amount (1−5 μg) of AtPRA1.F4 inhibited the vacuolar trafficking of Spo:GFP (Fig. 6B), indicating that the AtPRA1.F4 level is critical for its function in vacuolar trafficking.
Figure 6.
HA:AtPRA1.F4 overexpression and AtPRA1.F4 knockdown inhibit the trafficking of Golgi-dependent vacuolar proteins at the Golgi apparatus. A, Inhibition of vacuolar trafficking in atpra1.f4 plants. Spo:GFP was transformed into protoplasts from wild-type (WT) and atpra1.f4 plants, and their trafficking was examined at various time points by western-blot analysis using anti-GFP antibody. Trafficking efficiency was quantified from the ratio of the processed form versus the total amount of expressed protein. Error bars represent sd (n = 3). Asterisks indicate statistically significant differences at P ≤ 0.05 (*) between wild-type and atpra1.f4 plants. B, Complementation of the defect in vacuolar trafficking of Spo:GFP in atpra1.f4 protoplasts. Spo:GFP was cotransformed with different amounts of HA:AtPRA1.F4 (0.1, 1, and 5 μg) into atpra1.f4 protoplasts, and the trafficking of Spo:GFP was examined at 36 h after transformation by western-blot analysis using anti-GFP antibody. C and D, Inhibition of vacuolar trafficking in HA:AtPRA1.F4 OX plants. Spo:GFP (C) and AALP:GFP (D) were transformed into protoplasts from HA:AtPRA1.F4 OX plants, and their trafficking was examined at various time points by western-blot analysis using anti-GFP antibody. Trafficking efficiency was quantified from the ratio of the processed form versus the total amount of expressed protein. Error bars represent sd (n = 3 in C and n = 2 in D). Asterisks indicate statistically significant differences at P ≤ 0.05 (*) or P ≤ 0.01 (**) between R6 and HA:AtPRA1.F4. TE, Transient expression in protoplasts.
Next, we further examined the effect of HA:AtPRA1.F4 overexpression on the trafficking of vacuolar proteins using the reporter proteins Spo:GFP and AALP:GFP, a fusion protein of Arabidopsis aleurain-like protein (AALP) and GFP (Sohn et al., 2003). These constructs were cotransformed into wild-type protoplasts together with HA:AtPRA1.F4 or R6. Protein extracts from protoplasts were analyzed by western blotting using anti-GFP antibody. The trafficking efficiency of these proteins decreased significantly with coexpression of HA:AtPRA1.F4 compared with that of the control (Fig. 6, C and D), indicating that HA:AtPRA1.F4 overexpression inhibits the trafficking of vacuolar proteins. This result is similar to that of a previous study showing that overexpression of AtPRA1.B6, another AtPRA1 isoform, inhibits vacuolar trafficking (Lee et al., 2011). To corroborate this finding, we tested whether the trafficking of stably expressed AALP:GFP in transgenic plants is inhibited by HA:AtPRA1.F4 overexpression. We generated transgenic plants harboring AALP:GFP under the control of a dexamethasone-inducible promoter. Protoplasts from AALP:GFP-overexpressing plants were transformed with HA:AtPRA1.F4 or R6 and then treated with dexamethasone to induce AALP:GFP expression. Protein extracts were prepared at various time points after transformation, and AALP:GFP trafficking was examined by western-blot analysis using anti-GFP antibody. Once again, HA:AtPRA1.F4 overexpression reduced the trafficking efficiency of AALP:GFP compared with that of the controls (Supplemental Fig. S4).
Next, we examined the trafficking of membrane proteins to the tonoplast. AtβFruct4, VAM3, VAMP711, and TPK1 were fused to GFP or YFP, and the resulting constructs were examined for their trafficking in protoplasts under HA:AtPRA1.F4 overexpression. AtβFruct4:GFP is trafficked to the tonoplast as a membrane protein, where it is proteolytically processed to yield soluble GFP in the vacuolar lumen (Jung et al., 2011). Once again, HA:AtPRA1.F4 overexpression significantly reduced the trafficking of AtβFruct4:GFP compared with that of the control (Supplemental Fig. S5). In the cases of GFP:VAM3, GFP:VAMP711, and TPK1:YFP, trafficking was determined by GFP or YFP fluorescence patterns. In control protoplasts, these proteins produced ring patterns with occasional punctate foci, indicating that they are largely targeted to the tonoplast (Supplemental Fig. S6, A, E, and I). By contrast, in HA:AtPRA1.F4 OX protoplasts, these proteins produced heavily punctate staining patterns, indicating that HA:AtPRA1.F4 overexpression inhibits the trafficking of these proteins to the tonoplast. The punctate staining patterns may reflect protein accumulation in the Golgi apparatus. This was tested by cotransforming GFP:VAM3, GFP:VAMP711, or TPK1:YFP into protoplasts of HA:AtPRA1.F4 OX plants together with KAM1ΔC:mRFP and comparing the localization patterns. VAM3:GFP, VAMP711:GFP, and TPK1:YFP closely overlapped with KAM1ΔC:mRFP at the punctate foci. The same results were obtained with wild-type protoplasts when these constructs were cotransformed with the two additional constructs, HA:AtPRA1.F4 and KAM1ΔC:mRFP (Supplemental Fig. S6). These results indicate that stable or transient HA:AtPRA1.F4 expression in transgenic plants or protoplasts, respectively, inhibits the trafficking of vacuolar membrane proteins and causes them to accumulate at the Golgi apparatus. These results are consistent with the results showing that Na+/K+-ATPase activity is lower in HA:AtPRA1.F4 OX plants compared with that in control plants.
HA:AtPRA1.F4 Overexpression Differentially Affects the Trafficking of Trans-Golgi Network-Localized SNAREs
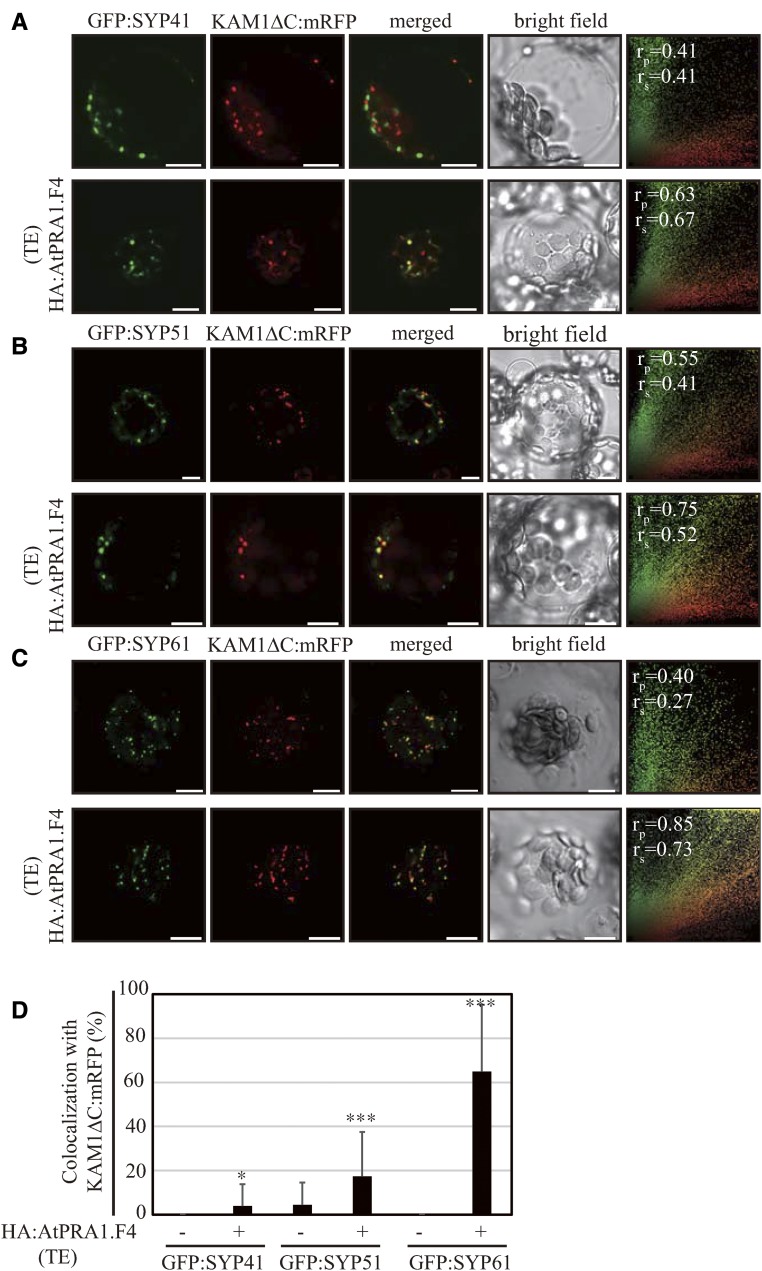
The results shown in Figure 6 and Supplemental Figures S4 to S6 prompted us to test the effect of AtPRA1.F4 overexpression on the trafficking of additional post-Golgi proteins, and we selected trans-Golgi network (TGN)-localized proteins because many SNARE proteins, such as SYP41, SYP51, and SYP61, localize to the TGN. These proteins were fused to GFP and transformed into wild-type protoplasts together with HA:AtPRA1.F4 or R6. The chimeric protein fusion construct KAM1ΔC:mRFP, which localized to the trans-Golgi (Tamura et al., 2005), was included in the transformation as a Golgi marker. In control protoplasts, all three GFP-fused SNAREs, GFP:SYP41, GFP:SYP51, and GFP:SYP61, produced punctate staining patterns. GFP:SYP41-positive and GFP:SYP61-positive punctate stains did not show any overlap with KAM1ΔC:mRFP-positive punctate stains. A small proportion of GFP:SYP51-positive punctate stains overlapped with KAM1ΔC:mRFP-positive punctate stains (Fig. 7, A–C, top rows). These results confirm the TGN localization of GFP-fused SYP41, SYP51, and SYP61. However, these TGN-localized SYP proteins showed varying degrees of colocalization with KAM1ΔC:mRFP in HA:AtPRA1.F4 OX protoplasts: GFP:SYP61, GFP:SYP51, and GFP:SYP41 showed greater than 65%, 17%, and 4% overlap with KAM1ΔC:mRFP at punctate stains, respectively (Fig. 7, A–C, bottom rows, and D). Colocalization of these SNAREs with KAM1ΔC:mRFP suggests that HA:AtPRA1.F4 overexpression differentially inhibits trafficking from the trans-Golgi to the TGN. To confirm these results, GFP:SYP61 and KAM1ΔC:mRFP were cotransformed into protoplasts from HA:AtPRA1.F4 OX plants. The number of punctate foci stained with both GFP:SYP61 and KAM1ΔC:mRFP was increased in HA:AtPRA1.F4 OX plants (Supplemental Fig. S7A) compared with controls (Fig. 7C, top row). Additionally, we determined the localization of endogenous SYP61 and ectopically expressed HA:AtPRA1.F4 in the roots of HA:AtPRA1.F4 OX plants using anti-SYP61 and anti-HA antibodies, respectively. SYP61 showed close colocalization with Golgi-localized HA:AtPRA1.F4 (Supplemental Fig. S7B), confirming that HA:AtPRA1.F4 overexpression inhibits SYP61 trafficking from the Golgi apparatus to the TGN, thereby causing its accumulation at the Golgi apparatus. However, the fact that the three SYP proteins showed varying degrees of overlap with KAM1ΔC:mRFP suggests that high levels of HA:AtPRA1.F4 differentially inhibit the exit of these TGN-localized SNAREs from the Golgi apparatus.
