The animal-like cryptochrome, which accumulates and is located differentially during the Chlamydomonas life cycle, controls germination in a positive and gametogenesis in a negative manner.
Abstract
Cryptochromes are known as flavin-binding blue light receptors in bacteria, fungi, plants, and insects. The animal-like cryptochrome (aCRY) of the green alga Chlamydomonas reinhardtii has extended our view on cryptochromes, because it responds also to other wavelengths of the visible spectrum, including red light. Here, we have investigated if aCRY is involved in the regulation of the sexual life cycle of C. reinhardtii, which is controlled by blue and red light at the steps of gametogenesis along with its restoration and germination. We show that aCRY is differentially expressed not only during the life cycle but also within the cell as part of the soluble and/or membrane-associated protein fraction. Moreover, localization of aCRY within the algal cell body varies between vegetative cells and the different cell types of gametogenesis. aCRY is significantly (early day) or to a small extent (late night) enriched in the nucleus in vegetative cells. In pregametes, gametes and dark-inactivated gametes, aCRY is localized over the cell body. aCRY plays an important role in the sexual life cycle of C. reinhardtii: It controls the germination of the alga, under which the zygote undergoes meiosis, in a positive manner, similar to the regulation by the blue light receptors phototropin and plant cryptochrome (pCRY). However, aCRY acts in combination with pCRY as a negative regulator for mating ability as well as for mating maintenance, opposite to the function of phototropin in these processes.
Light is an essential environmental factor for photosynthetic organisms, serving as a source of energy and signal information. To precisely perceive and respond to different wavelengths of the light spectrum, eukaryotic photosynthetic microorganisms and higher plants have developed different classes of light-sensitive receptors, including cryptochromes (CRYs), phototropins (PHOTs), phytochromes, UV-B resistance locus8, rhodopsins, and aureochromes (Christie, 2007; Hegemann, 2008; Kianianmomeni and Hallmann, 2014; Galvão and Fankhauser, 2015).
The green biflagellate alga Chlamydomonas reinhardtii (named Chlamydomonas hereafter) has served for many years not only as a model organism to study photosynthesis and flagella (Merchant et al., 2007), but has also been heavily explored with regard to photoperception. Chlamydomonas has a primitive visual system, the eyespot apparatus, where several photoreceptors related to photo-orientation are located. These include two channelrhodopsins (ChR1 and 2), at least one His kinase rhodopsin (HKR1), and one PHOT (Schmidt et al., 2006; Hegemann, 2008; Luck et al., 2012). ChR1 and 2 are microbial rhodopsins that act as light-gated ion channels and absorb light mainly in the green and blue region of the visible spectrum to mediate phototaxis and photophobic responses (Nagel et al., 2002, 2003; Sineshchekov et al., 2002; Govorunova et al., 2004). Recently, it was found that trafficking of these rhodopsins into the eyespot is even connected with the intraflagellar transport (Awasthi et al., 2016). Less is known about the His kinase rhodopsin HKR1, which is bimodally switched by absorbing UV-A and blue light, respectively, in vitro and has been suggested to mediate UV avoidance (Luck et al., 2012). Chlamydomonas PHOT is a well-studied blue light sensory photoreceptor. It was shown to be involved in the development of the eyespot size in vegetative cells and in the adjustment of the expression level of ChR1 (Trippens et al., 2012).
Photoreceptors play also important roles besides in the visual system of Chlamydomonas. PHOT is additionally present in the flagella and the cell body (Huang et al., 2004). It regulates the light-induced expression level of genes related to chlorophyll and carotenoid biosynthesis and light-harvesting complexes, both under blue and red light (Im et al., 2006) as well as the switch-off of chemotaxis toward nitrite in pregametes (Ermilova et al., 2004). In gametes, it regulates phototaxis (Trippens et al., 2012). Moreover, it participates in the regulation of light-dependent steps in the sexual life cycle as outlined below (Huang and Beck, 2003). Recently, it was shown that PHOT acts also as a photoreceptor that mediates the feedback regulation of photosynthesis for high light acclimation (Petroutsos et al., 2016). Besides PHOT, a functional UV-B resistance locus8 homolog was found to induce UV-B acclimation in Chlamydomonas by initiating the signaling pathway via interacting with COP1 in a UV-B dependent manner (Tilbrook et al., 2016).
As members of a photoreceptor family of emerging complexity, four CRYs are encoded in Chlamydomonas, an animal-like CRY (aCRY) and a plant CRY (pCRY, formerly known as Chlamydomonas photolyase homolog 1) as well as two so-called CRY-DASH (Drosophila, Arabidopsis, Synechocystis, human) proteins (Beel et al., 2012; Müller et al., 2017). CRYs bind flavin adenine dinucleotide and share sequence homology and structural similarity with photolyases, which are enzymes that repair DNA photolesions caused by UV-B light (Chaves et al., 2011). CRY photoreceptors usually lack DNA repair activity, but dual functions have been demonstrated for cryptochrome photolyase family1 members of the marine diatom Phaeodactylum tricornutum and the primitive green alga Oestreococcus tauri (summarized in Fortunato et al., 2015). CRYs share a specific nonconserved C-terminal extension used for signal transduction (Chaves et al., 2011). Most studies of Chlamydomonas CRYs have been performed on pCRY. In vitro analysis of the photolyase homology region of pCRY showed that blue light illumination induces the formation of the flavin neutral radical, autophosphorylation, and changes in conformation (Immeln et al., 2007; Immeln et al., 2010; Thöing et al., 2015). In vivo studies have suggested that pCRY is rapidly degraded by the proteasome pathway in a blue- and red-light-dependent manner (Reisdorph and Small, 2004). pcry knockdown lines with a reduced pCRY level down to approximately 11% revealed that pCRY is not only involved in entraining the Chlamydomonas circadian clock by blue light (Forbes-Stovall et al., 2014; Müller et al., 2017), but it seems also associated with the circadian oscillator (Müller et al., 2017). Moreover, pCRY is involved in the control of the sexual cycle (Müller et al., 2017).
Recently, aCRY was functionally characterized in Chlamydomonas using an insertional acry mutant with a strongly reduced level (down to approximately 20%) compared to wild type (100%). In response to blue and red light, the transcript levels of several genes of chlorophyll and carotenoid biosynthesis, light-harvesting complexes, nitrogen metabolism, the cell cycle, and the circadian clock were strongly altered in the wild type but significantly less changed in the acry mutant (Beel et al., 2012, 2013). Moreover, yellow but not far-red light caused comparable changes on the transcript level of selected genes. These data are in agreement with in vitro data showing that blue, yellow, and red light is absorbed by the neutral radical state of flavin in aCRY, but not far-red light. Thus, it was assumed that the neutral radical of the flavin chromophore acts as a dark form of the sensor, which absorbs in almost the entire visible spectrum (below 680 nm). A conversion by red light of the neutral radical state to the anionic fully reduced state was found by spectroscopic analyses to go hand in hand with conformational changes of aCRY (Spexard et al., 2014). In contrast, blue light illumination of the oxidized flavin did not elicit such change, albeit easily producing the neutral radical form (Spexard et al., 2014; Nohr et al., 2016). Moreover, an essential role of an unusually long-lived tyrosyl radical at position 373 of aCRY was found recently to be involved in the red light response (Oldemeyer et al., 2016). This residue is only conserved in close homologs, which points to the existence of a separate aCRY subfamily within the CRY family. Heterologously expressed and purified aCRY forms a dimer in the dark, while partial oligomerization was observed upon illumination via disulfide bridge formation at Cys 482 (Oldemeyer et al., 2016). Despite these advances at the molecular level, the in vivo functions of aCRY remain elusive to a large extent.
It is known that light as well as growth conditions influence the sexual life cycle of Chlamydomonas. Nitrogen depletion leads to the conversion of haploid vegetative cells to pregametes having mating type plus or minus (Huang and Beck, 2003; Goodenough et al., 2007). Illumination provokes the transition from pregametes to gametes, which achieve full mating ability (Beck and Acker, 1992). While some Chlamydomonas strains do not differentiate at all into gametes in the dark because they absolutely require light for this process, others can differentiate to gametes in the dark to a small extent, but need light for higher conversion rates (Saito et al., 1998). Gamete maintenance is also dependent on light. Consequent dark treatment of mature gametes leads to the loss of mating ability producing so called dark-inactivated gametes. Further light exposure restores the mating ability (reactivated gametes). The conversion from pregametes to gametes and from dark-inactivated gametes to reactivated gametes, also known as gametogenesis and restoration, is mainly influenced by blue light, but to some extent also by red light (Weissig and Beck, 1991; Pan et al., 1997), indicating the participation of blue and/or red light photoreceptors in these processes. When mature gametes of the plus and minus mating types are mixed, they will mate and form diploid zygotes. The germination process of the zygote into four haploid vegetative cells again requires light as well as nitrogen-containing medium. In darkness, zygotes become dormant cells (Gloeckner and Beck, 1995).
