The NAC transcription factor OsNAC2 plays an important role in ABA-dependent leaf senescence by targeting both abscisic acid biosynthetic and senescence-associated genes.
Abstract
It is well known that abscisic acid (ABA)-induced leaf senescence and premature leaf senescence negatively affect the yield of rice (Oryza sativa). However, the molecular mechanism underlying this relationship, especially the upstream transcriptional network that modulates ABA level during leaf senescence, remains largely unknown. Here, we demonstrate a rice NAC transcription factor, OsNAC2, that participates in ABA-induced leaf senescence. Overexpression of OsNAC2 dramatically accelerated leaf senescence, whereas its knockdown lines showed a delay in leaf senescence. Chromatin immunoprecipitation-quantitative PCR, dual-luciferase, and yeast one-hybrid assays demonstrated that OsNAC2 directly activates expression of chlorophyll degradation genes, OsSGR and OsNYC3. Moreover, ectopic expression of OsNAC2 leads to an increase in ABA levels via directly up-regulating expression of ABA biosynthetic genes (OsNCED3 and OsZEP1) as well as down-regulating the ABA catabolic gene (OsABA8ox1). Interestingly, OsNAC2 is upregulated by a lower level of ABA but downregulated by a higher level of ABA, indicating a feedback repression of OsNAC2 by ABA. Additionally, reduced OsNAC2 expression leads to about 10% increase in the grain yield of RNAi lines. The novel ABA-NAC-SAGs regulatory module might provide a new insight into the molecular action of ABA to enhance leaf senescence and elucidates the transcriptional network of ABA production during leaf senescence in rice.
Senescence is the last stage of leaf development. During this period, various changes occur at the physiological, biochemical, and molecular levels. For example, macromolecules including lipids, proteins, and nucleic acids are hydrolyzed, which leads to disassembly of mitochondria and nuclei, and to cell death (Buchanan‐Wollaston et al., 2005; Ulker et al., 2007). Although senescence is an active process to salvage nutrients from old tissues, precocious senescence will shorten the growth stage of crops and be unfavorable to agronomic production (Woo et al., 2013).
The most distinguishing feature in leaf senescence is the yellowing phenotype, which is a visible marker of the degradation of macromolecules (Kim et al., 2006). The chlorophyll degradation pathway is one of the most characterized ones for macromolecule degradation in plants (Hörtensteiner, 2006). Overexpressing NON-YELLOW COLORING1 (NYC1) or NYC1-like genes in rice (Oryza sativa) can induce degradation of chlorophyll (Sato et al., 2009). A pph (encoding pheophytinase) mutant is abnormal in chlorophyll degradation during senescence and therefore exhibits a stay-green phenotype (Schelbert et al., 2009). Mutation of the PAO (Pheophorbide a oxygenase) gene leads to retention of chlorophyll in leaves during dark-induced senescence in Arabidopsis (Arabidopsis thaliana; Pruzinská et al., 2005). Recently, the highly conserved STAY-GREEN (SGR) in higher plants has been identified to be chloroplast-localized dechelatase (Shimoda et al., 2016).
The senescence process is highly regulated by a range of important factors. It has been demonstrated that the transcriptional level of NAC (NAM, ATAF1, and CUC2) transcription factors are up-regulated and play a fundamental role in leaf senescence (Gregersen and Holm, 2007). The expression of senescence-associated genes (SAGs) is regulated by these senescence-related NAC transcription factors, such as ORESARA1 (ORE1) (Kim et al., 2009), Oresara1 sister 1 (ORS1) (Balazadeh et al., 2011), Phytochrome-interacting factor 4 (PIF4) and PIF5 (Sakuraba et al., 2014), ANAC046 (Oda-Yamamizo et al., 2016), and AtNAP (Guo and Gan, 2006). However, the molecular mechanism underlying NAC transcription factor (TF)-regulated leaf senescence remains poorly understood in rice. NTL4 promotes drought-induced senescence probably due to direct regulation of reactive oxygen species (ROS) synthesis (Lee et al., 2012). The expression of both OsNAC5 (Sperotto et al., 2009) and OsNAC6 (Nakashima et al., 2007) was increased during leaf senescence. NAC106 plays an important role in regulation of the aging process, and its overexpression defers senescence in rice (Sakuraba et al., 2015). Additionally, OsNAP was reported to control expression of genes involved in chlorophyll degradation by direct binding to the promoter regions (Liang et al., 2014).
In higher plants, the level of ABA is finely controlled by the balance between the rates of ABA biosynthesis and catabolism (Nambara and Marion-Poll, 2005). Key enzymes controlling ABA production include 9′-cis-epoxycarotenoid dioxygenases (NCEDs), which are involved in xanthophyll cleavage (Tan et al., 2003); zeaxanthin epoxidase (ZEP), which converts zeaxanthin to violaxanthin via antheraxanthin (Oliver et al., 2007); and ABSCISIC ALDEHYDE OXIDASE3 (AAO3), which is responsible for the final step in ABA biosynthesis (Yang et al., 2014). The oxidation of ABA to phaseic acid is catalyzed by ABA 8′-hydroxylase, which is encoded probably by three genes (OsABA8ox1, 2, and 3) in rice (Saika et al., 2007).
ABA has vital function in stress responses and regulates various plant developments, e.g. seed maturation and dormancy, organ abscission, and leaf senescence (Chandler and Robertson, 1994; Cutler et al., 2010). It has long been known that ABA participates in leaf senescence (Becker and Apel, 1993). ABA can induce the expression of some SAGs, e.g. NYC1 (Kusaba et al., 2007), SGR (Park et al., 2007), PPH (Schelbert et al., 2009), and PaO (Pruzinská et al., 2005), and then promotes plant senescence. The expression of NAC TFs, including VNI2 (Yang et al., 2011), SNAC-As (Takasaki et al., 2015), ORE1 (Kim et al., 2011), and OsNAP (Liang et al., 2014), is significantly up-regulated after ABA treatment by an unknown molecular mechanism. AtNAP could specifically bind to the promoter of AAO3, one of the key enzymes controlling ABA production, and promote transcription of chlorophyll degradation genes (Yang et al., 2014). Although the biosynthetic pathways that generate ABA and its downstream signaling mechanisms have been extensively studied, and ABA-induced leaf senescence has long been observed, the upstream transcriptional network that modulates its level during leaf senescence and connects its action to leaf senescence is less clear.
Previously, the transcription factor OsNAC2 was reported to promote shoot branching (Mao et al., 2007), and our previous study showed that OsNAC2 affects plant height and regulates abiotic stress tolerance in rice (Shen et al., 2017). Previous study of our lab showed that overexpression of OsNAC2 endowed rice with premature senility, which indicated OsNAC2 might play a role in the pathway of leaf senescence (Chen et al., 2015). However, the underlying molecular mechanisms of OsNAC2 regulating leaf senescence were poorly understood. Here, we show that OsNAC2 promotes leaf senescence in rice by induction of ABA biosynthetic genes (OsNCED3 and OsZEP1) and down-regulating ABA catabolic gene (OsABA8ox1) by targeting to their promoters, which leads to increased levels of ABA. OsNAC2 also directly regulated chlorophyll degradation genes, OsSGR and OsNYC3. Our data elucidate the transcriptional network of ABA production during leaf senescence in rice. The novel ABA-NAC-SAG regulatory module might provide a new insight into the molecular action of ABA to enhance leaf senescence and the function of NAC TFs in pathway ABA induce leaf senescence.
RESULTS
OsNAC2 Is Highly Expressed during Leaf Senescence and Positively Regulates Leaf Senescence in an Age-Dependent Manner
To study the role of OsNAC2 in leaf senescence, we examined the senescence symptoms of transgenic lines, including 35S::OsNAC2 lines (OsNAC2-OX, ON7, and ON11) and knockdown lines (OsNAC2-RNAi18, RNAi25, and RNAi31), which were previously reported (Chen et al., 2015). The third leaves of 6-week-old seedlings hydroponically cultivated were used for statistics of yellowing rate. It was evident that older leaves became yellow earlier in OsNAC2-OX lines but much later in RNAi lines than in the wild type (Fig. 1A). Consistent with this visible symptom, the yellowing rate of leaves from OsNAC2-OX was obviously higher compared with that in the wild type, whereas the OsNAC2-RNAi line had few yellow leaves in the 4-week-old rice plant (Fig. 1B). It is considered that stay-green phenotype in grain-filling stage leads to the increase of rice yield (Sakuraba et al., 2015). We subsequently examined the natural senescence phenotype of OsNAC2 transgenic lines during grain-filling stage and found that OsNAC2-RNAi exhibited a delayed-senescence phenotype, while OsNAC2-OX exhibited a precocious-senescence phenotype (Supplemental Fig. S1).
Figure 1.
OsNAC2 is upregulated during leaf senescence. A, Phenotype of the third leaves in 6-week-old wild-type and OsNAC2 transgenic plants. B, Quantitative analysis of the yellowing rate of the leaves shown in A. C, qPCR analysis of OsNAC2 expression in various ages and regions of rice leaves. LS, ES, NS, and YL mean late-senescing, early-senescing, no senescing, and young leaves, respectively. T, M, and B mean the top, middle, and base of the leaf. Relative mRNA level was calculated using the ΔΔCT method from triplicate data. OsActin was used as an internal control to normalize the different samples with the same amount of plant RNA. Data are mean ± se with three replicates. Asterisk indicates a significant difference between the wild type and OsNAC2 transgenic lines by t test: *P < 0.05.
We then investigated temporal and spatial expression patterns of OsNAC2 using transgenic plants (pOsNAC2-GUS) expressing β-glucuronidase (GUS) driven by the native promoter of OsNAC2, a 2-kb genomic DNA fragment upstream of the transcription start site. GUS staining analysis showed that GUS activity was obviously higher at the senescing area than at the no senescing area in the same leaves. Moreover, GUS was expressed specifically in coleoptiles in the course of germination (from 1 to 10 d; Supplemental Fig. S2). The coleoptile is known to protect true leaves against soil pressures and other physical constraints and is the first organ of rice plants to senescence after germination (Kawai and Uchimiya, 2000). Therefore, these data suggest that OsNAC2 is involved in the aging process.
To further assess the transcriptional level of OsNAC2 during leaf aging, we tested OsNAC2 expression in leaves at different developmental stages by qPCR. The OsNAC2 transcripts were higher in late-senescing and early-senescing leaves than in young leaves and nonsenescing leaves (Fig. 1C). Consistent with the results from GUS staining, OsNAC2 expression increased gradually from the tip to the base of a fully expanded leaf (Supplemental Fig. S2). Taken together, our results indicate that OsNAC2 plays an important role in leaf senescence.
