The C terminus of the oleuropein-specific β-glucosidase from olive encompasses a bifunctional role to assemble an active multimeric enzyme and to regulate nuclear localization, keeping it conditionally inaccessible.
Abstract
Oleuropein, a terpene-derived glycosylated secoiridoid biosynthesized exclusively by members of the Oleaceae family, is involved in a two-component defense system comprising a β-glucosidase that activates oleuropein into a toxic glutaraldehyde-like structure. Oleuropein and its deglycosylated derivatives have high pharmaceutical interest. In this study we determined that the in planta heterologous expressed OeGLU, an oleuropein-specific β-glucosidase from olive (Olea europaea), had enzymatic kinetics similar to the olive native enzyme. The C terminus encompassing the nuclear localization signal sequesters the enzyme in the nucleus, and predetermines the protein-protein recognition and homodimerization. Biochemical analysis revealed that OeGLU is a homomultimer with high Mr. In silico prediction modeling of the complex structure and bimolecular fluorescence complementation analyses revealed that the C terminus of OeGLU is essential for the proper assembly of an octameric form, a key conformational feature that determines the activity of the enzyme. Our results demonstrate that intrinsic characteristics of the OeGLU ensure separation from oleuropein and keep the dual-partner defensive system conditionally inactive. Upon cell destruction, the dual-partner defense system is activated and olive massively releases the arsenal of defense.
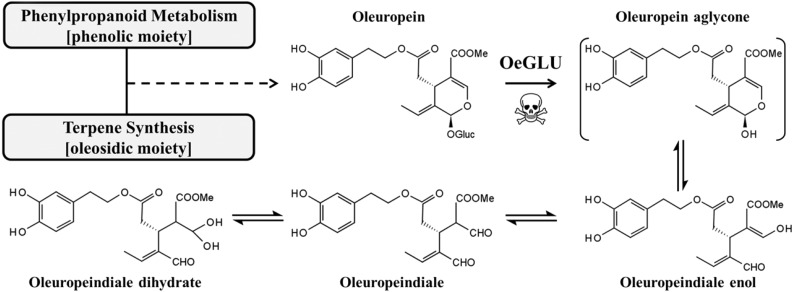
Secondary metabolism assists organisms to address (a)biotic challenges that will arise during their lifetime. A plethora of diverse secondary metabolites can be found in the plant kingdom, often restricted in certain plant families or even genera and species. Apart from the importance in plant ecophysiology, intermediates or final products of the secondary metabolic pathways have high nutritional or pharmaceutical value for humans. Over the last decade, many of the healthy advantages of the Mediterranean diet have been attributed to olives and olive oil (Visioli et al., 2002). Olea europaea (Oleaceae) produces a unique number of terpene-derived secoiridoids, known as oleosides, conjugates of glycosylated elenolic acid with a characteristic exocyclic 8,9-olefinic bond (Soler‐Rivas et al., 2000). The genus Caiophora (Loasaceae) and members of the Oleaceae family are the only known source of oleosides (Weigend et al., 2000; Jensen et al., 2002; Obied et al., 2008).
The dominant secoiridoid in olive is oleuropein, an ester of elenolic acid with 2′-(3′,4′-dihydroxyphenyl)ethanol (hydroxytyrosol), and accumulates up to 14% in young drupes and leaves (Amiot et al., 1986; Ortega-García and Peragón, 2010). Oleuropein is a metabolite of high interest for humans as it exhibits antioxidant, antiinflammatory, antiproliferative, antimicrobic and antiviral activity (Bulotta et al., 2013). Apart from a recently identified iridoid synthase (Alagna et al., 2016), other enzymes involved in the biosynthesis of oleuropein are unknown. This metabolite is stored in a glycosylated and thus inactive form, but upon cell disruption (i.e. by pathogen attack) the molecule becomes accessible to the physically separated oleuropein-specific β-glucosidase. The resulting aglycone form of oleuropein is unstable and rapidly converted to a highly reactive molecule with glutaraldehyde-like structure. This product covalently binds to amino acids and exhibits strong protein denaturing/cross-linking activities (Fig. 1; Konno et al., 1999; Uccella, 2000; Koudounas et al., 2015). The two components comprising a defensive glucoside and a specialized detonating β-glucosidase, usually separated in a distinct cellular compartment, are usually referred to as a “dual-partner defense system.” This strategic pluripotent defense is conserved in plants (Morant et al., 2008; Pentzold et al., 2014).
Figure 1.
Schematic representation of oleuropein and the main aglycone rearrangements after deglycosylation (Uccella, 2000).
β-Glucosidases (E.C. 3.2.1.21) catalyze the hydrolysis of glycosidic bonds from glucosides and oligosaccharides. Plants contain a large set of β-glucosidases involved in diverse biological functions ranging from phytohormone activation to defense (Katudat Cairns and Esen, 2010). Despite the pluralism of metabolites plants have evolved as chemical arsenal (i.e. alkaloids, glucosinolates, cyanogenic conjugates, saponins, benzoxazinoids, cardenolides and iridoids), glycosylation seems to be a conserved strategy that increases water solubility, reduces chemical reactivity and improves chemical stability (Jones and Vogt, 2001; Mithöfer and Boland, 2012; Piasecka et al., 2015). Defensive β-glucosidases have been diversified during evolution and acquired high specificity toward their respective substrates (Hösel and Conn, 1982; Verdoucq et al., 2004; Seshadri et al., 2009; Xia et al., 2012). To avoid any toxicity in the host cell, plants have invented a mechanism that involves discrete localization of the detonating enzyme and its respective metabolite/substrate. The decompartmentalization ensures that the toxic aglycone will be massively produced only after cell disruption. Well-documented examples are the glucosinolate/myrosinase system known as the “mustard oil bomb” (Brassicales), the avenacoside/avenacosidase system (Avena sativa), the DI(M)BOA/β-glucosidase system (Poaceae), and the cyanide bomb distributed in several plant families (Morant et al., 2008).
Olive oil is produced by crushing of olives followed by malaxation (coalescence of oil droplets through mixing of olive paste). During this process, oleuropein is exposed to peroxidases and polyphenoloxidases. Nevertheless it is widely accepted that the enzymatic activity of β-glucosidase on oleuropein is the dominant reaction to shape the composition of virgin olive oil in secoiridoid derivatives, defining therefore the oil’s nutritional and organoleptic properties (Obied et al., 2008; Romero-Segura et al., 2012; Hachicha Hbaieb et al., 2015). We have recently identified OeGLU, to our knowledge the first Glycoside Hydrolase Family 1 (GH1) β-glucosidase in Oleaceae able to activate oleuropein (Koudounas et al., 2015).
β-Glucosidases involved in the dual-partner defense systems exhibit highly diversified quaternary structures (Morant et al., 2008). A critical question is whether a quaternary assembly is necessary for the specificity and the activity of β-glucosidase enzyme. Recent advances have shown that agroinfiltration is a successful approach to comprehend specialized biochemical reactions by introducing the appropriate genes in tobacco (Nicotiana benthamiana) plants, especially when other heterologous expression systems fail to produce active enzymes (Miettinen et al., 2014; Lau and Sattely, 2015; Rajniak et al., 2015). Here we show that the oleuropein β-glucosidase (OeGLU) is sequestered in the nucleus due to a nuclear localization signal (NLS) present in the polypeptide. We further demonstrate that the C terminus of the polypeptide encompassing the NLS is decisive for protein/protein interaction. In the nucleus, the final subunit organization forms a high molecular mass structure. We further show that the high molecular mass structure necessitates the β-glucosidase activity. In silico analysis predicted that the enzyme could form a homopolymer composed of eight subunits, consistent with the estimation of the enzyme’s Mr.