Previous studies showed that PHOT and pCRY are blue light receptors involved in the light-regulated steps of gametogenesis and zygote germination (Huang and Beck, 2003; Müller et al., 2017). So far, there is no evidence for a red light-absorbing photoreceptor involved in the sexual life cycle of Chlamydomonas. Because phytochrome is not encoded in the Chlamydomonas genome (Mittag et al., 2005; Merchant et al., 2007), aCRY is a candidate for playing also a role in the process of gametogenesis and/or zygote germination in addition to PHOT and pCRY. Here, we characterized the expression level and the subcellular localization of aCRY at different stages of the Chlamydomonas sexual cycle and found significant differences during its life cycle. Moreover, we detected the involvement of aCRY in all light-regulated steps of the sexual cycle, either in a positive or negative manner.
RESULTS
aCRY Is Primarily Soluble and Is Found in Complexes in Vegetative Cells of Chlamydomonas
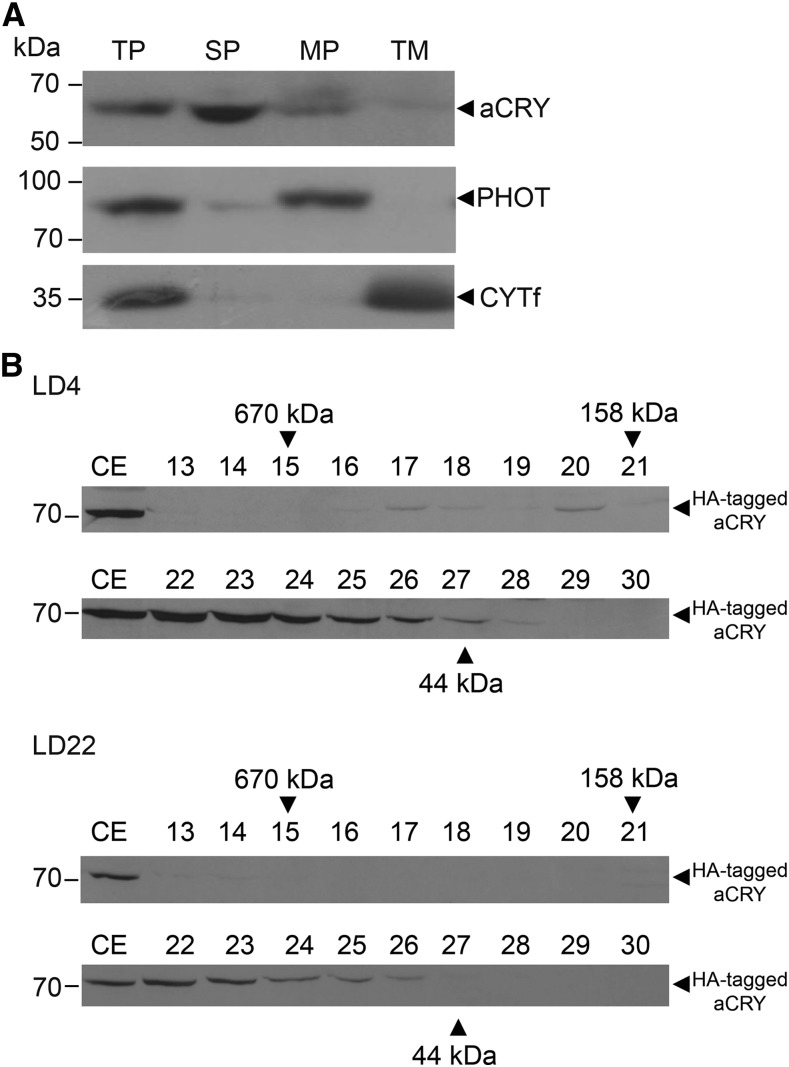
Soluble aCRY protein accumulates to its highest level at the beginning of the day in vegetative cells and is present at its lowest level at the beginning of the night (Beel et al., 2012). To investigate if aCRY may be associated also with membranes similar to the Chlamydomonas blue light receptor PHOT, we prepared subfractions of vegetative cells harvested during the day (LD4). For this purpose, total proteins were separated into fractions containing soluble, peripheral membrane, and transmembrane proteins (see “Materials and Methods”) and analyzed on immunoblots along with anti-aCRY antibodies. Anti-LOV1 (PHOT) and anti-cytochrome f (CYTf) antibodies were used as controls for a membrane-associated (PHOT) and a transmembrane (CYTf) protein, respectively. As shown previously (Huang et al., 2002), PHOT was strongly enriched in the peripheral membrane fraction, weakly present in the soluble fraction and missing in the transmembrane fraction (Fig. 1A). CYTf is known to be located in thylakoid membranes (Willey et al., 1984; Schulze et al., 2013) and appeared predominantly in the transmembrane fraction (Fig. 1A). aCRY was mainly present in the soluble fraction, but also detectable in the membrane-associated fraction to a smaller extent (Fig. 1A). It did not accumulate to any significant level in the transmembrane protein fraction, consistent with the theoretical prediction by the TMHMM Server (Sonnhammer et al., 1998) that it lacks transmembrane domains. As a result, aCRY is predominantly present as a soluble protein but also has the ability to associate with membranes in vegetative cells.
Figure 1.
aCRY is mainly present as soluble protein and forms complexes during day and night. A, Soluble proteins (SP), membrane-associated proteins (MP), and transmembrane proteins (TM) of total proteins (TP) were prepared from vegetative cells harvested at LD4 of the mt+ strain (see “Materials and Methods”). Thirty-microgram proteins of each fraction were separated by 9% SDS-PAGE and immunoblotted with anti-aCRY, anti-LOV1 (PHOT), and anti-CYTf antibodies, respectively (see “Materials and Methods”). B, Complex formation of aCRY in Chlamydomonas. Cells from a 3 × HA tagged-aCRY line (see “Materials and Methods”) were harvested at LD4 and LD22. Soluble proteins from a crude extract (CE) were separated by size exclusion chromatography (see “Materials and Methods”). Proteins from fractions 13 to 30 were separated by 9% SDS-PAGE and immunoblotted with anti-HA antibodies. Fraction 22 corresponds to a molecular mass range of 130 to 140 kD, while fractions 24 and 25 represent an apparent molecular mass of 70 to 80 kD, corresponding to monomeric aCRY. The molecular masses of the standard protein marker are shown with arrowheads.
It was shown previously that aCRY forms homomers in vitro (Oldemeyer et al., 2016). Here, we analyzed if aCRY is also present in complexes in vivo. For this purpose, we separated soluble proteins of crude extracts from cells harvested at LD4 or night (LD22) by size exclusion chromatography (see “Materials and Methods”). We used a transgenic line where acrymut (Beel et al., 2012) was complemented with a C-terminal HA-tagged aCRY gene under the control of the endogenous promotor (see “Materials and Methods” for details). Mr standards were used for size determination. The presence of aCRY in the different fractions was assayed on immunoblots using anti-HA antibodies, which recognize the tagged aCRY with a theoretical molecular mass of 69.7 kD (Supplemental Fig. S1; Fig. 1B). During both day and night, aCRY was detected to some extent as a monomer (Fig. 1B, fractions 24 and 25), but was mainly present in a complex of 130 to 140 kD (Fig. 1B, fraction 22). It seems likely that this complex represents a homodimer of aCRY as also detected in vitro (Oldemeyer et al., 2016). However, we cannot fully exclude the possibility that another partner may be present in this complex. Interestingly, aCRY also forms complexes of low abundance with a size of approximately 200 and 400 kD (Fig. 1B, fractions 17 and 20, respectively) in vivo during the day, which might represent larger oligomers of aCRY, as also found in vitro after illumination (Oldemeyer et al., 2016). These data suggest a signaling role of aCRY as part of homo- or heteromeric complexes in vegetative cells.
Soluble and Membrane-Associated aCRYs Accumulate Differentially during the Sexual Cycle
Light is involved in the process of gametogenesis, where primarily blue and to some extent red light are effective (Weissig and Beck, 1991). To examine if aCRY with its ability to absorb in the blue and red region of the visible spectrum (Beel et al., 2012) may have a function in the sexual life cycle (cartooned in Fig. 2), we first analyzed the expression status of aCRY at different stages of the Chlamydomonas life cycle. Soluble proteins from vegetative cells, pregametes, gametes, early zygotes, and late zygotes were prepared (see “Materials and Methods”) and analyzed on immunoblots with anti-aCRY antibodies. As shown previously (Beel et al., 2012), soluble aCRY accumulates in vegetative cells at the end of the night (LD24; Fig. 3A). In contrast, aCRY accumulation is strongly reduced in pregametes and gametes, both in cells of mating type plus and minus. The strong reduction was also found in early zygotes (Fig. 3A).
Figure 2.