To further analyze evolutionary relationships of OsNAC2 in the NAC family, a total of 234 NAC proteins, comprising 99 from Arabidopsis (TAIR, http://www.Arabidopsis.org/), and 135 from rice (Rice Genome Annotation Project, http://rice.plantbiology.msu.edu/), were used for construction of an phylogenetic tree, by MEGA5.1 software using the maximum likelihood method and the bootstrap test carried out with 1,000 replications. As illustrated in Supplemental Figure S3, the phylogenetic analysis classified the NACs into 14 groups, designated as Roman numeral families (NAC-I to NAC-XIV) based on tree topologies. The published senescence-associated NACs (Kim et al., 2016; Liang et al., 2014; Nakashima et al., 2007; Oda-Yamamizo et al., 2016; Sperotto et al., 2009) fall in groups I to IV. In detail, ANAC053, ANC016, ANAC017, and ANAC096 located in NAC II, ANAC083, and ONAC106 located in NAC III, while ANAC042, ONAC058 (OsNAP), AtNAP, ANAC047, ANAC019, AtNAC3, OsNAC5, ATAF1, and OsNAC5 located in NAC IV. OsNAC2 had high homology with AtNAC6 (ANAC092), together with ANAC046 and ANAC059 (ORS1), forming a distinct cluster NAC I (Supplemental Fig. S3), which was consistent with the previous reports (You et al., 2015; Hu et al., 2010).
Overexpression of OsNAC2 Causes Early Senescence of Rice and Tobacco Leaves
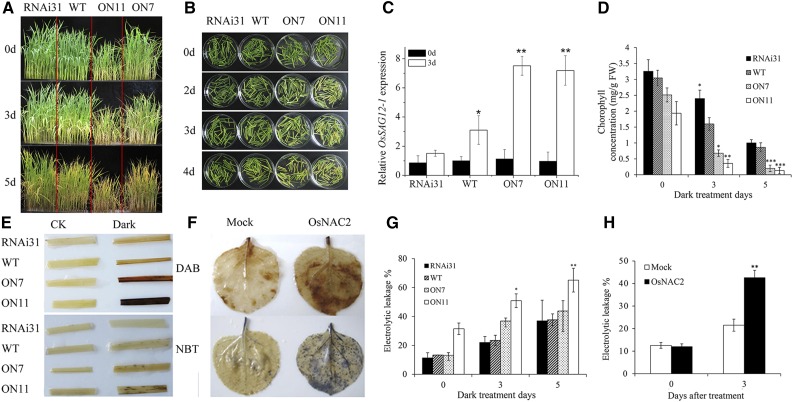
Dark treatment is used frequently as an effective method to simulate synchronous senescence (Kim et al., 2006; Kong et al., 2006). To gain a mechanistic understanding of the potential role of OsNAC2 in leaf senescence, rice plants were treated by darkness with various times. After 5-d dark treatment, OsNAC2-OX lines exhibited a more severe leaf senescent phenotype, while RNAi transgenic lines displayed weaker leaf senescence, compared with the wild type (Fig. 2A). Similarly, an accelerated yellowing phenotype was also observed in detached and 4-d dark-treated leaves from OsNAC2-OX plants (Fig. 2B). AtSAG12, an Arabidopsis gene encoding a Cys protease, is considered a molecular marker the study of developmental senescence (Weaver et al., 1998). In rice, OsSAG12-1 is the closest homolog of AtSAG12 and the expression of OsSAG12-1 is induced during senescence (Singh et al., 2013). Thus, we determined the mRNA level of OsSAG12-1 in detached leaves of OsNAC2 transgenic plants incubated 3 d in the darkness and found that OsSAG12-1 significantly upregulated in OsNAC2-OX plants (ON11 and ON7; Fig. 2C). Conformably, chlorophyll content in OsNAC2-OX leaves treated with darkness for 5 d was 0.2 mg/g fresh weight (FW), only 25% of that found in wild-type leaves (Fig. 2D). These results demonstrate that chlorophyll is degraded at a more rapid rate in OsNAC2-OX than in wild-type plants.
Figure 2.
Overexpression of OsNAC2 causes early senescence in rice and tobacco. A, Phenotypes of 4-week-old wild-type and OsNAC2-transgenic plants after 0, 3, and 5 d dark treatment. B, Phenotypes of detached leaves in 4-week-old wild-type and transgenic plants after 0, 2, 3, and 4 d dark treatment. C, Expression of OsSAG12-1 in OsNAC2-OX, the wild type, and OsNAC2-RNAi lines after 3 d dark treatment. D, Chlorophyll content of the leaves in OsNAC2 plants after 0, 3, and 5 d dark treatment. Values are means ± sd of five replicates. E, DAB and NBT staining of wild-type and transgenic seedlings leaves after 4 d dark treatment. F, DAB and NBT staining in tobacco leaves by a transient expression of OsNAC2 after 3 d dark treatment. G, Ion leakage analysis of leaves in 4-week-old wild-type and transgenic rice plants after 0, 3, and 5 d dark treatment. Values are means ± sd of five replicates. H, Ion leakage analysis of leaves in tobacco leaves with OsNAC2 transient expression after 3 d dark treatment. Values are means ± sd of five replicates. Asterisks indicate a significant difference between wild-type and OsNAC2 transgenic lines by t test: *P < 0.05 and **P < 0.01.
We also investigated ion leakage rates and ROS in dark-treated leaves. Consistently, OsNAC2-OX lines displayed significantly higher ion leakage (Fig. 2G), indicating that overexpressing OsNAC2 results in cell membrane damage in dark. In addition, ROS analysis showed an increased level of H2O2 and O−2 in ON7 and ON11 lines (Fig. 2E). Same results were obtained in a transient expression assay in tobacco (Nicotiana benthamiana) leaves (Fig. 2, F and H). These results suggest that OsNAC2 is positively regulatory factor for leaf senescence.
Overexpression of OsNAC2 Alters the Expression of Senescence-Related Genes
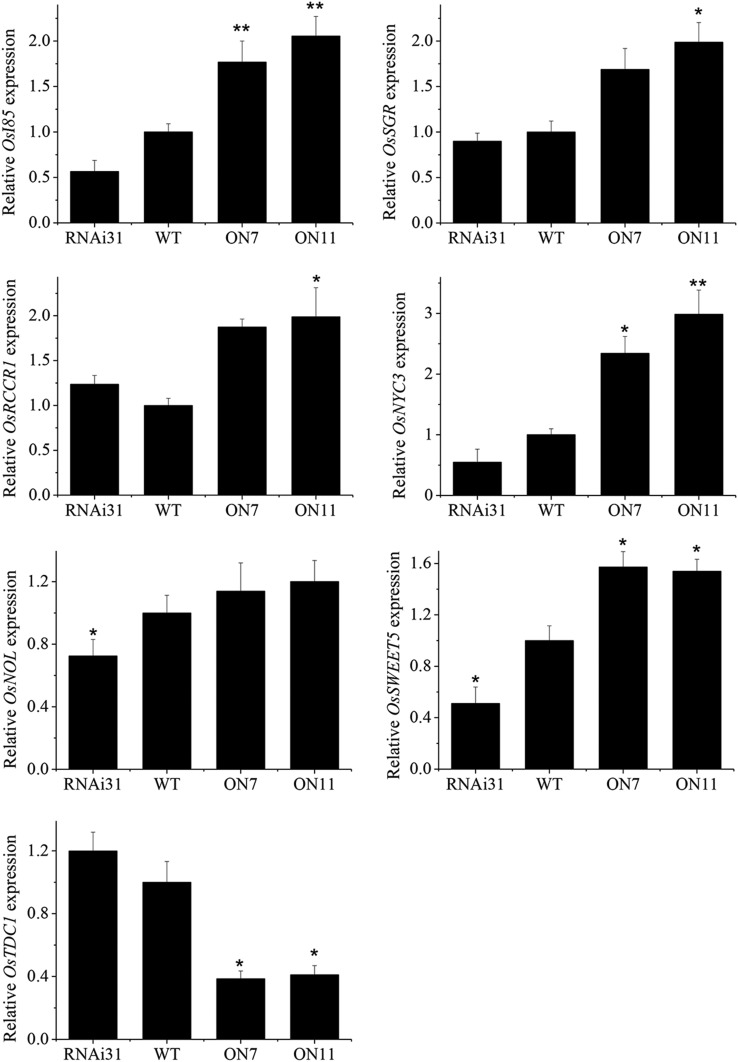
Based on the leaf aging phenotype mentioned above, we predicted that the expression of senescence-related genes would be significantly different between OsNAC2-overexpressing and RNAi lines. To understand the molecular mechanism of OsNAC2-regulated chlorophyll degradation, we profiled genome-wide gene expression in 2-week-old OsNAC2-OX and wild-type plants using rice microarray. Many differently expressed genes were involved in leaf senescence, such as chlorophyll breakdown genes OsNYC3, OsNOL, OsRCCR1, OsPAO, and OsSGR; serotonin biosynthesis gene, OsTDC1; glyoxylate cycle related gene, OsI85; and Gal transport gene, OsSWEET5 (Table I). The changes of some SAGs expression, e.g. OsSAG12-1, in OsNAC2-OX were not so significant by t test, probably because 2-week-old seedlings may be not in the procedure of senescence. However, there were still some SAGs showed significant changes in OsNAC2-OX, e.g. OsSGR, OsSWEET5, and OsTDC1, which gave information of probably candidate downstream targets of OsNAC2. Transcript levels of these senescence relative genes were further confirmed by qPCR. Significant increases in transcript levels of OsI85, OsNOL, OsSGR, OsRCCR1, OsSWEET5, and OsNYC3 and decrease in that of OsTDC1 were found in OsNAC2-OX compared with the wild type (Fig. 3). These results indicate that OsNAC2 plays a regulatory role, either direct or indirect, in expression of chlorophyll degradation genes during leaf senescence.
