AP-3-mediated vacuolar trafficking of PAT10 involves a subpopulation of COPII and the vacuolar tethering complex HOPS.
Abstract
Plant vacuoles are versatile organelles critical for plant growth and responses to environment. Vacuolar proteins are transported from the endoplasmic reticulum via multiple routes in plants. Two classic routes bear great similarity to other phyla with major regulators known, such as COPII and Rab5 GTPases. By contrast, vacuolar trafficking mediated by adaptor protein-3 (AP-3) or that independent of the Golgi has few recognized cargos and none of the regulators. In search of novel regulators for vacuolar trafficking routes and by using a fluorescence-based forward genetic screen, we demonstrated that the multispan transmembrane protein, Arabidopsis (Arabidopsis thaliana) PROTEIN S-ACYL TRANSFERASE10 (PAT10), is an AP-3-mediated vacuolar cargo. We show that the tonoplast targeting of PAT10 is mediated by the AP-3 complex but independent of the Rab5-mediated post-Golgi trafficking route. We also report that AP-3-mediated vacuolar trafficking involves a subpopulation of COPII and requires the vacuolar tethering complex HOPS. In addition, we have identified two novel mutant alleles of AP-3δ, whose point mutations interfered with the formation of the AP-3 complex as well as its membrane targeting. The results presented here shed new light on the vacuolar trafficking route mediated by AP-3 in plant cells.
Plant vacuoles are versatile organelles critical for plant growth and responses to the environment (Pacini et al., 2011). The functionality of vacuoles is fulfilled through proteins localized at the vacuolar membrane (i.e. the tonoplast) and those in the vacuolar lumen. Transmembrane proteins at the tonoplast mediate ion homeostasis and turgor regulation, whereas luminal proteins inside the vacuoles participate in nutrient recycling and signal termination.
Vacuolar proteins are synthesized at the endoplasmic reticulum (ER) and transported via multiple routes to their destination in plants (Bassham et al., 2008; Hwang, 2008; Pedrazzini et al., 2013; Uemura and Ueda, 2014). Four distinct vacuolar trafficking routes have been proposed in plant cells (Uemura and Ueda, 2014). A classic route bearing high similarity to yeast and metazoans utilizes COPII-mediated anterograde trafficking from the ER to the Golgi. It passes through the trans-Golgi network/early endosome (TGN/EE) to prevacuolar compartments (PVC)/multivesicular bodies (MVB) through sequential activation of the small GTPases Rab5 and Rab7 (Cui et al., 2014; Ebine et al., 2014; Singh et al., 2014) and finally reaches the vacuoles. The second route is slightly distinct from the first, such that Rab7 is not required (Ebine et al., 2014; Feng et al., 2017). The third route is mediated by Adaptor Protein-3 (AP-3), a heterotetrameric complex consisting of two large subunits (δ and β), a medium subunit (µ), and a small subunit (σ; Bassham et al., 2008; Feraru et al., 2010; Zwiewka et al., 2011). Finally, several vacuolar proteins have been reported to target to the tonoplast independent of the Golgi (i.e. directly from the ER to the vacuoles; Viotti et al., 2013). Many cargos and regulators have been identified for the first two routes (Bassham et al., 2008; Hwang, 2008; Pedrazzini et al., 2013; Uemura and Ueda, 2014). By contrast, few cargos have been identified for the AP-3-mediated trafficking route and the ER-to-vacuole direct route (Wolfenstetter et al., 2012; Viotti et al., 2013; Ebine et al., 2014). Regulators controlling these two vacuolar trafficking routes are obscure.
We have demonstrated previously that a multispan tonoplast protein Arabidopsis (Arabidopsis thaliana) PROTEIN S-ACYL TRANSFERASE10 (PAT10) mediates the S-acylation-dependent tonoplast association of CBL2, CBL3, and CBL6 (Zhou et al., 2013; Zhang et al., 2015). CBLs are calcineurin B-like proteins critical for Ca2+ signaling during plant growth and responses to abiotic stresses (Batistic et al., 2010; Tang et al., 2012). Although most CBLs are soluble proteins, they associate with either the plasma membrane (PM) or the tonoplast through PAT-mediated S-acylation (Batistic et al., 2010, 2012; Zhou et al., 2013). Therefore, the subcellular targeting of CBLs depends on the location of their upstream PATs. Interestingly, a previous study showed that the tonoplast association of CBL6 is independent of the Golgi (Bottanelli et al., 2011), implying that PAT10 is transported to the tonoplast via unconventional routes.
To determine the trafficking route of PAT10 to the tonoplast and identify novel regulatory components in vacuolar trafficking routes, we performed a fluorescence-based forward genetic screen. Here, we report that (1) the tonoplast targeting of PAT10 is mediated through the AP-3 complex but independent of Rab5; (2) the AP-3-mediated vacuolar trafficking of PAT10 involves a subpopulation of COPII; and (3) AP-3-mediated vacuolar trafficking requires the vacuolar tethering complex HOPS (homotypic fusion and vacuole protein sorting). In addition, we have identified two novel mutant alleles of AP-3δ, whose point mutations interfered with the formation of the AP-3 complex as well as its membrane targeting. The results presented here shed new light on the vacuolar trafficking route mediated by AP-3 in plant cells.
RESULTS
Targeting of PAT10 to the Tonoplast Is Independent of Rab5-Mediated Post-Golgi Trafficking Routes
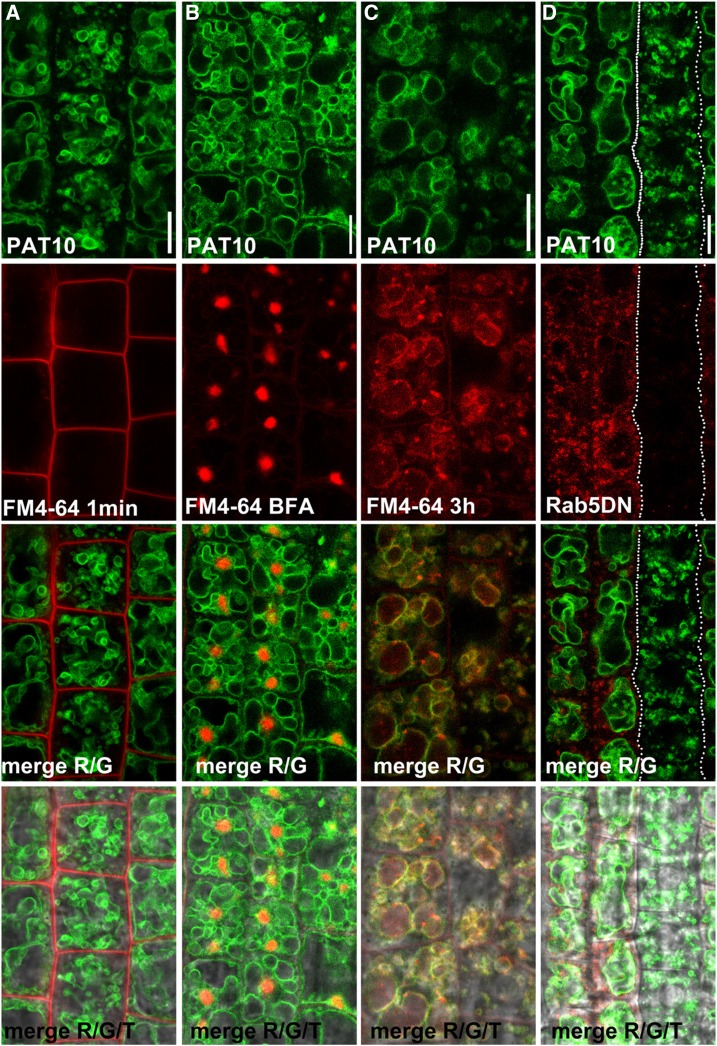
To test whether the vacuolar trafficking of PAT10 was independent of the classic post-Golgi trafficking route, we applied both genetic and pharmacological approaches. We first treated PAT10g-GFP;pat10-1 plants with the lipophilic dye FM4-64. Because the ectopic expression of PAT10 resulted in its mistargeting to the Golgi (Batistic, 2012; Qi et al., 2013), likely due to saturation of the ER sorting machinery, we analyzed the localization of PAT10 using PAT10g-GFP;pat10-1, in which all mutant phenotypes were rescued and the protein was targeted exclusively to the tonoplast (Zhou et al., 2013). As reported (Bolte et al., 2004), FM4-64 first labels the PM (Fig. 1A) and then internalizes to reach the tonoplast through endocytic trafficking (Fig. 1C). Treatment of the fungal toxin brefeldin A (BFA) resulted in the aggregation of internalized FM4-64 signals into so-called BFA compartments (Fig. 1B). However, the tonoplast association of PAT10 was not interfered with by BFA treatment (Fig. 1C), suggesting that the tonoplast targeting of PAT10 was independent of BFA-sensitive post-Golgi trafficking.