RESULTS
OeGLU Kinetic Parameters
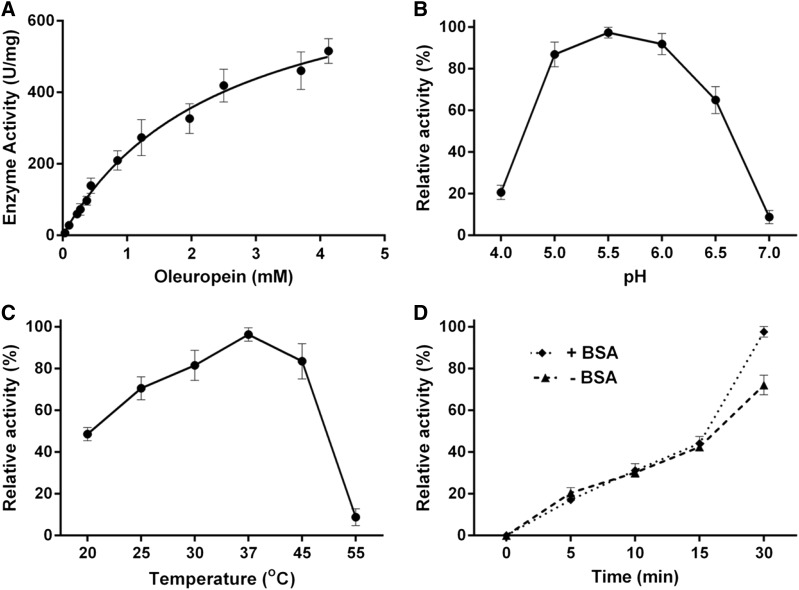
To determine whether the protein encoded by the heterologously expressed OeGLU gene has enzymatic characteristics similar to the native enzyme from olive, we performed three types of experiments. The heterologous synthesized OeGLU was assayed with the natural substrate oleuropein to determine the reaction kinetics. First, we estimated the Michaelis-Menten curve and showed that the Km of OeGLU for oleuropein is 2.48 mm (Fig. 2A). Then we determined the optimum pH of the reaction. The enzymatic activity exhibited a narrow optimum pH curve with highest activity at pH 5.5 and the relative activity showed a rapid decline beyond this pH value, reaching negligible β-glucosidase reactivity at pH 7.0 (Fig. 2B). Consistent with the ambient temperature of olive tree growth, a broad optimum temperature curve was obtained with highest activity at 37°C. The OeGLU retained greater than 50% of the highest enzymatic activity between 25°C and 45°C. β-Glucosidase activity was undetected at 55°C (Fig. 2C). The estimated enzymatic kinetic values of the heterologously expressed OeGLU gene were very similar to the ones determined using purified β-glucosidases from olive extracts (Romero-Segura et al., 2009; Gutierrez-Rosales et al., 2012).
Figure 2.
OeGLU kinetic parameters. Michaelis-Menten curve of OeGLU using oleuropein as substrate (A). Effect of pH (B), temperature (C), and the absence or presence of BSA in the reaction medium (D) on the OeGLU relative activity with oleuropein. Error bars, SD; n = 3 biological replicates.
Knowing that oleuropein aglycone exhibits strong protein-denaturing/protein-cross-linking properties (Koudounas et al., 2015), we asked whether the enzymatically produced oleuropein aglycone had any impact on the activity of the enzyme. Interestingly, no significant difference in the enzymatic activity could be observed between reactions incubated for 5, 10, or 15 min, in the presence or the absence of bovine serum albumin (BSA; i.e. providing another target for the oleuropein aglycone). In 30 min, OeGLU lost 25% of the original activity when incubated in the absence of BSA (Fig. 2D).
OeGLU Is Localized in the Nucleus
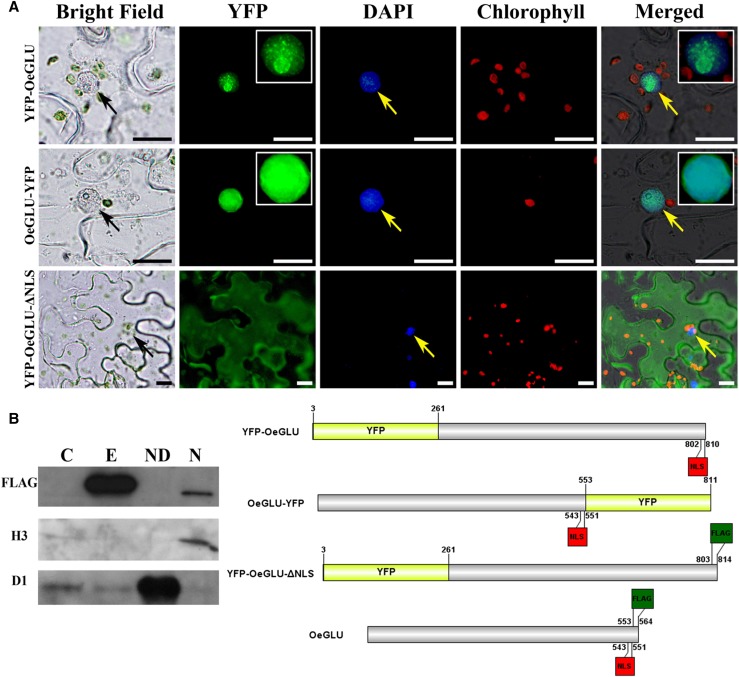
Oleuropein is localized in the vacuoles or cytosol of the cells (Konno et al., 1998; Bitonti et al., 2000). Accordingly, the OeGLU compartmentalization could allow the oleuropein/OeGLU dual-partner defense system to be inadequately active within olive cells where both OeGLU and oleuropein are highly accumulated (Amiot et al., 1986; Gutierrez-Rosales et al., 2012; Koudounas et al., 2015). In silico analysis of the OeGLU was predicted to contain a putative NLS at amino acid 542 to 550 (DRRKRLRGS) and no nuclear export signal (Supplemental Fig. S1). To rigorously test the location of the OeGLU, yellow fluorescent protein (YFP) fusion constructs were used. Although both OeGLU-YFP and YFP-OeGLU chimeric proteins clearly confirmed nuclear localization in transiently expressing leaf epidermal tobacco cells (Fig. 3A), the fluorescent pattern was different. When YFP was fused at the N terminus of the enzyme, discrete fluorescent foci were detected, whereas when YFP was fused at the C terminus of OeGLU, fluorescence was diffused throughout the nucleus (Fig. 3A). The distinctive fluorescent pattern has also been observed in transformants of the closely related strictosidine β-glucosidase from Madagascar periwinkle (Catharanthus roseus; Guirimand et al., 2010).
Figure 3.
OeGLU is localized in the nucleus. A, Epifluorescence microscopy of tobacco epidermal cells agroinfiltrated with YFP-fused constructs of OeGLU. OeGLU is localized in the nucleus as confirmed by both YFP-OeGLU and OeGLU-YFP constructs. Deletion of the predicted NLS (YFP-OeGLU-ΔNLS) resulted in a diffused pattern of fluorescence localized in the cytoplasm. Zoomed view (2×) of the nuclei of the YFP-OeGLU and OeGLU-YFP constructs under the YFP filter is depicted in insets. Nuclei were stained with DAPI. Scale bars, 10 μm. Arrows indicate nuclei. B, Western-blot analysis of crude protein extracts from tobacco plants agroinfiltrated with empty vector or with OeGLU. Nucleus-depleted and nuclear fractions of OeGLU. The protein samples were immunodetected either with anti-FLAG (sc-807), with antihistone H3 (nuclear marker), or with anti-D1 (chloroplastic marker) primary antibodies. Schematic representation of the constructs used. Numbers indicate the position of the amino acid residues. C, Empty vector; E, OeGLU; ND, nucleus-depleted; N, nuclear.
To further confirm the engagement of the NLS motif in nuclear targeting of the enzyme, we deleted the NLS sequence. Numerous observations of infiltrated leaf epidermal tobacco cells transiently expressing the YFP-OeGLU-ΔNLS construct revealed that fluorescence was localized solely in the cytoplasm (Fig. 3A). The integrity of the OeGLU (∼63 kD) fused with YFP (∼28 kD) chimeric polypeptides was confirmed by immunoblot analysis with anti-green fluorescent protein (anti-GFP) and as expected, a single band at ∼91 kD was detected (Supplemental Fig. S2). To validate the nuclear localization, immunoblot analysis of protein extracts from tobacco plants agroinfiltrated with OeGLU showed that the enzyme is detected only in the nuclear fraction and not in the nucleus-depleted fraction (Fig. 3B). These results confirmed the distinctive compartmentalization of the β-glucosidase enzyme in the nucleus.
Monomers of OeGLU Self-Recognize and Interact Autonomously
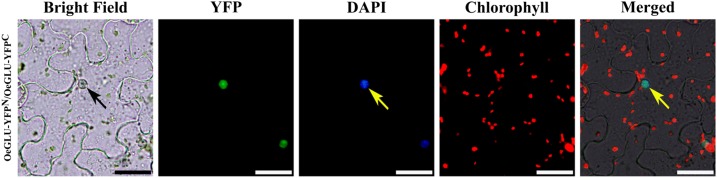
Defensive β-glucosidases from different plant species usually form oligomers of two or more subunits (Morant et al., 2008). To determine protein-protein recognition and interaction of OeGLU monomers, bimolecular fluorescence complementation (BiFC) analysis was applied. Transiently coexpressed OeGLU monomers fused to the C terminus with either YFPN or YFPC in tobacco leaf epidermal cells revealed a strong interaction and the reconstituted fluorescent signal was detected only in the nucleus (Fig. 4). As expected, no signal was observed in controls of coagroinfiltrated OeGLU-YFPN/C with the respective empty vectors (Supplemental Fig. S3). Consistent with OeGLU localization, protein-protein interaction pattern argues that monomers are rapidly deposited in the nucleus and then self-assembled.
