Arabidopsis SPA proteins are required for light-induced nuclear exclusion of the repressor of light signaling COP1 but have no effect on nuclear accumulation of COP1 in darkness.
Abstract
The Arabidopsis (Arabidopsis thaliana) COP1/SPA ubiquitin ligase is a central repressor that suppresses light signaling in darkness by targeting positive regulators of the light response, mainly transcription factors, for degradation. Light inactivates COP1/SPA, in part by excluding COP1 from the nucleus. SPA proteins are essential cofactors of COP1, but their exact role in the COP1/SPA complex is thus far unknown. To unravel a potential role of SPA proteins in COP1 nucleocytoplasmic partitioning, we monitored the subcellular localization of COP1 in a spa1234 quadruple mutant (spaQn). We analyzed a YFP-COP1-expressing transgenic line and endogenous COP1 after subcellular fractionation. In dark-grown seedlings, both YFP-COP1 and endogenous COP1 accumulated in the nucleus in the absence and presence of SPA proteins, indicating that SPA proteins are not required for nuclear localization of COP1 in darkness. In contrast, in white light-grown seedlings, spaQn mutants failed to relocalize COP1 from the nucleus to the cytoplasm. Hence, SPA proteins are necessary for the light-controlled change in COP1 subcellular localization. We conclude that SPA proteins have a dual role: (1) they are required for light-responsiveness of COP1 subcellular localization, and (2) they promote COP1 activity in darkness in a fashion that is independent of the nuclear import/nuclear retention of COP1.
Plants constantly monitor the quality, intensity, direction, and periodicity of light and adjust their growth and development accordingly, thereby ensuring their survival and reproductive success. To this end, they have evolved a set of photoreceptors, among them the red/far-red light-sensing phytochromes (phyA-E) and the UV-A/blue light-sensing cryptochromes (cry1, cry2), that perceive the ambient light environment. When activated by light, these photoreceptors promote a number of developmental responses, such as seedling de-etiolation, leaf growth, stomata formation, and the regulation of flowering time, collectively known as photomorphogenesis (Kami et al., 2010).
Downstream of the phytochromes and cryptochromes, the CONSTITUTIVELY PHOTOMORPHOGENIC 1/SUPPRESSOR OF PHYTOCHROME A-105 (COP1/SPA) complex acts as a central repressor of light signaling in the dark (Lau and Deng, 2012; Menon et al., 2016). COP1/SPA is a tetrameric complex consisting of two COP1 and two SPA proteins of the four-member SPA protein family (SPA1-SPA4; Zhu et al., 2008). The COP1/SPA complex is thought to act as the substrate recognition subunit of a CULLIN 4 (CUL4)/DAMAGED DNA BINDING PROTEIN 1 (DDB1) E3 ubiquitin ligase (Chen et al., 2010). It recognizes and polyubiquitinates photomorphogenesis-promoting transcription factors, such as ELONGATED HYPOCOTYL 5, LONG HYPOCOTYL IN FAR-RED 1, PRODUCTION OF ANTHOCYANIN PIGMENT1, PAP2, and CONSTANS, thereby targeting them for proteasomal degradation in dark-grown seedlings and plants (Lau and Deng, 2012; Menon et al., 2016). As a consequence, mutants lacking functional COP1 or all four SPA genes display strong constitutive photomorphogenesis, exhibiting features of light-grown plants in complete darkness (Deng et al., 1991; Laubinger et al., 2004; Ordoñez-Herrera et al., 2015). The four SPA genes have partially redundant but also distinct functions throughout plant development (Laubinger et al., 2004, 2006; Fittinghoff et al., 2006; Balcerowicz et al., 2011; Rolauffs et al., 2012; Chen et al., 2016).
Light activates phytochrome and cryptochrome photoreceptors that subsequently bind and inhibit the COP1/SPA E3 ubiquitin ligase, thus allowing the target transcription factors to accumulate and initiate photomorphogenesis. Light inhibits COP1/SPA activity by at least three mechanisms: (1) COP1 is excluded from the nucleus in the light and is thereby separated from its target proteins (von Arnim and Deng, 1994; Pacín et al., 2014). phyA, phyB, and cry1 promote COP1 nuclear exclusion in response to continuous far-red (FRc), red (Rc), and blue light (Bc), respectively (Osterlund and Deng, 1998), while simulated shade, that is, a low R:FR or B:FR ratio, promotes its nuclear accumulation (Pacín et al., 2013). (2) Phytochromes promote proteasomal degradation of SPA1 and SPA2 proteins upon light exposure in a COP1-dependent manner and thereby decrease SPA protein levels (Balcerowicz et al., 2011; Chen et al., 2015); and (3) phyA, phyB, and cry1 bind to SPA1 in a light-dependent manner, thereby reducing the COP1-SPA1 interaction (Lian et al., 2011; Liu et al., 2011; Lu et al., 2015; Sheerin et al., 2015).
Despite the fact that the light-induced nuclear exclusion of COP1 was the first of these processes to be described, its precise mechanism is still not well understood. The COP1 protein harbors a nuclear localization signal (NLS) and a cytoplasmic localization signal (CLS), both of which are essential for proper COP1 nucleocytoplasmic partitioning (Stacey et al., 1999, 2000; Subramanian et al., 2004). Within the nucleus, COP1 is targeted to discrete domains, referred to as speckles or photobodies, where it colocalizes with other factors of the light signaling pathway, including SPAs, cryptochromes, and phytochromes as well as target transcription factors (Van Buskirk et al., 2012). Both nuclear localization and speckle formation are required for proper COP1 function (Stacey et al., 1999, 2000; Subramanian et al., 2004).