Figure 7.
HA:AtPRA1.F4 overexpression differentially inhibits the trafficking of TGN-localized SNARE proteins at the Golgi apparatus. A to C, Effects of HA:AtPRA1.F4 overexpression on the localization of SYP41:GFP, SYP51:GFP, and SYP61:GFP. The indicated SNARE and KAM1ΔC:mRFP constructs were transformed into wild-type protoplasts together with HA:AtPRA1.F4 or R6 (empty vector), and the localization of SYP41:GFP (A), SYP51:GFP (B), or SYP61:GFP (C) was examined. For colocalization of SNAREs with KAM1ΔC:mRFP, images (n = 35 and 30 for SYP41, n = 35 and 45 for SYP51, and n = 21 and 30 for SYP61 without and with HA:AtPRA1.F4, respectively) were analyzed using the Pearson and Spearman correlation coefficient colocalization plug-in of ImageJ software. The results are shown as scatterplots (right-most images). Bars = 10 μm. D, Quantification of colocalization. The extent of colocalization of the indicated SNARE proteins with KAM1ΔC:mRFP was calculated and presented in relative values. Error bars represent sd (n = 37 and 53 for SYP41, n = 34 and 41 for SYP51, and n = 19 and 20 for SYP61 without and with HA:AtPRA1.F4, respectively). Asterisks indicate statistically significant differences at P ≤ 0.05 (*) or P ≤ 0.001 (***) between R6 and HA:AtPRA1.F4. TE, Transient expression in protoplasts.
HA:AtPRA1.F4 Overexpression Inhibits the Trafficking of Membrane Proteins to the PM But Does Not Inhibit the Trafficking of Secretory Proteins to the Apoplast
The Golgi apparatus plays a crucial role in the trafficking of proteins to the PM and protein secretion out of the cell. We examined the effect of HA:AtPRA1.F4 overexpression on these protein-trafficking pathways. In control protoplasts, AHA2:GFP, a fusion between PM-localized H+-ATPase and GFP, was efficiently targeted to the PM, as reported previously (Kim et al., 2001). However, HA:AtPRA1.F4 overexpression caused AHA2:GFP to produce a punctuate staining pattern (Fig. 8A), indicating that HA:AtPRA1.F4 overexpression inhibits the trafficking of proteins to the PM. AHA2:GFP colocalized with KAM1ΔC:mRFP at the punctate foci (Fig. 8B), indicating that AHA2:GFP accumulated at the Golgi apparatus. To further confirm this finding, we examined the trafficking of another PM protein, PMP:GFP-gly, a fusion protein containing the PM protein PMP and GFP with a C-terminal N-glycan site (Lee et al., 2011). In control protoplasts, PMP:GFP-gly produced the typical ring-type PM pattern with occasional punctate foci, but HA:AtPRA1.F4 overexpression caused a predominantly punctate staining pattern (Supplemental Fig. S8A). However, in both samples, the majority of PMP:GFP-gly contained Endo H-resistant N-glycans (Supplemental Fig. S8B), indicating that HA:AtPRA1.F4 overexpression does not affect the trafficking of AHA2:GFP into the Golgi apparatus but rather inhibits its exit from the Golgi apparatus.
Figure 8.
HA:AtPRA1.F4 overexpression inhibits the trafficking of PM proteins at the Golgi apparatus. AHA2:GFP was introduced into wild-type protoplasts together with R6 or HA:AtPRA1.F4 (A) or both HA:AtPRA1.F4 and KAM1ΔC:mRFP (B), and the localization of PM proteins was examined. TE, Transient expression in protoplasts. Bars = 10 μm.
Next, we examined the effect of HA:AtPRA1.F4 overexpression on the trafficking of secretory proteins. invertase:GFP, a fusion protein of apoplastic invertase and GFP, is efficiently secreted into the incubation medium of protoplasts (Kim et al., 2005). Transgenic plants harboring invertase:GFP were generated and crossed with HA:AtPRA1.F4 OX plants, and homozygous plants for both invertase:GFP and HA:AtPRA1.F4 were screened from the F2 generation. In these homozygous plants, GFP signals were detected at the apoplastic regions, similar to plants harboring only invertase:GFP (Fig. 9A), indicating that HA:AtPRA1.F4 overexpression does not inhibit invertase:GFP secretion into the apoplast. To further confirm this, invertase:GFP was cotransformed into wild-type protoplasts together with HA:AtPRA1.F4 or R6, and invertase:GFP secretion was examined. Protein extracts from protoplasts and the incubation medium were analyzed by western blotting using anti-GFP antibody. In both cotransformed samples, most of the invertase:GFP was detected in the incubation medium (Fig. 9B), confirming that HA:AtPRA1.F4 overexpression does not inhibit invertase:GFP secretion. HA:AtPRA1.F4 overexpression was confirmed by western blotting using anti-HA antibody. The fact that HA:AtPRA1.F4 was detected only in protoplast extracts excludes the nonspecific contamination of protoplast proteins in the medium.
Figure 9.
HA:AtPRA1.F4 overexpression does not inhibit the trafficking of secretory proteins. A and B, HA:AtPRA1.F4 overexpression does not inhibit invertase:GFP secretion. A, Plants harboring invertase:GFP were crossed with HA:AtPRA1.F4 OX plants, and homozygous plants were screened in the F2 population. Localization of invertase:GFP was examined in leaf tissue from homozygous plants expressing both invertase:GFP and HA:AtPRA1.F4. SE, Stable expression in transgenic plants; WT, wild type. B, invertase:GFP was cotransformed into wild-type protoplasts with HA:AtPRA1.F4 or R6, and protein extracts from protoplasts and incubation medium were analyzed by western blotting. C, Protoplasts; M, medium; TE, transient expression in protoplasts. C and D, HA:AtPRA1.F4 overexpression does not inhibit secGFP secretion. C, secGFP was transformed into HA:AtPRA1.F4 OX protoplasts alone or cotransformed into wild-type protoplasts together with HA:AtPRA1.F4 or R6. Subsequently, protein extracts from protoplasts and incubation medium were analyzed at 24 h after transformation by western blotting. Asterisks indicate statistically significant differences at P ≤ 0.001 (***) between R6 and Sar-1[H74L]. D, Protein extracts from protoplasts and incubation medium were prepared at 36 h after transformation, treated with Endo H, and analyzed by western blotting using anti-GFP and anti-HA antibodies. Gly, Glycosylated form; processed, processed form; un-gly, unglycosylated form.