Figure 4.
Monomers of OeGLU recognize and interact autonomously. In vivo self-interactions of OeGLU in leaf epidermal cells of N. benthamiana cotransformed with OeGLU fused with either YFPN or YFPC fragments using BiFC assays. Interaction between two OeGLU monomers allowed reconstitution of the YFP fluorochrome. Nuclei were stained with DAPI. Arrows indicate nucleus. Scale bars, 20 μm.
OeGLU Multimerizes
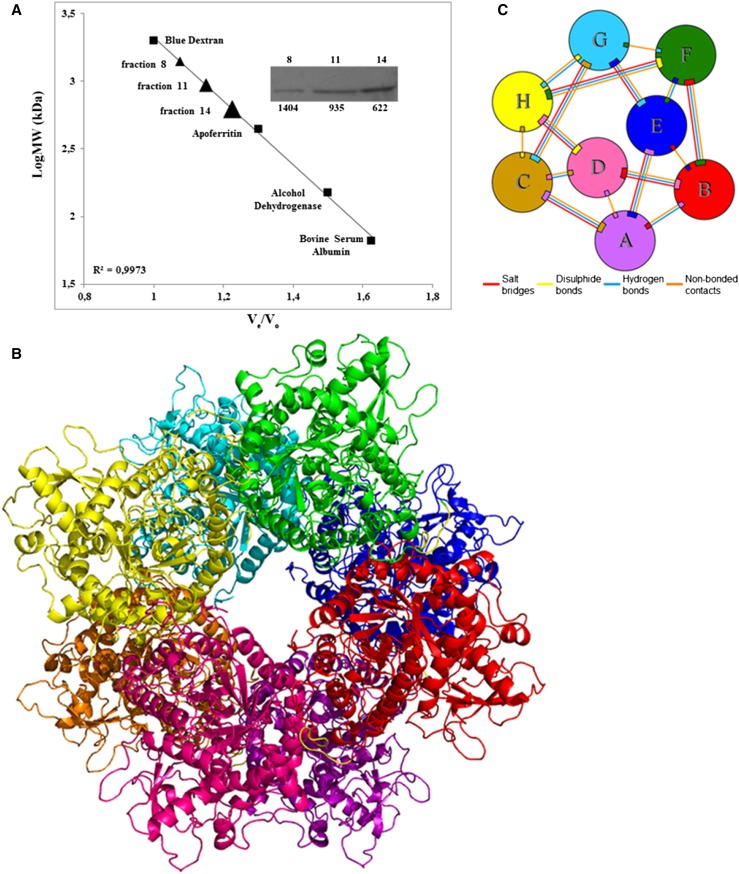
To determine the quaternary structure of the recombinant OeGLU, we used gel filtration chromatography of proteins extracted from tobacco plants transiently expressing the OeGLU. Immunoblot analysis of the fractions demonstrated that the native form of OeGLU multimerizes whereas a limited amount of the enzyme could be detected as an ∼63 kD monomer (Fig. 5A; Supplemental Fig. S4). OeGLU was mainly detected in the fraction of 622 kD and, to a lesser extent, in fractions of higher molecular mass. Taken together, OeGLU is a supra-Mr enzyme and the estimated size from gel filtration predicts that the native form of the enzyme acquires a higher than octamer conformation.
Figure 5.
OeGLU multimerizes. A, Calibration curve of the column used for gel filtration chromatography using the molecular weight marker proteins (squares). Protein extract from tobacco plants transiently expressing OeGLU, was applied to the column and the protein fractions obtained were immunodetected with anti-FLAG (Supplemental Fig. S4). OeGLU was mainly detected in the fractions indicated on the calibration curve (triangles). The numbers above the inset correspond to the protein fractions collected and the numbers below to the calculated molecular masses (measured in kD). B, Overall predicted quaternary structure of the octameric conformation of OeGLU based on in silico analysis. C, Schematic diagram of the interactions between eight monomers (A–H). Colored lines joining the polypeptide chain monomers represent a different type of interaction as indicated. Ve, elution volume; Vo, void volume; LogMW, log (molecular weights).
In Silico Prediction of the Quaternary Structure of OeGLU
Biochemical analysis revealed that the native form of the OeGLU is composed of more than eight subunits. To predict putative domains that contribute in the formation of a multimeric OeGLU, we performed an in silico analysis. Although the ab initio prediction of the quaternary structure of complexes is, as yet, a challenging field of computational biology, our pipeline successfully predicted amino acids of the C-terminal that possibly interact among the monomers in four (poses 1, 2, 3, and 5) out of the five suggested poses (Supplemental Fig. S5, A–G). Poses 1 to 4 had a high structural similarity score to well-known defensive plant β-glucosidases ranging from 0.9122 to 0.9273, and pose 5 had a lower similarity score (0.7745) with the structure of an archaeon β-glucosidase (Supplemental Fig. S5). Even though the tetramer (Supplemental Fig. S5G) was predicted to have a higher number of salt bridges than the octamer 28 versus 20 (Supplemental Fig. S5E), the latter was predicted to have 104 and the former 95 hydrogen bonds. Considering the high Mr of the native OeGLU enzyme as determined by chromatography analysis, we propose that the quaternary structure of the protein is composed of at least eight subunits (Figs. 5, B and C).
Deletion of the NLS Affects the Quaternary Structure of OeGLU, a Key Feature for the Enzymatic Activity
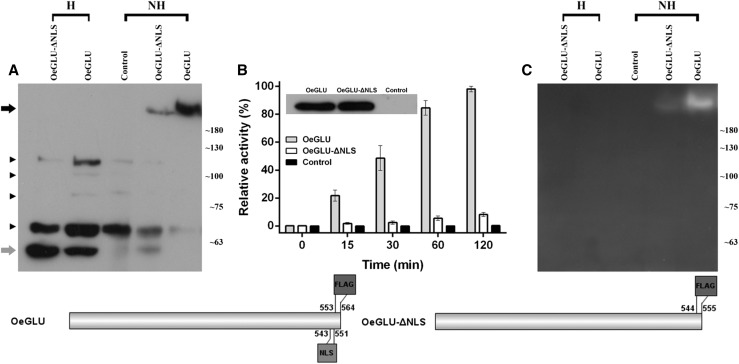
The repeatedly observed distinct pattern of fluorescence between the YFP-OeGLU and the OeGLU-YFP chimeric proteins in the nucleus (Fig. 3) has also been observed in strictosidine β-glucosidase from C. roseus (Guirimand et al., 2010). This feature prompted us to investigate the C-terminal engagement in the interaction of the monomers. We therefore asked whether deletion of the NLS would have an impact on the quaternary structure of OeGLU. Partially denatured conditions (without any heat treatment) of tobacco protein extracts separated on SDS-PAGE followed by immunoblot analysis with anti-FLAG, revealed that OeGLU was solely detected as a high-Mr multimer. Heat denaturation of the tobacco protein extracts revealed that the OeGLU was solely detected as a monomeric form (Fig. 6A). Surprisingly the transiently expressed OeGLU-ΔNLS in tobacco leaves showed an increased ratio of the monomeric over the multimeric form. Nevertheless, the multimeric form derived from OeGLU-ΔNLS had similar molecular mass to the one derived from OeGLU (Fig. 6A). The integrity of tobacco proteins was verified with Coomassie brilliant blue (CBB) staining (Supplemental Fig. S6). The deletion of the nine amino acids at the C terminus had a major impact on the stability of the OeGLU quaternary structure.
Figure 6.
Deletion of the NLS affects the quaternary structure of OeGLU, a key feature for the enzymatic activity. A, Western blot with anti-FLAG antibody (sc-166355) of crude protein extracts from tobacco plants transiently expressing OeGLU-ΔNLS, OeGLU, and Control (empty vector). Equal amounts of proteins (10 μg) were loaded on polyacrylamide gels (6%) and the samples were either heat treated at 90°C for 5 min, or not heat treated before electrophoresis. Black and gray arrows on the left indicate the multimeric and the monomeric forms of OeGLU, respectively. Arrowheads indicate unspecific cross reactivity of the antibody. Molecular mass markers are given on the right in kilodaltons. B, Relative enzymatic activity of the tobacco crude protein extracts expressing OeGLU, OeGLU-ΔNLS, or Control (empty vector) constructs using oleuropein as substrate was measured by HPLC in indicated time intervals. From this specific series of agroinfiltration, equal quantities of the transiently expressed constructs were verified by western blotting of heated (i.e. monomeric) protein samples (depicted in the inset) using anti-FLAG antibody (sc-807). Error bars, SD; n = 3 biological replicates. C, Zymogram of protein extracts as in (A) was developed with 1.5 mm MUGlc. Molecular mass markers are given on the right in kD. Schematic representation of the constructs used. Numbers indicate the position of the amino acid residues. H, heat treated; NH, non-heat treated.