Whether SPA proteins have a role in the subcellular localization of COP1 has yet not been investigated. It has been suggested that SPA proteins might be required to retain COP1 in the nucleus (Yi and Deng, 2005). This would agree with the reduced interaction between COP1 and SPA1 observed in the light compared to the dark (Lian et al., 2011; Liu et al., 2011; Lu et al., 2015; Sheerin et al., 2015). However, using a combination of confocal microscopy and cell fractionation experiments, we show that SPA proteins have little effect on the nuclear accumulation of COP1 in darkness, but instead are required for its proper nuclear exclusion in white light.
RESULTS
SPA Proteins Are Not Required for Nuclear Accumulation and Photobody Formation of YFP-COP1 in Dark-Grown Seedlings
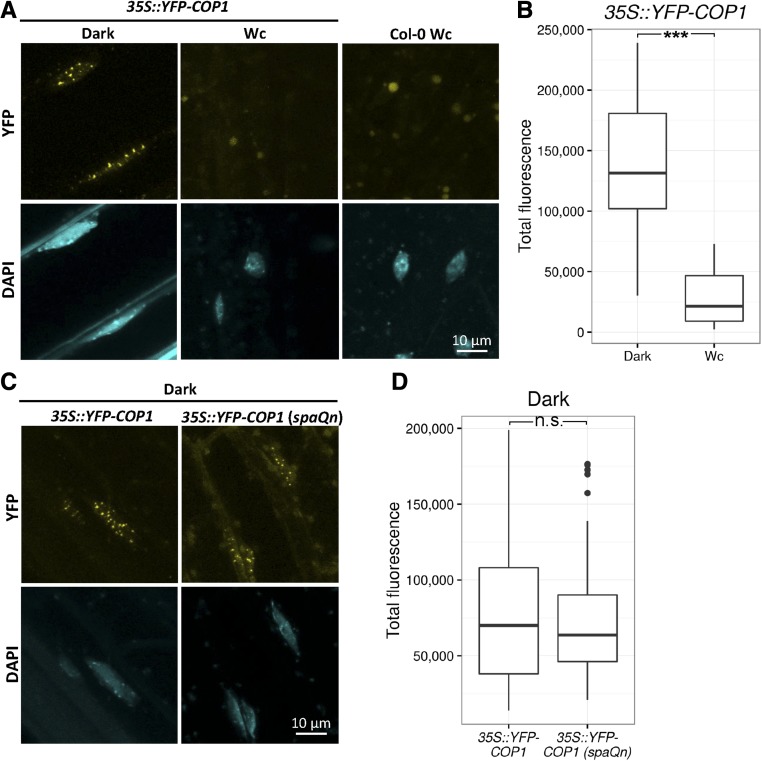
Since SPA proteins are essential for COP1 function in darkness, we asked whether SPA proteins are required for the nuclear localization of COP1 in dark-grown seedlings. To this end, we crossed the 35S::YFP-COP1 transgene (Oravecz et al., 2006) into the spaQn mutant background that carries null alleles of all four Arabidopsis SPA genes (Ordoñez-Herrera et al., 2015). As a control, we first analyzed YFP-COP1 localization in the progenitor line, that is, in a SPA wild-type background. Distinct YFP-COP1 photobodies were detected in the hypocotyl nuclei of dark-grown seedlings, whereas no such signal was detected in hypocotyls of seedlings grown in continuous white light (Wc; Fig. 1A). The weak, round-shaped signals localized outside the nucleus of light-grown seedlings were background signals, likely derived from chloroplasts, since they were equally found in nontransgenic wild-type seedlings (Fig. 1A). The disappearance of YFP-COP1 from the nucleus in Wc was not due to YFP-COP1 degradation, since total YFP-COP1 levels were not lowered by Wc treatment (Supplemental Fig. S1). In total, these results are in close agreement with previous findings (von Arnim and Deng, 1994; Stacey and von Arnim, 1999; Pacín et al., 2014).
Figure 1.
SPA proteins are not required for nuclear accumulation and photobody formation of YFP-COP1 in dark-grown seedlings. A, Representative images of hypocotyl nuclei of 4-d-old 35S::YFP-COP1 and Col-0 wild-type seedlings grown in darkness or 40 µmol m−2 s−1 Wc. B, Average nuclear YFP fluorescence per nucleus quantified from hypocotyl nuclei of 35S::YFP-COP1 seedlings grown under the same conditions as in A. A total of 20 or 53 nuclei in 8 to 10 seedlings was analyzed in darkness or Wc, respectively. Percentage of nuclei showing speckles was 100% in darkness and 0% in Wc. C and D, Representative images (C) and average nuclear YFP fluorescence (D) of hypocotyl nuclei of 4-d-old 35S::YFP-COP1 and 35S::YFP-COP1 (spaQn) seedlings grown in darkness. A total of 26 or 60 nuclei in 8 to 10 seedlings of 35S::YFP-COP1 or 35S::YFP-COP1 (spaQn), respectively, was analyzed. Percentage of nuclei showing speckles was 85% for 35S::YFP-COP1 and 91% for 35S::YFP-COP1. Upper and lower hinges of boxplot correspond to the first and third quartiles, respectively. Whiskers extend from the hinges to the highest/lowest value within 1.5 * interquartile range (IQR). Data points beyond this range are represented as dots. *** indicates that true difference between means is not equal to 0 at P < 0.001 in two-sample Student’s t test. n.s., not significant at P = 0.05.