Table I. List of senescence-related genes changed in OsNAC2-OX line by microarray analysis.
| Annotation | Probe Set_ID | Gene Symbol | Fold Change | References |
|---|---|---|---|---|
| Ospse1 | Os.35614.1.S1_at | Os01g0546800 | 0.721459 | Wu et al. (2013) |
| SPL28 | Os.38379.1.S1_at | Os01g0703600 | 0.689033* | Qiao et al. (2010) |
| OsSAG12-1 | Os.4181.1.S1_at | Os01g0907600 | 0.799077 | Singh et al. (2013) |
| NYC1 | Os.18818.1.S1_a_at | Os01g0227100 | 0.637737* | Kusaba et al. (2007) |
| OsWRKY80 | Os.20028.1.S1_at | Os09g0481700 | 1.545195* | Ricachenevsky et al. (2010) |
| OsSGR | Os.11666.1.S1_at | Os09g0532000 | 2.434277** | Park et al. (2007) |
| OsRCCR1 | Os.5164.1.S1_at | Os10g0389200 | 0.884418 | Tang et al. (2011) |
| OsNAC5 | Os.4385.1.S1_at | Os11g0184900 | 0.826093 | Sperotto et al. (2009) |
| NYC3 | Os.11724.2.S1_s_at | Os06g0354700 | 1.090994 | Morita et al. (2009) |
| NOL | Os.17490.1.A1_at | Os03g0654600 | 1.124019 | Sato et al. (2009) |
| OsI85 | Os.22277.1.S1_at | Os07g0529000 | 1.240426* | Pistelli et al. (1991) |
| PHYB | Os.4601.1.S1_a_at | Os03g0309200 | 0.598971* | Piao et al. (2015) |
| cZOGT1 | Os.46328.1.S1_at | Os04g0556500 | 0.706998* | Kudo et al. (2012) |
| OsSWEET5 | Os.6338.1.S1_at | Os05g0588500 | 1.333925* | Zhou et al. (2014) |
| TDC2 | Os.49621.1.S1_at | Os07g0437500 | 1.518717* | Kang et al. (2009) |
| TDC1 | Os.27876.1.S1_x_at | Os08g0140300 | 0.25689** | Kang et al. (2009) |
Figure 3.
qPCR confirmation of expression of senescence-related genes in the 2-week-old wild type and OsNAC2-OX. Relative mRNA level was calculated using the ΔΔCT method from triplicate data. OsActin was used as internal control to normalize the different samples with the same amount of plant RNA. Data are mean ± se with three replicates. Asterisks indicate a significant difference between wild-type and OsNAC2 transgenic lines by t test: *P < 0.05 and **P < 0.01.
OsNAC2 Directly Regulates Expression of OsNYC3 and OsSGR Genes
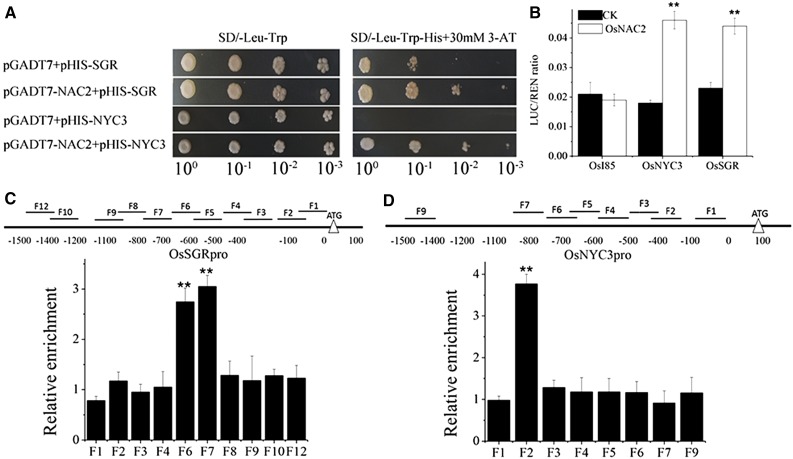
To further investigate whether OsNAC2 directly regulates expression of chlorophyll degradation genes, yeast one-hybrid assays were used to test the interaction of OsNAC2 with DNA. The full-length cDNA of OsNAC2 was fused in frame to the GAL4 activation domain in the pGADT7 vector. The promoter sequence regions of OsI85, OsSGR, and OsNYC3 were ligated into the pHIS vector. The yeast one-hybrid assays showed that OsNAC2 directly interacts with the promoter sequences of OsSGR and OsNYC3 (Fig. 4A).
Figure 4.
OsSGR and OsNYC3 are the direct target genes of OsNAC2. A, Interaction of OsNAC2 with the promoters of OsSGR and OsNYC3 by yeast one-hybrid assays. The yeast cells were grown on SD/-Leu/-Trp/-His/+30 mm 3-amino-1, 2,4-triazole medium. B, Interaction of OsNAC2 with the promoters of OsSGR and OsNYC3 by dual-luciferase promoter activation assays in tobacco. C and D, ChIP-qPCR assays. Total protein extracted from 35S:OsNAC2-mGFP transgenic plants hydroponically cultivated for 4 weeks were immunoprecipitated with an anti-GFP antibody. Fragmented genomic DNA was eluted from the protein-DNA complexes and subjected to qPCR analysis. The long black bars represent promoter regions for which we designed primers. The numbers under the bar show the distance from ATG start codon. Short bars represents for the corresponding region of each pair of primers on the promoter. Error bars are the se for three biological repeats: *P < 0.05 and **P < 0.01.
We also performed transient expression assays for transactivation in N. benthamiana. The promoter sequences of OsSGR and OsNYC3 were cloned into pGreenII 0800-LUC, and the open reading frame of OsNAC2 was cloned into pGreenII 62-SK. The highest activities were observed when OsNAC2 was coinfiltrated with the promoter of OsNYC3 and OsSGR (Fig. 4B).
To test whether endogenous OsNAC2 specifically binds to its target genes, chromatin immunoprecipitation (ChIP)-qPCR was performed using the anti-green fluorescent protein (anti-GFP) antibody and specific primers. Our results showed that specific fragments in the promoter of OsNYC3 and OsSGR were significantly enriched in the GFP antibody-immunoprecipitated DNA (Fig. 4, C and D). Taken together, the data indicate that OsNAC2 is a transcription factor specifically binding to its target genes, OsNYC3 and OsSGR.
ABA Participates in Leaf Senescence by Modulating OsNAC2 Expression
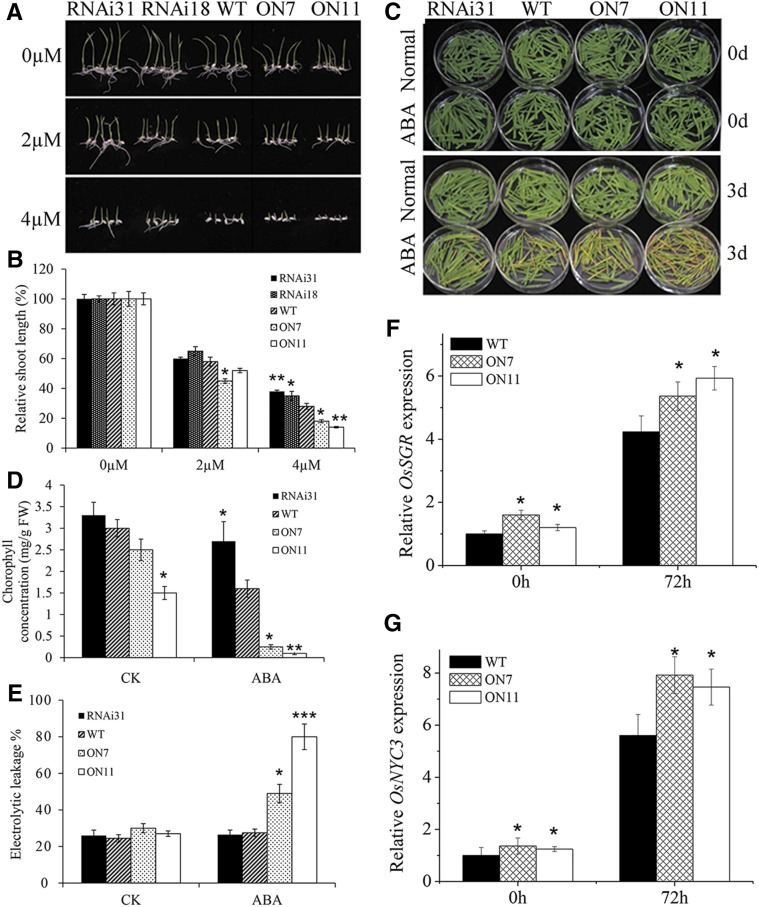
Leaf senescence can be modulated by a variety of phytohormones and environmental factors, and ABA may be one of the most important phytohormones regulating both leaf senescence and expression of NAC TFs (Liang et al., 2014). To determine whether ABA-regulated senescence is mediated by OsNAC2, exogenously ABA was applied on different transgenic lines. Our results showed that in the presence of ABA (either 2 or 4 µm), the shoot length of OsNAC2-OX lines was significantly shorter than that of the wild type, whereas shoot length of RNAi lines was longer than that of the wild type, indicating that ABA sensitivity of rice plants is positively correlated to the level of OsNAC2 expression (Fig. 5, A and B). Next, we examined the effect of exogenously applied ABA on chlorophyll degradation. Consistent with the visible phenotypes, among ABA-treated leaves, those excised from OsNAC2-OX showed the fastest yellowing while those from OsNAC2-RNAi exhibited a stay-green phenotype, compared with the wild type (Fig. 5C). Chlorophyll concentrations in OsNAC2-RNAi, wild-type, and OsNAC2-OX leaves treated with ABA for 3 d were 1.6, 0.8, and 0.1 µg/mg FW, respectively (Fig. 5D); the severity of ion leakage was the most in OsNAC2-OX, followed by the wild type and OsNAC2-RNAi (Fig. 5E). Moreover, expression of two OsNAC2-targeted SAGs, OsSGR and OsNYC3, was significantly upregulated 72 h after ABA treatment (Fig. 5, F and G). These results imply that OsNAC2 mediates ABA-induced leaf senescence.
Figure 5.
Effects of exogenous ABA on chlorophyll content and SAG gene transcript levels in wild-type, OsNAC2-OX, and OsNAC2-RNAi plants. A, ABA effect on seedling growth on MS media. B, Relative shoot length of wild-type, OsNAC2-OX, and OsNAC2-RNAi plants in the presence of different ABA concentrations. The value of shoot length without ABA treatment was set to 1. C, Phenotype of detached leaves from wild-type, OsNAC2-OX, and OsNAC2-RNAi plants in the presence of 20 µm ABA. D, Chlorophyll content of the leaves in wild-type, OsNAC2-OX, and OsNAC2-RNAi plants treated with 20 µm ABA for 3 d. E, Ion leakage analysis of leaves in 2-week-old wild-type, OsNAC2-OX, and OsNAC2-RNAi plants treated with 20 µm ABA for 3 d. F and G, Relative expression of OsSGR and OsNYC3 72 h after 20 µm ABA treatment. HAT, hours after treatment. Data are means ± se with at least three biological replicates. Asterisks represent statistically significant differences between wild-type and transgenic plants: *P < 0.05, **P < 0.01, and ***P < 0.001.