Figure 1.
Genetic interference of Rab5-dependent post-Golgi trafficking did not impair the tonoplast association of PAT10. A to C, Confocal laser scanning micrograph (CLSM) of root epidermal cells from 4-d-after-germination (DAG) seedlings of PAT10g-GFP;pat10-1 transgenic plants pulse labeled with FM4-64 for 1 min (A), for 5 min and then treated with BFA for 50 min (B), or for 3 h (C). D, CLSM of root epidermal cells from 4-DAG seedlings of PAT10g-GFP;pat10-1;ProCPC:RFP-Rab5DN transgenic plants. Regions between the dotted lines are H cells with no Rab5DN expression. Merges of the RFP/GFP channels (R/G) and merges of the RFP/GFP/transmission channels (R/G/T) are shown at the bottom. Bars = 10 µm.
Because the classic post-Golgi trafficking routes require the activity of Rab5 (Uemura and Ueda, 2014), we expressed a dominant negative (DN) Rab5 to inhibit Rab5-dependent post-Golgi trafficking (Sohn et al., 2003; Lee et al., 2004). We used the promoter of CARPRICE (ProCPC) that is specifically active in nontrichoblast cells (N cells; Wada et al., 2002), while GFP-labeled PAT10 is expressed in both N cells and H cells (trichoblast cells). Thus, targeting of the same transgene can be analyzed simultaneously in the same root under various conditions. The expression of Rab5DN also did not alter the tonoplast association of PAT10 (Fig. 1D). To verify that the genetic interference worked as it should, we examined the effect of Rab5DN on inositol transporter1 (INT1), whose tonoplast localization relies on Rab5-dependent vacuolar trafficking (Wolfenstetter et al., 2012). In contrast to the tonoplast association of INT1 in the wild type, both the PM and the tonoplast were labeled with INT1 signals in cells expressing Rab5DN (Supplemental Fig. S1). These results indicated that PAT10 did not take the classic Rab5-depedent vacuolar trafficking routes.
Fluorescence-Based Forward Genetic Screening to Identify Key Components Regulating Rab5-Independent Vacuolar Trafficking Routes
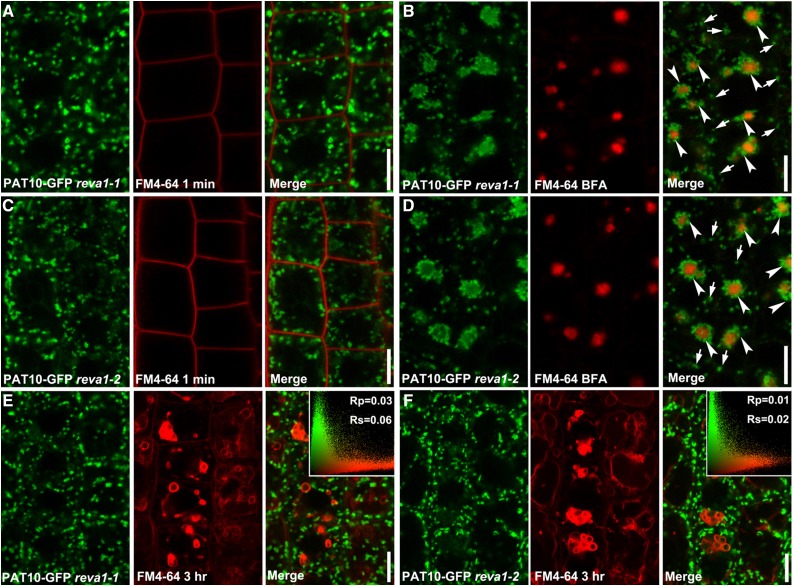
To determine the trafficking route of PAT10 to vacuoles and identify novel regulatory components in Rab5-independent trafficking routes, we performed fluorescence-based forward genetic screening. Because PAT10 is expressed in pollen and pollen tubes (Zhou et al., 2013), an in vitro pollen germination assay at the M1 generation would be able to detect mutations that resulted in the mistargeting of PAT10. We screened around 3,000 M1 plants from an ethyl methanesulfonate (EMS)-mutagenized PAT10g-GFP;pat10-1 population using fluorescence microscopy for pollen tubes that display aberrant targeting of PAT10-GFP. Two lines showed similar alterations at the subcellular distribution of PAT10-GFP (Supplemental Fig. S2). These two mutants are recessive, because PAT10-GFP was distributed to punctate vesicles in M2 roots but not in M1 roots. By crosses, we also verified that the two mutants were allelic, and thus named them reva1-1 and reva1-2 (for regulators of vacuolar trafficking1). Instead of the tonoplast distribution, PAT10-GFP was distributed as punctate vesicles (Fig. 2, A, C, E, and F). BFA treatment resulted in the accumulation of most PAT10-positive signals into a circle surrounding the FM4-64-labeled BFA cores (Fig. 2, B and D), indicative of its Golgi identity (Lam et al., 2009).
Figure 2.
The tonoplast association of PAT10 was interfered with in the reva1 mutants. CLSM of root epidermal cells from 4-DAG seedlings of reva1-1 (A, B, and E) or reva1-2 (C, D, and F). Roots were pulse labeled with FM4-64 for 1 min (A and C) or for 3 h (E and F) or were pulse labeled with FM4-64 and followed by BFA treatment for 50 min (B and D). Arrowheads in B and D point at BFA compartments. Arrows in B and D point at BFA-insensitive vesicles. Insets in E and F indicate the Pearson correlation coefficient (Rp) and the Spearman correlation coefficient (Rs). Results are representative of over 30 images. Bars = 10 µm.
The Tonoplast Targeting of PAT10 Is Mediated by AP-3
To identify the casual gene for the reva1 mutants, we combined map-based cloning and sequencing (Supplemental Fig. S2). Rough mapping using F2 progeny derived from an outcross with Landsberg erecta (Ler) indicated that mutations were on chromosome 1. By sequencing, we identified a base pair change in the coding sequence of At1g48760, resulting in an amino acid substitution (G94D for reva1-1 and E278K for reva1-2; Supplemental Fig. S3). This gene encodes the δ-subunit of AP-3 (AP-3δ). Because two mutants of AP-3δ, the EMS-mutagenized pat4-1 and the T-DNA insertional mutant pat4-2, were identified previously (Feraru et al., 2010; Zwiewka et al., 2011), we renamed reva1-1 and reva1-2 as pat4-3 and pat4-4, respectively.
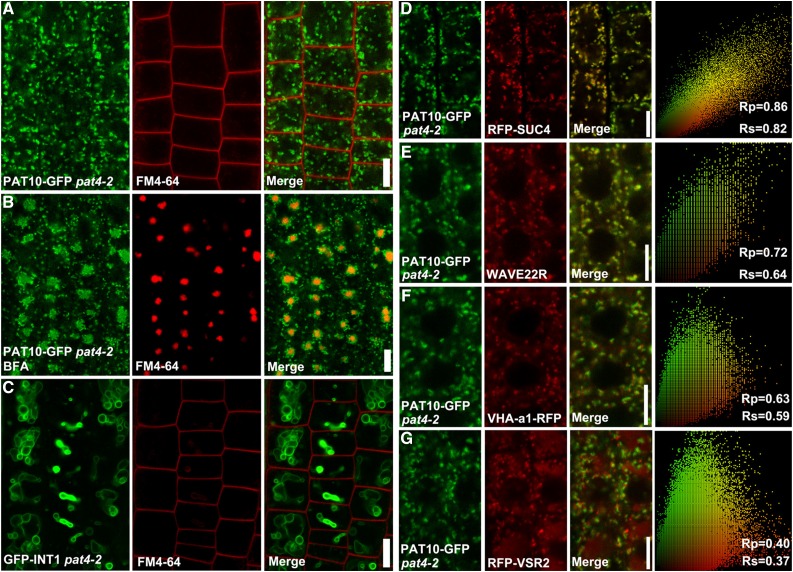
To verify that the tonoplast association of PAT10 depended on AP-3, we first analyzed its subcellular targeting in pat4-2, the null mutant allele of AP-3δ (Feraru et al., 2010). As in pat4-3 and pat4-4 (Fig. 2), PAT10-GFP was distributed in punctate vesicles (Fig. 3A) that formed a ring-shaped structure surrounding the FM4-64 core upon BFA treatment in pat4-2 (Fig. 3B). By contrast, INT1, whose tonoplast association is independent of AP-3 (Wolfenstetter et al., 2012), was distributed at a ring-shaped membrane (Fig. 3C), as typical of vacuolar morphology in AP-3 mutants (Feraru et al., 2010; Zwiewka et al., 2011). To provide further evidence, we introduced ProUBQ10:RFP-SUC4 into PAT10g-GFP; pat4-2. As an AP-3-mediated tonoplast cargo, SUC4 was relocalized into cis-Golgi stacks in AP-3 mutants (Wolfenstetter et al., 2012). Indeed, PAT10-GFP and RFP-SUC4 overlapped substantially at punctate vesicles in pat4-2 (Fig. 3D), supporting PAT10 as the other AP-3 cargo. We also introduced fluorescent markers for the Golgi apparatus (WAVE22R; Geldner et al., 2009), for the TGN/EE (VHA-a1-RFP; Dettmer et al., 2006), and for the PVC/MVB (RFP-VSR2; Wang et al., 2011). As expected, PAT10 showed the most substantial colocalization with the Golgi marker (Fig. 3E) but not that of TGN/EE (Fig. 3F) or PVC/MVB (Fig. 1G). These results demonstrated that the tonoplast association of PAT10 relies on AP-3 and that mutations at AP-3 resulted in the retention of PAT10 at the Golgi.