Because the NLS is not a part of the core (β/α)8 TIM-barrel fold and none of the predicted amino acids that constitute the active site of OeGLU are located in this motif (Koudounas et al., 2015), we questioned whether this structurally altered variant had an impact on the enzymatic activity of β-glucosidase. To test the activity, we performed an HPLC analysis with oleuropein as a substrate. Although both constructs were equally expressed as revealed by western blot, the relative activity of the OeGLU-ΔNLS mutant was <10% compared to the wild-type OeGLU (Fig. 6B). To exclude the possibility that the monomeric variant of OeGLU is enzymatically active but the activity had become deteriorated by the produced oleuropein aglycone, which exhibits protein-denaturing/protein-cross-linking properties, we performed a zymogram using the general substrate 4-methylumbelliferyl-β-d-glucopyranoside (MUGlc). The multimeric variant of OeGLU-ΔNLS and the wild-type OeGLU were able to deglycosylate MUGlc (Fig. 6C). No fluorescence could be detected in the monomeric form of OeGLU-ΔNLS or OeGLU, indicating that the monomers have negligible β-glucosidase activity. Taken together, the results strongly suggest that the assembly of the multimeric form of OeGLU is actively regulated by the C-terminal domain and is a prerequisite for the activity of the enzyme.
The C Terminus of OeGLU Is Necessary and Sufficient for the Interaction of the Monomers
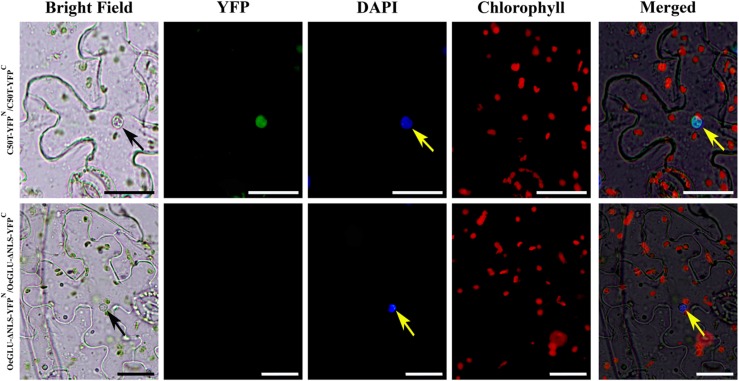
To validate and further examine the engagement of the C terminus in the protein-protein interaction of OeGLU monomers, we performed two sets of complementary experiments using BiFC assays. A protein-protein interaction of a construct lacking the NLS (OeGLU-ΔNLS) resulted in no signal. Despite numerous observations, no fluorescence could be detected in tobacco epidermal cells transiently expressing the OeGLU-ΔNLS-YFPN/C constructs (Fig. 7). On the other hand, fluorescence due to reconstitution of the YFP was observed in the nucleus of tobacco epidermal cells transiently expressing the C50T-YFPN/C constructs comprising the last 50 C-terminal amino acids of OeGLU (Fig. 7). In control tobacco leaf cells, no fluorescence was observed when C50T-YFPN or C50T-YFPC and OeGLU-ΔNLS-YFPN or OeGLU-ΔNLS-YFPC constructs were transiently coexpressed with the appropriate YFP domain (Supplemental Fig. S7). Taken together, the last 50 amino acids of OeGLU at C terminus of the polypeptide, encompassing the NLS, are sufficient to reconstitute a BiFC signal suggesting that the amino acids in this domain execute strong interactions of OeGLU monomers to form an active oleuropein β-glucosidase enzyme. Consistent with this conjecture, in silico analysis of the quaternary structure predicted the amino acids at the C terminus of the OeGLU to be highly engaged in bonding of monomers (Supplemental Fig. S5).
Figure 7.
The C terminus of OeGLU is necessary and sufficient for the interaction of the monomers. Agrobacterium tumefaciens suspensions harboring the C50T-YFPN/C or the OeGLU-ΔNLS-YFPN/C constructs were coinfiltrated in tobacco epidermal leaf cells. Interaction between two monomers allowed reconstitution of the YFP fluorochrome only in the case of C50T. Nuclei were stained with DAPI. Scale bars, 20 μm. Arrows indicate nuclei.
Detection of the Oleuropein β-Glucosidase-Specific Activity in Zymograms
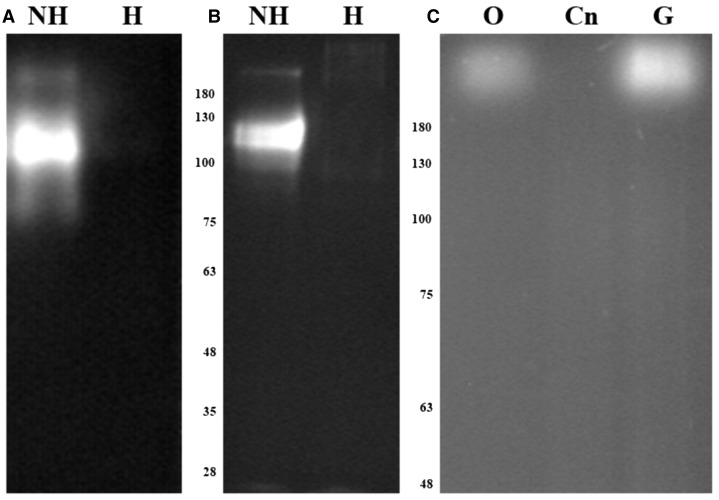
Oleuropein is a nonfluorescent metabolite (Obied et al., 2007), but HPLC coupled with fluorescence detector has been proven to be an efficient methodology to detect and quantify an isomer of oleuropein aglycone (Selvaggini et al., 2006). This difference in fluorescence between the glycosylated and aglycone forms of oleuropein has not been highlighted in the literature. We questioned whether we could detect the deglycosylation of oleuropein using zymography to compare OeGLU with other endogenous β-glucosidases of olive. Because almond (Prunus dulcis) β-glucosidase is often used to produce oleuropein aglycone (Capasso et al., 1997), we initially used this commercially available purified enzyme to perform in-gel activity assays (Fig. 8A). Surprisingly, using almond β-glucosidase, a similar fluorescent band was detected either when the gel was developed with the commonly used MUGlc as an unspecific substrate or when developed with oleuropein as substrate (Fig. 8B). No fluorescence could be detected when almond β-glucosidase was heat denatured, revealing that the fluorescent signal corresponds to enzymatic deglycosylation of oleuropein or MUGlc by almond β-glucosidase (Fig. 8, A and B).
Figure 8.
Deglycosylation of oleuropein detected on polyacrylamide gels. Zymograms were developed using MUGlc or oleuropein as substrates. Purified almond β-glucosidase (10 μg) was either non-heat or heat treated, resolved on 10% SDS-PAGE, and developed with 1.5 mm MUGlc (A) or 3 mm oleuropein (B). Proteins extracted from olive mesocarp (2.5 μg) or tobacco plants infiltrated with empty vector (25 μg), or expressing OeGLU (25 μg), were resolved on 7% SDS-PAGE and developed with 1.5 mm oleuropein (C). Molecular mass markers are given in kilodaltons. Cn, Empty vector; G, OeGLU; H, heat treated; NH, non-heat treated; O, olive mesocarp.
The same oleuropein staining approach was further applied on crude protein extracts from olive mesocarp along with extracts from tobacco plants transiently expressing OeGLU. Clearly the zymogram developed with oleuropein resulted in fluorescent bands derived from the specific production of oleuropein aglycone. The deglycosylation of oleuropein by the multimeric form of OeGLU from olive or tobacco extracts had similar molecular mass and pattern (Fig. 8C). Interestingly, the enzymatic activity of OeGLU was retained despite the addition of SDS and β-mercaptoethanol (Supplemental Fig. S8). Although this outcome seems to be contradictory, plant β-glucosidases are known to have a compact quaternary structure that remains active in concentrations of up to 3.2% SDS and 5% β-mercaptoethanol (Esen and Gungor, 1991, 1993) or even exhibit enhanced activity in the presence of reducing agents (Esen, 1992; Odoux et al., 2003). Because a single fluorescent band was detected in olive mesocarp protein extracts reacting with oleuropein that had a similar molecular mass compared to the transiently expressed olive β-glucosidase in tobacco cells, OeGLU is the major oleuropein-specific β-glucosidase in olive.