OsNAC2 Enhances ABA Concentration via Regulating Expression of ABA Metabolism Genes
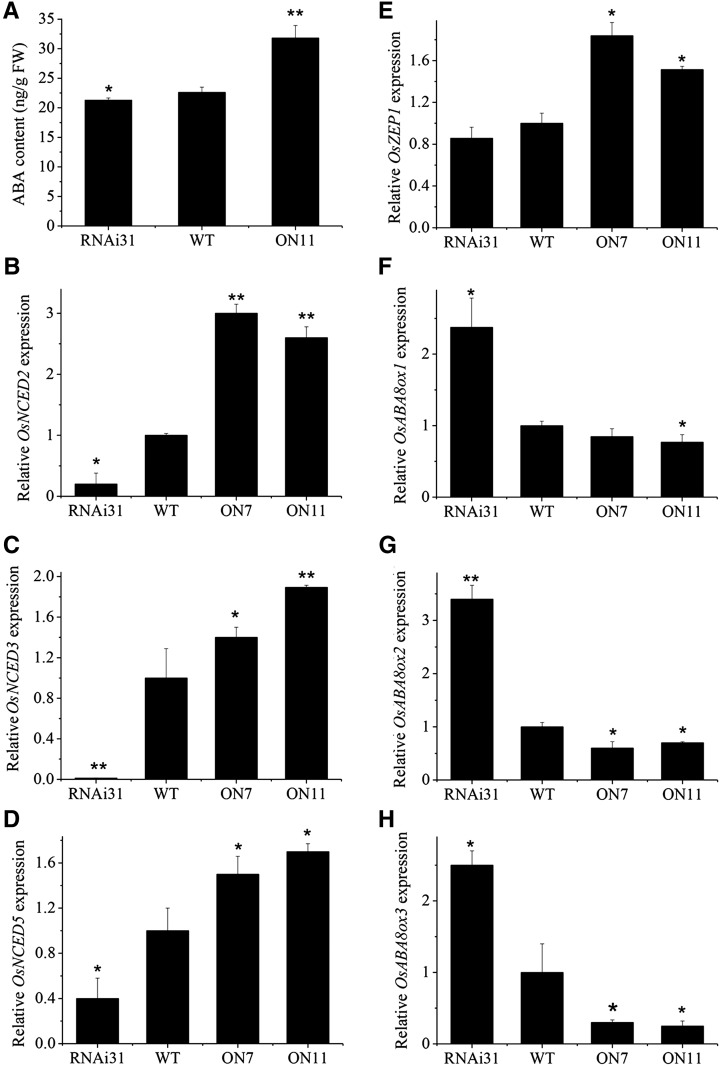
To test whether endogenous ABA production was affected by OsNAC2, we examined the ABA level in 2-week-old leaves of OsNAC2-OX, OsNAC2-RNAi, and wild-type plants. The ABA content of OsNAC2-OX was 31.81 ± 2.11 ng/g FW, which was significantly higher than that of wild-type plants (22.62 ± 0.88 ng/g FW; Fig. 6A). Combined with our previously reported data, we hypothesized that the expression of ABA metabolism-associated genes is affected by OsNAC2. qPCR analysis showed that transcription levels of key ABA biosynthetic genes, including OsNCED2 (Fig. 6B), OsNCED3 (Fig. 6C), OsNCED5 (Fig. 6D), and OsZEP1 (Fig. 6E), were significantly upregulated in OsNAC2-OX, whereas ABA catabolism genes, including OsABA8ox1 (Fig. 6F), OsABA8ox2 (Fig. 6G), and OsABA8ox3 (Fig. 6H), were down-regulated. Thus, it appears that OsNAC2 enhances the level of ABA via up-regulation of ABA biosynthetic genes and down-regulation of ABA catabolism genes as well.
Figure 6.
ABA content and expression of ABA metabolism-related genes in wild-type, OsNAC2-OX, and OsNAC2-RNAi plants. Data were means ± se with at least three biological replicates. Asterisks represent statistically significant differences between wild-type and transgenic plants: *P < 0.05 and **P < 0.01.
OsNAC2 Binds Directly to Promoters of ABA Metabolism-Related Genes
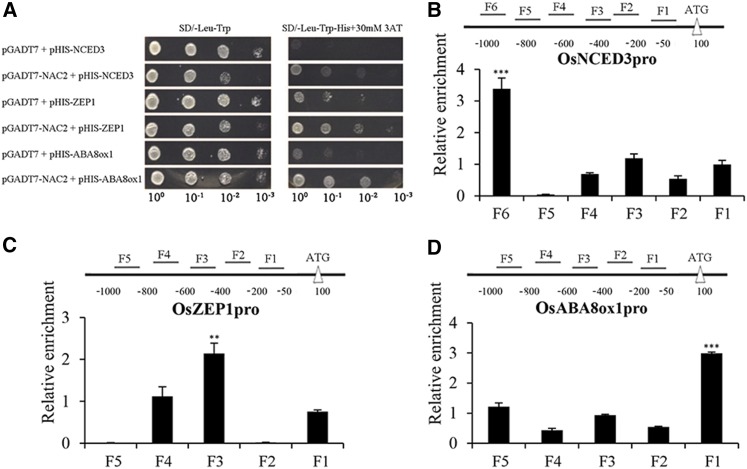
To investigate whether OsNAC2 directly regulates the expression of ABA metabolism-related genes, yeast one-hybrid analysis was performed to test direct interaction between OsNAC2 and DNA. Our results showed that the GAL4 transcriptional activation domain fused with OsNAC2 activates the HIS3 reporter gene driven by the promoter of OsNCED3, OsZEP1, or OsABA8ox1 (Fig. 7A). However, OsNAC2 did not bind directly to the promoters of OsNCED2, OsNCED5, OsABA8ox2, and OsABA8ox3 (Supplemental Fig. S4). ChIP-qPCR assays further confirmed that OsNAC2 indeed bound to the promoters of OsNCED3 (Fig. 7B), OsZEP1(Fig. 7C), and OsABA8ox1 (Fig. 7D). Thus, OsNAC2 is a transcription factor directly regulating expression of ABA metabolism-related genes.
Figure 7.
Interactions between OsNAC2 and the promoters of OsNCED3, OsZEP1, and OsABA8ox1. A, Interaction of OsNAC2 with the promoters of OsNCED3, OsZEP1, and OsABA8ox1 by yeast one-hybrid assays. The yeast cells were grown on SD/-Leu/-Trp/-His/+30 mm 3-amino-1, 2,4-triazole medium. B to D, ChIP-qPCR assays of OsNAC2 binding to the promoters of OsNCED3 (B), OsZEP1 (C), and OsABA8ox1 (D). Total protein extracted from 35S:OsNAC2-mGFP transgenic plants hydroponically cultivated for 4 weeks was immunoprecipitated with an anti-GFP antibody. Fragmented genomic DNA was eluted from the protein-DNA complexes and subjected to qPCR analysis. The long black bars represent for promoter region for which we designed primers. The numbers under the bar show the distance from ATG start codon. Short bars represents for the corresponding region of each pair of primers on the promoter. Error bars are the se for three biological repeats: *P < 0.05, **P < 0.01, and ***P < 0.001.
Previous work has defined the 4-bp core sequence of the NAC binding motif as CACG (Olsen et al., 2005). Thus, we checked whether CACG sequences exist in the promoters of these five target genes. The ChIP experiments identified the positive fragments, −500 ∼ −800, −150 ∼ −400, −850 ∼ −1,000, −450 ∼ −650, and −50 ∼ −250 in the promoters of target genes, OsSGR, OsNYC3, OsNCED3, OsZEP1, and OsABA8ox1, respectively (Fig. 7, B–D). Sequence searching by Clone Manage Software showed that all the five positive fragments contained the core sequence of NAC binding motif CACG (Supplemental Fig. S5), which indicated CACG might also be the binding motif of OsNAC2.
ABA Regulates OsNAC2 and Senescence-Related Gene Expression in a Dose-Dependent Manner But Temporally
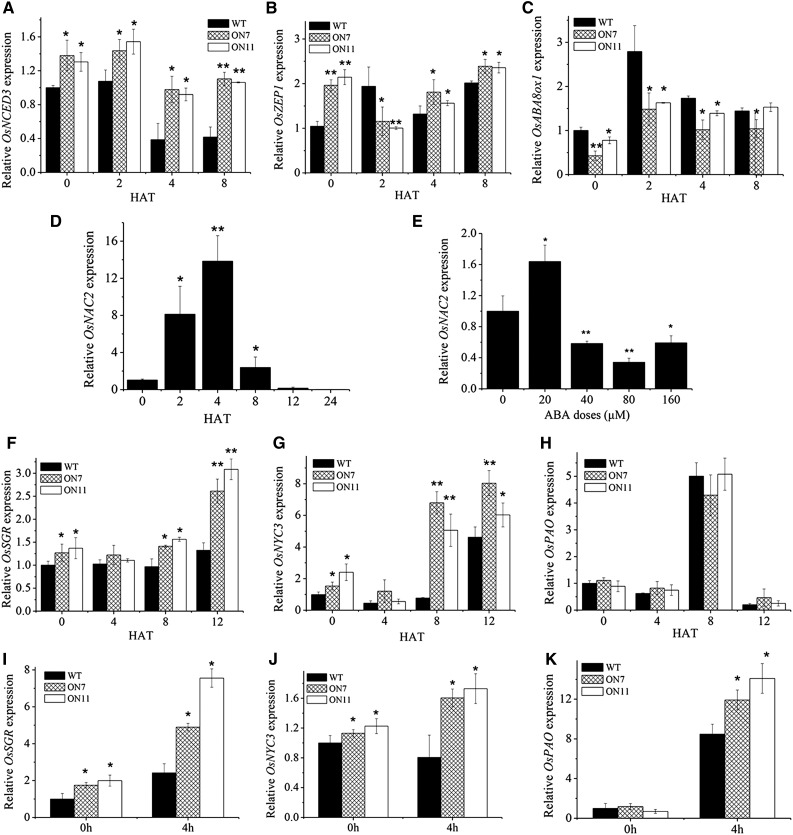
We then investigated whether ABA affects expression of OsNAC2. As shown in Figure 8D, the level of OsNAC2 transcripts increased up to 8- and 16-fold of the ABA-untreated seedlings at 2 and 4 h after 20 µm ABA treatment, respectively. Consistently, expression of OsNCED3 and OsZEP1 obviously increased 2 h after 20 µm ABA treatment and was generally higher in OsNAC2-OX plants than the wild type (Fig. 8, A and B), whereas OsABA8ox1 significantly downregulated in OsNAC2-OX, compared with the wild type (Fig. 8C). Thus, it seems that ABA-promoted expression of OsNCED3 and OsZEP1, and ABA-suppressed expression of OsABA8ox1 are mediated by OsNAC2. However, OsNAC2 transcription decreased rapidly 8 h after ABA treatment (Fig. 8D). We also examined the kinetic expression of OsNAC2 4 h after different concentrations of ABA. OsNAC2 was induced exclusively by 20 µm ABA (Fig. 8E). In contrast, ABA concentrations over 40 µm had an inhibitory action on the expression of OsNAC2 (Fig. 8E). Thus, ABA regulates expression of OsNAC2 either in a positive or a negative manner dependent on the dose of ABA. Interestingly, at 2 h of ABA treatment, the transcript level of OsABA8ox1 increased in the wild type, while that of OsZEP1 decreased in OsNAC2-OX (Fig. 8, B and C). ABA was the final production of OsZEP1 and the substrate of OsABA8ox1 (Oliver et al., 2007; Zhu et al., 2009). Therefore, the accumulation of ABA might feedback regulates expression of OsZEP1 and OsABA8ox1.