The life cycle of Chlamydomonas (modified from Huang and Beck, 2003; Müller et al., 2017). Haploid vegetative cells (V) perform asexual reproduction under optimal growth conditions. They turn into pG under nitrogen deprivation conditions in darkness. Light induces the formation of gametes (G). Gametes may lose their mating ability and turn to dark-inactivated gametes (dG) upon dark treatment. For simplicity, only the conversion of the plus strain is shown. When gametes of two different mating types are mixed, they will mate and fuse to a quadriflagellated cell that is called early zygote (eZ). The early zygotes convert to mature zygotes (Z) having a thick cell wall after exposure to 15 to 18 h light followed by 5 d in the dark (Jiang and Stern, 2009). In the absence of any light, zygotes will stay as dormant cells; when light and nitrogen are available, they will undergo meiosis and germinate into four vegetative cells, two plus and two minus.
Figure 3.
Accumulation of soluble and membrane-bound aCRY at different stages of the Chlamydomonas sexual cycle. A, The expression of soluble aCRY in vegetative cells (V), pG, and gametes (G) of mating type plus and minus (mt+ and mt−) as well as in early zygotes (eZ) and zygotes (Z). V, pG, G, and eZ were prepared as described in “Materials and Methods.” +N or −N represents the presence or the absence of a nitrogen source in the medium. Black bars on the top indicate darkness and white bars light. Seventy-five-microgram proteins of each sample were separated by 9% SDS-PAGE and immunoblotted with anti-aCRY antibodies. Unspecified protein bands from the PVDF membrane stained with Coomassie Brilliant Blue R250 after immunochemical detection were used as loading control (LC). B, Expression of aCRY in a soluble protein fraction of late zygotes under darkness (D), and 6 h (L6), 12 h (L12), and 24 h (L24) illumination, respectively. The modified version of aCRY (upper band in D, L6, L12, and L24) was quantified in combination with aCRY (indicated as dark plus light gray portion) as well as alone (light gray portion). C, The expression of aCRY in the membrane fractions of V, pG, and G of mt+ and mt− wild-type (WT) strains, as well as of early zygotes (eZ). D, The expression of aCRY in the membrane fraction of Z under darkness (D) as well as after 6 h (L6), 12 h (L12), and 24 h (L24) illumination. A to D, Quantified aCRY protein levels of three biological replicates relative to the protein level of V cells of the mt− strain, harvested at LD24 in the dark, are shown in each diagram with standard deviations.
In contrast to these results, soluble aCRY was visible under all tested conditions (under dark and after 6, 12, and 24 h illumination) in late zygotes, albeit it was more abundant after illumination (Fig. 3B). In this case, a modified form of aCRY was observed to a significant extent, whose nature was unclear (Fig. 3B). One possibility was that aCRY is phosphorylated during this stage of the sexual cycle. To check for this possibility, we treated proteins of a crude extract with λ protein phosphatase, but the potential modification of aCRY was not significantly changed (Supplemental Fig. S2), excluding phosphorylation. Thus, the nature of the modification in late zygotes is still unknown. Our data show that soluble aCRY is differentially expressed at different life stages of Chlamydomonas.
Since aCRY is also present in the fraction of membrane-associated proteins in vegetative cells to a certain extent, we checked whether the observed changes in the abundance of soluble aCRY during the life cycle can be also found with membrane-associated aCRY. Surprisingly, membrane-associated aCRY accumulates in pregametes and gametes of both mating types (Fig. 3C). Compared to vegetative cells, its level is only slightly reduced. Only in early zygotes, aCRY protein level is strongly decreased (Fig. 3C). Membrane-associated aCRY was also found in the membrane fraction of late zygotes, but no modified form was detected there (Fig. 3D). In contrast to soluble aCRY, membrane-associated aCRY is most abundant in the dark. These data suggest that the soluble and membrane-associated forms of aCRY are subject to different control mechanisms during the life cycle, aside from in early zygotes, where both are only present in small amounts.
The low amount of soluble and membrane-bound aCRY in some stages of the algal life cycle may be caused by degradation of aCRY by the proteasome pathway and/or by yet another mechanism of degradation, as found recently for pCRY in gametes (Müller et al., 2017). To check for this, the cell-permeable proteasome inhibitor (MG 132) was added prior to harvesting the cells. In this case, soluble and membrane-bound aCRY accumulate at higher rates in pregametes, gametes, and early zygotes (Fig. 4, A and B), suggesting that aCRY degradation includes the proteasome pathway in all investigated stages. Even in vegetative cells, soluble and membrane-bound aCRY appear to accumulate at a higher rate in the presence of the proteasome inhibitor, suggesting that aCRY undergoes degradation via the proteasome pathway even in these cells to a certain extent.
Figure 4.
Degradation of soluble and membrane-bound aCRY via the proteasome pathway. Samples were labeled and prepared as described in Fig. 3 and either treated with proteasome inhibitor MG 132 (+PI, carbobenzoxy-leucyl-leucyl-leucinal) 1 h before harvesting with a final concentration of 10 µm or not (−PI). A and B, Accumulation of aCRY in a soluble (A) or membrane-bound (B) protein fraction of V, pG, G, and eZ, respectively. The expression of aCRY in V (−PI) was used as a control. The protein level of aCRY was quantified based on three biological replicates; error bars show standard deviations.
Blue and Red Light Affect Germination, Which Is Positively Regulated by aCRY
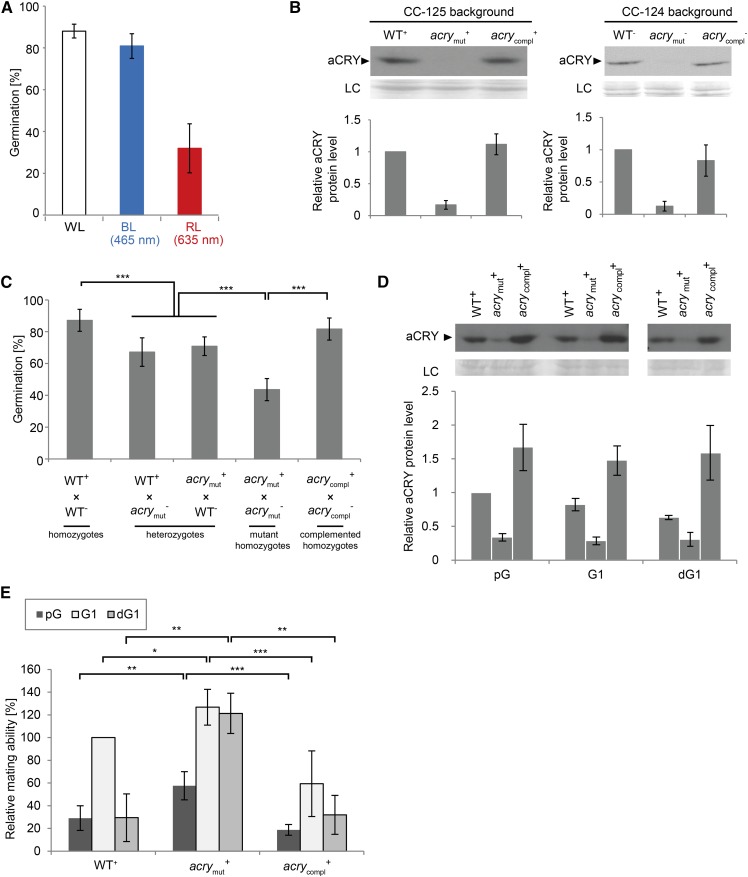
In the sexual life cycle of Chlamydomonas, both gametogenesis and zygote germination are light-dependent steps (Fig. 2; Treier et al., 1989; Gloeckner and Beck, 1995). In wild-type strains of Chlamydomonas, diploid zygotes can only undergo meiosis and germinate into haploid vegetative cells when illuminated in the presence of a nitrogen source. The role of different specific light qualities on germination had not been studied previously. Thus, we analyzed if blue and red light could influence germination, because they are both absorbed by the neutral radical form of aCRY (Beel et al., 2012). For this purpose, we performed tests with strains CC-125 (mt+) and CC-124 (mt−) as mating partners (see “Materials and Methods”), which are frequently used for this purpose (Suzuki and Johnson, 2002; Huang and Beck, 2003). Blue and red light were applied at 465 nm and 635 nm, respectively, according to the absorption peaks of aCRY (Beel et al., 2012). Blue light induced zygote germination nearly as effectively as white light (80%–90%; Fig. 5A), but red light also had a profound effect on germination (32.0% induction; Fig. 5A). As it is known that zygotes cannot germinate in the dark (Gloeckner and Beck, 1995), our results indicate that germination is largely affected by blue, but also significantly by red light. Thus, aCRY may be involved in germination, which was tested in the next step.