In spaQn 35S::YFP-COP1 seedlings grown in darkness, nuclear YFP-COP1 photobodies were observed that were very similar to those observed in the parental 35S::YFP-COP1 line (Fig. 1C). Thus, SPA proteins are not essential for nuclear accumulation and photobody formation of YFP-COP1.
While no visual change in YFP-COP1 photobody formation was detected between SPA wild-type and spaQn backgrounds, there might still be a quantitative difference in the nuclear YFP-COP1 accumulation in the dark. To investigate this possibility, nuclear fluorescence intensity was measured in hypocotyl nuclei of the YFP-COP1-expressing seedlings. To this end, we generated 3-dimensional images (z-stacks) from DAPI-stained hypocotyl cells of 35S::YFP-COP1 seedlings using confocal microscopy. Detecting both DAPI and YFP signals in these stacks allowed us to measure YFP-fluorescence intensities within the 3-dimensional space of a nucleus (see “Materials and Methods” for further details). In a SPA wild-type background, nuclear YFP-COP1 fluorescence intensity was strongly reduced in Wc-grown seedlings when compared to dark-grown seedlings (Fig. 1B), thus confirming the visual impression shown in Figure 1a. When comparing YFP-COP1 fluorescence intensity in nuclei of dark-grown 35S::YFP-COP1 and spaQn 35S::YFP-COP1 seedlings, no dramatic difference was observed (Fig. 1D). Especially considering that total YFP-COP1 levels were somewhat reduced in dark-grown spaQn when compared to the SPA-wild-type background (Supplemental Fig. S1), these results confirm that nuclear accumulation of YFP-COP1 is not impaired by the absence of SPA proteins.
SPA Proteins Are Essential for the White Light-Induced Reduction in Nuclear Accumulation and Photobody Formation of YFP-COP1
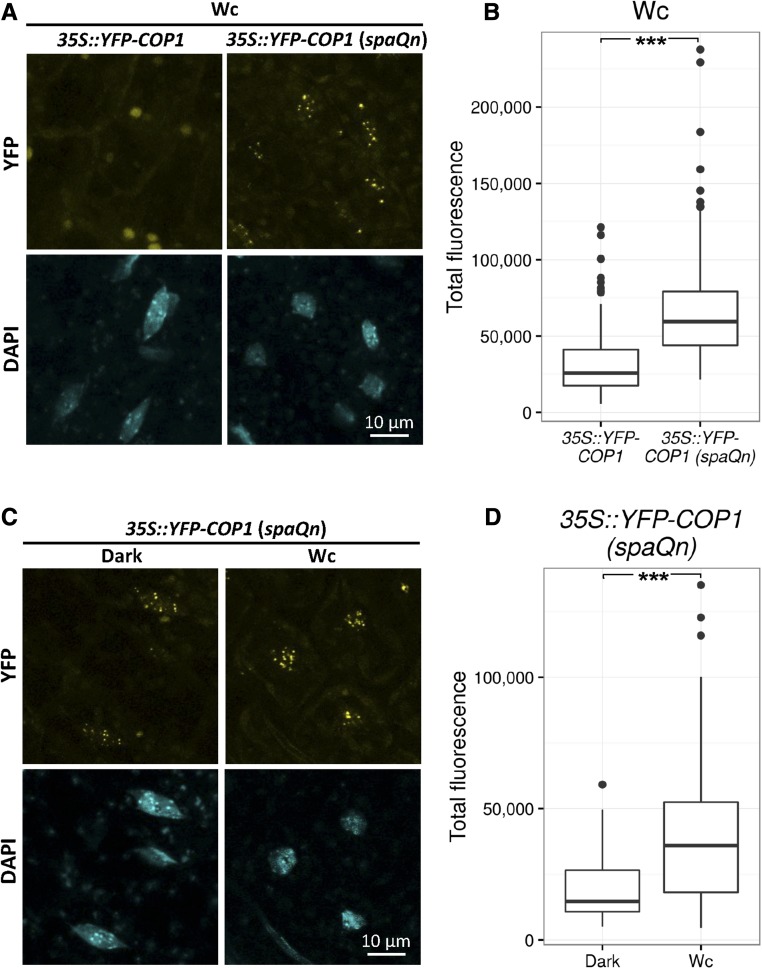
We subsequently assessed YFP-COP1 subcellular localization in hypocotyl cells of Wc-grown seedlings. As shown above, YFP-COP1 did not form detectable photobodies in a SPA wild-type background. However, notably, in the spaQn background, many nuclear-localized YFP-COP1 speckles were observed (Fig. 2A). Fluorescence intensity quantification confirmed that the nuclear YFP-COP1 signal was significantly higher in spaQn when compared to the SPA wild-type background (Fig. 2B). This increase in YFP-COP1 fluorescence in the nuclei of spaQn seedlings is not due to impaired YFP-COP1 protein degradation, because total levels of YFP-COP1 were lower, rather than higher, in Wc-grown spaQn than in SPA wild-type seedlings (Supplemental Fig. S1). When directly comparing nuclear YFP-COP1 fluorescence in dark- and Wc-grown spaQn seedlings, there was no detectable difference in photobody formation (Fig. 2C), and the nuclear YFP-COP1 signal intensity was even higher in Wc than in darkness (Fig. 2D). Taken together, these results suggest that light-regulated nucleocytoplasmic partitioning of YFP-COP1 is strongly impaired in the spaQn background.
Figure 2.