To obtain supporting evidence for this finding, we examined the trafficking of another secretory protein, secretory GFP-gly (secGFP-gly), which is an artificial protein generated by fusing the N-terminal ER-targeting signal of binding protein with GFP containing a C-terminal glycosylation site (Park et al., 2013). We introduced secGFP-gly into wild-type protoplasts together with HA:AtPRA1.F4 or R6. In addition, HA:AtPRA1.F4 OX protoplasts were transformed with secGFP-gly. The amount of secGFP-gly detected in the incubation medium was comparable among the three different conditions (Fig. 9C), indicating that neither transient nor stable HA:AtPRA1.F4 overexpression inhibits secGFP-gly secretion. The secretion efficiency ranged from 55% to 60% during a 24-h incubation under all three conditions. We confirmed that secGFP-gly is secreted through the Golgi apparatus. Trafficking of secGFP-gly was strongly inhibited at the ER by expression of the cotransformed Sar-1[H74L], a dominant-negative Sar-1 mutant (Fig. 9C; Takeuchi et al., 2000; Lee et al., 2011). Moreover, secGFP-gly protein in the incubation medium was resistant to Endo H (Fig. 9D), suggesting that secGFP-gly was transported through the Golgi apparatus.
Next, we examined the trafficking of secretory proteins in atpra1.f4 plants. invertase:GFP or secGFP-gly was cotransformed into wild-type and atpra1.f4 protoplasts, and their secretion was examined. Protein extracts from protoplasts and incubation medium were analyzed by western blotting using anti-GFP antibody. In both samples, most invertase:GFP was detected in the incubation medium (Supplemental Fig. S9A). Moreover, the amount of secGFP-gly detected in the incubation medium of atpra1.f4 protoplasts was comparable to that from wild-type protoplasts (Supplemental Fig. S9B), indicating that knockdown mutation of AtPRA1.F4 does not inhibit invertase:GFP and secGFP-gly secretion. Furthermore, secGFP-gly in the incubation medium was resistant to Endo H (Supplemental Fig. S9C), similar to that in the incubation medium of protoplasts transformed with HA:AtPRA1.F4, indicating that secGFP-gly is transported through the Golgi apparatus. These results raise the intriguing possibility that multiple pathways exist to transport PM and secretory proteins out of the Golgi apparatus.
HA:AtPRA1.F4 Overexpression Does Not Inhibit the Trafficking of Golgi-Independent Pathways
Two independent pathways exist for the trafficking of tonoplast intrinsic proteins (TIPs) to the tonoplast in Arabidopsis. GFP-TIP2;1 (GFP-δ-TIP) traffics to the tonoplast via Golgi-independent and Golgi-dependent mechanisms in hypocotyls and roots, respectively (Rivera-Serrano et al., 2012). In the case of TIP3;1, its trafficking to the tonoplast occurs in a Golgi-independent manner in leaf protoplasts (Park et al., 2004). We examined the effect of HA:AtPRA1.F4 overexpression on Golgi-independent trafficking pathways using δ-TIP (TIP2;1). Transgenic plants harboring YFP:δ-TIP were crossed with HA:AtPRA1.F4 OX plants. YFP:δ-TIP localization was examined in intact leaves and protoplasts of homozygous plants for both YFP:δ-TIP and HA:AtPRA1.F4. YFP:δ-TIP was detected at the tonoplast, similar to control plants not overexpressing HA:AtPRA1.F4 (Supplemental Fig. S10A). Next, we examined the trafficking of VHA-c3, which also is targeted to the tonoplast in a Golgi-independent manner (Seidel et al., 2013). VHA-c3:YFP localized to the tonoplast in both wild-type and HA:AtPRA1.F4 OX protoplasts (Supplemental Fig. S10B). These results indicate that proteins targeted to the vacuole by Golgi-independent pathways are not affected by HA:AtPRA1.F4 overexpression.
To corroborate these findings, we measured vacuolar pH because correct trafficking of vacuolar H+-ATPase subunits is crucial for the maintenance of proper vacuolar pH. The vacuolar H+-ATPase subunits Vha-a2 and Vha-a3 are targeted to the tonoplast via a Golgi-independent pathway (Viotti et al., 2013). The vacuolar pH in 6-d-old control and HA:AtPRA1.F4 OX seedlings was measured using the fluorescein-based ratiometric pH indicator BCECF-acetoxymethyl ester (Ozkan and Mutharasan, 2002; Krebs et al., 2010). The vacuolar pH in the roots of HA:AtPRA1.F4 OX plants was more acidic than that of control plants (Fig. 10A). To obtain supporting evidence for this finding, we measured the ATPase activity of vacuolar membrane fractions. Consistent with the vacuolar pH, the vacuolar ATPase activity was higher in HA:AtPRA1.F4 OX plants than in control plants (Fig. 10B). To confirm that the increased activity of vacuolar ATPase is the underlying cause of lower pH in the vacuole of HA:AtPRA1.F4 OX plants, wild-type and HA:AtPRA1.F4 OX seedlings were treated with concanamycin A (ConcA), a vacuolar ATPase inhibitor (Robinson and Pimpl, 2014; Kriegel et al., 2015). ConcA treatment significantly increased vacuolar pH to the same values in both wild-type and HA:AtPRA1.F4 OX plants (Fig. 10C), indicating that higher vacuolar ATPase activity contributes to the more acidic vacuolar pH in HA:AtPRA1.F4 OX plants. These results raise the possibility that protein trafficking to the TGN or tonoplast via Golgi-independent pathways may be facilitated by the inhibition of Golgi-dependent trafficking to these organelles, and this may be mediated by HA:AtPRA1.F4 expression.
Figure 10.
HA:AtPRA1.F4 OX plants display reduced vacuolar pH and higher levels of vacuolar ATPase activity. A, Vacuolar pH in root epidermal cells of 6-d-old seedlings was measured as described in “Materials and Methods.” Three independent experiments were performed with more than 50 cells from 15 seedlings in each experiment. The asterisk indicates a significant difference at P ≤ 0.05 (Student’s t test). Error bars represent sd (n = 202 in pBIB and n = 195 in HA:AtPRA1.F4 OX plants). B, Effect of AtPRA1.F4 overexpression on vacuolar ATPase activity. Vacuolar ATPase activity was measured using purified vacuolar proteins prepared from 17-d-old seedlings as described in “Materials and Methods.” Wild-type activity was set to 100%. The asterisk indicates a significant difference at P ≤ 0.05 calculated using Student’s t test. Error bars represent sd (n = 3). C, Effect of ConcA on vacuolar pH. Six-day-old seedlings of control plants and HA:AtPRA1.F4 OX plants were incubated in 1 μm ConcA or dimethyl sulfoxide (DMSO) for 20 h before measurement of pH. Asterisks indicate significant differences at P ≤ 0.05 (*) and P ≤ 0.01 (**) by Student’s t test. Error bars represent sd (n = 3).
Both HA:AtPRA1.F4 Overexpression and AtPRA1.F4 Knockdown Cause Vesiculation of the Golgi Apparatus
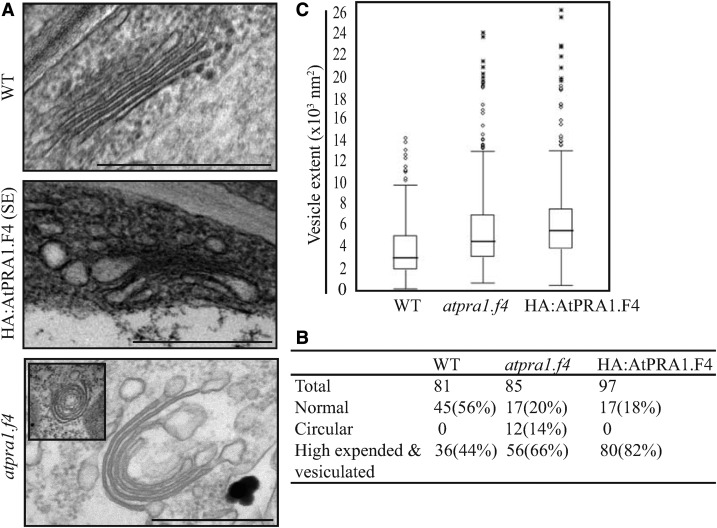
To elucidate the underlying cause of the defect in protein trafficking at the Golgi apparatus resulting from HA:AtPRA1.F4 overexpression or low levels of AtPRA1.F4, we examined whether HA:AtPRA1.F4 overexpression or AtPRA1.F4 knockdown has an effect on Golgi apparatus morphology. Ultrathin sections of leaf tissue were prepared, and the Golgi apparatus was examined by transmission electron microscopy (TEM). HA:AtPRA1.F4 OX and atpra1.f4 plants displayed abnormal Golgi apparatus, with a highly expanded and vesiculated or circular morphology, in contrast to the well-stacked Golgi apparatus in wild-type plants. Quantification revealed that 82% of the Golgi apparatus had a highly expanded and/or vesiculated morphology in HA:AtPRA1.F4 OX plants, and this figure was 80% in atpra1.f4 plants, whereas only 44% of the Golgi showed a small degree of expanded or vesiculated morphology in wild-type plants (Fig. 11). The two-dimensional sizes of vesicles associated with the Golgi apparatus in the TEM images were compared for HA:AtPRA1.F4 OX and wild-type plants. The average vesicle sizes in HA:AtPRA1.F4 OX, atpra1.f4, and wild-type plants were 5,444, 4,532, and 3,043 nm2, respectively. HA:AtPRA1.F4 OX and atpra1.f4 plants also contained extremely expanded vesicles compared with those of wild-type plants. These combined results indicate that both HA:AtPRA1.F4 overexpression and AtPRA1.F4 knockdown disrupt Golgi apparatus morphology.