Figure 5.
aCRY has a positive influence on the germination efficiency, which is supported by blue and red light, and a negative influence on mating ability and maintenance. A, The influence of blue and red light on germination. Blue (465 nm, BL), red (635 nm, RL) and white light (WL, see “Materials and Methods”) were used to illuminate wild-type (WT) zygotes during the germination process. About 50 zygotes were counted for each condition and each replicate (n = 3). Mean values and standard deviations are shown. B, Quantification of soluble aCRY in WT strain CC-125 (mating type plus) and CC-124 (mating type minus), acrymut strains and corresponding complemented strains. Seventy-five-microgram proteins of each sample were separated by 9% SDS-PAGE and immunoblotted with anti-aCRY antibodies. CC-125 (WT+) and CC-124 (WT−) were used for comparison and their levels were set to 100%. Unspecified protein bands from the PVDF membrane were used as loading control (LC). The quantifications of aCRY in these strains are based on three independent biological replicates and shown with standard deviations. C, Germination profiles of crosses between WT, acrymut, and acrycompl strains. Germination of zygotes of several crossing between WT and mutant strains, including WT homozygotes (WT+ × WT−), heterozygotes (WT+ × acrymut− or acrymut+ × WT−), and acry mutant homozygotes (acrymut+ × acrymut−) are shown. In addition, the germination rate of homozygotes of the complemented strains (acrycompl+ × acrycompl−) is presented. At least three biological replicates were performed. Error bars indicate standard deviations. Student’s t tests were performed (***P < 0.001). D, Quantification of aCRY in total (soluble and membrane-bound) proteins of the different cell types of gametogenesis. The cell types (see “Materials and Methods”) include (1) pG kept in the dark, (2) pG illuminated for 1 h with light resulting in G1 gametes (G1), and (3) G1 gametes that have been exposed to darkness for 1 h resulting in dG1 of WT, acrymut, and acrycompl mt+ strains. Fifty-microgram proteins of each sample were separated by 9% SDS-PAGE and immunoblotted with anti-aCRY antibodies. The expression level of aCRY in pG of WT was used for comparison. Three biological replicates were performed. Error bars indicate standard deviations. E, The influence of aCRY on mating ability and the maintenance of mating ability. Mean values and standard deviations of at least three biological replicates are shown. Student’s t tests were performed (***P < 0.001; **P < 0.01, *P < 0.05). Mating ability assays of WT, acrymut, and acrycompl strains. pG, G1, and dG1 cells (see D for explanations) were used for the mating ability tests. The mating ability of G1 of WT was set to 100% and used for comparison.
First, we backcrossed the acry mutant phenotype from strain CRMS101 (Beel et al., 2012) into CC-124 and CC-125 (see “Materials and Methods”; Supplemental Fig. S3; Figure 5B). It should be noted that the mutant acry phenotype is based on an insertion of the selection marker in an intron of aCRY, resulting in a knockdown, but not a knockout, of aCRY protein, as already mentioned in the introduction (Beel et al., 2012). The crossed mutant strains showed also a strong reduction of the aCRY level compared to wild type. One acry mutant strain (acrymut, mt+) had a reduction down to approximately 17% compared to wild type mt+, and the other one (acrymut, mt−) down to approximately 13% compared to wild type mt− (Fig. 5B). acrymut strains of both mating types with strongly reduced aCRY protein levels were used for the germination assay along with wild-type strains of both mating types (CC-125 and CC-124) as controls. Homozygotes of wild type (wild type+ × wild type−) showed a germination value of about 90% (Fig. 5C). Both heterozygotes of wild type and acrymut (wild type+ × acrymut−, acrymut+ × wild type−) exhibited a significant lower germination rate (approximately 70%; Fig. 5C). Furthermore, the germination of the homozygote of acrymut revealed a strongly reduced germination level of approximately 40% (Fig. 5C). In contrast, a complemented homozygote (acrycompl+ × acrycompl−) that has aCRY levels of 112% and 83%, respectively (Fig. 5B), showed again a significantly higher germination rate (82%; Fig. 5C). These data demonstrate that aCRY is indeed involved in the process of the zygote germination step, where it acts as a positive regulator in addition to PHOT and pCRY (Huang and Beck, 2003; Müller et al., 2017).
aCRY Is a Negative Regulator of Mating Ability and Mating Maintenance
In a next step, we tested whether aCRY participates in the gamete formation process, which is also under light control (Weissig and Beck, 1991). For the mating ability test, strain acrymut (mt+) was chosen (see “Materials and Methods”). The gamete formation of pregametes exposed for 1 h to light results in so-called G1 gametes. Incubation of mature gametes in the dark for 1 h (named dG1) leads to a loss of their mating ability (Beck and Acker, 1992; Huang and Beck, 2003). We analyzed the level of total (soluble and membrane-bound) aCRY in pregametes (pG) and gametes (G1), where the membrane-bound form of aCRY dominates, as well as in dark-inactivated gametes (dG1) in wild type (mt+), strain acrymut (mt+), and the complemented strain. The level of total aCRY was reduced to approximately 25% in the mutant strain and recovered to around 150% in the complemented strain (Fig. 5D).
For the mating ability test, wild-type strain (mt+) was set to 100% for the state of G1 gametes and used for comparison. As expected (Saito et al., 1998), wild-type pregametes exhibited only a low mating ability rate (29%; Fig. 5E) compared to wild-type G1 gametes, as shown previously (Beck and Acker, 1992), suggesting the involvement of photoreceptors. Dark treatment of G1 gametes resulted in a strong loss of mating ability in the wild-type strain (Fig. 5E), consistent with former studies (Beck and Acker, 1992; Pan et al., 1997; Huang and Beck, 2003). In acrymut, the mating abilities were significantly higher than that of wild type both in pregametes and G1 gametes (Fig. 5E). In the complemented strain of acrymut (see “Materials and Methods”), named acrycompl (∼150% compared to wild type; Fig. 5D), the mating abilities of pregametes and G1 gametes were again reduced and slightly lower than in wild type (Fig. 5E). These data suggest that aCRY is involved in mating ability and seems to exert a negative regulation in the mating pathway, because the reduction of the aCRY level causes an increase in the mating ability.
We also analyzed whether aCRY has an effect on the phototactic behavior of the cells after the transition from pregametes to G1 gametes. For that purpose, equal amounts of pregametes were evenly distributed in a flat culture flask with shaking under light for 1 h. The swimming behavior of the G1 cells was compared immediately after the stop of shaking (control, 0 min) and keeping the cells in a light gradient for 20 min (see “Materials and Methods”) and was also compared to vegetative cells. Cells from wild type and acrycompl accumulated to a large extent toward the higher light intensity, both in G1 and vegetative cells (Supplemental Fig. S4). The positive phototactic behavior toward the higher light intensity also occurred in acrymut, however at a significantly lower rate. Many cells were still widely distributed over the flask and were also present at the regions with low light intensities. These data suggest that aCRY contributes to the regulation of phototactic behavior in G1 gametes and vegetative cells.
As mentioned above, incubation of mature gametes in the dark leads to a loss of their mating ability. To test a possible role of aCRY in mating maintenance ability, the G1 gametes of wild type, acrymut, and acrycompl were incubated in the dark for 1 h. The dark treatment resulted in a strong loss of mating ability in the wild-type strain (Fig. 5E), consistent with former studies (Beck and Acker, 1992; Pan et al., 1997; Huang and Beck, 2003). Interestingly, dark treatment only provoked a minor—or nonexistent—loss of mating ability in acrymut, indicating that aCRY is an indispensable component for the loss of the mating ability pathway. acrycompl displayed a similar reduction in mating ability after dark treatment as wild type (Fig. 5E), which confirms the negative role of aCRY in the regulation of mating maintenance.
aCRY Shifts in Its Intracellular Localization during the Life Cycle
The change in abundance of soluble and membrane-bound aCRY in vegetative cells compared to pregametes and gametes might be associated with a change in the intracellular localization of aCRY. To check for this, we performed immunolocalization studies with a transgenic wild-type line expressing HA-tagged aCRY (Supplemental Fig. S1C; “Materials and Methods”) as well as with wild type (negative control) along with anti-HA antibodies that have been previously used for immunohistochemistry studies in C. reinhardtii (anti-HA, clone 3F10 Roche/Sigma; Niwa et al., 2013). Vegetative cells were taken at LD4 and LD22 when aCRY was shown to be accumulated at high levels in a diurnal cycle (Beel et al., 2012) but present in different complexes (Fig. 1B). Moreover, pG, G1 gametes, and dG1 were used for the experiments. Negative controls with wild-type cells of the different life cycle stages were performed under the same microscopic settings as with the transgenic line. They showed no significant signals with the anti-HA antibodies (Supplemental Fig. S5) in contrast to the transgenic wild-type line expressing HA-tagged aCRY (Fig. 6). In vegetative cells of the transgenic line, taken at LD4, aCRY was observed in a circular region below the flagella, which had been labeled with anti-tubulin antibodies, indicating that it may be enriched in the nucleus (Fig. 6A, 1). To examine whether this is indeed the case, we used antibodies against the nuclear pore complex as further control (Fig. 6A, 2). The pictures confirm nuclear localization of aCRY, as also visible in an overview picture (Fig. 6A, 3). aCRY is found to a significant extent in the nucleus during LD4. In contrast, aCRY appears to be present throughout the cell body during LD22. A representative cell is depicted in Figure 6A, 4. Only in a few cells, it is in addition strongly (Fig. 6A, 5) or partially (Fig. 6A, 6) enriched in the nucleus.
Figure 6.
(Figure continues on following page.)