Figure 3.
The tonoplast targeting of PAT10 is mediated by AP-3. A and B, CLSM of root epidermal cells from PAT10g-GFP pat4-2 transgenic plants upon 1 min of FM4-64 staining (A) or pulse labeled with FM4-64 and followed by BFA treatment for 50 min (B). C, CLSM of root epidermal cells from ProUBQ10:GFP-INT1;pat4-2 transgenic plants upon 1 min of FM4-64 staining. D to G, CLSM of root epidermal cells from PAT10g-GFP;ProUBQ10:RFP-SUC4;pat4-2 (D), PAT10g-GFP;WAVE22R;pat4-2 (E), PAT10g-GFP;VHA-a1-RFP;pat4-2 (F), and PAT10g-GFP;ProUBQ10:RFP-VSR2;pat4-2 (G) transgenic plants. The Pearson correlation coefficient (Rp) and the Spearman correlation coefficient (Rs) are indicated on the scatterplots at the right side of each micrograph. Bars = 10 µm.
To determine whether the tonoplast association of PAT10 was compromised in the mutants of other AP-3 subunits, we introduced PAT10-GFP into pat2-2 (a null mutant of AP-3β; Zwiewka et al., 2011), ap-3µ-2 (a mutant of AP-3µ; Kansup et al., 2013), and ap-3σ (a mutant of AP-3σ; Supplemental Fig. S4). Not surprisingly, all AP-3 mutants tested showed defective tonoplast targeting of PAT10-GFP (Supplemental Fig. S4). In addition, the expression of AP-3δ-RFP driven by ProUBQ10 fully restored the tonoplast targeting of PAT10-GFP in pat4-2 (Supplemental Fig. S4), confirming the AP-3-dependent trafficking of PAT10.
Gly-94 and Glu-278 of AP-3δ Are Key Residues Affecting AP-3 Complex Formation and Subcellular Targeting
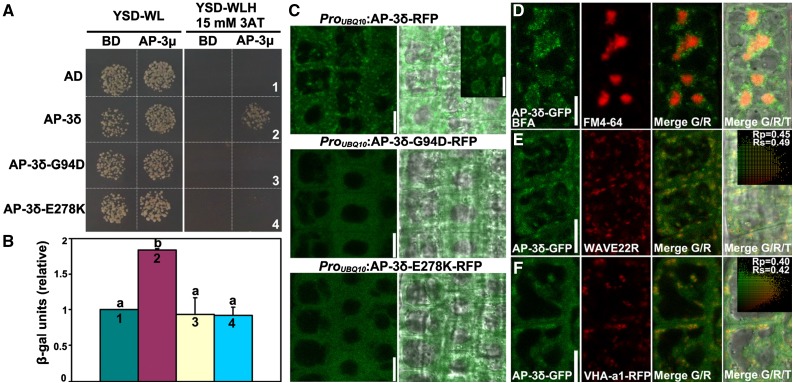
All F1 plants from crosses between pat4-2 and pat4-3 or pat4-4 showed the same PAT10-GFP distribution pattern as the parental homozygous plants (Fig. 4, D–G), suggesting that both pat4-3 and pat4-4 are null mutants of AP-3δ. Indeed, the Gly-94 and Glu-278 residues are evolutionarily conserved (Supplemental Fig. S3). To determine how these amino acid substitutions affected the function of AP-3, we first performed yeast two-hybrid (Y2H) assays between different AP-3 subunits. Coimmunoprecipitation showed that AP-3δ may interact directly with AP-3µ (Zwiewka et al., 2011). Using Y2H assays, we showed that the wild-type AP-3δ but not the two mutated versions interacted with AP-3µ (Fig. 4, A and B). Next, we expressed RFP translational fusions of AP-3δ and the two mutant variants in root epidermal cells to determine their subcellular targeting. The wild-type AP-3δ-RFP was localized at punctate vesicles, which accumulate into ring-shaped structure with faint signals also in the BFA core upon BFA treatment (Fig. 4, C and D), indicative of Golgi and TGN/EE identity. By contrast, the two AP-3δ mutants showed diffused cytoplasmic localization (Fig. 4C). To provide further evidence of the subcellular targeting of AP-3, we generated transgenic plants expressing both AP-3δ-GFP and RFP-fused organelle markers, including WAVE22R for the Golgi (Geldner et al., 2009) and VHA-a1-RFP for the TGN/EE (Dettmer et al., 2006). Colocalization with these fluorescence markers showed that AP-3 associates with the Golgi and partially with the TGN/EE (Fig. 4, E and F). These results demonstrated that the two residues, Gly-94 and Glu-278, are critical not only for the formation of AP-3 but also its subcellular targeting.
Figure 4.
Gly-94 and Glu-278 of AP-3δ are key residues affecting AP-3 complex formation and subcellular targeting. A, Y2H assays between AP-3μ and AP-3δ or its mutants (G94D and E278K). Yeast strains cotransformed with Binding domain (BD)-AP-3μ and Activation domain (AD)-AP-3δ fusion constructs were growing on nonselective plates (-WL) or on selective plates (-WLH/15 mm 3AT). BD or AD alone was used as the control, respectively. 3AT, 3-Amino-1,2,4-triazole. Results shown are representative images from five independent experiments. B, Quantification of β-galactosidase activity from the Y2H assay as shown in A. Results shown are means ± se (n = 5). Each number (1–4) corresponds to the yeast strain labeled in A. Different letters indicate significantly different groups (one-way ANOVA, Tukey-Kramer test, P < 0.01). C, CLSM of root epidermal cells expressing RFP translational fusions of AP-3δ, AP-3δ-G94D, or AP-3δ-E278K. The inset shows CLSM of root epidermal cells expressing AP-3δ-RFP treated with BFA for 50 min. Merges of fluorescence and transmission images are shown at the side. D to F, CLSM of root epidermal cells expressing AP-3δ-GFP, colabeled with FM4-64, and treated with BFA (D) or coexpressed with the Golgi marker WAVE22R (E) or with the TGN/EE marker VHA-a1-RFP (F). Merges of the RFP/GFP channels (R/G) and merges of the RFP/GFP/transmission channels (R/G/T) are shown at the right. The Pearson correlation coefficient (Rp) and the Spearman correlation coefficient (Rs) are shown on the scatterplots at the right. Bars = 10 µm.