SPA proteins are essential for light-induced reduction in YFP-COP1 nuclear accumulation and photobody formation. A and B, Representative images (A) and average nuclear YFP fluorescence (B) of hypocotyl nuclei of 4-d-old 35S::YFP-COP1 and 35S::YFP-COP1 (spaQn) seedlings grown in 40 µmol m−2 s−1 Wc. A total of 146 or 208 nuclei was analyzed in 8 to 10 seedlings of 35S::YFP-COP1 or 35S::YFP-COP1 (spaQn), respectively. Percentage of nuclei showing speckles was 0% for 35S::YFP-COP1 and 97% for 35S::YFP-COP1. C and D, Representative images (C) and average nuclear YFP fluorescence (D) of hypocotyl nuclei of 4-d-old 35S::YFP-COP1 (spaQn) seedlings grown in darkness or 40 µmol m−2 s−1 Wc. A total of 106 or 169 nuclei was analyzed in 8 to 10 seedlings grown in darkness or Wc, respectively. Percentage of nuclei showing speckles was 78% in darkness and 97% in Wc. Upper and lower hinges of boxplot correspond to the first and third quartiles, respectively. Whiskers extend from the hinges to the highest/lowest value within 1.5 * interquartile range (IQR). Data points beyond this range are represented as dots. *** indicate that true difference between means is not equal to 0 at P < 0.001 in two-sample Student’s t test.
Nucleocytoplasmic Partitioning of the Endogenous COP1 Protein Is Perturbed in spaQn
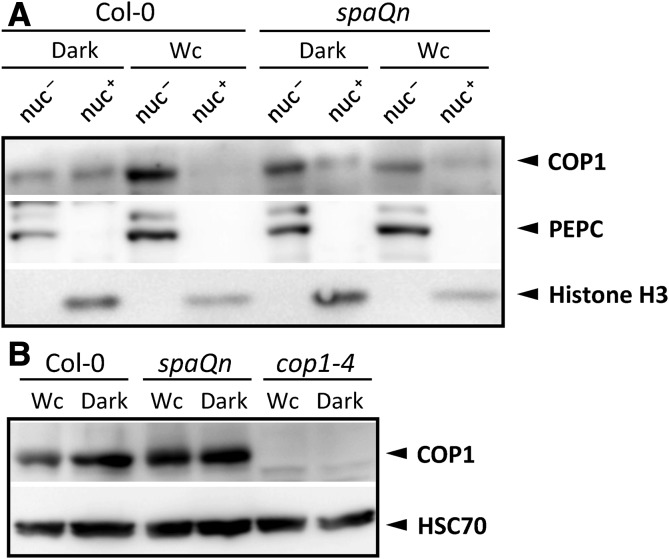
The observations made using the transgenic 35S::YFP-COP1 line suggest that SPA proteins are required for light-induced nuclear exclusion of COP1. We further tested this hypothesis by analyzing endogenous COP1 levels in nuclear and cytosolic fractions derived from biochemical fractionation experiments. In the wild type, the abundance of nuclear COP1 decreased in Wc-grown seedlings when compared to dark-grown seedlings, while concomitantly the cytosolic COP1 levels increased in Wc when compared to darkness (Fig. 3A). This finding is consistent with results obtained previously (Yu et al., 2013) and confirms that COP1 is excluded from the nucleus in the light. In the spaQn mutant, light did not cause a dramatic change in COP1 levels in the nuclear fraction or in the cytosolic fraction. Importantly, light exposure did not cause a reduction in nuclear COP1 levels. Similarly, cytosolic COP1 levels were not increased by light in spaQn (Fig. 3A; Supplemental Table S1). No major differences in total COP1 levels were detected between wild-type and spaQn seedlings (Fig. 3B; Supplemental Table S1), suggesting that differences observed in COP1 subcellular localization between wild type and spaQn are not due to differential COP1 protein stability. In total, these results indicate that nucleocytoplasmic partitioning of the COP1 protein was strongly impaired in light-grown spaQn mutant seedlings.
Figure 3.
COP1 nucleocytoplasmic partitioning is disturbed in the spaQn mutant. A and B, Immunodetection of COP1 protein in nuclei-depleted (nuc–) and nuclei-enriched (nuc+) protein fractions (A) or in total protein extracts (B) of 4-d-old Col-0 and spaQn seedlings grown in darkness or 40 µmol m−2 s−1 Wc. PEPC was used as a cytosolic marker and Histone H3 was used as a nuclear marker. HSC70 was used as a loading control for total protein preparations. Proteins were detected using anti-COP1, anti-PEPC, anti-Histone H3, and anti-HSC70 antibodies, respectively.
Taken together, these results agree with the observations made using the YFP-COP1-expressing seedlings and thus indicate that SPA proteins are not important for nuclear accumulation of COP1 in darkness, but are instead important for the light response of COP1, that is, for dissolving COP1 photobodies and for excluding COP1 from the nucleus in the light.