After shifting vegetative cells to the sexual cycle (cartooned in Fig. 2), aCRY was observed throughout the cell body in all investigated cell types (pG, G1 gametes, and dG1). It was never found in the nucleus in all these cell types (Fig. 6B). Representative cells are shown for the state of pregametes (Fig. 6B, 1 and 2), G1 gametes (Fig. 6B, 3), and dark-inactivated G1 gametes (Fig. 6B, 4). In summary, our data show a localization pattern of aCRY that is diurnal as well as sexually influenced.
DISCUSSION
Blue light and its absorbing receptors are known to control the cell and life cycles of so far investigated algae, including Chlamydomonas, the marine diatom Phaeodactylum tricornutum and the stramenopile alga Vaucheria frigida (Huang and Beck, 2003; Oldenhof et al., 2004; Takahashi et al., 2007; Huysman et al., 2013; Müller et al., 2017). To date, the receptors PHOT, pCRY, and aureochrome have been identified to be involved in these processes. PHOT and aureochrome have in common that they contain LOV (light, oxygen, or voltage) domains as a sensory unit binding flavin as a chromophore (Christie, 2007; Suetsugu and Wada, 2013). Flavin is also acting as chromophore for pCRY (see “Introduction”). Here, we have extended the spectrum of involved photoreceptors to include aCRY for life cycle control in Chlamydomonas. It has been shown previously that blue and to a smaller extent red light are involved in the process of gametogenesis along with its restoration in Chlamydomonas (Weissig and Beck, 1991; Pan et al., 1997). Our data demonstrate that blue and red light are also involved in the process of germination (Figs. 5A and 7). aCRY represented a premium candidate for controlling these processes in addition, because of its ability to absorb not only blue but also red light based on the flavin neutral radical (Beel et al., 2012; Spexard et al., 2014; Oldemeyer et al., 2016). As shown here, aCRY plays indeed a key role in the steps of gametogenesis and zygote germination. Using acry mutants of both mating types and the corresponding complemented strains, we found that aCRY is involved in three steps of the sexual cycle, (1) in mating ability, (2) in the loss of mating ability in the dark, and (3) in zygote germination (Fig. 7).
Figure 7.
Overview of the role of aCRY, pCRY, and PHOT in the Chlamydomonas sexual life cycle. Ellipses represent the aCRY, pCRY, and PHOT proteins with the colors indicating the absorbed light spectrum. The negative roles of aCRY and pCRY in gametogenesis seem not to be based on light and are thus shown with a gray background. For the light-dependent steps in the sexual life cycle, gametogenesis along with its restoration and germination, blue and red arrows indicate light qualities, affecting these pathways to different extents (as indicated by the length of the arrows).
The role of aCRY in the development of mating ability and its maintenance is, however, counterintuitive for a photoreceptor. In both functions, aCRY appears to act as a negative regulator in the dark (Figs. 5E and 7), which is similar to pCRY (Müller et al., 2017). To our knowledge, only the illuminated forms of the photoreceptors ChR1 and PHOT are active in promoting responses in the alga (Hegemann, 2008). In the dark, the activity of the ion channel ChR1 and the LOV/kinase domains of PHOT, respectively, are strongly reduced. In contrast, phytochrome has been demonstrated to be active in both functional states, Pr and Pfr, regulating different physiological pathways in plants (Rockwell et al., 2006). However, plant phytochrome is considered to be a bimodal photochromic switch, where in the dark the Pr form is accumulated only slowly. aCRY and pCRY, similar to other cryptochromes, are not known to exhibit any enzymatic function in the dark, which might drive the loss of mating ability for dark-exposed gametes (Fig. 5E). Therefore, a negative regulation by aCRY and pCRY in the dark might possibly be exerted by dissociation of any of them from an enzyme such as a phosphatase, thereby releasing an inhibitory interaction. It should be noted that such a simple scenario would not account for the increase in mating ability in the acry and pcry mutants as compared to wild type after illumination (Figs. 5E and 7). Clearly, a molecular elucidation of such bimodal activity of aCRY and pCRY in the dark as well as the light will be rewarding.
In this context, the differential expression pattern of aCRY in vegetative cells, pregametes, gametes and early as well as late zygotes is also of interest along with the differences in the abundance of soluble and membrane-associated aCRY forms in those cell types. In the case of pCRY, we do not yet know if a membrane-associated form exists, but it was shown recently that its soluble form is also subject to degradation in pregametes and gametes (Müller et al., 2017). In land plants and animals, CRYs seem to be present as soluble proteins and perform roles in the nucleus and/or cytosol (Chaves et al., 2011; Ahmad, 2016; Liu et al., 2016). Similarly, Chlamydomonas aCRY is present mostly as soluble protein in vegetative cells and associated to membranes to a smaller extent (Fig. 1A). The soluble aCRY seems to form a homodimer or even an oligomer during the day when performing its asexual functions, consistent with in vitro data showing that heterologously expressed full-length aCRY is present as a dimer in the dark and partially oligomerizes after light exposure (Oldemeyer et al., 2016). In pregametes and gametes of both mating types as well as in early zygotes, soluble aCRY is strongly degraded, apparently by the proteasome pathway as shown with inhibitor studies. In contrast, membrane-associated aCRY is rather stable in pregametes and gametes and only partially degraded by the proteasome (Figs. 3 and 4), indicating that individual functions of these forms may exist and pointing to the possibility that the membrane-associated form of aCRY in pregametes and gametes may have different functions and interaction partners. This separation may go hand in hand with the intracellular localization of aCRY, which is different in the cell types of gametogenesis compared to vegetative cells with regard to the complete lack of aCRY in the nucleus in the sexual cells (Fig. 6). It might be that the membrane association of aCRY in these cells is functionally connected to its distribution over the cell body and its absence in the nucleus. In vegetative cells, aCRY is clearly present in the nucleus, especially during day. In fact, aCRY bears a nuclear localization motif “RKKATAAAGGSSGAGRGKK” in its C-terminal region with the bipartite basic region known from other nuclear proteins (see underlined amino acid regions) such as in the constitutively nuclear CRY2 of Arabidopsis (Zuo et al., 2012).
In contrast to pregametes and gametes, membrane-associated aCRY is also strongly reduced in early zygotes. One possible explanation for this finding is that aCRY completes its role in the process of mating and is not needed afterward. In mature zygotes, both soluble and membrane-associated aCRY accumulate again to various extents. It would be interesting to see if this goes hand in hand with nuclear localization as observed in vegetative cells, where soluble aCRY is found. However, determination of the intracellular localization of aCRY in zygotes is practically not feasible by immunolocalization studies due to the thick and rigid zygote cell wall. Notably, at this stage of the life cycle, aCRY plays a role as a positive regulator for the light-dependent germination step (Figs. 5C and 7) during which the zygote undergoes meiosis, resulting in four vegetative cells. Thus, it exerts the same role as PHOT and pCRY in this process (Huang and Beck, 2003; Müller et al., 2017) and is acting complementarily with PHOT and pCRY, but with an extended spectral response.
Gametogenesis is also controlled by PHOT (Huang et al., 2002; Huang and Beck, 2003) and by pCRY. However, the roles of aCRY, pCRY, and PHOT are contrary in the gametogenesis step. In phot knockdown strains, the conversion of pregametes to mature gametes is reduced (Huang and Beck, 2003), suggesting a positive regulation for PHOT, while in the acry and pcry mutants, the mating ability of the partially mating-competent pregametes of strain CC-125 (Saito et al., 1998) as well as of gametes is significantly increased (Figs. 5E and 7; Müller et al., 2017). Moreover, there is a significant loss of mating ability of dark-exposed gametes in phot knockdowns similar to wild type, whereas PHOT is involved in the restoration of the mating ability of dark-inactivated gametes (Huang and Beck, 2003; Fig. 7). In contrast, in the acry and pcry mutants, high mating abilities are maintained upon dark exposure of mature gametes, opposite to wild type (Figs. 5E and 7). To obtain even more detailed knowledge of the overlapping functions of aCRY, pCRY, and PHOT and to distinguish them from the action of other potentially involved blue light-absorbing photoreceptors in Chlamydomonas (see “Introduction”), a triple acry/pcry/phot knockout mutant would be useful. However, while a well-characterized null mutant for PHOT exists and was used for functional characterizations (Zorin et al., 2009; Trippens et al., 2012; Petroutsos et al., 2016), it was generated in a flagella-less strain that does not cross. Moreover, the insertional acry and pcry mutants are only knockdown mutants, because the insertion is in an intron in both cases (Beel et al., 2012; Müller et al., 2017).
aCRY and PHOT may be interconnected within a functional network. Former data showed an influence of a red light receptor, most probably aCRY, on the expression of certain genes associated with photosynthetic function that require wild-type PHOT (Im et al., 2006). Moreover, it is intriguing that both aCRY and PHOT exist (also) as membrane-associated proteins, especially in pregametes, gametes, and late zygotes in the case of aCRY. This offers the possibility of a signal network in the vicinity of the plasma membrane. In the moss Physcomitrella patens, an interaction between PHOT and the red light photoreceptor phytochrome at the plasma membrane was found. In this case, phytochrome interacts with PHOT in its photoactivated state exclusively at the plasma membrane and activates a direct cytoplasmic signal to guide directional responses to light (Jaedicke et al., 2012). Because phytochrome is not encoded in the genome of Chlamydomonas (Mittag et al., 2005; Merchant et al., 2007), it is conceivable that membrane-associated aCRY may act in Chlamydomonas together with PHOT, in a direct or indirect manner.