CBL2 Targets to Both the Tonoplast and the PM in pat4-2 Depending on S-Acylation
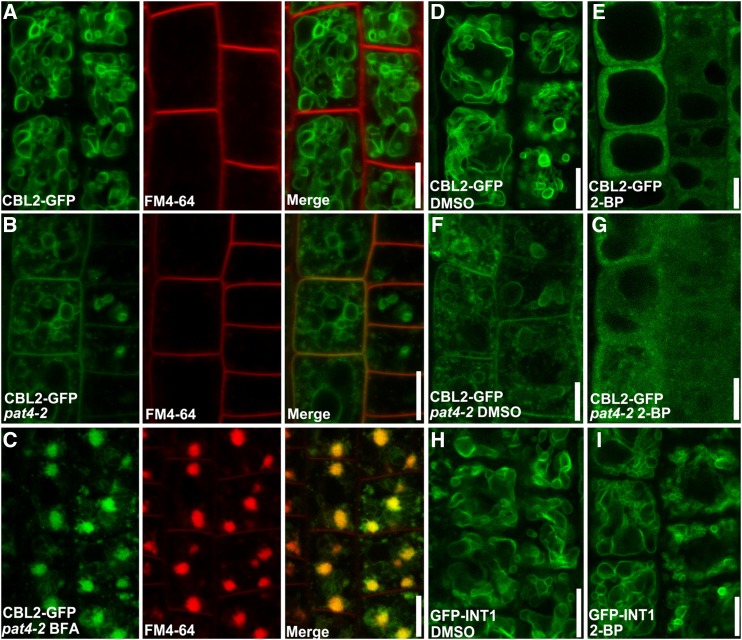
PAT10 loss of function resulted in pleiotropic growth defects (Zhou et al., 2013), which is inconsistent with the fact that functional loss of AP-3 subunits only showed subtle defects in plant growth (Niihama et al., 2009; Feraru et al., 2010; Zwiewka et al., 2011), considering that PAT10 is mistargeted in the ap-3 mutants. To determine the reason behind the discrepancy, we analyzed the localization of CBL2 in ap-3 mutants, because the function of PAT10 was mediated mainly through the tonoplast association of CBL2 and CBL3 (Tang et al., 2012; Zhou et al., 2013). Interestingly, CBL2 was detected both at the PM and at the tonoplast in pat4-2 (Fig. 5B) and pat2-2 (Supplemental Fig. S5), in contrast to its tonoplast localization in the wild type (Fig. 5A). BFA treatment in pat4-2 and pat2-2 resulted in the accumulation of most CBL2 signals in the BFA compartments (Fig. 5C, Supplemental Fig. S5), overlapping with those of FM4-64 (Fig. 5C), indicating mis-localization of CBL2 in the mutants. Upon BFA treatment, no PM-associated signals were detected for CBL2 (Fig. 5C). Treatment of the protein S-acylation inhibitor 2-bromopalmitate (2-BP; Hemsley and Grierson, 2008; Zhou et al., 2013; Zhang et al., 2015) in the wild type resulted in the redistribution of CBL2 from the tonoplast into the cytoplasm (Fig. 5E), in contrast to treatment with dimethyl sulfoxide (DMSO; Fig. 5D). The same 2-BP treatment did not disturb the tonoplast association of INT1 (Fig. 5, H and I). In pat4-2, 2-BP caused the redistribution of CBL2 from both the PM and the tonoplast to the cytoplasm (Fig. 5, F and G), indicating that both the PM-associated and the tonoplast-associated CBL2 were modified by S-acylation. These results suggested that, in pat4-2 and in pat2-2, where PAT10 was retained at the Golgi, CBL2 was nonselectively transported through vesicular trafficking to the PM and to the vacuoles after being S-acylated at the Golgi.
Figure 5.
CBL2 targets to both the tonoplast and the PM in pat4-2 depending on S-acylation. A and B, CLSM of root epidermal cells stained with FM4-64 for 1 min from ProUBQ10:CBL2-GFP (A) or ProUBQ10:CBL2-GFP;pat4-2 (B) transgenic plants. C, CLSM of root epidermal cells pulse labeled with FM4-64 and treated with BFA for 50 min from ProUBQ10:CBL2-GFP;pat4-2 transgenic plants. Note that BFA compartments contain both CBL2 and FM4-64 signals. D and E, CLSM of root epidermal cells from ProUBQ10:CBL2-GFP transgenic plants, which have been treated with either DMSO (D) or 2-BP (E). F and G, CLSM of root epidermal cells from ProUBQ10:CBL2-GFP;pat4-2 transgenic plants, which have been treated with either DMSO (F) or 2-BP (G). H and I, CLSM of root epidermal cells from ProUBQ10:GFP-INT1 transgenic plants, which have been treated with either DMSO (H) or 2-BP (I). Bars = 10 µm.
AP-3-Dependent Vacuolar Trafficking Involves a Subpopulation of COPII
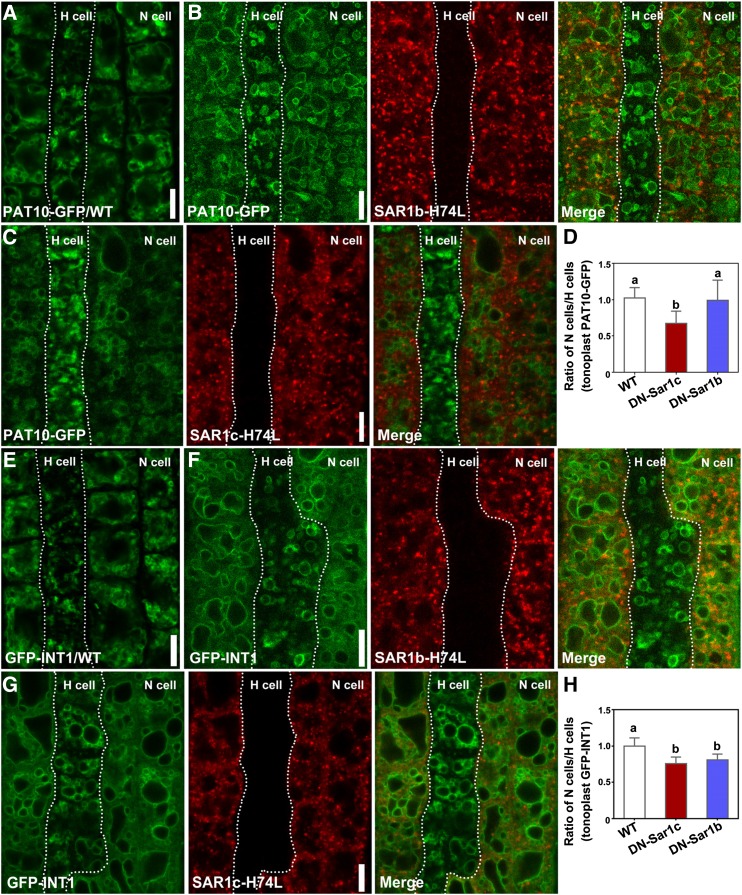
The tonoplast association of CBLs was reported to be independent of COPII based on genetic interference of Sar-1 or Sac12 (Batistic et al., 2010; Bottanelli et al., 2011). Sar-1 is a COPII-recruiting guanosine triphosphate-hydrolyzing protein (GTPase; Takeuchi et al., 2000) found at the ER exit site (ERES) but also over the ER network and mediating vesicle trafficking from the ER to the Golgi (Bassham et al., 2008; Chung et al., 2016). To determine whether the tonoplast targeting of PAT10 relies on COPII, we expressed a DN-Sar-1b specifically in N cells using ProCPC. We used the ratio of tonoplast-associated fluorescence intensities between N cells and H cells to quantify the difference, which not only excludes the influence of different vacuolar morphology between N cells and H cells but also excludes the influence of differential transgene expression among different roots. By measuring the fluorescence intensities of PAT10-GFP on the tonoplast in N cells and H cells either from wild-type or DN-Sar-1b plants, we found that DN-Sar-1b did not interfere with the tonoplast targeting of PAT10 (Fig. 6, A, B, and D; Supplemental Data Set S1). However, the expression of DN-Sar-1b did compromise the tonoplast targeting of INT1 (Fig. 6, E, F, and H; Supplemental Data Set S1), such that GFP-INT1 showed a significantly reduced association at the tonoplast in DN-Sar-1b-expressing cells (Fig. 6, F and H; Supplemental Data Set S1) than in those of the wild type (Fig. 6, A and H; Supplemental Data Set S1).
Figure 6.
AP-3-dependent vacuolar trafficking involves a subpopulation of COPII. A to C, CLSM of root epidermal cells from plants transformed with PAT10g-GFP (A), PAT10g-GFP;ProCPC:RFP-SAR1b-H74L (B), or PAT10g-GFP;ProCPC:RFP-SAR1c-H74L (C). D, Ratio of tonoplast-associated PAT10-GFP fluorescence intensity between N cells and H cells. E to G, CLSM of root epidermal cells from plants transformed with GFP-INT1 (E), GFP-INT1;ProCPC:RFP-SAR1b-H74L (F), or GFP-INT1;ProCPC:RFP-SAR1c-H74L (G). H, Ratio of tonoplast-associated GFP-INT1 fluorescence intensity between N cells and H cells. For B, C, F, and G, images from left to right are GFP channel, RFP channel, and the merge of GFP and RFP channels. Dotted lines illustrate the border between H cells and N cells. Results shown in D and H are means ± sd (n = 25). Means with different letters indicate significant differences (one-way ANOVA, Dunnett’s multiple comparisons test, P < 0.05). WT, Wild type. Bars = 10 µm.