Different Light Qualities Have Distinct Effects on YFP-COP1 Subcellular Localization
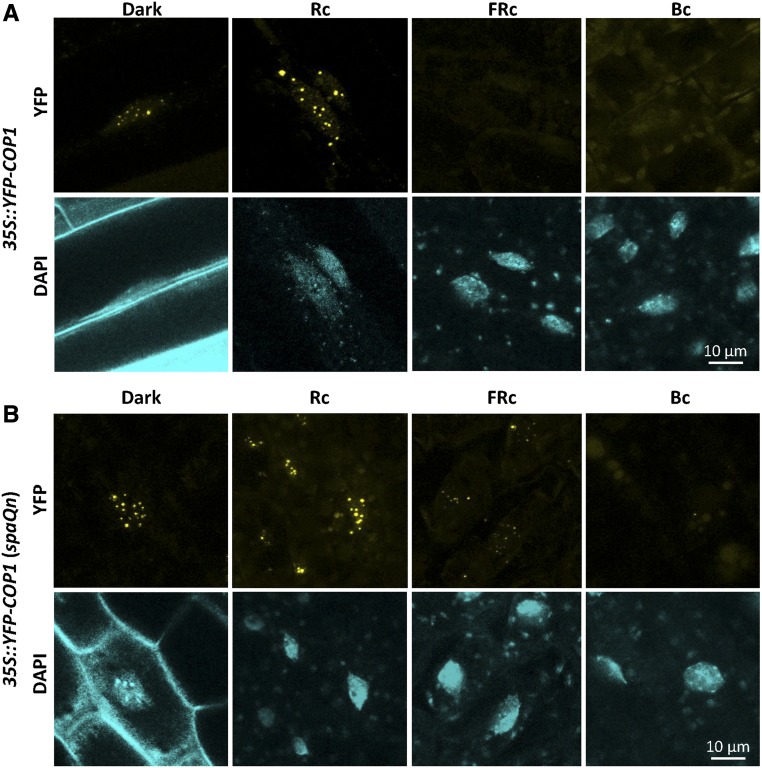
When analyzed in a GUS-COP1-expressing line, nuclear exclusion appeared to be strongest in white light, but was also induced to varying degrees by FRc, Rc, and Bc (Osterlund and Deng, 1998). These results, combined with the finding that YFP-COP1 still formed speckles in hypocotyl cells of Wc-grown seedlings in the spaQn background, motivated us to investigate the nuclear accumulation of YFP-COP1 in the spaQn mutant grown in monochromatic light. To this end, we grew seedlings expressing YFP-COP1 in the SPA wild-type and the spaQn mutant backgrounds in Rc, FRc, and Bc. In the SPA wild-type background, YFP-COP1 speckles were not detectable in both FRc and Bc, that is, FRc and Bc-grown seedlings appeared similar to Wc-grown seedlings (Fig. 4A). In contrast, we detected strong nuclear YFP-COP1 accumulation in Rc in this line, with photobody formation that was at least as strong as in darkness. Microscopy of spaQn YFP-COP1 seedlings grown in these light conditions revealed the formation of speckles in both Rc and FRc (Fig. 4B), while we were not able to detect YFP-COP1 speckles in Bc-grown seedlings. Hence, FRc exposure, similar to Wc exposure, abolished nuclear YFP-COP1 photobody formation in a SPA-dependent manner. In Rc, YFP-COP1 speckle formation did not change when compared to dark-grown seedlings, neither in the SPA wild-type nor in the spaQn mutant background, suggesting that Rc exposure was not sufficient to exclude YFP-COP1 from the nucleus in this transgenic line. We used the maximum fluence rate of Rc that can be produced by our LED light sources (60 µmol m−2 s−1), which should be sufficient to fully activate phyB. In Bc, YFP-COP1 speckles were absent in both the SPA wild-type and the spaQn mutant background, indicating that removal of YFP-COP1 from the nucleus in Bc is SPA-independent. Taken together, these results suggest a complex coaction of various photoreceptors in the control of nucleo-cytoplasmic partitioning of YFP-COP1.
Figure 4.
YFP-COP1 exhibits different degrees of nuclear accumulation in different light qualities. A and B, Representative images of hypocotyl nuclei of 4-d-old 35S::YFP-COP1 (A) and 35S::YFP-COP1 (spaQn) (B) seedlings grown in darkness, 60 µmol m−2 s−1 Rc, 3 µmol m−2 s−1 FRc, or 50 µmol m−2 s−1 Bc.
DISCUSSION
It has been established over the past two decades that the COP1/SPA complex acts as part of an E3 ubiquitin ligase that promotes the degradation of positive factors of light responses in dark-grown seedlings. The severe constitutive photomorphogenesis-phenotype found in cop1 and spa mutants clearly indicates that both COP1 and SPA proteins are necessary for the activity of the COP1/SPA E3 ubiquitin ligase (Deng et al., 1991; Laubinger et al., 2004; Ordoñez-Herrera et al., 2015). However, the specific role(s) of the SPA proteins within the COP1/SPA complex remain enigmatic. Since COP1 needs to be in the nucleus to exert its function (Stacey et al., 2000; Subramanian et al., 2004), we asked whether SPA proteins might be required for the nuclear accumulation of COP1. This idea was also driven by the overlap between the CLS and the SPA-interacting domain of COP1 that both map to the coiled-coil domain of COP1 (Stacey et al., 1999; Hoecker and Quail, 2001; Laubinger and Hoecker, 2003; Saijo et al., 2003; Chen et al., 2016). However, we found that the nuclear accumulation of COP1 in dark-grown seedlings was not impaired in a spa quadruple mutant that is devoid of all four SPA proteins. Both endogenous COP1 and a YFP-COP1 fusion protein showed normal accumulation of COP1 in the nucleus in the absence of SPA when seedlings were grown in darkness. Also, photobody formation of YFP-COP1 was not visibly impaired in dark-grown spaQn when compared to SPA wild type (Supplemental Table S2). These observations argue against the idea that SPA proteins have major functions in nuclear translocation/trapping of COP1 or its recruitment into photobodies. Hence, the NLS of COP1 appears to be sufficient to promote COP1 nuclear localization in darkness. Alternatively, COP1-interaction partners other than SPA proteins might be involved in this process. There is evidence that the COP9 signalosome (CSN) is involved in COP1 nuclear accumulation in dark-grown seedlings: nuclear localization of a GUS-COP1 fusion protein in darkness is lost in several csn mutants and subunit CSN1 interacts with and promotes nuclear accumulation of COP1 (Wang et al., 2009).