Figure 11.
HA:AtPRA1.F4 OX and atpra1.f4 plants display highly altered Golgi apparatus. A, Golgi morphology. Ultrathin sections of leaf tissues from wild-type (WT), atpra1.f4, and HA:AtPRA1.F4 OX plants were examined by TEM. The inset in the atpra1.f4 image shows a circular form of the Golgi apparatus. Bars = 0.5 μm. B, Quantification of the vesiculated Golgi apparatus. The indicated numbers of Golgi apparatuses were grouped into two categories based on morphology. One group displayed normal morphology (representative image shown at top in A); the other group displayed highly expanded and/or vesiculated structures (representative image shown at bottom in A). Two or more of the Golgi-associated vesicles were larger than 3,000 nm2 (the size of the vesicle) and were considered to be an expanded form in the quantification. C, Effect of HA:AtPRA1.F4 overexpression and AtPRA1.F4 knockdown mutation on vesicle size. The vesicle volume was calculated using ImageJ (values are shown in the box plot). The horizontal line in each box represents the median value of the distribution. The boundaries of the box represent the lower and upper quartile values. The whiskers extending vertically from the bottom and top portions of each box represent the extent of the rest of the data. The number of Golgi apparatuses examined was 81, 85, and 97, respectively, for control, atpra1.f4, and HA:AtPRA1.F4 OX plants.
DISCUSSION
We investigated the physiological role of AtPRA1.F4 using transgenic plants overexpressing HA-tagged AtPRA1.F4 and atpra1.f4 plants in which AtPRA1.F4 was knocked down. Both HA:AtPRA1.F4 OX and atpra1.f4 plants exhibited altered phenotypes at the cellular and whole-plant levels. Of particular note, hypersensitivity to high concentrations of NaCl and KCl, higher pH in the apoplast, and lower vacuolar Na+/K+-ATPase and PM ATPase activities were shared by the two types of plants. By contrast, other phenotypes, such as an increase in the number of secondary roots and root hairs and delayed flowering, were specific to HA:AtPRA1.F4 OX plants, while atpra1.f4 plants were smaller in stature, had shorter roots, and exhibited a reduced seed yield. The difference in the phenotype between the two types of plants, atpra1.f4 and HA:AtPRA1.F4 OX, might have stemmed from the ectopic expression of AtPRA1.F4:HA by the strong CaMV 35S promoter. In Arabidopsis, a large number of AtPRA1 isoforms exist with varying degrees of sequence identity. They show differential spatiotemporal expression patterns. Moreover, these proteins form homodimers or heterodimers depending on individual isoforms (Alvim Kamei et al., 2008). Thus, HA:AtPRA1.F4 that was expressed nonspecifically in many tissues might have been be involved in nonspecific interactions with other isoforms or AtPRA1-interacting proteins. Morphological alterations in growth-related phenotypes are not easily understood based on the proposed function of PRA1 proteins. The defective trafficking of various proteins could indirectly cause such developmental and growth abnormalities. However, hypersensitivity to high concentrations of NaCl and KCl and an increase in apoplastic pH may be more easily understood based on the proposed function of PRA1, which is the delivery of prenylated Rabs to specific organelles for proper trafficking through endomembrane compartments. Consistent with the enhanced sensitivity to salt stress, HA:AtPRA1.F4 OX plants displayed lower vacuolar Na+/K+-ATPase and PM ATPase activities, which might have been caused by the defect in protein trafficking resulting from high levels of AtPRA1.F4. Indeed, previous studies showed that overexpression of PRA1 isoforms leads to the inhibition of protein trafficking at the ER in both animals and plants (Ruggiero et al., 2008; Lee et al., 2011). However, atpra1.f4 plants that had lower levels of AtPRA1.F4 also displayed lower vacuolar Na+/K+-ATPase and PM ATPase activities, similar to HA:AtPRA1.F4 OX plants. Thus, these results strongly suggest that appropriate levels of AtPRA1.F4 are critical for its normal function in protein trafficking.
PRA1 proteins are involved in delivering prenylated Rabs to their specific target organelles (Abdul-Ghani et al., 2001; Sivars et al., 2003; Seabra and Wasmeier, 2004). Therefore, the correct localization of PRA1 isoforms should be one of the key factors that allow them to perform their physiological role. We showed that HA:AtPRA1.F4 localized to the Golgi apparatus when it was expressed either stably in transgenic plants or transiently in protoplasts. The Golgi localization of HA:AtPRA1.F4 is consistent with the phenotype of HA:AtPRA1.F4 OX plants, which showed defective trafficking to both the PM and tonoplast. Moreover, electron microscopy analysis revealed that the Golgi apparatus in HA:AtPRA1.F4 OX and atpra1.f4 plants was highly vesiculated and/or expanded. The exact reason why high or low levels of HA:AtPRA1.F4 cause vesiculation of the Golgi apparatus is not currently understood. It has been shown that proper trafficking through the Golgi apparatus is crucial for maintaining its structural integrity (Fujiwara et al., 1988; Ritzenthaler et al., 2002). In addition, in animal cells, vesiculation of the Golgi apparatus is caused by the suppression or overexpression of certain types of Golgi-localized proteins or loading the Golgi with high levels of cholesterol (Dahan et al., 2005; Irannejad and Wedegaertner, 2010; Sato et al., 2011), which subsequently affects protein trafficking. In the case of vesiculation induced by high levels of cholesterol, the cell surface-localized protein VSVG still can be transported into, but not out of, the vesiculated Golgi apparatus (Dahan et al., 2005). PRA1 proteins are thought to be involved in the recruitment of Rabs; therefore, Golgi-localized AtPRA1.F4 may be involved in the recruitment of Rabs to the Golgi apparatus.
Multiple Rab proteins are known to be targeted to the Golgi apparatus. In this study, we did not address which Rabs are recruited to the Golgi apparatus by AtPRA1.F4. It is likely that multiple Rabs may be affected by AtPRA1.F4 overexpression or knockdown. The observed morphological phenotypes and/or defects in trafficking in HA:AtPRA1.F4 OX and atpra1.f4 plants are likely to be direct or indirect downstream effects resulting from the perturbation of a number of Rab proteins in these plants. Biochemical analysis revealed that HA:AtPRA1.F4 in transgenic plants formed a high-Mr complex, most likely via homomeric interactions. Homomeric oligomerization of AtPRA1.F4 may be an important biochemical property of AtPRA1.F4 function. This idea is supported by results showing that both high and low levels of AtPRA1.F4 resulted in similar defects in vacuolar trafficking: high levels of HA:AtPRA1.F4 may result in excessive oligomerization, whereas low levels of AtPRA1.F4 may result in insufficient complex formation, which in turn may lead to a failure to recruit Rabs to the Golgi apparatus or to release recruited Rabs to allow them to interact with downstream effector proteins. Thus, in either case, abnormal levels of AtPRA1.F4 may hamper the normal trafficking of most, but not all, post-Golgi proteins at the trans-Golgi, which in turn leads to abnormal Golgi structures with bigger vesicles. Currently, how the abnormal levels of AtPRA1.F4 lead to the generation of abnormally big vesicles or the accumulation of vesicles around the Golgi is not understood. However, in this condition, anterograde trafficking from the ER to the trans-Golgi still occurs normally, as observed with trafficking of ST:GFP and Kam1ΔC:mRFP to the trans-Golgi. According to the cisternal maturation model, no vesicles are involved in trafficking from the trans-Golgi to the TGN (Bonfanti et al., 1998). By contrast, this model proposes that vesicles produced at the trans-Golgi are involved in the retrograde trafficking of trans-Golgi-localized proteins to earlier cisternae to give rise to the TGN (Glick and Luini, 2011). Thus, it is possible that AtPRA1.4 plays a role in the retrograde trafficking at the Golgi apparatus. However, this idea should be further tested in the future.