DISCUSSION
The interplay between plants and herbivores has armored the former with diversified glycosylated defensive metabolites activated upon specific β-glucosidases (Morant et al., 2008), providing mighty arsenals against a broad spectrum of herbivores. Accumulated data have highlighted a distinctive compartmentalization of the dual-partner defense system in different organelles to prevent the autotoxicity. Oleuropein is the principal secoiridoid in O. europaea and provides the chemical arsenal in members of the Oleaceae family after β-glucosidase activity (Konno et al., 1999; Soler‐Rivas et al., 2000; Koudounas et al., 2015). Recently, oleuropein and derivatives of the secoiridoid biochemical pathway have drawn attention as natural products with high pharmaceutical interest (Bulotta et al., 2013).
OeGLU is a GH1 β-glucosidase that activates oleuropein into a potent protein cross linker due to a glutaraldehyde-like structure (Koudounas et al., 2015). Here, we demonstrate that the activity of the enzyme is attributed to the formation of a supramolecular structure and in silico analysis predicted the formation of an octamer. Further, we delineate the importance of the C terminus, encompassing the NLS motif, to play a major role in protein-protein interaction of homodimer formation and multimerization. This process leads to the formation of an active enzyme only when the protein is assembled in the nucleus providing the fundamental properties to physically separate OeGLU from its substrate. We further determined the kinetic parameters of OeGLU and developed an in situ fluorescent oleuropein-specific assay to monitor the deglycosylation of oleuropein.
The purified enzyme from tobacco leaves transiently expressing the OeGLU exhibited a Km of 2.48 mm for oleuropein. This value is very similar to the reported 2.78 mm Km of the olive-purified β-glucosidase for the substrate oleuropein (Gutierrez-Rosales et al., 2012). The optimum pH and temperature of the transiently expressed enzyme activity were 5.5 and 37°C, respectively. These results coincide well with previous studies of the purified β-glucosidase enzymes from olive (Romero-Segura et al., 2009; Kara et al., 2011). Taken together, the heterologous expression of OeGLU gene in tobacco cells produced a metabolically active enzyme having similar biochemical characteristics and kinetics to the olive enzyme.
Three independent approaches (i.e. subcellular fractionation followed by western-blot analysis, imaging of YFP-fused constructs, and BiFC analysis) demonstrated that OeGLU is targeted to the nucleus and deletion of the predicted NLS, located in the C terminus, resulted in cytoplasmic localization. This result coincides with the intense in situ staining of β-glucosidase activity observed in the nucleus of cells surrounding olive fruit injuries (Spadafora et al., 2008). In addition, the closely related strictosidine and raucaffricine specific β-glucosidases and a β-glucosidase from barrel clover (Medicago truncatula) involved in flavonoids have nuclear localization (Naoumkina et al., 2007; Guirimand et al., 2010). Nuclear localization of enzymes involved in both anabolic (Saslowsky et al., 2005) and catabolic (Stavrinides et al., 2015) pathways of heterogeneous and diversified secondary metabolites (i.e. flavonoids and alkaloids) strongly indicate that metabolic channeling through the plant nucleus is an important but underestimated key characteristic in secondary metabolism. The reason why evolution has chosen this compartment as a metabolic hub and the impact in plant physiology remains to be addressed.
Although the tertiary structure of plant GH1 β-glucosidases is highly conserved, the quaternary structures of the active enzymes are exceedingly diversified (Morant et al., 2008). To get insights into the quaternary structure of OeGLU, we conducted gel filtration chromatography and BiFC analysis. Consistent with the fact that several defensive plant β-glucosidases are known to form supramolecular structures of up to 1500 kD (Fan and Conn, 1985; Nisius, 1988; Fieldes and Gerhardt, 1994; Geerlings et al., 2000), gel filtration chromatography analysis revealed that OeGLU assembles into high-molecular-mass aggregates, mainly a multimer of ∼622 kD.
Because OeGLU that was heterologously expressed in a non-oleuropein-producing plant had similar molecular mass compared to the native olive β-glucosidase, as revealed by zymography, we concluded that OeGLU homomultimerizes autonomously. Alternatively, common aggregation processes or factors present also in the tobacco nucleus could possess the specific activity to multimerize the OeGLU. A fluorescence signal detected in the nucleus of coagroinfiltrated N. benthamiana leaves, due to reconstitution of YFP in BiFC analysis, confirmed the physical interaction and the autonomous multimerization. Supporting this conclusion, heterologous expression of the closely related strictosidine β-glucosidase in a prokaryote resulted in an active multimeric form (Guirimand et al., 2010). It has been proposed that aggregated β-glucosidases exhibit enhanced enzymatic activity (Kim and Kim, 1998; Lee et al., 2006) and increased resistance against herbivore proteases (Gus-Mayer et al., 1994; Guirimand et al., 2010). Interestingly, oligomerization of a GH1 β-glucosidase from the psychrophilic prokaryote Exiguobacterium antarcticum B7 was correlated with cold adaptation (Zanphorlin et al., 2016). Taking into consideration that plants are obliged to recruit their defensive β-glucosidases in a wide temperature range, multimerization may also be a strategy to retain the activity at low temperatures. In line with this notion, the key feature of β-glucosidase supramolecular structure determines its activity. Oleuropein aglycone with glutaraldehyde-like structure covalently binds to amino acids and exhibits strong protein denaturing/cross-linking properties (Konno et al., 1999; Koudounas et al., 2015). However, OeGLU retains more than 75% of its initial activity in the absence of BSA. This shows that the supramolecular enzyme structure is not prone to oleuropein aglycone protein-denaturing/protein-cross-linking properties, suggesting that the enzyme compensates the denaturing properties of aglycone to continue the enzymatic reaction.
In-gel activity assay is a reliable technique used for detecting enzymatic reactions (Gabriel and Gersten, 1992), and when combined with a specialized substrate allows the direct detection of specific enzymes from crude extracts (Luijendijk et al., 1996). Oleuropein is a nonfluorescence substrate (Obied et al., 2007) but deglycosylation by specific β-glucosidase is a fluorogenic reaction, as revealed by zymography. To the best of our knowledge, this is reported for the first time and raises the possibility of performing highly specific zymograms to detect isozymes among members of the Oleaceae family. This scheme could be used during protein purification steps or to rapidly screen oleuropeinolytic microorganisms, a subject of intense interest for biological debittering of olives and biotechnological use (Ciafardini et al., 1994; Arroyo-López et al., 2012).
Surprisingly, the last nine amino acid residues at the C terminus, encompassing the NLS motif, had also a major impact in the quaternary structure of the native OeGLU form. Deletion of the nine amino acid residues (OeGLU-ΔNLS) resulted in an increased ratio of the monomeric over the multimeric form, suggesting that other domains of the polypeptide are engaged in the formation of the quaternary structure as predicted by in silico analysis. However, zymography and HPLC analysis confirmed that the quaternary structure assembly assisted by the NLS motif is required for OeGLU to exhibit enzymatic activity.
The NLS acts as an aggregating motif and the OeGLU-ΔNLS construct did not reconstitute the fluorescence, showing that the OeGLU-ΔNLS variant did not interact in protein/protein BiFC assays. This outcome could be attributed to either low affinity of self-recognition of the monomers or to the presence of half of the YFP at the C terminus constraining the recognition affinity. Further, the last 50 amino acids reconstituted the fluorescent signal in BiFC assays, revealing that the C terminus is responsible for the nuclear localization, and recognition and interaction of monomers to form the supramolecular active form of OeGLU in the nucleus. Consistent with this result, when YFP was fused with OeGLU at the C terminus (OeGLU-YFP), the pattern of the nuclear fluorescent signal was distinct from that observed when YFP was fused to the N terminus (YFP-OeGLU) of the enzyme. The latter showed that the fluorescent signal had a speckle appearance, indicative of enzymatic foci and aggregation to form a supramolecular enzyme structure in the nucleus. In silico prediction of the quaternary structure of OeGLU agreed that four out of five computed poses engage amino acid residues at the C terminus to form a self-assembly suprastructure. The size of the predicted octameric structure is very close to the experimentally estimated molecular mass of OeGLU.
It has been suggested that the C terminus of the closely related strictosidine-specific β-glucosidase has to be accessible for proper conformation (Guirimand et al., 2010). Interestingly, substitution of a single amino acid in the C terminus of the dimeric AsGLU2 β-glucosidase from oat with the corresponding residue from the multimeric AsGLU1, resulted in a homomultimeric AsGLU2 that exhibited enhanced enzymatic activity (Kwak et al., 2009). Further, substitution of 25 amino acids in the N terminus of the hexameric DIMBOA β-glucosidase from wheat (Triticum aestivum) with the corresponding residues from the dimeric DIMBOA β-glucosidase from maize (Zea mays) converted the enzyme in a dimeric form with barely detected enzymatic activity (Sue et al., 2006). Taken together, N- or C-terminal domains actively participate and precisely regulate the quaternary structure of the enzymatically active forms of plant GH1 β-glucosidases.