Light exposure causes exclusion of COP1 from the nucleus, thereby inhibiting COP1/SPA activity (von Arnim and Deng, 1994; Pacín et al., 2014). The biochemical mechanism regulating this process is thus far unknown. Our results show that nuclear exclusion of COP1 in Wc is strongly impaired in the absence of SPA proteins. Both endogenous COP1 and the YFP-COP1 fusion protein were retained in the nucleus in Wc-grown spaQn seedlings but not in SPA wild-type seedlings. Also, YFP-COP1 photobodies, which dissolve in Wc in a SPA wild-type background, were retained in a spaQn mutant background. Taken together, the subcellular localization of COP1 was not responsive to Wc in the absence of SPA. The reason for the insensitivity of COP1 to light in spaQn is so far unknown. It is possible that SPA proteins are necessary for the observed light-induced association of photoreceptors with the COP1/SPA complex (Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011; Lu et al., 2015; Sheerin et al., 2015). Alternatively, SPA proteins might be necessary to induce or mediate conformational changes in the COP1 protein that lead to nuclear exclusion of COP1 in Wc. This may include shielding of the NLS and/or exposure of the CLS of COP1 after light exposure. A third possibility is that SPA proteins are not directly affecting COP1 subcellular localization but that they act indirectly via a feedback mechanism, that is, by affecting the expression, stability, or activity of components of the so-far-unknown COP1 import/export machinery. Such feedback control of nuclear accumulation of light signaling components has been reported previously: PHYTOCHROME-INTERACTING FACTORs (PIFs) facilitate phyB nuclear import upon light exposure (Pfeiffer et al., 2012). Since active phyB causes PIF degradation (Leivar and Quail, 2011), phyB effectively reduces the PIFs’ positive effect on its own nuclear accumulation during prolonged irradiation. A similar feedback mechanism occurs during phyA signaling: phyA requires the shuttle proteins FAR-RED ELONGATED HYPOCOTYL 1 (FHY1) and FHY1-LIKE (FHL) for its nuclear import upon FR-irradiation (Hiltbrunner et al., 2005; Hiltbrunner et al., 2006), yet once in the nucleus, active phyA reduces expression of FHY1 and FHL and thereby limits the pool of these transport proteins (Li et al., 2010). Eventually, phyB and SPA1 have been reported to act together to desensitize phyA signaling in FR by promoting each other’s nuclear accumulation (Zheng et al., 2013).
Since Wc activates several classes of photoreceptors, we also analyzed the subcellular localization of COP1 in pure Rc, FRc, and Bc, which induce only a subset of photoreceptors, respectively (Supplemental Table S2). In FRc, similar results were obtained as in Wc, that is, nuclear exclusion of YFP-COP1 was observed in FRc-grown seedlings and this YFP-COP1 exclusion was dependent on SPA proteins. This indicates that phyA is sufficient for nuclear exclusion of YFP-COP1, in agreement with previous data (Osterlund and Deng, 1998). Furthermore, these results show that SPA proteins are required for the phyA-mediated nuclear exclusion of YFP-COP1 in FRc. In Rc, however, we did not observe a change in the subcellular localization of YFP-COP1, which disagrees with previous data obtained with a GUS-COP1-expressing line (Osterlund and Deng, 1998). We do not know the reason for this discrepancy and therefore can only speculate that it may be due to the respective transgenic line used. When observing endogenous COP1 protein levels in nuclear and cytoplasmic fractions, no difference was detected between seedlings grown in darkness and seedlings exposed to Rc for 24 h (Jang et al., 2010), which is consistent with our observations made using YFP-COP1. Hence, a potential role of SPA in possible Rc-controlled COP1 localization remains unclear. Due to the very limited seed production of the severely dwarfed spaQn plants, extensive cell fractionation experiments are unfortunately not possible. In Bc, similar to Wc, YFP-COP1 was excluded from the nucleus. However, Bc-induced nuclear exclusion of YFP-COP1 was not SPA dependent. This suggests that the subcellular localization of COP1 is controlled by a complex, nonadditive interplay among different photoreceptors, likely phytochromes and cryptochromes, in Wc-grown seedlings that cannot be mimicked by activating individual photoreceptors. Synergism between R and B was also observed previously (Sellaro et al., 2009).
In total, our results indicate that SPA proteins are necessary for the responsiveness of the COP1 subcellular localization to white light, that is, for transmitting the light signal to COP1. However, this is clearly not the only function of SPA proteins in the COP1/SPA complex: the failure of spaQn mutants to exclude COP1 from the nucleus in the light does not appear to enhance COP1 activity in the nucleus, since spaQn mutants exhibit a very short hypocotyl in Wc (Ordoñez-Herrera et al., 2015). This indicates that the nuclear COP1 in Wc-grown spaQn is largely inactive, similar to nuclear COP1 in dark-grown spaQn mutants. Hence, SPA proteins appear to have a dual role: they confer light-responsiveness to COP1 in Wc-grown seedlings and they are required for COP1 function per se, mainly in darkness. It remains to be determined which activity SPA proteins confer to the COP1/SPA complex in darkness. Based on our results, we can exclude the possibility that SPA proteins confer nuclear localization of COP1 in darkness. Hence, SPA proteins might be required for the assembly of a CUL4-DDB1-RBX1COP1/SPA complex, for binding of substrate transcription factors, and/or for COP1/SPA E3 ligase activity.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The genotypes spa1-100 spa2-2 spa3-1 spa4-3 (spaQn) and 35S::YFP-COP1 (cop1-4) were described previously (Oravecz et al., 2006; Ordoñez-Herrera et al., 2015). spaQn plants expressing 35S::YFP-COP1 were generated by crossing 35S::YFP-COP1 (cop1-4) to a spa1 spa2 spa3 mutant segregating for the spa4-3 allele. 35S::YFP-COP1 spaQn plants were identified by PCR using primers distinguishing between the respective mutant and wild-type alleles and fluorescence microscopy for the presence of the 35S::YFP-COP1 transgene.