The Golgi apparatus plays a crucial role in protein trafficking to various endomembrane compartments. Vesiculation of the Golgi apparatus resulting from HA:AtPRA1.F4 overexpression or knockdown may be the underlying cause of inhibition in various post-Golgi trafficking pathways. Indeed, similar to a previous finding in animal cells (Dahan et al., 2005), proteins could still be transported into, but not out of, a vesiculated Golgi apparatus. Intriguingly, the failure of proteins to exit from the vesiculated Golgi apparatus appeared to vary with the type of cargo. For example, of the TGN-localized SNAREs tested in this study (SYP41, SYP51, and SYP61), trafficking of SYP61 from vesiculated Golgi was essentially completely inhibited but that of SYP51 and SYP41 was only moderately inhibited and not inhibited, respectively. The trafficking of both soluble and membrane vacuolar proteins and PM proteins from vesiculated Golgi was strongly inhibited. However, the trafficking of secretory proteins was not inhibited. It is generally thought that secretory vesicles generated at the TGN carry both the PM and secretory proteins. After the fusion of these secretory vesicles to the PM, the secretory proteins within the vesicles will end up at the apoplast and the PM proteins on the vesicle membranes at the PM (Gu et al., 2001). Thus, one possibility is that the differential effects were exerted at the trans-Golgi but not at the TGN; secretory proteins were somehow transported to the TGN and then to the apoplast, whereas the trafficking of PM proteins was blocked at the trans-Golgi. In fact, ap1m2 plants, which have a mutation at the gene encoding one isoform of the two μ-adaptins of adaptor protein complex 1, also have an abnormal Golgi structure. These plants show a strong defect in the trafficking of PM proteins but a rather mild inhibition in the trafficking of secretory proteins (Park et al., 2013).
The mechanism underlying the differential inhibition of trafficking at the Golgi by HA:AtPRA1.F4 overexpression is intriguing. It is generally accepted that most proteins targeted to post-Golgi compartments are delivered from the ER to the Golgi apparatus and then sorted at the TGN, depending on their final destination (Nakatsu and Ohno, 2003; Bowers and Stevens, 2005; Newell-Litwa et al., 2007). The TGN, therefore, is the crucial sorting place for trafficking to post-Golgi compartments. However, it is not clearly understood how proteins are transported from the trans-Golgi cisternae to the TGN. In general, the Golgi is closely associated with the TGN, and they are transported together in the cell (Dettmer et al., 2006). One hypothesis is that the TGN arises from the trans-Golgi cisternae via cisternal maturation. Consistent with this hypothesis, Staehelin and Kang (2008) used electron tomography to show that the TGN in plants appears to arise from the trans-Golgi cisternae via cisternal peeling. If this is the case, no specific transport mechanism is necessary to transport cargo from the Golgi apparatus to the TGN. However, plant cells also contain Golgi-independent TGN (Uemura et al., 2014), which may require a specific mechanism for the trafficking of cargo from the Golgi apparatus to the TGN. A recent study showed that the majority of GNOM ARF-GEFs localize with GNL1 to distinct but partially overlapping Golgi subdomains (Naramoto et al., 2014), raising the possibility that the Golgi apparatus may consist of distinct domains that are involved in differential cargo trafficking. Our results here provide compelling evidence that trafficking from the trans-Golgi cisternae to the TGN may occur via multiple mechanisms. We found evidence for both AtPRA1.F4-dependent and AtPRA1.F4-independent mechanisms. Indeed, the sorting of specific cargo destined for the protein storage vacuole in plants starts at the cis-Golgi and continues throughout the Golgi apparatus (Hillmer et al., 2001; Hinz et al., 2007). Some secretory cargo molecules such as pectins and the cellulose synthase complex appear to exit the Golgi stack before reaching the TGN (Moore et al., 1991; Zhang and Staehelin, 1992; Crowell et al., 2009). However, it is not clearly understood what underlies these differential targeting pathways at the Golgi apparatus. Specific sorting signals are required for targeting vacuolar proteins to the vacuole (Matsuoka and Neuhaus, 1999), and PM proteins also contain a specific targeting signal (Barfield et al., 2009; Parmar et al., 2014), whereas secretory proteins do not contain any specific sorting signals and are transported out of the cell by the bulk-flow mechanism (Wieland et al., 1987; Denecke et al., 1990; Phillipson et al., 2001). One hypothesis is that the sorting signals of cargo proteins may play a role in the differential trafficking from the Golgi apparatus to the TGN, consistent with our results in plants overexpressing HA:AtPRA1.F4. However, we cannot rule out other possibilities. One of them is that a defect in the normal pathway may lead to alternative or abnormal pathways of trafficking from the trans-Golgi to the downstream compartments, which depends on the types of cargo. In fact, this has been observed in previous studies (Walworth et al., 1992; Geng et al., 2010; van der Vaart et al., 2010). Further studies are necessary to test the validity of these hypotheses and to determine the specific mechanism in the future.
MATERIALS AND METHODS
Plant Materials and Culture
Arabidopsis (Arabidopsis thaliana ecotype Columbia) was grown on B5 plates in a culture room under 70% relative humidity and a 16-h-light/8-h-dark cycle. Leaf tissues obtained from 15- to 18-d-old plants were used immediately for protoplast preparation (Jin et al., 2001).
Construction of 35S::HA:AtPRA1.F4 and Generation of Transgenic Plants Expressing HA:AtPRA1.F4
Full-length AtPRA1.F4 (At3G13710) cDNA was isolated by PCR using specific primers (5′-CGCGGATCCGAATGGCGAACAACGACGAGATT-3′ and 5′-CCGCTCGAGTTAAGACGAAGGGTGTGA-3′) and ligated into a pUC vector containing the CaMV 35S promoter, an HA tag, a multiple cloning site, and a NOS terminator to generate pUC-35S::HA:AtPRA1.F4. To generate transgenic plants constitutively overexpressing AtPRA1.F4, the HA:AtPRA1.F4 fragment from the pUC-35S::HA:AtPRA1.F4 construct was inserted into a pBIB binary vector using XbaI and KpnI restriction endonuclease sites. The resulting construct was introduced into Agrobacterium tumefaciens and used to generate transgenic plants by floral dipping (Clough and Bent, 1998).
Transient Expression, Immunofluorescence Staining, and Microscopy
Plasmid DNA was purified using Qiagen columns and introduced into Arabidopsis leaf cell protoplasts by polyethylene glycol-mediated transformation (Jin et al., 2001; Kim et al., 2001). Protoplasts were prepared from fresh leaf tissues (Lee et al., 2002; Jung et al., 2011). The expression of these constructs was monitored at various time points after transformation. Images of GFP fluorescence were obtained from intact protoplasts placed on glass slides and covered with a coverslip. For immunostaining, protoplasts on coverslips were fixed with 4% (v/v) paraformaldehyde (Park et al., 2004). Fixed protoplasts were labeled with anti-HA, anti-γ-COP, anti-AtRabF2a, or anti-SYP61 antibody as the primary antibody (Pimpl et al., 2000) followed by incubation with TRITC-labeled anti-rat or anti-rabbit IgG (Molecular Probes) for 3 h. Immunostained protoplasts were mounted in medium (120 mm Tris, pH 8.4, and 30% glycerol) containing Mowiol 4-88 (Calbiochem).
Images were captured using a Zeiss LSM 510 META laser scanning confocal microscope and processed using the LSM5 image browser and Adobe Photoshop 7. The excitation wavelength/emission filters used for mesophyll protoplasts and root tissues were 488 nm (argon ion laser)/505 to 530 bandpass for GFP/FITC and 543 nm/560 to 615 bandpass for RFP/TRITC. Images were obtained using a cooled CCD camera and a Zeiss Axioplan fluorescence microscope. The filter sets used for GFP/FITC and RFP/TRITC were XF116 (exciter, 474AF20; dichroic, 500DRLP; emitter, 510AF23) and XF33/E (exciter, 535DF35; dichroic, 570DRLP; emitter, 605DF50 [Omega]), respectively. Data were processed using Adobe Photoshop software, and images were rendered in pseudocolor.
Measurement of PM ATPase Activity
Protoplasts were prepared from 14-d-old plants (Jin et al., 2001) suspended in 5 mL of homogenizing buffer (330 mm Suc, 50 mm Tris-HCl, pH 7, 5 mm EDTA, 5 mm DTT, 0.5 mm phenylmethylsulfonyl fluoride, and 0.2% BSA) in the presence of 0.6% (w/v) polyvinylpyrrolidone and gently homogenized by passing through two layers of nylon filters (11-μm pore size; Millipore) three times. Extracts were centrifuged at 10,000g for 20 min. Supernatants were ultracentrifuged at 100,000g for 1 h, and the resulting precipitates were resuspended in suspension buffer (330 mm Suc, 5 mm potassium phosphate, pH 7.5, 5 mm KCl, 1 mm DTT, and 0.1 mm EDTA). The suspension (2 mL) was added to 10 mL of an aqueous two-phase mixture with a final composition of 6.5% (w/v) Dextran T500, 6.5% (w/v) PEG3350, 330 mm Suc, 5 mm potassium phosphate, pH 7.8, and 5 mm KCl. After centrifugation at 1,000g for 5 min, the resulting upper phase was mixed with dilution buffer (330 mm Suc, 5 mm Tris-Cl, pH 7.5, and 5 mm KCl) and centrifuged at 100,000g for 1 h. The pellet (containing PM) was resuspended in reaction buffer A (30 mm imidazole-HCl, pH 7.5, 50 mm KCl, 4 mm MgCl2, and 0.05% Brij 58).