Figure 8.
Expression of ABA metabolism and senescence-related genes after different concentration of ABA treatments. A to D and F to H, Treatments with 20 µm ABA. I to K, Treatments with 80 µm ABA. Relative mRNA level was calculated using the ΔΔCT method from triplicate data. OsActin was used as an internal control to normalize the different samples with the same amount of plant RNA. Data are mean ± se with three replicates. Asterisks indicate a significant difference between wild-type and OsNAC2 transgenic lines by t test: *P < 0.05, **P < 0.01, and ***P < 0.001.
To gain a mechanistic understanding of the effect of ABA on rice senescence, transcript levels of some SAGs were measured by qPCR. There was no obvious change in the expression of OsSGR and OsNYC3 4 h after 20 µm ABA treatment. Then, significant up-regulation of OsSGR and OsNYC3 in OsNAC2-OX line was observed at 8 and 12 h of ABA treatment (Fig. 8, F and G). Instead, the expression of ABA metabolism-related genes varied 2 or 4 h after ABA treatment (Fig. 8, A–C), which suggested the temporality of OsNAC2 regulating ABA metabolism-related genes and SAGs. Treatment of leaves with ABA resulted in no significant change of OsPAO transcript levels between OsNAC2-OX and the wild type at all three time points: 4, 8, and 12 h (Fig. 8H), which suggested that ABA modulated the expression of OsPAO in an OsNAC2-independent manner.
On the contrary, 80 µm ABA led to significant increase in transcript levels of both target (OsSGR and OsNYC3) and nontarget (OsPAO) genes in OsNAC2-OX line 4 h after treatment, compared with the wild type. In detail, expression of OsSGR and OsNYC3 increased 7.6- and 1.72-fold, respectively, obviously lower than that of OsPAO (14.1-fold; Fig. 8, I–K), which was consistent with the pervious finding that high dose of ABA had a feedback regulation of OsNAC2 and then inhibited the expression of target-genes (Fig. 8E). So, it’s interesting that high dose of ABA might mediate SAGs expression probably in both OsNAC2-dependent and OsNAC2-independent pathway.
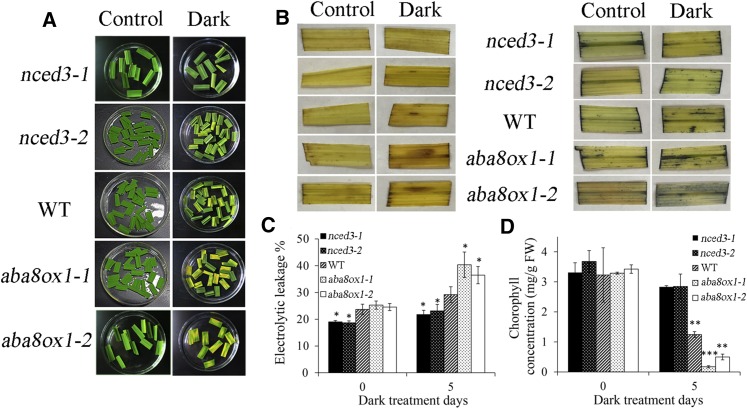
nced3 and aba8ox1 Mutants Exhibit a Stay-Green and Senescence Phenotype, Respectively
To test whether OsNCED3 and OsABA8ox1 are involved in leaf senescence, we obtained the tilling mutants of OsNCED3 (nced3-1,-2) and OsABA8ox1 (aba8ox1-1,-2; Supplemental Fig. S6). After 5 d of dark treatment, nced3-1,-2 leaves showed a stay-green phenotype similar to that of OsNAC2-RNAi plants, while aba8ox1-1,-2 showed a senescent phenotype similar to that of OsNAC2-OX plants (Fig. 9A). Consistent with this, ion leakage rates, ROS levels, and chlorophyll concentrations were significantly higher in aba8ox1 -1,-2 leaves but lower in nced3-1,-2 leaves than in wild-type leaves after dark treatment (Fig. 9, B–D). Taken together, these data indicate that OsNCED3 and OsABA8ox1 are involved in OsNAC2-regulated plant senescence.
Figure 9.
Effect of darkness on detached leaves of 8-week-old wild-type, nced3-1,2, and aba8ox1-1,2 plants. A, Phenotype of wild-type, nced3-1,2, and aba8ox1-1,2 leaves after 5 d dark treatment. B, DAB and NBT staining of 6-week-old wild-type, nced3-1,2, and aba8ox1-1,2 leaves treated with dark for 5 d. C, Ion leakage analysis of leaves in 6-week-old wild-type, nced3-1,2, and aba8ox1-1,2 leaves after 5 d dark treatment. D, Chlorophyll content in leaves prior to and after dark treatment. Values are means ± sd of five measurements. Asterisks indicate a significant difference between wild-type and OsNAC2 transgenic lines by t test: *P < 0.05, **P < 0.01, and ***P < 0.001.
Down-Regulation of OsNAC2 Leads to Increased Grain Yield
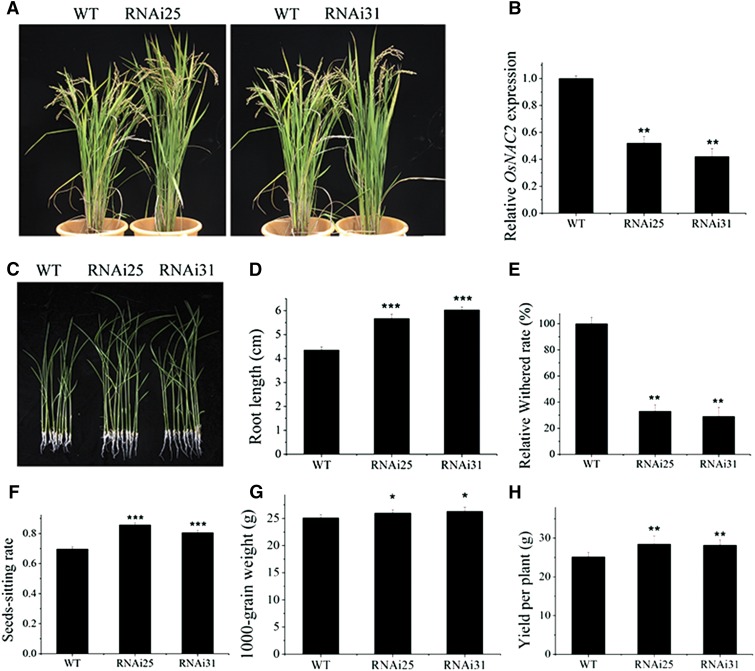
Our RNAi transgenic lines displayed distinctly delayed leaf senescence (Fig. 10A), which was consistent with the observed decline in OsNAC2 expression (Fig. 10B). So, we next investigated whether grain yield is improved when OsNAC2-RNAi lines were grown in the field. As expected, the root length of OsNAC2-RNAi lines was significantly longer than that of the wild type (Fig. 10, C and D). After 150 mm NaCl treatment, OsNAC2-RNAi lines showed a lower withered rate compared with the wild type (Fig. 10E). Furthermore, an examination of the agronomic traits related to grain yield in two OsNAC2-RNAi lines, RNAi25 and RNAi31, showed a 16.1% and 10.9% increase in seed-setting ratio and a 3.6% and 4.6% increase in 1,000-grain weight, respectively, compared with nontransgenic controls (Fig. 10, F and G). These changes resulted in increased grain yields of 11.6% and 10.6% in the RNAi25 and RNAi31 lines, respectively (Fig. 10H). Thus, regulating the expression of OsNAC2 should be a useful strategy for improving rice yield in the future.
Figure 10.
Agronomic traits of OsNAC2-RNAi transgenic lines. A, Morphology of 4-month-old mature OsNAC2-RNAi lines grown in field conditions. B, Expression of OsNAC2 in RNAi plants. C, Root phenotype of 2-week-old wild-type and OsNAC2-RNAi lines. D, Root length of 2-week-old OsNAC2-RNAi plants compared with the wild type. E, Relative withered rate of 2-week-old wild-type and OsNAC2-RNAi line 3 d after 150 mm NaCl treatment. F to H, Seed-setting rate (F), 1,000-grain weight (G), and grain yield per plant (H) in the RNAi lines. Values are means ± sd of 10 measurements. Asterisks indicate a significant difference between wild-type and OsNAC2 transgenic lines by t test: *P < 0.05, **P < 0.01, and ***P < 0.001.
DISCUSSION
Leaf senescence is a complex and highly programmed process, including down-regulation of photosynthesis, disintegration of chloroplasts, degradation of nucleic acid, protein, and lipid, and recycling of nutrients. Although significant progress has been made toward our understanding of leaf senescence in Arabidopsis, it remains largely unknown how leaf senescence is regulated in rice, where chlorophyll degradation is the most characterized, e.g. RLS1 (Jiao et al., 2012), OsDOS (Kong et al., 2006), and NYC1 (Sato et al., 2009). The NAC family is one of the most important transcriptional factors involved in leaf senescence (Gregersen and Holm, 2007). Previous reports have shown that NAC transcription factors, NTL4 (Lee et al., 2012), OsNAC5 (Sperotto et al., 2009), OsNAC6 (Nakashima et al., 2007), OsORE1 (Kim et al., 2014), ONAC106 (Sakuraba et al., 2015), and OsNAP (Liang et al., 2014), participated in aging process of rice. Among them, OsNAP was identified to directly control expression of SAGs, OsSGR, OsNYC1, OsNYC3, OsRCCR1 and OsI57, which function in chlorophyll degradation and glyoxylate cycle (Liang et al., 2014). Additionally, ONAC106 also directly controls the expression of OsSGR and OsNYC1 (Sakuraba et al., 2015). In this study, we identified and characterized OsNAC2, which acts as an important regulator in leaf senescence. OsNAC2 is upregulated during leaf senescence (Fig. 1), and overexpression of OsNAC2 can alter the expression of SAGs (Fig. 3). Furthermore, OsNAC2 can directly interact with the promoter sequences of OsSGR and OsNYC3 (Fig. 4). Although OsNAP and OsNAC2 play a similar role in modulating expression of SAGs, we found that expression of OsNAC2 and OsNAP was not affected in OsNAP-OX and OsNAC transgenic plants, respectively (Supplemental Fig. S7). These results suggest that OsNAC2 accelerates leaf senescence in an OsNAP-independent manner.