The life cycle of Chlamydomonas is functionally closely connected with the survival strategy of the alga. Gametogenesis is induced by removal of the nitrogen source and further driven by light (Fig. 2). In nature, nitrogen-deprived environmental conditions can easily occur in freshwater as well as in moist soil, both habitats where Chlamydomonas lives. The formation of the zygotes ensures that the alga survives such adverse conditions for a long time in this “dormant” stage. However, the alga is flexible to reprogram gamete formation, if the natural conditions change and it is reexposed suddenly to a nitrogen source such as nitrate (Pozuelo et al., 2000, and literature therein). To allow such a reprogramming, positive and negative elements are needed at the switch from vegetative cells to gametes. aCRY and pCRY represent such negative elements, while PHOT acts as the positive element. Once the zygotes are formed, a process back to gametes is not available. Meiosis is now needed to produce vegetative cells that can divide quickly. Thus, it makes sense to accumulate positive elements with different spectral responses (aCRY, pCRY, and PHOT) at this stage of the sexual cycle that promote this process in the presence of nitrogen and blue as well as red light.
MATERIALS AND METHODS
Strains and Culture Conditions
Chlamydomonas reinhardtii wild-type strains CC-125 (nit1, nit2, agg1+, mt+) and CC-124 (nit1, nit2, agg1, mt−) were routinely used for life-cycle-related experiments unless otherwise indicated. They are frequently used strains for life-cycle analysis (Suzuki and Johnson, 2002; Huang and Beck, 2003). CC-124 was used as mating partner to test the mating ability of plus strains. For studies in the background of mutant acry, the paromomycin resistant acrymut strain CRMS101 with D66 background (Beel et al., 2012) was backcrossed into CC-124 and CC-125 (see Supplemental Fig. S3). Two of the paromomycin-resistant progenies (one mt+ and one mt−) were chosen to generate complemented strains (acrycompl) as described below. Moreover, a HA-tagged aCRY line expressed in the background of acrymut that had been backcrossed to wild-type strain SAG73.72 (Beel et al., 2012) was used for size exclusion chromatography and a HA-tagged line in the background of SAG73.72 for the immunolocalization studies (see also below). Strains were cultured under 12 h light/12 h dark (LD12/12) cycle with a light intensity of 75 μmol∙m−2∙s−1 in Tris-acetate-phosphate (TAP) medium (Harris, 1989), unless otherwise indicated. Cells were harvested at the indicated LD time points, where LD0 represents the beginning of the day and LD12 represents the beginning of the night.
Complementation of acrymut by Transformation with pKP39
pKP39, a vector containing the full-length aCRY gene of Chlamydomonas (Beel et al., 2012), was used for complementing the acry mutant strains (Supplemental Fig. S3). For this purpose, the acrymut strains with CC-124 or CC-125 background were transformed with KpnI-linearized pKP39 using the autolysin method (Iliev et al., 2006). pKP39 confers hygromycin resistance to the transgenic lines that were selected on TAP agar medium containing 20 µg/mL hygromycin B.
Generation of HA-Tagged aCRY Lines
Vectors pKP20-Hyg and pKP20 (Supplemental Fig. S1) containing both the full-length genomic DNA of the aCRY gene along with its putative promotor region and the sequence for a 3 × HA-tag at the end of its last exon together with the RBCS2 3′ UTR as well as the hygromycin B (pKP20-Hyg) resistance gene from pHyg3 or the APVIII-based paromomycin resistance (pKP20) for selection in C. reinhardtii (Berthold et al., 2002) were used for transformation. The mutant strain SAG73.72:acry1A (Beel et al., 2012) and wild-type strain SAG73.72 were transformed with AhdI-linearized pKP20-Hyg and pKP20, respectively. Cells were grown on TAP agar medium containing 20 μg/mL hygromycin B and 50 µg/mL paromomycin, respectively. The selected strains with HA-tagged aCRY expression were used as described above.
Size Exclusion Chromatography
Size exclusion chromatography was performed as recently described for pCRY (Müller et al., 2017) using an Äkta FPLC (GE Healthcare) with a Superdex 200 Increase 10/300 GL column (GE Healthcare) at 4°C with the following details. Extracts of soluble proteins were made with the elution buffer containing protease inhibitor cocktail (cOmplete Protease Inhibitor Cocktail, Roche) according to Iliev et al. (2006) and treated as described (Müller et al., 2017). Five-hundred-microliter fractions were collected and proteins denatured; 100 µL of these aliquots were separated by 9% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by immunoblots (see below) with anti-HA antibodies (dilution 1:2000). Standard marker proteins (Bio-Rad) were used to determine the apparent molecular mass of the sample by calibration.
Sample Preparation of Pregametes, Gametes, Early Zygotes, and Late Zygotes
Vegetative cells were transferred to TAP without NH4+ at LD12 and kept for 14 h in darkness to induce pregametes. Afterward, the pregametes were exposed to white light of 75 μmol∙m−2∙s−1 for 6 h for mature gamete formation. Mature gametes (plus and minus) were mixed to allow mating for 4 h without stirring, and zygote pellicles (early zygotes) formed in the medium (Wegener et al., 1989) were harvested.
For late zygote formation, early zygote pellicles in the liquid medium were transferred to TAP plates using an inoculation loop, incubated for 16 to 18 h in the light (75 μmol∙m−2∙s−1) and then kept in the dark for 5 d for maturation. Zygotes were harvested according to Wegener et al. (1989) with minor modifications. In detail, the plates with mature zygotes were exposed to chloroform vapors for 90 s to kill unmated cells. The zygotes were scraped off the plates with a razor blade and suspended in 4 mL TAP medium under safety far-red light (Müller et al., 2017). To further destroy unmated cells, 30 s sonication steps were performed two times with a power of 25 to 30 W using a Sonicator (Bioblock Scientific-Vibra Cell 72405) equipped with a microtip. After sonication, the zygotes were centrifuged at 1500g for 5 min, and the pellet was washed with 20 mL fresh TAP medium. The centrifugation and the washing steps were repeated another two times to remove most of the fraction of unmated cells (Wegener et al., 1989). Then one-fourth of the remaining pure zygotes were harvested in the dark. The remaining zygotes were illuminated for 6, 12, or 24 h and then harvested.
Crude Protein Extracts and Immunoblots
For crude extraction of total proteins (comprising soluble, membrane-associated, and transmembrane proteins), cells were boiled in 2 × SDS buffer according to Schulze et al. (2013). Soluble proteins of the crude extraction were obtained using a glass bead procedure according to Zhao et al. (2004). After removal of the soluble protein, the remaining pellets were resolved in 2 × SDS-sample buffer (Schulze et al., 2013) for obtaining proteins of the membrane fraction (comprising membrane associated and transmembrane proteins). In order to break the hard cell wall of the late zygotes for protein extraction, samples were ground in liquid nitrogen using a mortar and pestle (Aoyama et al., 2014) instead of the glass bead method (Zhao et al., 2004).
For the separation of membrane-associated proteins and transmembrane proteins, the protein pellet containing the membrane fraction (see above) was washed with ice-cold 1 × phosphate-buffered saline (PBS; 10 mm Na2HPO4, 1.8 mm KH2PO4, 2.7 mm KCl, 137 mm NaCl, pH 7.4) two times. Then the pellet was incubated with 0.1 m sodium carbonate buffer (pH 11.8) at 4°C with gentle agitation for 1 h (Fujiki et al., 1982). After ultracentrifugation at 93,000g, the remaining pellet containing the transmembrane proteins was washed with 30 mL 0.1 m sodium carbonate buffer and resolved in 2 × SDS-sample buffer. The supernatant fraction containing the membrane-associated proteins was precipitated with 100% (w/v) trichloroacetic acid (TCA) overnight at 4°C and spun down at 5000g for 10 min. The pellets were washed three times with ice-cold acetone, then dried and resolved directly in 2 × SDS-sample buffer (Schulze et al., 2013).
Protein concentrations were determined by the Neuhoff method (Neuhoff et al., 1979). The immunoblots for the detection of aCRY, PHOT, and CYTf, respectively, were performed with the same procedure as described previously (Beel et al., 2012). The antibody dilution for detecting PHOT was 1:2000, and for CYTf 1:15000. Anti-LOV1 (PHOT) antibodies were kindly provided by Peter Hegemann, Berlin, Germany. The antibodies for detecting CYTf were purchased from Agrisera.
Protein levels were quantified using ImageJ 1.46r (Wayne Rasband, National Institutes of Health).