ATPase activity was measured in samples containing 10 to 20 μg of PM proteins by generating inside-out vesicles using 0.05% Brij 58 (Sigma-Aldrich; Johansson et al., 1995). The vanadate-sensitive ATPase activity was calculated by subtracting the background signal in reaction buffer B (30 mm imidazole-HCl, pH 7.5, 50 mm KCl, 4 mm MgCl2, 2 mm sodium orthovanadate, and 0.05% Brij 580) from that in reaction buffer A. Sodium orthovanadate stock (100 mm) was prepared as described previously (Surowy and Sussman, 1986). The reaction mixture (1 mL) was incubated on ice for 1 h in the dark, and the reaction was initiated by adding 4 mm Tris-ATP (Tris hydroxymethyl aminomethane salt of ATP; Sigma-Aldrich). After a 10-min incubation at 23°C, the reaction was stopped by adding 100 μL of 20% SDS. To measure the activity, the reaction mixture (500 μL) was mixed with 500 μL of reagent A (3% ascorbic acid, 0.5 m HCl, and 0.5% ammonium molybdate) and incubated on ice for 10 min to enhance the color. Subsequently, 500 μL of reagent B (2% sodium meta-arsenite and 2% trisodium citrate) was added to the reaction and incubated at 37°C for 10 min. The activity was measured spectrophotometrically based on the A850. A850 values were converted to ATPase activity using a standard curve.
Measurement of Vacuolar Na+/K+-ATPase and Vacuolar ATPase Activities
Vacuoles were prepared from protoplasts according to a previously published method (Kim et al., 2005). Briefly, protoplasts were loaded onto a Ficoll step gradient consisting of 12% and 15% Ficoll in 0.6 m mannitol and 20 mm HEPES, pH 7.7, and subjected to ultracentrifugation at 150,000g for 2 h. The uppermost and pellet fractions containing intact vacuoles and nonvacuolar protoplasts, respectively, were collected separately. Vacuoles were pelleted from the top fraction by centrifugation at 19,000g for 10 min and diluted in buffer (5 mm HEPES-KOH, pH 7.5, 160 mm NaCl, and 1 mm EDTA). Purified vacuoles were used for vacuolar Na+/K+-ATPase activity assays or subjected to TCA precipitation for protein gel-blot analysis.
Vacuolar Na+/K+-ATPase activity was assayed according to a previously published method (Sarkar, 2002) with minor modifications. Vacuolar proteins (20 μg) were added to reaction buffer (30 mm imidazole-HCl, pH 7.4, 130 mm NaCl, 20 mm KCl, and 4 mm MgCl2) with or without 1 mm ouabain. The reaction mixture was incubated on ice in the dark for 60 min, and the reaction was initiated by adding 4 mm Tris-ATP (Sigma-Aldrich), followed by incubation at 37°C for 10 min. The total reaction volume was 1 mL. The reaction was stopped by adding 100 μL of 20% SDS. For inorganic phosphate estimation, 500 μL of reaction mixture (1.1-mL total volume) was quickly mixed with 500 μL of reagent A (3% ascorbic acid in 0.5 m HCl and 0.5% ammonium molybdate solution), and tubes were incubated on ice for 10 min. Subsequently, 500 μL of reaction mixture was mixed with 500 μL of reagent B (2% sodium meta-arsenite, 2% trisodium citrate, and 2% acetic acid) and incubated at 37°C. The A850 was measured at 10 min after the start of the incubation using a Beckman DU-800 spectrophotometer. Na+/K+-ATPase activity was calculated from the difference in inorganic phosphate content between reaction mixtures with or without 1 mm ouabain.
Vacuolar ATPase activity was measured as described for PM H+-ATPase activity measurement using vacuolar proteins. Brij 58, which was used to generate inside-out vesicles, was omitted from the buffer for measurement of PM ATPase activity.
Measurement of Vacuolar pH
Seedlings (5−6 d old) grown on one-half-strength MS agar plates supplemented with 1% Suc (pH 5.8) were transferred to liquid one-half-strength MS medium (pH 5.8) supplemented with 1% Suc and 10 μm BCECF in the presence of 0.02% pluronic acid and then incubated for 1 h at 22°C in the dark. Plants were treated with ConcA (1 μm) or the equivalent amount of DMSO (ConcA stock solution was prepared in DMSO) for the indicated times at room temperature before BCECF application.
Measurement of Apoplastic pH
Apoplastic pH was measured according to a previously published method (Cho et al., 2012; Villiers and Kwak, 2013). Leaves from 4- to 5-week-old plants were collected and immersed in 50 mL of water and then vacuumed for 5 min. The vacuum treatment was repeated four times. Subsequently, the leaf tissues were quickly dried with a paper towel and placed in a syringe without a plunger. The syringe was placed into a 1.5-mL conical tube, and the whole assembly was placed in a 50-mL conical tube and centrifuged at 1,000g for 10 min at 4°C. Apoplastic fluids were collected, and 160 µL was placed on a 96-well plate and mixed with 40 µL of 100 mg mL−1 8-hydroxypyrene-1,3,6-trisulfonic acid trisodium salt solution (Invitrogen). The fluorescence signal was measured at 510 nm after excitation at 460 nm using a plate reader (Tecan). The solution pH was determined from a standard curve generated using Britton-Robinson universal buffer (0.05 m H3BO3, 0.05 m H3PO4, and 0.05 m CH3COOH) adjusted to pH values ranging from 4 to 6 (in 0.5-unit increments) with 1 m NaOH.
Measurement of Proton Extrusion from Root Tissues (Rhizosphere pH)
To measure H+ efflux, plants were grown vertically on one-half-strength MS agar plates for 10 d and then transferred to one-half-strength MS liquid medium containing the pH indicator Bromocresol Purple (0.005%). At 2 d after the start of the incubation, the absorbance of the incubation medium was measured at 590 nm. The pH of the solution was determined from a standard curve (pH range ∼5.2–5.8).
Endo H Digestion
For Endo H treatments, protein extracts were denatured by adding 100 µL of 2× denaturation buffer (1% SDS and 2% β-mercaptoethanol) and boiled for 10 min. After cooling to 22°C, 100 µL of 2× reaction buffer (100 mm sodium citrate, pH 5.5) was added. The sample (100 µL) was mixed with 1 µL of Endo H (1 unit µL−1). The reaction was incubated at 37°C for 1 h and terminated by adding 1× SDS-PAGE loading buffer. Control samples were treated in the same way but without Endo H.
Electron Microscopy
Leaf tissue from 5-d-old seedlings was fixed with 2% paraformaldehyde- and 2% glutaraldehyde-containing sodium cacodylate buffer (50 mm sodium cacodylate, pH 7.2) at 4°C for 4 h. Samples were washed three times with sodium cacodylate buffer followed by postfixation with 1% osmium tetroxide in sodium cacodylate buffer at 4°C for 2 h. After washing briefly with distilled water, samples were treated with 0.5% uranyl acetate at 4°C for 30 min, followed by dehydration with a graded series of ethanol. Dehydrated samples were infiltrated with a graded series of ethanol and Spurr’s resin mixture before final embedding in 100% Spurr’s resin. Infiltrated samples were polymerized at 4°C for 24 h. Ultrathin sections were prepared with an ultramicrotome (MT-X; RMC), stained with 2% uranyl acetate and Reynolds’ lead citrate, and examined with a transmission electron microscope (JEM-1010; JEOL) at 80 kV. The two-dimensional size of vesicles was calculated using ImageJ software, and data were processed to yield a box plot with Fast Statistic version 2.0.4.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AtPRA1.F4 (At3G13710), AtPRA1.F3 (At3G13720), Actin2 (At3G18780).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of atpra1.f4 plants.
Supplemental Figure S2. HA:AtPRA1.F4 forms highly stable high-Mr complexes.
Supplemental Figure S3. AtPRA1.F4 OX plants show alterations in root hair development and flowering time.
Supplemental Figure S4. Trafficking of stably expressed AALP:GFP is inhibited by HA:AtPRA1.F4 overexpression.
Supplemental Figure S5. Trafficking of AtβFruct4:GFP is inhibited by HA:AtPRA1.F4 overexpression.
Supplemental Figure S6. HA:AtPRA1.F4 overexpression inhibits the Golgi-dependent trafficking of vacuolar membrane proteins at the Golgi apparatus.
Supplemental Figure S7. GFP:SYP61 accumulates at the Golgi apparatus in HA:AtPRA1.F4 OX plants.
Supplemental Figure S8. HA:AtPRA1.F4 inhibits PMP:GFP-gly trafficking.
Supplemental Figure S9. AtPRA1.F4 knockdown does not inhibit the trafficking of secretory proteins.
Supplemental Figure S10. HA:AtPRA1.F4 overexpression does not affect the Golgi-independent trafficking of vacuolar proteins.
Supplemental Table S1. Nucleotide sequences of primers used for qRT-PCR.