A recent study indicated that, although different Sar-1s in Arabidopsis share high sequence homology, their function might be distinct (Zeng et al., 2015). Real-time quantitative PCR showed that Sar-1b and Sar-1c are constitutively expressed, while the transcript level of Sar-1a is low in all tissues examined (Supplemental Fig. S6). We also confirmed that Sar-1c is expressed in root epidermal cells, where PAT10-GFP localization was analyzed (Supplemental Fig. S6). Thus, it was likely that only a subpopulation of COPII was involved in the transport of PAT10. To test this hypothesis, we generated ProCPC:DN-Sar-1c and analyzed its effect on the targeting of PAT10. Indeed, the expression of DN-Sar-1c substantially reduced the tonoplast association of both PAT10 (Fig. 6, C and D; Supplemental Data Set S1) and INT1 (Fig. 6, G and H; Supplemental Data Set S1) compared with the wild type (Fig. 6, A and G; Supplemental Data Set S1). These results suggested that the AP-3-mediated vacuolar trafficking of PAT10 was selectively transported at the ER by a subpopulation of COPII in which Sar-1c is a key component.
The HOPS Complex Is Required for AP-3-Mediated Vacuolar Trafficking
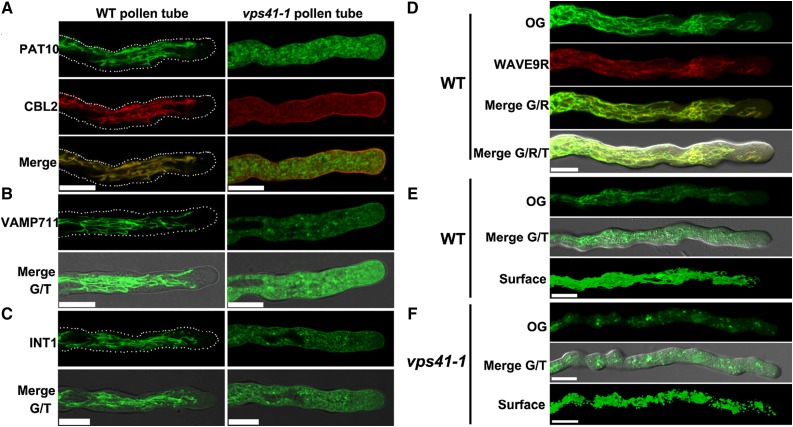
In yeast, the HOPS complex controls membrane fusion at the vacuoles (Nickerson et al., 2009). Interestingly, mutations either at AP-3 subunits or VPS41, a key component of HOPS, were able to suppress the zigzag growth of vti11 (Niihama et al., 2009). We thus considered the possibility of whether the function of HOPS was required for AP-3-mediated tonoplast targeting. It was reported recently that the functional loss of VPS41 resulted in male gametophytic lethality by interfering with in vivo pollen tube growth (Hao et al., 2016). Therefore, we decided to use the in vitro pollen tube growth system to examine the effect of VPS41 functional loss on the tonoplast targeting of AP-3 cargos. To this purpose, we introduced PAT10g-GFP into vps41-1/+, a null mutant for VPS41 (Hao et al., 2016). In wild-type pollen tubes, PAT10 and CBL2 were colocalized to filamentous and tubule structures, excluding from the apical region (Fig. 7A), which is typical for tonoplast distribution (Hicks et al., 2004). By contrast, PAT10 was distributed into small cytoplasmic vesicles in vps41-1 pollen tubes (Fig. 7A). In comparison, CBL2 was detected mostly at the PM, and to a lesser extent, cytoplasmic vesicles in vps41-1 (Fig. 7A). To exclude the possibility that the PAT10-positive vesicles were deformed vacuoles due to VPS41 loss of function, we stained pollen tubes with Oregon Green (OG). OG labels the dynamic tubular vacuoles of wild-type pollen tubes (Fig. 7, D and E), colocalizing with the tonoplast fluorescent marker WAVE9R (Geldner et al., 2009). By contrast, vacuoles of vps41-1 pollen tubes, as indicated by OG labeling, showed numerous granules (Fig. 7F), larger than the punctate vesicles where PAT10 was targeted to vps41-1 pollen tubes (Fig. 7A). These results indicated that the tonoplast association of PAT10 requires functional VPS41.
Figure 7.
The HOPS complex is required for AP-3-mediated vacuolar trafficking. A, CLSM of a wild-type (WT) or vps41-1 pollen tube coexpressing PAT10g-GFP and CBL2-RFP. Merges of the GFP and RFP channels are shown at the bottom. B and C, CLSM of a wild-type or vps41-1 pollen tube expressing GFP-VAMP711 (B) or GFP-INT1 (C). Merges of the GFP and transmission channels are shown at the bottom. Dotted lines illustrate the silhouettes of pollen tubes. D, CLSM of a wild-type pollen tube expressing WAVE9R (red) and stained with OG (green). E and F, CLSM of a wild-type (E) or vps41-1 pollen tube (F) stained with OG. Three-dimensional surface rendering images showing vacuolar structures are at the bottom. Bars = 10 µm.
To determine whether it was the case for other AP-3 cargos, we also introduced ProUBQ10:GFP-VAMP711 in vps41-1. VAMP711 is a close homolog of the known AP-3 cargo VAMP713 (Ebine et al., 2011). We demonstrated that VAMP711 also relies on AP-3 for its tonoplast targeting by introducing ProUBQ10:GFP-VAMP711 in pat4-2 (Supplemental Fig. S7). The tonoplast association of VAMP711 was compromised in pat4-2 such that signals were detected at the PM, cytoplasmic vesicles, and faintly at the tonoplast (Supplemental Fig. S7). Treatment of BFA resulted in the disappearance of GFP signals from the PM into BFA compartments, although a few deformed vacuoles in pat4-2 were still labeled with GFP-VAMP711 signals (Supplemental Fig. S7). These results suggested that VAMP711 relies mainly on AP-3 for its tonoplast targeting. Indeed, VAMP711 was relocalized to cytoplasmic vesicles as well as the apical PM in vps41-1, instead of the tonoplast (Fig. 7B). Interestingly, INT1, the cargo of the Rab5-mediated vacuolar trafficking route, also was mistargeted in vps41-1 pollen tubes (Fig. 7C), suggesting that the VPS41-participating HOPS complex is crucial for the vacuolar consumption of both AP-3-mediated and Rab5-mediated vesicles.
DISCUSSION
The Vacuolar Targeting of PAT10 Depends on AP-3
Several lines of evidence indicate that the multispan transmembrane protein PAT10 is transported to the tonoplast from the ER through AP-3-mediated trafficking (Fig. 8). First, the genetic interference of Rab5 did not affect the tonoplast association of PAT10 (Fig. 1), excluding the involvement of Rab5-mediated vacuolar trafficking. Second, interfering with the formation of Sar-1c-positive COPII by expressing DN-Sar-1c caused the reduced signal of PAT10 at the tonoplast (Fig. 6), indicating the passage of PAT10 through the Golgi during its transport. Third, PAT10 was retained at the Golgi in all AP-3 mutants tested (Figs. 2 and 3), similar to SUC4, another multispan transmembrane protein identified as an AP-3 cargo (Wolfenstetter et al., 2012). Interestingly, unlike SUC4 and PAT10, the tail-anchored tonoplast protein VAMP711, a close homolog of the AP-3 cargo VAMP713 (Ebine et al., 2011), was not retained at the Golgi in AP-3 mutants (Supplemental Fig. S7). Instead, it was mistargeted to the PM in addition to the tonoplast (Supplemental Fig. S7), suggesting that different AP-3 cargos, once reaching the Golgi and in the absence of AP-3, are either retained at the Golgi or transported nonselectively through vesicular trafficking to the PM and the tonoplast.