LED light sources and growth conditions were described previously (Baumgardt et al., 2002; Laubinger et al., 2004).
Confocal Laser-Scanning Microscopy
Four-day-old seedlings expressing 35S::YFP-COP1 were used to analyze YFP-COP1 subcellular localization. For identification of nuclei, seedlings were stained with 1 µg/mL DAPI solution for 15 to 20 min at room temperature, rinsed in 70% ethanol, and mounted in water. Seedlings were observed under a Leica SP8 confocal laser-scanning microscope. DAPI fluorescence was excited with a Diode 405 laser and observed between 443 and 488 nm, while YFP fluorescence was excited with an Argon 514 laser and observed between 520 and 580 nm.
Quantification of Nuclear YFP Fluorescence
Hyperstacks obtained from confocal laser-scanning microscopy were analyzed in 3D-ImageJ Suite (Schindelin et al., 2012, 2015; Ollion et al., 2013). First, 3D thresholding of DAPI z-stacks was achieved with the tool “3D Hysteresis Thresholding.” Thresholded images were then segmented with the tool “3D segmentation” to receive coherent, 3-dimensional regions of interest (RoIs). Nuclear volume was measured by capturing the volume of RoIs with “Measure 3D.” Finally, RoIs were superimposed on 3D-filtered YFP z-stacks to capture fluorescence intensity of nuclei with “Quantif 3D.” For each genotype and condition, 8 to 10 seedlings were analyzed in each experiment. For each seedling, the fluorescence of 2 to 20 nuclei was quantified, as indicated in the legends of the presented experiments. Since fluorescence values differed significantly between experiments performed on different days, only fluorescence values obtained from the same experiment were directly compared in this study. The observations made in the experiments described above were robust in at least two independent replicate experiments.
Total Protein Extraction, Nuclear Fractionation, and Immunoblot Analysis
Total protein was extracted from approximately 100 mg seedling tissue as described (Chen et al., 2015). Nuclei-enriched and nuclei-depleted protein fractions were prepared from approximately 3 g seedling tissue using the Cell Lytic Plant Nuclear Extraction Kit (Sigma-Aldrich) with modifications according to Jang et al. (2010). Equal amounts of total protein and equal volumes of nuclei-enriched and nuclei-depleted protein were used for immunoblot analysis. Proteins were detected using anti-Histone H3 (Abcam), anti-PEPC (Rockland), anti-HSC70 (Stressgen), and anti-COP1 (Balcerowicz et al., 2011) primary antibodies in combination with antirabbit IgG-HRP and antimouse IgG-HRP (Sigma-Aldrich) secondary antibodies, respectively. HRP activity was detected using the SuperSignal West Femto Maximum Sensitivity kit (Thermo Scientific) and visualized by a LAS-4000 Mini bioimager (GE Healthcare Life Sciences). Protein bands were quantified using the ImageJ Gel Analyzer plugin.
Accession Numbers
COP1 (At2g32950), SPA1 (At2g46340), SPA2 (At4g11110), SPA3 (At3g15354), SPA4 (At1g53090).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. YFP-COP1 protein levels in 35S::YFP-COP1 and 35S::YFP-COP1 spaQn.
Supplemental Table S1. Quantification of total and nuclear COP1 levels in extracts of wild-type and spaQn Arabidopsis seedlings grown in darkness or continuous white light for 4 d.
Supplemental Table S2. Summary of COP1 subcellular localization obtained by fluorescence microscopy of transgenic 35S::YFP-COP1 seedlings and subcellular fractionation of wild-type seedlings
Acknowledgments
We are grateful to Roman Ulm for the 35S::YFP-COP1 line and to Dmitry Lapin and Jane Parker for the gift of the anti-PEPC antibody. We thank Klaus Menrath, his greenhouse staff, and many undergraduate students for expert care of our plants.