In conclusion, OeGLU is transferred into the nucleus by its NLS and multimerizes, due to the C-terminal domain, into a supramolecular active enzyme, therein preventing any potential cytoplasmic leakage. This compartmentalization ensures separation from oleuropein, and keeps the dual-partner defensive system conditionally inactive, avoiding the production of the potent cytotoxic aglycone. This physical separation is lost when the olive cell is destroyed or invaded by herbivores, bringing the dual-partner defense system in close proximity and activating the enzymatic reaction, thus the olive massively releases the arsenal of defense.
MATERIALS AND METHODS
OeGLU Purification and Kinetics
Crude proteins from Nicotiana benthamiana leaves agroinfiltrated with the pGPTV/35S-OeGLU-FLAG construct were extracted at 6 d postinfiltration (dpi) as described by Koudounas et al. (2015). OeGLU was purified by affinity chromatography using the FLAG Immunoprecipitation Kit (cat. no. FLAGIPT1; Sigma-Aldrich) in accordance with the manufacturer’s instructions. The kinetic parameters of the purified OeGLU (e.g. Km) were determined by incubating 20 ng of enzyme with oleuropein (Extrasynthese) in the range of 0.01 to 4.50 mm and measuring the deglycosylation degree by HPLC as described by Koudounas et al. (2015). The Michaelis-Menten curve was generated using the GraphPad Prism program (GraphPad Software). Optimum pH of the enzymatic reaction was determined by incubating 20 μg crude extract of tobacco plants expressing OeGLU with 5 mm oleuropein in the pH range of 4 to 7 at 37°C for 30 min, using sodium acetate and P buffers (150 mm). Optimum temperature was determined in the range of 20 to 55°C in 150 mm sodium acetate buffer (pH 5.5) for 30 min. The inhibitory effect of the produced oleuropein aglycone on OeGLU activity was examined during enzyme incubation in 150 mm sodium acetate buffer (pH 5.5) at 37°C, in the presence or absence of 0.05% (w/v) BSA. The enzyme activity was monitored at regular time intervals. All reactions were performed at least three times. Two technical replicates from each of the three biological replicates were assayed.
Isolation of Nuclear Fraction
Young N. benthamiana leaves agroinfiltrated with the pGPTV/35S-OeGLU-FLAG construct were frozen in liquid nitrogen and ground into powder using a mortar and pestle. Approximately 2 g of the powder was resuspended in 30 mL of ice-cold Nuclei Extraction buffer 1 (0.4 m Suc, 10 mm Tris-HCl pH 8.0, 10 mm MgCl2, 5 mm β-mercaptoethanol, and 1 mm PMSF). The solution was filtered through Miracloth, an aliquot of the filtrate was collected as input material, and the remaining was centrifuged for 20 min at 1,900g at 4°C. The supernatant was gently removed, the pellet was resuspended in 1 mL Nuclei extraction buffer 2 (0.25 m Suc, 10 mm Tris-HCl pH 8.0, 10 mm MgCl2, 0.5% Triton X-100, 5 mm β-mercaptoethanol, and 1 mm PMSF), and then centrifuged for 20 min at 1,900g at 4°C. The supernatant was collected and referred to as “nucleus-depleted fraction.” The pellet was resuspended in 0.3 mL nuclei extraction buffer 2 (containing 0.15% Triton X-100), slowly overlayed on 0.5 mL nuclei extraction buffer 3 (1.7 m Suc, 10 mm Tris-HCl pH 8.0, 2 mm MgCl2, 0.15% Triton X-100, 5 mm β-mercaptoethanol, and 1 mm PMSF), and centrifuged for 45 min at 16,000g at 4°C. This step was repeated until the pellet appeared white. Finally the pellet was resuspended in 0.06 mL nuclei extraction buffer 1 without Suc and is referred to as “nuclear fraction.”
Immunoblotting Analysis
Protein samples were quantified by the Bradford assay (Bradford, 1976) and subjected to SDS-PAGE as described by Koudounas et al. (2015). Gels were stained with CBB or analyzed by western blot using two anti-FLAG antibodies (cat. nos. sc-807 and sc-166355; Santa Cruz Biotechnology), antihistone H3 (cat. no. 4499; Cell Signaling Technology), or anti-D1 (Zhang et al., 1999) as primary antibodies, and horseradish peroxidase-conjugated goat antirabbit IgG as a secondary antibody (cat. no. sc-2004; Santa Cruz Biotechnology) according to standard protocols.
YFP Imaging
For subcellular localization analysis, the cDNA of OeGLU was amplified by RT-PCR and cloned into the SmaI site of pUC18 using the primers ForKozak and RevMluIStop, or ForKozakMluI and RevStop to introduce an MluI restriction site at the C or N terminus, respectively (Supplemental Table S1). After verification by sequencing, the Citrine-YFP reporter gene was introduced into the MluI site, as described by Daras et al. (2014), to generate the in-frame fused constructs OeGLU-YFP and YFP-OeGLU, respectively. The constructs were directly cloned into the pGPTV-HPT binary vector, carrying the 35S cauliflower mosaic virus promoter using the XbaI and SacI restriction sites. The same cloning strategy was applied for the preparation of the YFP-OeGLU-ΔNLS construct using the primers ForKozakMluI and ΔNLS-FLAG (Supplemental Table S1) to delete the predicted NLS at amino acids 542 to 550 (DRRKRLRGS). The resulting vectors were introduced into Agrobacterium tumefaciens strain C58C1 RifR and infiltrated into the abaxial side of 2- to 4-week-old N. benthamiana leaves using a syringe without a needle as described by Koudounas et al. (2015). Leaf epidermal cells (3 dpi) were incubated with 5 μm 4',6-diamidino-2-phenylindole (DAPI) for 3 min and examined under a model no. BX-50 epifluorescence microscope (Olympus) equipped with the HBO 100 W/2 mercury short-arc lamp (Osram). YFP fluorescence was visualized using a cat. no. 41017 Endow GFP Bandpass filter set (excitation filter of 450–490 nm, beamsplitter of 495-nm long-pass, emission filter of 500–550 nm; Chroma Technology). Chlorophyll autofluorescence was visualized using the U-MSWG filter set (excitation filter of 480–550 nm, dichroic mirror of 570 nm, emission filter of >590 nm; Olympus) and DAPI fluorescence was visualized using the U-MWU filter set (excitation filter of 330–385 nm, dichroic mirror of 400 nm, emission filter of >420 nm; Olympus). Images were captured with a model no. DP71 camera (Olympus) using Cell^A (Olympus Soft Imaging Solutions). Merged images were processed with Adobe Photoshop CS5 (version 12.0; Adobe Systems). The integrity of the fused proteins was additionally verified with immunoblot analysis using anti-GFP (cat. no. sc8334; Santa Cruz Biotechnology) as primary antibody that recognizes the GFP variants including YFP.
BiFC Studies of OeGLU Interactions
For BiFC analysis, the cDNA of OeGLU was amplified by RT-PCR and cloned into the SmaI site of pUC18 using the primers ForKozak or ForMluIC50T and RevSPLT or RevSPLT-ΔNLS (Supplemental Table S1). The products were verified by sequencing and were directly cloned into the pSPYNE and pSPYCE vectors using the XbaI and XhoI restriction sites to generate in-frame fusions with YFPN (amino acids 1–155), and YFPC (amino acids 156–239), respectively. The resulting vectors were introduced in A. tumefaciens strain C58C1 RifR and infiltrated into the abaxial side of 2- to 4-week-old N. benthamiana leaves. Specific BiFC protein-protein interactions were examined by epifluorescence microscopy using a model no. BX-50 epifluorescence microscope (Olympus) and the filter sets described.
Gel Filtration Chromatography
Crude proteins from N. benthamiana leaves agroinfiltrated with the pGPTV/35S-OeGLU-FLAG construct were extracted at 6 dpi, as described by Koudounas et al. (2015). Ammonium sulfate was added to the extract and the pellet obtained at 55% saturation was collected. The pellet was dissolved in the initial buffer and the concentrated protein was used for gel filtration chromatography. Gel filtration was performed at 4°C using a Sephacryl S-300 HR column (GE Healthcare Bio-Sciences) precalibrated with BSA (66 kD), alcohol dehydrogenase (150 kD), apoferritin (443 kD), and blue-dextran (2,000 kD; Sigma-Aldrich). Fractions of 1 mL were collected, concentrated five times using the methanol/chloroform/water precipitation protocol (Wessel and Flügge, 1984), and evaluated using anti-FLAG immunoblot analysis.