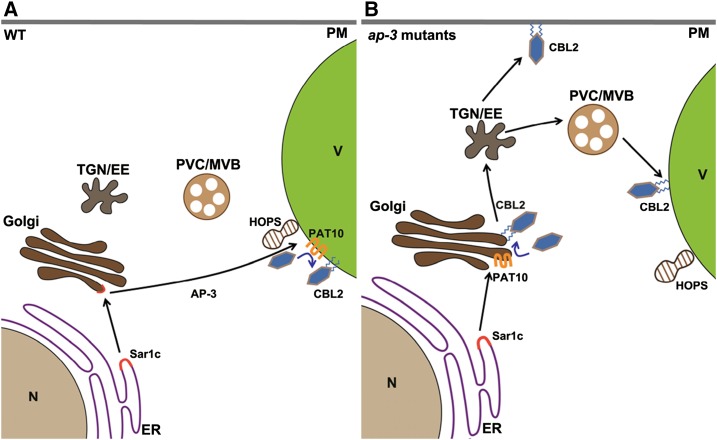
Figure 8.
Cartoon model showing the AP-3-dependent PAT10 trafficking. A, In the wild type (WT), PAT10 was selectively packed into the Sar-1c-dependent COPII complex at the ER and destined for the Golgi. AP-3 mediates the transport of PAT10 from the Golgi to the vacuoles together with the HOPS complex, where cytosolic CBL2 is S-acylated and anchored to the tonoplast. B, In the mutants of AP-3, PAT10 was retained at the Golgi, where cytosolic CBL2 is S-acylated and anchored to the Golgi. Due to active vesicular trafficking at the Golgi, S-acylated CBL2 follows Rab5-dependent trafficking from the Golgi to the TGN/EE, where it targets to the PM or the tonoplast nonselectively.
PAT10-Mediated Membrane Anchorage of CBL2 Is Independent of Membrane Context
We at first excluded the possibility that AP-3 was involved in the tonoplast targeting of PAT10 for the simple reason that mutations in PAT10 resulted in strong developmental defects (Zhou et al., 2013) while ap-3 mutants were only mildly affected (Niihama et al., 2009; Feraru et al., 2010; Zwiewka et al., 2011). However, to our surprise, although PAT10 was retained at the Golgi (Figs. 2 and 3), its substrate CBL2 was targeted to the tonoplast and the PM in an S-acylation-dependent way (Fig. 5). The partial distribution of CBL2 at the tonoplast may be the reason that ap-3 mutants did not show the defects of pat10, because PAT10 functions through tonoplast targeting of CBL2 and CBL3 (Zhou et al., 2013). In ap-3 mutants, PAT10 was retained mostly at the Golgi (Figs. 2 and 3). Thus, S-acylation of CBL2 must have happened at the Golgi by the function of Golgi-retained PAT10. This would indicate that S-acylation by PAT10 is independent of its membrane context (i.e. either at the Golgi or at the tonoplast) and that PAT10 is able to mediate the membrane anchorage of cytoplasmic CBL2. Because the Golgi is an endomembrane compartment constantly undergoing vesicle trafficking, the membrane-anchored CBL2 presumably followed vesicle flow nonselectively to the PM or the tonoplast through the TGN/EE (Figs. 5 and 7). Consistently, the tail-anchored tonoplast protein, VAMP711, also was nonselectively transported to the PM and the tonoplast through the TGN/EE in the absence of AP-3 (Supplemental Fig. S7). Indeed, ours (Supplemental Fig. S1) and previous results (Ebine et al., 2014) showed that inhibiting the Rab5-mediated vacuolar trafficking route resulted in mistargeting of tonoplast proteins to the PM, supporting the nonselective transport of Golgi-retained proteins in the case of disrupted vacuolar trafficking.
AP-3-Specific Vacuolar Trafficking of PAT10 Relies on the Sar-1c-Positive COPII Complex
COPII is an evolutionarily conserved complex mediating anterograde vesicular trafficking at the ER (Chung et al., 2016). It was proposed recently that different Sar-1 paralogs encoded in plant genomes may have distinct functions (Chung et al., 2016). Indeed, an amino acid difference determines the separate function of two Sar-1s (Zeng et al., 2015). The tonoplast association of CBL6 was unaffected by interfering with the formation of Sar-1b-positive COPII (Batistic et al., 2010), which is also true for PAT10 (Fig. 6). However, the fact that PAT10 is transported via AP-3 indicates its passage through the Golgi. Consistently, we have demonstrated that manipulating the activity of Sar-1c but not Sar-1b substantially compromised the tonoplast targeting of PAT10 (Fig. 6). How the specificity between different Sar-1s and cargos is determined requires further investigation.
The Tethering Complex HOPS Is Essential for AP-3- and Rab5-Mediated Vacuolar Trafficking
Compared with extensive studies on the trafficking routes vacuolar proteins take, little is known of the mechanism regulating the final arrival of vesicles at the tonoplast. We show here that both PAT10 and INT1, cargos mediated by AP-3 and Rab5, respectively, were mislocalized in vps41 (Fig. 7). In addition, functional loss of AP-3 resulted in the mistargeting of CBL2 to the PM and the tonoplast (Fig. 5), while CBL2 was detected only at the PM in vps41 pollen tubes (Fig. 7). These results indicate that VPS41 is critical for the vacuolar targeting of both AP-3- and Rab5-mediated vesicles. The essential requirement of HOPS to vacuolar fusion is consistent with the fact that functional loss of VPS41, and by inference the HOPS complex, resulted in male gametophytic lethality (Hao et al., 2016).
MATERIALS AND METHODS
Plant Materials and Growth Condition
Arabidopsis (Arabidopsis thaliana) plants were grown and transformed as described (Zhou et al., 2013). The Columbia-0 ecotype was used as the wild type. Transgenic plants were selected on one-half-strength Murashige and Skoog (MS) medium supplemented with 30 µg mL−1 Basta salts (Sigma-Aldrich) or 50 µg mL−1 hygromycin (Roche). The T-DNA insertion lines, SALK_076372 (vps41-1; Hao et al., 2016), SAIL_1258_G03 (pat2-2; Feraru et al., 2010), SALK_069881C (pat4-2; Zwiewka et al., 2011), SALK_064486 (ap-3µ-2; Kansup et al., 2013), and SAIL_269_F04 (ap-3σ), were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org). The T-DNA insertion line SALK_024964 (pat10-1) was described (Zhou et al., 2013). The pat4-3/reva1-1 and pat4-4/reva1-2 mutant alleles were isolated from an M2 population of PAT10g-GFP;pat10-1 seeds mutagenized with EMS, which resulted in a nucleotide substitution at AP-3δ (i.e. G281A or G832A, respectively). Fluorescent marker lines for colabeling experiments, including WAVE22R (Geldner et al., 2009), WAVE9R (Geldner et al., 2009), and VHA-a1-RFP (Dettmer et al., 2006), were described previously.
DNA Manipulation
All constructs were generated using Gateway technology (Invitrogen) except where noted. Entry vectors for genes or fragments of interest were generated with the following primer pairs: ZP929/ZP930 for Sar-1b, ZP4302/ZP4303 for Sar-1c, ZP5324/ZP5325 for ProSAR1C, ZP1845/ZP1846 for INT1, ZP3601/ZP3602 for VAMP711, ZP4639/ZP4640 for SUC4, ZP4427/ZP4428 for AP-3δ (without stop codon), ZP4427/ZP4606 for AP-3δ (with stop codon), and ZP4210/ZP4608 for AP-3μ. Dominant negative mutants of Rab GTPases were generated using the Phusion site-directed mutagenesis kit according to the manufacturer’s instructions from corresponding entry vectors with the following primer pairs: ZP1280/ZP1281 for Sar-1b-H74L, ZP4304/ZP4305 for Sar-1c-H74L, and ZP1275/ZP1276 for ARA7-S24N (Rab5DN). All entry clones were generated in the pENTR/D/TOPO vector (Invitrogen). All PCR amplifications used Phusion hot-start high-fidelity DNA polymerase with the annealing temperature and extension times recommended by the manufacturer. All entry vectors were sequenced, and sequences were analyzed using Vector NTI (Invitrogen). The Bioneer PCR purification kit and Spin mini prep kit were used for PCR product recovery and plasmid DNA extraction, respectively.