Mating Ability, Mating Maintenance, and Germination Assays
The mating ability, mating maintenance, and germination assays were performed as previously described (Müller et al., 2017). The light intensities used for the germination assays were 30 μmol∙m−2∙s−1 for white light, 30 μmol∙m−2∙s−1 for blue light (SuperFlux LED from Lumitronix LED-Technik, peak at 465 nm, full width at half maximum of 18.5 nm) and 41 μmol∙m−2∙s−1 for red light (SuperFlux LED from Lumitronix LED-Technik, peak at 635 nm, full width at half maximum of 17 nm).
Phototactic Behavior Test
G1 (in TAP medium lacking a nitrogen source) as well as vegetative cells (in TAP medium) were used for phototactic behavior tests. For this purpose, the cells were put into the dark. Light was introduced from one side with a fluence of 30 μmol∙m−2∙s−1, and pictures were taken from above.
Immunolocalization Studies
For immunolocalization studies, 2 mL cells from mid log-phase cultures were harvested at the indicated time points by centrifugation at 500g for 10 min and washed with 2 mL microtubule-stabilizing buffer (50 mm Piperazine-N,N′-bis-(2-ethanesulphonic acid), 5 mm EGTA, 1 mm MgSO4, pH 7.4, with NaOH), named MTSB. After an additional centrifugation step, the cells were resuspended in 2 mL MTSB and spotted on poly-l-lysine-coated microscope slides and allowed to settle down for 20 min at room temperature in the dark to avoid photoaccumulation. Slides were washed once for 5 min in 1 × PBS (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.7 mm KH2PO4) and then fixed for 5 min with freshly mixed fixation buffer A (100 mm sodium phosphate buffer, pH 7.4) and B (8% paraformaldehyde, 10 mm NaOH, pH 7.4). After two additional 1 × PBS washing steps for 5 min, the permeabilization step was performed for 30 s in −20°C cold methanol with two more following washing steps. Blocking (1 × PBS, 3% bovine serum albumin, 0.1% Tween 20) was performed for 60 min at 37°C in a humidity incubation chamber, followed by a washing step in 1× PBS. The first antibody was incubated for 1 h at 37°C with a dilution of 1:250 in case of the anti-HA antibody (Sigma, clone 3F10 produced in rat) and 1:5000 for the anti-nuclear pore complex (Biolegend, clone MAb414 produced in mouse) and the anti-acetylated tubulin antibodies (Sigma, clone 6-11B-1, produced in mouse), respectively. The cells were washed five times in 1 × PBS for 5 min each, then blocked again for 30 min at 37°C and incubated with the labeled second antibody for 1 h at 37°C, followed by five washing steps with 1 × PBS, 5 min each. The slides were cover slipped with ProLong Gold Antifade Reagent (Invitrogen). All antibodies were diluted in blocking buffer without Tween 20. The goat anti-rat antibody was labeled with AlexaFluor488 (Life Technologies) and used in a 1:2000 dilution. The goat anti-mouse antibody was labeled with AlexaFluor555 (Life Technologies) and used in a 1:5000 dilution. All dark samples were prepared under far-red safety light (according to Müller et al., 2017) until the permeabilization step. Confocal microscopy was performed with Zeiss LSM 780 and a 40×/1.30 NA EC Plan-Neofluar objective. For antibody visualization, 488 and 561 nm laser with 0.2% intensity were used and the signals from 490 nm to 553 nm (AlexaFluor488) and 570 nm to 641 nm (AlexaFluor555) were measured with a Gallium Arsenide Phosphide Detector. The image calculation and finalization were performed with Zen 2012 program (Zeiss).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequences of pKP20-Hyg and pKP20, respectively, and scheme of pKP20-Hyg.
Supplemental Figure S2. The expression of aCRY in late zygotes after protein phosphatase treatment.
Supplemental Figure S3. Genetic background of the strains used in this work.
Supplemental Figure S4. Phototactic behavior of gametes and vegetative cells of wild type, acrymut, and acrycompl.
Supplemental Figure S5. Negative controls for immunofluorescence micrographs of wild-type cells using anti-HA antibodies.
Acknowledgments
We thank Peter Hegemann for anti-PHOT antibodies and Anne Herbst for her help in the preparation of the protocol for immunolocalization studies.
Footnotes
This work was supported by the German Research Foundation within Research Group FOR1261 (grants Mi373/12-2 to M.M. and Ko3580/1-2 to T.K.), by a Heisenberg Fellowship to T.K. (KO3580/4-1), and by the Jena School for Microbial Communication (JSMC). Y.Z. acknowledges a fellowship from the JSMC.
Current address: Max Planck Institute for Chemical Ecology, 07745 Jena, Germany.
References
- Ahmad M. (2016) Photocycle and signaling mechanisms of plant cryptochromes. Curr Opin Plant Biol 33: 108–115 [DOI] [PubMed] [Google Scholar]
- Aoyama H, Saitoh S, Kuroiwa T, Nakamura S (2014) Comparative analysis of zygospore transcripts during early germination in Chlamydomonas reinhardtii. J Plant Physiol 171: 1685–1692 [DOI] [PubMed] [Google Scholar]
- Awasthi M, Ranjan P, Sharma K, Veetil SK, Kateriya S (2016) The trafficking of bacterial type rhodopsins into the Chlamydomonas eyespot and flagella is IFT mediated. Sci Rep 6: 34646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CF, Acker A (1992) Gametic differentiation of Chlamydomonas reinhardtii: Control by nitrogen and light. Plant Physiol 98: 822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel B, Müller N, Kottke T, Mittag M (2013) News about cryptochrome photoreceptors in algae. Plant Signal Behav 8: e22870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel B, Prager K, Spexard M, Sasso S, Weiss D, Müller N, Heinnickel M, Dewez D, Ikoma D, Grossman AR, et al. (2012) A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell 24: 2992–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold P, Schmitt R, Mages W (2002) An engineered Streptomyces hygroscopicus aph 7" gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153: 401–412 [DOI] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GTJ, Batschauer A, Ahmad M (2011) The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62: 335–364 [DOI] [PubMed] [Google Scholar]
- Christie JM. (2007) Phototropin blue-light receptors. Annu Rev Plant Biol 58: 21–45 [DOI] [PubMed] [Google Scholar]
- Ermilova EV, Zalutskaya ZM, Huang K, Beck CF (2004) Phototropin plays a crucial role in controlling changes in chemotaxis during the initial phase of the sexual life cycle in Chlamydomonas. Planta 219: 420–427 [DOI] [PubMed] [Google Scholar]
- Forbes-Stovall J, Howton J, Young M, Davis G, Chandler T, Kessler B, Rinehart CA, Jacobshagen S (2014) Chlamydomonas reinhardtii strain CC-124 is highly sensitive to blue light in addition to green and red light in resetting its circadian clock, with the blue-light photoreceptor plant cryptochrome likely acting as negative modulator. Plant Physiol Biochem 75: 14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato AE, Annunziata R, Jaubert M, Bouly J-P, Falciatore A (2015) Dealing with light: The widespread and multitasking cryptochrome/photolyase family in photosynthetic organisms. J Plant Physiol 172: 42–54 [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: Application to endoplasmic reticulum. J Cell Biol 93: 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão VC, Fankhauser C (2015) Sensing the light environment in plants: Photoreceptors and early signaling steps. Curr Opin Neurobiol 34: 46–53 [DOI] [PubMed] [Google Scholar]
- Gloeckner G, Beck CF (1995) Genes involved in light control of sexual differentiation in Chlamydomonas reinhardtii. Genetics 141: 937–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Lin H, Lee JH (2007) Sex determination in Chlamydomonas. Semin Cell Dev Biol 18: 350–361 [DOI] [PubMed] [Google Scholar]
- Govorunova EG, Jung KH, Sineshchekov OA, Spudich JL (2004) Chlamydomonas sensory rhodopsins A and B: Cellular content and role in photophobic responses. Biophys J 86: 2342–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Sourcebook. Academic Press, San Diego, CA [Google Scholar]
- Hegemann P. (2008) Algal sensory photoreceptors. Annu Rev Plant Biol 59: 167–189 [DOI] [PubMed] [Google Scholar]
- Huang K, Beck CF (2003) Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 100: 6269–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Kunkel T, Beck CF (2004) Localization of the blue-light receptor phototropin to the flagella of the green alga Chlamydomonas reinhardtii. Mol Biol Cell 15: 3605–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Merkle T, Beck CF (2002) Isolation and characterization of a Chlamydomonas gene that encodes a putative blue-light photoreceptor of the phototropin family. Physiol Plant 115: 613–622 [DOI] [PubMed] [Google Scholar]
- Huysman MJJ, Fortunato AE, Matthijs M, Costa BS, Vanderhaeghen R, Van den Daele H, Sachse M, Inzé D, Bowler C, Kroth PG, et al. (2013) AUREOCHROME1a-mediated induction of the diatom-specific cyclin dsCYC2 controls the onset of cell division in diatoms (Phaeodactylum tricornutum). Plant Cell 25: 215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev D, Voytsekh O, Schmidt EM, Fiedler M, Nykytenko A, Mittag M (2006) A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol 142: 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im CS, Eberhard S, Huang K, Beck CF, Grossman AR (2006) Phototropin involvement in the expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii. Plant J 48: 1–16 [DOI] [PubMed] [Google Scholar]