ABA is generally considered as a senescence initiator (Cutler et al., 2010). Recently, OsNAP, a transcriptional activator of senescence in rice, is found to repress ABA biosynthesis. ABA concentration is lower in rice leaves of a gain-of-function ps1-D mutant while higher in those of OsNAP-RNAi plants, compared with the wild type. ABA biosynthetic genes, such as OsNCED1, OsNCED3, OsNCED4, and OsZEP, appeared to be downregulated in the ps1-D mutant (Liang et al., 2014). However, how OsNAP regulates ABA biosynthetic genes, and how ABA accelerates senescence remains unclear in rice. In this study, we found that ABA content was significantly higher in OsNAC2-OX transgenic plants than in the wild-type plants (Fig. 6A). Four ABA biosynthetic genes were upregulated, while three ABA degradation genes were downregulated in OsNAC2-OX plants (Fig. 6). These findings indicate that OsNAC2 regulates rice leaf senescence in a different way from OsNAP. Further analysis revealed that OsNAC2 can directly bind to the promoter of OsNCED3, OsZEP1, and OsABA8ox1 (Fig. 7). In Arabidopsis leaves, NAP was reported to promote chlorophyll degradation by directly binding the promoter of ABA biosynthetic gene, AAO3, which enhances the transcription of AAO3 and leads to increased levels of ABA (Yang et al., 2014). The enzymatic steps of ABA biosynthesis, from zeaxanthin to active ABA, and catabolism have been elucidated to be potential points of control to regulate endogenous ABA concentrations (Seo et al., 2009). Our work provided evidence to support that OsNCED3, OsZEP1, and OsABA8ox1 are key points to control ABA levels. OsNCED3 is one of the five NCED genes encoding 9-cis epoxy-carotenoid dioxygenase, which is involved in xanthophyll cleavage (Hwang et al., 2010), while OsZEP1 encodes a zeaxanthin epoxidase responsible for the first step in ABA biosynthesis (Oliver et al., 2007). OsABA8ox1 is a key gene in ABA catabolism and has ABA 8′-hydroxylase activity in rice (Zhu et al., 2009). Importantly, nced3 leaves showed a stay-green phenotype similar to that of OsNAC2-RNAi plants, while aba8ox1 showed premature senility similar to that of OsNAC2-OX plants (Fig. 9A). It was probably that OsNAC2 might not activate the expression of downstream genes without targeting OsNCED3 and OsABA8ox1, which indicated that OsNCED3 and OsABA8ox1 were the targets of OsNAC2 directly. The hybridization of nced3 or aba8ox1 with OsNAC2-OX line or OsNAC2 RNAi line should be further conducted for genetic analysis. These data indicate that OsNCED3, OsZEP1, and OsABA8ox1 act downstream of OsNAC2, consistent with the conclusion that OsNAC2 accelerates senescence via induction of ABA biosynthesis in rice.
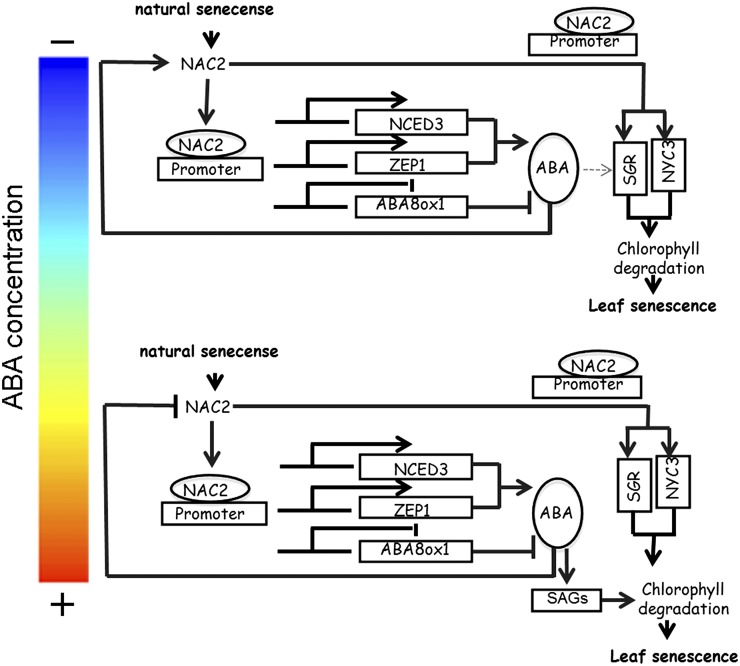
Based on the above results, we proposed a following model for the role of OsNAC2 in leaf senescence (Fig. 11). The plant hormone ABA, together with others including ethylene, jasmonic acid, and salicylic acid, is the major regulator for senescence. In our research, lower concentrations of ABA induce the transcription of OsNAC2 (Fig. 8E), which activates expression of OsNCED3 and OsZEP1, but inhibits expression of OsABA8ox1 by binding to a specific sequence in their promoters (Figs. 6 and 7). The subsequent increase in OsNCED3 and OsZEP1 activity and decrease in OsABA8ox1 activity enhance ABA accumulation in rice leaves (Fig. 6A). Furthermore, OsNAC2 can also directly bind to the promoters of OsSGR and OsNYC3, and increase their expression (Figs. 3 and 4), resulting in chlorophyll degradation and leaf senescence (Figs. 2 and 3). In contrast, although OsNAC2 increases its own expression by induction of ABA biosynthesis, it appears that high level of ABA may have a feedback repression of the expression of OsNAC2 (Fig. 8E) and then slow down ABA synthesis, which might be one of the mechanisms to finely regulate plant growth and development. The resulting also increases the transcription of chlorophyll degradation genes, OsPAO, OsSGR, and OsNYC3, via unknown intermediates and leads to leaf senescence. Thus, the NAC transcription factor OsNAC2 acts as a key regulator linking ABA and leaf senescence processes, which might provide a new vision of NAC TFs function in plant development.
Figure 11.
Proposed model of OsNAC2 role in plant leaf senescence of rice.
The CUC subfamily has provided information about the roles in plant development, such as the formation of the cotyledon boundary, the shoot apical meristem (Vroemen et al., 2003), gynoecium, and ovule (Ishida et al., 2000). NAC proteins, which belong to the CUC subfamily, have been demonstrated to play a pivotal part in plant development (Olsen et al., 2005), stress responses (Puranik et al., 2012), and senescence (Gregersen and Holm, 2007). ANAC096 (Xu et al., 2013), ANAC019 (Jensen et al., 2010), ANAC055, ANAC072/RD26, ANAC002/ATAF1, ANAC081/ATAF2, ANAC102, and ANAC032 (Takasaki et al., 2015) function as positive regulators of ABA signaling. ORE1/ANAC092/NAC2 (Balazadeh et al., 2010), ORS1 (Balazadeh et al., 2011), and AtNAP (Guo and Gan, 2006) are positive regulators of senescence. ANAC016 also has an important role in ABA-inducible leaf senescence by directly promoting AtNAP (Kim et al., 2013). Transcription factors act as not only activators but also repressors in plants. For instance, ATAF1 functions as a negative regulator in drought signaling pathways through modulation of osmotic stress-responsive gene expression (Lu et al., 2007). ATAF2, in the same clade with ATAF1, and ANAC019/ANAC055 were recently reported to negatively regulate the expression of pathogenesis-related genes (Bu et al., 2008). JUB1 is a negative regulator of senescence (Wu et al., 2012). Although, in general, we proposed OsNAC2 accelerates leaf senescence in rice, it is novel that OsNAC2 positively regulated OsSGR, OsNYC3, OsNCED3, and OsZEP1, and negatively regulated OsABA8ox1 by directly binding to their promoters (Figs. 4 and 7). Similarly, Chen et al. (2015) reported that OsNAC2 repressed the expression of GA biosynthetic genes, OsKO2, and enhanced the expression of OsEATB, which encodes a repressor of GA biosynthesis (Chen et al., 2015). Additionally, MYC2, a basic helix-loop-helix domain-containing TF, acts as both activator and repressor to modulate the expression jasmonic acid-responsive genes in Arabidopsis (Dombrecht et al., 2007). The recognition site of MYC2 appears to function as both positive and negative regulator of the expression of the MYC2 promoter-GUS fusion gene (Abe et al., 1997). Recently, CONSTANS, the eponymous member of the family of CONSTANS-like transcriptional regulators, mediates photoperiodic flowering by physically interacting with microproteins, miP1a and miP1b (Graeff et al., 2016).Thus, TFs play a multiple and complicated role in development and stress responses of plants. Future work on the interaction of OsNAC2 with other proteins might help us better understand the regulatory mechanism of transcriptional activation in TFs.
MATERIALS AND METHODS
Plant Materials and Treatments
Plant material used in this study was Nipponbare (Oryza sativa japonica cv Nipponbare). The vector in which 1,500-bp full-length promoter of OsNAC2 is connected with GUS was as described by Shen et al. (2017). Overexpression and RNAi transgenic lines were constructed before and the details are described by Chen et al. (2015). The seeds of OsNAC2-OX, wild-type, and OsNAC2-RNAi plants were germinated in petri dishes with wetted filter paper at 37°C under dark condition. After 48 h incubation, uniformly germinated seeds were selected and cultivated in a beaker containing half-strength Kimura B solution as described previously (Chu and Lee, 1990). The hydroponically cultivated seedlings were grown in a phytotron in a 16/8-h light/dark cycle, 28°C, 300 W/m2 light intensity, and 40% humidity regime. Leaves were used in experiments and all experiments were repeated three times independently.
For dark-induced leaf senescence, 4-week-old seedlings (cultivated as described above) were treated with 0, 3, and 5 d of darkness to observe their phenotypic characters. The leaves were also used for chlorophyll and ion leakage rates measurements. The detached leaves were treated with 0, 2, 3, and 4 d of darkness to obtain the phenotype in vitro. For ABA-induced leaf senescence, rice seeds were germinated on Murashige and Skoog medium containing 0, 2, and 4 µm ABA, and the sensitivity index of seed germination and shoot length to ABA was evaluated. Detached leaves (2 weeks old) were placed in a petri dish wetted with 20 µm ABA and kept under dim light (40 mmol/s/m2). Chlorophyll and ion leakage rates were also measured. The 20 and 80 µm ABA-treated leaves were collected at designated time intervals for total RNA extraction. Additionally, treatment of ABA at 0, 20, 40, 80, and 160 µm was conducted for OsNAC2 expression pattern.