Glossary
- ER
endoplasmic reticulum
- CaMV
cauliflower mosaic virus
- PVC
prevacuolar compartment
- PM
plasma membrane
- TGN
trans-Golgi network
- ConcA
concanamycin A
- TEM
transmission electron microscopy
- TRITC
tetramethylrhodamine
- FITC
fluorescein isothiocyanate
- MS
Murashige and Skoog
- DMSO
dimethyl sulfoxide
Footnotes
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, Information & Communication Technology (ICT) & Future Planning (2016R1E1A1A02922014).
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ010953012017)” Rural Development Administration, Republic of Korea (PJ010953012017).
References
- Abdul-Ghani M, Gougeon PY, Prosser DC, Da-Silva LF, Ngsee JK (2001) PRA isoforms are targeted to distinct membrane compartments. J Biol Chem 276: 6225–6233 [DOI] [PubMed] [Google Scholar]
- Alvim Kamei CL, Boruc J, Vandepoele K, Van den Daele H, Maes S, Russinova E, Inzé D, De Veylder L (2008) The PRA1 gene family in Arabidopsis. Plant Physiol 147: 1735–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barfield RM, Fromme JC, Schekman R (2009) The exomer coat complex transports Fus1p to the plasma membrane via a novel plasma membrane sorting signal in yeast. Mol Biol Cell 20: 4985–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfanti L, Mironov AA Jr, Martínez-Menárguez JA, Martella O, Fusella A, Baldassarre M, Buccione R, Geuze HJ, Mironov AA, Luini A (1998) Procollagen traverses the Golgi stack without leaving the lumen of cisternae: evidence for cisternal maturation. Cell 95: 993–1003 [DOI] [PubMed] [Google Scholar]
- Bowers K, Stevens TH (2005) Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1744: 438–454 [DOI] [PubMed] [Google Scholar]
- Calero M, Winand NJ, Collins RN (2002) Identification of the novel proteins Yip4p and Yip5p as Rab GTPase interacting factors. FEBS Lett 515: 89–98 [DOI] [PubMed] [Google Scholar]
- Chavrier P, Goud B (1999) The role of ARF and Rab GTPases in membrane transport. Curr Opin Cell Biol 11: 466–475 [DOI] [PubMed] [Google Scholar]
- Cho D, Villiers F, Kroniewicz L, Lee S, Seo YJ, Hirschi KD, Leonhardt N, Kwak JM (2012) Vacuolar CAX1 and CAX3 influence auxin transport in guard cells via regulation of apoplastic pH. Plant Physiol 160: 1293–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colicelli J. (2004) Human RAS superfamily proteins and related GTPases. Sci STKE 2004: RE13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Höfte H, Vernhettes S (2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan S, Anderson KL, Weller S, Krueger E, McNiven MA (2005) Agonist-induced vesiculation of the Golgi apparatus in pancreatic acinar cells. Gastroenterology 129: 2032–2046 [DOI] [PubMed] [Google Scholar]
- Denecke J, Botterman J, Deblaere R (1990) Protein secretion in plant cells can occur via a default pathway. Plant Cell 2: 51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP (2008) Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc 3: 619–628 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y (1988) Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem 263: 18545–18552 [PubMed] [Google Scholar]
- Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ (2010) Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell 21: 2257–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS, Luini A (2011) Models for Golgi traffic: a critical assessment. Cold Spring Harb Perspect Biol 3: a005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glomset JA, Farnsworth CC (1994) Role of protein modification reactions in programming interactions between ras-related GTPases and cell membranes. Annu Rev Cell Biol 10: 181–205 [DOI] [PubMed] [Google Scholar]
- Gu F, Crump CM, Thomas G (2001) Trans-Golgi network sorting. Cell Mol Life Sci 58: 1067–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes CR, Brandizzi F, Andreeva AV (1999) Endomembranes and vesicle trafficking. Curr Opin Plant Biol 2: 454–461 [DOI] [PubMed] [Google Scholar]
- Heo JB, Bang WY, Kim SW, Hwang SM, Son YS, Im CH, Acharya BR, Kim CW, Kim SW, Lee BH, et al. (2010) OsPRA1 plays a significant role in targeting of OsRab7 into the tonoplast via the prevacuolar compartment during vacuolar trafficking in plant cells. Planta 232: 861–871 [DOI] [PubMed] [Google Scholar]
- Hillmer S, Movafeghi A, Robinson DG, Hinz G (2001) Vacuolar storage proteins are sorted in the cis-cisternae of the pea cotyledon Golgi apparatus. J Cell Biol 152: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz G, Colanesi S, Hillmer S, Rogers JC, Robinson DG (2007) Localization of vacuolar transport receptors and cargo proteins in the Golgi apparatus of developing Arabidopsis embryos. Traffic 8: 1452–1464 [DOI] [PubMed] [Google Scholar]
- Huang Z, Andrianov VM, Han Y, Howell SH (2001) Identification of Arabidopsis proteins that interact with the cauliflower mosaic virus (CaMV) movement protein. Plant Mol Biol 47: 663–675 [DOI] [PubMed] [Google Scholar]
- Irannejad R, Wedegaertner PB (2010) Regulation of constitutive cargo transport from the trans-Golgi network to plasma membrane by Golgi-localized G protein betagamma subunits. J Biol Chem 285: 32393–32404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I (2001) A new dynamic-like proetin, ADL6, is involved in trafficking from the trans-Gogi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson F, Olbe M, Sommarin M, Larsson C (1995) Brij 58, a polyoxyethylene acyl ether, creates membrane vesicles of uniform sidedness: a new tool to obtain inside-out (cytoplasmic side-out) plasma membrane vesicles. Plant J 7: 165–173 [DOI] [PubMed] [Google Scholar]
- Jung CJ, Lee MH, Min MK, Hwang I (2011) Localization and trafficking of an isoform of the AtPRA1 family to the Golgi apparatus depend on both N- and C-terminal sequence motifs. Traffic 12: 185–200 [DOI] [PubMed] [Google Scholar]
- Kim DH, Eu YJ, Yoo CM, Kim YW, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I (2001) Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13: 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Park M, Kim SJ, Hwang I (2005) Actin filaments play a critical role in vacuolar trafficking at the Golgi complex in plant cells. Plant Cell 17: 888–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T. (2000) Clathrin. Annu Rev Biochem 69: 699–727 [DOI] [PubMed] [Google Scholar]
- Kotzer AM, Brandizzi F, Neumann U, Paris N, Moore I, Hawes C (2004) AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J Cell Sci 117: 6377–6389 [DOI] [PubMed] [Google Scholar]
- Krebs M, Beyhl D, Görlich E, Al-Rasheid KAS, Marten I, Stierhof YD, Hedrich R, Schumacher K (2010) Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc Natl Acad Sci USA 107: 3251–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel A, Andrés Z, Medzihradszky A, Krüger F, Scholl S, Delang S, Patir-Nebioglu MG, Gute G, Yang H, Murphy AS, et al. (2015) Job sharing in the endomembrane system: vacuolar acidification requires the combined activity of V-ATPase and V-PPase. Plant Cell 27: 3383–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar T, Götte M, Gallwitz D (1997) Vesicular transport: how many Ypt/Rab-GTPases make a eukaryotic cell? Trends Biochem Sci 22: 468–472 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Sohn EJ, Lee MH, Hwang I (2004) The Arabidopsis rab5 homologs rha1 and ara7 localize to the prevacuolar compartment. Plant Cell Physiol 45: 1211–1220 [DOI] [PubMed] [Google Scholar]
- Lee MH, Jung C, Lee J, Kim SY, Lee Y, Hwang I (2011) An Arabidopsis prenylated Rab acceptor 1 isoform, AtPRA1.B6, displays differential inhibitory effects on anterograde trafficking of proteins at the endoplasmic reticulum. Plant Physiol 157: 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Min MK, Lee YJ, Jin JB, Shin DH, Kim DH, Lee KH, Hwang I (2002) ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiol 129: 1507–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Veeraprame H, Bayan N, Li G (2004) The C-terminus of prenylin is important in forming a dimer conformation necessary for endoplasmic-reticulum-to-Golgi transport. Biochem J 380: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HP, Wu CC, Chang YS (2006) PRA1 promotes the intracellular trafficking and NF-kappaB signaling of EBV latent membrane protein 1. EMBO J 25: 4120–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Neuhaus JM (1999) Cis-elements of protein transport to the vacuoles. J Exp Bot 50: 165–174 [Google Scholar]
- Moore PJ, Swords KM, Lynch MA, Staehelin LA (1991) Spatial organization of the assembly pathways of glycoproteins and complex polysaccharides in the Golgi apparatus of plants. J Cell Biol 112: 589–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F, Ohno H (2003) Adaptor protein complexes as the key regulators of protein sorting in the post-Golgi network. Cell Struct Funct 28: 419–429 [DOI] [PubMed] [Google Scholar]