- Immeln D, Pokorny R, Herman E, Moldt J, Batschauer A, Kottke T (2010) Photoreaction of plant and DASH cryptochromes probed by infrared spectroscopy: The neutral radical state of flavoproteins. J Phys Chem B 114: 17155–17161 [DOI] [PubMed] [Google Scholar]
- Immeln D, Schlesinger R, Heberle J, Kottke T (2007) Blue light induces radical formation and autophosphorylation in the light-sensitive domain of Chlamydomonas cryptochrome. J Biol Chem 282: 21720–21728 [DOI] [PubMed] [Google Scholar]
- Jaedicke K, Lichtenthäler AL, Meyberg R, Zeidler M, Hughes J (2012) A phytochrome-phototropin light signaling complex at the plasma membrane. Proc Natl Acad Sci USA 109: 12231–12236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Stern D (2009) Mating and tetrad separation of Chlamydomonas reinhardtii for genetic analysis. J Vis Exp 12: 1274. 10.3791/1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kianianmomeni A, Hallmann A (2014) Algal photoreceptors: In vivo functions and potential applications. Planta 239: 1–26 [DOI] [PubMed] [Google Scholar]
- Liu B, Yang Z, Gomez A, Liu B, Lin C, Oka Y (2016) Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana. J Plant Res 129: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck M, Mathes T, Bruun S, Fudim R, Hagedorn R, Tran Nguyen TM, Kateriya S, Kennis JTM, Hildebrandt P, Hegemann P (2012) A photochromic histidine kinase rhodopsin (HKR1) that is bimodally switched by ultraviolet and blue light. J Biol Chem 287: 40083–40090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag M, Kiaulehn S, Johnson CH (2005) The circadian clock in Chlamydomonas reinhardtii. What is it for? What is it similar to? Plant Physiol 137: 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller N, Wenzel S, Zou Y, Künzel S, Sasso S, Weiß D, Prager K, Grossman AR, Kottke T, Mittag M (March 30, 2017) A plant cryptochrome controls key features of the Chlamydomonas circadian clock and its life cycle. Plant Physiol 174: 185–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P (2002) Channelrhodopsin-1: A light-gated proton channel in green algae. Science 296: 2395–2398 [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E (2003) Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA 100: 13940–13945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhoff V, Philipp K, Zimmer HG, Mesecke S (1979) A simple, versatile, sensitive and volume-independent method for quantitative protein determination which is independent of other external influences. Hoppe Seylers Z Physiol Chem 360: 1657–1670 [DOI] [PubMed] [Google Scholar]
- Niwa Y, Matsuo T, Onai K, Kato D, Tachikawa M, Ishiura M (2013) Phase-resetting mechanism of the circadian clock in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 110: 13666–13671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohr D, Franz S, Rodriguez R, Paulus B, Essen L-O, Weber S, Schleicher E (2016) Extended electron-transfer in animal cryptochromes mediated by a tetrad of aromatic amino acids. Biophys J 111: 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldemeyer S, Franz S, Wenzel S, Essen L-O, Mittag M, Kottke T (2016) Essential role of an unusually long-lived tyrosyl radical in the response to red light of the animal-like cryptochrome aCRY. J Biol Chem 291: 14062–14071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenhof H, Bisová K, van den Ende H, Zachleder V (2004) Effect of red and blue light on the timing of cyclin-dependent kinase activity and the timing of cell division in Chlamydomonas reinhardtii. Plant Physiol Biochem 42: 341–348 [DOI] [PubMed] [Google Scholar]
- Pan JM, Haring MA, Beck CF (1997) Characterization of blue light signal transduction chains that control development and maintenance of sexual competence in Chlamydomonas reinhardtii. Plant Physiol 115: 1241–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroutsos D, Tokutsu R, Maruyama S, Flori S, Greiner A, Magneschi L, Cusant L, Kottke T, Mittag M, Hegemann P, et al. (2016) A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature 537: 563–566 [DOI] [PubMed] [Google Scholar]
- Pozuelo M, Merchán F, Macías MI, Beck CF, Galván A, Fernández E (2000) The negative effect of nitrate on gametogenesis is independent of nitrate assimilation in Chlamydomonas reinhardtii. Planta 211: 287–292 [DOI] [PubMed] [Google Scholar]
- Reisdorph NA, Small GD (2004) The CPH1 gene of Chlamydomonas reinhardtii encodes two forms of cryptochrome whose levels are controlled by light-induced proteolysis. Plant Physiol 134: 1546–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Su Y-S, Lagarias JC (2006) Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Inoue M, Yamada M, Matsuda Y (1998) Control of gametic differentiation and activity by light in Chlamydomonas reinhardtii. Plant Cell Physiol 39: 8–15 [Google Scholar]
- Schmidt M, Gessner G, Luff M, Heiland I, Wagner V, Kaminski M, Geimer S, Eitzinger N, Reissenweber T, Voytsekh O, et al. (2006) Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell 18: 1908–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze T, Schreiber S, Iliev D, Boesger J, Trippens J, Kreimer G, Mittag M (2013) The heme-binding protein SOUL3 of Chlamydomonas reinhardtii influences size and position of the eyespot. Mol Plant 6: 931–944 [DOI] [PubMed] [Google Scholar]
- Sineshchekov OA, Jung KH, Spudich JL (2002) Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 99: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6: 175–182 [PubMed] [Google Scholar]
- Spexard M, Thöing C, Beel B, Mittag M, Kottke T (2014) Response of the Sensory animal-like cryptochrome aCRY to blue and red light as revealed by infrared difference spectroscopy. Biochemistry 53: 1041–1050 [DOI] [PubMed] [Google Scholar]
- Suetsugu N, Wada M (2013) Evolution of three LOV blue light receptor families in green plants and photosynthetic stramenopiles: Phototropin, ZTL/FKF1/LKP2 and aureochrome. Plant Cell Physiol 54: 8–23 [DOI] [PubMed] [Google Scholar]
- Suzuki L, Johnson CH (2002) Photoperiodic control of germination in the unicell Chlamydomonas. Naturwissenschaften 89: 214–220 [DOI] [PubMed] [Google Scholar]
- Takahashi F, Yamagata D, Ishikawa M, Fukamatsu Y, Ogura Y, Kasahara M, Kiyosue T, Kikuyama M, Wada M, Kataoka H (2007) AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proc Natl Acad Sci USA 104: 19625–19630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöing C, Oldemeyer S, Kottke T (2015) Microsecond deprotonation of aspartic acid and response of the α/β subdomain precede C-terminal signaling in the blue light sensor plant cryptochrome. J Am Chem Soc 137: 5990–5999 [DOI] [PubMed] [Google Scholar]
- Tilbrook K, Dubois M, Crocco CD, Yin R, Chappuis R, Allorent G, Schmid-Siegert E, Goldschmidt-Clermont M, Ulm R (2016) UV-B perception and acclimation in Chlamydomonas reinhardtii. Plant Cell 28: 966–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier U, Fuchs S, Weber M, Wakarchuk WW, Beck CF (1989) Gametic differentiation in Chlamydomonas reinhardtii—light dependence and gene-expression patterns. Arch Microbiol 152: 572–577 [Google Scholar]
- Trippens J, Greiner A, Schellwat J, Neukam M, Rottmann T, Lu Y, Kateriya S, Hegemann P, Kreimer G (2012) Phototropin influence on eyespot development and regulation of phototactic behavior in Chlamydomonas reinhardtii. Plant Cell 24: 4687–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener D, Treier U, Beck CF (1989) Procedures for the generation of mature Chlamydomonas reinhardtii zygotes for molecular and biochemical analyses. Plant Physiol 90: 512–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissig H, Beck CF (1991) Action spectrum for the light-dependent step in gametic differentiation of Chlamydomonas reinhardtii. Plant Physiol 97: 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey DL, Auffret AD, Gray JC (1984) Structure and topology of cytochrome f in pea chloroplast membranes. Cell 36: 555–562 [DOI] [PubMed] [Google Scholar]
- Zhao B, Schneid C, Iliev D, Schmidt EM, Wagner V, Wollnik F, Mittag M (2004) The circadian RNA-binding protein CHLAMY 1 represents a novel type heteromer of RNA recognition motif and lysine homology domain-containing subunits. Eukaryot Cell 3: 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorin B, Lu Y, Sizova I, Hegemann P (2009) Nuclear gene targeting in Chlamydomonas as exemplified by disruption of the PHOT gene. Gene 432: 91–96 [DOI] [PubMed] [Google Scholar]
- Zuo ZC, Meng YY, Yu XH, Zhang ZL, Feng DS, Sun SF, Liu B, Lin CT (2012) A study of the blue-light-dependent phosphorylation, degradation, and photobody formation of Arabidopsis CRY2. Mol Plant 5: 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]