In Silico Analysis of OeGLU
Subcellular localization predictions were performed using cNLS Mapper (Kosugi et al., 2009) or WoLF PSORT (Horton et al., 2007). The tertiary protein structure prediction of OeGLU (NCBI accession no. AAL93619) was generated ab initio with the Phyre2 application (Kelley et al., 2015) using the intensive modeling mode. The quaternary protein structure of OeGLU was predicted by submitting the tertiary model to the GalaxyGemini server (Lee et al., 2013). The resulting poses were analyzed with the software PyMOL (DeLano, 2002) and protein-protein interactions were identified with the software PDBsum (Laskowski, 2001).
Oleuropein Activity Staining in Polyacrylamide Gels
For the in-gel oleuropein activity staining, 10 μg of purified almond β-glucosidase (Sigma-Aldrich) were subjected to 10% SDS-PAGE under seminative conditions as described in Koudounas et al. (2015) and incubated with either 1.5 mm MUGlc or 3 mm oleuropein for 10 min. The fluorescent bands corresponding to β-glucosidase activity were photographed under UV light. The same staining method (developed with 1.5 mm oleuropein) was used for crude protein extracts, subjected to seminative 7% SDS-PAGE, from tobacco plants (25 μg) infiltrated with either the pGPTV/35S-OeGLU-FLAG or the pGPTV/35S (empty vector) construct, along with crude protein extracts (2.5 μg) from 17 weeks after flowering olive (Olea europaea) mesocarp.
Accession Numbers
The full-length cDNA sequence of OeGLU can be found in the GenBank/EMBL/DDBJ databases under accession no. AY083162.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence alignment of the proteins OeGLU and CrSGD.
Supplemental Figure S2. Protein extracts from tobacco plants expressing different Oe-GLU variants.
Supplemental Figure S3. Controls of BiFC assays.
Supplemental Figure S4. Western-blot analysis of fractions obtained after gel-filtration chromatography.
Supplemental Figure S5. In silico analysis and prediction of the quaternary structure of OeGLU.
Supplemental Figure S6. CBB staining of crude protein extracts from tobacco plants transiently expressing OeGLU or OeGLU-ΔNLS.
Supplemental Figure S7. Controls of OeGLU dimerization assays.
Supplemental Figure S8. Effect of reducing agents and SDS on specific enzymatic activity of OeGLU.
Supplemental Table S1. List of primers and constructs used in this study.
References
- Alagna F, Geu-Flores F, Kries H, Panara F, Baldoni L, O’Connor SE, Osbourn A (2016) Identification and characterization of the iridoid synthase involved in oleuropein biosynthesis in olive (Olea europaea) fruits. J Biol Chem 291: 5542–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiot MJ, Fleuriet A, Macheix JJ (1986) Importance and evolution of phenolic compounds in olive during growth and maturation. J Agric Food Chem 34: 823–826 [Google Scholar]
- Arroyo-López FN, Romero-Gil V, Bautista-Gallego J, Rodríguez-Gómez F, Jiménez-Díaz R, García-García P, Querol A, Garrido-Fernández A (2012) Yeasts in table olive processing: desirable or spoilage microorganisms? Int J Food Microbiol 160: 42–49 [DOI] [PubMed] [Google Scholar]
- Bitonti M, Chiappetta A, Innocenti A, Uccella N (2000) Biomolecular characterisation and histological distribution of biophenols in green mature fruit of Olea europaea Cassanese cv. In C Vitagliano, GP Martelli, eds, IV International Symposium on Olive Growing, Valenzano, Italy. ISHS Acta Horticulturae, Leuven, Belgium 586, pp 515–519 [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bulotta S, Oliverio M, Russo D, Procopio A (2013) Biological activity of oleuropein and its derivatives. In Ramawat KG, Mérillon J-M, eds, Natural Products: Phytochemistry, Botany and Metabolism of Alkaloids, Phenolics and Terpenes. Springer, Berlin, Germany, pp 3605–3638 [Google Scholar]
- Capasso R, Evidente A, Visca C, Gianfreda L, Maremonti M, Greco G (1997) Production of glucose and bioactive aglycone by chemical and enzymatic hydrolysis of purified oleuropein from Olea europea. Appl Biochem Biotechnol 61: 365–377 [Google Scholar]
- Ciafardini G, Marsilio V, Lanza B, Pozzi N (1994) Hydrolysis of oleuropein by Lactobacillus plantarum strains associated with olive fermentation. Appl Environ Microbiol 60: 4142–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daras G, Rigas S, Tsitsekian D, Zur H, Tuller T, Hatzopoulos P (2014) Alternative transcription initiation and the AUG context configuration control dual-organellar targeting and functional competence of Arabidopsis Lon1 protease. Mol Plant 7: 989–1005 [DOI] [PubMed] [Google Scholar]
- DeLano WL. (2002) The PyMOL Molecular Graphics System. Delano Scientific, San Carlos, CA [Google Scholar]
- Esen A. (1992) Purification and partial characterization of maize (Zea mays L.) β-glucosidase. Plant Physiol 98: 174–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esen A, Gungor G (1991) Detection of β-glucosidase activity on sodium dodecyl sulphate-polyacrylamide gels. Appl Theor Electrophor 2: 63–69 [PubMed] [Google Scholar]
- Esen A, Gungor G (1993) Stability and activity of plant and fungal β-glucosidases under denaturing conditions. In Esen A, ed, β-Glucosidases, Vol 533 American Chemical Society, Washington, DC, pp 214–239 [Google Scholar]
- Fan TWM, Conn EE (1985) Isolation and characterization of two cyanogenic β-glucosidases from flax seeds. Arch Biochem Biophys 243: 361–373 [DOI] [PubMed] [Google Scholar]
- Fieldes MA, Gerhardt KE (1994) An examination of the β-glucosidase (linamarase) banding pattern in flax seedlings using Ferguson plots and sodium dodecyl sulphate-polyacrylamide gel electrophoresis. Electrophoresis 15: 654–661 [DOI] [PubMed] [Google Scholar]
- Gabriel O, Gersten DM (1992) Staining for enzymatic activity after gel electrophoresis, I. Anal Biochem 203: 1–21 [DOI] [PubMed] [Google Scholar]
- Geerlings A, Ibañez MM, Memelink J, van der Heijden R, Verpoorte R (2000) Molecular cloning and analysis of strictosidine β-D-glucosidase, an enzyme in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. J Biol Chem 275: 3051–3056 [DOI] [PubMed] [Google Scholar]
- Guirimand G, Courdavault V, Lanoue A, Mahroug S, Guihur A, Blanc N, Giglioli-Guivarc’h N, St-Pierre B, Burlat V (2010) Strictosidine activation in Apocynaceae: towards a “nuclear time bomb”? BMC Plant Biol 10: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gus-Mayer S, Brunner H, Schneider-Poetsch HAW, Rüdiger W (1994) Avenacosidase from oat: purification, sequence analysis and biochemical characterization of a new member of the BGA family of β-glucosidases. Plant Mol Biol 26: 909–921 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Rosales F, Romero MP, Casanovas M, Motilva MJ, Mínguez-Mosquera MI (2012) β-Glucosidase involvement in the formation and transformation of oleuropein during the growth and development of olive fruits (Olea europaea L. cv. Arbequina) grown under different farming practices. J Agric Food Chem 60: 4348–4358 [DOI] [PubMed] [Google Scholar]
- Hachicha Hbaieb R, Kotti F, García-Rodríguez R, Gargouri M, Sanz C, Pérez AG (2015) Monitoring endogenous enzymes during olive fruit ripening and storage: correlation with virgin olive oil phenolic profiles. Food Chem 174: 240–247 [DOI] [PubMed] [Google Scholar]
- Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hösel W, Conn EE (1982) The aglycone specificity of plant β-glycosidases. Trends Biochem Sci 7: 219–221 [Google Scholar]
- Jensen SR, Franzyk H, Wallander E (2002) Chemotaxonomy of the Oleaceae: iridoids as taxonomic markers. Phytochemistry 60: 213–231 [DOI] [PubMed] [Google Scholar]
- Jones P, Vogt T (2001) Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta 213: 164–174 [DOI] [PubMed] [Google Scholar]
- Kara HE, Sinan S, Turan Y (2011) Purification of β-glucosidase from olive (Olea europaea L.) fruit tissue with specifically designed hydrophobic interaction chromatography and characterization of the purified enzyme. J Chromatogr B Analyt Technol Biomed Life Sci 879: 1507–1512 [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketudat Cairns JR, Esen A (2010) β-Glucosidases. Cell Mol Life Sci 67: 3389–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YW, Kim IS (1998) Subunit composition and oligomer stability of oat β-glucosidase isozymes. Biochim Biophys Acta 1388: 457–464 [DOI] [PubMed] [Google Scholar]