GUS Staining
The native promoter of OsNAC2, a 2-kb genomic DNA fragment upstream of the transcription start site, was cloned and transcriptionally fused to the upstream of the GUS-coding sequence, and the OsNAC2 promoter::GUS construct (pOsNAC2-GUS) was transformed into wild-type rice plants (Nipponbare) plants. Seeds of transgenic rice (T1) harboring an OsNAC2 Pro::GUS construct was used for GUS staining from 1 to 10 d. Primers for amplifying OsNAC2 promoter are shown in Supplemental Table S1. β-GUS staining was performed as described previously (Woo et al., 2001). Images were taken directly or with a stereomicroscope (Leica ZOOM 2000).
Histochemical Staining
O2− and H2O2 were determined using nitro blue tetrazolium (NBT) and 3,3′-diaminobenzidine (DAB) histochemical staining, respectively. Leaves after different treatment were detached from different lines and submerged in NBT solution (1 mg/mL NBT plus 10 mm NaN3 solution in 10 mm potassium phosphate buffer, pH 7.8) or DAB solution (1 mg/mL, pH 5.5). After leaves were stained for 40 min (NBT) or 2 h (DAB), leaves were boiled in 95% ethanol for 15 min to remove the chlorophyll completely and stored in 60% glycerol for observation and imaging.
Chlorophyll Measurements
Chlorophyll was extracted from 50 mg of leaf tissue with different treatment, and its content was determined by measuring the A663 and 645 nm using a visspectro photometer [Jinghua (Shanghai) Co.] as described previously (Kim et al., 2006): total Chl (mg/L) = Chla + Chlb = 20.2A645 + 8.02A663.
Measurement of Ion Leakage Rates
Ion leakage rates were measured as described previously (Lee et al., 2015). The conductivity of the solution was measured using an FG3-ELK conductivity meter (Mettler Toledo Instruments). The rate of ion leakage is expressed as the percentage of initial conductivity divided by the total conductivity.
Rice Microarray Analysis
For microarray analysis, rice seedlings were grown in basal nutrient solution under the condition of a 16-h-light/8-h-dark photoperiod at 28°C. Total RNA was extracted from the leaves of 2-week-old wild-type and OsNAC2-OX (ON11) plants according to the manufacturer’s instructions, and biotin-labeled cRNA was obtained by GeneChip 3′IVT Express Kit (Affymetrix). Array hybridization and wash were then conducted with the GeneChip Hybridization, Wash and Stain Kit (Affymetrix) in Hybridization Oven 645 (Affymetrix) and Fluidics Station 450 (Affymetrix). Slides were scanned by GeneChip Scanner 3000 and Command Console software 3.1 (Affymetrix) with default settings. Three independent biological replicates were conducted for the wild-type and OsNAC2-OX (ON11) plants. Raw data were normalized by the robust multiarray average algorithm, and fold change and pfp values were calculated by rank product method using R (3.0.0). The differently expressed genes in wild-type and OsNAC2-OX plants were classified functionally using the biological process category of Rice Gene Ontology (www.geneontology.com). Significant interactors were determined using a two-sample analysis (t test).
Endogenous ABA Determination
Two-week-old leaves of OsNAC2-OX, wild type, and OsNAC2-RNAi lines were collected, weighed, and immediately frozen in liquid nitrogen. Frozen leaves were pulverized and ABA was extracted as described previously (Chen et al., 2012). Quantitative determination of endogenous ABA was performed on a UPLC-MS/MS system consisting of an AB SCIEX 4500 triple quadrupole mass spectrometer and a Shimadzu LC-30AD UPLC system.Three independent biological repeats were performed.
qPCR Analysis
Untreated and treated leaf tissue obtained from different lines about 2 weeks old was used as a source of total RNA. Total RNAs were extracted with RNAiso reagent (TaKaRa code no. 9752A). The total RNAs were purified and reversed to cDNA with the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa code no. RR047A). The qPCR analysis was performed using SYBR Premix EX Taq (TaKaRa code no. RR420A) on a MyiQ2 real-time PCR detection system (Bio-Rad) according to the manufacturer’s instructions. PCR efficiency (95–105%) was verified. Transcript levels of target genes were normalized to that of the housekeeping gene OsActin using the equation of 2−△△CT, where CT is the threshold cycle for each gene in every sample. Primer sequences are listed in Supplemental Table S1.
Constructions for Yeast One-Hybrid Assay
For yeast one-hybrid assays, the coding sequence of OsNAC2 was amplified using thermostable DNA polymerase from Thermococcus kodakarensis (KOD) polymerase (Toyobo) and was subcloned into the EcoRI and XhoI sites of pGADT7 vector. To generate OsSGRp::HIS3, OsI85p::HIS3, OsNYC3p::HIS3, OsNCED2p::HIS3, OsNCED3p::HIS3, OsNCED5p::HIS3, OsZEP1p::HIS3, OsABA8ox1p::HIS3, OsABA8ox2p::HIS3, and OsABA8ox3p::HIS3 reporter constructs, the promoter fragments were amplified by PCR and then cloned into pHIS2.1 vector through EcoRI-SpeI, EcoRI- SpeI, SacI-SpeI, EcoRI-MluI, EcoRI-MluI, MluI- SpeI, EcoRI-MluI, EcoRI-MluI, EcoRI-MluI, and EcoRI-MluI sites, respectively. All primers used for cloning these constructs are listed in Supplemental Table S1. These vectors and empty vector were transformed into yeast strain AH109 by the PEG/LiAc method, and yeast cells were plated onto SD/–His/–Trp/–Leu + 30 mm 3-amino-1, 2,4-triazole medium for stringent screening of the possible interactions, according to the protocol of Matchmaker GAL4 One-Hybrid System (Clontech).
Dual-Luciferase Assay of Transiently Transformed Tobacco Leaves
To generate the construct of LUC reporter for the dual-luciferase assays, the coding sequence of OsNAC2 was cloned into pGreenII 62-SK vector. The promoter sequences of OsSGR and OsNYC3 were amplified from rice genome and inserted into pGreenII0800-LUC through KpnI-NcoI sites, respectively. Primers are listed in Supplemental Table S1.
The Nicotiana benthamiana leaves were used for dual-luciferase assays as described previously (Hellens et al., 2005). The effector constructs (p35S-OsNAC2 or p35S) and the above reporter constructs harboring the luciferase (LUC) gene were introduced into Agrobacterium tumefaciens strain GV3101, respectively. After cotransformation, the tobacco plants were placed under 23°C for 3 d. The firefly LUC and Renilla luciferase (REN) activities were measured using the Dual-Luciferase Reporter Assay System (Promega).The LUC activity was normalized to the REN activity.
ChIP
ChIP was performed based on a previous report (Gendrel and Colot, 2005) with OsNAC2-OX transgenic seedlings, in which a GFP coding sequence was fused in frame to the 3′end of the OsNAC2 gene. About 4 g of 4-week-old OsNAC2-OX seedlings was treated and fixed with 1.0% formaldehyde for 10 min subsequently. Antibodies against mGFP (Santa Cruz Biotechnology) were used for immunoprecipitation. Protein-A-agarose beads were blocked with salmon sperm DNA and used to pull down the protein-DNA complex. The DNA fragments of the ChIP were used for qPCR. Primers were selected in the promoter regions of each selected gene. The ChIP experiments were repeated three times with the similar data. Primer pairs for qPCR were listed in Supplemental Table S1.
Field Cultivation of Rice
The rice plants were grown in a standard paddy field at the Experimental Station of Fudan University in Taicang Jiangshu province. The field planting was with three replicates according to a randomized complete-block design; the spacing of the transplanted plants was 20 × 20 cm. The area per plot was designed as 4 m2. These plants were then grown under the conditions of conventional cultivation. For agronomic traits measurement, at least 10 plants randomly obtained from the center of the plot were used to avoid any irregularities at the margins of the plot.
Tilling Mutant and Verification
The tilling mutant of OsABA8ox1 and OsNCED3 bought from Liu Lab at Key Laboratory of Plant Molecular Physiology, CAS, was germinated and hydroponically cultivated as described previously (Chu and Lee, 1990). These tilling mutants were planted in the paddy field at Fudan University. For mutant verification, 2-week-old leaf tissue of each mutant line was used for DNA extracting by Plant Genomic DNA Kit (Tiangen; code DP305). The primers, aba8ox1-F (5′-CTAACTCCTCTTCCCACTCTGCTT-3′), aba8ox1-R (5′-ATTTCAGTTAAGGAACCAGTCTGC-3′), nced3-F (5′-TCTCTCTCTCGGCAGAAACACACC-3′), and nced3-R (5′-CGGGTCGAGGAGGCCGCAGGCGGC-3′), were provided by Liu Lab at Key Laboratory of Plant Molecular Physiology, CAS, for amplifying the fragments of OsABA8ox1 and OsNCED3. PCR was carried out using PrimerSTAR Max DNA Polymerase (TaKaRa code no. R045A) according to the manufacturer’s instruction. The obtained fragments were sequenced by Sangon Biotech (Shanghai) Co. Additionally, leaves of the mutants collected from the heading stage were treated with 5 d of darkness to observe their phenotypes. Chlorophyll and ion leakage rates were also measured.
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL databases under the following accession numbers: OsNAC2 (Os04g0460600), OsNAP (Os03g0327800), OsSGR (Os09g0532000), OsNYC3 (Os06g0354700), OsI85 (Os07g0529000), OsRCCR1 (Os10g0389300), OsPaO (Os03g0805900), OsNCED2 (Os12g0435200), OsNCED3 (Os03g0645900), OsNCED5 (Os12g0617400), OsABA8ox1 (Os02g0703600), OsABA8ox2 (Os08g0472800), OsABA8ox3 (Os09g0457100), OsZEP1 (Os04g0448900), OsActin (Os10g0510000), OsNOL (Os03g0654600), OsSWEET5 (Os05g0588500), OsTDC1 (Os08g0140300), and OsNAP (Os03g0327800).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Natural senescence phenotype of OsNAC2 transgenic lines during grain-filling stage in field condition.
Supplemental Figure S2. Histochemical staining of a transgenic line expressing pOsNAC2-GUS in 6-week-old wild-type leaf and seeds 1 to 10 d after germination.
Supplemental Figure S3. Phylogenetic tree among NACs in Arabidopsis and rice.
Supplemental Figure S4. Yeast one-hybrid assays between OsNAC2 and the promoter of senescence and ABA metabolism-related genes.
Supplemental Figure S5. NAC binding motif CACG searching.
Supplemental Figure S6. Sequence alignment of mutant aba8ox1 and nced3 with the wild type.
Supplemental Figure S7. Expression of OsNAC2 in OsNAP-OX (OX11 and OX17) and OsNAP-SRDX (SRDX4, SRDX6, and SRDX7) lines and OsNAP in OsNAC2-OX and OsNAC2-RNAi lines.
Supplemental Table S1. Primers used for the sequencing of different genes in rice.