Destination vectors for protein expression in planta, ProUBQ10:GW-GFP and ProUBQ10:GFP-GW, were modified by replacing the Pro35S from Pro35S:GW-GFP and Pro35S:GFP-GW (Karimi et al., 2002), respectively, with ProUBQ10 (amplified with the primer pair ZP510/ZP511) using HindIII/SpeI double digestion. A 1,252-bp sequence upstream of the CAPRICE start codon was amplified with ZP1257/ZP1258 and digested with SacI/SpeI. The resulting fragment replaced the Pro35S in Pro35S:RFP-GW and Pro35S:GW-RFP (Karimi et al., 2002) to generate the destination vectors ProCPC:RFP-GW and ProCPC:GW-RFP. The destination vector GW:GUS was described (Zhou et al., 2013). The destination vectors for BD and AD fusions in Y2H assays were pDEST32 and pDEST22, respectively (Invitrogen). Expression vectors were generated by LR reactions using LR Clonase II (Invitrogen). The expression vectors PAT10g-GFP and ProUBQ10:CBL2-RFP were described previously (Zhou et al., 2013; Zhang et al., 2015). All primers are listed in Supplemental Table S1.
Genotyping PCR, Reverse Transcription-PCR, and Quantitative Real-Time PCR
Primers used to verify T-DNA insertions at the DNA level are as follows: ZP4408/ZP4409 for the wild-type copy and ZP4408/ZP1 for the mutant copy of VPS41 in SALK_076372 (vps41-1); ZP4702/ZP4703 for the wild-type copy and ZP4703/ZP4 for the mutant copy of AP-3β in SAIL_1258_G03 (pat2-2); ZP3639/ZP3640 for the wild-type copy and ZP3640/ZP1 for the mutant copy of AP-3δ in SALK_069881C (pat4-2); ZP4704/ZP4705 for the wild-type copy and ZP4705/ZP1 for the mutant copy of AP-3µ in SALK_064486 (ap-3µ-2); and ZP4706/ZP4707 for the wild-type copy and ZP4707/ZP4 for the mutant copy of AP-3σ in SAIL_269_F04 (ap-3σ). Primers used to verify the mutant allele of ap-3σ at the RNA level by reverse transcription-PCR are ZP4430/ZP4431. Primers used to perform quantitative real-time PCR are as follows: ZP5304/ZP5305 for Sar-1a, ZP5306/ZP5307 for Sar-1b, and ZP5308/ZP5309 for Sar-1c. Total RNA extraction, reverse transcription-PCR, and quantitative real-time PCR were performed as described (Zhou et al., 2013). All primers are listed in Supplemental Table S1.
EMS Mutagenesis and Map-Based Cloning
About 10,000 PAT10g-GFP;pat10-1 seeds were incubated with 25 mL of 0.2% (v/v) EMS in a 50-mL Falcon tube on a tube rotator for 15 h. The seeds were rinsed with water 10 times before being dried under a fume cupboard. In total, 3,000 M1 plants were analyzed by in vitro germination assay (Boavida and McCormick, 2007). Plants whose pollen tubes showed altered PAT10-GFP localization were selected for outcrosses. The mapping populations were generated by a cross between Ler and the corresponding mutant. We roughly mapped both pat4-3 and pat4-4 loci on chromosome 1 using an F2 mapping population (38 F2 progeny for pat4-3 and 30 F2 progeny for pat4-4). We used the simple sequence length polymorphism markers that can recognize polymorphisms between Columbia-0 and Ler based on information provided by TAIR.
Protein Interaction Assays
Y2H assays were performed using the Pro-Quest system (Invitrogen). YSD medium supplemented with all amino acids except Trp and Leu was used to select diploids. YSD medium supplemented with 15 mm 3-amino-1,2,4-triazole and all amino acids except Trp, Leu, and His was used to select colonies showing positive interactions. Quantification of β-galactosidase activity from Y2H experiments was performed as described previously (Huang et al., 2013).
Pharmacological Treatment
Stock solutions of various pharmacological drugs were prepared using DMSO as the solvent at the following concentrations: 20 mm 2-BP, 35 mm BFA, and 4 mm FM4-64. Stock solutions were diluted and added to one-half-strength MS medium at the following designated final concentrations, 20 μm 2-BP, 50 μm BFA, and 4 μm FM4-64. DMSO was equally diluted as the controls. All experiments were repeated at least three times. The 4-DAG seedlings were dipped in liquid one-half-strength MS medium supplemented with 4 μm FM4-64 (Invitrogen) for 5 min (except for the 1-min assay) at room temperature. Seedlings were then washed three times with liquid one-half-strength MS medium and treated with 50 μm BFA for 50 min prior to confocal fluorescence imaging. In the 2-BP treatment assay, 4-DAG seedlings were dipped in liquid one-half-strength MS medium supplemented with 20 μm 2-BP (Sigma) or DMSO overnight before imaging.
OG Staining of Growing Pollen Tubes
Fluorescence labeling of vacuoles in growing pollen tubes with the fluorescent cell-permeant dye OG 488 carboxylic acid diacetate (Invitrogen) was performed as follows: pollen grains were germinated on pollen germination medium (PGM) for 2.5 h, and then 100 μL of liquid PGM supplemented with 10 µm OG from a 10 mm stock solution in DMSO was dropped on in vitro growing pollen tubes for 50 min at 28°C in the dark. The pollen tubes were then washed with liquid PGM without OG before being imaged.
Fluorescence Microscopy
Fluorescence images were captured using a Zeiss LSM880 laser scanning microscope with a 40/1.3 oil objective. GFP-RFP double-labeled plant materials were captured alternately using line switching with the multitrack function (488 nm for GFP and 561 nm for RFP). Fluorescence was detected using a 505- to 550-nm band-pass filter for GFP and a 575- to 650-nm band-pass filter for RFP. Image processing was performed with the Zeiss LSM image-processing software. The quantification of PAT10-GFP was presented using the tonoplast fluorescence intensity of N cells with Sar-1DN expression versus those without. The average signal intensity of regions of interest at the tonoplast was measured using ImageJ.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: At5g55990 for CBL2; At2g46410 for CPC; At2g43330 for INT1; At3g51390 for PAT10; At4g19640 for RAB5; At1g09180 for Sar-1a; At1g56330 for Sar-1b; At4g02080 for Sar-1c; At4g32150 for VAMP711; At1g08190 for VPS41; At1g09960 for SUC4; At1g48760 for AP-3δ; At3g55480 for AP-3β; At1g56590 for AP-3μ; and At3g50860 for AP-3σ.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The expression of Rab5DN interfered with the vacuolar targeting of INT1.
Supplemental Figure S2. Identification of the reva1 mutants by EMS mutagenesis and cloning.
Supplemental Figure S3. The Gly-94 and Glu-278 residues of AP-3δ are evolutionarily conserved.
Supplemental Figure S4. The tonoplast targeting of PAT10 is mediated by AP-3.
Supplemental Figure S5. The tonoplast association of CBL2 depends on AP-3- but not Rab5-mediated post-Golgi trafficking.
Supplemental Figure S6. Expression of Sar-1 homologs in Arabidopsis.
Supplemental Figure S7. The tonoplast-targeted VAMP711 is another AP-3 cargo.
Supplemental Table S1. Oligonucleotides used in this study.
Supplemental Data Set S1. Source images and raw quantification data for Figure 6, D and G.
Acknowledgments
We thank Takashi Ueda for the ARA7/Rab5 entry vector and the Arabidopsis Biological Resource Center for the mutant seeds.
Glossary
- ER
endoplasmic reticulum
- TGN/EE
trans-Golgi network/early endosome
- PVC
prevacuolar compartments
- MVB
multivesicular bodies
- PM
plasma membrane
- BFA
brefeldin A
- EMS
ethyl methanesulfonate
- Y2H
yeast two-hybrid
- 2-BP
2-bromopalmitate
- DMSO
dimethyl sulfoxide
- OG
Oregon Green
- MS
Murashige and Skoog
- PGM
pollen germination medium
- CLSM
confocal laser scanning micrograph
- DAG
days after germination
Footnotes
This work was supported by the Major Research Plan (grant no. 2013CB945102) from the Ministry of Science and Technology of China, the National Natural Science Foundation of China (grant nos. 31261160490, 31271578, 31471304, and 31625003), and the Tai-Shan Scholar Program of the Shandong Provincial Government.
Articles can be viewed without a subscription.