- Konno K, Hirayama C, Yasui H, Nakamura M (1999) Enzymatic activation of oleuropein: a protein crosslinker used as a chemical defense in the privet tree. Proc Natl Acad Sci USA 96: 9159–9164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno K, Yasui H, Hirayama C, Shinbo H (1998) Glycine protects against strong protein-denaturing activity of oleuropein. A phenolic compound in privet leaves. J Chem Ecol 24: 735–751 [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA 106: 10171–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudounas K, Banilas G, Michaelidis C, Demoliou C, Rigas S, Hatzopoulos P (2015) A defence-related Olea europaea β-glucosidase hydrolyses and activates oleuropein into a potent protein cross-linking agent. J Exp Bot 66: 2093–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S-N, Kim S-Y, Choi S-R, Kim I-S (2009) Assembly and function of AsGlu2 fibrillar multimer of oat β-glucosidase. Biochim Biophys Acta 1794: 526–531 [DOI] [PubMed] [Google Scholar]
- Laskowski RA. (2001) PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res 29: 221–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau W, Sattely ES (2015) Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science 349: 1224–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Park H, Ko J, Seok C (2013) GalaxyGemini: a web server for protein homo-oligomer structure prediction based on similarity. Bioinformatics 29: 1078–1080 [DOI] [PubMed] [Google Scholar]
- Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ, Hwang I (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Luijendijk T, Stevens L, Verpoorte R (1996) Reaction for the localization of strictosidine glucosidase activity on polyacrylamide gels. Phytochem Anal 7: 16–19 [Google Scholar]
- Miettinen K, Dong L, Navrot N, Schneider T, Burlat V, Pollier J, Woittiez L, van der Krol S, Lugan R, Ilc T, et al. (2014) The seco-iridoid pathway from Catharanthus roseus. Nat Commun 5: 3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithöfer A, Boland W (2012) Plant defense against herbivores: chemical aspects. Annu Rev Plant Biol 63: 431–450 [DOI] [PubMed] [Google Scholar]
- Morant AV, Jørgensen K, Jørgensen C, Paquette SM, Sánchez-Pérez R, Møller BL, Bak S (2008) β-Glucosidases as detonators of plant chemical defense. Phytochemistry 69: 1795–1813 [DOI] [PubMed] [Google Scholar]
- Naoumkina M, Farag MA, Sumner LW, Tang Y, Liu CJ, Dixon RA (2007) Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc Natl Acad Sci USA 104: 17909–17915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisius A. (1988) The stromacentre in Avena plastids: an aggregation of β-glucosidase responsible for the activation of oat-leaf saponins. Planta 173: 474–481 [DOI] [PubMed] [Google Scholar]
- Obied HK, Bedgood DR Jr, Prenzler PD, Robards K (2007) Chemical screening of olive biophenol extracts by hyphenated liquid chromatography. Anal Chim Acta 603: 176–189 [DOI] [PubMed] [Google Scholar]
- Obied HK, Prenzler PD, Ryan D, Servili M, Taticchi A, Esposto S, Robards K (2008) Biosynthesis and biotransformations of phenol-conjugated oleosidic secoiridoids from Olea europaea L. Nat Prod Rep 25: 1167–1179 [DOI] [PubMed] [Google Scholar]
- Odoux E, Chauwin A, Brillouet JM (2003) Purification and characterization of vanilla bean (Vanilla planifolia Andrews) β-D-glucosidase. J Agric Food Chem 51: 3168–3173 [DOI] [PubMed] [Google Scholar]
- Ortega-García F, Peragón J (2010) HPLC analysis of oleuropein, hydroxytyrosol, and tyrosol in stems and roots of Olea europaea L. cv. Picual during ripening. J Sci Food Agric 90: 2295–2300 [DOI] [PubMed] [Google Scholar]
- Pentzold S, Zagrobelny M, Rook F, Bak S (2014) How insects overcome two-component plant chemical defence: plant β-glucosidases as the main target for herbivore adaptation. Biol Rev Camb Philos Soc 89: 531–551 [DOI] [PubMed] [Google Scholar]
- Piasecka A, Jedrzejczak-Rey N, Bednarek P (2015) Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol 206: 948–964 [DOI] [PubMed] [Google Scholar]
- Rajniak J, Barco B, Clay NK, Sattely ES (2015) A new cyanogenic metabolite in Arabidopsis required for inducible pathogen defence. Nature 525: 376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Segura C, García-Rodríguez R, Sánchez-Ortiz A, Sanz C, Pérez AG (2012) The role of olive β-glucosidase in shaping the phenolic profile of virgin olive oil. Food Res Int 45: 191–196 [Google Scholar]
- Romero-Segura C, Sanz C, Perez AG (2009) Purification and characterization of an olive fruit β-glucosidase involved in the biosynthesis of virgin olive oil phenolics. J Agric Food Chem 57: 7983–7988 [DOI] [PubMed] [Google Scholar]
- Saslowsky DE, Warek U, Winkel BSJ (2005) Nuclear localization of flavonoid enzymes in Arabidopsis. J Biol Chem 280: 23735–23740 [DOI] [PubMed] [Google Scholar]
- Selvaggini R, Servili M, Urbani S, Esposto S, Taticchi A, Montedoro G (2006) Evaluation of phenolic compounds in virgin olive oil by direct injection in high-performance liquid chromatography with fluorometric detection. J Agric Food Chem 54: 2832–2838 [DOI] [PubMed] [Google Scholar]
- Seshadri S, Akiyama T, Opassiri R, Kuaprasert B, Cairns JK (2009) Structural and enzymatic characterization of Os3BGlu6, a rice β-glucosidase hydrolyzing hydrophobic glycosides and (1→3)- and (1→2)-linked disaccharides. Plant Physiol 151: 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler‐Rivas C, Espín JC, Wichers HJ (2000) Oleuropein and related compounds. J Sci Food Agric 80: 1013–1023 [Google Scholar]
- Spadafora A, Mazzuca S, Chiappetta FF, Parise A, Perri E, Innocenti AM (2008) Oleuropein-specific-β-glucosidase activity marks the early response of olive fruits (Olea europaea) to mimed insect attack. Agric Sci China 7: 703–712 [Google Scholar]
- Stavrinides A, Tatsis EC, Foureau E, Caputi L, Kellner F, Courdavault V, O’Connor SE (2015) Unlocking the diversity of alkaloids in Catharanthus roseus: nuclear localization suggests metabolic channeling in secondary metabolism. Chem Biol 22: 336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sue M, Yamazaki K, Yajima S, Nomura T, Matsukawa T, Iwamura H, Miyamoto T (2006) Molecular and structural characterization of hexameric β-D-glucosidases in wheat and rye. Plant Physiol 141: 1237–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uccella N. (2000) Olive biophenols: biomolecular characterization, distribution and phytoalexin histochemical localization in the drupes. Trends Food Sci Technol 11: 315–327 [Google Scholar]
- Verdoucq L, Morinière J, Bevan DR, Esen A, Vasella A, Henrissat B, Czjze M (2004) Structural determinants of substrate specificity in family 1 β-glucosidases: novel insights from the crystal structure of sorghum dhurrinase-1, a plant β-glucosidase with strict specificity, in complex with its natural substrate. J Biol Chem 279: 31796–31803 [DOI] [PubMed] [Google Scholar]
- Visioli F, Poli A, Gall C (2002) Antioxidant and other biological activities of phenols from olives and olive oil. Med Res Rev 22: 65–75 [DOI] [PubMed] [Google Scholar]
- Weigend M, Kufer J, Müller AA (2000) Phytochemistry and the systematics and ecology of Loasaceae and Gronoviaceae (Loasales). Am J Bot 87: 1202–1210 [PubMed] [Google Scholar]
- Wessel D, Flügge UI (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Xia L, Ruppert M, Wang M, Panjikar S, Lin H, Rajendran C, Barleben L, Stöckigt J (2012) Structures of alkaloid biosynthetic glucosidases decode substrate specificity. ACS Chem Biol 7: 226–234 [DOI] [PubMed] [Google Scholar]
- Zanphorlin LM, de Giuseppe PO, Honorato RV, Tonoli CCC, Fattori J, Crespim E, de Oliveira PSL, Ruller R, Murakami MT (2016) Oligomerization as a strategy for cold adaptation: structure and dynamics of the GH1 β-glucosidase from Exiguobacterium antarcticum B7. Sci Rep 6: 23776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Paakkarinen V, van Wijk KJ, Aro EM (1999) Co-translational assembly of the D1 protein into photosystem II. J Biol Chem 274: 16062–16067 [DOI] [PubMed] [Google Scholar]