Glossary
- Bc
continuous blue light
- CLS
cytoplasmic localization signal
- FRc
continuous far-red light
- NLS
nuclear localization signal
- Rc
continuous red light
- RoIs
regions of interest
- Wc
continuous white light
References
- Balcerowicz M, Fittinghoff K, Wirthmueller L, Maier A, Fackendahl P, Fiene G, Koncz C, Hoecker U (2011) Light exposure of Arabidopsis seedlings causes rapid de-stabilization as well as selective post-translational inactivation of the repressor of photomorphogenesis SPA2. Plant J 65: 712–723 [DOI] [PubMed] [Google Scholar]
- Baumgardt R-L, Oliverio KA, Casal JJ, Hoecker U (2002) SPA1, a component of phytochrome A signal transduction, regulates the light signaling current. Planta 215: 745–753 [DOI] [PubMed] [Google Scholar]
- Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee J-H, Zhu D, Deng XW (2010) Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Lory N, Stauber J, Hoecker U (2015) Photoreceptor Specificity in the Light-Induced and COP1-Mediated Rapid Degradation of the Repressor of Photomorphogenesis SPA2 in Arabidopsis. PLoS Genet 11: e1005516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wirthmueller L, Stauber J, Lory N, Holtkotte X, Leson L, Schenkel C, Ahmad M, Hoecker U (2016) The functional divergence between SPA1 and SPA2 in Arabidopsis photomorphogenesis maps primarily to the respective N-terminal kinase-like domain. BMC Plant Biol 16: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Caspar T, Quail PH (1991) cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev 5: 1172–1182 [DOI] [PubMed] [Google Scholar]
- Fittinghoff K, Laubinger S, Nixdorf M, Fackendahl P, Baumgardt R-L, Batschauer A, Hoecker U (2006) Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J 47: 577–590 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A, Tscheuschler A, Viczián A, Kunkel T, Kircher S, Schäfer E (2006) FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol 47: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A, Viczián A, Bury E, Tscheuschler A, Kircher S, Tóth R, Honsberger A, Nagy F, Fankhauser C, Schäfer E (2005) Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr Biol 15: 2125–2130 [DOI] [PubMed] [Google Scholar]
- Hoecker U, Quail PH (2001) The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J Biol Chem 276: 38173–38178 [DOI] [PubMed] [Google Scholar]
- Jang I-C, Henriques R, Seo HS, Nagatani A, Chua N-H (2010) Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Fittinghoff K, Hoecker U (2004) The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in arabidopsis. Plant Cell 16: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Hoecker U (2003) The SPA1-like proteins SPA3 and SPA4 repress photomorphogenesis in the light. Plant J 35: 373–385 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Le Gourrierec J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133: 3213–3222 [DOI] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Gao S, Martinez C, He G, Zhou Z, Huang X, Lee J-H, Zhang H, Shen Y, et al. (2010) Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H-L, He S-B, Zhang Y-C, Zhu D-M, Zhang J-Y, Jia K-P, Sun S-X, Li L, Yang H-Q (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X-D, Zhou C-M, Xu P-B, Luo Q, Lian H-L, Yang H-Q (2015) Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant 8: 467–478 [DOI] [PubMed] [Google Scholar]
- Menon C, Sheerin DJ, Hiltbrunner A (2016) SPA proteins: SPAnning the gap between visible light and gene expression. Planta 244: 297–312 [DOI] [PubMed] [Google Scholar]
- Ollion J, Cochennec J, Loll F, Escudé C, Boudier T (2013) TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29: 1840–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Adám E, Schäfer E, Nagy F, Ulm R (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordoñez-Herrera N, Fackendahl P, Yu X, Schaefer S, Koncz C, Hoecker U (2015) A cop1 spa mutant deficient in COP1 and SPA proteins reveals partial co-action of COP1 and SPA during Arabidopsis post-embryonic development and photomorphogenesis. Mol Plant 8: 479–481 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Deng X-W (1998) Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J 16: 201–208 [DOI] [PubMed] [Google Scholar]
- Pacín M, Legris M, Casal JJ (2013) COP1 re-accumulates in the nucleus under shade. Plant J 75: 631–641 [DOI] [PubMed] [Google Scholar]
- Pacín M, Legris M, Casal JJ (2014) Rapid decline in nuclear costitutive photomorphogenesis1 abundance anticipates the stabilization of its target elongated hypocotyl5 in the light. Plant Physiol 164: 1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Nagel M-K, Popp C, Wüst F, Bindics J, Viczián A, Hiltbrunner A, Nagy F, Kunkel T, Schäfer E (2012) Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc Natl Acad Sci USA 109: 5892–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolauffs S, Fackendahl P, Sahm J, Fiene G, Hoecker U (2012) Arabidopsis COP1 and SPA genes are essential for plant elongation but not for acceleration of flowering time in response to a low red light to far-red light ratio. Plant Physiol 160: 2015–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Rueden CT, Hiner MC, Eliceiri KW (2015) The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev 82: 518–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellaro R, Hoecker U, Yanovsky M, Chory J, Casal JJ (2009) Synergism of red and blue light in the control of Arabidopsis gene expression and development. Curr Biol 19: 1216–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof Y-D, Huq E, Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Hicks SN, von Arnim AG (1999) Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1. Plant Cell 11: 349–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Kopp OR, Kim T-H, von Arnim AG (2000) Modular domain structure of Arabidopsis COP1. Reconstitution of activity by fragment complementation and mutational analysis of a nuclear localization signal in planta. Plant Physiol 124: 979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, von Arnim AG (1999) A novel motif mediates the targeting of the Arabidopsis COP1 protein to subnuclear foci. J Biol Chem 274: 27231–27236 [DOI] [PubMed] [Google Scholar]
- Subramanian C, Kim B-H, Lyssenko NN, Xu X, Johnson CH, von Arnim AG (2004) The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: mutational analysis by bioluminescence resonance energy transfer. Proc Natl Acad Sci USA 101: 6798–6802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk EK, Decker PV, Chen M (2012) Photobodies in light signaling. Plant Physiol 158: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng X-W (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Wang X, Li W, Piqueras R, Cao K, Deng XW, Wei N (2009) Regulation of COP1 nuclear localization by the COP9 signalosome via direct interaction with CSN1. Plant J 58: 655–667 [DOI] [PubMed] [Google Scholar]
- Yi C, Deng XW (2005) COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Yu Y, Wang J, Zhang Z, Quan R, Zhang H, Deng XW, Ma L, Huang R (2013) Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PLoS Genet 9: e1004025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Wu S, Zhai H, Zhou P, Song M, Su L, Xi Y, Li Z, Cai Y, Meng F, et al. (2013) Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell 25: 115–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Maier A, Lee J-H, Laubinger S, Saijo Y, Wang H, Qu L-J, Hoecker U, Deng XW (2008) Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]