References
- Bassham DC, Brandizzi F, Otegui MS, Sanderfoot AA (2008) The secretory system of Arabidopsis. The Arabidopsis Book 6: e0116, doi/10.1199/tab.0116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O. (2012) Genomics and localization of the Arabidopsis DHHC-cysteine-rich domain S-acyltransferase protein family. Plant Physiol 160: 1597–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O, Rehers M, Akerman A, Schlücking K, Steinhorst L, Yalovsky S, Kudla J (2012) S-Acylation-dependent association of the calcium sensor CBL2 with the vacuolar membrane is essential for proper abscisic acid responses. Cell Res 22: 1155–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O, Waadt R, Steinhorst L, Held K, Kudla J (2010) CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J 61: 211–222 [DOI] [PubMed] [Google Scholar]
- Boavida LC, McCormick S (2007) Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J 52: 570–582 [DOI] [PubMed] [Google Scholar]
- Bolte S, Talbot C, Boutte Y, Catrice O, Read ND, Satiat-Jeunemaitre B (2004) FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J Microsc 214: 159–173 [DOI] [PubMed] [Google Scholar]
- Bottanelli F, Foresti O, Hanton S, Denecke J (2011) Vacuolar transport in tobacco leaf epidermis cells involves a single route for soluble cargo and multiple routes for membrane cargo. Plant Cell 23: 3007–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KP, Zeng Y, Jiang L (2016) COPII paralogs in plants: functional redundancy or diversity? Trends Plant Sci 21: 758–769 [DOI] [PubMed] [Google Scholar]
- Cui Y, Zhao Q, Gao C, Ding Y, Zeng Y, Ueda T, Nakano A, Jiang L (2014) Activation of the Rab7 GTPase by the MON1-CCZ1 complex is essential for PVC-to-vacuole trafficking and plant growth in Arabidopsis. Plant Cell 26: 2080–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K, Fujimoto M, Okatani Y, Nishiyama T, Goh T, Ito E, Dainobu T, Nishitani A, Uemura T, Sato MH, et al. (2011) A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol 13: 853–859 [DOI] [PubMed] [Google Scholar]
- Ebine K, Inoue T, Ito J, Ito E, Uemura T, Goh T, Abe H, Sato K, Nakano A, Ueda T (2014) Plant vacuolar trafficking occurs through distinctly regulated pathways. Curr Biol 24: 1375–1382 [DOI] [PubMed] [Google Scholar]
- Feng QN, Zhang Y, Li S (2017) Tonoplast targeting of VHA-a3 relies on a Rab5-mediated but Rab7-independent vacuolar trafficking route. J Integr Plant Biol 59: 230–233 [DOI] [PubMed] [Google Scholar]
- Feraru E, Paciorek T, Feraru MI, Zwiewka M, De Groodt R, De Rycke R, Kleine-Vehn J, Friml J (2010) The AP-3 β adaptin mediates the biogenesis and function of lytic vacuoles in Arabidopsis. Plant Cell 22: 2812–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Liu J, Zhong S, Gu H, Qu LJ (2016) AtVPS41-mediated endocytic pathway is essential for pollen tube-stigma interaction in Arabidopsis. Proc Natl Acad Sci USA 113: 6307–6312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley PA, Grierson CS (2008) Multiple roles for protein palmitoylation in plants. Trends Plant Sci 13: 295–302 [DOI] [PubMed] [Google Scholar]
- Hicks GR, Rojo E, Hong S, Carter DG, Raikhel NV (2004) Geminating pollen has tubular vacuoles, displays highly dynamic vacuole biogenesis, and requires VACUOLESS1 for proper function. Plant Physiol 134: 1227–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GQ, Li E, Ge FR, Li S, Wang Q, Zhang CQ, Zhang Y (2013) Arabidopsis RopGEF4 and RopGEF10 are important for FERONIA-mediated developmental but not environmental regulation of root hair growth. New Phytol 200: 1089–1101 [DOI] [PubMed] [Google Scholar]
- Hwang I. (2008) Sorting and anterograde trafficking at the Golgi apparatus. Plant Physiol 148: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansup J, Tsugama D, Liu S, Takano T (2013) The Arabidopsis adaptor protein AP-3μ interacts with the G-protein β subunit AGB1 and is involved in abscisic acid regulation of germination and post-germination development. J Exp Bot 64: 5611–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Lam SK, Cai Y, Tse YC, Wang J, Law AH, Pimpl P, Chan HY, Xia J, Jiang L (2009) BFA-induced compartments from the Golgi apparatus and trans-Golgi network/early endosome are distinct in plant cells. Plant J 60: 865–881 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Sohn EJ, Lee MH, Hwang I (2004) The Arabidopsis rab5 homologs rha1 and ara7 localize to the prevacuolar compartment. Plant Cell Physiol 45: 1211–1220 [DOI] [PubMed] [Google Scholar]
- Nickerson DP, Brett CL, Merz AJ (2009) Vps-C complexes: gatekeepers of endolysosomal traffic. Curr Opin Cell Biol 21: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niihama M, Takemoto N, Hashiguchi Y, Tasaka M, Morita MT (2009) ZIP genes encode proteins involved in membrane trafficking of the TGN-PVC/vacuoles. Plant Cell Physiol 50: 2057–2068 [DOI] [PubMed] [Google Scholar]
- Pacini E, Jacquard C, Clément C (2011) Pollen vacuoles and their significance. Planta 234: 217–227 [DOI] [PubMed] [Google Scholar]
- Pedrazzini E, Komarova NY, Rentsch D, Vitale A (2013) Traffic routes and signals for the tonoplast. Traffic 14: 622–628 [DOI] [PubMed] [Google Scholar]
- Qi B, Doughty J, Hooley R (2013) A Golgi and tonoplast localized S-acyl transferase is involved in cell expansion, cell division, vascular patterning and fertility in Arabidopsis. New Phytol 200: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MK, Krüger F, Beckmann H, Brumm S, Vermeer JE, Munnik T, Mayer U, Stierhof YD, Grefen C, Schumacher K, et al. (2014) Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB-vacuole fusion. Curr Biol 24: 1383–1389 [DOI] [PubMed] [Google Scholar]
- Sohn EJ, Kim ES, Zhao M, Kim SJ, Kim H, Kim YW, Lee YJ, Hillmer S, Sohn U, Jiang L, et al. (2003) Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15: 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Ueda T, Sato K, Abe H, Nagata T, Nakano A (2000) A dominant negative mutant of sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J 23: 517–525 [DOI] [PubMed] [Google Scholar]
- Tang RJ, Liu H, Yang Y, Yang L, Gao XS, Garcia VJ, Luan S, Zhang HX (2012) Tonoplast calcium sensors CBL2 and CBL3 control plant growth and ion homeostasis through regulating V-ATPase activity in Arabidopsis. Cell Res 22: 1650–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Ueda T (2014) Plant vacuolar trafficking driven by RAB and SNARE proteins. Curr Opin Plant Biol 22: 116–121 [DOI] [PubMed] [Google Scholar]
- Viotti C, Krüger F, Krebs M, Neubert C, Fink F, Lupanga U, Scheuring D, Boutté Y, Frescatada-Rosa M, Wolfenstetter S, et al. (2013) The endoplasmic reticulum is the main membrane source for biogenesis of the lytic vacuole in Arabidopsis. Plant Cell 25: 3434–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K (2002) Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129: 5409–5419 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhuang XH, Hillmer S, Robinson DG, Jiang LW (2011) Vacuolar sorting receptor (VSR) proteins reach the plasma membrane in germinating pollen tubes. Mol Plant 4: 845–853 [DOI] [PubMed] [Google Scholar]
- Wolfenstetter S, Wirsching P, Dotzauer D, Schneider S, Sauer N (2012) Routes to the tonoplast: the sorting of tonoplast transporters in Arabidopsis mesophyll protoplasts. Plant Cell 24: 215–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Chung KP, Li B, Lai CM, Lam SK, Wang X, Cui Y, Gao C, Luo M, Wong KB, et al. (2015) Unique COPII component AtSar1a/AtSec23a pair is required for the distinct function of protein ER export in Arabidopsis thaliana. Proc Natl Acad Sci USA 112: 14360–14365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL, Li E, Feng QN, Zhao XY, Ge FR, Zhang Y, Li S (2015) Protein palmitoylation is critical for the polar growth of root hairs in Arabidopsis. BMC Plant Biol 15: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LZ, Li S, Feng QN, Zhang YL, Zhao X, Zeng YL, Wang H, Jiang L, Zhang Y (2013) Protein S-ACYL TRANSFERASE10 is critical for development and salt tolerance in Arabidopsis. Plant Cell 25: 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiewka M, Feraru E, Möller B, Hwang I, Feraru MI, Kleine-Vehn J, Weijers D, Friml J (2011) The AP-3 adaptor complex is required for vacuolar function in Arabidopsis. Cell Res 21: 1711–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]