Glossary
- SAG
senescence-associated gene
- TF
transcription factor
- ROS
reactive oxygen species
- FW
fresh weight
- ChIP
chromatin immunoprecipitation
- NBT
nitro blue tetrazolium
- DAB
3,3′-diaminobenzidine
Footnotes
Articles can be viewed without a subscription.
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller-Roeber B (2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4: 346–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62: 250–264 [DOI] [PubMed] [Google Scholar]
- Becker W, Apel K (1993) Differences in gene expression between natural and artificially induced leaf senescence. Planta 189: 74–79 [Google Scholar]
- Bu Q, Jiang H, Li CB, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C (2008) Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res 18: 756–767 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, Leaver CJ (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Chandler PM, Robertson M (1994) Gene expression regulated by abscisic acid and its relation to stress tolerance. Annu Rev Plant Biol 45: 113–141 [Google Scholar]
- Chen ML, Fu XM, Liu JQ, Ye TT, Hou SY, Huang YQ, Yuan BF, Wu Y, Feng YQ (2012) Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J Chromatogr B Analyt Technol Biomed Life Sci 905: 67–74 [DOI] [PubMed] [Google Scholar]
- Chen X, Lu S, Wang Y, Zhang X, Lv B, Luo L, Xi D, Shen J, Ma H, Ming F (2015) OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. Plant J 82: 302–314 [DOI] [PubMed] [Google Scholar]
- Chu C, Lee TM (1990) The relationship between ethylene biosynthesis and chilling tolerance in seedlings of rice (Oryza sativa). Bot Bull Acad Sin 30: 263–273 [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, Kazan K (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Colot V (2005) Arabidopsis epigenetics: when RNA meets chromatin. Curr Opin Plant Biol 8: 142–147 [DOI] [PubMed] [Google Scholar]
- Graeff M, Straub D, Eguen T, Dolde U, Rodrigues V, Brandt R, Wenkel S (2016) Microprotein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in Arabidopsis. PLoS Genet 12: e1005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen PL, Holm PB (2007) Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotechnol J 5: 192–206 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S. (2006) Chlorophyll degradation during senescence. Annu Rev Plant Biol 57: 55–77 [DOI] [PubMed] [Google Scholar]
- Hu R, Qi G, Kong Y, Kong D, Gao Q, Zhou G (2010) Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol 10: 145 10.1186/1471-2229-10-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Chen HC, Huang WY, Chu YC, Choutou S, Cheng WH (2010) Ectopic expression of rice OsNCED3 in Arabidopsis increases ABA level and alters leaf morphology. Plant Sci 178: 12–22 [Google Scholar]
- Ishida T, Aida M, Takada S, Tasaka M (2000) Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant Cell Physiol 41: 60–67 [DOI] [PubMed] [Google Scholar]
- Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O’Shea C, Skriver K (2010) The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem J 426: 183–196 [DOI] [PubMed] [Google Scholar]
- Jiao BB, Wang JJ, Zhu XD, Zeng LJ, Li Q, He ZH (2012) A novel protein RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice. Mol Plant 5: 205–217 [DOI] [PubMed] [Google Scholar]
- Kang K, Kim YS, Park S, Back K (2009) Senescence-induced serotonin biosynthesis and its role in delaying senescence in rice leaves. Plant Physiol 150: 1380–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Uchimiya H (2000) Coleoptile senescence in rice (Oryza sativa L.). Ann Bot (Lond) 86: 405–414 [Google Scholar]
- Kim HJ, Chung KM, Woo HR (2011) Three positive regulators of leaf senescence in Arabidopsis, ORE1, ORE3 and ORE9, play roles in crosstalk among multiple hormone-mediated senescence pathways. Genes Genomics 33: 373–381 [Google Scholar]
- Kim YS, Sakuraba Y, Han SH, Yoo SC, Paek NC (2013) Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol 54: 1660–1672 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Hong SH, Kim YW, Lee IH, Jun JH, Phee BK, Rupak T, Jeong H, Lee Y, Hong BS, et al. (2014) Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J Exp Bot 65: 4023–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Nam HG, Lim PO (2016) Regulatory network of NAC transcription factors in leaf senescence. Curr Opin Plant Biol 33: 48–56 [DOI] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG (2009) Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kong Z, Li M, Yang W, Xu W, Xue Y (2006) A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol 141: 1376–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T, Makita N, Kojima M, Tokunaga H, Sakakibara H (2012) Cytokinin activity of cis-zeatin and phenotypic alterations induced by overexpression of putative cis-Zeatin-O-glucosyltransferase in rice. Plant Physiol 160: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, Tanaka A (2007) Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19: 1362–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Seo PJ, Lee HJ, Park CM (2012) A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J 70: 831–844 [DOI] [PubMed] [Google Scholar]
- Lee SH, Sakuraba Y, Lee T, Kim KW, An G, Lee HY, Paek NC (2015) Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J Integr Plant Biol 57: 562–576 [DOI] [PubMed] [Google Scholar]
- Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J, Chu C (2014) OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA 111: 10013–10018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PL, Chen NZ, An R, Su Z, Qi BS, Ren F, Chen J, Wang XC (2007) A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol 63: 289–305 [DOI] [PubMed] [Google Scholar]
- Mao C, Ding W, Wu Y, Yu J, He X, Shou H, Wu P (2007) Overexpression of a NAC-domain protein promotes shoot branching in rice. New Phytol 176: 288–298 [DOI] [PubMed] [Google Scholar]
- Morita R, Sato Y, Masuda Y, Nishimura M, Kusaba M (2009) Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J 59: 940–952 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51: 617–630 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Oda-Yamamizo C, Mitsuda N, Sakamoto S, Ogawa D, Ohme-Takagi M, Ohmiya A (2016) The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Sci Rep 6: 23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SN, Dennis ES, Dolferus R (2007) ABA regulates apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in rice. Plant Cell Physiol 48: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al. (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19: 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao W, Kim EY, Han SH, Sakuraba Y, Paek NC (2015) Rice phytochrome B (OsPhyB) negatively regulates dark- and starvation-induced leaf senescence. Plants (Basel) 4: 644–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistelli L, De Bellis L, Alpi A (1991) Peroxisomal enzyme activities in attached senescing leaves. Planta 184: 151–153 [DOI] [PubMed] [Google Scholar]
- Pruzinská A, Tanner G, Aubry S, Anders I, Moser S, Müller T, Ongania KH, Kräutler B, Youn JY, Liljegren SJ, Hörtensteiner S (2005) Chlorophyll breakdown in senescent Arabidopsis leaves. Characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol 139: 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17: 369–381 [DOI] [PubMed] [Google Scholar]
- Qiao Y, Jiang W, Lee J, Park B, Choi MS, Piao R, Woo MO, Roh JH, Han L, Paek NC, Seo HS, Koh HJ (2010) SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol 185: 258–274 [DOI] [PubMed] [Google Scholar]
- Ricachenevsky FK, Sperotto RA, Menguer PK, Fett JP (2010) Identification of Fe-excess-induced genes in rice shoots reveals a WRKY transcription factor responsive to Fe, drought and senescence. Mol Biol Rep 37: 3735–3745 [DOI] [PubMed] [Google Scholar]
- Saika H, Okamoto M, Miyoshi K, Kushiro T, Shinoda S, Jikumaru Y, Fujimoto M, Arikawa T, Takahashi H, Ando M, et al. (2007) Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol 48: 287–298 [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Jeong J, Kang MY, Kim J, Paek NC, Choi G (2014) Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun 5: 4636. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Piao W, Lim JH, Han SH, Kim YS, An G, Paek NC (2015) Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol 56: 2325–2339 [DOI] [PubMed] [Google Scholar]
- Sato Y, Morita R, Katsuma S, Nishimura M, Tanaka A, Kusaba M (2009) Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J 57: 120–131 [DOI] [PubMed] [Google Scholar]
- Schelbert S, Aubry S, Burla B, Agne B, Kessler F, Krupinska K, Hörtensteiner S (2009) Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21: 767–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Nambara E, Choi G, Yamaguchi S (2009) Interaction of light and hormone signals in germinating seeds. Plant Mol Biol 69: 463–472 [DOI] [PubMed] [Google Scholar]
- Shen J, Lv B, Luo L, He J, Mao C, Xi D, Ming F (2017) The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci Rep 7: 40641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda Y, Ito H, Tanaka A (2016) Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 28: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Giri MK, Singh PK, Siddiqui A, Nandi AK (2013) Down-regulation of OsSAG12-1 results in enhanced senescence and pathogen-induced cell death in transgenic rice plants. J Biosci 38: 583–592 [DOI] [PubMed] [Google Scholar]
- Sperotto RA, Ricachenevsky FK, Duarte GL, Boff T, Lopes KL, Sperb ER, Grusak MA, Fett JP (2009) Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta 230: 985–1002 [DOI] [PubMed] [Google Scholar]
- Takasaki H, Maruyama K, Takahashi F, Fujita M, Yoshida T, Nakashima K, Myouga F, Toyooka K, Yamaguchi-Shinozaki K, Shinozaki K (2015) SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J 84: 1114–1123 [DOI] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Tang Y, Li M, Chen Y, Wu P, Wu G, Jiang H (2011) Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. J Plant Physiol 168: 1952–1959 [DOI] [PubMed] [Google Scholar]
- Ulker B, Shahid Mukhtar M, Somssich IE (2007) The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226: 125–137 [DOI] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC (2003) The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15: 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37: 455–469 [DOI] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG (2001) ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Kim HJ, Nam HG, Lim PO (2013) Plant leaf senescence and death - regulation by multiple layers of control and implications for aging in general. J Cell Sci 126: 4823–4833 [DOI] [PubMed] [Google Scholar]
- Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor MI, Asensi-Fabado MA, Munné-Bosch S, Antonio C, Tohge T, et al. (2012) JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24: 482–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HB, Wang B, Chen Y, Liu YG, Chen L (2013) Characterization and fine mapping of the rice premature senescence mutant ospse1. Theor Appl Genet 126: 1897–1907 [DOI] [PubMed] [Google Scholar]
- Xu ZY, Kim SY, Hyeon Y, Kim DH, Dong T, Park Y, Jin JB, Joo SH, Kim SK, Hong JC, Hwang D, Hwang I (2013) The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25: 4708–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Worley E, Udvardi M (2014) A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26: 4862–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SD, Seo PJ, Yoon HK, Park CM (2011) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Zhang L, Song B, Qi X, Chan Z (2015) Systematic analysis and identification of stress-responsive genes of the NAC gene family in Brachypodium distachyon. PLoS One 10: e0122027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu L, Huang W, Yuan M, Zhou F, Li X, Lin Y (2014) Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS One 9: e94210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Ye N, Zhang J (2009) Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol 50: 644–651 [DOI] [PubMed] [Google Scholar]