- Naramoto S, Otegui MS, Kutsuna N, de Rycke R, Dainobu T, Karampelias M, Fujimoto M, Feraru E, Miki D, Fukuda H, Nakano A, Friml J (2014) Insights into the localization and function of the membrane trafficking regulator GNOM ARF-GEF at the Golgi apparatus in Arabidopsis. Plant Cell 26: 3062–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Litwa K, Seong E, Burmeister M, Faundez V (2007) Neuronal and non-neuronal functions of the AP-3 sorting machinery. J Cell Sci 120: 531–541 [DOI] [PubMed] [Google Scholar]
- Novick P, Zerial M (1997) The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol 9: 496–504 [DOI] [PubMed] [Google Scholar]
- Ozkan P, Mutharasan R (2002) A rapid method for measuring intracellular pH using BCECF-AM. Biochim Biophys Acta 1572: 143–148 [DOI] [PubMed] [Google Scholar]
- Park M, Kim SJ, Vitale A, Hwang I (2004) Identification of the protein storage vacuole and protein targeting to the vacuole in leaf cells of three plant species. Plant Physiol 134: 625–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Song K, Reichardt I, Kim H, Mayer U, Stierhof YD, Hwang I, Jürgens G (2013) Arabidopsis μ-adaptin subunit AP1M of adaptor protein complex 1 mediates late secretory and vacuolar traffic and is required for growth. Proc Natl Acad Sci USA 110: 10318–10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar HB, Barry C, Kai F, Duncan R (2014) Golgi complex-plasma membrane trafficking directed by an autonomous, tribasic Golgi export signal. Mol Biol Cell 25: 866–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC (2000) The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol 301: 1077–1087 [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Aivazian D (2004) Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol 5: 886–896 [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. (2001) Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol 11: 487–491 [DOI] [PubMed] [Google Scholar]
- Phillipson BA, Pimpl P, daSilva LLP, Crofts AJ, Taylor JP, Movafeghi A, Robinson DG, Denecke J (2001) Secretory bulk flow of soluble proteins is efficient and COPII dependent. Plant Cell 13: 2005–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpl P, Movafeghi A, Coughlan S, Denecke K, Hillmer S, Robinson DG (2000) In situ localization and in vitro induction of plant COPI-coated vesicles. Plant Cell 12: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzenthaler C, Nebenführ A, Movafeghi A, Stussi-Garaud C, Behnia L, Pimpl P, Staehelin LA, Robinson DG (2002) Reevaluation of the effects of brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell 14: 237–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Serrano EE, Rodriguez-Welsh MF, Hicks GR, Rojas-Pierce M (2012) A small molecule inhibitor partitions two distinct pathways for trafficking of tonoplast intrinsic proteins in Arabidopsis. PLoS ONE 7: e44735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DG, Pimpl P (2014) Clathrin and post-Golgi trafficking: a very complicated issue. Trends Plant Sci 19: 134–139 [DOI] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT (1996) Protein sorting by transport vesicles. Science 272: 227–234 [DOI] [PubMed] [Google Scholar]
- Ruggiero AM, Liu Y, Vidensky S, Maier S, Jung E, Farhan H, Robinson MB, Sitte HH, Rothstein JD (2008) The endoplasmic reticulum exit of glutamate transporter is regulated by the inducible mammalian Yip6b/GTRAP3-18 protein. J Biol Chem 283: 6175–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito-Nakano Y, Loftus BJ, Hall N, Nozaki T (2005) The diversity of Rab GTPases in Entamoeba histolytica. Exp Parasitol 110: 244–252 [DOI] [PubMed] [Google Scholar]
- Sarkar PK. (2002) A quick assay for Na+-K+-AtPase specific activity. Z Naturforsch C 57: 562–564 [DOI] [PubMed] [Google Scholar]
- Sato M, Saegusa K, Sato K, Hara T, Harada A, Sato K (2011) Caenorhabditis elegans SNAP-29 is required for organellar integrity of the endomembrane system and general exocytosis in intestinal epithelial cells. Mol Biol Cell 22: 2579–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R, Orci L (1996) Coat proteins and vesicle budding. Science 271: 1526–1533 [DOI] [PubMed] [Google Scholar]
- Seabra MC, Goldstein JL, Südhof TC, Brown MS (1992) Rab geranylgeranyl transferase: a multisubunit enzyme that prenylates GTP-binding proteins terminating in Cys-X-Cys or Cys-Cys. J Biol Chem 267: 14497–14503 [PubMed] [Google Scholar]
- Seabra MC, Wasmeier C (2004) Controlling the location and activation of Rab GTPases. Curr Opin Cell Biol 16: 451–457 [DOI] [PubMed] [Google Scholar]
- Segev N. (2001) Ypt and Rab GTPases: insight into functions through novel interactions. Curr Opin Cell Biol 13: 500–511 [DOI] [PubMed] [Google Scholar]
- Seidel T, Siek M, Marg B, Dietz KJ (2013) Energization of vacuolar transport in plant cells and its significance under stress. Int Rev Cell Mol Biol 304: 57–131 [DOI] [PubMed] [Google Scholar]
- Sivars U, Aivazian D, Pfeffer S (2005) Purification and properties of Yip3/PRA1 as a Rab GDI displacement factor. Methods Enzymol 403: 348–356 [DOI] [PubMed] [Google Scholar]
- Sivars U, Aivazian D, Pfeffer SR (2003) Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature 425: 856–859 [DOI] [PubMed] [Google Scholar]
- Sohn EJ, Kim ES, Zhao M, Kim SJ, Kim H, Kim YW, Lee YJ, Hillmer S, Sohn U, Jiang L, et al. (2003) Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15: 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somsel Rodman J, Wandinger-Ness A (2000) Rab GTPases coordinate endocytosis. J Cell Sci 113: 183–192 [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Kang BH (2008) Nanoscale architecture of endoplasmic reticulum export sites and of Golgi membranes as determined by electron tomography. Plant Physiol 147: 1454–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surowy TK, Sussman MR (1986) Immunological cross-reactivity and inhibitor sensitivities of the plasma membrane H+-ATPase from plants and fungi. Biochim Biophys Acta 848: 24–34 [Google Scholar]
- Takeuchi M, Ueda T, Sato K, Abe H, Nagata T, Nakano A (2000) A dominant negative mutant of sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J 23: 517–525 [DOI] [PubMed] [Google Scholar]
- Tamura K, Shimada T, Kondo M, Nishimura M, Hara-Nishimura I (2005) KATAMARI1/MURUS3 is a novel Golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17: 1764–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmapuram J, Duan H, Liu L, Schuler MA (2005) Bicistronic and fused monocistronic transcripts are derived from adjacent loci in the Arabidopsis genome. RNA 11: 128–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Suda Y, Ueda T, Nakano A (2014) Dynamic behavior of the trans-Golgi network in root tissues of Arabidopsis revealed by super-resolution live imaging. Plant Cell Physiol 55: 694–703 [DOI] [PubMed] [Google Scholar]
- Ueda T, Nakano A (2002) Vesicular traffic: an integral part of plant life. Curr Opin Plant Biol 5: 513–517 [DOI] [PubMed] [Google Scholar]
- van der Vaart A, Griffith J, Reggiori F (2010) Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell 21: 2270–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol 131: 1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villiers F, Kwak JM (2013) Rapid apoplastic pH measurement in Arabidopsis leaves using a fluorescent dye. Plant Signal Behav 8: e22587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti C, Krüger F, Krebs M, Neubert C, Fink F, Lupanga U, Scheuring D, Boutté Y, Frescatada-Rosa M, Wolfenstetter S, et al. (2013) The endoplasmic reticulum is the main membrane source for biogenesis of the lytic vacuole in Arabidopsis. Plant Cell 25: 3434–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth NC, Brennwald P, Kabcenell AK, Garrett M, Novick P (1992) Hydrolysis of GTP by Sec4 protein plays an important role in vesicular transport and is stimulated by a GTPase-activating protein in Saccharomyces cerevisiae. Mol Cell Biol 12: 2017–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MG, Hughson FM (2000) Membrane tethering and fusion in the secretory and endocytic pathways. Traffic 1: 588–597 [DOI] [PubMed] [Google Scholar]
- Wee EGT, Sherrier DJ, Prime TA, Dupree P (1998) Targeting of active sialyltransferase to the plant Golgi apparatus. Plant Cell 10: 1759–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland FT, Gleason ML, Serafini TA, Rothman JE (1987) The rate of bulk flow from the endoplasmic reticulum to the cell surface. Cell 50: 289–300 [DOI] [PubMed] [Google Scholar]
- Yang X, Matern HT, Gallwitz D (1998) Specific binding to a novel and essential Golgi membrane protein (Yip1p) functionally links the transport GTPases Ypt1p and Ypt31p. EMBO J 17: 4954–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, McBride H (2001) Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2: 107–117 [DOI] [PubMed] [Google Scholar]
- Zhang GF, Staehelin LA (1992) Functional compartmentation of the Golgi apparatus of plant cells: immunocytochemical analysis of high-pressure frozen- and freeze-substituted sycamore maple suspension culture cells. Plant Physiol 99: 1070–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, Acar M, Hoskins RA, Bellen HJ, Scott MP (2007) Thirty-one flavors of Drosophila rab proteins. Genetics 176: 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]